Abstract
The STR Sequencing Project (STRSeq) was initiated to facilitate the description of sequence-based alleles at the Short Tandem Repeat (STR) loci targeted in human identification assays. This international collaborative effort, which has been endorsed by the ISFG DNA Commission, provides a framework for communication among laboratories. The initial data used to populate the project are the aggregate alleles observed in targeted sequencing studies across four laboratories: National Institute of Standards and Technology (N=1786), Kings College London (N=1043), University of North Texas Health Sciences Center (N=839), and University of Santiago de Compostela (N=944), for a total of 4612 individuals. STRSeq data are maintained as GenBank records at the U.S. National Center for Biotechnology Information (NCBI), which participates in a daily data exchange with the DNA DataBank of Japan (DDBJ) and the European Nucleotide Archive (ENA). Each GenBank record contains the observed sequence of a STR region, annotation (“bracketing”) of the repeat region and flanking region polymorphisms, information regarding the sequencing assay and data quality, and backward compatible length-based allele designation. STRSeq GenBank records are organized within a BioProject at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/380127), which is sub-divided into: commonly used autosomal STRs, alternate autosomal STRs, Y-chromosomal STRs, and X-chromosomal STRs. Each of these categories is further divided into locus-specific BioProjects. The BioProject hierarchy facilitates access to the GenBank records by browsing, BLAST searching, or ftp download. Future plans include user interface tools at strseq.nist.gov, a pathway for submission of additional allele records by laboratories performing population sample sequencing and interaction with the STRidER web portal for quality control (http://strider.online).
1. Introduction
As the forensic DNA community evaluates the potential of sequencing applications for Short Tandem Repeat (STR) loci, it is imperative to define the allelic diversity in these regions of the human genome. Large-scale sequencing projects within the broader genomics community may use shorter read chemistries (e.g. 100 bp) and may not describe repetitive regions due to their complexity and non-conformity to typical alignment parameters [1]. Additionally, knowledge of the forensic literature is needed to report STR sequences in the same manner established by the forensic community.
Even within forensic sequencing studies, there are differences in the reporting of sequence-based STR alleles. Names of convenience such as 20 (a) [2] or FL1X20 [3] have not been standardized and may create confusion about the specific allele being reported. There may be differences in format for the compression or “bracketing” of STR sequences, such as ATAG[9] [4, 5] or [ATAG]9 [6] or [ATAG]9 [7]. More importantly, there may be differences in strand reporting where choice of the forward strand will match the reference sequence direction, and choice of the reverse strand aligns the sequence in the opposite direction. The DNA Commission of the ISFG on minimal nomenclature requirements in 2016 recommended reporting all sequences in the forward strand orientation [8]. However, some loci were historically reported on the reverse strand [9]. In particular, STRs for which the reported strand has changed over time may differ in reporting where the repeat region begins. This can result in shifted (different) allele number designations for the same sequence [8]. Lastly, the recovery and reporting of varying lengths of flanking regions (and hence flanking region variants) is inherent to differences in kit designs and bioinformatic pipelines.
The international forensic DNA community continues to develop guidance on STR sequence nomenclature, and additional resources for quality control of STR sequence data are being developed [10]. However, the need for standardization is immediate. A 2016 survey was recently published by the European Network of Forensic Science Institutes (ENFSI) DNA Working Group [11], in which over half of the 33 responding laboratories have already purchased at least one sequencing instrument. Additionally, the respondents (primarily composed of government forensic laboratories across 25 countries) reported lack of nomenclature and reporting standards as the highest ranking scientific and legal challenge for the implementation of new sequencing technologies in forensic genetics. Also in 2016, the Applied Genetics Group of the U.S. National Institute of Standards and Technology (NIST) queried forensic laboratories to assess the utility of STR reference sequences for loci of forensic interest. The feedback received from 22 laboratories (representing 11 countries) mirrored the ENSFI survey with strong support for the development of STR sequence nomenclature resources.
In response to this need, NIST partnered with the U.S. National Center for Biotechnology Information (NCBI), leveraging NIST’s over 20-year history supporting the forensic STR typing community [12] and NCBI’s extensive infrastructure for accepting, maintaining and serving DNA sequence data. Through this partnership, the STR Sequencing Project (STRSeq) has been initiated to facilitate the description of sequence-based alleles at the STRs targeted in human identification assays. This resource consists of a curated catalog of sequence diversity at forensic STR loci, along with the key elements of nomenclature conforming to current guidelines [8], and will serve as the data backbone during this time of transition, as well as a stable resource for the future.
