Abstract
Background
Model organisms are at the core of life science research. Notable examples include the mouse as a model for humans, baker’s yeast for eukaryotic unicellular life and simple genetics, or the enterobacteria phage λ in virology. Plant research was an exception to this rule, with researchers relying on a variety of non-model plants until the eventual adoption of Arabidopsis thaliana as primary plant model in the 1980s. This proved to be an unprecedented success, and several secondary plant models have since been established. Currently, we are experiencing another wave of expansion in the set of plant models.
Scope
Since the 2000s, new model plants have been established to study numerous aspects of plant biology, such as the evolution of land plants, grasses, invasive and parasitic plant life, adaptation to environmental challenges, and the development of morphological diversity. Concurrent with the establishment of new plant models, the advent of the ‘omics’ era in biology has led to a resurgence of the more complex non-model plants. With this review, we introduce some of the new and fascinating plant models, outline why they are interesting subjects to study, the questions they will help to answer, and the molecular tools that have been established and are available to researchers.
Conclusions
Understanding the molecular mechanisms underlying all aspects of plant biology can only be achieved with the adoption of a comprehensive set of models, each of which allows the assessment of at least one aspect of plant life. The model plants described here represent a step forward towards our goal to explore and comprehend the diversity of plant form and function. Still, several questions remain unanswered, but the constant development of novel technologies in molecular biology and bioinformatics is already paving the way for the next generation of plant models.
Keywords: Plant biology, model organisms, plant models, non-model plant models, Cardamine hirsuta, Eutrema salsugineum, Marchantia polymorpha, Phragmites australis, Pisum, sativum, Setaria viridis, Striga hermonthica
INTRODUCTION
Model organisms (MOs) are used in research to study certain scientific questions (Ankeny and Leonelli, 2011). They can either function as a representative for a whole group of organisms (such as plants, mammalians or prokaryotes), or act as a ‘stand-in’ for specific organisms of interest that cannot be easily studied, such as mice instead of humans for example for ethical reasons. There are two main reasons to use MOs. First, they are typically simple, both biologically and in handling. MOs are generally small, can be easily grown in a lab, have short life cycles, produce sufficient offspring, have small and simple genomes, and can be easily transformed, mutated and crossed. Second, to study every aspect of a given organism’s life, sophisticated methods, techniques and equipments are typically required (Ankeny and Leonelli, 2011). Their development, production, acquisition and maintenance can be expensive, time-consuming and laborious. Therefore, it is more practical to focus on specific MOs for the initial development and production of such technologies, instead of studying countless different organisms, each with individual requirements. Eventually, knowledge gained with an MO can be extrapolated to the actual organisms of interest, allowing researchers to limit experimentation on these organisms to a few targeted and well-established final tests.
In the plant field, Arabidopsis thaliana was only established as a universal MO in the 1980s (Somssich, 2018). One reason for this relatively late adoption of a plant MO was that plant-specific aspects of development, morphology and physiology were typically studied directly in established crops, thereby eliminating the usual final step of extrapolating the knowledge from the model to the crops (Koornneef and Meinke, 2010). These plants are now considered part of the ‘non-model plant models’ group, meaning that they are established MOs, without actually carrying the typical characteristics of MOs. Relying on such non-model plant models in plant research became problematic with the advent of modern genetics and molecular biology (Koornneef and Meinke, 2010). When these fields became more important, the work with non-model plant models became technically impractical, slow and inefficient. As a result, plant biologists eventually recognized the need to adopt one specific model as a means of advancing the plant science field, resulting in A. thaliana becoming the universal MO (Koornneef and Meinke, 2010). Since then, the field of plant biology, and specifically plant molecular biology and genetics, has expanded enormously and produced a wealth of knowledge and understanding of plant biology (Somerville and Koornneef, 2002).
In a first wave of expansion, mostly in the late 1990s, the plant community adopted a set of ‘second-generation’ plant models. These were chosen to represent individual groups of plants that were too distantly related to A. thaliana to be studied in this primary model (Chang et al., 2016). Among those adopted, Brachypodium distachyon was chosen as a grass (monocot) model, Physcomitrella patens to represent the mosses, Medicago truncatula to cover the legumes and Populus trichocarpa to study trees (Cook, 1999; Draper et al., 2001; Cove, 2005; Jansson and Douglas, 2007; Chang et al., 2016). At the same time, A. thaliana is still far from being ‘solved’, and A. thaliana research will remain at the forefront of plant science (Provart et al., 2016). As such, it will continue to produce new insights at an ever-increasing molecular detail, while providing a basis for the development of new techniques (Provart et al., 2016). Notably, the field of A. thaliana research has seen its own expansion with the emerging research area of natural variation (Weigel, 2012).
More recently, some third-generation model plants have been proposed to cover research areas such as the early evolution of land plants from aquatic ancestors, plant parasitism, the formation of complex organs, tissue forms and shapes, and specific adaptations to environmental conditions. Concurrently, the group of non-model plant models is also experiencing a resurgence since plant science entered the genomics (or generally ‘omics’) era (Rowan et al., 2011). New genomics techniques such as high-throughput whole genome sequencing, the CRISPR/Cas9 system for precise genome editing, new cloning techniques that make it easier than ever to clone and express genes of interest, de novo gene synthesis, or the modern high- and super-resolution fluorescence microscopy techniques have advanced molecular biology research even for these highly complex plants, allowing them a comeback into modern molecular biology labs (Rowan et al., 2011; Borrill, 2020). Philippa Borrill has recently written an insightful article on the ‘blurring of the boundaries between cereal crops and model plants’ (Borrill, 2020).
With this review, we will introduce some of these emerging third-generation plant models. More precisely, we will discuss Marchantia polymorpha as a model to study land plant evolution, Setaria viridis as a model for C4 photosynthesis and biomass recalcitrance, Phragmites australis for invasive plants, Striga hermonthica for plant parasitism, Eutrema salsugineum for salt tolerance and Cardamine hirsuta for comparative developmental studies. Furthermore, we will discuss Pisum sativum, a member of the non-model plant model group that is currently experiencing a resurgence as a model for legume crops. The scientific and biological relevance of these species are discussed, and the tools and resources available for the scientific community are highlighted.
THE NEW PLANT MODELS TO STUDY
Land plant evolution: introducing Marchantia polymorpha (common liverwort)
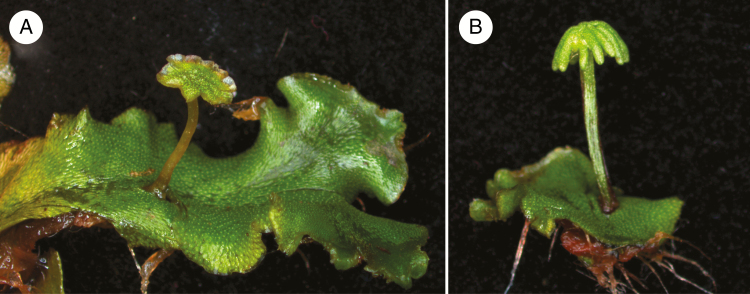
The conquering of land by plants ~470 million years ago was a major step in evolution (Bowman et al., 2016b). Fossil and phylogenetic evidence suggest that land plants evolved from a common charophycean algal ancestor with a haplobiontic life cycle, meaning a dominant multicellular gametophyte (n), while the diploid phase only includes a fertilized unicellular zygote that immediately undergoes meiosis (Bowman et al., 2016b). In land plants, both the gametophyte (n) and the sporophyte (2n) produce multicellular bodies (Bowman et al., 2016b). The relative dominance of these two multicellular phases has shifted during land plant evolution: the haploid phase is dominant in basal land plants while the sporophyte is only short lived and determinate, thereby mosre closely resembling the charophycean algae (Bowman et al., 2016b). In vascular plants, the diploid phase became dominant over the haploid phase, causing morphological diversity of vascular plants to reside in the sporophyte, while the gametophyte was reduced to a few cells that produce male and female gametes (Bowman et al., 2016b). In this context it is an open question whether the genetic programme underlying the development of two multicellular bodies, and the genetic programme that enabled the increasing complexity of the sporophyte, already pre-existed in the algal ancestor, or if they evolved de novo. To address this, it is of major importance to study the development of a basal land plant, as well as the relationships of this basal plant to its ancestors, charophycean algae and its descendants, vascular plants. Bryophytes are a group of basal land plants that include the non-vascular liverworts, mosses and hornworts (Mishler and Churchill, 1984). Marchantia polymorpha (Fig. 1) is a complex thalloid liverwort with a well-studied taxonomy and morphology (Bowman, 2016). Liverworts have experienced a low rate of chromosomal and molecular evolution, and thus the genetic makeup of M. polymorpha is probably more similar to that of the common ancestor of all land plants, making it a versatile model to study land plant origin and evolution (Bowman et al., 2016a).
Fig. 1.
Marchantia polymorpha. Marchantia polymorpha produces a haploid thallus with either (A) a male antheridiophore containing antheridia with flagellated sperm, or (B) a female archegoniophore with archegonia holding an egg. Upon fertilization, the diploid sporophyte undergoes mitosis followed by meiotic divisions of the sporogenous tissues to produce haploid spores. Photo credit Tom Dierschke (Monash University).
The predominant and persisting generation of the M. polymorpha life cycle is the gametophyte. This haploid dominance makes genetic analysis faster compared to diploid-dominant plants, as it eliminates the need of heterozygosity, allowing mutant and transgenic phenotypes to be studied in their isolated generation. Marchantia polymorpha can reproduce sexually through flagellated sperm and egg cells, which are produced in the gametophores (antheridia and archegonia) (Fig. 1A, B) (Shimamura, 2016). Antheridia produce sperm, while archegonia produce eggs (Shimamura, 2016). Marchantia polymorpha can also reproduce asexually via small, disc-shaped propagules called gemmae that are formed in gemmae cups on the dorsal side of the haploid thallus and remain dormant until dispersed (Eklund et al., 2015). These two modes of reproduction allow genetic crossings and the establishment and propagation of individual isogenic lines from a spore or gemma, derived from a single cell (Ishizaki et al., 2016). The gametophytic generation can be cultured and maintained under sterile conditions or stored at ultra-low temperatures, and cryopreservation of fertile M. polymorpha spermatozoa has been reported (Ishizaki et al., 2016; Togawa et al., 2018). These techniques provide the opportunity to reliably preserve M. polymorpha lines. Further advantages include a short generation time of ~3 months, 2–3 weeks for asexual reproduction and a small genome size (225.8 Mb, nine chromosomes) (Ishizaki et al., 2016; Bowman et al., 2017). The apparent absence of ancient polyploidization and the lack of gene duplication also account for a low functional redundancy (Ishizaki et al., 2016; Bowman et al., 2017).
An assembled M. polymorpha genome sequence was generated using the natural accessions Takaragaike-1 (Tak-1, male) and Takaragaike-2 (Tak-2, female), which were isolated in Kyoto, Japan (Okada et al., 2000; Ishizaki et al., 2008; Bowman et al., 2017). However, as M. polymorpha is a cosmopolitan species distributed globally from tropical to arctic climates, other natural accessions have been collected as laboratory strains and used for experimental research. At present, no comprehensive collection of all accessions exists in the research community. The first genetic transformations of M. polymorpha were achieved by particle bombardment, but practical high-frequency Agrobacterium-mediated transformation protocols are now available as well (Takenaka et al., 2000; Ishizaki et al., 2008; Tsuboyama and Kodama, 2014; Tsuboyama-Tanaka and Kodama, 2015). Common binary vectors such as pCAMBIA, pPZP and pGWBs can be used for transformations, and a gene targeting procedure via homologous recombination has also been adopted for M. polymorpha (Terada et al., 2002; Ishizaki et al., 2013a, 2015). In addition, CRISPR/Cas9-based targeted mutagenesis has been demonstrated to work efficiently; however, the haploid dominancy of the M. polymorpha life cycle limits the ability to isolate mutants of essential genes, and as such, null mutations are potentially lethal (Sugano et al., 2018). To overcome this issue, knockdown strategies such as inducible artificial microRNA (amiR)-mediated gene silencing or the Cre/loxP site-specific recombination system, combined with heat-shock- and DEX-controlled gene expression, were established (Flores-Sandoval et al., 2016; Nishihama et al., 2016). Constitutive overexpression can be achieved using the CaMV 35S or the endogenous ELONGATION FACTOR 1a (MpEF1a) promoter (Althoff et al., 2014). Both are capable of driving strong expression, but there are significant differences in terms of spatial distribution (Kajikawa et al., 2003; Althoff et al., 2014; Kubota et al., 2014; Sugano et al., 2014; Eklund et al., 2015; Flores-Sandoval et al., 2015; Kato et al., 2015). For gene expression studies, RNA in situ hybridization protocols and reporter genes such as β-glucuronidase (GUS) and fluorescent proteins have been tested and used successfully (Ishizaki et al., 2012, 2013b; Althoff et al., 2014; Komatsu et al., 2014; Kubota et al., 2014). Due to its low genetic redundancy, M. polymorpha is also highly suitable for forward genetic approaches. For instance, a T-DNA tagging strategy to generate mutants has been successfully employed, as has physical mutagenesis using X-ray irradiation (Miller et al., 1962; Ueda et al., 2012; Ishizaki et al., 2013b, 2016). In addition to the nuclear and organelle genome sequences, microRNA (miRNA) profiles and their targets, as well as DNA methylation profiles for different developmental stages and tissues are available (Lin et al., 2016; Tsuzuki et al., 2016; Bowman et al., 2017; Schmid et al., 2018). Recently, a whole suite of molecular biology and genetics tools, protocols and resources for the work with M. polymorpha has been made available as the OpenPlant toolkit (Sauret-Güeto et al., 2020).
Recent findings have shed some light on the basic principle of how sporophyte-specific gene expression is initiated in land plants. The core regulatory network controlling this genetic switch involves the interaction, translocation and subsequent regulatory action of a BELL-LIKE (BELL) and a KNOTTED 1 LIKE HOMEOBOX (KNOXI) transcription factor (Lee et al., 2008; Bowman et al., 2016b; Dierschke et al., 2020). This mechanism probably evolved in unicellular green algae, such as Chlamydomonas reinhardtii, and then diversified to activate sporophytic gene expression in land plants (Floyd and Bowman, 2007; Bowman et al., 2016b; Horst et al., 2016; Frangedakis et al., 2017). However, less is known about the genetic programmes that enabled sporophytic multicellularity and three-dimensional (3-D) growth. One underlying feature of multicellular life is the network of signalling pathways by which cells communicate (Bowman et al., 2017). Analysis of the M. polymorpha genome demonstrated that all necessary components for most land-plant signalling pathways are also encoded in the M. polymorpha genome, but reduced to the minimum number of components (Bowman et al., 2017). In the case of the auxin signalling pathway, the network in M. polymorpha is simple but functional, with all components existing as single orthologues (Kato et al., 2015). Phylogenetic analyses suggest that this feature is shared with the last common ancestor of land plants, and that M. polymorpha has probably retained this ancestral condition (Flores-Sandoval et al., 2015). Studies have also shown that auxin is required for cell patterning during transition from 2-D to 3-D growth in the M. polymorpha gametophyte (Flores-Sandoval et al., 2015). Similarly, it was found that the jasmonate signalling network in M. polymorpha consists of some ancient components and others that arose through duplication and neofunctionalization of algal genes (Han, 2017; Monte et al., 2018; Bowman et al., 2019). More recently, M. polymorpha has also been adopted as a model for evolutionary molecular plant–microbe interaction studies, with the first bacterial, fungal and oomycete pathogens being described (Carella et al., 2019; Gimenez-Ibanez et al., 2019; Matsui et al., 2019). In all cases, infection with the pathogen results in the activation of typical hallmarks of plant immunity, demonstrating that the plant is also a suitable model to study defensive mechanisms without the redundancy present in vascular plants (Carella et al., 2019; Gimenez-Ibanez et al., 2019; Matsui et al., 2019). With regard to cell-wall biology, the evolution of the highly complex cellulose synthesis machinery has recently been analysed (Lampugnani et al., 2019). At the core of this machinery are the members of the CELLULOSE SYNTHASE (CesA) family, which form multimeric complexes in vascular plants, making it complicated to study their function (Lampugnani et al., 2019). It was found that these key components already exist in M. polymorpha, but that this early land plant only has two CesA genes, compared to ten in A. thaliana (Lampugnani et al., 2019).
These studies have revealed that components of the different pathways often consist of a combination of pre-existing algal genes and/or genes that have undergone neofunctionalization (Bowman et al., 2017). Accordingly, genetic regulators that were considered specific to land plants have since been found in the charophycean algae (Catarino et al., 2016; Bowman et al., 2017; Wilhelmsson et al., 2017; Vries and Archibald, 2018). Hence, a number of developmental innovations relevant to land plant evolution can be traced back to the common ancestor of land plants. This includes UVB-tolerance through UVR8-mediated flavonoid induction and flavonoid production in response to abiotic stress, the genetic control of vegetative reproduction, photoperiodic control for the transition from vegetative to reproductive growth, and germ cell differentiation (Kubota et al., 2014; Koi et al., 2016; Albert et al., 2018; Clayton et al., 2018; Hiwatashi et al., 2018; Yamaoka et al., 2018). Moreover, these basic mechanisms were first acquired in the gametophytic generation, then co-opted between the generations, and finally diversified to pattern the sporophyte (Bowman et al., 2019).
C 4 photosynthesis and biomass recalcitrance: introducing Setaria viridis (green foxtail)
Historically, most research on grass genetics and genomics has been carried out in agriculturally important crops, such as maize, wheat or rice. These plants are not ideal MOs because of particular intrinsic difficulties, such as long life cycles, large plant size and lack of efficient transformation protocols (Li and Brutnell, 2011; Brutnell et al., 2015). The temperate grass Brachypodium distachyon was later adopted as a model grass at a remarkably rapid rate due to several biological attributes, such as small stature, short life cycle, simple growth requirements and amenability to genetic transformation. Despite its major contributions to research, B. distachyon lacks perhaps one of the most economically important traits generally found in grasses: the mechanism of C4 photosynthesis (Schuler et al., 2016). The productivity of several grasses used for food and bioenergy is driven by C4 photosynthesis, which confers improved radiation, nitrogen and water-use efficiencies when compared to C3 photosynthesis, while reducing losses caused by photorespiration (Schuler et al., 2016). Therefore, engineering C4 photosynthesis into C3 crops is a major objective for crop improvement, but such a strategy has been hampered by the lack of a complete list of genes and their corresponding functions required to support the trait (Weber and Bar-Even, 2019).
Setaria viridis (Fig. 2) is a C4 grass belonging to the subfamily Panicoideae, tribe Paniceae, which is sister to the tribe Andropogoneae, one of the most economically important groups of plants that includes maize, sorghum and sugarcane (Huang et al., 2016). Setaria viridis shows several desirable attributes for an MO, such as a short life cycle (6–8 weeks), self-fertility, small stature (15–30 cm), large seed yield (~13 000 seeds per plant), simple growth requirements and a small diploid genome (510 Mb). Boosted by its remarkable capacity to invade, colonize and adapt to local environments, Se. viridis has spread from its centre of origin in Eurasia to a wide range of habitats, becoming the most widely distributed weed in the world (Li and Brutnell, 2011; Zhu et al., 2017). Significant phenotypic variation is observed among different natural populations, including differences in inflorescence architecture, plant height, seed morphology and flowering time (Li and Brutnell, 2011). However, the genetic diversity underlying those traits is apparently low and distributed in subpopulations, suggesting strong local adaptation (Li and Brutnell, 2011; Brutnell et al., 2015). It is largely accepted that Se. viridis is the wild ancestor of the cereal crop Setaria italica (common name: foxtail millet) (Li and Brutnell, 2011; Brutnell et al., 2015; Huang et al., 2016). Although genetically similar, Se. italica shows distinct morphological and physiological traits compared to its wild ancestor, including larger stature, enlarged inflorescence, reduced vegetative branching, synchrony of flowering and loss of seed dormancy. These differences are thought to be part of the ‘domestication syndrome’ caused by artificial selection made by humans during foxtail millet domestication (Li and Brutnell, 2011). Regarding its photosynthetic apparatus, Se. viridis is a C4 plant employing an NADP-dependent malic enzyme (NADP-ME subtype) as the decarboxylating enzyme located in the bundle sheath, similar to important food and bioenergy crops such as maize, sorghum and sugarcane (Danila et al., 2016). These facts make Se. viridis an excellent model for studying plant domestication, to understand the molecular mechanisms underlying several aspects of C4 photosynthesis and to validate biotechnological strategies aimed at boosting plant yields.
Fig. 2.
Setaria viridis. Representative picture of Se. viridis, an emerging model for C4 grasses.
A wider adoption of Se. viridis as a universal grass model will be facilitated by the continuing development of novel resources and protocols. The foundation for genetic, genomic and functional studies has already been created with its published genome (Bennetzen et al., 2012), which is in its second annotated version and available at Phytozome (https://phytozome.jgi.doe.gov). To further exploit the genetics of S. viridis, an optimized protocol for genetic crosses has been developed, involving panicle pruning followed by emasculation using hot water treatment to kill viable pollen (Jiang et al., 2013). This method was reported to yield one to seven cross-pollinated seeds per panicle (Jiang et al., 2013). Another major breakthrough was the establishment of various Agrobacterium tumefaciens-mediated transformation protocols, including tissue culture-based and floral-dip methods (Martins et al., 2015a, b). Although the latter method is more straightforward, it typically shows very low transformation efficiencies (~0.6–0.8 %), whereas the more laborious and time-consuming tissue culture-based method presents a transformation frequency as high as 29 % (Martins et al., 2015a, b). These methods provide a valuable tool for gene discovery and functional studies, and for rapid validation of biotechnological strategies before their translation to dedicated crops. Reliable reference genes for expression analysis via quantitative real-time PCR have been identified and validated for a wide range of experimental conditions, including different plant developmental stages and diverse stress conditions (Martins et al., 2016). Standard phenotyping protocols have also been developed, with detailed descriptions of multiple growth and developmental assays under controlled conditions, and in response to phytohormone treatment and abiotic stresses (Acharya et al., 2017). These assays will be particularly important for mutant screens, for large-scale phenotyping, and for the characterization of transgenic lines during functional studies. Growth in a hydroponic system allows the uniform production of robust seedlings that can be used to assess plant responses to a wide range of chemicals in highly reproducible experiments (Monte-Bello et al., 2018).
Setaria viridis is being employed in efforts to address three major ‘biological problems’: (1) plant domestication, (2) C4 photosynthesis and (3) biomass recalcitrance. Although some genes involved in plant domestication might be conserved among species, no differences were found in the coding sequences of candidate domestication genes between Se. viridis and its cereal crop descendant, Se. italica, suggesting that a different set of genes or regulatory mechanisms was involved in foxtail millet domestication (Bennetzen et al., 2012). Still, several quantitative trait loci of key domestication traits were mapped and partially characterized (with candidate genes often identified), such as those related to shattering, plant height, plant branching, flowering time and photoperiod sensitivity. These data are nicely compiled and discussed in the excellent review from H. Hu et al. (2018). Setaria viridis has also been widely employed to study C4 photosynthesis (Brutnell et al., 2015; Huang and Brutnell, 2016; Zhu et al., 2017). In addition to its use in comparative transcriptomic studies with C3 and intermediate C3–C4 species to select novel candidate genes related to the C4 mechanism, the availability of efficient transformation approaches has allowed Se. viridis to become a platform to functionally validate those targets, and consequently, to provide new insights into several aspects of C4 photosynthesis (Boyd et al., 2015; Alonso-Cantabrana et al., 2018). Transgenic Se. viridis depleted in carbonic anhydrase (CA) was generated to address the role of CA in C4 photosynthesis (Osborn et al., 2017). CA is localized in the cytosol of mesophyll cells, where it catalyses the hydration of CO2 to HCO3−, which is further used by phosphoenolpyruvate carboxylase (PEPC) in the first step of C4 photosynthesis (Osborn et al., 2017). It was shown that under normal atmospheric conditions, CA activity was not rate-limiting for C4 photosynthesis in Se. viridis, whereas under conditions that result in lower intercellular CO2 concentrations (such as drought), mesophyll conductance may pose a greater limitation than CA activity (Osborn et al., 2017). Therefore, increasing mesophyll conductance may be an interesting strategy to boost CO2 assimilation in a scenario of global warming and limited water availability. Silencing of PEPC in Se. viridis resulted in reduced cell wall thickness and increased plasmodesmata (PD) density at the mesophyll–bundle sheath interface, leading to an intriguing speculation that PD development might be responsive to changes in C4 photosynthetic flux (Alonso-Cantabrana et al., 2018). These are only a few examples demonstrating the potential of Se. viridis as a model for molecular manipulation of the C4 photosynthetic pathway.
Setaria viridis has also been suggested as a model for lignocellulosic biomass crops, based on its phylogenetic proximity to potential feedstock, such as sugarcane, Miscanthus spp. and switchgrass (Brutnell et al., 2010, 2015; Li and Brutnell, 2011). Plant biomass is mainly composed of secondary cell walls (SCWs), whose major components are polysaccharides that can potentially be converted into fermentable sugars for the production of biofuels and biomaterials. However, the complex chemical compositions and rigid structure of SCWs hinder the efficient processing of plant biomass in biorefineries, an issue known as biomass recalcitrance (Marriott et al., 2016). Therefore, the production of optimized bioenergy crops with reduced recalcitrance requires a deep characterization of several aspects of SCW deposition. Above-ground biomass of Se. viridis was shown to be similar to that of other panicoid bioenergy crops in terms of cellulose and lignin content and cell wall polysaccharide composition (Petti et al., 2013). In addition, the characteristics of the CELLULOSE SYNTHASE gene superfamily and the accumulation and distribution of (1,3;1,4)-β-glucans, polysaccharides that are typical of grass cell walls, were shown to be similar between Se. viridis and other C4 grasses (Ermawar et al., 2015). The core set of biosynthetic genes potentially involved in developmental lignification and lignin-related laccases were identified using a combination of comparative phylogenetic studies, high-throughput expression analysis and quantitative RT-PCR analysis (Ferreira et al., 2019; Simões et al., 2020). Regarding gene discovery, only one SCW-related gene has been functionally characterized in Se. viridis. Souza et al. (2018) showed that the BAHD acyl-CoA transferase SvBAHD01 has a key role in arabinoxylan (AX) feruloylation in Se. viridis, as down-regulation of this gene resulted in a 60 % decrease in AX feruloylation in stems without affecting biomass accumulation. Notably, biomass saccharification efficiency was increased by ~40–60 %, which not only demonstrates that AX feruloylation is a promising target for reducing biomass recalcitrance, but also confirms Se. viridis as a platform to validate biotechnological strategies.
The development of diverse resources and tools for Se. viridis is rapidly advancing, although various challenges and opportunities are predicted for the Setaria community. Despite an efficient tissue culture-based transformation protocol being available, a more robust spike dip protocol is urgently needed to boost functional studies and gene discovery efforts. The use of CRISPR/Cas9 technology to generate null mutants will also greatly increase the possibilities for functional studies. The close genetic relationship between Se. italica and Se. viridis, in addition to the continuing development of new genome technologies, will probably facilitate the identification of the set of genes responsible for the phenotypic variation occurring during the domestication process. Finally, deeper knowledge on SCW biology in Se. viridis is essential to understand the molecular basis of biomass recalcitrance prior to the development of biotechnological strategies to generate optimized crops for biorefineries.
Invasive plants: introducing Phragmites australis (common reed)
Exploration and globalization have rapidly increased since the industrial revolution, and natural barriers that typically restrict species’ ranges have largely dissolved (Hulme, 2009). Many species previously confined to certain geographical regions have been introduced to non-native locations through human activity, leading to invasion events, where the introduced species establishes itself in a new habitat and outcompetes native species due to diverse ecological factors (Kolar and Lodge, 2001). Examples of such factors include a lack of natural predators, exploitation of eutrophic conditions and plant-specific characteristics, such as shoot and leaf-area allocation, fitness, growth rate and size (Keane and Crawley, 2002; van Kleunen et al., 2010; Mozdzer and Megonigal, 2012; Liu et al., 2018). Invasive species negatively impact global species’ diversity and ecosystem health and pose a global economic burden. Invasive plants cost the USA ~$35 billion annually, mainly associated with negative impacts to agricultural operations (Pimentel et al., 2005; Dogra et al., 2010). The scientific community would benefit from an established plant model to continue driving research in the field of invasion biology. The well-studied species Phragmites australis (common reed) is emerging as a promising model candidate (Meyerson et al., 2016b; Packer et al., 2017a).
Phragmites australis (Fig. 3) is a cosmopolitan perennial grass that spans all continents except Antarctica (Clevering and Lissner, 1999; Packer et al., 2017b). It is associated with widespread growth in wetland habitats, particularly in marshes and along the shores of freshwater and brackish water bodies (Chambers et al., 1999; Packer et al., 2017b). Often found growing in dense patches (so-called ‘stands’; Fig. 3), it typically propagates vegetatively through rhizome and stolon growth (Lambertini et al., 2008). Phragmites australis displays a high degree of phenotypic plasticity, a range of salt tolerances and, in some cases, the ability to grow in arid environments (Saltonstall et al., 2010; Achenbach et al., 2013; Holmes et al., 2016; Packer et al., 2017b). Sexual reproduction is facilitated through wind pollination of its inflorescences, which bear thousands of hermaphrodite florets capable of producing ~1000 seeds with long hairs to facilitate wind dispersal (McKee and Richards, 1996; Saltonstall et al., 2010). Cross-pollination by hand drastically increases seed set, which is typically quite low due to partial self-incompatibility (Ishii and Kadono, 2002; Lambert and Casagrande, 2007b; Kettenring et al., 2011). Although flowering can take several years following germination, vegetative propagation via a rhizome is a simple, rapid way to produce genetic clones (Ali et al., 2002; Saltonstall et al., 2010). Phragmites australis has been present in North America for at least 40 000 years, and during the 1800s it was an uncommon plant with distribution gaps across the continent; however, it now spans the entirety of the USA and into Canada (Hansen, 1978; Saltonstall, 2002). Anthropogenic habitat disturbance and seed dispersal are probably major promoters of the surge in population and range. Recently, chloroplast DNA sequencing has revealed that an invasive European haplotype heavily contributed to this expansion (Saltonstall, 2002; Meyerson and Cronin, 2013). This rapid, cryptic colonization has been the subject of many studies surrounding invasion biology, including phenotypic plasticity, genetic diversity, hybridization with native plants, predation and nutrient foraging, among others (Clevering and Lissner, 1999; Vretare et al., 2001; Meyerson et al., 2010; Mozdzer et al., 2010; Saltonstall et al., 2014; Allen et al., 2015).
Fig. 3.
Phragmites australis. A mature, flowering stand of Ph. australis (left) growing next to immature plants (right).
Global phylogenetic analyses using conserved chloroplast DNA sequences have identified 27 haplotypes, with 11 being native to North America (Saltonstall, 2002). Furthermore, by analysing variable nuclear markers, high rates of genetic diversity were found among the invasive European ‘M’ haplotype across North America, suggesting a series of multiple introductions from Europe (Kirk et al., 2011; Plut et al., 2011). A second European haplotype, referred to as ‘L1’, was identified among two stands in Quebec, Canada, although whether it is invasive was not confirmed (Meyerson and Cronin, 2013). As there are 14 European haplotypes, it is important to investigate the differing traits between M and other haplotypes, to understand the factors that give rise to its invasive nature.
There is no genome sequence available for Ph. australis; however, a full plastid genome is available on the NCBI website (accession PRJNA174737), and a transcriptome dataset can be found at the NCBI Sequence Read Archive (accessions SRR3233385–SRR3233398, GenBank BioProject accession PRJNA314710, Shotgun Assembly accession GEKX00000000) (Holmes et al., 2016). Transcriptomics-based studies have aimed to identify genes involved in salinity tolerance and rhizome growth, with 124 450 unique transcripts assembled and 1280 non-redundant proteins identified using mass spectrometry (He et al., 2012; Eller et al., 2014; Holmes et al., 2016). Tools have been developed to determine chloroplast haplotypes and nuclear genotypes, including chloroplast DNA markers, restriction fragment length polymorphisms, amplified fragment length polymorphisms and microsatellite DNA markers (Saltonstall 2002, 2003a, b; Lambertini et al., 2006). Denmark hosts a garden containing 188 genotypes of Ph. australis, acting as a living library of plants that can be used for physiological and genetic analyses (Lambertini et al., 2012). PhragNet is a network of individuals overseeing the management of 209 Ph. australis stands from 16 states spanning the USA and the Canadian province of Ontario, established to crowdsource ecological and genetic investigations of native and non-native haplotypes (Hunt et al., 2017). Phragmites australis can grow under standard glasshouse conditions in soil or hydroponics, with rhizome cuttings growing up to 2 m within 5 months, allowing rapid tissue production for growth assays and sampling (Ali et al., 2002; Vasquez et al., 2005). As seeds exhibit varying degrees of dormancy, protocols have been developed to increase germination efficiency, involving diurnal temperature fluctuations and high-intensity lighting (Kettenring and Whigham, 2009; Saltonstall et al., 2010). For genetic manipulations, an optimized protocol has been established to generate stable Ph. australis transformants using agrobacterium-mediated transformation of callus tissue (Kim et al., 2013). Additionally, Ph. australis has been successfully propagated by somatic embryogenesis (Lauzer et al., 2000).
The ‘large genome constraint hypothesis’ suggests that species with smaller genome sizes are more amenable to adapting a larger range of physiological traits and expanded ecological distributions, allowing them to exploit more extreme environments (Knight et al., 2005). Invasive species often have smaller genome sizes compared to non-invasive species; indeed, the genome size of the invasive European haplotype is 6.9 % smaller than native North American Ph. australis (Bennett et al., 1998; Pandit et al., 2014; Pyšek et al., 2018). The European haplotype displays traits favouring invasive species characteristics, including resistance to aphid predation, low C : N ratio, long rhizomes, and an abundance of early emerging shoots, which may be linked to its smaller genome (Pyšek et al., 2018). Phragmites australis exhibits a diversity of karyotypes, with cells containing 12 chromosomes and individuals displaying a range of euploidy and aneuploidy (Clevering and Lissner, 1999). Compared to octoploids, Ph. australis tetraploids grow taller with an increased abundance of stems, exhibit stronger chemical defence mechanisms, increased water content in leaves, and support more aphids (Meyerson et al., 2016a). By taking advantage of variations in ploidy and genome sizes, Ph. australis has been proposed as a model to study the relationship between genome size, ploidy and invasion potential, whereas these variations, coupled with long flowering periods, make it less suitable for classical genetics and more so for population genetics (Suda et al., 2015).
The ‘enemy-release hypothesis’ suggests species introduced outside of their native range are less vulnerable to predation due to lack of co-evolved natural enemies (Keane and Crawley, 2002). Native North American Ph. australis exhibits significantly higher rates of herbivory by gallflies and aphids compared to the invasive haplotype, leading to delayed flowering time, stem chlorosis and, in some cases, whole plant death (Lambert et al., 2007; Lambert and Casagrande, 2007a; Park and Blossey, 2008; Allen et al., 2015). The preference for herbivores to target native Ph. australis is interesting from a genetics perspective to investigate potential genes that may influence herbivory and defence.
The invasive European haplotype resists higher salinity levels compared to native Ph. australis, which may be linked to its invasive nature (Vasquez et al., 2005). It was shown that ploidy does not affect salinity tolerance, but rather there exists a partial correlation with geographical origin, suggesting localized adaptation (Achenbach et al., 2013). Furthermore, transcriptomics studies on salinity tolerance identified numerous differentially regulated genes, including the HIGH AFFINITY K+ TRANSPORTER (HAK/HAT) gene family expressed in salt-tolerant reed plants (Eller et al., 2014; Holmes et al., 2016). Yeast that express the Ph. australis gene PhaHAK2 or PhaHAK5 exhibit decreased potassium uptake in the presence of sodium chloride, and increased sodium permeability (Takahashi et al., 2007a, b), suggesting an importance in potassium/sodium balance for salinity tolerance.
Differences in nutrient requirements, nutrient use efficiencies and biomass allocation may give invasive species competitive advantages over native species (Kroons and Hutchings, 1995; Zedler and Kercher, 2004). As nutrients are absorbed at the root–soil interface, root morphology and root system architecture play important roles in nutrient foraging, absorption and transport (Fitter et al., 2002; Giehl and Wirén, 2014). Native North American Ph. australis develops thin, compact rhizomes with an abundance of lateral roots, whereas the invasive haplotype develops thick, long rhizomes with fewer lateral roots but increased root hair abundance (Holdredge et al., 2010). Under nutrient-limited conditions, both native and invasive Ph. australis develop the same above-ground and below-ground biomass; however, under nutrient-rich/eutrophic conditions, the invasive haplotype grows significantly faster, with a doubling of rhizome biomass and length, and significant increase in above-ground biomass (Holdredge et al., 2010). The invasive haplotype is associated with growth in soils containing higher nitrite/nitrate and ammonium, and in the native haplotypes, nitrogen assimilation is decreased at a higher rate under increasing salinity (Mozdzer et al., 2010; Hunt et al., 2017). These findings support the hypothesis that invasive Ph. australis is capable of exploiting eutrophic habitats under a wider range of environmental conditions compared to native haplotypes (Mozdzer et al., 2010). Investigations into plant nutrition between haplotypes using molecular biology approaches may elucidate important genetic determinants for classifying and predicting invasive species.
Phragmites australis has been studied in thousands of publications from physiology and ecology perspectives; however, advances in technology and techniques now facilitate molecular and genetic investigations. It will be important to sequence and assemble the genomes of native, invasive and ancestral European haplotypes to undertake broader ‘omics’ studies and to study single genes. The establishment of mutant libraries would greatly benefit the study of Ph. australis, and researchers could take advantage of natural variation among the many haplotypes. The influence of epigenetics is increasingly being investigated in the field of invasion biology, including in Ph. australis, to help understand the invasion rates of species with low genetic diversity and monoclonal growth (Prentis et al., 2008; Spens and Douhovnikoff, 2016). In North America, it was confirmed that introduced Ph. australis has interbred with native haplotypes, producing hybrids that maintain invasive traits (Meyerson et al., 2010; Williams et al., 2019). This provides another area of research into how introgression influences species invasion.
Plant parasitism: introducing Striga hermonthica (witchweed)
The other species discussed in this review are green, autotrophic plants that produce carbohydrates through photosynthesis, although parasitic plants live heterotrophically and survive from water and nutrients from host plants (Musselman, 1980). Parasites of the genus Striga (witchweed) cause annual losses of 293 000 tons (the equivalent of US$117 million) in milled rice production (Rodenburg et al., 2016). The dodder plant Cuscuta is another species that is increasingly being used to study plant parasitism (Vogel et al., 2018). While Cuscuta infects eudicotyledonous crop plants, such as sugar beet, potato and tomato, Striga targets monocotyledonous plants, including the main cereal crops, such as sorghum, millet, rice and maize; therefore, we will focus our review on this parasite (Musselman, 1980; Mishra, 2009; Vogel et al., 2018). Striga crop infestation occurs worldwide, but primarily in Africa, India, China, Indonesia and the USA (Musselman, 1980; Doggett 1987). To date, these root parasitic plants are mainly controlled by herbicide applications and by breeding host plants for resistance (Samejima and Sugimoto, 2018). However, to efficiently combat crop infestation by parasitic plants and to reduce the use of herbicides and other harmful chemicals, a better understanding of the development and physiology of parasitic plants, and especially their host plant invasion mechanisms, is urgently needed. Nevertheless, research in this area is hampered by the lack of an established model organism.
Striga’s trivial name ‘witchweed’ originated from the belief of early farmers that a Striga-infested host plant must be ‘bewitched’ when it exhibited drought-like symptoms for no apparent reason, as the Striga plant was still below ground and therefore invisible (Musselman, 1980; Runo and Kuria, 2018). The most studied Striga species are those with highest economic importance, including Striga asiatica, Striga gesneroides and Striga hermonthica, which are the focus of this chapter (Fig. 4 shows St. hermonthica) (Musselman, 1980). Recently, the life cycle and parasitic characteristics have been reviewed by Runo and Kuria (2018). Striga seeds in the soil only germinate in the presence of a potential host plant, which they sense through a germination-inducing signal secreted by host roots (Musselman, 1980). This germination stimulant has been identified as strigolactones: a group of terpenoid lactones considered phytohormones (Cook et al., 1972; Umehara et al., 2008). Germination is first visible by the outgrowth of the radicle, often referred to as the ‘germ tube’ (Musselman, 1980). A specialized organ is then formed at the tip of the radicle, known as the haustorium, which attaches to and penetrates the host root, subsequently forming a xylem connection with the host plant (Yoshida et al., 2016). After attachment, the Striga seedling grows below ground and develops its first leaves. During this time, most of the damage to the host plant occurs, resulting in symptoms that resemble drought and nutrient deficiency and ultimately cause severe stunting of the host plant (Berner et al., 1995). After emerging from the soil, Striga produces chlorophyll and begins to photosynthesize, completing its life cycle with flowering and the production of new seeds (Fig. 4) (Berner et al., 1995). Striga hermonthica exhibits several favourable characteristics of a model plant. It can be grown in growth chambers and glasshouses due to its low height of around 30 cm, and has a short life cycle of 3–4 months, consisting of 4–7 weeks below ground, 4 weeks from emergence to flowering, and 4 weeks to seed maturation. Striga hermonthica produces a high number of seeds (up to 42 000 per plant) that remain viable for over 20 years, and its attachment to host plants can be carried out in the lab in rhizotrons, as well as on agar plates (Doggett, 1987; Berner et al., 1995; Yoshida and Shirasu, 2009; Mohamed et al., 2010). Furthermore, because St. hermonthica is able to invade established plant models, such as rice, maize and sorghum, mutant and natural variation collections from these crops can be exploited to analyse potential mechanisms of resistance (Cissoko et al., 2011; Mbuvi et al., 2017). The model eudicot A. thaliana is also susceptible to St. hermonthica, but the parasite fails to invade the vessel elements; therefore, it may be used to study vessel element resistance and help to differentiate between attack and actual infection (Yoshida and Shirasu, 2009). Furthermore, as some Striga species, such as St. hermonthica, are cross-fertilizing, whereas others are self-fertilizing (e.g. St. asiatica), parasitic invasion strategies can also be studied in a context of adaptation and speciation (Safa et al., 1984).
Fig. 4.
Striga hermonthica. Mature St. hermonthica (white arrowhead) next to its host plant (grey arrowhead). Photo credit Boubacar Kountche (KAUST).
A reference genome for St. asiatica is published, and the parasitic plant genome project has generated large-scale transcriptomic datasets for St. hermonthica, providing a comprehensive developmental expression atlas (Westwood et al., 2012; Yang et al., 2015; Yoshida et al., 2019). This includes expression data for different developmental stages, during host plant attack, and from different tissues of the adult plant. Additionally, housekeeping genes for quantitative PCR experiments have been established (Fernández-Aparicio et al., 2013). These tools can aid to identify targets for putative herbicides. To genetically manipulate St. hermonthica, a virus-induced gene silencing system was established, in which Agrobacterium transformation is used to introduce a virus-based T-DNA that activates post-transcriptional gene silencing in order to reduce expression of a target gene (Kirigia et al., 2014). For Agrobacterium transformation, both leaf transformation and the Agro-drench method can be used, which involves applying the Agrobacterium solution directly onto the soil adjacent to the crown part of the 3- to 4-week-old St. hermonthica seedling (Kirigia et al., 2014).
The recent sequencing of the St. asiatica genome has contributed to understanding the evolution of parasitic plants (Yoshida et al., 2019). One of the main insights supports the hypothesis that transition from autotrophic to parasitic life includes three stages: (1) Neofunctionalization of existing genes and pathways to develop the distinct parasitic organs. Striga asiatica has undergone at least two whole-genome duplications, allowing for the recruitment of genes for new functions, where genes for lateral root development were recruited for haustorium formation, which could be specifically useful for the formation of new xylem connections. (2) The establishment of host-dependence, which goes along with a loss of gene functions involved in photosynthesis and hormone responses. (3) The establishment of a cellular transport machinery that facilitates the transport of host resources to the parasite (Yoshida et al., 2019). Next to these findings, genome sequencing has also uncovered evidence for horizontal gene transfer, specifically of retrotransposons, indicating gene flow from hosts to the parasite (Yoshida et al., 2019).
Another milestone in understanding Striga infection was the discovery of the substance exudated by host plants that induces Striga germination, which was later found to be the phytohormone strigolactone (Cook et al., 1972; Umehara et al., 2008). Genome analysis revealed that the family of strigolactone receptors is highly expanded in the Striga genome, and many of the receptors were found to be highly expressed at the seedling stage, probably to facilitate the detection of host plants (Yoshida et al., 2019). A useful tool to identify or test strigolactone receptors is the fluorescent substrate Yoshimulactone Green (YLG) (Tsuchiya et al., 2015). In several plants, including A. thaliana, rice, and petunia, α/β-hydrolase-fold enzymes have been identified as strigolactone receptors. These proteins bind strigolactones and subsequently hydrolyse them into two fragments. YLG takes advantage of this by structurally mimicking a strigolactone, but its breakdown products include one fragment that becomes fluorescent following cleavage. This visible readout can be used to further test the putative strigolactone receptors identified in the genome, which can then be targeted by blocking agents that bind to Striga but not to the host’s strigolactone receptors, thereby suppressing Striga germination (Tsuchiya et al., 2015; Shahul Hameed et al., 2018). Synthetic strigolactones can also be potentially utilized for so-called ‘suicidal germination’, in which germination stimulants are applied to the soil before planting the target crop. This causes the parasitic plant’s seeds to germinate and die due to the lack of nutrients before crops are planted (Uraguchi et al., 2018; Kountche et al., 2019). Conversely, the engineering of crops with reduced strigolactone exudation should impair Striga germination, thereby reducing infection efficiency. Indeed, mutations at the LOW GERMINATION STIMULANT 1 (LGS1) locus in sorghum caused a reduction in exudation of a highly active form of strigolactone, resulting in lower germination rates of Striga in proximity to the host plant (Gobena et al., 2017).
Besides manipulating strigolactone exudation, host plants also form mechanical barriers to block the formation of a vascular connection between host and parasite (Yoshida and Shirasu, 2009). Some plants can inhibit cell wall degradation by the parasite prior to haustorium attack on the root, while others prevent penetration by accumulating blocking substances, such as the deposition of lignin that was found in the St. hermonthica-resistant rice cultivar ‘Nipponbare’ (Mutuku et al., 2019). Finally, establishment of the vascular connection fails in various plant species, but the mechanism remains unknown (Yoshida and Shirasu, 2009).
A better understanding of the growth and development, as well as the physiology and invasion strategies, of parasitic plants would aid in developing better strategies for combating these agricultural pests to reduce yield losses. Because A. thaliana is resistant to St. hermonthica invasion, the library of established marker lines available for A. thaliana could be tested for their role in St. hermonthica resistance, including markers for developmental genes, resistance genes or genes involved in cell wall integrity sensing. Furthermore, natural variation among Arabidopsis, but also susceptible host crops, can be exploited to find accessions with enhanced or reduced tolerance, which might be correlated with changes in the genome or epigenome, and could help to get a better understanding of naturally evolved resistance. From a developmental perspective, it will be exciting to analyse the organ formation of the parasite, because it is unclear how the transition from a root-like organ to a haustorium takes place. This transition is crucial for the xylem connection to the host plant that provides water and nutrients to the parasite. Interestingly, St. hermonthica does not respond to the phytohormone absicic acid (ABA), which controls stomata closure, and is thereby able to maintain a high transpiration rate also under drought conditions, favouring its parasitic behaviour (Fujioka et al., 2019). Because other members of the Orobanchaceae are sensitive to ABA, this attribute makes Striga outstanding even among other root parasitic plants and interesting as a subject to study physiological questions.
Salt tolerance: introducing Eutrema salsugineum (salt cress)
Soil salinity, the contamination of otherwise fertile soil with salt cations, is a major problem for agriculture worldwide (Shabala, 2013). Soil salinity is now estimated to affect ~50 % of irrigated land, resulting in massive losses in agricultural production (Shabala, 2013). To combat this problem, research has focused on improving the salt tolerance of crop plants; however, most research in understanding the molecular basis of salt tolerance is conducted on the model plant A. thaliana, which is a glycophyte (meaning that it is salt-sensitive; Bressan et al., 2001). To fully understand salt tolerance, a halophyte model is needed (a plant that has already evolved salt tolerance) allowing researchers to study and learn from this plant’s adaptation to saline environments (Bressan et al., 2001). To this end, the salt cress Eutrema salsugineum (formerly Thellungiella salsuginea or Thellungiella halophila) was suggested as a new model plant (Bressan et al., 2001).
