Abstract
Hepatocellular carcinoma (HCC) is among the leading causes of cancer-related death worldwide. Patients with hepatitis B virus (HBV) pre-S mutants in liver tissues or blood have been regarded as a high-risk population for HCC development and recurrence. Detection of pre-S mutants in clinical specimens is thus important for early diagnosis and prognosis of HCC to improve patient survival. Recently, we have developed a next-generation sequencing (NGS)-based platform that can quantitatively detect pre-S mutants in patient plasma with superior sensitivity and accuracy. In this study, we compared the pre-S genotyping results from plasma by the NGS-based analysis with those from liver tissues by the immunohistochemistry (IHC)-based analysis in 30 HBV-related HCC patients. We demonstrated that the detection rate of pre-S mutants was significantly higher by NGS- than by IHC-based analysis. There was a moderate to good agreement between both analyses in detection of pre-S mutants. Compared with the IHC, the NGS-based detection of pre-S mutants in patient plasma could determine the patterns of pre-S mutants in liver tissues more efficiently in a noninvasive manner. Our data suggest that the NGS-based platform may represent a promising approach for detection of pre-S mutants as biomarkers of HBV-related HCC in clinical practice.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and lethal human cancers worldwide, causing approximately 700,000 deaths per year [1–3]. Although liver transplantation and surgical resection are widely regarded as potentially curative treatments for HCC patients, the former is limited by the scarcity of donor livers and the latter is not suitable for every patient [4, 5]. Moreover, recurrence after curative resection of HCC is a frequent event, leading to poor patient survival [6, 7]. Currently available chemotherapeutic or molecular targeted drugs provide only limited survival benefit for HCC patients [8, 9]. Therefore, a reliable biomarker and its detection method are urgently needed for early diagnosis and prognosis of HCC to improve patient survival.
Chronic hepatitis B virus (HBV) infection is one of the major risk factors for HCC development, accounting for over 50% of total cases worldwide [10–12]. Our previous studies have identified that two different types of ground glass hepatocytes (GGH, designated types I and II) in liver tissues contain pre-S mutants harboring deletions in the pre-S1 (pre-S1 mutant) and pre-S2 (pre-S2 mutant) gene segments of HBV large surface antigen, respectively [13, 14]. As important HBV oncoproteins, pre-S mutants can induce multiple oncogenic signaling pathways, contributing to hepatocyte proliferation, genomic instability, and eventually HCC formation in vitro and in vivo [15–18]. Moreover, patients carrying pre-S mutants in liver tissues or blood have a 5-fold higher risk to develop HCC [19, 20], and represent a high-risk population for HCC recurrence after curative surgical resection even under antiviral treatment [21–23]. As a result, pre-S mutants have emerged as powerful biomarkers for HBV-related HCC.
Two methods have been commonly used to detect the presence of pre-S mutants in chronic HBV carriers and HBV-related HCC patients, one based on immunohistochemistry (IHC) staining of HBV surface antigen (HBsAg) for GGH visualization in liver tissues [14, 21, 22], and another TA cloning following polymerase chain reaction (PCR) amplification of pre-S gene for DNA sequencing in serum/plasma samples [13, 19, 20]. Unlike these two methods providing only qualitative and semi-quantitative results, we have recently developed a next-generation sequencing (NGS)-based platform for quantitative detection of pre-S mutants in patient plasma [24]. In contrast to the TA cloning-based method, the NGS-based platform can detect the presence of pre-S mutants (not only types but also levels) in patient plasma with higher sensitivity and accuracy [24]. However, it remains unclear whether the pre-S genotyping results by the NGS-based platform from the plasma of patients may correspond with those by the IHC-based method from the liver tissues.
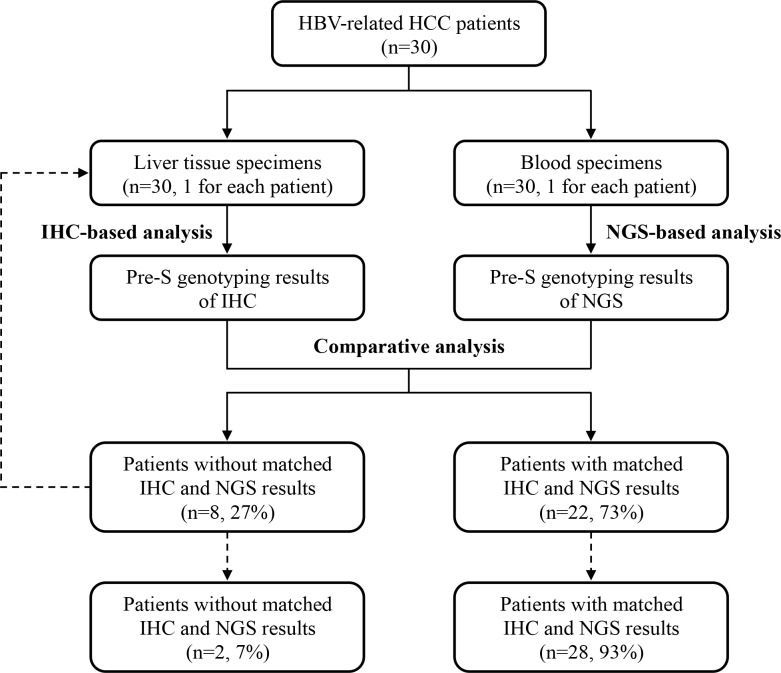
To clarify this issue, in this study, we collected liver tissue and plasma specimens from 30 HBV-related HCC patients for detection of pre-S mutants by IHC- and NGS-based analyses, respectively (Fig 1). Furthermore, the pre-S genotyping results of IHC and NGS were comparatively analyzed to determine the percentage of patients with or without consistent patterns of pre-S mutants between liver tissues and plasma.
Fig 1. Flowchart for the comparison of pre-S genotyping results between liver tissues and plasma in this study.
Liver tissue and plasma specimens collected from 30 HBV-related HCC patients (1 for each patient) were analyzed by IHC and NGS for detection of pre-S mutants, respectively. Comparative analysis of the pre-S genotyping results revealed that 22 out of 30 (73%) patients had matched IHC and NGS results but the other 8 (27%) did not. Further analysis of another 1 liver tissue specimen obtained from each of the 8 patients by IHC increased the number of the patients with matched results up to 28 (93%) (as indicated by dashed lines). The Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; NGS, next-generation sequencing.
Materials and methods
Human specimens
The liver tissue and plasma specimens obtained during and before surgery, respectively, were collected retrospectively from 30 HBV-related HCC patients who underwent surgical treatment at China Medical University Hospital (Taichung, Taiwan), from Jan 2006 to Jul 2017, under the approval of the China Medical University & Hospital Research Ethics Committee (protocol No. CMUH107-REC1-080). The clinicopathological records of the patients were also collected.
IHC-based pre-S genotyping analysis
The IHC-based detection of pre-S mutants was performed as previously described [21]. Briefly, liver tissue sections were incubated with the primary antibody anti-HBsAg (MA5-13059; Thermo Fisher Scientific, Rockford, IL, USA), followed by a horseradish peroxidase-conjugated secondary antibody. Subsequently, the sections were visualized with the aminoethyl carbazole substrate kit (Zymed Laboratories, San Francisco, CA, USA) and counterstained with hematoxylin. Finally, the types and scores of GGH were assessed by 2 pathologists (HWT and IJS). The percentage of each type of GGH was scored from 0 to 4 corresponding to 0%, <5%, 5% to 9%, 10% to 29%, and ≥30% of hepatocytes in liver tissues, respectively.
NGS-based pre-S genotyping analysis
The NGS-based detection of pre-S mutants was performed as previously described [24]. Briefly, DNA isolated from plasma specimens was used as template for PCR amplification of the pre-S gene. Next, the pre-S gene PCR products were subjected to NGS sequencing analysis (Illumina, San Diego, CA, USA). Finally, the deletion types, regions, and levels of pre-S mutants were determined by using our customized scripts. The level of each type of pre-S mutant was defined as the number of pre-S gene DNA with deletion divided by the total number of pre-S gene DNA and then multiplied by 100.
Statistical analysis
The difference between the pre-S genotyping results by IHC- and NGS-based analyses was compared by the McNemar’s paired proportion test, where a P value<0.05 was considered to indicate statistical significance. The agreement between both analyses in pre-S genotyping was evaluated by calculating the simple kappa coefficient with 95% CI, where κ = 1 denotes perfect agreement; κ>0.80, excellent; κ>0.60, good; κ>0.40, moderate; κ>0.20, fair; and κ>0 indicates poor agreement [25].
