Abstract
Silk has a long track record of clinical use in the human body, and new formulations, including silk nanoparticles, continue to reveal the promise of this natural biopolymer for healthcare applications. Native silk fibroin can be isolated directly from the silk gland, but generating sufficient material for routine studies is difficult. Consequently, silk fibroin, typically extracted from cocoons, serves as the source for nanoparticle formation. This silk requires extensive processing (e.g., degumming, dissolution, etc.) to yield a hypoallergenic aqueous silk stock, but the impact of processing on nanoparticle production and characteristics is largely unknown. Here, manual and microfluidic-assisted silk nanoparticle manufacturing from 60- and 90-min degummed silk yielded consistent particle sizes (100.9–114.1 nm) with low polydispersity. However, the zeta potential was significantly lower (P < 0.05) for microfluidic-manufactured nanoparticles (−28 to −29 mV) than for manually produced nanoparticles (−39 to −43 mV). Molecular weight analysis showed a nanoparticle composition similar to that of the silk fibroin starting stock. Reducing the molecular weight of silk fibroin reduced the particle size for degumming times ≤30 min, whereas increasing the molecular weight polydispersity improved the nanoparticle homogeneity. Prolonged degumming (>30 min) had no significant effect on particle attributes. Overall, the results showed that silk fibroin processing directly impacts nanoparticle characteristics.
Keywords: silk fibroin, nanoparticles, molecular weight, biopolymer, biomaterials
Introduction
For millennia, humans have exploited silk for biomedical applications.1 The outstanding mechanical properties,2 biocompatibility, and biodegradation1,3 of silk have been its critical quality attributes. Silk is produced by various insects and spiders. However, the unique ability to farm Bombyx mori silkworms (i.e., sericulture) enables the industrial production of the common silk. The highly dependable supply chain for B. mori silk fibroin has supported its widespread use in humans. B. mori silk is the only silk type approved for routine clinical use both as a fiber (i.e., sutures and surgical mesh, SERI Surgical Scaffold) and as a hydrogel (Silk Voice for vocal fold augmentation, Sofregen Medical inc, Medford, MA, U.S.A.).1 The B. mori silk fibroin protein consists of a light (≈26 kDa)4 and a heavy chain (≈391 kDa)5,6 linked by a single disulfide bond.7 Unlike the light chain, the heavy chain has a highly repetitive block copolymer structure consisting of short hydrophilic amorphous domains and dominating crystallizable hydrophobic blocks. These hydrophobic blocks typically contain glycine-X (GX) repeats, where X is alanine (A) (65%), serine (S) (23%), or tyrosine (Y) (9%) and form extensive β-sheet structures.5,6 The β-sheet structures are not only important for the mechanical properties of silk fibroin but also contribute to the excellent biocompatibility1,3 of silk while protecting silk from rapid proteolytic degradation.8
The processing of common silk fibers into a regenerated silk fibroin protein stock is a well-established practice;9 this aqueous stock is similar, but not identical, to the native one present in the silk gland for B. mori silkworms.10 Native silk fibroin was therefore included in the present study as a control. Our ability to reverse-engineer the B. mori silk cocoon has enabled the development of new silk formats.9 Examples include films, scaffolds, hydrogels, and micro- or nanoparticles.11 Silk has persistently shown biocompatibility, biodegradation, and mechanical robustness across many different biomedical applications.1,3 For example, silk nanoparticles are emerging as interesting drug-delivery platforms12,13 and have been earmarked for systemic applications, including active solid tumor targeting.14 A broad range of silk nanoparticle manufacturing methods have been explored (reviewed in15), for example, emulsification (170 nm),16 poly(vinylalcohol) blending (300 nm to 20 μm),17 salting out (480 nm to 1.2 μm),18 supercritical CO2 (50–100 nm),19,20 and nanoprecipitation using organic solvents (50–300 nm).21−23 Nanoprecipitation is an easy and robust method of manual-based nanoparticle manufacturing, although microfluidic manufacturing routes are now being explored to optimize process control, reduce wastage, and enhance product quality and consistency.24 Microfluidics has been used to generate recombinant spider silk microparticles (e.g., ref (25)). We24 as well as others18,26,27 have recently applied microfluidic-based approaches to tune both recombinant and silk fibroin nanoparticle manufacturing. Silk fibroin nanoparticles have demonstrated a desirable performance profile, including low blood coagulation,28 metabolic rewiring,29 and differential macrophage activation.30 Silk nanoparticles also improved drug delivery into drug-resistant cancer cells via endocytosis-mediated uptake22 and default trafficking, resulting in lysosomotrophic drug release31 in response to low pH and lysosomal enzymes.8 Now, in silico modeling32 and wet lab experiments33 are unraveling drug-silk interactions. Our ability to process silk fibroin under mild conditions and exploit silk self-assembly, thus eliminating the need for chemical cross-linkers, is important for the design of silk nanomedicines.15,34 The capacity of silk fibroin to protect liable payloads from degradation singles this biopolymer out from many other (bio)polymers.35,36
Formation of the regenerated silk fibroin protein stock requires sericin removal (i.e., degumming),9 which is an essential step to yield hypoallergenic silk fibroin.1 The most common degumming method is the use of alkaline solutions at high temperatures.9 Next, the extracted silk fibers are dissolved using a chaotropic agent (e.g., LiBr), or as recently published, in melted ionic liquids in combination with harsh ultrasonification37 to dismantle higher-order silk fibroin structures.9 During degumming and, to a lesser extent, during dissolution in chaotropic agents, fragmentation of the silk protein backbone occurs,38 reducing the molecular weight and subsequently increasing the polydispersity of the biopolymer,39,40 with studies reporting the consequences on mechanics, self-assembly,40 degradation,41 and drug release.42 However, the impact of silk fibroin stock heterogeneity on nanoparticle characteristics remains poorly understood, and the molecular weight composition of silk nanoparticles is unknown. The influence of the silk fibroin stock during manual or microfluidic desolvation also requires investigation to determine if this influences the physical properties of the resulting silk nanoparticles. Therefore, this study addressed these issues by tuning the silk stock processing parameters and the method of desolvation to assess the impact on the silk nanoparticle secondary structure, zeta potential, size, composition, morphology, and cytotoxicity.
Experimental Section
Extraction of B. mori Silk and Silk Solution Preparation
Reverse-Engineered Silk Fibroin
Silk from B. mori cocoons was extracted as described elsewhere;9 a video format has been reported previously.43 Briefly, B. mori cocoons were cut and boiled in 0.02 M Na2CO3 solution for either 10, 30, 60, or 90 minutes. The degummed silk was washed with ultrapure water, air-dried, and dissolved in 9.3 M LiBr solution for up to 4 h at 60 °C. The resulting solution was dialyzed (molecular weight cutoff 3500 Da, Thermo Fisher Scientific, Waltham, MA, U.S.A.) against ultrapure water for 72 h to remove the LiBr salt.