Eutrema salsugineum is thought to have originated in the Shandong province of China, from where it spread to north-east Asia, across the Bering Strait to north-west Canada, and then along the Rocky Mountains into the USA (Wang et al., 2015; German and Koch, 2017). In the lab, work has been done with plants originating from Yukon, Canada (Fig. 5A), and Shandong, China (Fig. 5B) (Koch and German, 2013). Eutrema salsugineum was identified through its ability to thrive under extreme conditions, such as drought, salinity and frost, as well as by its morphological similarity to A. thaliana (Fig. 5) (Bressan et al., 2001). It has a short life cycle of ~2–3 months, is self-fertile, produces around 4000–8000 seeds, can be efficiently transformed using the floral-dip method, and can be ethyl methanesulfonate (EMS)-mutagenized (Bressan et al., 2001; Inan et al., 2004). It also has a small genome (~260 Mb, double that of A. thaliana), consisting of seven chromosomes with an average coding sequence identity of ~92 % to A. thaliana (Bressan et al., 2001; Inan et al., 2004; Wu et al., 2012). However, in contrast to A. thaliana, E. salsugineum supposedly has an obligate vernalization requirement of ~3 weeks in order to flower, which was confirmed for the Yukon accession, whereas the Shandong accession flowered without a vernalization step (Bressan et al., 2001; Guo et al., 2012; M. Somssich et al., unpubl. data). Furthermore, E. salsugineum is able to withstand a salinity shock of up to 500 mm NaCl, whereas A. thaliana is already sensitive to 100 mm (Bressan et al., 2001; Inan et al., 2004). To do so, E. salsugineum has evolved several morpho-physiological mechanisms: stomata in E. salsugineum leaves are present at higher density when compared to those of A. thaliana, but their conductance is lower and they respond to salt stress by closing more tightly, leading to lower transpiration rates (Inan et al., 2004). The leaves are also more succulent-like, with a second layer of palisade mesophyll cells, and they are frequently shed during extreme salt stress (Inan et al., 2004). The roots develop additional layers of endodermis and cortex cells in order to restrict ion movement towards the vasculature, thereby limiting salt uptake during salt exposure (Inan et al., 2004). Curiously, germination is actually impaired in E. salsugineum when grown on high-salt medium, compared to A. thaliana, probably to delay germination during unfavourable conditions (Inan et al., 2004). In addition to this increased salt tolerance, E. salsugineum also has a higher cold tolerance, being able to survive a cold shock of −15 °C, and is also more tolerant to phosphate starvation (Inan et al., 2004; Velasco et al., 2016).
Fig. 5.
Eutrema salsugineum. Eutrema salsugineum plants of the Yukon (A) and Shandong (B) natural accessions.
Because E. salsugineum was suggested as a potential model for salt tolerance, several labs have focused on ‘omics’ approaches to characterize the plant, resulting in several datasets that are now available to the community. Two draft genomes (using Sanger and Illumina sequencing) and the chloroplast genome are available (Wu et al., 2012; Yang et al., 2013; Guo et al., 2016). Microarray and expressed sequence tag transcriptomes, and proteome datasets from non-stressed plants and plants that were exposed to cold, drought and salt stress have also been published (Wong et al., 2006; Zhang et al., 2008; Pang et al., 2010). Genome-wide characterization of miRNAs was performed using high-throughput sequencing, and genes differentially regulated after salt stress were identified (Zhang et al., 2013). Metabolomic datasets are available for control plants and plants that were exposed to osmotic stress alongside an A. thaliana metabolome for comparison (Lugan et al., 2010). Metabolomics and transcriptomics data were also generated for the Yukon accession with salt-stressed plants grown in growth chambers under a controlled environment or in their natural habitat (Guevara et al., 2012). RNA-sequencing datasets were generated for a comparative study of the Yukon and Shandong accessions (Champigny et al., 2013). Two studies describe the identification and expression analysis of aquaporin family proteins that regulate water conductivity and could be important for the salt tolerance of E. salsugineum (Qian et al., 2019; Qin et al., 2019). To identify shoot- or root-derived signals that are important for salt tolerance, grafting experiments between A. thaliana and E. salsugineum have also been successfully performed ( Y. Li et al., 2019). Finally, the methylome of E. salsugineum is also available (Bewick et al., 2016). Several of these resources, especially protocols and genome datasets, were made available early on the thellungiella.org webpage and via the plant genomics portal Phytozome.
Salt stress is a combination of ionic and osmotic stress (Lugan et al., 2010). Successful adaptation to these conditions involves four interacting basic signal perception–response systems: ion homeostasis, osmotic adjustments, injury avoidance and growth changes (Zhu, 2001). Data for E. salsugineum give some indications on how this species has adapted to such conditions. On the genetic level, several candidate genes potentially involved in salt stress adaptation were identified. Interestingly, some EMS mutants of E. salsugineum with decreased salt tolerance follow a single-locus genetic segregation pattern, indicating that individual loci can contribute significantly to salt tolerance (Inan et al., 2004). Two examples that were studied in closer detail are the LATE EMBRYOGENESIS ABUNDANT PROTEIN 1 (LEA1) and the MOLYBDENUM COFACOR SULFURASE 1 (Mcsu1) genes (Zhang et al., 2012; Zhou et al., 2015). LEA1 was upregulated under salt stress conditions, and ectopic overexpression of the E. salsugineum LEA1 gene in A. thaliana and yeast was shown to increase the salt tolerance of both organisms (Zhang et al., 2012). Similarly, overexpression of E. salsugineum Mcsu1 increased drought tolerance in transgenic alfalfa plants in an ABA-dependent manner (Zhou et al., 2015). Several genes that are known to be salt stress-associated in A. thaliana are constitutively expressed at higher levels in E. salsugineum, and are further induced under stress (Inan et al., 2004). Interestingly, when comparing the transcriptomes of Yukon E. salsugineum plants grown in their natural Yukon habitat or under controlled conditions in a growth chamber, there was a difference in both gene expression and phenotype (Guevara et al., 2012). Furthermore, there was comparatively little overlap in gene activation in response to natural occurring drought and drought treatment (Guevara et al., 2012). The transcriptomes of the Yukon and Shandong accessions grown in their natural environment did not display drastic differences; however, among the differentially regulated genes were several stress-related genes, which could help to differentiate between genes involved in salt- and cold-stress adaptation, because the latter would be required primarily in Yukon plants (Champigny et al., 2013). Concerning osmotic stress, a comparison of the metabolomes of A. thaliana and E. salsugineum did not reveal any major differences in activated pathways, but rather quantitative differences (Lugan et al., 2010). Overall, E. salsugineum seems to cope better with dehydration, for example through stabilization of the shoot to soil water gradient, or through adjustments in water solubility and polarity of their metabolites (Lugan et al., 2010). Furthermore, two proteins of the dehydrin family were implicated to be involved in cytoskeleton-stabilization during drought stress, to improve dehydration tolerance (Rahman et al., 2011).
Regarding ion homeostasis under salt stress conditions, halophytes are typically classified as either ion excluders or accumulators (Hasegawa et al., 2000); however, the ability to tightly regulate the uptake and distribution of salt ions within the plant seems to be a key attribute of halophytes (Hasegawa et al., 2000). Importantly, E. salsugineum can discriminate between sodium and potassium ions during salt stress, and has two barriers to control salt uptake: one at the root–soil interface, and another particularly strong one, at the site of xylem-loading, preventing salt entry and transport into the shoot and above-ground organs (Volkov et al., 2004; Volkov and Amtmann, 2006). At the site of xylem-loading, sodium and potassium translocation is negatively correlated in several plants, meaning loading of sodium into the xylem was paralleled by unloading of potassium (Volkov et al., 2004). This connection seems to be lost in E. salsugineum, where potassium can be translocated independently of sodium (Volkov et al. 2004). One of the main sites for sodium deposition under salt stress conditions are the old leaves of E. salsugineum, which appear to act as a salt sink (Vera-Estrella et al., 2005).
The large amount of ‘omics’ data available for E. salsugineum provide several starting points for new research projects, and the close relationship to A. thaliana should allow the use of standard molecular tools, such as fluorescent reporters. One tool that is lacking is a mutant plant collection, such as the T-DNA collections for A. thaliana, although the two draft genomes in combination with the CRISPR/Cas9 system may allow the generation and study of specific mutants. Accordingly, interesting candidate genes, identified by mining of the available ‘omics’ datasets, could be easily tested. Such candidates could then be expressed in A. thaliana to test if they can improve the salt tolerance of this glycophyte, before moving on to crop plants. However, with these large-scale datasets readily available, it appears that an integrated systems biology approach would be an especially interesting way to characterize salt tolerance on a whole system level. While manipulating individual genes can already cause specific effects, to really engineer salt-tolerant crop plants it must be assumed that the plant has to be comprehensively reprogrammed.
Comparative development: introducing Cardamine hirsuta (hairy bittercress)
Over the course of the last two decades, comparative development studies between different Brassica species have become a useful tool to uncover molecular mechanisms underlying morphological variability. While the success of A. thaliana as the main model system to research plant development is apparent, the study of developmental mechanisms governing morphological traits, such as compound leaf development, formation of multiple cortical cell layers or explosive pod shattering, cannot be performed in this species. Therefore, close relatives of A. thaliana that have evolved these distinct morphological or ecological features have been adopted as new models to allow for comparative analyses (Hay and Tsiantis, 2016). Cardamine hirsuta was among the earliest plants adopted for this reason (Fig. 6) (Hay and Tsiantis, 2016). Initially chosen to uncover the molecular mechanisms controlling leaf shape variability, and more precisely the evolution of complex leaves from simple leaves, C. hirsuta has since proven to be an interesting model for several developmental processes, thereby making it a complementary development model next to A. thaliana (Hay and Tsiantis, 2016; di Ruocco et al., 2018b). Cardamine hirsuta is endemic to Europe and North Africa, but several populations are also found on Atlantic islands and in North America, although these populations were only recently introduced (Hay et al., 2014). The genus name Cardamine is derived from the Greek ‘Kardamon’ (Nasturtium), owing to its similar taste, whereas the species name hirsuta is derived from the Latin word for ‘hairy’, due to the massive presence of trichomes and root hairs on the plant. Cardamine hirsuta is a close relative of A. thaliana, but it exhibits morphologically divergent traits from its famous relative, such as compound leaves, pod shattering, and altered root anatomy and trichome morphology. Studies on fossils estimated that the lineages of C. hirsuta and A. thaliana diverged roughly 14 million years ago, a moderately short time in terms of species divergence (Beilstein et al., 2008, 2010; Couvreur et al., 2010). Among the A. thaliana relatives, C. hirsuta stands out because it shows the important characteristics of a model system, including a small diploid genome (196 Mb) on eight chromosomes, being self-compatible, possible clonal propagation, a short life cycle of 3–4 months, and with abundant seed set (Hay et al., 2014). Furthermore, the availability of the complete genome sequence allows studies on large-scale genomic rearrangements, which have driven the evolution of specific traits (Monniaux et al., 2018). Production of transgenics is also simple, as C. hirsuta can be transformed by the Agrobacterium tumefaciens-based floral dip method, albeit with a lower efficiency (~35 %) when compared to A. thaliana (Clough and Bent, 1998; Hay et al., 2014). All of these characteristics make C. hirsuta a suitable counterpart to A. thaliana for exhaustive and unbiased parallel genetic studies of intraspecific phenotypic variability. The C. hirsuta genome has recently been sequenced and annotated, simplifying genetic analysis, genome-wide characterization studies and cloning (Hay and Tsiantis, 2006; Barkoulas et al., 2008; Gan et al., 2016). In conjunction with this, it is now possible to easily perform tissue/organ-specific RNA sequencing (Gan et al., 2016). Transcriptome data of leaf, fruit and simulated shade-treated plants are available on the C. hirsuta genome assembly website (http://chi.mpipz.mpg.de/assembly.html). The ease of genetic tractability in C. hirsuta enables agile genetic screens and gene expression analyses. Several mutant lines for genes involved in root, leaf and flower development are available, as well as fluorescent markers, such as the auxin signalling marker DR5::3XVENUS and the cortical marker CO2::3xVENUS (di Ruocco et al., 2018a). Moreover, the use of artificial miRNA or engineered nucleic molecules targeting endogenous miRNA have also been established in C. hirsuta to knock down gene activity or miRNAs, respectively (Schwab et al., 2010; Todesco et al., 2010; Rubio-Somoza et al., 2014). The relative recent divergence of A. thaliana and C. hirsuta not only allows the utilization of most molecular biology and genetics tools developed for A. thaliana, but also permits clonal analysis experiments in a comparative context. Methodologies to acquire high-resolution images of cellular organization in C. hirsuta organs have been developed for in silico analysis, cell tracking and growth quantification via specialized software such as Morphographix (Vlad et al., 2014; Barbier de Reuille et al., 2015; Kierzkowski et al., 2019). Furthermore, the availability of several different natural C. hirsuta accessions permits quantitative trait loci analyses, an important tool to study the basis of intraspecific morphological diversity (Hay et al., 2014; Cartolano et al., 2015). While there are several morphologically divergent traits separating C. hirsuta and A. thaliana, research has predominantly focused on leaf shape, pod shattering and root anatomy. While A. thaliana exhibits simple leaf morphology, C. hirsuta carries compound leaves, which develop a lamina dissected into discrete units called leaflets (Fig. 6) (Hay and Tsiantis, 2016). It was found that several C. hirsuta orthologues of meristem-specific A. thaliana genes are expressed in C. hirsuta leaves (Blein et al., 2008; Hasson et al., 2010; Rast-Somssich et al., 2015). This includes members of the Class I and II KNOX, PLETHORA and CUP SHAPED COTYLEDON gene families (Blein et al., 2008; Hasson et al., 2010; Rast-Somssich et al., 2015). Indeed, knock down of those genes in C. hirsuta leads to leaf simplification, whereas their ectopic expression in A. thaliana leaves enhances leaf complexity (Hay and Tsiantis, 2006; Blein et al., 2008; Rast-Somssich et al., 2015; Gan et al., 2016). Another fundamental regulator for compound leaf development identified in C. hirsuta is the REDUCED LEAF COMPLEXITY (RCO) transcription factor, whuch is a paralogue of the A. thaliana LATE MERISTEM IDENTITY (LMI) (Vlad et al., 2014). RCO is derived from an LMI gene duplication event in an ancestor of C. hirsuta and is conserved in all brassicas with compound leaves (Vlad et al., 2014). The RCO gene was lost in more recent species with simple leaves, such as A. thaliana (Vlad et al., 2014). LMI and RCO show complementary expression domains in A. thaliana and C. hirsuta, where LMI is expressed in terminal and lateral leaflet margins. Conversely, RCO is expressed only at the base of terminal and lateral leaflets, where it locally represses growth, thereby dissecting the leaf and allowing the leaflet to form (Vlad et al., 2014; Vuolo et al., 2018; Kierzkowski et al., 2019). It was recently shown in A. thaliana that LMI controls leaf growth via regulation of endoreduplication timing. In the future, it will be interesting to understand whether RCO represses growth at the margin of the leaflet, controlling cell endoreduplication via LMI1, or whether RCO controls other pathways to repress growth.
Fig. 6.
Cardamine hirsute. Four-week-old C. hirsuta plant. Scale bar = 1 cm.
More recently, C. hirsuta was adopted to study the genetic differences underlying seed dispersal mechanisms (Hofhuis et al., 2016). Cardamine hirsuta disperses its seeds through explosive pod shattering, a mechanism used by some angiosperm species to launch seeds far from the parent (Hofhuis et al., 2016). Using C. hirsuta as a model, it was shown that explosive pod shattering depends on the asymmetrical deposition of lignin in the secondary walls of cells in the silique’s endocarp, in combination with an increase in turgor pressure (Hofhuis et al., 2016). Rapid expansion of the exocarp cells, followed by an increase in turgor, and the inflexibility of the endocarp cells induce a coiling of the valves and launching of the seeds (Hofhuis et al., 2016).
Work on C. hirsuta has also expanded our understanding of the genetic basis underlying the differences in root anatomy (di Ruocco et al., 2018a, b). The cortex is a fundamental root tissue for plant life as its secondary growth helps plants to cope with different environmental conditions, such as wet lands or cold weather (di Ruocco et al., 2018b). The number of cortical layers can range from one to several, representing a paradigmatic example of interspecific anatomical variability (di Ruocco et al., 2018a). Cardamine hirsuta roots have two cortical layers (an outer and an inner one) whereas A. thaliana roots have only one (di Ruocco et al., 2018a, b). Comparing cortical development of A. thaliana and C. hirsuta allows for studying the basis of these anatomical differences. The cortex and endodermis of A. thaliana roots emerge through an asymmetric cell division of a stem cell daughter, called the cortex and endodermis initial (CEI) (di Ruocco et al., 2018a; di Mambro et al., 2018). This patterning mechanism is partially based on the miRNA165- and miRNA166- (miR165/6) dependent exclusion of HOMEODOMAIN LEUCINE ZIPPER III (HD-ZIPIII) transcription factor expression in the CEI, cortex and endodermis cells (Carlsbecker et al., 2010). In C. hirsuta, miR165/6 activity is confined to the cortex and endodermis, but is absent from the CEI, resulting in the CEI giving rise to a cortex cell and a cell with mixed cortex and endodermis identity, called CEM (di Ruocco et al., 2018a, b). The CEM cells undergo a second asymmetric division, producing the endodermal layer and an inner cortical cell layer (di Ruocco et al., 2018a, b). Hence, a differential distribution of miR165/6 activity underlies the variability of cortical cell layers between A. thaliana and C. hirsuta. It will now be interesting to understand how this diverse distribution of miR165/6 is generated and how HD-ZIPIIIs regulate the asymmetric cell divisions.
