Abstract
Outbreaks associated with rapidly growing mycobacteria (RGM) have been increasingly reported worldwide, including in Brazil. Among the RGM, the Mycobacterium abscessus complex is the most pathogenic and related to multidrug resistance. The aim of this study was to evaluate the antimicrobial susceptibility and molecular profile of RGM isolates involved in new postsurgical infection outbreaks in Brazil since 2007. Of the 109 cases reported in the state of Rio Grande do Sul between 2007 and 2011, 43 (39 %) had confirmed mycobacterial growth in culture. Clinical isolates were obtained from biopsy specimens or abscess aspirates. PRA-hsp65 restriction pattern identified the isolates as M. abscessus type 2, and partial rpoB sequencing confirmed the identification as M. abscessus subsp. bolletii. All isolates were susceptible to amikacin and resistant to ciprofloxacin, doxycycline, sulfamethoxazole, moxifloxacin and tobramycin. Most isolates (72 %) were fully susceptible to cefoxitin but six isolates (14 %) were fully resistant to clarithromycin. The latter differed from the susceptibility profiles of the previously described BRA100 clone from other Brazilian regions. Nevertheless, pulsed-field gel electrophoresis analysis revealed that these isolates belonged to a single BRA100 clone. In conclusion, our study reports the persistence of an emergent single and highly resistant clone of M. abscessus subsp. bolletii for several years even after national implementation of infection control measures.
Introduction
The nontuberculous mycobacteria (NTM) species most commonly associated with localized infections of the skin and subcutaneous tissues include Mycobacterium marinum and M. haemophilum and the rapidly growing mycobacteria (RGM) (M. abscessus, M. massiliense, M. chelonae, M. fortuitum) (Kasperbauer & Huitt, 2013). RGM species are frequently involved in infections after traumatic injuries or surgery, and are of particular concern due to their tendency to dissemination in immunocompromised patients (Falkinham, 2013; Kothavade et al., 2013; Redelman-Sidi & Sepkowitz, 2010).
There is a debate about the actual taxonomy of the genus; although there is evidence suggesting the presence of three subspecies of the M. abcessus complex, most studies in Brazil use only two subspecies, M. abscessus subsp. bolletii (which includes M. massiliense and M. bolletii) and M. abscessus subsp. abscessus (which includes M. abscessus) (Euzéby, 2014; Sampaio, 2010). The M. chelonae-abscessus group has emerged as an important cause of both community and hospital-acquired infections in humans (Koh et al., 2011). Several cases of infection due to this RGM have been reported in many countries, including Brazil (Cardoso et al., 2008; Cheng et al., 2013; Duarte et al., 2009; Leão et al., 2010; Monego et al., 2011; Viana-Niero et al., 2008).
The first outbreak of M. abscessus subsp. bolletii reported in Brazil occurred in 2004 and involved 68 patients who had undergone laparoscopic surgery (Viana-Niero et al., 2008). A series of nosocomial outbreaks, all of them associated with invasive procedures, have since been reported in several Brazilian states (ANVISA, 2011). Representative isolates proved to be genetically related and were identified as a single clone, denoted BRA100 (Duarte et al., 2009). The BRA100 strain is known to be resistant to high glutaraldehyde (GTA) concentrations (up to 7 %, after 15 to 30 min of exposure), which proves that this product is non-effective against specific strains of RGM (Duarte et al., 2009; Leão et al., 2010; Lorena et al., 2010).
The epidemiological situation in Brazil has made the government surveillance system adopt containment and control actions. These include analysis of sanitizing products in relation to the RGM susceptibility, deployment of a rapid response system for outbreak reporting, support of appropriate laboratory diagnosis of RGM infections, and an emphasis on the application of National Health Surveillance Agency guidelines (ANVISA, 2011).
Although RGM has already been reported in several Brazilian regions, scarce data are available from the state of Rio Grande do Sul (Leão et al., 2010). Thus, the aim of this study was to evaluate the susceptibility and molecular profile of RGM isolates involved in postsurgical infection outbreaks occurring in Rio Grande do Sul after the largest Brazilian epidemic (2006–2007).
Methods
Isolates and microbiological procedures.
