Abstract
Active self-propelled colloidal populations induce time-dependent three-dimensional fluid flows, which alter the rheological (viscoelastic) properties of their fluidic media. Researchers have also studied passive colloids mixed with bacterial suspensions to understand the hydrodynamic coupling between active and passive colloids. With recent developments in biological cell-driven biohybrid microswimmers, different type biological microswimmer (e.g., bacteria and algae) populations need to interact fluidically with each other in the same fluidic media, while such interactions have not been studied experimentally yet. Therefore, we report the swimming behavior of two opposite types of biological microswimmer (active colloid) populations: Chlamydomonas reinhardtii (C. reinhardtii) algae (puller-type microswimmers) population in coculture with Escherichia coli (E. coli) bacteria (pusher-type microswimmers) population. We observed noticeable fluidic coupling deviations from the existing understanding of passive colloids mixed with bacterial suspensions previously studied in the literature. The fluidic coupling among puller- and pusher-type microswimmers led to nonequilibrium fluctuations in the fluid flow due to their opposite swimming patterns. Such coupling could be the main reason behind the shift in motility behaviors of these two opposite-type swimmer populations suspended in the same fluidic media.
Introduction
Recently, bacteria-driven1−6 (pusher-type) and algae-driven7,8 (puller-type) biohybrid microswimmers have been proposed to be used in potentially high-impact environmental remediation, lab-on-a-chip, and active in vivo targeted drug delivery applications.9−12 On the other hand, researchers have mixed passive colloids with bacterial suspensions to study the hydrodynamic coupling between active and passive colloidal particles.13 These experiments showed noticeable deviations from the existing understanding of active colloids. The nonequilibrium fluctuations in the fluid flow due to the swimming patterns of bacteria are considered to be the main reason behind the emergence of collective motion in passive colloids suspended in the active bacterial culture media.14−16 However, in the natural ecological niches and future potential medical and environmental remediation application scenarios, opposite-type bacteria- and algae-driven biohybrid microswimmer populations need to interact with each other. For example, bacteria and microalgae populations can interact and exist together in their complex natural habitat. Such complex interactions can develop spontaneous fluidic flows in the absence of imposed gradients by using the energy supplied by the active microswimmers.8 The instability of an isotropic suspension of active microswimmers involves the coupling of swimmer orientation, fluidic flow, and swimming-induced stresses.13 There is no study yet describing how these opposite-type microswimmers would hydrodynamically couple if they coexist in the same fluidic media.
In this report, we present an in vitro model to investigate whether there is any hydrodynamic interaction between the active pusher- and puller-type microswimmers (colloids) suspended in the same fluidic media. This will enable us to understand how such opposite-type active microswimmers maintain their dynamic interactions with their neighbors while simultaneously defending their hydrodynamic territory to perform functional tasks, such as reproduction, biofilm formation, and colonization. We hypothesize that the dynamic viscosity of one of the growing active colloid population (e.g., E. coli) may influence the onset of long-range velocity correlations of the other population (e.g., C. reinhardtii). The occurrence of such long-range fluid velocity correlations at different bacterial concentrations can be determined if such correlations are measured in the same coculture of pusher- and puller-type microswimmer populations. It is conceivable that the details of bacterial sensing and motility can be mapped to various future potential biohybrid microswimmer applications. Moreover, we postulate that the dynamics of density increase in one population, while keeping the other one constant, might influence the motility and directionality of the other population in the close vicinity. In this context, we investigated how hydrodynamic coupling between E. coli and C. reinhardtii populations influences the motion of the C. reinhardtii by tracking their three-dimensional (3D) swimming in a mixed fluidic medium.
Materials and Methods
Mixed Culture of Prokaryotic and Eukaryotic Cells
The experiments were conducted with living C. reinhardtii (strain 11-32), obtained from the culture collection of algae at Göttingen University (SAG), Germany. The microalgae were cultivated in BG-11 medium at room temperature and under a 12:12 light–dark cycle (Philips Master TL-D 58W/840 Super 80 Weiss) in 250 mL Erlenmeyer flasks, sealed with a permeable membrane to prevent contamination but allow ventilation. All cultures including the references were adjusted to pH 4.8 every day as previously described.17 Initially, all cultures had populations of about 5.5 × 105 cells/mL. On the first, third, and seventh days of the cultures, the number of cells was counted with a hemocytometer (Marienfeld, Lauda-Königshofen) to investigate the development of the cultures.
The bacterial strain was grown overnight at 37 °C in lysogeny broth (LB) medium. The inoculum for microswimmer assays was prepared from actively growing organisms (logarithmic phase). The concentration of bacteria suspension was determined by measuring their optical density at 600 nm (OD600). The effect of microalgae mixed culture on bacterial growth kinetics was investigated by measuring OD600 with a spectrophotometer (Synergy HTX, Biotek, Germany). For growth kinetic study, E. coli strains were cultured in LB medium and incubated at 37 °C on an orbital shaker (Gyromax, Amerex Instruments, Inc., Germany) at a rotation speed of 200 rpm for ∼6 h. Then, the bacterial suspension was diluted with the fresh medium until it corresponded to an OD600 of 0.1. 50 μL of the bacterial suspension (initial concentration of 105–106 CFU mL–1) was then added to each well of a microliter plate (Eppendorf, VWR, Germany) along with phosphate-buffered saline (PBS) (20 μL) as control. 50 μL of microalgae suspension was then added to respective wells and again incubated at 37 °C on the orbital shaker at a rotation speed of 200 rpm. The bacterial viability was monitored at regular intervals by taking absorbance at 600 nm. All studies were done in triplicates.
Evaluation of the Steering Properties of the Mixed C. reinhardtii and E. coli Populations
By keeping C. reinhardtii (puller-type microswimmer) density constant, we varied E. coli (pusher-type microswimmer) density in a high throughput manner using an 8-well microchamber (Micro-Slide, ibidi, Germany) and recorded time-lapse videos for 9 h. C. reinhardtii can be effectively tracked in 3D because of their intrinsic autofluorescence properties, with live imaging to extract various quantitative information. The 3D trajectories were generated by using a Nikon NIS 3D object tracking module. The motions of the mixed microswimmer populations were examined inside a custom-made microscope chamber slide harboring 8 wells (bottom thickness = 150 μm) for high-throughput analysis. The microscopy investigation was performed with a Zeiss Axio Observer A1 inverted microscope with an Axiocam 503 CCD camera and a 40× (numerical aperture = 0.6) objective lens. The microalgae and bacteria cocultures were illuminated by using a red filter with an emission peak at 655 nm, and a bandwidth of 15 nm (655/15 BrightLine HC, AHF Analysentechnik, Tübingen, Germany), to prevent phototactic biased motion of the microalgae and thus to avoid false-positive results.15
Viscosity Measurements to Determine Microrheology
The microrheology measurements of E. coli–C. reinhardtii suspensions were performed by using a precise computer-controlled viscometer (Hybrid Rheometer HR 10, TA Instruments, Germany) with a 40 mm titanium-coated parallel plate (titanium has higher sensitivity than a steel plate). This enabled us to quantify the storage (elastic) and loss (viscous) moduli as G′ and G″ components, respectively. The microrheological measurements on a mixed culture of microalgae and bacteria are technically tricky since the plate gap shall not exceed the size of a single cell (in this case ∼3–5 μm) and the area of measurement shall not be more than 15 cm2 since these cells are very soft and enable the rheometer to resolve the torques. To make the plates parallel, special polished glass plates, coated with a thin titanium layer (∼5 nm) and a root-mean-square surface roughness of 250 nm, were used in a modular compact rheometer from Anton-Paar GmbH (Berlin, Germany). Titanium coating exhibits higher sensitivity (∼nN range) compared with the conventional steel plates (∼μN range). Also, the surface roughness further improves the measurements by enhancing the surface stickiness of the cell mixture between the plates to eliminate the slip flow between the adjacent plates. 200 μL of unmixed/mixed culture suspensions with a high cell density (∼106 cells in each) was used for the microrheology experiments. After fixing the dry and clean top plate with a gap of ∼250 μm with the bottom plate, we carefully positioned the cell suspension in between the plate space using a micropipet. The capillary forces allow us to fill the gap between the top and bottom plate with the cell suspensions. Subsequently, a step shear strain of about 10% was applied for harmonic oscillation experiments to record the frequency and amplitude sweeps.
Viability of C. reinhardtii Incubated with E. coli
The sample from the incubator (Professional 3500, VWR, Germany) was stained by using 10 μM SYTOX Green (Molecular Probes, USA) in TAP media for 10 min at room temperature. Live cells in the sample were observed at 488 nm (Ar laser)/505–530 nm (excitation/emission) and dead cells were observed at 543 nm (HeNe laser)/560 nm (excitation/emission) by using a confocal laser microscope (LSM 510 META, Carl Zeiss, Germany). The ZEN 2009 Light Edition software (Carl Zeiss, Germany) was used to merge dual fluorescence images. About 500 cells in a random field at 100× magnification were utilized to calculate the viability of C. reinhardtii.
Time-Lapse Video Microscopy and Optical Flow Measurement
The dishes (ibidi, Germany) were placed on TCS-SP2 AOBS (Leica) at 60× with proper phase contrast filters and equipped with an incubation chamber (H301-EC-BL, Okolab) to keep the cells at 37 °C. Just before recording time-lapse videos, we observed freshly harvested C. reinhardtii at low (1 × 105 cells/mL), medium (5 × 105 cells/mL), and high (1 × 106 cells/mL) concentrations on the inverted optical microscope for routine motility and swimming characterizations. Movies were recorded close to the bottom of the focal plane in a range of ∼1–5 μm culture dish and then 50 μm away from the bottom to observe whether the two opposite microswimmers were performing 3D motion by moving in and out of the focal plane. Bright-field phase-contrast images were collected every 5 min overnight by a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ), connected to the Nikon Eclipse Ti confocal microscope with a Yokogawa CSU-W1 spinning disk. Observed areas were randomly chosen for the time-lapse acquisitions from the entire population.
Particle image velocimetry (PIV) analyses were performed by using the ImageJ18,19 plugin20 to characterize the fluid flows created by the microswimmers. In brief, a pair of images were divided into smaller regions (cross-correlation interrogation windows). The cross-correlation between these image subregions measured the optic flow (displacement or velocity of the microalgae) within the assigned image pair. By progressively decreasing the interrogation window size as 128 × 64 × 32 (in pixels), we obtained a better PIV resolution. The template matching with the normalized correlation coefficient algorithm was implemented for the image processing, where the interrogation windows were compared initially against a larger search window. The results of the PIV analysis were displayed as a vector plot and saved in plain text tabular format containing all the analysis results as .txt extension. The PIV ImageJ plugin is used for the visualization of the vector and the magnitude data from PIV analysis.
Statistical Analysis
The experiments were performed in triplicates. The results were expressed as the mean and the standard deviation of the values that were obtained from at least three independent experiments. Differences in the mean values between control experiments (without mixed culture) and bacteria/microalgae/tracer beads treated mixed culture experiments were analyzed by Student’s t test using GraphPad Prism 5 software. The significance level was set to 0.05 (p < 0.05) for all statistical analyses.
Results and Discussion
Biological microswimmers are active micrometer-scale colloids that self-propel by moving their flagella or cilia using chemical energy (ATP). According to the type of far-field fluid flow that they induce, they are classified as pushers and pullers. While pushers force fluid outward along their swimming direction and draw fluid into the sides of their bodies, pullers force the fluid inward along their swimming direction and repel fluid from their sides. Such opposite fluidic force dipoles have important consequences for the fluidic interactions among the microswimmers. To understand the hydrodynamic coupling between two opposite-type microswimmer populations, we mixed bacteria and microalgae in their natural culture and motility media and studied their hydrodynamic interactions in detail (Figure 1a).
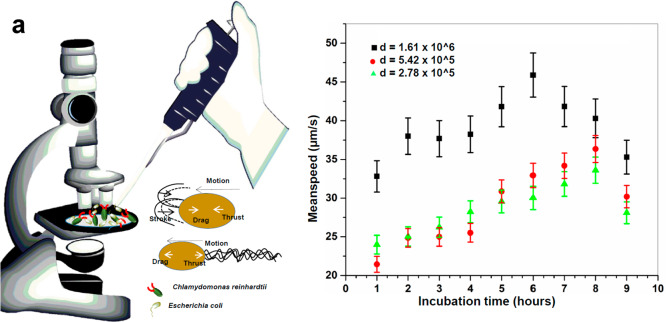
Figure 1.
(a) Schematics showing experimental procedure with diverse two opposite-type microswimmer population mixture characteristics. (b) Increase in mean speed of microalgae as a function of coculture incubation time. A constant-density C. reinhardtii motion was investigated against three different E. coli densities (d = initial density at each time point). Error bars show mean value ± standard deviation (SD) from three different experiments.
The mean speed of algae is plotted in Figure 1b as a function of incubation time for three different initial densities of bacteria (the density of bacteria itself is a function of coculture incubation time). As observed, the mean speed of C. reinhardtii (pullers) increased with the increasing density of E. coli (pushers), while C. reinhardtii density remained constant up to 6 h of mixed culture incubation. Subsequently, we observed a decrease in the mean speeds, which might arise due to the preparation and onset of microalgae division and arrests in motilities.21 The doubling time of C. reinhardtii is 7–8 h, whereas it is 45–60 min for E. coli. Therefore, we assume that microalgae density remained constant during the experiment. The imaging was completed within this time period. The increase in the population size of E. coli hindered their own mean speed via mechanical coupling among themselves. We hypothesize that the hydrodynamic coupling, based upon diverse swimming patterns of pushers and pullers, may not be as strong as the mechanical coupling mainly due to their size differences (E. coli ∼1–3 μm and C. reinhardtii ∼7–10 μm in diameter) and their motion confinements in two-dimensional (2D) and 3D space, respectively.
The mechanical coupling refers to coupling arising between microswimmers and extracellular polymeric substances, and the hydrodynamic coupling refers to fluid coupling arising due to the collective swimming of microalgae and bacteria.22 As qualitatively shown in Movie S1, after tracking autofluorescent C. reinhardtii (chlorophyll-bearing algae), it exhibited 3D swimming patterns as evident from the changes in directions at differential time points compared to E. coli (stained with SYTO 9 dye), which largely swim in the 2D plane. Therefore, the mechanical coupling may play an important role in swimming speed changes of the opposite-type microswimmers in a mixed culture. We would also like to mention that possibilities of other kinds of couplings, like chemical or any other kind of signaling, cannot be ruled out in the presented experimental study. It is very difficult to prove their presence or absence by using the current techniques. The focus of this work is to understand the complex interplay of the hydrodynamic and the mechanical coupling between two opposite-type active microswimmers.
When we traced E. coli and C. reinhardtii separately, we found that the mean speed of microalgae increases in the presence of bacteria; however, the mean speed of bacteria decreases as shown in Table 1. At early time points, the mixed algae–bacteria coculture chamber with the highest density demonstrated the highest mean speeds. As time advanced, E. coli grew in the chamber (E. coli vs C. reinhardtii doubling time ∼1 to ∼8 h), and thus, bacterial density increased. Up to a certain density, the mean speed of the microalgae increased, but close to 7–9 h, the speed further decreased irrespective of the cell density.
Table 1. Mean Speeds (in μm/s) of Microalgae, Bacteria, and Their Cocultures Measured for Initial Cell Density of d = 1 × 106 Cells/mL.
| microalgae | microalgae–bacteria mixture | bacteria | |
|---|---|---|---|
| microalgae | 41.3 ± 5.4 | 53.0 ± 4.9 | |
| bacteria | 24.1 ± 2.2 | 29.0 ± 1.7 |
This result could be due to the following reasons: we observed that when C. reinhardtii and E. coli cells were mixed together, bacterial cells preferably stayed at the bottom of the vessel (Figure S1a,b) irrespective of their prior mixing before adding to the microscopic imaging dish. Nevertheless, we performed movie recordings close to the bottom wall because there we observed a significant amount of moving C. reinhardtii for the analyses, whereas going away from the bottom we saw fewer moving cells due to preferable 2D motion of the E. coli.
In these conditions, we believe that bacterial cells found at the bottom acted as a carpet adding to an enhanced diffusion culture medium near the carpet of swimming E. coli.1 Therefore, moving fluid with a bacterial carpet enhanced the C. reinhardtii mean swimming speed. Furthermore, E. coli created the local force on the surrounding medium away from the swimming directions (Figure 1a), which might add to hydrodynamic-enhanced flow to C. reinhardtii.23
To understand increase (microalgae) versus decrease (bacteria) in swimming mean speeds, we examined relevant parameters such as viscosity variation, dynamic cluster formation, activity/motility induced phase separation, and individual flow fields of the microswimmers. We noticed that the E. coli–C. reinhardtii suspension with the highest initial bacterial density (Figure 1b) showed a maximum increase in microalgae mean speed. It is usually assumed that E. coli in dilute suspensions largely exhibits hydrodynamic coupling and less mechanical coupling. The mechanical coupling, however, evolves and starts dominating hydrodynamic coupling with the increasing cell population and contributes to long-range correlated motion.22 This could indirectly lead to an increase in diffusion and hence an increase in mean speed. We believe that in such a complex suspension of microswimmers with the opposite flow fields (pushers versus pullers in Figure 1a) extracellular polymer materials and bacterial debris act as matrices for the mechanical coupling. This leads to large-scale collective oscillation in C. reinhardtii in dense bacterial suspensions and counterintuitive weak synchronization among E. coli(24) (Movie S2).
The diffusion coefficient and mean speed measurements showed the active interaction between C. reinhardtii and E. coli, which increases the mean speed of the microalgae. To validate that the observed results were due to active swimming dynamics, we added passive polystyrene tracer beads, which have a similar size of microalgae, into the suspensions. We did not notice an equivalent diffusion and mean speed enhancement or changes in diffusion of tracer beads (Figure 2), indicating the active nature of the interaction. Upon mixing the tracer beads with the two opposite-type active microswimmers together, C. reinhardtii demonstrated significantly high mean speeds (t = 0.045, p ≤ 0.05), followed by E. coli and tracer beads. Irrespective of the cell density, when C. reinhardtii was mixed with E. coli and tracer beads separately, we saw maximum mean speed increase only when microalgae were mixed with bacteria. It gives a strong indication of complex mechanical coupling between puller- and pusher-type microswimmers (Figure S3).
Figure 2.
(a, b) Diffusion coefficient alterations in a bacteria–algae–beads mixed cocultures (a) and individual microswimmers (b). (c) High-density coculture of active microswimmers: (i) microalgae, (ii) bacteria, (iii) tracer beads, and (iv) overlay of microalgae, bacteria, and beads. Scale bar is 20 μm.
Figure 2c(i–iv) demonstrates fluorescent images of individual cultures of C. reinhardtii, E. coli, and tracer beads with their overlay (from left to right, respectively). Figure S2 demonstrates low, medium, and high density of C. reinhardtii, E. coli, tracer beads, and their overlay fluorescent images.
In the real scenario of microswimmer hydrodynamic coupling, the complex interplay among the passive viscoelastic biomaterial properties (swimmer cells), the fluidic stresses, and the internal micro/nanomotor active forces produces the configurational changes that constitute the versatile swimming patterns. The enhanced mean speed of the microalgae and the reduced mean speed of the bacteria could be further related to the physical origin of the effect of elasticity on their swimming speeds since favorable stroke asymmetry and swimmer elasticity contribute to a speed-up,17 and substantial boost happens when they work synergistically.
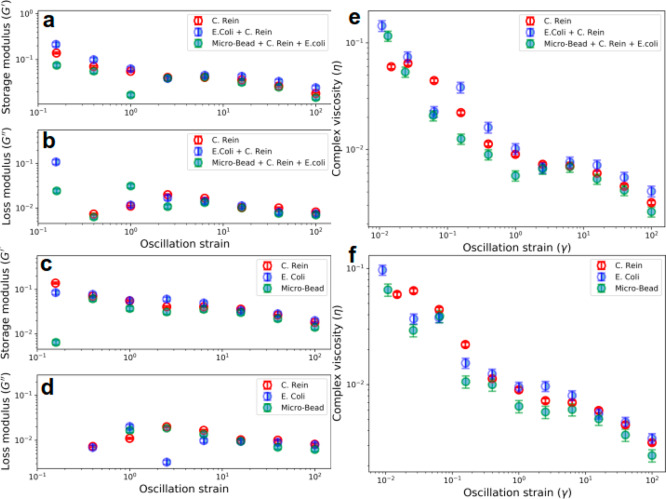
Active and passive rheology measurements were utilized to quantify the viscoelastic responses from the medium, which contained the active suspension of two opposite-type microswimmers. The results are shown in Figures 3a and 3c. The storage modulus or elastic component (G′) of mixed cells (blue half-filled circles in Figure 3a) is higher than the C. reinhardtii alone (red empty circles). The storage modulus represents the energy stored in the elastic structure of the sample. In monocultures (Figure 3a), we observed the high elastic component (G′) conspicuously only at low oscillation frequencies. With the increasing oscillation frequency, G′ for E. coli showed a slight dominance, and then an overlapping trend was observed among all three monocultures. This might be due to the enhanced coupling between the bacterial cell’s typical flagellar appendages resembling a large dangling chain as shown in complex polymer with advancing oscillation frequency, which contributes to the elastic component to Young’s moduli.25 The loss moduli (G″) or more precisely viscous moduli for the mixed and individual cultures were less predictable than the elastic component (G′) at lower oscillation frequencies. However, with the increasing oscillation frequency, the viscous component for C. reinhardtii was slightly higher than the other cultures as represented by red circles in Figure 3b,d.
Figure 3.
Quantification of microrheology changes in the microswimmer cocultures. (a, b) Storage modulus (G′) and loss modulus (G′′) of the mixed cultures. (c, d) Storage modulus (G′) and loss modulus (G′′) of individual cells and tracer beads, respectively. (e, f) Complex viscosity determination of the mixed- and monocultures of the microswimmers.
The loss moduli of the individual microswimmer cultures remained similar to that of mixed cultures (compare Figures 3b and 3d). Nevertheless, a general trend appeared that rheological properties, G′ (elastic or storage modulus) and G′′ (viscous or loss modulus), vary after mixing two opposite microswimmers. Passive rheology could be contributed by proteins and sugars secreted by growing E. coli. They are also known to contribute to such mechanical coupling involving complex dynamic rheology.22,26 This passive rheology may add to the viscosity of medium when microalgae and bacteria are mixed together, and this increasing viscosity speeds up self-propulsion.27,28 The puller-type microswimmer (C. reinhardtii) seems to swim counterintuitively faster when the viscosity of its surrounding fluid is increased, whereas the pusher-type microswimmer (E. coli) slows down (Table 1 and Figure 3). To confirm that passive rheology may contribute to the mechanical coupling and the mean speed increase, we enzymatically treated the mixed microalgae–bacteria culture with the DNase I and proteinase K which digest extracellular polymeric substances (EPS).29 We observed an increase in mean speeds via EPS-mediated reduced coupling, indicating the roles of extracellular matrices into the mechanical coupling (Figure 4a,b).
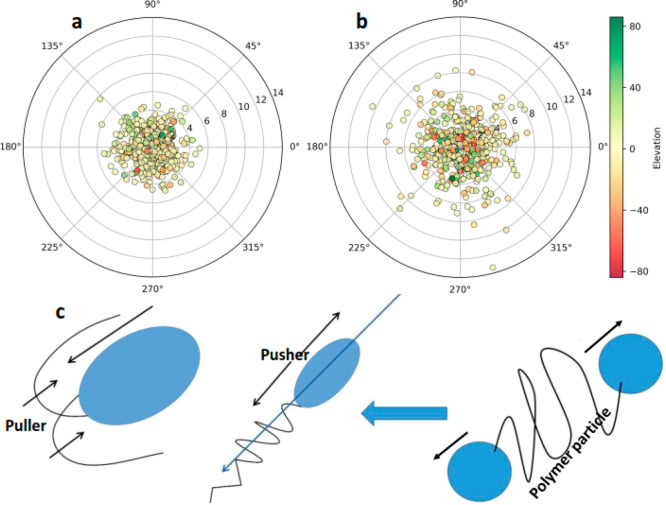
Figure 4.
Polar graph showing enhanced 3D motion of the microalgae in the coculture (a) after DNase/Proteinase K treatment compared with proteolytic enzyme untreated samples (b) as an influence of mechanical coupling. The dots represent the angular position of C. reinhardtii at different time points, angles in this plot represent the heading direction, numbers 0–14 on the polar graph show the segmentation lengths of swimmers, and colors of dots denote the elevation angle [in deg] recorded in 3D object tracking module. (c) Schematics showing polar stretching along the swimming axis of the bacteria create polymer-like microrheology, which contributes to the increase in viscosity of the medium.
We also compared the changes in G′ and G″ when microalgae were mixed with the tracer beads (Figure 3, left panel). The results indicate that the bacterial extracellular matrix in growth medium contributes significantly to the elasticity of mixed culture. The frequency responses observed for E. coli with and without mixing the microalgae and tracer beads were typical of weak viscoelastic liquid which is composed of untethered polymers.30 Furthermore, in dense bacterial suspensions, the mechanical coupling dominates over the hydrodynamic coupling because the length scale between the cells is below the detectable effective hydrodynamic distance (<10 μm).22 The changes in the complex viscosity as a function of strain are also plotted for mixed cultures and monocultures (Figure 3e,f). Long-range coupling between the opposite-type microswimmers can be explained via screening the force field arising due to the collective swimming of the bacteria/microalgae or the hydrodynamic effects. Like complex polymers, we cannot undermine long-range hydrodynamic interactions originating due to the growth of an extracellular matrix of bacteria toward a biofilm in our experiment.
Also, large-scale cooperative effects in bacteria at high cell densities, such as transient jet and vortex patterns appearing from the random switching transition, cause turbulent collective behavior among the cells, which are known mediators of reducing the medium viscosity.31 On the contrary, in low-density suspensions, E. coli might experience and share contacts as well as hydrodynamic interactions with their neighbors. Bacteria with their numerous extensions and appendages (e.g., flagella, fimbriae, lipopolysaccharides, pili, and lipoproteins) are effectively a larger and equivalent object like C. reinhardtii than the cell body itself. Therefore, size-based differences contributing to the mechanical and hydrodynamic coupling could be negated. We hypothesize that pusher versus puller complex swimming gait might lead to varying mean speeds for the two opposite microswimmers presented herein. The basic coupling mechanism among active microswimmers at high-density suspensions in the absence of shear flow and the effect of microswimmer activity on effective microrheology can be understood by their force dipole as explained in the earlier section. In the case of C. reinhardtii as an active puller-type microswimmer, the local flow induces a force dipole that tends to resist elongation along the swimming direction. On the contrary, in the high-density microalgae–bacteria suspensions the force dipole due to swimming is of the opposite sign for E. coli pushers, which favors stretching along the swimming direction analogous to polymer-like microrheology as shown in Figure 4c.
Complex Dynamics and Pattern Formation in Active Suspensions of Self-Propelled Microswimmers
Next, we investigated bacteria–microalgae suspensions in context with phase behavior and kinetics in the mixture of two opposite-type active microswimmers (pusher versus puller) behavior. We observed an activity-induced phase separation and self-assembly in mixtures of active bacteria–microalgae suspensions. The phase separation in self-propelled artificial phoretic particles is a positive accumulation feedback triggered by a slowdown of particles due to random collisions. Contrary to artificial particles, active bacteria or microalgae colloids could involve a different mechanism as they avoid crossing and colliding with the neighboring active particles.32 We observed that C. reinhardtii formed microclusters in the mixed coculture suspensions with orientation preferences as shown with the arrow direction in Figure 5a and the overlay of autofluorescent microalgae (as seen with the gray dots in Figure 5b and Movie S3). We found that, compared to the dynamics of the one type of active colloid population cases, the dynamics of two opposite-type active colloid mixtures show enhanced fluctuations with frequent fission and fusion of clusters. We also note that the overall dynamics of active-passive mixtures are very different from that of the corresponding purely active systems.5 Most importantly, the dynamics of phase separating mixtures is much more evident with clusters constantly moving, splitting, and merging (Movie S3).
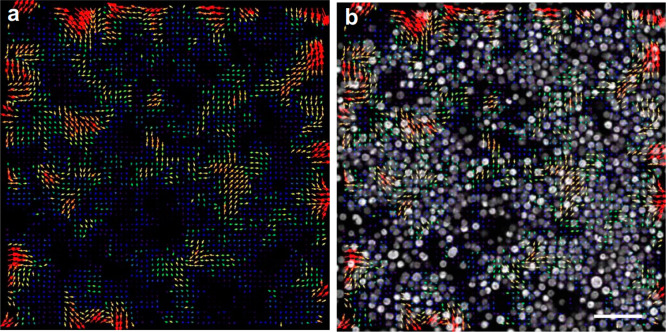
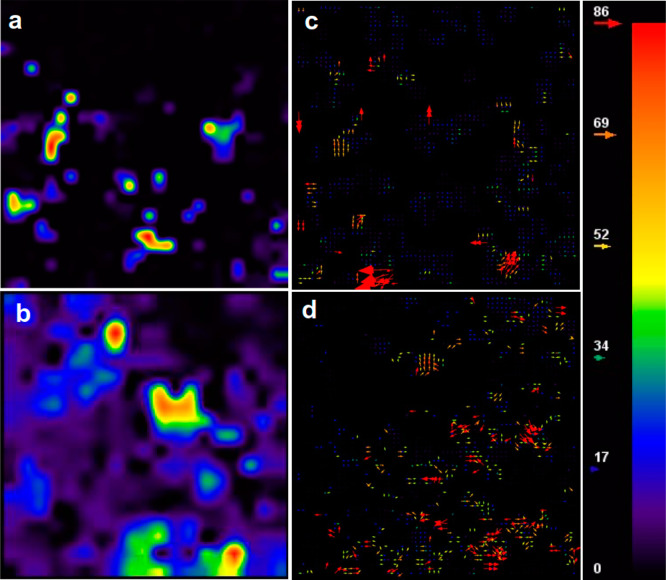
Figure 5.
Particle image velocimetry (PIV) analysis of fluidic coupling in the bacteria–microalgae cocultures. (a) Microalgae concentration field. Arrows exhibit the direction or orientation of the microalgae clusters compared with the previous frame of the image. (b) Overlay of the PIV color-coded vector plot with time-lapse sequence from mixed E. coli and C. reinhardtii coculture (autofluorescence tracked) showing the microalgae displacement field overlaid with concentration field. From one time frame to the next, the directional motion, microalgae cluster splitting, and formation can be compared. Being smaller size and no fluorescent, bacterial cells can be barely seen in (b). Scale bar is 20 μm.
At lower density and early time points of coculture, the evolution of the C. reinhardtii cluster distribution is mainly characterized by short-scale fluctuations. Both the concentration field and the cluster flow field become smoother. The mean concentration and the velocity field also change quite drastically and quickly and become very smooth and correlated over scales of the order of the cluster size. At high density and longer time scales, both the velocity and concentration field begin to develop and exhibit strong fluctuations, as seen in Figure 6a–d and Movie S3. The velocity fields remain correlated over large length scales. The strong fluctuations are not steady with time. The position and shape of the fluctuations keep evolving in time, with dense regions constantly reorganizing, merging, and breaking up, while their magnitude stabilizes as a result of diffusion. Similar behavior is reported for single species (pushers) and the coculture results presented herein.33 It appears that pusher-type microswimmers transmit these hydrodynamic fluctuations to pullers. None of these dynamic mixing, instabilities, and pattern formation is observed in active suspensions of puller-type microswimmers alone grown in isolations.
Figure 6.
Velocity magnitude map and velocity vector plot for (a, c) low-density coculture and (b, d) high-density coculture.
Conclusion
There are several unexplained phenomena in microbiology regarding the understanding of bacterial native milieu, e.g., the inception and diffusion of signaling molecules in quorum sensing, the osmotic flow of nutrients via a viscoelastic medium, and the differential efficacy of antibiotic actions at low- and high-density bacterial populations.34 Understanding the early viscoelastic extracellular matrix and their influences on the neighboring microswimmer community presented in this study may provide essential background for the future in such investigations. Furthermore, puller- versus pusher-type microswimmer behavior in lieu of their mutual interactions and hydrodynamic/mechanical coupling in the fluidic media presented herein could be used to design biohybrid microrobotic systems powered by both E. coli and C. reinhardtii in a single microdevice.35,36 An important next step is utilizing both the control (phototaxis and autonomous fluorescence properties of C. reinhardtii) and sensing capabilities (optimum pH sensing of E. coli)37 of these diverse microswimmers, which will expand the potential applications of biohybrid microswimmers for targeted drug delivery and environmental remediation in the future.
Acknowledgments
A.V.S. thanks the Max Planck Institute for Intelligent Systems for the Grassroots project grants M10335 and M10338. This work was funded by the Max Planck Society.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.9b03665.
Author Contributions
A.V.S. and V.K. contributed equally to this work. A.V.S., V.K., and M.S. proposed the research. A.V.S., G.Sa., and V.K. designed the experiments. A.V.S., O.Y., G.Sa., and V.K. performed the experiments. A.V.S. G.Sa., and V.K. analyzed the data. A.V.S and V.K. produced the videos. M.S. and J.B. supervised the research. A.V.S., G.Sa., V.K., O.Y., J.B., and M.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Darnton N.; Turner L.; Breuer K.; Berg H. C. Moving fluid with bacterial carpets. Biophys. J. 2004, 86 (3), 1863–1870. 10.1016/S0006-3495(04)74253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alapan Y.; Yasa O.; Schauer O.; Giltinan J.; Tabak A. F.; Sourjik V.; Sitti M. Soft erythrocyte-based bacterial microswimmers for cargo delivery. Science Robotics 2018, 3 (17), eaar4423 10.1126/scirobotics.aar4423. [DOI] [PubMed] [Google Scholar]
- Singh A. V.; Sitti M. Patterned and Specific Attachment of Bacteria on Biohybrid Bacteria-Driven Microswimmers. Adv. Healthcare Mater. 2016, 5 (18), 2325–2331. 10.1002/adhm.201600155. [DOI] [PubMed] [Google Scholar]
- Singh A. V.; Hosseinidoust Z.; Park B.-W.; Yasa O.; Sitti M. Microemulsion-based soft bacteria-driven microswimmers for active cargo delivery. ACS Nano 2017, 11 (10), 9759–9769. 10.1021/acsnano.7b02082. [DOI] [PubMed] [Google Scholar]
- Park B.-W.; Zhuang J.; Yasa O.; Sitti M. Multifunctional bacteria-driven microswimmers for targeted active drug delivery. ACS Nano 2017, 11 (9), 8910–8923. 10.1021/acsnano.7b03207. [DOI] [PubMed] [Google Scholar]
- Mostaghaci B.; Yasa O.; Zhuang J.; Sitti M. Bioadhesive bacterial microswimmers for targeted drug delivery in the urinary and gastrointestinal tracts. Advanced Science 2017, 4 (6), 1700058. 10.1002/advs.201700058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasa O.; Erkoc P.; Alapan Y.; Sitti M. Microalga-Powered Microswimmers toward Active Cargo Delivery. Adv. Mater. 2018, 30 (45), 1804130. 10.1002/adma.201804130. [DOI] [PubMed] [Google Scholar]
- Santomauro G.; Singh A. V.; Park B.-W.; Mohammadrahimi M.; Erkoc P.; Goering E.; Schütz G.; Sitti M.; Bill J. Incorporation of Terbium into a Microalga Leads to Magnetotactic Swimmers. Advanced Biosystems 2018, 2 (12), 1800039. 10.1002/adbi.201800039. [DOI] [Google Scholar]
- Erkoc P.; Yasa I. C.; Ceylan H.; Yasa O.; Alapan Y.; Sitti M. Mobile Microrobots for Active Therapeutic Delivery. Advanced Therapeutics 2019, 2 (1), 1800064. 10.1002/adtp.201800064. [DOI] [Google Scholar]
- Singh A. V.; Ansari M. H. D.; Laux P.; Luch A. Micro-nanorobots: important considerations when developing novel drug delivery platforms. Expert Opin. Drug Delivery 2019, 16 (11), 1259–1275. 10.1080/17425247.2019.1676228. [DOI] [PubMed] [Google Scholar]
- Hosseinidoust Z.; Mostaghaci B.; Yasa O.; Park B.-W.; