Abstract
High-field asymmetric waveform ion mobility spectrometry (FAIMS) enables the separation of ions on the basis of their differential mobility in an asymmetric oscillating electric field. We, and others, have previously demonstrated the benefits of FAIMS for the analysis of peptides and denatured proteins. To date, FAIMS has not been integrated with native mass spectrometry of folded proteins and protein complexes, largely due to concerns over the heating effects associated with the high electric fields employed. Here, we demonstrate the newly introduced cylindrical FAIMS Pro device coupled with an Orbitrap Eclipse enables analysis of intact protein assemblies up to 147 kDa. No evidence for dissociation was detected suggesting that any field heating is insufficient to disrupt the noncovalent interactions governing these assemblies. Moreover, the FAIMS device was integrated into native liquid extraction surface analysis (LESA) MS of protein assemblies directly from thin tissue sections. Intact tetrameric hemoglobin (64 kDa) and trimeric reactive intermediate deiminase A (RidA, 43 kDa) were detected. Improvements in signal-to-noise of between 1.5× and 12× were observed for these protein assemblies on integration of FAIMS.
In native mass spectrometry (MS), noncovalent interactions that were present in solution-phase are maintained in the gas-phase, enabling structural information to be inferred.1 Ion mobility spectrometry (IMS) has played an important role in establishing native mass spectrometry as a powerful tool in the analysis of protein complexes.2−6 In most cases, IMS-MS offers the ability to derive collision cross sections (CCSs), delivering insight into tertiary and quaternary structure, in addition to providing information regarding stoichiometry in the case of protein assemblies or complexes. The structural information gleaned from native MS is complementary to that obtained from established biophysical techniques such as X-ray crystallography.4 Classical drift tube IMS (DTIMS),7−9 and the commercially available traveling wave ion mobility spectrometry (TWIMS)5,10 and trapped ion mobility spectrometry (TIMS)11,12 all offer the ability to measure CCS. The latter has been evaluated for native MS analysis using ubiquitin.13
IMS also offers improvements in signal-to-noise (S/N) by enabling ions of differing mobility to be separated from one another prior to mass analysis. High-field asymmetric waveform ion mobility spectrometry (FAIMS)14 has shown particular promise in this regard. FAIMS separates gas-phase ions on the basis of their differential mobilities in high and low electric fields. The ions are passed by a carrier gas between parallel electrodes to which an asymmetric waveform is applied. The amplitude of the waveform is the dispersion voltage (DV) (giving rise to the dispersion field (DF)). As a consequence of their differential mobility, ions stray from their original trajectory, an occurrence which is corrected for by applying a dc compensation voltage (CV). By tuning the CV, it is possible to selectively transmit ions through the FAIMS device. FAIMS offers a more orthogonal mode of separation to mass spectrometry than other ion mobility techniques.15,16
To date, it has not been possible to derive CCS from FAIMS measurements; however, as mentioned above, FAIMS offers significant advantages in terms of improved S/N. Much of our current interest in FAIMS derives from the improved S/N of protein signals detected following direct sampling of biological substrates, including dried blood spots, bacterial colonies, and tissue.17−21 In those works, FAIMS MS was coupled with liquid extraction surface analysis (LESA) using denaturing solvents, i.e., the proteins detected were intact but unfolded. Improvements to S/N have been demonstrated with both FAIMS and TWIMS for desorption electrospray ionization (DESI) imaging of proteins.22,23 Kelleher and co-workers have also recently demonstrated the benefits of FAIMS for the analysis of intact (but unfolded) proteins.24
More recently, we have been developing native LESA MS in which folded proteins and protein assemblies are extracted directly from thin tissue sections and dried blood spots.25−27 A key benefit of LESA is the reduced (often absent) requirements for sample preparation. In the case of native LESA, the benefits include the potential for integrated structural and spatial information. An obvious question to pose is: Can the benefits of FAIMS be applied to native MS and native LESA MS? Native LESA MS, in particular, is characterized by noisy mass spectra, a consequence of the omission of washing protocols in order to avoid disrupting protein structure and use of aqueous extraction solvents. Improvements in S/N would therefore be most welcome.