Results
Patient profile and clinicopathological data
The clinicopathological characteristics of the 30 HBV-related HCC patients enrolled in this study are summarized in Table 1. Among all patients, 26 (87%) were men, 4 (13%) were women, and the median age was 54 years (range, 28 to 78). 27 patients (90%) were HBsAg positive and 26 patients (87%) were HBV e antigen (HBeAg) negative. 24 patients (80%) had genotype B and 6 patients (20%) had genotype C HBV infection. HBV DNA was detected in 20 (67%) patients at a median of 6.8×104 copies/mL (range, 30.1 to 1.5×108). Tumor size was recorded in 29 (97%) patients at a median of 4.0 cm (range, 1.5 to 35.0).
Table 1. Clinicopathological characteristics of the 30 HBV-related HCC patients enrolled in this study.
| Characteristicsa | No. of Patients | Median (Range) |
|---|---|---|
| Age (years) | 30 | 54 (28−78) |
| Gender (men/women) | 26/4 | |
| Smoking (yes/no) | 10/20 | |
| Alcohol (yes/no) | 7/23 | |
| HBsAg (positive/negative/NA) | 27/0/3 | |
| HBeAg (positive/negative) | 4/26 | |
| HBV genotype (B/C) | 24/6 | |
| HBV DNA (copies/mL) (20−1.7×108/<20/NA)b | 20/2/8 | 6.8×104 (30.1−1.5×108) |
| Albumin (g/dL) | 30 | 3.7 (2.4−4.9) |
| AST (U/L) | 30 | 53.5 (14.0−290.0) |
| ALT (U/L) | 29 | 56 (13−292) |
| AFP (ng/mL) (≤54000/>54000)c | 26/4 | 35.7 (2.4−4550.0) |
| Tumor size (cm) | 29 | 4.0 (1.5−35.0) |
| Tumor encapsulation (yes/no) | 22/7 | |
| Lymph node involvement (yes/no) | 4/26 | |
| Portal vein thrombosis (yes/no) | 0/30 | |
| Satellite nodule (yes/no) | 6/24 | |
| Vascular invasion (yes/no) | 14/16 | |
| Distant metastasis (yes/no) | 3/27 | |
| Steatosis grade (0/1/2/3) | 18/5/0/0 | |
| Metavir inflammation score (0/1/2/3) | 5/19/1/0 | |
| Ishak fibrosis score (0/1/2/3/4/5/6) | 1/5/5/8/5/1/4 | |
| Child-Pugh cirrhosis score (A/B/C) | 21/6/2 | |
| CLIP score (0/1/2/3/4/5/6) | 10/11/5/2/1/0/0 | |
| BCLC stage (A/B/C/D) | 18/6/4/2 | |
| AJCC TNM stage (I/II/IIIA/IIIB/IIIC/IVA/IVB) | 11/12/3/0/3/0/1 |
aOnly patients with available data were analyzed.
bHBV DNA was measured with a range of 20 to 1.7×108 copies/mL.
cAFP was measured with the highest detection limit of 54000 ng/mL.
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HBsAg; hepatitis B surface antigen; NA, not available; HBeAg, hepatitis B e antigen; AST, aspartate aminotransferase; ALT, alanine aminotransferase; AFP, alpha-fetoprotein; CLIP, Cancer of the Liver Italian Program; BCLC, Barcelona Clinic Liver Cancer; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis.
IHC-based pre-S genotyping in liver tissues
By IHC-based pre-S genotyping analysis, we first detected pre-S mutants in liver tissues of the 30 HBV-related HCC patients. As shown in Table 2 and S1 Table, either or both of types I and II GGH were detected in 19 out of 30 (63%) patients, among whom 4 (13%) patients had only type I GGH (score 1–4), 6 (20%) patients had only type II GGH (score 1–4), and 9 (30%) patients had both type I GGH (score 1–4) and type II GGH (score 1–4). Additionally, 13 (43%) patients had type I GGH (score 1–4) without/with type II GGH, and 15 (50%) patients had type II GGH (score 1–4) without/with type I GGH.