Cardamine hirsuta has been useful in shedding light on developmental questions that could not be answered utilizing only A. thaliana as the sole plant development model. The use of the CRISPR/Cas9 system, together with the generation of ad hoc suppressor screens, will probably allow the discovery of additional genetic networks underlying the development of species-specific morphological traits. Nowadays, several molecular mechanisms governing characteristic morphological traits of C. hirsuta are starting to be unveiled. In the future, it will be interesting to understand whether the knowledge acquired from C. hirsuta can be extrapolated to phylogenetically distant species having similar morphological traits.
Legume crops: reintroducing Pisum sativum (pea)
The broad genetic diversity within the family Fabaceae offers a wealth of material to optimize crops for the changing climate. Intraspecific (gene pool) diversity allows optimization through breeding, whereas diversity in environmental tolerances between species may help by giving options for alternative crop traits. However, compared with cereals, legumes have been largely neglected by gene technology (Considine et al., 2017). Pisum sativum (pea) is the oldest ‘model’ legume, but comparatively little investment has been made toward pea research. This is expected to change due to the recently published genome, which will bring pea into the genomic era (Kreplak et al., 2019). To some degree, this recent lack of investment has to do with pea being part of the non-model plant model group, which does not carry the typical characteristics of a good model system. Due to pea’s agricultural importance and the fact that humans have been optimizing it for centuries through breeding and research makes it more applicably relevant than traditional model plants. Pea and several other classical models became problematic to work with once the era of molecular genetics arrived, for several reasons. The pea genome is large (~4.45 Gb) and highly complex, with up to 97 % being repetitive DNA composed of transposable elements (Macas et al., 2007; Kreplak et al., 2019). This presented too great a challenge for early genome sequencing and assembly approaches for pea, and eventually resulted in the adoption of Medicago truncatula and Lotus japonica as model legume species (Barker et al., 1990; Cook, 1999; Stougaard, 2014). There has long been a battle between the two systems to be the universally accepted legume model. Work on both persists (especially for symbiosis genetics); however, they both have their practical disadvantages and have proven to be difficult plants to work with in the lab. Medicago truncatula and L. japonica do not have the century-old background of research that pea possesses, and unlike pea, are not seed-crop plants. With the advent of ‘next-generation’ techniques, such as advanced whole-genome sequencing approaches and modern cloning techniques, the problems that hampered pea research since the emergence of molecular biology and genetics in the 1980s have now been overcome (Smýkal et al., 2012; Kreplak et al., 2019). Due to these developments, Pi. sativum, one of the first plants studied by geneticists, has finally arrived in the genome era of plant science, and has become the most well-characterized legume used in plant biochemistry and physiology (Meisrimler et al., 2016).
Pea (Fig. 7) has a long history of scientific investigation that dates back to its use by Thomas Andrew Knight in the 1790s, and more famously by Gregor Johann Mendel in the 1860s in early studies of inheritance (Mendel, 1865; Shull and Fisher Stanfield, 1939). Ellis et al. (2011) nicely illustrate the molecular nature of some of Mendel’s results (also reviewed by Reid and Ross, 2011). Pea was prominent early on as a genetic biochemical model, particularly for seed embryo biology and hormonal control of plant growth, differentiation, and plant architecture, due to its predictable, well-characterized growth habit and developmental staging (Marinos, 1970; Knott, 1987; Wang and Hedley, 1991; Sauer et al., 2006; Gomez-Roldan et al., 2008; Balla et al., 2011). In more recent times, pea has proven valuable for studying morphological and developmental processes, such as flowering time control and circadian rhythms (Hecht et al., 2007; Weller and Ortega, 2015). In addition, the high agronomical relevance of nitrogen-fixation in the root nodules of legumes is an area of great interest due to the reduction in fertilizer requirement (Hirsch, 1992; Beckie and Brandt, 1997; Scharff et al., 2003). The pea diploid genome is roughly 10× larger than that of Medicago truncatula, but when discounting the repetitive DNA sequences, the exomic component of the pea genome is actually smaller than that of Medicago truncatula, with an estimated 45 000 and 62 000 genes, respectively (Macas et al., 2007; Tang et al., 2014; Sudheesh et al., 2015; Kreplak et al., 2019). The large structure of the pea flower makes for easy emasculation and crossing without the magnification aid required for A. thaliana or Medicago truncatula. Flowers remain closed, and efficient self-fertilization thus occurs without the need for a pollinator species. This also makes the flowers ideal for controlled, manual cross-pollination, as the unopened flower buds have receptive stigmas and undehisced anthers that are easy to remove. Newly opened flowers provide an abundance of brightly coloured, self-adhering pollen for crossings. Dwarf varieties can be employed in a research setting for cultivation in small cabinets and glasshouses, using only simple tying or staking to manage individuals (Ross and Reid, 1991). The pea life cycle from germination to harvest takes from 8 to 12 weeks (Mobini and Warkentin, 2016). Most common laboratory varieties are domesticated forms that have indehiscent pods, allowing fruit and seed to be left to desiccate on the plant and easily collected (Weeden et al., 2002). Pea has a significant advantage over typical lab models such as A. thaliana, or field-suitable models such as maize, as pea is suitable for growth in the field, glasshouse, growth chambers and tissue culture environments.
Fig. 7.
Pisum sativum. Pisum sativum can be easily maintained and studied in a controlled, laboratory or glasshouse setting. Simple tying and twisting of the plants as they grow allows for easy comparison of their physiology.
A fully annotated and assemble genome sequence for Pi. sativum ‘Caméor’ has been published recently, thanks to the rapid evolution of next-generation sequencing technologies, bridging the gap between classical ‘model’ plants and crop plants (Kreplak et al., 2019). A large number of pea gene-based molecular markers have been designed and a comprehensive map of key trait-associated genes in the pea genome has been constructed using molecular markers and cDNA cloned for comparative mapping studies. Kulaeva et al. (2017) have combined the molecular pea markers into one user-friendly online tool: the Pea Marker Database (PMD). With the published genome, opportunities for gene-discovery, characterization of known and unknown mutants, and genomic-assisted crop improvement are now immense. Pea seeds are amenable to EMS mutagenesis, and extensive collections of TILLING mutants of both ‘Caméor’ and ‘Terese’ Pi. sativum cultivars with phenotypic and sequence data are available through UTILLdb (Triques et al., 2007; Dalmais et al., 2008; Sharma et al., 2009). Pea transcriptomes and proteomes are published and annotated using the genomes of Medicago truncatula and other sequenced model species (Schiltz, 2004; Bourgeois et al., 2009; Franssen et al., 2011; Alves-Carvalho et al., 2015). The pea chloroplast genome has also been sequenced, which provides information that can be used for both evolutionary and transgenic applications (Magee et al., 2010). Worldwide germplasm collections provide a wealth of diverse genetic material for crop breeding and optimization, with over 6000 accessions being listed on the USDA National Plant Germplasm System (Smýkal et al., 2011; United States Department of Agriculture et al., 2019). Pea is amenable to genetic transformation using Agrobacterium-mediated transformation of different sources of initial explants, such as protoplasts, lateral cotyledonary meristems, or segments of nodes, epicotyls and embryonic axis, but like many other legumes, optimization of transformation efficiency remains a challenge due to recalcitrance to post-transformation regeneration (Puonti-Kaerlas et al., 1990; Kathen and Jacobsen, 1993; Schroeder et al., 1993; Bean et al., 1997; Grant et al., 1998; Grant and Cooper, 2003). With pea growing in interest as a favourable legume research species, more research should be invested in improving transformation as a genetic tool. Particular interest is being paid to reduce the length of the breeding cycle in pea (Mobini and Warkentin, 2016). Termed ‘speed breeding’, this research aims to overcome the longer life cycles of typical crop plants through the manipulation of growth conditions and hormonal application, and has been proven to work efficiently for pea (Watson et al., 2018).
Pea was one of the first plants to be domesticated. This brings the benefits of thousands of years of selection for favourable traits of a crop plant, which also benefits its candidature as a strong model plant (Mikić et al., 2014). For example, beneficial traits include high-yielding seed pods that all mature around the same time and do not shatter, and a predictable growth habit and determinate growth (Weeden, 2018). Pea provides biological information not accessible with other models such as A. thaliana. The well-characterized life-cycle stages and caulescent habit (cf. the rosette of A. thaliana) can make many types of physiological manipulations easier, and allows for detailed physiological measurements, such as studying shoot branching, axillary bud formation, compound leaf development and coiling of tendrils (Jaffe and Galston, 1966; Ingram et al., 1984; Knott, 1987; Beveridge, 2000; Yaxley et al., 2001). Pea produces a compound inflorescence consisting of lateral secondary inflorescences, making it an interesting plant from a floral development perspective (Ferrándiz et al., 1999). The ability to graft and extract phloem and xylem sap provides a platform to study whole-plant physiological processes, such as nutrient uptake, long-distance communication through hormones, mRNA and protein signals, and even epigenetic control (Urquhart and Joy, 1981, 1982; Lexa and Cheeseman, 1997; Beveridge et al., 1997; Kabir et al., 2013). Pea provides the benefits of researching a legume seed crop and having direct agronomic application for seed crops, without the issues of genome duplication that has occurred in soybean (Schmutz et al., 2010). Pea offers insight into the 18 000+ other legume species, many of which we rely on for food and pasture (Graham and Vance, 2003). Finally, legumes play a pivotal role in crop rotation, with the symbiotic bacteria in the nitrogen-fixing root nodules providing bio-available nitrogen, thereby minimizing fertilizer requirements and the associated cost and environmental impact (Courty et al., 2015). The wealth of historical research, combined with the recently published genome waiting to be fully utilized, means pea promises a breadth of information vital for key biological processes that have applications for yield, fruit set and low-input farming systems, thereby contributing to food security and improving sustainable agricultural practices.
OUTLOOK AND CONCLUSIONS
The new models discussed here have been around for several years and are well established. There are, however, several more plant species that have either already been established or were proposed as new models to answer even more specific scientific questions. In the final part of this work, we would like to give a brief mention to some of the fascinating plants that were not included here, mainly due to space constraints, but that could be part of a future wave of plant models. Boechera (rockcress) and Erythranthe guttata (yellow monkeyflower) allow the study of genotypic and phenotypic trait variations among natural populations, while Silene latifolia (white campion) is an interesting model to study the evolution of sexual plant systems (Bernasconi et al., 2009; Rushworth et al., 2011; Yuan, 2018). Azolla and Ceratopteris have been suggested as model ferns, as have the duckweeds Lemna minor and Spirodela polyrhiza for aquatic plant life and phytoremediation (Gupta and Prakash, 2013; Sessa et al., 2014). Capsella rubella has also been studied for some time, and is used to investigate plant reproductive biology (Guo et al., 2009). Hibiscus trionum (Venice mallow) is an interesting new model to study pollinator attraction, while Utricularia gibba (floating bladderwort) is an interesting model for the evolution of carnivorous plant life and three-dimensional plant form, as well as genome biology (Vignolini et al., 2015; Renner et al., 2018; Whitewoods et al., 2020).
There are also some very interesting recent developments regarding other non-model plant models. Similar to the case of Pi. sativum that we have described in this paper, research on Triticum aestivum has also been hampered by the enormous complexity of the plant’s hexaploid genome. On top of that, the space and time required to grow wheat over multiple generations have proven to be significantly problematic in carrying out research. The past year has seen two giant leaps taken to improve these conditions. First, the speed breeding technique has accelerated plant growth speed, thereby decreasing generation time and accelerating research (Ghosh et al., 2018; Watson et al., 2018). Additionally, the publication of the annotated wheat genome has provided the basis for full genetic and genomic work [International Wheat Genome Sequencing Consortium (IWGSC) 2018]. Of all the non-model plant models, rice is probably the most developed one to date, although a major problem that persists is the propagation of such a big plant in the confined space of a research laboratory. Publication of the ‘Xiaowei’ germplasm now aims to eliminate this issue (S. Hu et al., 2018). ‘Xiaowei’ is a dwarf mutant of the japonica and indica rice varieties, which is 30 % smaller than the wild type varieties and exhibits a shorter growth period, lower biomass and improved space utilization (S. Hu et al., 2018). As such, it should be suitable for large-scale indoor experiments before moving on to the standard rice varieties and field studies. Finally, Nature Plants has recently announced the return of the snapdragon, referring to the genus Antirrhinum, which has been a very important plant model throughout the 20th century to specifically study flower development (Schwarz-Sommer et al., 2003; Nature Plants, 2019). The recent publication of its genome might reignite interest to study Antirrhinum majus as a model for flower development and genome architecture (Schwarz-Sommer et al., 2003; M. Li et al., 2019).
In conclusion, the expansions in the set of available plant models represents a paradigm shift in plant research. The 19th and 20th centuries were mostly defined by the use of non-model plant models to study agriculturally relevant or phenotypically interesting traits. Following the adoption of A. thaliana as the primary plant model, plant science entered the era of molecular biology and genetics, in which traits could be studied at the molecular level. With the availability of new ‘omics’ tools, new plant models are added to our collection at an unprecedented speed, and old non-model plant models are, in many regards, elevated to proper model system status. With these recent developments, we will draw closer to eventually understanding plant life with all its different aspects and facets.
FUNDING
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (BIOEN Young Investigators Awards research grant 2015/02527–1 to I.C.); the SPRINT Project (2016/50189-0 to I.C.); Conselho Nacional de Desenvolvimento Científico e Tecnológico (research fellowship 302927/2018–2 to I.C.); an FIRB (Futuro In Ricerca) research grant (to R.D.I.), the Melbourne-Potsdam PhD Program, and the Natural Sciences and Engineering Research Council of Canada (to M.S.O.); the Australian Government Research Training Program (to K.L.P.); and the Deutsche Forschungsgemeinschaft (DFG) (projects 329720419 to M.I.R-S. and 344523413 to M.S.).
ACKNOWLEDGEMENTS
The authors would like to thank Staffan Persson, James L. Weller and John L. Bowman for comments on the manuscript, as well as Tom Dierschke and Boubacar Kountche for providing photographs of M. polymorpha and St. hermonthica, respectively.