In addressing the above question, a key consideration is field heating. That is, FAIMS employs high electric fields meaning that ions experience collisional heating. This phenomenon was first considered by Purves et al., who calculated that the average increase in temperature experienced by ubiquitin ions was ∼7 K when the DV was 4.4 kV.28 Later work by Shvartsburg et al., in which they argued that the maximum increase in temperature was more relevant when considering the effects of field heating, found the maximum temperature increase to be ∼50 K for ubiquitin ions (DV = 4 kV).16 Field heating is inversely proportional to the square of CCS,29 and has been shown to be enhanced when lighter carrier gases such as He or H2 are employed.30−33 Robinson et al. have shown that field heating effects may be mitigated by cooling the carrier gas.15
Field heating clearly has the potential to be detrimental to the structural analysis of protein ions by causing unfolding. Nevertheless, Shvartsburg et al. have demonstrated the transmission of noncovalent dimers of bovine serum albumin through a FAIMS device.34 Bovine serum albumin typically exists as a monomer but is known to undergo aggregation under stress conditions. The dimers of BSA observed in that experiment cannot therefore be considered “native”, despite the maintenance of noncovalent interactions. In other work, conformers of monomeric β2-microglobulin have been studied by FAIMS MS and correlated with circular dichroism spectroscopy and NMR spectroscopy.35 These studies suggest that FAIMS may be compatible with native MS. To address this issue, here we have considered the FAIMS MS of well-established protein standards including Zn-bound carbonic anyhydrase and the homotetramers concanavalin A and alcohol dehydrogenase. We have also coupled FAIMS MS with native LESA of heterotetrameric hemoglobin and homotrimeric reactive intermediate deiminase A (RidA) from mouse and rat kidney tissue. In all cases, the intact protein complex or protein assemblies were transmitted through the FAIMS device suggesting that field heating does not disrupt noncovalent interactions governing quaternary structure and ligand binding or at least not to the extent that dissociation occurs. The results also demonstrate improvements in S/N for these protein assemblies.
Experimental Section
Detailed descriptions of the methods are provided in the Supporting Information. In brief, samples of protein standards carbonic anhydrase (CAH), concanavalin A (ConA), and yeast alcohol dehydrogenase (ADH) were prepared in ammonium acetate solution. Thin tissue sections of mouse and rat kidney were prepared at 10 μm thickness and thaw mounted onto glass slides. Experiments were performed on an Orbitrap Eclipse mass spectrometer (Thermo Fisher, San Jose, CA) equipped with a FAIMS Pro device and the extended mass range option allowing detection of ions up to m/z 8000. FAIMS Pro features optimized gas flows for improved ion transmission and an electrode gap of 1.5 mm, which shortens ion residence time versus previous FAIMS devices.36 Samples of CAH, ConA, and ADH were introduced by nanoESI and thin tissue sections were sampled by liquid extraction surface analysis (LESA), both using the Triversa Nanomate platform (Advion, Ithaca, NY). The dispersion voltage (DV) was fixed at −5 kV (corresponding to a dispersion field of −33.3 kV/cm). Compensation voltage (CV) scans for optimization of transmission of each analyte were performed for the range −60 V to −10 V in steps of 1 V per scan. Subsequent static FAIMS analyses were performed, in which the optimum CV (chosen for transmission of charge states observed in the absence of FAIMS) was applied. For mass spectra acquired with FAIMS voltages off, the DV and CV were set to 0 V. All other settings were unchanged.
Results and Discussion
Samples of carbonic anhydrase (CAH), concanavalin A (ConA), and alcohol dehydrogenase (ADH) in ammonium acetate solution were analyzed by direct infusion nESI FAIMS mass spectrometry. As described below, each protein sample exhibited narrow charge state distributions, indicative of retention of intramolecular interactions, and quaternary structure was maintained (as evidenced by m/z values corresponding to protein–ligand complexes or protein assemblies). In each case, the optimum CV for transmission was determined by stepping through the CV range −60 V to −10 V at a rate of 1 V per scan. Extracted ion chromatograms revealed both the range of CVs over which the protein complexes were transmitted and the optimum CV for transmission. Subsequent static FAIMS analysis, employing the optimum CV, were performed.