Native Silk Fibroin
Silk fibroin subjected to minimal processing was directly isolated from the silk gland. Briefly, B. mori silkworms in the fourth instar were purchased (Silkwormstore Ltd., London, U.K.) and reared on reconstituted mulberry leaves at 32 °C with a 12 h day–night cycle. Native silk was isolated from the gland of B. mori silkworms in the fifth instar that had started to spin.44 Silkworms were anesthetized by placing them on ice; they were decapitated, and the gland was removed. The gland section between the sericin-free posterior and the posterior median section was cut and transferred into ice-cold ultrapure water. Next, the epithelium was peeled off under a stereomicroscope with forceps. The contents were washed three times for 45 min with ultrapure water to remove residual sericin, diluted in 1 mL ultrapure water, and kept at 4 °C overnight. This silk stock was denoted as native silk. The silk concentration was determined gravimetrically by lyophilizing a known volume of the solution and measuring the resulting dry weight.
Preparation of Silk Nanoparticles
Silk nanoparticles were produced by nanoprecipitation using either a manual process22 or a microfluidic setup24 (Figure 1a). First, the silk solutions from B. mori cocoons were diluted to 3% w/v with ultrapure water. Native silk solutions had a high viscosity and were, therefore, diluted to 0.25% w/v to allow handling during nanoparticle production. For the manual setup, the silk solution was added dropwise to isopropanol (99.5%, Ph. Eur) at a volume ratio of 5:1 (i.e., final isopropanol to silk solution ratio). The microfluidic-assisted setup was a NanoAssemblrTM benchtop instrument (version 1.5, model number: SN: NA-1.5-16) (NanoAssemblrTM, Precision Nanosystems Inc. Vancouver, Canada) equipped with a microfluidic cartridge (product code: NIT0012) (Precision Nanosystems). A total flow rate of 1 mL min–1 at a volume ratio of 5:1 of isopropanol to silk was used. The volumes were kept equal for the manual preparation process. To ensure proper mixing and nanoparticle formation, 350 μL at the beginning and 50 μL at the end of the process were discarded. The resulting nanoparticle suspensions were centrifuged at 48,400g for 2 h; the supernatant was aspirated, and the residue was resuspended in ultrapure water by sonicating twice for 30 seconds at 30% amplitude (Sonoplus HD 2070 ultrasonic probe, Bandelin, Berlin, Germany). These steps were repeated twice. After the final centrifugation, a concentrated silk nanoparticle suspension was prepared by resuspending the residue in 4 mL of water. The particles were stored at 4 °C until use.
Figure 1.
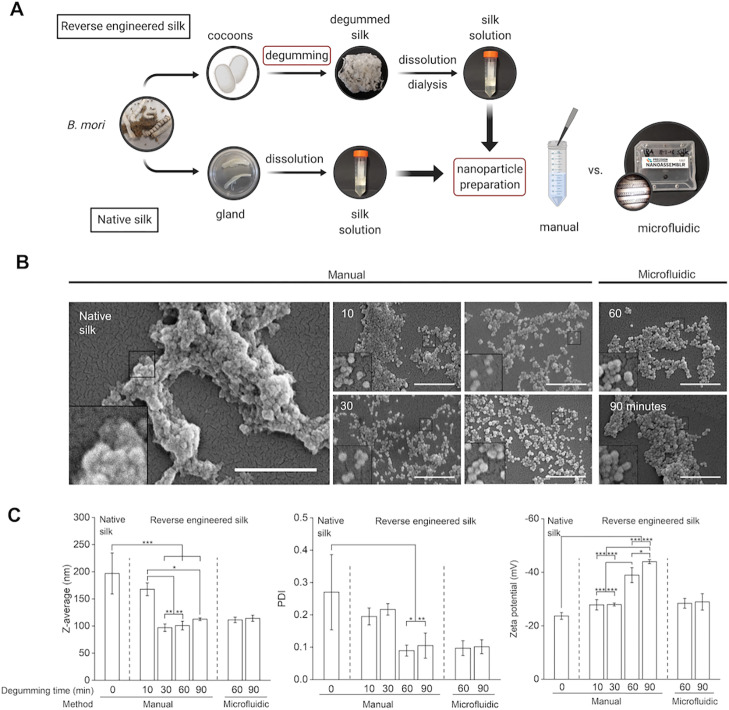
Impact of degumming time and manufacturing method on nanoparticle characteristics. (A) Schematic overview of the silk nanoparticle manufacturing process with either the manual or microfluidic setup. Top row: steps for reverse-engineered silk solution. Bottom row: native silk extraction and preparation of stock solution. Red boxes indicate that the process parameters varied. This panel was created with BioRender.com. (B) Qualitative assessment of nanoparticles with SEM (scale bar: 1 μm). (C) Quantitative nanoparticle assessment of size, polydispersity, and surface charge (n ≥ 3).
Characterization of Silk Nanoparticles
The yield of nanoparticles was determined by generating a standard curve of absorbance to mass for each formulation. A dilution series for each nanoparticle suspension was prepared, and the absorbance was measured at 600 nm (POLARstar Omega, BMG Labtech GmbH, Germany). Next, a known volume of silk suspension was lyophilized, and the mass of the dried residue was determined gravimetrically to relate the measured absorbance to mass. By taking into account the concentration and volume of the silk solution used, the yield of nanoparticles was calculated. The particle size and zeta potential were studied as detailed elsewhere.45 Briefly, the particles were diluted in ultrapure water and measured by dynamic light scattering (DLS, Zetasizer Nano-ZS Malvern Instrument, Worcestershire, U.K.) using refractive indices of 1.33 and 1.45 for ultrapure water and protein, respectively.
Scanning Electron Microscopy
Scanning electron microscopy (SEM) was used to visualize the morphology of the nanoparticles. Lyophilized silk nanoparticles were resuspended in distilled water to a concentration of 1 mg mL–1. The samples were then pipetted onto a silicon wafer and lyophilized overnight. The specimens were sputter-coated with 15 nm of gold using an ACE200 low vacuum sputter coater (Leica Microsystems, Wetzlar, Germany) and analyzed with an FE-SEM SU6600 (Hitachi High Technologies, Krefeld, Germany) at 5 kV. The SEM images were processed using ImageJ v1.52n (National Institutes of Health, Bethesda, MD, U.S.A.).
Silk Primary Structure Analysis
The silk protein size distribution was assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Lyophilized silk nanoparticles were dissolved in 9.3 M LiBr solution at 60 °C and dialyzed (molecular weight cutoff 3500 Da) against ultrapure water for 24 h to remove the LiBr salt. Laemmli sample buffer and β-mercaptoethanol (Bio-Rad Laboratories, Inc., Hercules, CA, U.S.A.) were added, and the samples were reduced for 10 min at 70 °C and loaded onto 4 to 20% Mini-PROTEAN TGX precast gels (Bio-Rad Laboratories). The gels were run with trisglycine SDS running buffer under reducing conditions. Spectra Multicolor High Range Protein Ladder (Invitrogen, Carlsbad, CA, U.S.A.) was run as a molecular standard. The gels were stained with a SilverXpress Silver Staining Kit (Thermo Fisher Scientific, Waltham, MA, USA). Images were acquired with an 8 megapixel Wileyfox Swift 2 plus phone, and the pixel density was analyzed by ImageJ v1.52n (National Institutes of Health, Bethesda, MD, U.S.A). The vertical profile was plotted against the distance in pixels using OriginPro2018b software (OriginLab Corporation, Northampton, MA, U.S.A.).