2. Samples and submission strategy
The initial data used to populate STRSeq are the aggregate alleles observed in targeted sequencing studies of single source samples across four laboratories: NIST, Kings College London (KCL), University of North Texas Health Science Center (UNT), and University of Santiago de Compostela (USC), for a total of 4612 individuals. The number of alleles aggregated differs by locus due to variable multiplex performance and quality requirements described in Section 3. Since only aggregate alleles are displayed, the source of the alleles is anonymized. The targeted sequence data used in STRSeq either have been, or are expected to be published by the submitting laboratory ([6, 13], additional manuscripts in preparation). Records will be added to the STRSeq BioProject in sets, largely coinciding with associated publications, as follows:
NIST: N=1786 samples from multiple sources: 1) N=665 liquid blood samples purchased from Interstate Blood Bank (Memphis, TN) and Millennium Biotech, Inc. (Ft. Lauderdale, FL) with self-declared ancestries from three different U.S. population groups: Caucasian, African American, and Hispanic; 2) N=781 buccal swabs provided by DNA Diagnostics Center (Fairfield, OH) from paternity testing samples with self-declared ancestries from four different US population groups: Caucasian, African American, East Asian and Hispanic; 3) N=297 buccal swabs collected from anonymous volunteers of self-reported, diverse ancestries, provided by the George Washington University; and 4) N=43 control samples and reference materials. All samples have been sequenced with the ForenSeq system (Illumina) and a subset (>600 samples) has overlapping sequence data from the PowerSeq Auto-Y assay (Promega). In addition, for the majority of these samples, capillary electrophoresis (CE) data have been published previously at all sequenced STR loci [14, 15].
KCL: N=1043 samples were obtained from consenting adult volunteers resident in the UK. The samples relate to six different UK population groups with self-declared ancestries of: White British, West African, North East African, South Asian, Chinese and Middle Eastern. All samples have been sequenced with the ForenSeq system (Illumina) and additionally genotyped with at least two commonly available CE kits.
UNT: N=839 samples which have been described in associated sequence-based allele frequency publications and were sequenced with the ForenSeq system (Illumina) [6, 13].
USC: N=944 samples from the HGDP-CEPH diversity panel cell-line DNAs from 51 diverse populations were sequenced with the ForenSeq system (Illumina).
Initially, STRSeq records will be created for the STR loci targeted in the aforementioned assays; additional records will be created as samples are sequenced with other available commercial assays, e.g. Precision ID GlobalFiler NGS STR Panel (Thermo Fisher Scientific). If new STR loci (see [16]) are targeted in commercially available assays launched in the future, additional records will be created.
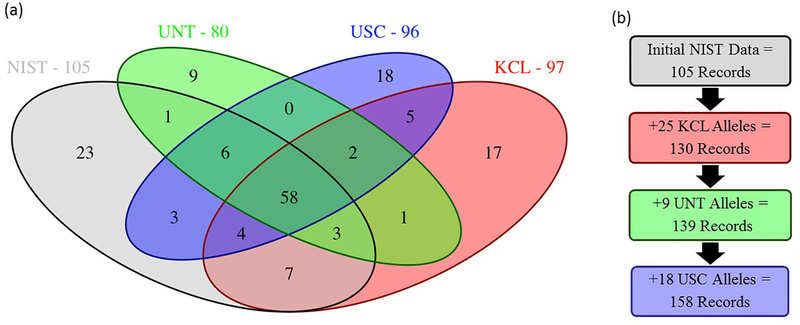
A single laboratory will be indicated as having submitted each record. The association of a submitting laboratory with a record does not imply “discovery” of a sequence variant; rather the designation is simply the organization that initially provided the sequence and maintains the supporting data. For the initial data set, NIST will be the submitting laboratory of all sequences generated at NIST and the other laboratories will be the submitting laboratory of those sequences generated at that specific laboratory for which records do not already exist in the database. Duplicate records will not be created, which will generally result in a decreasing number of new sequence records as successive sample sets are added. Fig. 1 outlines an example submission strategy of non-duplicate allele records that might be expected from a typical highly polymorphic STR such as D12S391.