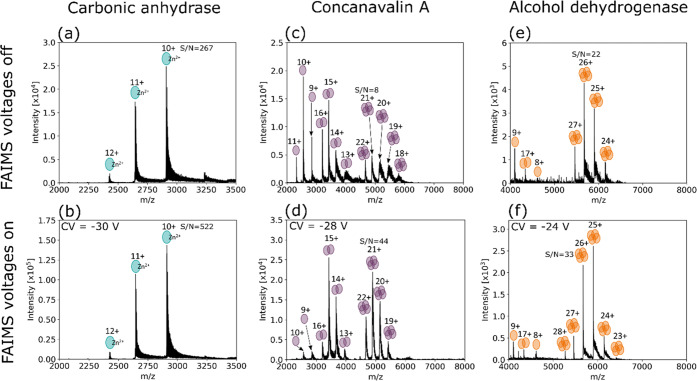
Figure 1a,b shows the mass spectra obtained with FAIMS voltages off and at CV = −30 V for CAH. CAH features a Zn2+ cofactor under native conditions (see Protein Data Bank entry 1v9e).37,38 The Zn2+ cofactor remained bound to CAH (approximately 29.1 kDa, z = 9+ to 12+) when transmitted by FAIMS. The S/N of the 10+ charge state of the holo-protein improved by ∼2-fold. The holo-protein was transmitted over the CV range −50 to −25 V in charge states 9+ to 12+. The apoprotein was not observed at any CV; see Figure S1, Supporting Information. If the protein was unfolded as a result of heating in the FAIMS device, it would likely not be observed under the static conditions optimized for the holo-protein, the so-called FAIMS self-cleaning mechanism; however, it might be observed at another CV. Figure S1c shows the total mass spectrum obtained following a scan of CVs and the apoprotein is not detected.
Figure 1.
(a) Mass spectrum of carbonic anhydrase acquired with FAIMS voltages off; (b) FAIMS mass spectrum of carbonic anhydrase acquired with a static CV of −30 V; (c) mass spectrum of concanavalin A acquired with FAIMS voltages off; (d) FAIMS mass spectrum of concanavalin A acquired with a static CV of −28 V. Monomer, homodimer, and homotetramer ions were detected both with and without FAIMS; (e) mass spectrum of alcohol dehydrogenase acquired with FAIMS voltages off; (f) FAIMS mass spectrum of alcohol dehydrogenase acquired with a static CV of −24 V. Monomer, homodimer, and homotetramer ions were detected both with and without FAIMS.
ConA (PDB entry 1gkb) was detected as the monomer (approximately 25.7 kDa, z = 9+ to 11+), homodimer (approximately 51.5 kDa, z = 13+ to 16+) and homotetramer (approximately 103 kDa, z = 19+ to 22+), the result of a pH-dependent equilibrium.39 The mass spectrum obtained with the FAIMS voltages off is shown in Figure 1c. CV optimization revealed transmission of tetramer charge states 19+ to 22+ over the CV range −44 V to −15 V (Figure S2, Supporting Information). The FAIMS mass spectrum shown in Figure 1d was obtained with a static CV of −28 V, providing balanced transmission for the four charge states. At this CV, the S/N of the 21+ charge state of the tetramer improved by over 5-fold. Monomer units with a narrow charge state distribution were also detected both with and without FAIMS voltages, indicative of their presence in solution rather than a gas-phase dissociation product. At the static CV of −28 V, the monomer ions were poorly transmitted; monomer transmission was optimal at a CV of approximately −36 V.