Secondary Structure Analysis of Silk Nanoparticles
The secondary structure of silk nanoparticles was examined using Fourier transform infrared (FTIR) spectroscopy. Silk nanoparticles were lyophilized before measurement to remove water. Native silk solution was air-dried prior to measurement. As reference controls, silk films were used that were either air-dried (low β-sheet content) or autoclaved to induce high β-sheet content. For all measurements, 128 scans were acquired at a resolution of 4 cm–1 over a 400 to 4000 cm–1 wavenumber range using a PerkinElmer Spectrum 100 instrument (PerkinElmer, Waltham, MA, U.S.A.). The measured interferogram was transformed into a spectrum by Fourier transformation. The spectra were normalized and analyzed with OriginPro2018b software by peak-fitting the amide I region as shown elsewhere.46 Briefly, the baseline was subtracted, and the full peak width at half-maximum was fixed at a constant value to resolve overlapping bands and avoid overfitting. The amide I region was identified (1595–1705 cm–1) and fitted by using 11 peaks centered around 1611 cm–1 as a tyrosine side chain, 1619, 1924, 1630, and 1698 cm–1 as a β-sheet structure, 1640 cm–1 as a random coil structure, 1659 cm–1 as an α-helical structure, and 1666, 1680, and 1691 cm–1 as a β-turn structure. The relative areas of the fitted peaks were used to determine the composition of the secondary structure (Figure S3).
In Vitro Cytotoxicity of Silk Nanoparticles
The mouse macrophage cell line RAW264.7 (ATCC, Manassas, VA, U.S.A.) was cultured in Dulbecco’s modified Eagle’s medium (4.5 g glucose, 110 mg sodium pyruvate) supplemented with 10% v/v fetal bovine serum, 50 U mL–1 penicillin, and 50 μg mL–1 streptomycin (Thermo Fisher Scientific, U.K.). Cultures were maintained in a humidified 5% CO2 atmosphere at 37 °C and cultured on tissue culture-treated polystyrene (Corning, New York, NY, U.S.A.). The cells were subcultured every 2–3 days (80% confluency) by scraping. For cytotoxicity assays, the cells were seeded into 96-well plates at a density of 1.5 × 104 cells per cm2 and allowed to recover overnight. Next, the medium was removed and replaced with a fresh medium containing silk nanoparticles (concentration range 2.5–250 μg mL–1). The cells were incubated for 48 h, and then 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromid (5 mg mL–1 in phosphate-buffered saline) was added to each well, and the cells were incubated for an additional 5 h. The supernatant was aspirated, the formazan crystals were dissolved in 100 μL dimethyl sulfoxide, and the absorbance was measured at 570 nm. Cell viability was calculated as a percentage by normalizing to the untreated control population (i.e., zero nanoparticle dose).
Statistical Analyses
Data were analyzed using OriginPro2018b. One-way analysis of variance followed by Bonferroni’s post hoc test was conducted between multiple groups. Sample pairs were analyzed by either a paired or unpaired t-Test. Normality and homogeneity of variances were assumed. Asterisks denote statistical significance as follows: *P < 0.05, **P < 0.01, and ***P < 0.001. All data are presented as mean values ±standard deviation, and the number of independent experiments (n) is noted in each figure legend.
Results
Nanoparticle Morphology, Size, and Surface Charge
Silk nanoparticle morphology was analyzed using SEM. All particles prepared from degummed silk had a spherical shape (Figure 1b). Stocks of 60- or 90-min degummed silk resulted in the most uniform particles independent of the manufacturing approach. These results correlated well with quantitative measurements using DLS (detailed below). The use of native silk for particle formulation led to irregularly shaped particles that appeared to aggregate and showed wide size distributions as quantified with DLS. The largest nanoparticles (196.8 nm) were obtained when using native silk (i.e., 0 min of degumming). The particle size decreased with 10 min of degumming time (167.7 nm) and ranged between 96.9 and 114.1 nm when using longer degumming times (30–90 min) (Figure 1b). However, polydispersity was the highest for 10- and 30-min degumming times (0.195–0.217), whereas the particles showed narrower size distribution for degumming times of 60 and 90 min with a polydispersity index of 0.088 and 0.107, respectively (Figure 1c). Microfluidic manufacturing, compared to the manual method, showed no substantial differences in size or polydispersity, whereas the surface charge decreased from −39 and −43 mV to less negative values (−28 and −29 mV) for 60- and 90-min degummed silks, respectively (Figure 1c). Across all processing conditions and silk stocks, the particle size ranged from 96.9 to 196.8 nm with a polydispersity index between 0.088 and 0.270. All particle formulations showed a negative surface charge, ranging from −24 to −43 mV (Figure 1c). Overall, yields ranged from approximately 8% to 25%; higher yields were observed for longer degummed silks (Figure S2). For 10-min degummed silk, both manual and microfluidic manufacturing had low yields (approximately 8%). The microfluidic-based method showed a significantly improved yield for 30-min degummed silk when compared to the manual method (8 vs 17%). For both 60- and 90-min degummed silk, yields were further improved (ranging between 19 and 25%) but was the best for the manual method.
Secondary Structure Analysis of Silk Films and Nanoparticles
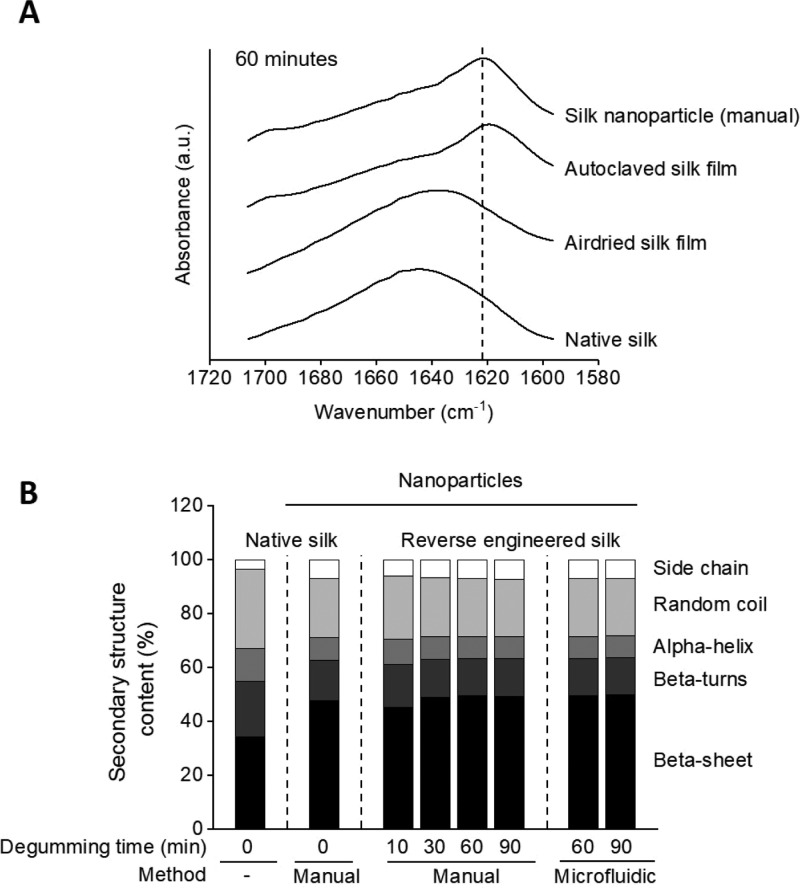
The secondary structure was analyzed using FTIR spectroscopy and peak fitting of the amide I region. Native silk obtained from the silkworm gland and air-dried silk (60-min degumming time) showed a characteristic peak at higher wavenumbers, indicating higher content of random coil structures (Figure 2a). In contrast, autoclaved silk and nanoparticles made with 60-min degummed silk showed a band between 1622 and 1627 cm–1, which is characteristic of the antiparallel β-sheet structure. These results correlated well with the secondary structure analysis after peak fitting (Figure 2b). Silk nanoparticles typically showed higher β-sheet (45–50%) and lower random coil content (21–23%) compared to native silk (34% β-sheet and 29% random coil). Using 10-min degummed silk generated nanoparticles with lower β-sheet content (45%) compared to the other nanoparticle formulations. All other degumming times showed no substantial differences between the samples (Figures 2 and S4). There was no significant difference in the secondary structure for batch or microfluidic manufactured silk nanoparticles.
Figure 2.
Secondary structure determination of silk nanoparticles. (A) FTIR absorbance spectra in the amide I region of nanoparticles manufactured from 60-min degummed silk with the manual method. Controls include native silk as well as air-dried and autoclaved silk film. The dashed line marks the absorption band between 1622 and 1627 cm–1, which is characteristic of the antiparallel β-sheet structure. (B) Secondary structure content of native silk and silk nanoparticles. Native silk fibroin was obtained from the silk gland with minimal processing. Silk nanoparticles were manufactured either manually or by microfluidics.
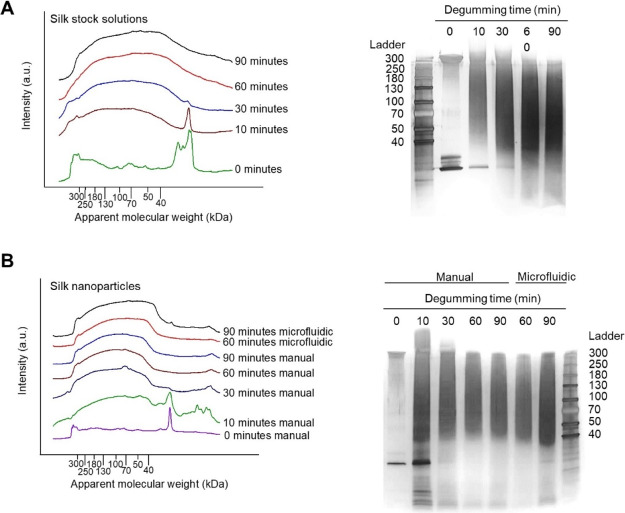
Fragmentation of the silk fibroin during the degumming process was analyzed using SDS-PAGE. The isolated native silk stock (i.e., 0 min of degumming) showed two major bands likely indicating the heavy chain (360 kDa) and light chain (26 kDa) of the silk fibroin (Figure 3a). With increasing degumming time, the clear bands at 360 and 26 kDa disappeared and were no longer visible at degumming times above 60 min. Furthermore, all degummed samples showed protein smearings that were shifted toward lower molecular weights with increasing degumming time. These trends were confirmed using densitometry gel analysis. Here, the absorbance intensity at 26 kDa decreased, and the overall intensity broadened with increasing degumming times. The assembly of silk fibroin fragments into nanoparticles was analyzed by redissolving silk nanoparticles and subjecting the extracted silk fibroin to the SDS-PAGE analysis (Figure 3b). Particles manufactured from native silk showed two main bands similar to those observed for the native silk fibroin stock solution. The analysis of nanoparticles from 10-min degummed silk showed a molecular weight distribution similar to what was observed in the respective stock solutions. At degumming times above 30 min, the silk fibroin band at 360 kDa disappeared completely from the silk fibroin stock; disassembly of the respective nanoparticles and analysis of the silk fibroin content showed no substantial differences between the starting stock and the incorporated silk fibroin fragments. There were no apparent differences in the silk incorporated into the nanoparticles generated using either manual or microfluidic-assisted manufacturing.
Figure 3.
Analysis of silk fibroin fragmentation and assembly into nanoparticles. SDS-PAGE (right) and densitometry analysis (left) of (A) silk stock solutions prepared from native and degummed B. mori silk and (B) analysis of the silk incorporated into nanoparticles manufactured manually or by microfluidics.
In Vitro Cytotoxicity of Silk Nanoparticles
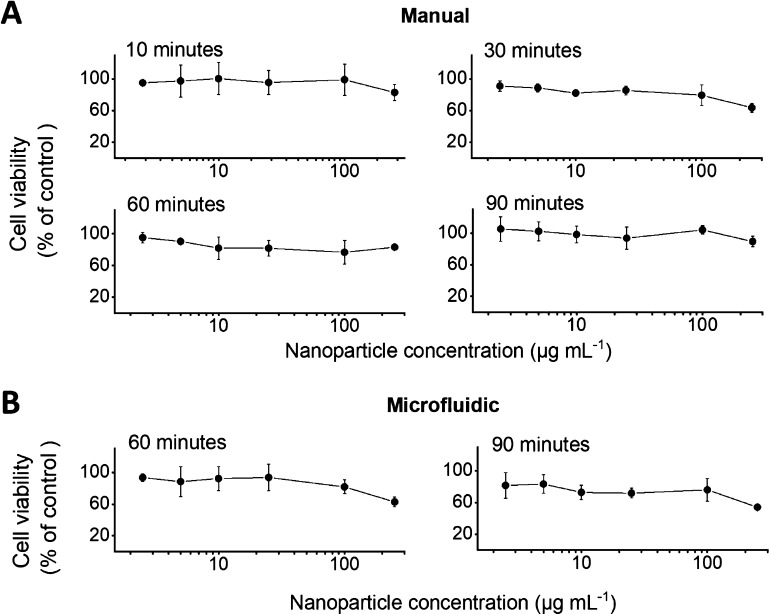
Cell viability of RAW 264.7 cells was analyzed using nanoparticle concentrations ranging from 2.5 to 250 μg mL–1 (Figure 4). The half-maximal inhibitory concentration (IC50) was >250 μg mL–1 for all formulations. Up to a concentration of 100 μg mL–1, the degumming time and the manufacturing method had no substantial effect on the cell viability. Overall, all formulations showed good biocompatibility in RAW 264.7 cells up to a concentration of 100 μg mL–1 (cell viability > 75%).
Figure 4.
In vitro cytotoxicity of silk nanoparticles in macrophages. RAW 264.7 cells were exposed for 48 h to 2.5–250 μg mL–1 nanoparticles prepared from 10-, 30-, 60-, or 90-min degummed silk. Nanoparticles were manufactured using either a (A) manual or (B) microfluidic method. (n = 3).
Discussion
Silk is now making inroads as a biopolymer for drug delivery across different material formats, including particles.34 A wide spectrum of manufacturing methods has been reported yielding silk particles ranging from the nano- to the microscale.12,15 For example, exploring the innovative use of ionic liquids.37 While the optimum particle size is application-dependent, particles in the nanometer size range permit first- to third-order targeting. For example, nanoparticles administered systemically have the potential to exploit leaky blood vessels that, in turn, facilitate the exit of nanoparticles from circulation and accumulation in the extracellular space and the subsequent entry of the nanoparticles into cells through endocytosis and lysosomal trafficking.13,47 The potential of tissue targeting of the payload and particle-mediated changes of cellular uptake mechanisms is exciting because they now provide a route to overcome delivery barriers for a broad range of payloads.48 However, the manufacture of these particles to a clinical standard is challenging, and improved production routes are needed.49,50 For example, the pharmaceutical industry is currently shifting from batch to continuous manufacturing in an effort to reduce manufacturing error, reduce contamination risk, and maximize production yields.51 Microfluidics is one such tool that is being adopted by the industry and hence included in the present study.
Our knowledge of silk processing parameters and the consequences on particle attributes is limited. One fundamental factor is silk degumming. To reflect current practices, we selected the commonly used sodium carbonate degumming process exploring a 10- to 90-min degumming time followed by the lithium-bromide silk fibroin dissolution protocol.9 Nanoparticles of similar sizes were obtained (96.9–122.4 nm) for a degumming time of 30–90 min using both microfluidic and manual-based production processes. This particle size is in good agreement with previous reports,22,43,45 although we observed a comparatively large particle size for 10-min degummed silk (167.7 nm) and increased polydispersity for the 10- and 30-min degummed samples. To differentiate, if this was due to silk fibroin stock attributes or the particle production process, we conducted pilot studies using our microfluidic setup (Figure S1, Supporting Information). Using microfluidics, the 10-min degummed silk fibroin yielded the largest silk nanoparticles (219.8 nm) with a comparatively high polydispersity. We therefore concluded that the silk fibroin stock critically influenced the size and sought to extract monodispersed silk fibroin directly from the glands of B. mori silkworms to be used in manual nanoparticle manufacturing. Native silk fibroin generated large silk nanoparticles (196.8 nm) with the highest polydispersity (0.270) and a significantly (P < 0.001) reduced surface charge when compared to 60-min degummed silk (−24 vs −39 mV). The native silk fibroin stock was diluted to reduce solution viscosity and improve handling. However, attempts to use this stock with our microfluidic setup resulted in catastrophic chip fouling; therefore, no preliminary data are available. Therefore, we decided not to further explore the native silk in the microfluidic setup because of the processing challenges but instead focus on reverse-engineered silk. We note that Holland and co-workers have successfully used native silk fibroin feedstocks in combination with microfluidics to create a microparticle library.44 We believe that microfluidic geometries and processing conditions are critical when working with native silk fibroin.
All nanoparticles manufactured in the present study were subjected to surface charge measurements. All current baseline measurements were conducted in water to minimize cofounding effects (e.g., aggregation). However, future studies exposing silk nanoparticles to buffered solutions could provide useful information to uncover further structural details. Manual manufacturing using 60- or 90-min degummed silk resulted in the most negative zeta potential values (−39 to −43 mV) similar to previous data.22,43,45 However, 10- and 30-min degummed silk yielded significantly (P < 0.001) lower surface charges (−28 and −28 mV, respectively). These values were closest to the zeta potential of native silk fibroin nanoparticles. Switching from manual to microfluidic-assisted production significantly (P < 0.05) reduced the zeta potential of 60- and 90-min degummed silk nanoparticles to values similar to those of 10- or 30-min degummed silk (Figure 1d), an important observation. These data suggest that the microfluidic setup generated nanoparticles that had a different silk fibroin packing geometry with more of the ionizable acidic functional groups hidden from the nanoparticle surface, albeit with a similar secondary structure (discussed below). This observation is surprising, and we speculate that the herringbone structure of the microfluidic chip allows for chaotic and rapid mixing,52 yielding silk nanoparticles with different surface characteristics. However, the exact mechanism of silk nanoparticle formation is currently unknown. Imaging showed that all processed samples contained nanoparticles, albeit with differences in appearance. Native silk fibroin nanoparticles showed a globular configuration with extensive cohesion and a “sticky” appearance that was less prominent for 10-min degummed samples and absent for all other nanoparticles. Degumming for 30 min and more resulted in a narrow particle size distribution and an appearance identical to those reported previously. Nanoparticle yields for identical degumming times were similar for both manual and microfluidic processed particles (Figure S2). A notable exception was 30-min degummed silk with microfluidic processing doubling the yield; the underlying reason for this is currently not known.
Secondary structure analysis revealed that both microfluidic and manually produced silk fibroin nanoparticles had a similar conformation; there were little differences in the secondary structure across the entire silk nanoparticle library. Both the production methods exposed silk nanoparticles to isopropanol. Isopropanol crystallizes silk, which is thereby expected to maximize the amount of β-sheets present in the secondary structure (Figure 2). However, in the absence of such a solvent, it is plausible to speculate that nanoparticles with an amorphous structure can be generated (e.g., ref (53)) by exploiting the silk fibroin natural micellar conformation. However, amorphous silk nanoparticles are predicted to have a poor shelf life and are thus of limited value for pharmaceutical applications. The native silk stock showed substantially lower crystallinity than samples exposed to isopropanol. However, we observed a β-sheet content of >30% which is higher than that reported in the literature (e.g., ref (54)). Secondary structure composition of native silk fibroin is easily influenced not only by its environment such as surrounding pH and salt concentration but also by mechanics such as shear stress and drying.55,56 This responsiveness can complicate the sample assessment but can also make silk fibroin a versatile material that is amenable to precise secondary structure tuning.57 Further evidence of process-dependent secondary structure tuning comes from experiments with the native silk fibroin stock. Here, different micrometer-sized silk constructs were manufactured using microfluidics; some of these shapes were highly amorphous.44
Contrary to our assumption that a narrow silk fibroin polydispersity would lead to a narrow particle size distribution, particle size control was the best at the 60- and 90-min degumming times. In contrast, a relatively large nanoparticle polydispersity was observed for the native silk fibroin stock despite the stock being essentially monodispersed. Seminal work by Carissimi and coworkers also suggests that a reduction in the molecular weight results in more uniform silk fibroin nanoparticles.58 Besides particle uniformity, the particle size was also monitored. Silk degummed for only 10 min yielded the largest nanoparticles (167.7 nm) and were most similar to those generated from the native silk fibroin stock (largest nanoparticles reported in this study). In contrast, degumming times of 30 min and longer consistently yielded silk nanoparticles in the 100 nm size range. This observation is in agreement with previous studies where a reduced silk fibroin molecular weight yielded smaller particles;59 our preliminary studies22 and work by Carissimi58 have corroborated these findings.
Commonly used silk degumming and dissolution processes38 damage the silk fibroin structure, resulting in a reduction in the molecular weight and increased polydispersity. We used sodium carbonate because it is widely used, effectively removes sericin, and results in up to 40% greater molecular weight reduction than sodium bicarbonate38 or urea.39 SDS-PAGE analysis indicated boiling time-dependent silk fibroin fragmentation of both light and heavy chains, confirming earlier reports.40 It is reasonable to speculate that the silk fibroin molecule does not undergo random molecular weight reduction except at key locations that are most susceptible. For example, the completely amorphous nature of a silk light chain renders this site most vulnerable to degradation as also the single disulfide bond between the light and heavy chains.7 The amorphous segments within a silk heavy chain are also expected to be damaged during degumming, whereas the silk crystalline domains are expected to last. SDS-PAGE analysis supports this sequence of events. SDS-PAGE analysis of 10-min degummed silk fibroin showed a prominent band at approximately 25 kDa, which was essentially absent at 30 min. It is plausible that this band is the liberated silk fibroin light chain. An alternative explanation could be that this band is the p25 glycoprotein fibrohexamerin (it has a molecular weight very similar to that of the silk fibroin light chain).5 Irrespective of the nature of this particular band, longer degumming times clearly lead to its fragmentation (i.e., disappearance). Therefore, the presence of this 25 kDa protein could impact particle formation and ultimately dictate the size. Although SDS-PAGE analysis of the silk fibroin stocks is important, in isolation, it provides little information about the actual composition of the silk nanoparticles. Therefore, silk nanoparticles were disassembled and analyzed with SDS-PAGE. Across all silk fibroin samples, the nanoparticle composition resembled the starting stock. There was no apparent preferential incorporation of protein subsets into the respective nanoparticles. However, silk nanoparticles generated from 10-min degummed silk had larger particle diameter (167.7 nm) and polydispersity (0.195) while containing a 25 kDa protein. One might speculate that the presence of this 25 kDa protein impacts nanoparticle characteristics (Figure 3b). Unfortunately, our experimental design did not allow us to prove this, and hence further studies are required to extract this 25 kDa band to fully elucidate its role. In addition to the spectrum of silk nanoparticle characteristics assessed here, the initial biological response toward macrophages was assessed too. All silk nanoparticles were well tolerated with similar results reported previously.29
Conclusions
Overall, this study demonstrates that silk fibroin processing directly impacts nanoparticle attributes. We employed both manual and microfluidic-assisted silk nanoparticle manufacturing routes and showed that neither the particle size nor the secondary structure was affected by the selected production methods. However, the zeta potential was significantly affected, and we speculate that microfluidics reduced the surface-accessible acidic functional groups. SDS-PAGE analysis of the nanoparticles showed that their composition resembled the silk fibroin starting stock. Overall, this study provides novel insight into the processing parameters influencing silk fibroin nanoparticle performance.
Acknowledgments
J.I.S. thanks ERASMUS + traineeship supported by the European Commission for funding this work. J.D.T. is supported by an EPSRC Doctoral Prize Award (EP/R513349/1). The authors acknowledge that the work was carried out in part at the EPSRC Future Manufacturing Research Hub for Continuous Manufacturing and Advanced Crystallisation (CMAC) (EP/P006965/1), supported by a UK Research Partnership Fund award from the Higher Education Funding Council for England (grant HH13054). The authors would like to acknowledge that the electron scanning microscopy work was carried out at the Advanced Materials Research Laboratory housed within the University of Strathclyde.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsbiomaterials.0c00202.
DLS and SEM of continuous manufactured nanoparticles from 10- and 30-min degummed silk; silk nanoparticle yields and exemplary band fitting of FTIR absorbance spectra; band assignment in the amide I region; FTIR absorbance spectra of silk film and nanoparticles from 10-, 30- and 90-min degummed silk; and secondary structure content of air-dried and autoclaved silk films (PDF)
Author Contributions
J.I.S. designed, analyzed, and interpreted the data and generated the manuscript draft. J.D.T and T.W. provided training, advised on experimental design, and contributed to the interpretation of the results. All authors discussed the results and/or provided advice on the experimental analysis. F.P.S. conceived the study and supervised the project. F.P.S. wrote and content-edited the manuscript with support from the other authors.
The authors declare no competing financial interest.
Notes
All data supporting this research are openly available from https://doi.org/10.15129/7387b6dc-f207-4c2f-8f97-94b3bd651468.
Supplementary Material
References
- Holland C.; Numata K.; Rnjak-Kovacina J.; Seib F. P. The Biomedical Use of Silk: Past, Present, Future. Adv. Healthcare Mater. 2019, 8, 1800465. 10.1002/adhm.201800465. [DOI] [PubMed] [Google Scholar]
- Omenetto F. G.; Kaplan D. L. New opportunities for an ancient material. Science 2010, 329, 528–531. 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber A. E.; Omenetto F. G.; Kaplan D. L. In vivo bioresponses to silk proteins. Biomaterials 2015, 71, 145–157. 10.1016/j.biomaterials.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K.; Kikuchi Y.; Takagi T.; Kikuchi A.; Oyama F.; Shimura K.; Mizuno S. Primary structure of the silk fibroin light chain determined by cDNA sequencing and peptide analysis. J. Mol. Biol. 1989, 210, 127–139. 10.1016/0022-2836(89)90295-7. [DOI] [PubMed] [Google Scholar]
- Inoue S.; Tanaka K.; Arisaka F.; Kimura S.; Ohtomo K.; Mizuno S. Silk Fibroin ofBombyx moriIs Secreted, Assembling a High Molecular Mass Elementary Unit Consisting of H-chain, L-chain, and P25, with a 6:6:1 Molar Ratio. J. Biol. Chem. 2000, 275, 40517–40528. 10.1074/jbc.M006897200. [DOI] [PubMed] [Google Scholar]
- Zhou C.-Z.; Confalonieri F.; Jacquet M.; Perasso R.; Li Z.-G.; Janin J. Silk fibroin: structural implications of a remarkable amino acid sequence. Proteins 2001, 44, 119–122. 10.1002/prot.1078. [DOI] [PubMed] [Google Scholar]
- Tanaka K.; Kajiyama N.; Ishikura K.; Waga S.; Kikuchi A.; Ohtomo K.; Takagi T.; Mizuno S. Determination of the site of disulfide linkage between heavy and light chains of silk fibroin produced by Bombyx mori. Biochim. Biophys. Acta 1999, 1432, 92–103. 10.1016/s0167-4838(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Wongpinyochit T.; Johnston B. F.; Seib F. P. Degradation Behavior of Silk Nanoparticles-Enzyme Responsiveness. ACS Biomater. Sci. Eng. 2018, 4, 942–951. 10.1021/acsbiomaterials.7b01021. [DOI] [PubMed] [Google Scholar]
- Rockwood D. N.; Preda R. C.; Yücel T.; Wang X.; Lovett M. L.; Kaplan D. L. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 2011, 6, 1612–1631. 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greving I.; Dicko C.; Terry A.; Callow P.; Vollrath F. Small angle neutron scattering of native and reconstituted silk fibroin. Soft Matter 2010, 6, 4389–4395. 10.1039/c0sm00108b. [DOI] [Google Scholar]
- Wenk E.; Merkle H. P.; Meinel L. Silk fibroin as a vehicle for drug delivery applications. J. Controlled Release 2011, 150, 128–141. 10.1016/j.jconrel.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Mottaghitalab F.; Farokhi M.; Shokrgozar M. A.; Atyabi F.; Hosseinkhani H. Silk fibroin nanoparticle as a novel drug delivery system. J. Controlled Release 2015, 206, 161–176. 10.1016/j.jconrel.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Seib F. P. Silk nanoparticles—an emerging anticancer nanomedicine. AIMS Bioeng. 2017, 4, 239–258. 10.3934/bioeng.2017.2.239. [DOI] [Google Scholar]
- Tian Y.; Jiang X.; Chen X.; Shao Z.; Yang W. Doxorubicin-loaded magnetic silk fibroin nanoparticles for targeted therapy of multidrug-resistant cancer. Adv. Mater. 2014, 26, 7393–7398. 10.1002/adma.201403562. [DOI] [PubMed] [Google Scholar]
- Crivelli B.; Perteghella S.; Bari E.; Sorrenti M.; Tripodo G.; Chlapanidas T.; Torre M. L. Silk nanoparticles: from inert supports to bioactive natural carriers for drug delivery. Soft Matter 2018, 14, 546–557. 10.1039/c7sm01631j. [DOI] [PubMed] [Google Scholar]
- Myung S. J.; Kim H.-S.; Kim Y.; Chen P.; Jin H.-J. Fluorescent Silk Fibroin Nanoparticles Prepared Using a Reverse Microemulsion. Macromol. Res. 2008, 16, 604–608. 10.1007/bf03218567. [DOI] [Google Scholar]
- Wang X.; Yucel T.; Lu Q.; Hu X.; Kaplan D. L. Silk nanospheres and microspheres from silk/pva blend films for drug delivery. Biomaterials 2010, 31, 1025–1035. 10.1016/j.biomaterials.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel A. S.; Hu X.; Park S.-H.; Kaplan D. L.; Scheibel T. R. Controlling silk fibroin particle features for drug delivery. Biomaterials 2010, 31, 4583–4591. 10.1016/j.biomaterials.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.-Q.; Kankala R. K.; He G.-Y.; Yang D.-Y.; Li G.-P.; Wang P.; Wang S.-B.; Zhang Y. S.; Chen A.-Z. Supercritical Fluid-Assisted Fabrication of Indocyanine Green-Encapsulated Silk Fibroin Nanoparticles for Dual-Triggered Cancer Therapy. ACS Biomater. Sci. Eng. 2018, 4, 3487–3497. 10.1021/acsbiomaterials.8b00705. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Li Y.; Chen A.-Z.; Zheng Z.-J.; Hu J.-Y.; Li J.-S.; Li G. Generation of Silk Fibroin Nanoparticles via Solution-Enhanced Dispersion by Supercritical CO2. Ind. Eng. Chem. Res. 2013, 52, 3752–3761. 10.1021/ie301907f. [DOI] [Google Scholar]
- Kundu J.; Chung Y.-I.; Kim Y. H.; Tae G.; Kundu S. C. Silk fibroin nanoparticles for cellular uptake and control release. Int. J. Pharm. 2010, 388, 242–250. 10.1016/j.ijpharm.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Seib F. P.; Jones G. T.; Rnjak-Kovacina J.; Lin Y.; Kaplan D. L. pH-dependent anticancer drug release from silk nanoparticles. Adv. Healthcare Mater. 2013, 2, 1606–1611. 10.1002/adhm.201300034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Q.; Shen W.-D.; Xiang R.-L.; Zhuge L.-J.; Gao W.-J.; Wang W.-B. Formation of silk fibroin nanoparticles in water-miscible organic solvent and their characterization. J. Nanopart. Res. 2007, 9, 885–900. 10.1007/s11051-006-9162-x. [DOI] [Google Scholar]
- Wongpinyochit T.; Totten J. D.; Johnston B. F.; Seib F. P. Microfluidic-assisted silk nanoparticle tuning. Nanoscale Adv. 2019, 1, 873–883. 10.1039/c8na00208h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel A.; Schwab M.; Slotta U.; Winter G.; Scheibel T. Processing conditions for the formation of spider silk microspheres. ChemSusChem 2008, 1, 413–416. 10.1002/cssc.200800030. [DOI] [PubMed] [Google Scholar]
- Florczak A.; Jastrzebska K.; Bialas W.; Mackiewicz A.; Dams-Kozlowska H. Optimization of spider silk sphere formation processing conditions to obtain carriers with controlled characteristics. J. Biomed. Mater. Res., Part A 2018, 106, 3211–3221. 10.1002/jbm.a.36516. [DOI] [PubMed] [Google Scholar]
- Mitropoulos A. N.; Perotto G.; Kim S.; Marelli B.; Kaplan D. L.; Omenetto F. G. Synthesis of silk fibroin micro- and submicron spheres using a co-flow capillary device. Adv. Mater. 2014, 26, 1105–1110. 10.1002/adma.201304244. [DOI] [PubMed] [Google Scholar]
- Maitz M. F.; Sperling C.; Wongpinyochit T.; Herklotz M.; Werner C.; Seib F. P. Biocompatibility assessment of silk nanoparticles: hemocompatibility and internalization by human blood cells. Nanomedicine 2017, 13, 2633–2642. 10.1016/j.nano.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Saborano R.; Wongpinyochit T.; Totten J. D.; Johnston B. F.; Seib F. P.; Duarte I. F. Metabolic Reprogramming of Macrophages Exposed to Silk, Poly(lactic-co-glycolic acid), and Silica Nanoparticles. Adv. Healthcare Mater. 2017, 6, 1601240. 10.1002/adhm.201601240. [DOI] [PubMed] [Google Scholar]
- Totten J. D.; Wongpinyochit T.; Carrola J.; Duarte I. F.; Seib F. P. PEGylation-Dependent Metabolic Rewiring of Macrophages with Silk Fibroin Nanoparticles. ACS Appl. Mater. Interfaces 2019, 11, 14515–14525. 10.1021/acsami.8b18716. [DOI] [PubMed] [Google Scholar]
- Totten J. D.; Wongpinyochit T.; Seib F. P. Silk nanoparticles: proof of lysosomotropic anticancer drug delivery at single-cell resolution. J. Drug Targeting 2017, 25, 865–872. 10.1080/1061186X.2017.1363212. [DOI] [PubMed] [Google Scholar]
- Wongpinyochit T.; Vassileiou A. D.; Gupta S.; Mushrif S. H.; Johnston B. F.; Seib F. P. Unraveling the Impact of High-Order Silk Structures on Molecular Drug Binding and Release Behaviors. J. Phys. Chem. Lett. 2019, 10, 4278–4284. 10.1021/acs.jpclett.9b01591. [DOI] [PubMed] [Google Scholar]
- Coburn J. M.; Na E.; Kaplan D. L. Modulation of vincristine and doxorubicin binding and release from silk films. J. Controlled Release 2015, 220, 229–238. 10.1016/j.jconrel.2015.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomeh M. A.; Hadianamrei R.; Zhao X. Silk Fibroin as a Functional Biomaterial for Drug and Gene Delivery. Pharmaceutics 2019, 11, 494. 10.3390/pharmaceutics11100494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard E. M.; Dennis P. B.; Omenetto F.; Naik R. R.; Kaplan D. L. Physical and chemical aspects of stabilization of compounds in silk. Biopolymers 2012, 97, 479–498. 10.1002/bip.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel T.; Lovett M. L.; Kaplan D. L. Silk-based biomaterials for sustained drug delivery. J. Controlled Release 2014, 190, 381–397. 10.1016/j.jconrel.2014.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Pérez A. A.; Montalbán M. G.; Aznar-Cervantes S. D.; Cragnolini F.; Cenis J. L.; Víllora G. Production of silk fibroin nanoparticles using ionic liquids and high-power ultrasounds. J. Appl. Polym. Sci. 2014, 132, 41702. 10.1002/app.41702. [DOI] [Google Scholar]
- Wang Q.; Chen Q.; Yang Y.; Shao Z. Effect of various dissolution systems on the molecular weight of regenerated silk fibroin. Biomacromolecules 2013, 14, 285–289. 10.1021/bm301741q. [DOI] [PubMed] [Google Scholar]
- Yamada H.; Nakao H.; Takasu Y.; Tsubouchi K. Preparation of undegraded native molecular fibroin solution from silkworm cocoons. Mater. Sci. Eng., C 2001, 14, 41–46. 10.1016/s0928-4931(01)00207-7. [DOI] [Google Scholar]
- Wray L. S.; Hu X.; Gallego J.; Georgakoudi I.; Omenetto F. G.; Schmidt D.; Kaplan D. L. Effect of processing on silk-based biomaterials: reproducibility and biocompatibility. J. Biomed. Mater. Res., Part B 2011, 99B, 89–101. 10.1002/jbm.b.31875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Wang B. Biodegradation of silk biomaterials. Int. J. Mol. Sci. 2009, 10, 1514–1524. 10.3390/ijms10041514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard E. M.; Hu X.; Finley V.; Kuo C. K.; Kaplan D. L. Effect of silk protein processing on drug delivery from silk films. Macromol. Biosci. 2013, 13, 311–320. 10.1002/mabi.201200323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongpinyochit T.; Johnston B. F.; Seib F. P. Manufacture and Drug Delivery Applications of Silk Nanoparticles. J. Visualized Exp. 2016, 116, e54669 10.3791/54669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanovich U.; Ruggeri F. S.; De Genst E.; Adamcik J.; Barros T. P.; Porter D.; Müller T.; Mezzenga R.; Dobson C. M.; Vollrath F.; Holland C.; Knowles T. P. J. Silk micrococoons for protein stabilisation and molecular encapsulation. Nat. Commun. 2017, 8, 15902. 10.1038/ncomms15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongpinyochit T.; Uhlmann P.; Urquhart A. J.; Seib F. P. PEGylated Silk Nanoparticles for Anticancer Drug Delivery. Biomacromolecules 2015, 16, 3712–3722. 10.1021/acs.biomac.5b01003. [DOI] [PubMed] [Google Scholar]
- Hu X.; Kaplan D.; Cebe P. Determining Beta-Sheet Crystallinity in Fibrous Proteins by Thermal Analysis and Infrared Spectroscopy. Macromolecules 2006, 39, 6161–6170. 10.1021/ma0610109. [DOI] [Google Scholar]
- Duncan R.; Richardson S. C. W. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Mol. Pharm. 2012, 9, 2380–2402. 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]
- Wahlich J.; Desai A.; Greco F.; Hill K.; Jones A. T.; Mrsny R. J.; Pasut G.; Perrie Y.; Seib F. P.; Seymour L. W.; Uchegbu I. F. Nanomedicines for the Delivery of Biologics. Pharmaceutics 2019, 11, 210. 10.3390/pharmaceutics11050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R.; Gaspar R. Nanomedicine(s) under the microscope. Mol. Pharm. 2011, 8, 2101–2141. 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- Torchilin V. P. Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat. Rev. Drug Discovery 2014, 13, 813–827. 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K. P.; Groh J. M.; Johnson M. D.; Burcham C. L.; Campbell B. M.; Diseroad W. D.; Heller M. R.; Howell J. R.; Kallman N. J.; Koenig T. M.; May S. A.; Miller R. D.; Mitchell D.; Myers D. P.; Myers S. S.; Phillips J. L.; Polster C. S.; White T. D.; Cashman J.; Hurley D.; Moylan R.; Sheehan P.; Spencer R. D.; Desmond K.; Desmond P.; Gowran O. Kilogram-scale prexasertib monolactate monohydrate synthesis under continuous-flow CGMP conditions. Science 2017, 356, 1144–1150. 10.1126/science.aan0745. [DOI] [PubMed] [Google Scholar]
- Stroock A. D.; Dertinger S. K.; Ajdari A.; Mezic I.; Stone H. A.; Whitesides G. M. Chaotic mixer for microchannels. Science 2002, 295, 647–651. 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Lu G.; Lu Q.; Kaplan D. L. Direct Formation of Silk Nanoparticles for Drug Delivery. ACS Biomater. Sci. Eng. 2016, 2, 2050–2057. 10.1021/acsbiomaterials.6b00457. [DOI] [PubMed] [Google Scholar]
- Laity P. R.; Gilks S. E.; Holland C. Rheological behaviour of native silk feedstocks. Polymer 2015, 67, 28–39. 10.1016/j.polymer.2015.04.049. [DOI] [Google Scholar]
- Chen X.; Shao Z.; Knight D. P.; Vollrath F. Conformation transition kinetics of Bombyx mori silk protein. Proteins 2007, 68, 223–231. 10.1002/prot.21414. [DOI] [PubMed] [Google Scholar]
- Laity P. R.; Baldwin E.; Holland C. Changes in silk feedstock rheology during cocoon construction: the role of calcium and potassium ions. Macromol. Biosci. 2019, 19, 1800188. 10.1002/mabi.201800188. [DOI] [PubMed] [Google Scholar]
- Hu X.; Shmelev K.; Sun L.; Gil E.-S.; Park S.-H.; Cebe P.; Kaplan D. L. Regulation of silk material structure by temperature-controlled water vapor annealing. Biomacromolecules 2011, 12, 1686–1696. 10.1021/bm200062a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimi G.; Lozano-Pérez A. A.; Montalbán M. G.; Aznar-Cervantes S. D.; Cenis J. L.; Víllora G. Revealing the Influence of the Degumming Process in the Properties of Silk Fibroin Nanoparticles. Polymers 2019, 11, 2045. 10.3390/polym11122045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J.; Park Y. H. Morphology of Regenerated Silk Fibroin: Effects of Freezing Temperature, Alcohol Addition, and Molecular Weight. J. Appl. Polym. Sci. 2001, 81, 3008–3021. 10.1002/app.1751. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.