Figure 1.
(a) Venn diagram demonstrating the overlap of D12S391 sequence-based alleles observed among the four laboratories, and the total number of unique sequence-based alleles observed within each laboratory. (b) Submission strategy for 158 unique sequence-based alleles observed at the D12S391 locus. The 105 unique alleles generated at NIST form the basis of STRSeq records. Subsequent submissions from KCL, UNT, and USC will add records for sequences generated at each laboratory for which records do not already exist (25, 9, and 18 records, respectively).
3. BioProject hierarchy and record format
The BioProject hierarchy serves to organize the GenBank records (Table 1). The highest-level STRSeq umbrella project contains four sub-umbrella projects: (a) Commonly Used Autosomal STR Loci, (b) Alternate Autosomal STR Loci, (c) Y-Chromosomal STR Loci, and (d) X-Chromosomal STR Loci. These sub-umbrella projects are divided further into locus-specific data-level projects which contain the GenBank sequence record data. Each umbrella and data-level project has a corresponding accession number, e.g. PRJNA380127 is the STRSeq umbrella project, PRJNA380345 is the Commonly Used Autosomal STR Loci sub-umbrella project, and PRJNA380554 is the TPOX Sequence-Based Alleles project (the common PRJNA prefix identifies the six-digit number as a BioProject). Entering one of these accession numbers at https://www.ncbi.nlm.nih.gov/bioproject allows direct access to the umbrella or data-level project of interest. Each BioProject page contains additional links for up, down, and cross navigation. Table 1 contains direct links to STRSeq umbrella and data-level projects.
Table 1.
STRSeq BioProject hierarchy, accession numbers, and direct links to all levels. The highest-level of organization is the STRSeq umbrella project (PRJNA380127, http://ncbi.nlm.nih.gov/bioproject/380127), containing four sub-umbrella projects: (a) Commonly Used Autosomal STR Loci, (b) Alternate Autosomal STR Loci, (c), Y-Chromosomal STR Loci and (d) X-Chromosomal STR Loci. Each of these contains locus-specific sub-projects, which are the data-level projects containing GenBank sequence records.
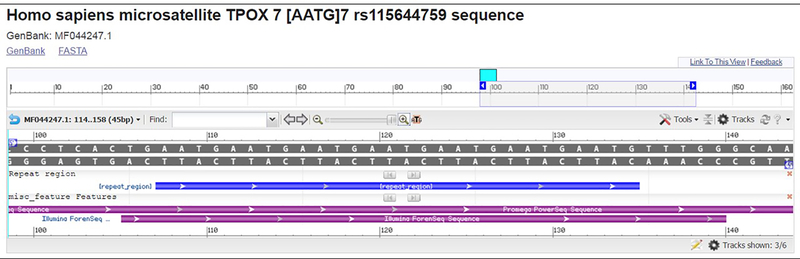
The sequence records in GenBank are flat files of specified format that can be downloaded and parsed en masse (see Fig. 2 for an example record for the TPOX locus). Starting from the bottom of the record, in a section labeled ORIGIN, users will find the full sequence that was reported by the submitting laboratory. The length of reported sequence is dependent upon the assay and the quality of the flanking sequence data, but generally will be consistent with the assay-specific configuration files published in [17]. Above the sequence is the FEATURES table, which includes the position of the repeat region within the sequence, the position and dbSNP rs number of variations in the flanking regions (when applicable), and the subset of sequence that was observed with different commercial assays (when applicable). Each feature can be selected in order to highlight the appropriate region in the sequence string. SNP rs numbers are hyperlinked to dbSNP, allowing users to navigate and access frequency information quickly. If the polymorphism has not been assigned a dbSNP reference number, the GRCh38 coordinate is given, and the field will be updated if an rs number is assigned later or if the assembly is updated.
Figure 2.
Example STRSeq GenBank record, available online at https://www.ncbi.nlm.nih.gov/nuccore/1197990967.