Table 2. Summary of the pre-S genotyping results by IHC-based analysis in 30 HBV-related HCC patients.
| Summary of The Pre-S Genotyping Resultsa | IHC-Based Analysis |
|---|---|
| Total patients (n) (%) | 30 (100) |
| Patients without type I and type II GGH (n) (%) | 11 (37) |
| Patients with type I and/or type II GGH (n) (%) | 19 (63) |
| Patients with type I GGH (score 1–4) and type II GGH (score 0) (n) (%) | 4 (13) |
| Patients with type I GGH (score 0) and type II GGH (score 1–4) (n) (%) | 6 (20) |
| Patients with type I GGH (score 1–4) and type II GGH (score 1–4) (n) (%) | 9 (30) |
| Patients with type I GGH (score 1–4) and type II GGH (score 0–4) (n) (%) | 13 (43) |
| Patients with type II GGH (score 1–4) and type I GGH (score 0–4) (n) (%) | 15 (50) |
aThe percentage of each type of GGH was scored from 0 to 4 corresponding to 0%, <5%, 5% to 9%, 10% to 29%, and ≥30% of hepatocytes in liver tissues, respectively.
Abbreviations: IHC, immunohistochemistry; GGH, ground glass hepatocytes; n, number; del, deletion.
NGS-based pre-S genotyping in plasma
By NGS-based pre-S genotyping analysis, we next detected pre-S mutants in plasma of the 30 HBV-related HCC patients. As shown in Table 3 and S1 Table, 25 out of 30 (83%) patients had pre-S deletion, among whom 4 (13%) patients had only pre-S1 deletion, 7 (23%) patients had only pre-S2 deletion, 3 (10%) patients had both pre-S1 and pre-S2 deletion, 1 (3%) patient had both pre-S2 and pre-S1+pre-S2 deletion, and 10 (34%) patients had all three types of pre-S deletion. Moreover, 18 (60%) and 21 (70%) patients had deletion spanning the pre-S1 and pre-S2 gene segments, respectively.
Table 3. Summary of the pre-S genotyping results by NGS-based analysis in 30 HBV-related HCC patients.
| Summary of The Pre-S Genotyping Results | NGS-Based Analysis |
|---|---|
| Total patients (n) (%) | 30 (100) |
| Patients without pre-S del (n) (%) | 5 (17) |
| Patients with pre-S del (n) (%) | 25 (83) |
| Patients with only pre-S1 del (n) (%) | 4 (13) |
| Patients with only pre-S2 del (n) (%) | 7 (23) |
| Patients with only pre-S1+pre-S2 del (n) (%) | 0 (0) |
| Patients with both pre-S1 and pre-S2 del (n) (%) | 3 (10) |
| Patients with both pre-S1 and pre-S1+pre-S2 del (n) (%) | 1 (3) |
| Patients with both pre-S2 and pre-S1+pre-S2 del (n) (%) | 0 (0) |
| Patients with all three types of pre-S del (n) (%) | 10 (34) |
| Patients with deletion spanning pre-S1 gene segment (n) (%) | 18 (60) |
| Patients with deletion spanning pre-S2 gene segment (n) (%) | 21 (70) |
Abbreviations: NGS, next-generation sequencing; n, number; del, deletion.
Detection of pre-S mutants by NGS in plasma determined their patterns in liver tissues with higher sensitivity and accuracy than by IHC
Comparative analysis of the pre-S genotyping results of IHC and NGS showed that 22 out of 30 (73%) patients had consistent results although the other 8 (27%) did not (S1 Table). In total 30 patients, pre-S mutants were detected in 19 (63%) and 25 (83%) patients by IHC and NGS, respectively (Tables 2 and 3). Six patients were negative for pre-S mutants by IHC-based analysis but conversely positive by NGS. The detection of pre-S mutants was significantly more sensitive by NGS- than by IHC-based analysis (McNemar’s paired proportion test, P value = 0.0143) (Table 4). The simple kappa value for the agreement between both analyses in detection of pre-S mutants was moderate at 0.51 (95% confidence interval (CI), 0.20 to 0.81) (Table 4). Similar results were observed when comparing both analyses in identifying patients with pre-S1 mutant (McNemar’s paired proportion test, P value = 0.0253; simple kappa coefficient, kappa value (κ) = 0.67 (95% CI, 0.42 to 0.92)) or pre-S2 mutant (McNemar’s paired proportion test, P value = 0.0143; simple kappa coefficient, κ = 0.60 (95% CI, 0.33 to 0.86)) (Table 4).