From 2007 to 2010, four RGM outbreaks occurred in different cities, and one single case was reported in 2011, in the southernmost region of Brazil, all due to infections secondary to invasive procedures. During this period, a total of 109 clinical specimens (biopsy or aspiration of abscess fluids) obtained from patients with signs of postsurgical infections were sent to the Instituto de Pesquisas Biológicas–Laboratório Central de Saúde Pública (IPB-LACEN/RS) for investigation. The clinical specimens were processed for acid-fast bacillus (AFB) smears (Ziehl–Neelsen stain) and cultured on Ogawa-Kudoh (OK) solid medium for up to 4 weeks at 35–37 °C. Ultimately, only 43 specimens exhibited mycobacterial growth in culture, and were evaluated further.
Species identification by PCR-restriction enzyme analysis (PRA).
DNA was extracted from colonies on OK and suspended in 250 µl of 1× Tris-EDTA (TE), boiled for 20 min, incubated in an ultrasonic water bath for 15 min and frozen at −20 °C. All 43 isolates were initially analysed using PRA of the hsp65 gene (PRA-hsp65), as described by Telenti et al. (1993), with modifications. The primers Tb11 (5′-ACCAACGATGGTGTGTCCAT) and Tb12 (5′-CTTGTCGAACCGCATACCCT) were used to generate a 441 bp fragment of the hsp65 gene. The PCR mixture consisted of 20 ng of mycobacterial DNA, 25 pmol of Tb11 and Tb12 primers, 10× buffer (20 mM Tris/HCl at pH 8.0, 50 mM KCl), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 10 % glycerol and 1.5 U GoTaq Hot Start Polymerase (Promega) in a 50 µl reaction volume. A 5 µl volume of PCRx Enhancer System (Invitrogen) was used to optimize the amplification reaction. Amplification was achieved with a MaxyGene Therm 1000 gradient thermal cycler (Axygen) under the following conditions: 45 cycles at 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min, with a final extension step at 72 °C for 7 min. Amplicons were digested with BstEII and HaeIII (Promega) in different reactions, and restriction fragments were analysed after 3 % agarose gel (Invitrogen) electrophoresis and staining with GelRed. The patterns observed were compared to those deposited in PRASITE (http://app.chuv.ch/prasite).
Species identification by DNA sequencing of the rpoB gene.
Identification was also performed by partial sequencing of the rpoB gene, as previously described (Adekambi et al., 2003). The PCR products were purified using the polyethylene glycol method (Jolley et al., 2004), and sequenced using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). The sequences obtained were analysed in Chromas version 2.3. Sequence alignment was performed in the dnastar Lasergene 7 Genomics Suite, and homology analysis was performed by comparison of the consensus sequence obtained from each isolate with those deposited in GenBank, using the blast (Basic Local Alignment Search Tool, http://www.ncbi.nlm.nih.gov/BLAST) algorithm.
Antimicrobial susceptibility testing.
Susceptibility to amikacin, cefoxitin, clarithromycin, ciprofloxacin, doxycycline, moxifloxacin, trimethoprim-sulfamethoxazole and tobramycin was tested by the broth microdilution method as recommended by the Clinical Laboratory Standards Institute (CLSI, 2011). Staphylococcus aureus ATCC 29213 was used as a quality control strain.
Pulsed-field gel electrophoresis (PFGE) analysis.
Isolates were typed by PFGE, as described by Leão et al. (2010). Briefly, DNA was isolated from the plug moulds, digested with DraI (Promega), and loaded into 1 % pulsed-field certified agarose (Bio-Rad). Electrophoresis, using thiourea in the running buffer, was carried out in a CHEF-DR III system (Bio-Rad) at 14 °C for 24 h at 6 V cm−1 with a switch time of 1.6–21.3 s. Bacteriophage lambda ladder PFG Marker (New England Biolabs) was used as a molecular size standard. PFGE patterns were analysed using BioNumerics v. 6.5 (Applied Maths). Dendrograms were constructed using the Dice coefficient and the unweighted pair-group method with arithmetic mean clustering (UPGMA), based on 2 % optimization and position tolerance.
The CRM-0019 strain, implicated in the outbreak that occurred in Rio de Janeiro from 2006 to 2007, and five other unrelated strains (CRM-0639, CRM-0640, CRM-0642, CRM-0803 and CRM-0836) from Rio de Janeiro were used for comparison to the molecular profile of our isolates.