Above the FEATURES table is the structured comments section (offset with ##humanSTR-START## and ##humanSTR-END##), which contains field-based information relevant to STRSeq records. The given Bracketed repeat is intended to be consistent with the guidance of the ISFG Nomenclature Commission [8]. Specific to STRSeq records is the lower-case formatting of selected bases within the Bracketed repeat, which highlights sequence tracts that are not counted toward the length-based allele designation (when applicable, e.g. D19S433 14 allele will be presented as: [AAGG] aaag [AAGG] tagg [AAGG]12). The Sequencing technology field lists the commercial assay(s) and instrument(s) used to generate the sequence data. The Coverage field lists the minimum threshold of reads observed for the reported sequence. The current threshold for STRSeq record creation is >30X. This is consistent with the default minimum “interpretation threshold” implemented in one commercial software, corresponding to the only relevant commercial assay with a published developmental validation [18] at the time of writing. This threshold will continue to be evaluated in the future as additional developmental validations are published. The Length-based tech. field lists the assay and instrument used to generate the Length-based allele given. Often a sequence will have been observed in multiple samples. The length-based information in each record indicates that, for at least one sample, the specified length-based allele was generated with the given length-based technology. This approach is not meant to be comprehensive; variation in the length-based allele among individuals or assays can result from indels in flanking regions. In some instances, length-based allele confirmation may not be possible, such as the lack of a CE assay for STRs targeted by commercial sequencing assays but not previously in common use. When a length-based allele confirmation has not been performed, the Length-based allele field will indicate e.g. “7 (Inferred from sequence)” and the Length-based tech. field will contain “Not reported”. The remaining information in the structured comments section orients the sequence on the chromosome and will be updated along with the reference sequence assembly.
Above the structured comments section is the COMMENT block, which is identical across records and recapitulates this paper. Above the COMMENT block are references. REFERENCE 1 will be this paper and REFERENCE 2 identifies the submitting laboratory. The remaining top-most fields contain information for GenBank record organization. The ACCESSION and VERSION number is the GenBank sequence identifier (e.g. MF044256.1 in Fig. 2). If future commercial assay typing provides additional flanking sequence, the updated sequence will become e.g. MF044256.2 (coexisting with MF044256.1). If the additional flanking sequence reveals a polymorphism, the additional sequence consistent with the reference sequence becomes e.g. MF044256.2 and a new record is created for the additional sequence which differs from the reference sequence.
The DEFINITION line near the top of the record is the descriptor present in a list of sequences (see https://www.ncbi.nlm.nih.gov/nuccore/?term=strseq+tpox), and will uniquely identify each allele with components of the record itself. In addition, the top of each record contains hyperlinks to the FASTA sequence, which can be downloaded, and a Graphics view (Fig. 3). This graphical display presents an interactive version of the sequence (displaying forward and reverse strands) and the features identified in the GenBank record: the repeat region, the region(s) reported from each available sequencing technology, and any associated flanking region polymorphisms. The information shown in Graphics view is dependent on the Tracks selected in the viewer. All available information for the record is displayed simultaneously by selecting both the Sequence and Aggregate features Track. More information and tutorials on the NCBI Sequence Viewer can be found at https://www.ncbi.nlm.nih.gov/tools/sviewer.
Figure 3.
Example Graphics view of STRSeq Genbank record, available and interactive online at https://www.ncbi.nlm.nih.gov/nuccore/1197990967?report=graph.
4. Typical use cases
Several use cases for STRSeq have been identified based on feedback from the forensic community:
1. As a teaching tool to explore STR sequences
The STRSeq BioProject is expected to be useful to forensic operational, academic, and commercial laboratories interested in sequencing STRs as it allows the viewing and downloading of repeat region motifs, flanking region polymorphisms, and commercial assay overlap.
2. As the data backbone for software development
This catalog of sequences with associated forensic formatting and stable links to GenBank records facilitates development of STR sequencing methods and bioinformatic pipelines that conform to agreed variant data frameworks.