ADH (PDB entry 4w6z) was detected in its homotetramer conformation (approximately 147 kDa, z = 23+ to 28+) over the CV range −40 V to −15 V (Figure S3, Supporting Information). Figure 1e shows the mass spectrum obtained with the FAIMS voltages off, and Figure 1f shows the static FAIMS mass spectrum obtained with CV = −24 V (selected for transmission of tetramers over a range of charge states). The S/N of the 26+ charge state of the tetramer increased by 50% at this CV. Ions with m/z corresponding to the homodimeric conformation (z = 16+ to 18+) were detected with FAIMS voltages on and off, suggestive of a degree of in-solution or in-source dissociation unrelated to FAIMS voltages.
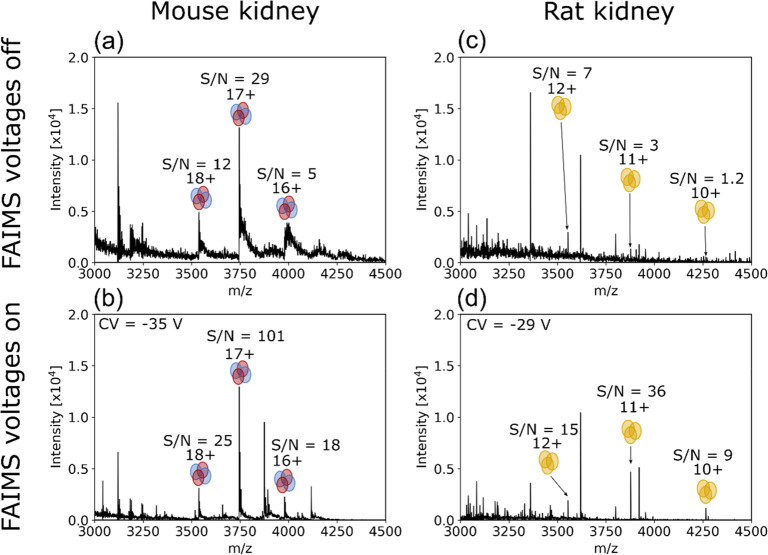
Subsequent experiments focused on integration of native LESA mass spectrometry with FAIMS. To assess the potential of native LESA FAIMS MS for analyzing protein assemblies directly from tissue, sections of mouse and rat kidney were sampled. The renal pelvis (see Figure S4a, Supporting Information) of the mouse kidney section, which is characterized by abundant vasculature, was sampled using ammonium acetate solution (200 mM) + 5% MeOH. These sampling conditions are known to be suitable for detection of the hemoglobin heterotetramer (αβ2H)2 (PDB entry 3hrw).25,27 Intact hemoglobin tetramer (αβ2H)2 (63.6 kDa) ions were observed, i.e., were successfully transmitted by the FAIMS device. (Protein assignment was based on mass measurement.) The CV was optimized for transmission of 17+ ions of the tetramer (see Figure S5, Supporting Information), and found to be −35 V. The LESA mass spectra with FAIMS voltages off and static FAIMS voltages on (CV = −35 V) are shown in Figure 2a,b, respectively. (αβ2H)2 ions were also detected in charge states 16+ and 18+. S/N was improved by approximately 2–3-fold for each charge state using the static CV while absolute signal intensities remained similar. In addition, heterodimer ions (αβ2H, 10+) were detected over the CV range −40 to −16 V and holo-alpha subunit (αH, 5+) ions were detected over the CV range −45 to −11 V, both also remaining bound to the heme ligand(s) (see Figure S5, Supporting Information). Heterodimers and heme-bound monomers are commonly observed in native LESA experiments25,40,41 (in the absence of FAIMS) and are the consequence of either physiological or in solution dissociation.42,43
Figure 2.
(a) LESA MS spectrum from a location on the mouse kidney section showing hemoglobin tetramer ions (16+ to 18+) detected with FAIMS voltages off. (b) With FAIMS voltages on (static CV = −35 V), the S/N for each Hb charge state was increased 2–3-fold with an obvious reduction in baseline signal intensity. Mass spectra are the average of 1 min of acquired data. (c) LESA MS spectrum of a location in the cortex of rat kidney section with FAIMS voltages off. Peaks corresponding to the homotrimeric RidA complex in the 10+ to 12+ charge states were detected. (d) LESA FAIMS mass spectrum (CV = −29 V) of an adjacent location in the cortex. S/N was increased and baseline signals reduced for the three charge states with FAIMS voltages on. Spectra are the average of 94 scans with a maximum injection time of 500 ms.