LITERATURE CITED
- Acharya BR, Roy Choudhury S, Estelle AB, et al. 2017. Optimization of phenotyping assays for the model monocot Setaria viridis. Frontiers in Plant Science 8: 2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achenbach L, Eller F, Nguyen LX, Brix H. 2013. Differences in salinity tolerance of genetically distinct Phragmites australis clones. AoB PLANTS 5: 1–19. [Google Scholar]
- Albert NW, Thrimawithana AH, McGhie TK, et al. 2018. Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. The New Phytologist 218: 554–566. [DOI] [PubMed] [Google Scholar]
- Ali NA, Bernal MP, Ater M. 2002. Tolerance and bioaccumulation of copper in Phragmites australis and Zea mays. Plant and Soil 239: 103–111. [Google Scholar]
- Allen WJ, Young RE, Bhattarai GP, et al. 2015. Multitrophic enemy escape of invasive Phragmites australis and its introduced herbivores in North America. Biological Invasions 17: 3419–3432. [Google Scholar]
- Alonso-Cantabrana H, Cousins AB, Danila F, et al. 2018. Diffusion of CO2 across the mesophyll-bundle sheath cell interface in a C4 plant with genetically reduced PEP carboxylase activity. Plant Physiology 178: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff F, Kopischke S, Zobell O, et al. 2014. Comparison of the MpEF1α and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Research 23: 235–244. [DOI] [PubMed] [Google Scholar]
- Alves-Carvalho S, Aubert G, Carrère S, et al. 2015. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. The Plant Journal 84: 1–19. [DOI] [PubMed] [Google Scholar]
- Ankeny RA, Leonelli S. 2011. What’s so special about model organisms? Studies in History and Philosophy of Science 42: 313–323. [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. The Plant Journal 65: 571–577. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska A-L, Kierzkowski D, et al. 2015. MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife 4: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, Blondon F, et al. 1990. Medicago truncatula, a model plant for studying the molecular genetics of theRhizobium-legume symbiosis. Plant Molecular Biology Reporter 8: 40–49. [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. 2008. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nature Genetics 40: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Bean SJ, Gooding PS, Mullincaux PM, Davies DR. 1997. A simple system for pea transformation. Plant Cell Reports 16: 513–519. [DOI] [PubMed] [Google Scholar]
- Beckie HJ, Brandt SA. 1997. Nitrogen contribution of field pea in annual cropping systems. 1. Nitrogen residual effect. Canadian Journal of Plant Science 77: 311–322. [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. 2008. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. American Journal of Botany 95: 1307–1327. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 107: 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. 1998. DNA amounts in two samples of weeds. Annals of Botany 82: 121–134. [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria. Nature Biotechnology 30: 555–561. [DOI] [PubMed] [Google Scholar]
- Bernasconi G, Antonovics J, Biere A, et al. 2009. Silene as a model system in ecology and evolution. Heredity 103: 5–14. [DOI] [PubMed] [Google Scholar]
- Berner DK, Kling JG, Sing BB. 1995. Striga research and control - A perspective from Africa. Plant Disease 79: 652–660. [Google Scholar]
- Beveridge CA. 2000. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regulation 32: 193–203. [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. 1997. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiology 115: 1251–1258. [Google Scholar]
- Bewick AJ, Ji L, Niederhuth CE, et al. 2016. On the origin and evolutionary consequences of gene body DNA methylation. Proceedings of the National Academy of Sciences of the United States of America 113: 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, et al. 2008. A conserved molecular framework for compound leaf development. Science (New York, N.Y.) 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Borrill P. 2020. Blurring the boundaries between cereal crops and model plants. New Phytologist. doi:10.1111/nph.16229. [DOI] [PubMed] [Google Scholar]
- Bourgeois M, Jacquin F, Savois V, et al. 2009. Dissecting the proteome of pea mature seeds reveals the phenotypic plasticity of seed protein composition. Proteomics 9: 254–271. [DOI] [PubMed] [Google Scholar]
- Bowman JL. 2016. A brief history of marchantia from Greece to genomics. Plant & Cell Physiology 57: 210–229. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Araki T, Kohchi T. 2016a Marchantia: past, present and future. Plant & Cell Physiology 57: 205–209. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Briginshaw LN, Fisher TJ, Flores-Sandoval E. 2019. Something ancient and something neofunctionalized-evolution of land plant hormone signaling pathways. Current Opinion in Plant Biology 47: 64–72. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304.e15. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Sakakibara K, Furumizu C, Dierschke T. 2016b Evolution in the cycles of life. Annual Review of Genetics 50: 133–154. [DOI] [PubMed] [Google Scholar]
- Boyd RA, Gandin A, Cousins AB. 2015. Temperature response of C4 photosynthesis: biochemical analysis of rubisco, phosphoenolpyruvate carboxylase and carbonic anhydrase in Setaria viridis. Plant Physiology 169: 00586.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK. 2001. Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiology 127: 1354–1360. [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Bennetzen JL, Vogel JP. 2015. Brachypodium distachyon and Setaria viridis: model genetic systems for the grasses. Annual Review of Plant Biology 66: 465–485. [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, et al. 2010. Setaria viridis: a model for C4 photosynthesis. The Plant Cell 22: 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella P, Gogleva A, Hoey DJ, et al. 2019. Conserved biochemical defenses underpin host responses to oomycete infection in an early-divergent land plant lineage. Current Biology 29: 2282–2294. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartolano M, Pieper B, Lempe J, et al. 2015. Heterochrony underpins natural variation in Cardamine hirsuta leaf form. Proceedings of the National Academy of Sciences of the United States of America 112: 10539–10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino B, Hetherington AJ, Emms DM, Kelly S, Dolan L. 2016. The stepwise increase in the number of transcription factor families in the Precambrian predated the diversification of plants on land. Molecular Biology and Evolution 33: 2815–2819. [DOI] [PubMed] [Google Scholar]
- Chambers RM, Meyerson LA, Saltonstall K. 1999. Expansion of Phragmites australis into tidal wetlands of North America. Aquatic Botany 64: 261–273. [Google Scholar]
- Champigny MJ, Sung WW, Catana V, et al. 2013. RNA-Seq effectively monitors gene expression in Eutrema salsugineum plants growing in an extreme natural habitat and in controlled growth cabinet conditions. BMC Genomics 14: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bowman JL, Meyerowitz EM. 2016. Field guide to plant model systems. Cell 167: 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissoko M, Boisnard A, Rodenburg J, Press MC, Scholes JD. 2011. New Rice for Africa (NERICA) cultivars exhibit different levels of post-attachment resistance against the parasitic weeds Striga hermonthica and Striga asiatica. The New Phytologist 192: 952–963. [DOI] [PubMed] [Google Scholar]
- Clayton WA, Albert NW, Thrimawithana AH, et al. 2018. UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. The Plant Journal 96: 503–517. [DOI] [PubMed] [Google Scholar]
- Clevering OA, Lissner J. 1999. Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquatic Botany 64: 185–208. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Considine MJ, Siddique KHM, Foyer CH. 2017. Nature’s pulse power: legumes, food security and climate change. Journal of Experimental Botany 68: 1815–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR. 1999. Medicago truncatula–a model in the making! Current Opinion in Plant Biology 2: 301–304. [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Wall ME, et al. 1972. Germination stimulants. II. Structure of strigol - A potent seed germination stimulant for witchweed (Striga lutea Lour.). Journal of the American Chemical Society 94: 6198–6199. [Google Scholar]
- Courty PE, Smith P, Koegel S, Redecker D, Wipf D. 2015. Inorganic nitrogen uptake and transport in beneficial plant root-microbe interactions. Critical Reviews in Plant Sciences 34: 4–16. [Google Scholar]
- Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution 27: 55–71. [DOI] [PubMed] [Google Scholar]
- Cove D. 2005. The moss Physcomitrella patens. Annual Review of Genetics 39: 339–358. [DOI] [PubMed] [Google Scholar]
- Dalmais M, Schmidt J, Le Signor C, et al. 2008. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biology 9: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila FR, Quick WP, White RG, Furbank RT, von Caemmerer S. 2016. The metabolite pathway between bundle sheath and mesophyll: quantification of plasmodesmata in leaves of C3 and C4 monocots. The Plant Cell 28: 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierschke T, Flores-Sondoval E, Rast-Somssich MI, Althoff F, Zachgo S, Bowman JL. 2020. Gamete-specific expression of TALE class HD genes activates the diploid sporophyte program in Marchantia polymorpha. BioRxiv doi:10.1101/2020.04.06.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett H. 1987. Striga. A parasitic witchweed. BioEssays 7: 135–138. [Google Scholar]
- Dogra KS, Sood SK, Dobhal PK, Sharma S. 2010. Alien plant invasion and their impact on indigenous species diversity at global scale: a review. Journal of Ecology and The Natural Environment 2: 175–186. [Google Scholar]
- Draper J, Mur LA, Jenkins G, et al. 2001. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology 127: 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ishizaki K, Flores-Sandoval E, et al. 2015. Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. The Plant Cell 27: 1650–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller F, Lambertini C, Nielsen MW, Radutoiu S, Brix H. 2014. Expression of major photosynthetic and salt-resistance genes in invasive reed lineages grown under elevated CO2 and temperature. Ecology and Evolution 4: 4161–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TH, Hofer JM, Timmerman-Vaughan GM, Coyne CJ, Hellens RP. 2011. Mendel, 150 years on. Trends in Plant Science 16: 590–596. [DOI] [PubMed] [Google Scholar]
- Ermawar RA, Collins HM, Byrt CS, et al. 2015. Genetics and physiology of cell wall polysaccharides in the model C4 grass, Setaria viridis spp. BMC Plant Biology 15: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M, Huang K, Wafula EK, et al. 2013. Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Molecular Biology Reports 40: 3395–3407. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Navarro C, Gomez MD, Canas LA, Beltran JP. 1999. Flower development in Pisum sativum: from the war of the whorls to the battle of the common primordia. Developmental Genetics 25: 280–290. [DOI] [PubMed] [Google Scholar]
- Ferreira SS, Simões MS, Carvalho GG, de Lima LGA, Svartman RMA, Cesarino I. 2019. The lignin toolbox of the model grass Setaria viridis. Plant Molecular Biology 101: 235–255. [DOI] [PubMed] [Google Scholar]
- Fitter A, Williamson L, Linkohr B, Leyser O. 2002. Root system architecture determines fitness in an Arabidopsis mutant in competition for immobile phosphate ions but not for nitrate ions. Proceedings of the Royal Society of London. Series B: Biological Sciences 269: 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sandoval E, Dierschke T, Fisher TJ, Bowman JL. 2016. Efficient and inducible use of artificial MicroRNAs in Marchantia polymorpha. Plant & Cell Physiology 57: 281–290. [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL. 2015. A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLOS Genetics 11: e1005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. 2007. The ancestral developmental tool kit of land plants. International Journal of Plant Sciences 168: 1–35. [Google Scholar]
- Frangedakis E, Saint-Marcoux D, Moody LA, Rabbinowitsch E, Langdale JA. 2017. Nonreciprocal complementation of KNOX gene function in land plants. The New Phytologist 216: 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber AP. 2011. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka H, Samejima H, Suzuki H, Mizutani M, Okamoto M, Sugimoto Y. 2019. Aberrant protein phosphatase 2C leads to abscisic acid insensitivity and high transpiration in parasitic Striga. Nature Plants 5: 258–262. [DOI] [PubMed] [Google Scholar]
- Gan X, Hay A, Kwantes M, et al. 2016. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nature Plants 2: 16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German DA, Koch MA. 2017. Eutrema salsugineum (Cruciferae) new to Mexico: a surprising generic record for the flora of Middle America. PhytoKeys 76: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watson A, Gonzalez-Navarro OE, et al. 2018. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nature Protocols 13: 369512. [DOI] [PubMed] [Google Scholar]
- Giehl RFH, Wirén Von N. 2014. Root nutrient foraging. Plant physiology 166: 509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Zamarreño AM, García-Mina JM, Solano R. 2019. An evolutionarily ancient immune system governs the interactions between Pseudomonas syringae and an early-diverging land plant lineage. Current Biology 29: 2270–2281. [DOI] [PubMed] [Google Scholar]
- Gobena D, Shimels M, Rich PJ, et al. 2017. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proceedings of the National Academy of Sciences of the United States of America 114: 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP. 2003. Legumes: importance and constraints to greater use. Plant Physiology 131: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JE, Cooper PA. 2003. Genetic transformation in pea. In: Jaiwal PK, Singh RP, eds. Applied Genetics of leguminosae biotechnology. Focus on biotechnology, Vol. 10B. Dordrecht: Springer, 23–34. [Google Scholar]
- Grant JE, Cooper PA, Gilpin BJ, et al. 1998. Kanamycin is effective for selecting transformed peas. Plant Science 139: 159–164. [Google Scholar]
- Guevara DR, Champigny MJ, Tattersall A, et al. 2012. Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biology 12: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Bechsgaard JS, Slotte T, et al. 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proceedings of the National Academy of Sciences of the United States of America 106: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Hao G, Ma T. 2016. The complete chloroplast genome of salt cress (Eutrema salsugineum). Mitochondrial DNA 27: 2862–2863. [DOI] [PubMed] [Google Scholar]
- Guo Y, Wang D, Jia W, Song J, Yang J, Wang B. 2012. Effects of seed vernalisation and photoperiod on flowering induction in the halophyte Thellungiella halophila. Australian Journal of Botany 60: 743. [Google Scholar]
- Gupta C, Prakash D. 2013. Duckweed: an effective tool for phyto-remediation. Toxicological & Environmental Chemistry 95: 1256–1266. [Google Scholar]
- Han GZ. 2017. Evolution of jasmonate biosynthesis and signaling mechanisms. Journal of Experimental Botany 68: 1323–1331. [DOI] [PubMed] [Google Scholar]
- Hansen RM. 1978. Shasta ground sloth food habits, Rampart Cave, Arizona. Paleobiology 4: 302–319. [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51: 463–499. [DOI] [PubMed] [Google Scholar]
- Hasson A, Blein T, Laufs P. 2010. Leaving the meristem behind: the genetic and molecular control of leaf patterning and morphogenesis. Comptes Rendus Biologies 333: 350–360. [DOI] [PubMed] [Google Scholar]
- Hay AS, Pieper B, Cooke E, et al. 2014. Cardamine hirsuta: a versatile genetic system for comparative studies. The Plant Journal 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nature Genetics 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2016. Cardamine hirsuta: a comparative view. Current Opinion in Genetics & Development 39: 1–7. [DOI] [PubMed] [Google Scholar]
- He R, Kim MJ, Nelson W, et al. 2012. Next-generation sequencing-based transcriptomic and proteomic analysis of the common reed, Phragmites australis (Poaceae), reveals genes involved in invasiveness and rhizome specificity. American Journal of Botany 99: 232–247. [DOI] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, et al. 2007. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiology 144: 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM. 1992. Developmental biology of legume nodulation. New Phytologist 122: 211–237. [DOI] [PubMed] [Google Scholar]
- Hiwatashi T, Quan KL, Yasui Y, et al. 2018. The RopGEF KARAPPO is essential for the initiation of vegetative reproduction in Marchantia. bioRxiv doi:10.1101/385682. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Moulton D, Lessinnes T, et al. 2016. morphomechanical innovation drives explosive seed dispersal. Cell 166: 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdredge C, Bertness MD, Von Wettberg E, Silliman BR. 2010. Nutrient enrichment enhances hidden differences in phenotype to drive a cryptic plant invasion. Oikos 119: 1776–1784. [Google Scholar]
- Holmes GD, Hall NE, Gendall AR, Boon PI, James EA. 2016. Using transcriptomics to identify differential gene expression in response to salinity among Australian Phragmites australis clones. Frontiers in Plant Science 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Katz A, Pereman I, Decker EL, Ohad N, Reski R. 2016. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nature Plants 2: 15209. [DOI] [PubMed] [Google Scholar]
- Hu S, Hu X, Hu J, et al. 2018. Xiaowei, a new rice germplasm for large-scale indoor research. Molecular Plant 11: 1418–1420. [DOI] [PubMed] [Google Scholar]
- Hu H, Mauro-Herrera M, Doust AN. 2018. Domestication and improvement in the model C4 grass, Setaria. Frontiers in Plant Science 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Brutnell TP. 2016. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Current Opinion in Plant Biology 31: 91–99. [DOI] [PubMed] [Google Scholar]
- Huang P, Shyu C, Coelho CP, Cao Y, Brutnell TP. 2016. Setaria viridis as a model system to advance millet genetics and genomics. Frontiers in Plant Science 7: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme PE. 2009. Trade, transport and trouble: managing invasive species pathways in an era of globalization. Journal of Applied Ecology 46: 10–18. [Google Scholar]