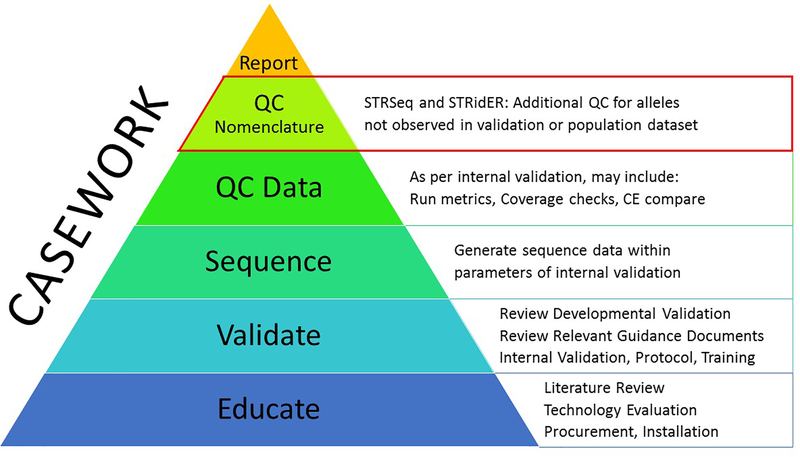
3. To provide a quality control function for the evaluation of rare sequences
When a sequence is observed in forensic casework that was not observed in initial validation studies or in the implemented allele frequency database, a STRSeq BLAST search determines if a similar or identical sequence has been recorded. When a link to previous data is identified, STRSeq provides nomenclature information and leads the analyst to published allele frequency data (see Fig. 4).
Figure 4.
Outline of the anticipated STRSeq use cases for evaluation of rare alleles in forensic casework, integrated into an overall quality assurance system.
5. Future directions for STRSeq
As previously described, sample sets and STRs will be added iteratively, allowing the BioProject to be built further and records to be released in phases. Once created, the GenBank records are expected to be stable but STRSeq should be viewed as a dynamic resource.
Some users will be familiar with NCBI interfaces and will quickly adapt their workflows to access, search, and download records contained in the STRSeq BioProject. While many tutorials exist to facilitate access to NCBI resources (see https://www.ncbi.nlm.nih.gov/guide/all/#howtos), it is likely that most users will prefer customized interface tools specific to this BioProject. Future plans include the development of such tools at strseq.nist.gov, in order to streamline BLAST searches and batch record downloads from the BioProject.
Additionally, we aim to provide a pathway for submission of new sequence records from laboratories performing population sample sequencing. We anticipate an integrated, seamless process whereby users upload population sample sequencing data to the STRidER web portal (http://strider.online) [10] for quality control, and STRidER queries STRSeq for a matching sequence accession number. In cases where the STRidER query finds no match in STRSeq, a process could be initiated to evaluate the sequence and then aim to create a new GenBank record. Such a process would strengthen the STRidER quality control function and expand STRSeq, while harmonizing nomenclature between both resources. This is particularly important for novel sequence variants likely to be encountered as population studies extend their geographic scope or sample numbers.
Acknowledgements
The authors express gratitude to the NCBI staff who have facilitated development of the BioProject: Drs. Lori Black, Melissa Landrum, Ilene Mizrachi, Kim Pruitt, George Riley, and Steven Sherry. The authors also acknowledge the input of the European Commission project DNASEQEX (HOME/2014/ISFP/AG/LAWX/4000007135) and the support of the ENFSI DNA Working Group and thank the many practitioners and researchers who provided valuable feedback.
NIST funding sources and disclaimers
This work was funded in part by the National Institute of Justice (NIJ) interagency agreement 1609-602-18NIJ: “Forensic DNA Applications of Next Generation Sequencing”. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Departments of Commerce or Justice. Certain commercial equipment, instruments and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that any of the materials, instruments or equipment identified are necessarily the best available for the purpose.
UNT funding sources and disclaimers
This work was supported in part by award no. 2015-DN-BX- K067, awarded by the National Institute of Justice, Office of Justice Programs, U.S. Department of Justice. The opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect those of the U.S. Department of Justice.