LESA sampling of the rat kidney tissue was performed in the cortex region (see Figure S4b, Supporting Information) using an extraction solvent comprising ammonium acetate solution (10 mM) containing 0.125% C8E4 detergent. This combination of sampling conditions and location has previously been shown to result in the detection of ions corresponding to the homotrimer reactive intermediate deiminase A (RidA).26 The homotrimer ions of RidA (42.6 kDa, PDB entry 1qah) were successfully transmitted through the FAIMS device and observed in charge states 9+ through 12+ (protein assignment was based on mass measurement). Extracted ion chromatograms for charge states 10+, 11+, and 12+ are shown in Figure S6, Supporting Information. The optimum CVs for transmission of these charge states were CV = −24 V, −27 V, and −32 V, respectively. A CV of −29 V was selected for subsequent LESA static FAIMS analysis to enable transmission of multiple charge states. The LESA mass spectra obtained from the rat kidney cortex with FAIMS voltages off and on (CV = −29 V) are shown in Figure 2c,d. For the 12+ charge state, S/N improved ∼2-fold with the FAIMS on and the absolute signal intensities were similar. For the 11+ and 10+ charge states, S/N improved between 7- and 12-fold with concomitant increases in absolute signal intensities.
For all the species described above, both the protein standards and the endogenous proteins sampled directly from tissue, the detection of the intact protein complexes in their native states, i.e., their established oligomeric forms in the case of the protein assemblies (homotetrameric ADH and ConA, heterotetrameric Hb, and homotrimeric RidA) and Zn-bound CAH, confirm that the FAIMS Pro is suitable for integration in native mass spectrometry. Any FAIMS-associated field heating is insufficient to cause dissociation of the protein complexes. The ions considered here are much larger than those previously considered in field heating experiments and, with the exception of CAH, have higher charge states. For ADH and ConA, monomer and dimer subunits were observed both with and without FAIMS indicating that they are present in solution rather than a consequence of heating in the gas-phase. For CAH, the holo-protein was observed exclusively. A scan of CV values designed to address the possibility of self-cleaning revealed no evidence for the apoprotein.
Conclusions
Intact protein complexes can be selectively transmitted by FAIMS prior to mass analysis, as evidenced by detection of protein assemblies up to 147 kDa. Improvements in S/N following integration of FAIMS were observed for all proteins studied here. Transmission through the FAIMS device suggests that any field heating induced by the device is insufficient to disrupt noncovalent interactions governing quaternary structure and ligand binding. Additionally, this is the first demonstration of native LESA FAIMS-MS for intact protein complexes sampled directly from tissue. Hemoglobin tetramer ions remained assembled and were separated from dimer and monomer units on the basis of their differential mobility. Trimeric RidA ions also remained intact. For complex samples analyzed with minimal sample preparation, such as thin tissue sections, FAIMS now presents an opportunity for improving native protein ion signals by reduction of chemical noise, an advantage previously shown for denaturing LESA MS.18,20,21
Acknowledgments
H.J.C. is an EPSRC Established Career Fellow (Grant EP/S002979/1). O.J.H. is funded by EPSRC (Grant EP/S002979/1). E.I.-T. is funded by EPSRC (Grant EP/R018367/1). The Orbitrap Eclipse mass spectrometer and FAIMS Pro device used in this work were funded by BBSRC (Grant BB/S019456/1).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c00649.
Experimental details; Figure S1, expanded m/z range showing the 10+ ion of holo-CAH obtained with the FAIMS voltages off, under static FAIMS conditions (CV = −30 V), and following a CV sweep from −60 V to −10 V; Figure S2, extracted ion chromatograms for ConA tetramers; Figure S3, extracted ion chromatograms for ADH tetramers; Figure S4, photographs of mouse and rat kidney sections; Figure S5, optimization of CV for transmission of hemoglobin ions; and Figure S6, optimization of CV for transmission of rat RidA ions (PDF)
The authors declare no competing financial interest.