Glutaraldehyde tolerance assay.
A set of randomly selected isolates were evaluated for their ability to survive after 30 min of exposure to 2.0 % commercial glutaraldehyde solutions using the suspension method, as recommended by the manufacturers for high-level disinfection and sterilization (Collins & Montalbine, 1976). The method was performed according to Lorena et al. (2010). The tolerance was defined as the ability of GTA to inhibit the visible mycobacterial growth.
Results
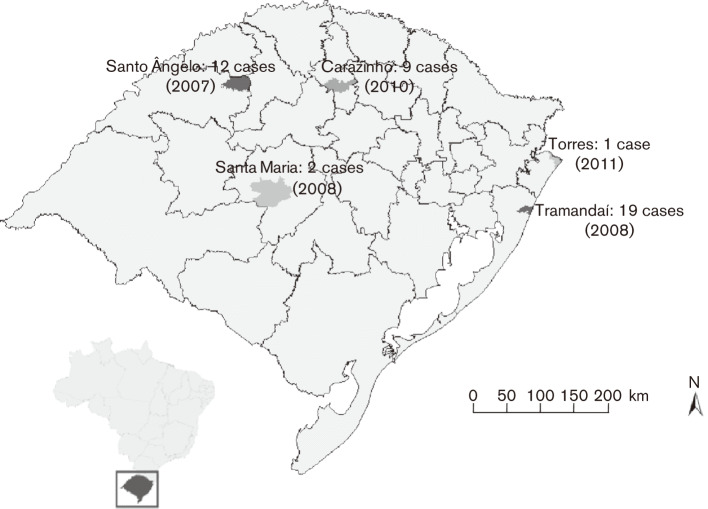
One hundred and nine cases of postsurgical infections were reported in Rio Grande do Sul from 2007 to 2011. There were 34 in 2007 in the municipality of Santo Ângelo; 48 in 2008 in the municipalities of Tramandaí (n = 44) and Santa Maria (n = 4); 26 in 2010 in the municipality of Carazinho; and one case in 2011 in the municipality of Torres (Fig. 1). Whereas only 13 clinical specimens (12 %) were positive on AFB staining, 43 (39 %) grew RGM in OK medium. All were identified as M. abscessus type 2 by PRA-hsp65 and were further confirmed as M. abscessus subsp. bolletii by rpoB gene sequencing (GenBank accession number KF360857).
Fig. 1.
Confirmed cases of M. abscessus subsp. bolletii from postsurgical infections in the state of Rio Grande do Sul, Brazil, 2007–2011.
The isolates were 100 % susceptible to amikacin but resistant to ciprofloxacin, doxycycline, moxifloxacin, trimethoprim-sulfamethoxazole and tobramycin. Furthermore, 31 (72 %) isolates were fully susceptible to cefoxitin, and six isolates (14 %) were fully resistant to clarithromycin (Table 1).
Table 1.
Susceptibility profile of 43 M. abscessus subsp. bolletii from southern Brazil
| Drug | Resistant [n (%)] | Intermediate [n (%)] | Susceptible [n (%)] |
| AMK | 0 | 0 | 43 (100) |
| FOX | 0 | 12 (28) | 31 (72) |
| CLR | 6 (14) | 0 | 37 (86) |
| CIP | 43 (100) | 0 | 0 |
| DOX | 43 (100) | 0 | 0 |
| SXT | 43 (100) | 0 | 0 |
| MXF | 43 (100) | 0 | 0 |
| TOB | 43 (100) | 0 | 0 |
AMK, Amikacin; CIP, ciprofloxacin; CLR, clarithromycin; DOX, doxycycline; FOX, cefoxitin; MXF, moxifloxacin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin.
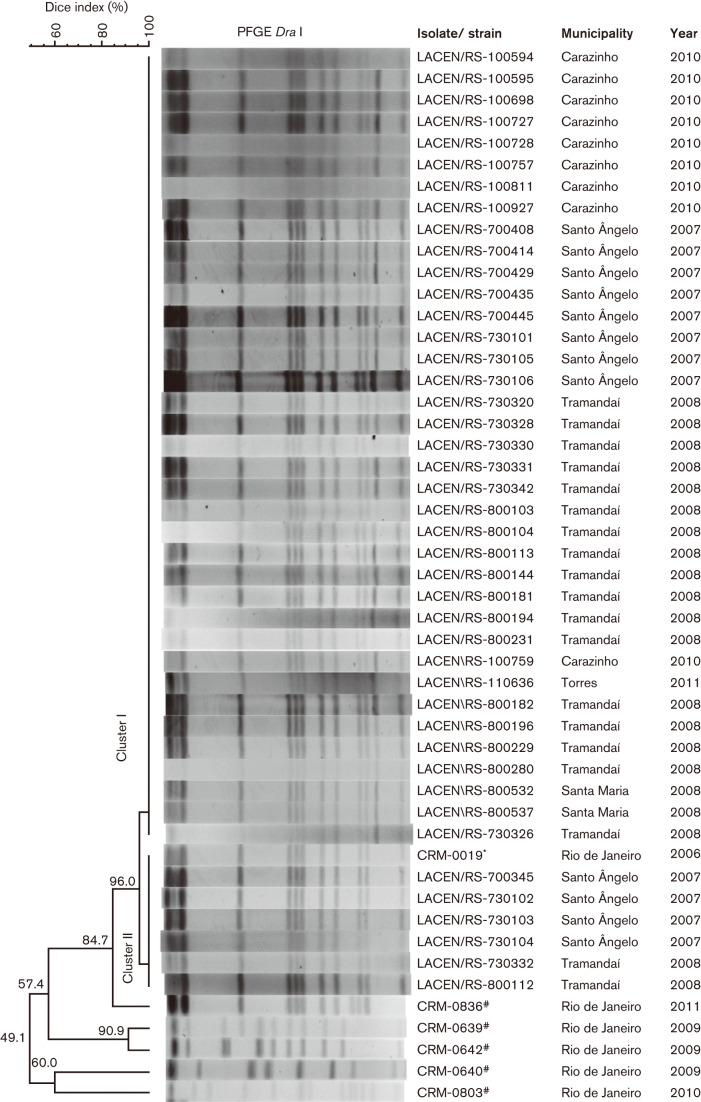
All 43 M. abscessus subsp. bolletii specimens generated interpretable patterns (12 or 13 bands) on PFGE and proved to be clonally related (>85 % of similarity). To evaluate the relationship of this clone with the outbreaks previously described in Brazil (Duarte et al., 2009), we compared the PFGE pattern of isolate CRM-0019 (from the 2006–2007 Rio de Janeiro outbreak), which belongs to the BRA100 clone, with the pattern of our isolates, and they also proved to be clonally related. Although all isolates were clonally related, they did not have identical PFGE patterns; the majority of isolates (n = 37) presented 13 bands (cluster I), but six isolates presented 12 bands (cluster II). The dendrogram is shown in Fig. 2.
Fig. 2.
Dendrogram of DraI PFGE patterns of isolates from 43 patients in five cities in southern Brazil. CRM-0019* isolate from the outbreak in Rio de Janeiro. CRM-0639#, CRM-0640#, CRM-0642#, CRM-0803# and CRM-0836# isolates from unrelated outbreaks in Brazil.
To evaluate the discriminatory power of PFGE, we evaluated the patterns of five epidemiologically unrelated M. abscessus subsp. bolletii, and all were distinguishable.
We selected a set of representative isolates to be tested for susceptibility to 2 % GTA. There were five isolates from Santo Angelo, seven isolates from Tramandaí, one isolate from Santa Maria, four from Carazinho and the only isolate from Torres. In total, we tested 18 isolates that represented 42 % of the total isolates obtained (n = 43). Eight isolates (44 %) were susceptible to 2 % GTA after 30 min exposure (isolates LACEN/RS-730320, LACEN/RS-800112, LACEN/RS-800229, LACEN/RS-700429, LACEN/RS-730101, LACEN/RS-730102, LACEN/RS-730104 and LACEN/RS-730106) and 10 isolates (56 %) were resistant to 2 % GTA after 30 min exposure (isolates LACEN/RS-730342, LACEN/RS-800103, LACEN/RS-800113, LACEN/RS-800196, LACEN/RS-800532, LACEN/RS-100594, LACEN/RS-100595, LACEN/RS-100698, LACEN/RS-100727 and LACEN/RS-110636).
Discussion
This study investigated 109 patients with postsurgical infections reported from 2007 to 2011 in five different municipalities of the state of Rio Grande do Sul, Brazil. Only 43 of the collected specimens presented mycobacterial growth in the OK medium, possibly due to the poor quality of the specimen, to storage conditions, or to the low sensitivity of the culture method for diagnosis of biopsy specimens and surgical wound aspirates, especially during antimicrobial treatment. In fact, the American Thoracic Society (Griffith et al., 2007) issued diagnostic criteria in 2007, which highlight the importance of drawing culture specimens from sterile body sites when attempting to establish true infection by NTM.
Both methods used here for species identification presented identical results for all isolates. Although PRA-hsp65 can be used as a quick and reliable method to identify NTM in the clinical laboratory (Chimara et al., 2008), a more complete characterization was obtained by sequencing of the rpoB gene, allowing subspecies identification.
Interestingly, we found 14 % resistance to clarithromycin among the isolates studied, demonstrating a different susceptibility profile compared with the BRA100 clone from other Brazilian regions (Duarte et al., 2009; Leão et al., 2009; Monego et al., 2011). This result is of particular relevance because macrolide-based regimens are often used for treatment of M. abscessus disease (Brown-Elliott et al., 2012; Griffith et al., 2007). Moreover, clarithromycin is usually the option for therapy combined with amikacin (Griffith et al., 2007). As resistance to clarithromycin is usually induced (Brown-Elliott et al., 2012), increased use of this antibiotic may have led to the emergence of resistant RGM. The susceptibility profiles observed in our isolates for amikacin, ciprofloxacin, doxycycline, trimethoprim-sulfamethoxazole and tobramycin were the same susceptibility pattern as previously reported for the BRA100 clone (Duarte et al., 2009; Monego et al., 2011). However, it is of note that most isolates of our study were susceptible to cefoxitin in contrast to the BRA100 from Rio de Janeiro, which tended to be resistant to this antibiotic (Duarte et al., 2009).
Our results confirmed the discriminatory power of PFGE, considering that distinct molecular profiles were observed among the isolates unrelated to the outbreaks. Although all isolates were clonally related, they did not exhibit identical PFGE patterns, resulting in two clusters with only one band (~50 kb) of difference between them. The band is thought to reflect the presence of a pMAB01-like plasmid but this was not confirmed experimentally.
Interestingly, the 43 strains (cluster I, 13 bands and cluster II, 12 bands) exhibited the same profile of the BRA100 clone, as previously reported in Brazilian outbreaks. The spread of a single clone of M. abscessus subsp. bolletii in different regions of the country seems to be related to a common source of infection, reinforcing the concept that the BRA100 clone is an emergent pathogen in the Brazilian hospital environment (Leão et al., 2010). Furthermore, recent studies indicated a molecular relationship between the BRA100 clone and strains from Taiwan, Malaysia, UK and USA, demonstrating that this clone is disseminated worldwide (Cheng et al., 2013; Davidson et al., 2013; Aitken et al., 2012). Some aspects that may explain the persistence of BRA100 in the environment are its virulence and biofilm formation, which is particularly associated with M. abscessus (Donlan, 2001; Duarte et al., 2009; Shang et al., 2011; Simoes et al., 2005). The main factor related specifically to Brazilian outbreaks was tolerance to 2 % GTA (Duarte et al., 2009; Lorena et al., 2010), although several mitigating measures have been recommended by the National Health Surveillance Agency, as mentioned above. We found that 56 % (10/18) of the BRA100 clone isolates survive abundantly after 30 min of exposure to 2 % GTA, which may have contributed to the possible dissemination of this biocide-tolerant micro-organism in different hospitals.
In conclusion, our study has shown the persistence of a single clone of M. abscessus subsp. bolletii, BRA100, which was responsible for outbreaks occurring in distinct periods from 2007 to 2011 in Rio Grande do Sul. It should be stressed that this is the first report of clarithromycin resistance among isolates belonging to BRA100, which indicates an important change in the susceptibility profile of this clone. M. abscessus subsp. bolletii BRA100 has persisted despite the control measures adopted by the Brazilian public health surveillance system, indicating that these actions alone are not being effective in preventing dissemination or that they are not being implemented rigorously enough by health institutions. Therefore, our study contributes to the knowledge of the epidemiological profile and the magnitude of infections attributable to BRA100 in Brazil.
Acknowledgements
This study received financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico, FIPE/HCPA 09-0653, ICOHRTA 5 U2R TW006883-02 and ENSP-011-LIV-10-2-3 (MCT/CNPq; Edital Universal, process no. 480789/2010-0 MCT/CNPq 14/2010). The authors would like to thank the staff from Seção de Micobactérias, IPB/LACEN–FEPPS and Laboratório de Micobactérias, Instituto de Microbiologia–UFRJ, for their assistance with sample processing and culture management; Marilina Bercini of Divisão de Vigilância Epidemiológica; and Ana Luiza Rammé of Divisão de Vigilância Sanitária, Centro Estadual de Vigilância em Saúde/RS.
Footnotes
Abbreviations: AFB, acid-fast bacillus; GTA, glutaraldehyde; NTB, nontuberculous mycobacteria; RGM, rapidly growing mycobacteria.
References
- Adékambi T., Colson P., Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003;41:5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken M. L., Limaye A., Pottinger P., Whimbey E., Goss C. H., Tonelli M. R., Cangelosi G. A., Dirac M. A., Olivier K. N.& other authors (2012Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center Am J Respir Crit Care Med 185231–232. 10.1164/ajrccm.185.2.231 [DOI] [PubMed] [Google Scholar]
- ANVISA. Relatório descrito de investigação de casos de infecções por micobactérias não tuberculosas de crescimento rápido (MCR) no Brasil no período de 1998 a 2009. 2011. Gerência Geral de Tecnologia em Serviços de Saúde. http://www.anvisa.gov.br/hotsite/hotsite_micobacteria/relatorio_descrito_mcr_16_02_11.pdf.
- Brown-Elliott B. A., Nash K. A., Wallace R. J., Jr Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev. 2012;25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso A. M., Martins de Sousa E., Viana-Niero C., Bonfim de Bortoli F., Pereira das Neves Z. C., Leão S. C., Junqueira-Kipnis A. P., Kipnis A. Emergence of nosocomial Mycobacterium massiliense infection in Goiás, Brazil. Microbes Infect. 2008;10:1552–1557. doi: 10.1016/j.micinf.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Cheng A., Liu Y. C., Chen M. L., Hung C. C., Tsai Y. T., Sheng W. H., Liao C. H., Hsueh P. R., Chen Y. C., Chang S.-C. Extrapulmonary infections caused by a dominant strain of Mycobacterium massiliense (Mycobacterium abscessus subspecies bolletii) Clin Microbiol Infect. 2013;19:E473–E482. doi: 10.1111/1469-0691.12261. [DOI] [PubMed] [Google Scholar]
- Chimara E., Ferrazoli L., Ueky S. Y., Martins M. C., Durham A. M., Arbeit R. D., Leão S. C. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008;8:48. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. Approved Standard, 2nd edn, CLSI document M24-A2. [PubMed] [Google Scholar]
- Collins F. M., Montalbine V. Mycobactericidal activity of glutaraldehyde solutions. J Clin Microbiol. 1976;4:408–412. doi: 10.1128/jcm.4.5.408-412.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. M., Hasan N. A., de Moura V. C., Duarte R. S., Jackson M., Strong M. Phylogenomics of Brazilian epidemic isolates of Mycobacterium abscessus subsp. bolletii reveals relationships of global outbreak strains. Infect Genet Evol. 2013;20:292–297. doi: 10.1016/j.meegid.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis. 2001;33:1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Duarte R. S., Lourenço M. C., Fonseca Lde. S., Leão S. C., Amorim Ede. L., Rocha I. L., Coelho F. S., Viana-Niero C., Gomes K. M.& other authors (2009Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol 472149–2155. 10.1128/JCM.00027-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euzéby J. P. LPSN list of prokaryotic names with standing in nomenclature. 2014. http://www.bacterio.net/ [DOI] [PMC free article] [PubMed]
- Falkinham J. O., III Ecology of nontuberculous mycobacteria–where do human infections come from? Semin Respir Crit Care Med. 2013;34:095–102. doi: 10.1055/s-0033-1333568. [DOI] [PubMed] [Google Scholar]
- Griffith D. E., Aksamit T., Brown-Elliott B. A., Catanzaro A., Daley C., Gordin F., Holland S. M., Horsburgh R., Huitt G.& other authors (2007An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases Am J Respir Crit Care Med 175367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- Jolley K. A., Chan M. S., Maiden M. C. mlstdbNet – distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperbauer S., Huitt G. Management of extrapulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34:143–150. doi: 10.1055/s-0033-1333576. [DOI] [PubMed] [Google Scholar]
- Koh W. J., Jeon K., Lee N. Y., Kim B. J., Kook Y. H., Lee S. H., Park Y. K., Kim C. K., Shin S. J.& other authors (2011Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med 183405–410. 10.1164/rccm.201003-0395OC [DOI] [PubMed] [Google Scholar]
- Kothavade R. J., Dhurat R. S., Mishra S. N., Kothavade U. R. Clinical and laboratory aspects of the diagnosis and management of cutaneous and subcutaneous infections caused by rapidly growing mycobacteria. Eur J Clin Microbiol Infect Dis. 2013;32:161–188. doi: 10.1007/s10096-012-1766-8. [DOI] [PubMed] [Google Scholar]
- Leão S. C., Viana-Niero C., Matsumoto C. K., Lima K. V., Lopes M. L., Palaci M., Hadad D. J., Vinhas S., Duarte R. S.& other authors (2010Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil Future Microbiol 5971–980. 10.2217/fmb.10.49 [DOI] [PubMed] [Google Scholar]
- Lorena N. S., Pitombo M. B., Côrtes P. B., Maya M. C., Silva M. G., Carvalho A. C., Coelho F. S., Miyazaki N. H., Marques E. A.& other authors (2010Mycobacterium massiliense BRA100 strain recovered from postsurgical infections: resistance to high concentrations of glutaraldehyde and alternative solutions for high level disinfection Acta Cir Bras 25455–459. 10.1590/S0102-86502010000500013 [DOI] [PubMed] [Google Scholar]
- Monego F., Duarte R. S., Nakatani S. M., Araújo W. N., Riediger I. N., Brockelt S., Souza V., Cataldo J. I., da Silva Dias R. C., Biondo A. W. Molecular identification and typing of Mycobacterium massiliense isolated from postsurgical infections in Brazil. Braz J Infect Dis. 2011;15:436–441. doi: 10.1016/S1413-8670(11)70224-0. [DOI] [PubMed] [Google Scholar]
- Redelman-Sidi G., Sepkowitz K. A. Rapidly growing mycobacteria infection in patients with cancer. Clin Infect Dis. 2010;51:422–434. doi: 10.1086/655140. [DOI] [PubMed] [Google Scholar]
- Sampaio J. L. Prokaryotic taxonomy rules and nomenclature changes in the Mycobacterium chelonae-abscessus group. Future Microbiol. 2010;5:1457. doi: 10.2217/fmb.10.111. [DOI] [PubMed] [Google Scholar]
- Shang S., Gibbs S., Henao-Tamayo M., Shanley C. A., McDonnell G., Duarte R. S., Ordway D. J., Jackson M. Increased virulence of an epidemic strain of Mycobacterium massiliense in mice. PLoS ONE. 2011;6:e24726. doi: 10.1371/journal.pone.0024726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões M., Pereira M. O., Vieira M. J. Effect of mechanical stress on biofilms challenged by different chemicals. Water Res. 2005;39:5142–5152. doi: 10.1016/j.watres.2005.09.028. [DOI] [PubMed] [Google Scholar]
- Telenti A., Marchesi F., Balz M., Bally F., Böttger E. C., Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Niero C., Lima K. V., Lopes M. L., da Silva Rabello M. C., Marsola L. R., Brilhante V. C., Durham A. M., Leão S. C. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol. 2008;46:850–855. doi: 10.1128/JCM.02052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]