References
- [1].Willems TZ.; Yuan J.;Gordon A.; Gymrek M; and Erlich Y, Genome-wide profiling of heritable and de novo STR variations, Nat Methods 14(6) (2017) 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Butler JM, Advanced Topics in Forensic DNA Typing: Methodology, Elsevier, USA, 2012. [Google Scholar]
- [3].Van Neste C, Van Criekinge W, Deforce D, Van Nieuwerburgh F, Forensic Loci Allele Database (FLAD): Automatically generated, permanent identifiers for sequenced forensic alleles, Forensic Sci Int Genet 20 (2016) e1–3. [DOI] [PubMed] [Google Scholar]
- [4].Gelardi C, Rockenbauer E, Dalsgaard S, Borsting C, Morling N, Second generation sequencing of three STRs D3S1358, D12S391 and D21S11 in Danes and a new nomenclature for sequenced STR alleles, Forensic Sci Int Genet 12 (2014) 38–41. [DOI] [PubMed] [Google Scholar]
- [5].van der Gaag KJ, de Leeuw RH, Hoogenboom J, Patel J, Storts DR, Laros JF, de Knijff P, Massively parallel sequencing of short tandem repeats-Population data and mixture analysis results for the PowerSeq system, Forensic Sci Int Genet 24 (2016) 86–96. [DOI] [PubMed] [Google Scholar]
- [6].Novroski NM, King JL, Churchill JD, Seah LH, Budowle B, Characterization of genetic sequence variation of 58 STR loci in four major population groups, Forensic Sci Int Genet 25 (2016) 214–226. [DOI] [PubMed] [Google Scholar]
- [7].Gettings KB, Kiesler KM, Faith SA, Montano E, Baker CH, Young BA, Guerrieri RA, Vallone PM, Sequence variation of 22 autosomal STR loci detected by next generation sequencing, Forensic Sci Int Genet 21 (2016) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Parson W, Ballard D, Budowle B, Butler JM, Gettings KB, Gill P, Gusmao L, Hares DR, Irwin JA, King JL, Knijff P, Morling N, Prinz M, Schneider PM, Neste CV, Willuweit S, Phillips C, Massively parallel sequencing of forensic STRs: Considerations of the DNA commission of the International Society for Forensic Genetics (ISFG) on minimal nomenclature requirements, Forensic Sci Int Genet 22 (2016) 54–63. [DOI] [PubMed] [Google Scholar]
- [9].Gettings KB, Aponte RA, Vallone PM, Butler JM, STR allele sequence variation: Current knowledge and future issues, Forensic Sci Int Genet 18 (2015) 118–30. [DOI] [PubMed] [Google Scholar]
- [10].Bodner M, Bastisch I, Butler JM, Fimmers R, Gill P, Gusmao L, Morling N, Phillips C, Prinz M, Schneider PM, Parson W, Recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG) on quality control of autosomal Short Tandem Repeat allele frequency databasing (STRidER), Forensic Sci Int Genet 24 (2016) 97–102. [DOI] [PubMed] [Google Scholar]
- [11].Alonso A, Muller P, Roewer L, Willuweit S, Budowle B, Parson W, European survey on forensic applications of massively parallel sequencing, Forensic Sci Int Genet 29 (2017) e23–e25. [DOI] [PubMed] [Google Scholar]
- [12].Ruitberg CM, Reeder DJ, and Butler JM, STRBase: A short tandem repeat DNA database for the human identity testing community, Nucleic acids research 29(1) (2001) 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wendt FR, King JL, Novroski NM, Churchill JD, Ng J, Oldt RF, McCulloh KL, Weise JA, Smith DG, Kanthaswamy S, Budowle B, Flanking region variation of ForenSeq DNA Signature Prep Kit STR and SNP loci in Yavapai Native Americans, Forensic Sci Int Genet 28 (2017) 146–154. [DOI] [PubMed] [Google Scholar]
- [14].Hill CR, Duewer DL, Kline MC, Coble MD, Butler JM, population data for US 29 autosomal STR loci, Forensic Sci Int Genet 7(3) (2013) e82–3. [DOI] [PubMed] [Google Scholar]
- [15].Hill CR, Kline MC, Coble MD, Butler JM, Characterization of 26 MiniSTR Loci for Improved Analysis of Degraded DNA Samples, J Forensic Sci 53(1) (2008) 73–80. [DOI] [PubMed] [Google Scholar]
- [16].Phillips C, A genomic audit of newly-adopted autosomal STRs for forensic identification, Forensic Sci Int Genet 29 (2017) 193–204. [DOI] [PubMed] [Google Scholar]
- [17].Woerner AE, King JL, Budowle B, Fast STR allele identification with STRait Razor 3.0, Forensic Science International: Genetics (2017). [DOI] [PubMed] [Google Scholar]
- [18].Jager AC, Alvarez ML, Davis CP, Guzman E, Han Y, Way L, Walichiewicz P, Silva D, Pham N, Caves G, Bruand J, Schlesinger F, Pond SJ, Varlaro J, Stephens KM, Holt CL, Developmental validation of the MiSeq FGx Forensic Genomics System for Targeted Next Generation Sequencing in Forensic DNA Casework and Database Laboratories, Forensic Sci Int Genet 28 (2017) 52–70. [DOI] [PubMed] [Google Scholar]