Table 4. Comparison of the pre-S genotyping results by IHC- and NGS-based analyses in 30 HBV-related HCC patients.
| Patient groups by pre-S genotyping | McNemar’s paired proportion test | Simple Kappa Coefficient | ||||
|---|---|---|---|---|---|---|
| NGS (−) | NGS (+) | P value | κ value | 95% CI | ||
| Patients without (−)/with (+) any type of GGH/pre-S del (n) (%) | IHC (−) | 5 (17)a | 6 (20)b | 0.0143 | 0.51 | 0.20–0.81 |
| IHC (+) | 0 (0)c | 19 (63)d | ||||
| Patients without (−)/with (+) type I GGH (score 1–4) and type II GGH (score 0–4)/deletion spanning pre-S1 gene segment (n) (%) | IHC (−) | 12 (40) | 5 (17) | 0.0253 | 0.67 | 0.42–0.92 |
| IHC (+) | 0 (0) | 13 (43) | ||||
| Patients without (−)/with (+) type II GGH (score 1–4) and type I GGH (score 0–4)/deletion spanning pre-S2 gene segment (n) (%) | IHC (−) | 9 (30) | 6 (20) | 0.0143 | 0.60 | 0.33–0.86 |
| IHC (+) | 0 (0) | 15 (50) | ||||
aThe number of patients without the indicated GGH and pre-S deletion by both IHC- and NGS-based analyses.
bThe number of patients with the indicated pre-S deletion by NGS- but not IHC-based analysis.
cThe number of patients with the indicated pre-S deletion by IHC- but not NGS-based analysis.
dThe number of patients with the indicated pre-S deletion by both IHC- and NGS-based analyses.
Abbreviations: NGS, next-generation sequencing; GGH, ground glass hepatocytes; IHC, immunohistochemistry; del, deletion; n, number.
Considering that the IHC results were originally obtained from only 1 liver tissue specimen (a part of liver tissues) from each of the 30 HBV-related HCC patients, it could be reasonably speculated that the results might be insufficient to reflect the patterns of pre-S mutants in whole liver tissues. To further confirm the patterns of pre-S mutants in liver tissues, another 1 liver tissue specimen (a different part of liver tissues) was collected from each of the 8 patients with inconsistent IHC and NGS results for additional IHC-based analysis. As shown in Table 5, based on the IHC results from the original specimen, 6 out of the 8 patients did not have any type of GGH, and the other 2 had only type I GGH. Further analysis of another 1 specimen showed that 4 out of the 6 patients without any type of GGH as well as the 2 patients with only type I GGH became detectable for the types of GGH corresponding with the pre-S genotyping results of NGS (Table 5). Overall, the proportion of patients with matched IHC and NGS results could be increased from 22 of 30 (73%) patients by analyzing 1 liver tissue specimen from each patient up to 28 of 30 (93%) by including 2 liver tissue specimens in analysis.
Table 5. The pre-S genotyping results by additional IHC-based analysis in selected HBV-related HCC patients.
| Patient No. | IHC Result (GGH Type (Score))a,b | NGS Result (Pre-S Deletion Type (%))d |
|---|---|---|
| 4 | 1. type I GGH (0), type II GGH (0) | 1. pre-S1 del (86.404)e |
| 2. type I GGH (1), type II GGH (0)c | 2. wild-type (12.695) | |
| 3. pre-S2 del (0.737) | ||
| 4. pre-S1+pre-S2 del (0.163) | ||
| 7 | 1. type I GGH (3), type II GGH (0) | 1. pre-S1+pre-S2 del (46.237) |
| 2. type I GGH (3), type II GGH (3) | 2. pre-S2 del (26.927) | |
| 3. pre-S1 del (14.368) | ||
| 4. wild-type (12.467) | ||
| 9 | 1. type I GGH (0), type II GGH (0) | 1. pre-S2 del (56.155) |
| 2. type I GGH (0), type II GGH (0) | 2. wild-type (42.610) | |
| 3. pre-S1+pre-S2 del (0.718) | ||
| 4. pre-S1 del (0.516) | ||
| 13 | 1. type I GGH (0), type II GGH (0) | 1. wild-type (94.701) |
| 2. type I GGH (1), type II GGH (0) | 2. pre-S1 del (5.086) | |
| 3. pre-S2 del (0.137) | ||
| 4. pre-S1+pre-S2 del (0.077) | ||
| 14 | 1. type I GGH (1), type II GGH (0) | 1. wild-type (69.001) |
| 2. type I GGH (1), type II GGH (3) | 2. pre-S1 del (20.530) | |
| 3. pre-S2 del (9.463) | ||
| 4. pre-S1+pre-S2 del (1.006) | ||
| 18 | 1. type I GGH (0), type II GGH (0) | 1. pre-S2 del (41.477) |
| 2. type I GGH (1), type II GGH (4) | 2. pre-S1+pre-S2 del (39.126) | |
| 3. wild-type (12.348) | ||
| 4. pre-S1 del (7.048) | ||
| 20 | 1. type I GGH (0), type II GGH (0) | 1. wild-type (30.944) |
| 2. type I GGH (1), type II GGH (1) | 2. pre-S2 del (30.409) | |
| 3. pre-S1+pre-S2 del (29.105) | ||
| 4. pre-S1 del (9.542) | ||
| 26 | 1. type I GGH (0), type II GGH (0) | 1. wild-type (30.973) |
| 2. type I GGH (0), type II GGH (0) | 2. pre-S1+pre-S2 del (27.774) | |
| 3. pre-S1 del (27.161) | ||
| 4. pre-S2 del (14.091) |
aThe percentage of each type of GGH was scored from 0 to 4 corresponding to 0%, <5%, 5% to 9%, 10% to 29%, and ≥30% of hepatocytes in liver tissues, respectively.
bThe results of two liver tissue specimens (the original one and another one) from each patient were shown in descending order.
cThe result of IHC in correspondence with that of NGS was shown in bold.
dThe total percentage of pre-S gene DNA in each type of pre-S deletion was shown in descending order.
eThe pre-S deletion type above the cut-off percentage (5.049) was shown in bold.
Abbreviations: IHC, immunohistochemistry; NGS, next-generation sequencing; GGH, ground glass hepatocytes; del, deletion.
Discussion
Despite substantial progress in treatment, high morbidity and mortality of HCC in patients with chronic HBV infection are still a significant health problem [26–28]. Therefore, early diagnosis and prognosis of HCC by reliable biomarkers remain a key goal for improving patient survival. The presence of pre-S mutants in liver tissues or blood has emerged as a valuable biomarker for HBV-related HCC [19–23]. Recently, we have developed a NGS-based platform for quantitative detection of pre-S mutants in patient plasma with high sensitivity and accuracy using easy and relatively less invasive methods [24]. In this study, we further demonstrated that the NGS-based pre-S genotyping in plasma could also efficiently determine the patterns of pre-S mutants in liver tissues in HBV-related HCC patients.
The presence of pre-S mutants in chronic HBV carriers and HBV-related HCC patients has been commonly detected from the serum/plasma and liver tissue specimens by the TA cloning- and IHC-based analyses, respectively [13, 14, 19–22]. However, these two methods provide only qualitative and semi-quantitative results and remain to be optimized in practical operation. The TA cloning-based analysis is mainly dependent on cloning and sequencing of pre-S gene PCR bands that are clearly separated and visualized in agarose gel; the unseparated or invisible PCR bands may be omitted during the procedure, thus leading to lower detection sensitivity [24]. Also, the cloning procedure is somewhat time-consuming. In addition, the IHC-based analysis relies majorly on staining and visualization of GGH in sections of liver tissue specimens that are parts of whole liver tissues; the results from partial liver tissues may insufficiently represent those from whole liver tissues, thus resulting in false-negative results. In supportive of this notion, in this study we showed that the detection rate of GGH in total 2 liver tissue specimens was considerably higher than that in only 1 liver tissue specimen in the same patient. Furthermore, another limitation for the IHC-based analysis is the need of invasive liver biopsy that may have potential adverse effects on some patients [29]. The IHC-based analysis is somewhat time- and manpower-consuming, requiring skillful surgeons for the operation, technicians for tissue preparation process then IHC staining, and experienced pathologists for data interoperation.
To optimize the methodology for detection of pre-S mutants in patient specimens, we have recently developed a NGS-based platform that can quantitatively detect the presence of each type of pre-S mutant with higher sensitivity and accuracy than the TA cloning-based analysis in the plasma of HBV-related HCC patients [24]. In this study, we further compared the pre-S genotyping results of the NGS-based analysis in plasma with those of the IHC-based analysis in liver tissues (1 specimen for each patient) and showed a moderate to good agreement between both analyses in detection of pre-S mutants. Although the NGS exhibited significantly higher sensitivity than the IHC, the detection rate of pre-S mutants by IHC could be remarkably improved by including another 1 liver tissue specimen from each patient in analysis, leading to a higher level of consistency with the results of NGS-based analysis in plasma. Therefore, our data suggest that the NGS-based detection of pre-S mutants in plasma of patients can also determine the presence and pattern of pre-S mutants in liver tissues with better sensitivity and accuracy than the IHC-based analysis. Moreover, the detection procedure of the NGS-based analysis is somewhat more safe and efficient because there is no need for invasive liver biopsy to obtain more than one liver tissue specimens for IHC analysis; the NGS-based analysis only requires some plasma and technicians for a NGS run that should be able to get the results within 24 hours if the workflow is well established. Not only the NGS-based analysis saves time, but also providing a nonbiased overall assessment for pre-S mutant detection with better accuracy and safety.
Conclusions
In this study we demonstrated that the NGS-based pre-S genotyping in plasma could serve as an alternative approach to determine the presence and pattern of pre-S mutants in liver tissues in HBV-related HCC patients. Considering the advantages of high sensitivity and noninvasive procedure in detection of pre-S mutants, the NGS-based platform may have great promise for future clinical application in patients with chronic HBV infection or HBV-related HCC.
Supporting information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the China Medical University, Taichung, Taiwan (Grant Number CMU107-N-09) (to CFT). The funder had no role in study design, data collection and analysis, decision to publish, or preparation the manuscript.
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–55. 10.1016/S0140-6736(11)61347-0 . [DOI] [PubMed] [Google Scholar]
- 2.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15 Suppl 4:5–13. 10.1634/theoncologist.2010-S4-05 . [DOI] [PubMed] [Google Scholar]
- 3.Cheng KC, Lin WY, Liu CS, Lin CC, Lai HC, Lai SW. Association of different types of liver disease with demographic and clinical factors. Biomedicine (Taipei). 2016;6(3):16 10.7603/s40681-016-0016-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wall WJ, Marotta PJ. Surgery and transplantation for hepatocellular cancer. Liver Transpl. 2000;6(6 Suppl 2):S16–22. 10.1053/jlts.2000.19010 . [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008. J Hepatol. 2008;48 Suppl 1:S20–37. 10.1016/j.jhep.2008.01.022 . [DOI] [PubMed] [Google Scholar]
- 6.Marin-Hargreaves G, Azoulay D, Bismuth H. Hepatocellular carcinoma: surgical indications and results. Crit Rev Oncol Hematol. 2003;47(1):13–27. 10.1016/s1040-8428(02)00213-5 . [DOI] [PubMed] [Google Scholar]
- 7.Poon RT, Fan ST, Lo CM, Liu CL, Wong J. Intrahepatic recurrence after curative resection of hepatocellular carcinoma: long-term results of treatment and prognostic factors. Ann Surg. 1999;229(2):216–22. Epub 1999/02/19. 10.1097/00000658-199902000-00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llovet JM, Bruix J. Early diagnosis and treatment of hepatocellular carcinoma. Baillieres Best Pract Res Clin Gastroenterol. 2000;14(6):991–1008. Epub 2001/01/05. 10.1053/bega.2000.0143 . [DOI] [PubMed] [Google Scholar]
- 9.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. 10.1056/NEJMoa0708857 . [DOI] [PubMed] [Google Scholar]
- 10.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28(5):753–70. 10.1016/j.bpg.2014.08.007 . [DOI] [PubMed] [Google Scholar]
- 11.Beasley RP, Hwang LY. Hepatocellular carcinoma and hepatitis B virus. Semin Liver Dis. 1984;4(2):113–21. 10.1055/s-2008-1040651 . [DOI] [PubMed] [Google Scholar]
- 12.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362(9399):1907–17. Epub 2003/12/12. 10.1016/S0140-6736(03)14964-1 . [DOI] [PubMed] [Google Scholar]
- 13.Fan YF, Lu CC, Chen WC, Yao WJ, Wang HC, Chang TT, et al. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology. 2001;33(1):277–86. Epub 2000/12/22. 10.1053/jhep.2001.21163 . [DOI] [PubMed] [Google Scholar]
- 14.Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163(6):2441–9. 10.1016/S0002-9440(10)63599-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su IJ, Wang HC, Wu HC, Huang WY. Ground glass hepatocytes contain pre-S mutants and represent preneoplastic lesions in chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1169–74. 10.1111/j.1440-1746.2008.05348.x . [DOI] [PubMed] [Google Scholar]
- 16.Teng CF, Wu HC, Shyu WC, Jeng LB, Su IJ. Pre-S2 Mutant-Induced Mammalian Target of Rapamycin Signal Pathways as Potential Therapeutic Targets for Hepatitis B Virus-Associated Hepatocellular Carcinoma. Cell Transplant. 2017;26(3):429–38. 10.3727/096368916X694382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng YC, Neo JC, Wu JC, Chen YF, Kao CH, Tsai TF. Expression of a hepatitis B virus pre-S2 deletion mutant in the liver results in hepatomegaly and hepatocellular carcinoma in mice. J Pathol. 2017;241(4):463–74. Epub 2016/11/22. 10.1002/path.4850 . [DOI] [PubMed] [Google Scholar]
- 18.Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61(2):408–17. Epub 2014/05/08. 10.1016/j.jhep.2014.04.041 . [DOI] [PubMed] [Google Scholar]
- 19.Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, et al. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133(5):1466–74. 10.1053/j.gastro.2007.09.002 . [DOI] [PubMed] [Google Scholar]
- 20.Sinn DH, Choi MS, Gwak GY, Paik YH, Lee JH, Koh KC, et al. Pre-s mutation is a significant risk factor for hepatocellular carcinoma development: a long-term retrospective cohort study. Dig Dis Sci. 2013;58(3):751–8. Epub 2012/10/12. 10.1007/s10620-012-2408-9 . [DOI] [PubMed] [Google Scholar]
- 21.Tsai HW, Lin YJ, Lin PW, Wu HC, Hsu KH, Yen CJ, et al. A clustered ground-glass hepatocyte pattern represents a new prognostic marker for the recurrence of hepatocellular carcinoma after surgery. Cancer. 2011;117(13):2951–60. 10.1002/cncr.25837 . [DOI] [PubMed] [Google Scholar]
- 22.Tsai HW, Lin YJ, Wu HC, Chang TT, Wu IC, Cheng PN, et al. Resistance of ground glass hepatocytes to oral antivirals in chronic hepatitis B patients and implication for the development of hepatocellular carcinoma. Oncotarget. 2016;7(19):27724–34. 10.18632/oncotarget.8388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen CJ, Ai YL, Tsai HW, Chan SH, Yen CS, Cheng KH, et al. Hepatitis B virus surface gene pre-S2 mutant as a high-risk serum marker for hepatoma recurrence after curative hepatic resection. Hepatology. 2018. 10.1002/hep.29790 . [DOI] [PubMed] [Google Scholar]
- 24.Teng CF, Huang HY, Li TC, Shyu WC, Wu HC, Lin CY, et al. A Next-Generation Sequencing-Based Platform for Quantitative Detection of Hepatitis B Virus Pre-S Mutants in Plasma of Hepatocellular Carcinoma Patients. Sci Rep. 2018;8(1):14816 Epub 2018/10/06. 10.1038/s41598-018-33051-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74. . [PubMed] [Google Scholar]
- 26.Kubo S, Takemura S, Tanaka S, Shinkawa H, Nishioka T, Nozawa A, et al. Management of hepatitis B virus infection during treatment for hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21(27):8249–55. 10.3748/wjg.v21.i27.8249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamor PJ, deLemos AS, Russo MW. Viral hepatitis and hepatocellular carcinoma: etiology and management. J Gastrointest Oncol. 2017;8(2):229–42. 10.21037/jgo.2017.03.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HM, Banini BA. Updates on Chronic HBV: Current Challenges and Future Goals. Curr Treat Options Gastroenterol. 2019;17(2):271–91. Epub 2019/05/12. 10.1007/s11938-019-00236-3 . [DOI] [PubMed] [Google Scholar]
- 29.Thampanitchawong P, Piratvisuth T. Liver biopsy:complications and risk factors. World J Gastroenterol. 1999;5(4):301–4. Epub 2002/01/31. 10.3748/wjg.v5.i4.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.