- Hunt VM, Fant JB, Steger L, et al. 2017. PhragNet: crowdsourcing to investigate ecology and management of invasive Phragmites australis (common reed) in North America. Wetlands Ecology and Management 25: 607–618. [Google Scholar]
- Inan G, Zhang Q, Li P, et al. 2004. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiology 135: 1718–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, Murfet IC, Gaskin P, Willis CL, Macmillan J. 1984. Internode length in Pisum. Planta 160: 455–463. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Ishii J, Kadono Y. 2002. Factors influencing seed production of Phragmites australis. Aquatic Botany 72: 129–141. [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. 2008. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant & Cell Physiology 49: 1084–1091. [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Johzuka-Hisatomi Y, Ishida S, Iida S, Kohchi T. 2013a Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Scientific Reports 3: 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Mizutani M, Shimamura M, Masuda A, Nishihama R, Kohchi T. 2013b Essential role of the E3 ubiquitin ligase nopperabo1 in schizogenous intercellular space formation in the liverwort Marchantia polymorpha. The Plant Cell 25: 4075–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Nishihama R, Ueda M, et al. 2015. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One 10: e0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Nishihama R, Yamato KT, Kohchi T. 2016. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant & Cell Physiology 57: 262–270. [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T. 2012. Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. Journal of Plant Research 125: 643–651. [DOI] [PubMed] [Google Scholar]
- Jaffe MJ, Galston AW. 1966. Physiological studies on pea tendrils. I. Growth and coiling following mechanical stimulation. Plant Physiology 41: 1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ. 2007. Populus: a model system for plant biology. Annual Review of Plant Biology 58: 435–458. [DOI] [PubMed] [Google Scholar]
- Jiang H, Barbier H, Brutnell T. 2013. Methods for performing crosses in Setaria viridis, a new model system for the grasses. Journal of Visualized Experiments doi:10.3791/50527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir AH, Paltridge NG, Roessner U, Stangoulis JC. 2013. Mechanisms associated with Fe-deficiency tolerance and signaling in shoots of Pisum sativum. Physiologia Plantarum 147: 381–395. [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Yamaoka S, Yamato KT, et al. 2003. Functional analysis of a beta-ketoacyl-CoA synthase gene, MpFAE2, by gene silencing in the liverwort Marchantia polymorpha L. Bioscience, Biotechnology, and Biochemistry 67: 605–612. [DOI] [PubMed] [Google Scholar]
- de Kathen A, Jacobsen HJ. 1993. Transformation in pea (Pisum sativum L.) Bajaj YPS, ed. Plant Protoplasts and Genetic Engineering IV. Biotechnology in agriculture and forestry, Vol. 23. Berlin, Heidelberg: Springer, 331–347. [Google Scholar]
- Kato H, Ishizaki K, Kouno M, et al. 2015. Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genetics 11: e1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane RM, Crawley MJ. 2002. Exotic plant invasions and the enemy release hypothesis. Trends in Ecology & Evolution 17: 164–170. [Google Scholar]
- Kettenring KM, McCormick MK, Baron HM, Whigham DF. 2011. Mechanisms of Phragmites australis invasion: feedbacks among genetic diversity, nutrients, and sexual reproduction. Journal of Applied Ecology 48: 1305–1313. [Google Scholar]
- Kettenring KM, Whigham DF. 2009. Seed viability and seed dormancy of non-native Phragmites australis in suburbanized and forested watersheds of the Chesapeake Bay, USA. Aquatic Botany 91: 199–204. [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, et al. 2019. A growth-based framework for leaf shape development and diversity. Cell 177: 1405–1418.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-G, Sharmin SA, Alam I, et al. 2013. Agrobacterium-mediated transformation of reed (Phragmites communis Trinius) using mature seed-derived calli. GCB Bioenergy 5: 73–80. [Google Scholar]
- Kirigia D, Runo S, Alakonya A. 2014. A virus-induced gene silencing (VIGS) system for functional genomics in the parasitic plant Striga hermonthica. Plant Methods 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk H, Paul J, Straka J, Freeland JR. 2011. Long-distance dispersal and high genetic diversity are implicated in the invasive spread of the common reed, Phragmites australis (Poaceae), in northeastern North America. American Journal of Botany 98: 1180–1190. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Weber E, Fischer M. 2010. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecology Letters 13: 235–245. [DOI] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott CM. 1987. A key for stages of development of the pea (Pisum sativum). Annals of Applied Biology 111: 233–245. [Google Scholar]
- Koch MA, German DA. 2013. Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Frontiers in Plant Science 4: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koi S, Hisanaga T, Sato K, et al. 2016. An evolutionarily conserved plant RKD factor controls germ cell differentiation. Current Biology: CB 26: 1775–1781. [DOI] [PubMed] [Google Scholar]
- Kolar CS, Lodge DM. 2001. Progress in invasion biology: predicting invaders. Trends in Ecology & Evolution 16: 199–204. [DOI] [PubMed] [Google Scholar]
- Komatsu A, Terai M, Ishizaki K, et al. 2014. Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort Marchantia polymorpha. Plant Physiology 166: 411–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Meinke D. 2010. The development of Arabidopsis as a model plant. The Plant Journal 61: 909–921. [DOI] [PubMed] [Google Scholar]
- Kountche BA, Jamil M, Yonli D, et al. 2019. Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub‐Saharan Africa. Plants, People, Planet 1: 107–118. [Google Scholar]
- Kreplak J, Madoui MA, Cápal P, et al. 2019. A reference genome for pea provides insight into legume genome evolution. Nature Genetics 51: 1411–1422. [DOI] [PubMed] [Google Scholar]
- Kroons de H, Hutchings MJ. 1995. Morphological plasticity in clonal plants: the foraging concept reconsidered. Society 83: 143–152. [Google Scholar]
- Kubota A, Kita S, Ishizaki K, Nishihama R, Yamato KT, Kohchi T. 2014. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nature Communications 5: 3668. [DOI] [PubMed] [Google Scholar]
- Kulaeva OA, Zhernakov AI, Afonin AM, et al. 2017. Pea Marker Database (PMD) - A new online database combining known pea (Pisum sativum L.) gene-based markers. PLoS One 12: e0186713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AM, Casagrande RA. 2007a Susceptibility of native and non-native common reed to the non-native mealy plum aphid (Homoptera: Aphididae) in North America. Environmental Entomology 36: 451–457. [DOI] [PubMed] [Google Scholar]
- Lambert AM, Casagrande RA. 2007b Characteristics of a successful estuarine invader: evidence of self-compatibility in native and non-native lineages of Phragmites australis. Marine Ecology Progress Series 337: 299–301. [Google Scholar]
- Lambert AM, Winiarski K, Casagrande RA. 2007. Distribution and impact of exotic gall flies (Lipara sp.) on native and exotic Phragmites australis. Aquatic Botany 86: 163–170. [Google Scholar]
- Lambertini C, Gustafsson MHG, Frydenberg J, Lissner J, Speranza M, Brix H. 2006. A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Systematics and Evolution 258: 161–182. [Google Scholar]
- Lambertini C, Gustafsson MHG, Frydenberg J, Speranza M, Brix H. 2008. Genetic diversity patterns in Phragmites australis at the population, regional and continental scales. Aquatic Botany 88: 160–170. [Google Scholar]
- Lambertini C, Sorrell BK, Riis T, Olesen B, Brix H. 2012. Exploring the borders of European Phragmites within a cosmopolitan genus. Aob PLANTS 2012: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Flores-Sandoval E, Tan QW, Mutwil M, Bowman JL, Persson S. 2019. Cellulose synthesis – central components and their evolutionary relationships. Trends in Plant Science 24: 402–412. [DOI] [PubMed] [Google Scholar]
- Lauzer D, Dallaire S, Vincent G. 2000. In vitro propagation of reed grass by somatic embryogenesis. Plant Cell, Tissue and Organ Culture 60: 229–234. [Google Scholar]
- Lee JH, Lin H, Joo S, Goodenough U. 2008. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840. [DOI] [PubMed] [Google Scholar]
- Lexa M, Cheeseman JM. 1997. Growth and nitrogen relations in reciprocal grafts of wild-type and nitrate reductase-deficient mutants of pea (Pisum sativum L. var. Juneau). Journal of Experimental Botany 48: 1241–1250. [Google Scholar]
- Li P, Brutnell TP. 2011. Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. Journal of Experimental Botany 62: 3031–3037. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun W, Liu F, et al. 2019. Methods for grafting Arabidopsis thaliana and Eutrema salsugineum. Plant Methods 15: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang D, Gao Q, et al. 2019. Genome structure and evolution of Antirrhinum majus L. Nature Plants: 443515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Lu CW, Shen BN, et al. 2016. Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-Seq and degradome analyses. Plant & Cell Physiology 57: 339–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yang YB, Zhu ZH. 2018. Elevated nitrogen allows the weak invasive plant Galinsoga quadriradiata to become more vigorous with respect to inter-specific competition. Scientific Reports 8: 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugan R, Niogret MF, Leport L, et al. 2010. Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. The Plant Journal 64: 215–229. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilová A. 2007. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee AM, Aspinall S, Rice DW, et al. 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Research 20: 1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Mambro R, Sabatini S, Dello Ioio R. 2018. Patterning the axes: a lesson from the root. Plants 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinos NG. 1970. Embryogenesis of the pea (Pisum sativum). Protoplasma 71: 227–233. [Google Scholar]
- Marriott PE, Gómez LD, McQueen-Mason SJ. 2016. Unlocking the potential of lignocellulosic biomass through plant science. The New Phytologist 209: 1366–1381. [DOI] [PubMed] [Google Scholar]
- Martins PK, Mafra V, de Souza WR, et al. 2016. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Scientific Reports 6: 28348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins PK, Nakayama TJ, Ribeiro AP, et al. 2015. a Setaria viridis floral-dip: a simple and rapid Agrobacterium-mediated transformation method. Biotechnology Reports (Amsterdam, Netherlands) 6: 61–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins PK, Ribeiro AP, Cunha BADBD, Kobayashi AK, Molinari HBC. 2015b A simple and highly efficient Agrobacterium-mediated transformation protocol for Setaria viridis. Biotechnology Reports (Amsterdam, Netherlands) 6: 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Iwakawa H, Hyon G-S, et al. 2019. Isolation of natural fungal pathogens from Marchantia polymorpha reveals antagonism between salicylic acid and jasmonate during liverwort-fungus interactions. Plant and Cell Physiology 0: 1–11. [DOI] [PubMed] [Google Scholar]
- Mbuvi DA, Masiga CW, Kuria E, et al. 2017. Novel sources of Witchweed (Striga) resistance from wild sorghum accessions. Frontiers in Plant Science 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKEE J, Richards AJ. 1996. Variation in seed production and germinability in common reed (Phragmites australis) in Britain and France with respect to climate. The New Phytologist 133: 233–243. [DOI] [PubMed] [Google Scholar]
- Meisrimler CN, Wienkoop S, Lyon D, Geilfus CM, Lüthje S. 2016. Long-term iron deficiency: Tracing changes in the proteome of different pea (Pisum sativum L.) cultivars. Journal of Proteomics 140: 13–23. [DOI] [PubMed] [Google Scholar]
- Mendel G. 1865. Experiments in plant hybridization. Verhandlungen des naturforschenden Vereines in Brünn 4: 3–47. [Google Scholar]
- Meyerson LA, Cronin JT. 2013. Evidence for multiple introductions of Phragmites australis to North America: detection of a new non-native haplotype. Biological Invasions 15: 2605–2608. [Google Scholar]
- Meyerson LA, Cronin JT, Bhattarai GP, et al. 2016. a Do ploidy level and nuclear genome size and latitude of origin modify the expression of Phragmites australis traits and interactions with herbivores? Biological Invasions 18: 2531–2549. [Google Scholar]
- Meyerson LA, Cronin JT, Pyšek P. 2016b Phragmites australis as a model organism for studying plant invasions. Biological Invasions 18: 2421–2431. [Google Scholar]
- Meyerson LA, Viola D V., Brown RN. 2010. Hybridization of invasive Phragmites australis with a native subspecies in North America. Biological Invasions 12: 103–111. [Google Scholar]
- Mikić A, Medović A, Jovanović Ž, Stanisavljević N. 2014. Integrating archaeobotany, paleogenetics and historical linguistics may cast more light onto crop domestication: the case of pea (Pisum sativum). Genetic Resources and Crop Evolution 61: 887–892. [Google Scholar]
- Miller MW, Garber ED, Voth PD. 1962. Biosynthetic pathways in nutritionally deficient mutants of Marchantia polymorpha L. Nature 195: 1220–1221. [Google Scholar]
- Mishler BD, Churchill SP. 1984. A cladistic approach to the phylogeny of the ‘Bryophytes’. Brittonia 36: 406. [Google Scholar]
- Mishra JS. 2009. Biology and management of Cuscuta species. Indian Journal of Weed Sciences 41: 1–11. [Google Scholar]
- Mobini SH, Warkentin TD. 2016. A simple and efficient method of in vivo rapid generation technology in pea (Pisum sativum L.). In Vitro Cellular & Developmental Biology - Plant 52: 530–536. [Google Scholar]
- Mohamed AH, Housley TL, Ejeta G. 2010. An in vitro technique for studying specific Striga resistance mechanisms in sorghum. African Journal of Agricultural Research 5: 1868–1875. [Google Scholar]
- Monniaux M, Pieper B, McKim SM, et al. 2018. The role of APETALA1 in petal number robustness. eLife 7: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte I, Ishida S, Zamarreño AM, et al. 2018. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nature Chemical Biology 14: 480–488. [DOI] [PubMed] [Google Scholar]
- Monte-Bello CC, Araujo EF, Martins MCM, et al. 2018. A flexible low cost hydroponic system for assessing plant responses to small molecules in sterile conditions. Journal of Visualized Experiments 138 (2018 08 25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdzer TJ, Megonigal JP. 2012. Jack-and-master trait responses to elevated CO2 and N: a comparison of native and introduced Phragmites australis. PLoS One 7: e42794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdzer TJ, Zieman JC, McGlathery KJ. 2010. Nitrogen uptake by native and invasive temperate coastal macrophytes: importance of dissolved organic nitrogen. Estuaries and Coasts 33: 784–797. [Google Scholar]
- Musselman LJ. 1980. The biology of Striga, Orobanche, and other root-parasitic weeds. Annual Review of Phytopathology 18: 463–489. [Google Scholar]
- Mutuku JM, Cui S, Hori C, et al. 2019. The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiology 179: 1796–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nature Plants 2019. Return of the snapdragon. Nature Plants 5: 121–121. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Ishida S, Urawa H, Kamei Y, Kohchi T. 2016. Conditional gene expression/deletion systems for marchantia polymorpha using its own heat-shock promoter and Cre/loxP-Mediated site-specific recombination. Plant & Cell Physiology 57: 271–280. [DOI] [PubMed] [Google Scholar]
- Okada S, Fujisawa M, Sone T, et al. 2000. Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. The Plant Journal 24: 421–428. [DOI] [PubMed] [Google Scholar]
- Osborn HL, Alonso-Cantabrana H, Sharwood RE, et al. 2017. Effects of reduced carbonic anhydrase activity on CO2 assimilation rates in Setaria viridis: a transgenic analysis. Journal of Experimental Botany 68: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer JG, Meyerson LA, Richardson DM, et al. 2017. a Global networks for invasion science: benefits, challenges and guidelines. Biological Invasions 19: 1081–1096. [Google Scholar]
- Packer JG, Meyerson LA, Skálová H, Pyšek P, Kueffer C. 2017b Biological Flora of the British Isles: Phragmites australis. Journal of Ecology 105: 1123–1162. [Google Scholar]
- Pandit MK, White SM, Pocock MJ. 2014. The contrasting effects of genome size, chromosome number and ploidy level on plant invasiveness: a global analysis. The New Phytologist 203: 697–703. [DOI] [PubMed] [Google Scholar]
- Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X. 2010. Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. Journal of Proteome Research 9: 2584–2599. [DOI] [PubMed] [Google Scholar]
- Park MG, Blossey B. 2008. Importance of plant traits and herbivory for invasiveness of Phragmites australis (Poaceae). American Journal of Botany 95: 1557–1568. [DOI] [PubMed] [Google Scholar]
- Petti C, Shearer A, Tateno M, et al. 2013. Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and Panicoid crop species. Frontiers in Plant Science 4: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics 52: 273–288. [Google Scholar]
- Plut K, Paul J, Ciotir C, Major M, Freeland JR. 2011. Origin of non-native Phragmites australis in North America, a common wetland invader. Fundamental and Applied Limnology 179: 121–129. [Google Scholar]
- Prentis PJ, Wilson JR, Dormontt EE, Richardson DM, Lowe AJ. 2008. Adaptive evolution in invasive species. Trends in Plant Science 13: 288–294. [DOI] [PubMed] [Google Scholar]
- Provart NJ, Alonso J, Assmann SM, et al. 2016. 50 years of Arabidopsis research: highlights and future directions. The New Phytologist 209: 921–944. [DOI] [PubMed] [Google Scholar]
- Puonti-Kaerlas J, Eriksson T, Engström P. 1990. Production of transgenic pea (Pisum sativum L.) plants by Agrobacterium tumefaciens-mediated gene transfer. Theoretical and Applied Genetics 80: 246–252. [DOI] [PubMed] [Google Scholar]
- Pyšek P, Skálová H, Čuda J, et al. 2018. Small genome separates native and invasive populations in an ecologically important cosmopolitan grass. Ecology 99: 79–90. [DOI] [PubMed] [Google Scholar]
- Qian W, Yang X, Li J, Luo R, Yan X, Pang Q. 2019. Genome-wide characterization and expression analysis of aquaporins in salt cress (Eutrema salsugineum). PeerJ 7: e7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Liu Y, Han Y, et al. 2019. Aquaporins and their function in root water transport under salt stress conditions in Eutrema salsugineum. Plant Science 287: 110199. [DOI] [PubMed] [Google Scholar]
- Rahman LN, Smith GS, Bamm VV, et al. 2011. Phosphorylation of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 facilitates cation-induced conformational changes and actin assembly. Biochemistry 50: 9587–9604. [DOI] [PubMed] [Google Scholar]
- Rast-Somssich MI, Broholm S, Jenkins H, et al. 2015. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes & Development 29: 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Ross JJ. 2011. Mendel’s genes: toward a full molecular characterization. Genetics 189: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner T, Lan T, Farr KM, et al. 2018. Carnivorous plant genomes. Oxford: Oxford University Press. [Google Scholar]
- Rodenburg J, Demont M, Zwart SJ, Bastiaans L. 2016. Parasitic weed incidence and related economic losses in rice in Africa. Agriculture, Ecosystems & Environment 235: 306–317. [Google Scholar]
- Ross JJ, Reid JB. 1991. Internode length in Pisum: le5839 is a less severe allele than Mendel’s le. Pisum Genetics 23: 29–34. [Google Scholar]
- Rowan BA, Weigel D, Koenig D. 2011. Developmental genetics and new sequencing technologies: the rise of nonmodel organisms. Developmental Cell 21: 65–76. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, et al. 2014. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Current Biology: CB 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Runo S, Kuria EK. 2018. Habits of a highly successful cereal killer, Striga. PLOS Pathogens 14: e1006731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Ruocco G, Bertolotti G, Pacifici E, et al. 2018. Differential spatial distribution of miR165/6 determines variability in plant root anatomy. Development 145: dev153858. [DOI] [PubMed] [Google Scholar]
- di Ruocco G, di Mambro R, Dello Ioio R. 2018. Building the differences: a case for the ground tissue patterning in plants. Proceedings of the Royal Society B: Biological Sciences 285: 20181746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth CA, Song BH, Lee CR, Mitchell-Olds T. 2011. Boechera, a model system for ecological genomics. Molecular Ecology 20: 4843–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa SB, Jones BMG, Musselman LJ. 1984. Mechanisms favouring outbreeding in Striga hermonthica (Scrophulariaceae). New Phytologist 96: 299–305. [Google Scholar]
- Saltonstall K. 2002. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences of the United States of America 99: 2445–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K. 2003a A rapid method for identifying the origin of North American Phragmites populations using RFLP analysis. Wetlands 23: 1043–1047. [Google Scholar]
- Saltonstall K. 2003b Microsatellite variation within and among North American lineages of Phragmites australis. Molecular Ecology 12: 1689–1702. [DOI] [PubMed] [Google Scholar]
- Saltonstall K, Castillo HE, Blossey B. 2014. Confirmed field hybridization of native and introduced Phragmites australis (Poaceae) in North America. American Journal of Botany 101: 211–215. [DOI] [PubMed] [Google Scholar]
- Saltonstall K, Lambert A, Meyerson LA. 2010. Genetics and reproduction of common (Phragmites australis) and giant reed (Arundo donax). Invasive Plant Science and Management 3: 495–505. [Google Scholar]
- Samejima H, Sugimoto Y. 2018. Recent research progress in combatting root parasitic weeds. Biotechnology & Biotechnological Equipment 32: 221–240. [Google Scholar]
- Sauer M, Balla J, Luschnig C, et al. 2006. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes & Development 20: 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauret-Güeto S, Frangedakis E, Silvestri L, et al. 2020. Systematic tools for reprogramming plant gene expression in a simple model, Marchantia polymorpha. ACS Synthetic Biology 9: 864–882. [DOI] [PubMed] [Google Scholar]
- Scharff AM, Egsgaard H, Hansen PE, Rosendahl L. 2003. Exploring symbiotic nitrogen fixation and assimilation in pea root nodules by in vivo 15N nuclear magnetic resonance spectroscopy and liquid chromatography-mass spectrometry. Plant Physiology 131: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz S. 2004. Proteome reference maps of vegetative tissues in pea. An investigation of nitrogen mobilization from leaves during seed filling. Plant Physiology 135: 2241–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW, Giraldo-Fonseca A, Rövekamp M, Smetanin D, Bowman JL, Grossniklaus U. 2018. Extensive epigenetic reprogramming during the life cycle of Marchantia polymorpha. Genome Biology 19: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schroeder HE, Schotz AH, Wardley-Richardson T, Spencer D, Higgins T. 1993. Transformation and regeneration of two cultivars of pea (Pisum sativum L.). Plant Physiology 101: 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler ML, Mantegazza O, Weber AP. 2016. Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. The Plant Journal 87: 51–65. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Warthmann N, Weigel D. 2010. Directed gene silencing with artificial MicroRNAs. In: Meyers B, Green P, eds. Methods in Molecular Biology, Vol. 592. Totowa, NJ: Humana Press, 71–88. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Davies B, Hudson A. 2003. An everlasting pioneer: the story of Antirrhinum research. Nature Reviews Genetics 4: 655–664. [DOI] [PubMed] [Google Scholar]
- Sessa EB, Banks JA, Barker MS, et al. 2014. Between two fern genomes. Gigascience 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. 2013. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of Botany 112: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahul Hameed U, Haider I, Jamil M, et al. 2018. Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Reports 19: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Plaha P, Rathour R, Katoch V, Singh Y, Khalsa GS. 2009. Induced mutagenesis for improvement of garden pea. International Journal of Vegetable Science 16: 60–72. [Google Scholar]
- Shimamura M. 2016. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant & Cell Physiology 57: 230–256. [DOI] [PubMed] [Google Scholar]
- Shull CA, Fisher Stanfield J. 1939. Thomas Andrew Knight - in memoriam. Plant Physiology 14: 1–8. [Google Scholar]
- Simões MS, Carvalho GG, Ferreira SS, Hernandes-Lopes J, de Setta N, Cesarino I. 2020. Genome-wide characterization of the laccase gene family in Setaria viridis reveals members potentially involved in lignification. Planta 251: 46. [DOI] [PubMed] [Google Scholar]
- Smýkal P, Aubert G, Burstin J, et al. 2012. Pea (Pisum sativum L.) in the genomic era. Agronomy 2: 74–115. [Google Scholar]
- Smýkal P, Kenicer G, Flavell AJ, et al. 2011. Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genetic Resources 9: 4–18. [Google Scholar]
- Somerville C, Koornneef M. 2002. A fortunate choice: the history of Arabidopsis as a model plant. Nature Reviews. Genetics 3: 883–889. [DOI] [PubMed] [Google Scholar]
- Somssich M. 2018. A short history of Arabidopsis thaliana (L.) Heynh. Columbia-0. PeerJ Preprints e26931v3: 1–7. [Google Scholar]
- Souza de WR, Martins PK, Freeman J, et al. 2018. Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytologist 218: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spens AE, Douhovnikoff V. 2016. Epigenetic variation within Phragmites australis among lineages, genotypes, and ramets. Biological Invasions 18: 2457–2462. [Google Scholar]
- Stougaard J. 2014. Background and history of the Lotus japonicus model legume system. In: Tabata S, Stougaard J, eds. The Lotus japonicus Genome. Compendium of plant Genomes. Berlin, Heidelberg: Springer, 3–8. [Google Scholar]
- Suda J, Meyerson LA, Leitch IJ, Pyšek P. 2015. The hidden side of plant invasions: the role of genome size. The New Phytologist 205: 994–1007. [DOI] [PubMed] [Google Scholar]
- Sudheesh S, Sawbridge TI, Cogan NO, Kennedy P, Forster JW, Kaur S. 2015. De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genomics 16: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Nishihama R, Shirakawa M, et al. 2018. Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS One 13: e0205117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shirakawa M, Takagi J, et al. 2014. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant & Cell Physiology 55: 475–481. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Nishio T, Ichizen N, Takano T. 2007. Cloning and functional analysis of the K+ transporter, PhaHAK2, from salt-sensitive and salt-tolerant reed plants. Biotechnology Letters 29: 501–506. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Nishio T, Ichizen N, Takano T. 2007. High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Reports 26: 1673–1679. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Yamaoka S, Hanajiri T, et al. 2000. Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L. Transgenic Research 9: 179–185. [DOI] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, et al. 2014. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. 2002. Efficient gene targeting by homologous recombination in rice. Nature Biotechnology 20: 1030–1034. [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. 2010. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genetics 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Adachi T, Harada D, et al. 2018. Cryopreservation of Marchantia polymorpha spermatozoa. Journal of Plant Research 131: 1047–1054. [DOI] [PubMed] [Google Scholar]
- Triques K, Sturbois B, Gallais S, et al. 2007. Characterization of Arabidopsis thaliana mismatch specific endonucleases: application to mutation discovery by TILLING in pea. The Plant Journal 51: 1116–1125. [DOI] [PubMed] [Google Scholar]
- Tsuboyama S, Kodama Y. 2014. AgarTrap: a simplified Agrobacterium-mediated transformation method for sporelings of the liverwort Marchantia polymorpha L. Plant & Cell Physiology 55: 229–236. [DOI] [PubMed] [Google Scholar]
- Tsuboyama-Tanaka S, Kodama Y. 2015. AgarTrap-mediated genetic transformation using intact gemmae/gemmalings of the liverwort Marchantia polymorpha L. Journal of Plant Research 128: 337–344. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y, et al. 2015. PARASITIC PLANTS. probing strigolactone receptors in Striga hermonthica with fluorescence. Science (New York, N.Y.) 349: 864–868. [DOI] [PubMed] [Google Scholar]
- Tsuzuki M, Nishihama R, Ishizaki K, et al. 2016. Profiling and characterization of small RNAs in the liverwort, Marchantia polymorpha, belonging to the first diverged land plants. Plant & Cell Physiology 57: 359–372. [DOI] [PubMed] [Google Scholar]
- Ueda M, Kuniyoshi T, Yamamoto H, et al. 2012. Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. The Plant Journal 72: 683–693. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- United States Department of Agriculture, Agricultural Research Service, National Plant Germplasm System 2019. Germplasm Resources Information Network (GRIN-Taxonomy). Beltsville, MD: National Germplasm Resources Laboratory. [Google Scholar]
- Uraguchi D, Kuwata K, Hijikata Y, et al. 2018. A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science (New York, N.Y.) 362: 1301–1305. [DOI] [PubMed] [Google Scholar]
- Urquhart AA, Joy KW. 1981. Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiology 68: 750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AA, Joy KW. 1982. Transport, metabolism, and redistribution of xylem-borne amino acids in developing pea shoots. Plant Physiology 69: 1226–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez EA, Glenn EP, Brown JJ, Guntenspergen GR, Nelson SG. 2005. Salt tolerance underlies the cryptic invasion of North American salt marshes by an introduced haplotype of the common reed Phragmites australis (Poaceae). Marine Ecology Progress Series 298: 1–8. [Google Scholar]
- Velasco VM, Mansbridge J, Bremner S, et al. 2016. Acclimation of the crucifer Eutrema salsugineum to phosphate limitation is associated with constitutively high expression of phosphate-starvation genes. Plant, Cell & Environment 39: 1818–1834. [DOI] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, García-Ramírez L, Pantoja O. 2005. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiology 139: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignolini S, Moyroud E, Hingant T, et al. 2015. The flower of Hibiscus trionum is both visibly and measurably iridescent. The New Phytologist 205: 97–101. [DOI] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science (New York, N.Y.) 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vogel A, Schwacke R, Denton AK, et al. 2018. Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nature Communications 9: 2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V, Amtmann A. 2006. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. The Plant Journal 48: 342–353. [DOI] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A. 2004. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant, Cell and Environment 27: 1–14. [Google Scholar]
- Vretare V, Weisner SEB, Strand JA, Granéli W. 2001. Phenotypic plasticity in Phragmites australis as a functional response to water depth. Aquatic Botany 69: 127–145. [Google Scholar]
- Vries de J, Archibald JM. 2018. Plant evolution: landmarks on the path to terrestrial life. New Phytologist 217: 1428–1434. [DOI] [PubMed] [Google Scholar]
- Vuolo F, Kierzkowski D, Runions A, et al. 2018. LMI1 homeodomain protein regulates organ proportions by spatial modulation of endoreduplication. Genes & Development 32: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Hedley CL. 1991. Seed development in peas: knowing your three ‘r’s’ (or four, or five). Seed Science Research 1: 3–14. [Google Scholar]
- Wang XJ, Shi DC, Wang XY, Wang J, Sun YS, Liu JQ. 2015. Evolutionary migration of the Disjunct Salt Cress Eutrema salsugineum (= Thellungiella salsuginea, Brassicaceae) between Asia and North America. PLoS One 10: e0124010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A, Ghosh S, Williams MJ, et al. 2018. Speed breeding is a powerful tool to accelerate crop research and breeding. Nature Plants 4: 23–29. [DOI] [PubMed] [Google Scholar]
- Weber APM, Bar-Even A. 2019. Improving the efficiency of photosynthetic carbon reactions. Plant Physiology 179: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF. 2018. Domestication of pea (Pisum sativum L.): the case of the Abyssinian Pea. Frontiers in Plant Science 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF, Brauner S, Przyborowski JA. 2002. Genetic analysis of pod dehiscence in pea (Pisum sativum L.). Cellular & Molecular Biology Letters 7: 657–663. [PubMed] [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiology 158: 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ortega R. 2015. Genetic control of flowering time in legumes. Frontiers in Plant Science 6: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, DePamphilis CW, Das M, et al. 2012. The parasitic plant genome project: new tools for understanding the biology of orobanche and striga. Weed Science 60: 295–306. [Google Scholar]
- Whitewoods CD, Gonçalves B, Cheng J, et al. 2020. Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science (New York, N.Y.) 367: 91–96. [DOI] [PubMed] [Google Scholar]
- Wilhelmsson PKI, Mühlich C, Ullrich KK, Rensing SA. 2017. Comprehensive genome-wide classification reveals that many plant-specific transcription factors evolved in streptophyte algae. Genome Biology and Evolution 9: 3384–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Lambert AM, Long R, Saltonstall K. 2019. Does hybrid Phragmites australis differ from native and introduced lineages in reproductive, genetic, and morphological traits? American Journal of Botany 106: 29–41. [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, et al. 2006. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiology 140: 1437–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, et al. 2012. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proceedings of the National Academy of Sciences of the United States of America 109: 12219–12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Nishihama R, Yoshitake Y, et al. 2018. Generative cell specification requires transcription factors evolutionarily conserved in land plants. Current Biology 28: 479–486.e5. [DOI] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, et al. 2013. The reference genome of the halophytic plant Eutrema salsugineum. Frontiers in Plant Science 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, et al. 2015. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Molecular Biology and Evolution 32: 767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JL, Jablonski W, Reid JB. 2001. Leaf and flower development in pea (Pisum sativum L.): mutants cochleata and unifoliata. Annals of Botany 88: 225–234. [Google Scholar]
- Yoshida S, Cui S, Ichihashi Y, Shirasu K. 2016. The haustorium, a specialized invasive organ in parasitic plants. Annual Review of Plant Biology 67: 643–667. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, et al. 2019. Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Current Biology 29: 3041–3052.e4. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K. 2009. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. The New Phytologist 183: 180–189. [DOI] [PubMed] [Google Scholar]
- Yuan Y-W. 2018. Monkeyflowers (Mimulus): new model for plant developmental genetics and evo-devo. New Phytologist 222: 694–700. [DOI] [PubMed] [Google Scholar]
- Zedler JB, Kercher S. 2004. Causes and consequences of invasive plants in wetlands: opportunities, opportunists, and outcomes. Critical Reviews in Plant Sciences 23: 431–452. [Google Scholar]
- Zhang Y, Lai J, Sun S, et al. 2008. Comparison analysis of transcripts from the halophyte Thellungiella halophila. Journal of Integrative Plant Biology 50: 1327–1335. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li Y, Lai J, et al. 2012. Ectopic expression of a LEA protein gene TsLEA1 from Thellungiella salsuginea confers salt-tolerance in yeast and Arabidopsis. Molecular Biology Reports 39: 4627–4633. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhao C, Li M, et al. 2013. Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Biology 13: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Ma ZY, Zhu L, Guo JS, Zhu J, Wang JF. 2015. Overexpression of EsMcsu1 from the halophytic plant Eutrema salsugineum promotes abscisic acid biosynthesis and increases drought resistance in alfalfa (Medicago sativa L.). Genetics and Molecular Research: GMR 14: 17204–17218. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2001. Plant salt tolerance. Trends in Plant Science 6: 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu C, Yang J, Shyu C. 2017. Setaria comes of age: meeting report on the second International Setaria Genetics conference. Frontiers in Plant Science 8: 854–857. [DOI] [PMC free article] [PubMed] [Google Scholar]