Notes
Supplementary data supporting this research is openly available from the University of Birmingham data archive at DOI: 10.25500/edata.bham.00000476.
Supplementary Material
References
- Leney A. C.; Heck A. J. Native Mass Spectrometry: What is in the Name?. J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. 10.1007/s13361-016-1545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K.; Wildgoose J. L.; Langridge D. J.; Campuzano I. A method for direct measurement of ion mobilities using a travelling wave ion guide. Int. J. Mass Spectrom. 2010, 298, 10–16. 10.1016/j.ijms.2009.10.008. [DOI] [Google Scholar]
- Scarff C. A.; Patel V. J.; Thalassinos K.; Scrivens J. H. Probing hemoglobin structure by means of traveling-wave ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2009, 20, 625–631. 10.1016/j.jasms.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Scarff C. A.; Thalassinos K.; Hilton G. R.; Scrivens J. H. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun. Mass Spectrom. 2008, 22, 3297–3304. 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- Ruotolo B. T.; Benesch J. L.; Sandercock A. M.; Hyung S. J.; Robinson C. V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3, 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Michaelevski I.; Kirshenbaum N.; Sharon M. T-wave ion mobility-mass spectrometry: basic experimental procedures for protein complex analysis. J. Visualized Exp. 2010, 1985. 10.3791/1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmer D. E.; Hudgins R. R.; Jarrold M. F. Naked Protein Conformations: Cytochrome c in the Gas Phase. J. Am. Chem. Soc. 1995, 117, 10141–10142. 10.1021/ja00145a037. [DOI] [Google Scholar]
- Hudgins R. R.; Woenckhaus J.; Jarrold M. F. High resolution ion mobility measurements for gas phase proteins: correlation between solution phase and gas phase conformations. Int. J. Mass Spectrom. Ion Processes 1997, 165–166, 497–507. 10.1016/S0168-1176(97)00182-1. [DOI] [Google Scholar]
- Shelimov K. B.; Clemmer D. E.; Hudgins R. R.; Jarrold M. F. Protein Structurein Vacuo: Gas-Phase Conformations of BPTI and Cytochromec. J. Am. Chem. Soc. 1997, 119, 2240–2248. 10.1021/ja9619059. [DOI] [Google Scholar]
- Giles K.; Pringle S. D.; Worthington K. R.; Little D.; Wildgoose J. L.; Bateman R. H. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004, 18, 2401–2414. 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lima F.; Kaplan D. A.; Suetering J.; Park M. A. Gas-phase separation using a trapped ion mobility spectrometer. Int. J. Ion Mobility Spectrom. 2011, 14, 93. 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabelica V.; Shvartsburg A. A.; Afonso C.; Barran P.; Benesch J. L. P.; Bleiholder C.; Bowers M. T.; Bilbao A.; Bush M. F.; Campbell J. L.; Campuzano I. D. G.; Causon T.; Clowers B. H.; Creaser C. S.; De Pauw E.; Far J.; Fernandez-Lima F.; Fjeldsted J. C.; Giles K.; Groessl M.; et al. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom. Rev. 2019, 38, 291–320. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. C.; Kirk S. R.; Bleiholder C. On the structural denaturation of biological analytes in trapped ion mobility spectrometry - mass spectrometry. Analyst 2016, 141, 3722–3730. 10.1039/C5AN02399H. [DOI] [PubMed] [Google Scholar]
- Guevremont R. High-field asymmetric waveform ion mobility spectrometry: a new tool for mass spectrometry. J. Chromatogr A 2004, 1058, 3–19. 10.1016/S0021-9673(04)01478-5. [DOI] [PubMed] [Google Scholar]
- Robinson E. W.; Shvartsburg A. A.; Tang K.; Smith R. D. Control of ion distortion in field asymmetric waveform ion mobility spectrometry via variation of dispersion field and gas temperature. Anal. Chem. 2008, 80, 7508–7515. 10.1021/ac800655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsburg A. A.; Li F.; Tang K.; Smith R. D. Distortion of ion structures by field asymmetric waveform ion mobility spectrometry. Anal. Chem. 2007, 79, 1523–1528. 10.1021/ac061306c. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L.; Dexter A.; Creese A. J.; Cooper H. J. Liquid extraction surface analysis field asymmetric waveform ion mobility spectrometry mass spectrometry for the analysis of dried blood spots. Analyst 2015, 140, 6879–6885. 10.1039/C5AN00933B. [DOI] [PubMed] [Google Scholar]
- Kocurek K. I.; May R. C.; Cooper H. J. Application of High-Field Asymmetric Waveform Ion Mobility Separation to LESA Mass Spectrometry of Bacteria. Anal. Chem. 2019, 91, 4755–4761. 10.1021/acs.analchem.9b00307. [DOI] [PubMed] [Google Scholar]
- Sarsby J.; Griffiths R. L.; Race A. M.; Bunch J.; Randall E. C.; Creese A. J.; Cooper H. J. Liquid Extraction Surface Analysis Mass Spectrometry Coupled with Field Asymmetric Waveform Ion Mobility Spectrometry for Analysis of Intact Proteins from Biological Substrates. Anal. Chem. 2015, 87, 6794–6800. 10.1021/acs.analchem.5b01151. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L.; Creese A. J.; Race A. M.; Bunch J.; Cooper H. J. LESA FAIMS Mass Spectrometry for the Spatial Profiling of Proteins from Tissue. Anal. Chem. 2016, 88, 6758–6766. 10.1021/acs.analchem.6b01060. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L.; Simmonds A. L.; Swales J. G.; Goodwin R. J. A.; Cooper H. J. LESA MS Imaging of Heat-Preserved and Frozen Tissue: Benefits of Multistep Static FAIMS. Anal. Chem. 2018, 90, 13306–13314. 10.1021/acs.analchem.8b02739. [DOI] [PubMed] [Google Scholar]
- Towers M. W.; Karancsi T.; Jones E. A.; Pringle S. D.; Claude E. Optimised Desorption Electrospray Ionisation Mass Spectrometry Imaging (DESI-MSI) for the Analysis of Proteins/Peptides Directly from Tissue Sections on a Travelling Wave Ion Mobility Q-ToF. J. Am. Soc. Mass Spectrom. 2018, 29, 2456–2466. 10.1007/s13361-018-2049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza K. Y.; Feider C. L.; Klein D. R.; Rosenberg J. A.; Brodbelt J. S.; Eberlin L. S. Desorption Electrospray Ionization Mass Spectrometry Imaging of Proteins Directly from Biological Tissue Sections. Anal. Chem. 2018, 90, 7785–7789. 10.1021/acs.analchem.8b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melani R. D.; Srzentic K.; Gerbasi V. R.; McGee J. P.; Huguet R.; Fornelli L.; Kelleher N. L. Direct measurement of light and heavy antibody chains using ion mobility and middle-down mass spectrometry. MAbs 2019, 11, 1351–1357. 10.1080/19420862.2019.1668226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. J.; Griffiths R. L.; Edwards R. L.; Cooper H. J. Native Liquid Extraction Surface Analysis Mass Spectrometry: Analysis of Noncovalent Protein Complexes Directly from Dried Substrates. J. Am. Soc. Mass Spectrom. 2015, 26, 1320–1327. 10.1007/s13361-015-1152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. L.; Konijnenberg A.; Viner R.; Cooper H. J. Direct Mass Spectrometry Analysis of Protein Complexes and Intact Proteins up to > 70 kDa from Tissue. Anal. Chem. 2019, 91, 6962–6966. 10.1021/acs.analchem.9b00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. L.; Sisley E. K.; Lopez-Clavijo A. F.; Simmonds A. L.; Styles I. B.; Cooper H. J. Native mass spectrometry imaging of intact proteins and protein complexes in thin tissue sections. Int. J. Mass Spectrom. 2019, 437, 23–29. 10.1016/j.ijms.2017.10.009. [DOI] [Google Scholar]
- Purves R. W.; Barnett D. A.; Ells B.; Guevremont R. Elongated conformers of charge states + 11 to + 15 of bovine ubiquitin studied using ESI-FAIMS-MS. J. Am. Soc. Mass Spectrom. 2001, 12, 894–901. 10.1016/S1044-0305(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Baker E. S.; Clowers B. H.; Li F.; Tang K.; Tolmachev A. V.; Prior D. C.; Belov M. E.; Smith R. D. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J. Am. Soc. Mass Spectrom. 2007, 18, 1176–1187. 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsburg A. A.; Danielson W. F.; Smith R. D. High-resolution differential ion mobility separations using helium-rich gases. Anal. Chem. 2010, 82, 2456–2462. 10.1021/ac902852a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsburg A. A.; Smith R. D. Accelerated high-resolution differential ion mobility separations using hydrogen. Anal. Chem. 2011, 83, 9159–9166. 10.1021/ac202386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsburg A. A.; Smith R. D. High-resolution differential ion mobility spectrometry of a protein. Anal. Chem. 2013, 85, 10–13. 10.1021/ac3029129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsburg A. A. Ultrahigh-resolution differential ion mobility separations of conformers for proteins above 10 kDa: onset of dipole alignment?. Anal. Chem. 2014, 86, 10608–10615. 10.1021/ac502389a. [DOI] [PubMed] [Google Scholar]
- Shvartsburg A. A.; Noskov S. Y.; Purves R. W.; Smith R. D. Pendular proteins in gases and new avenues for characterization of macromolecules by ion mobility spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6495–6500. 10.1073/pnas.0812318106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysik A. J.; Read P.; Little D. R.; Bateman R. H.; Radford S. E.; Ashcroft A. E. Separation of beta2-microglobulin conformers by high-field asymmetric waveform ion mobility spectrometry (FAIMS) coupled to electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2229–2234. 10.1002/rcm.1613. [DOI] [PubMed] [Google Scholar]
- Prasad S.; Belford M. W.; Dunyach J.-J.; Purves R. W. On an Aerodynamic Mechanism to Enhance Ion Transmission and Sensitivity of FAIMS for Nano-Electrospray Ionization-Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2014, 25, 2143–2153. 10.1007/s13361-014-0995-8. [DOI] [PubMed] [Google Scholar]
- Saito R.; Sato T.; Ikai A.; Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 A resolution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 792–795. 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- Yin S.; Loo J. A. Top-Down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int. J. Mass Spectrom. 2011, 300, 118–122. 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeri Erba E.; Barylyuk K.; Yang Y.; Zenobi R. Quantifying protein-protein interactions within noncovalent complexes using electrospray ionization mass spectrometry. Anal. Chem. 2011, 83, 9251–9259. 10.1021/ac201576e. [DOI] [PubMed] [Google Scholar]
- Mikhailov V. A.; Griffiths R. L.; Cooper H. J. Liquid extraction surface analysis for native mass spectrometry: Protein complexes and ligand binding. Int. J. Mass Spectrom. 2017, 420, 43–50. 10.1016/j.ijms.2016.09.011. [DOI] [Google Scholar]
- Yan B.; Taylor A. J.; Bunch J. Cryo-LESA Mass Spectrometry-a Step Towards Truly Native Surface Sampling of Proteins. J. Am. Soc. Mass Spectrom. 2019, 30, 1179–1189. 10.1007/s13361-019-02178-7. [DOI] [PubMed] [Google Scholar]
- Griffith W. P.; Kaltashov I. A. Highly Asymmetric Interactions between Globin Chains during Hemoglobin Assembly Revealed by Electrospray Ionization Mass Spectrometry. Biochemistry 2003, 42, 10024–10033. 10.1021/bi034035y. [DOI] [PubMed] [Google Scholar]
- Schaer D. J.; Buehler P. W.; Alayash A. I.; Belcher J. D.; Vercellotti G. M. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2013, 121, 1276–1284. 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.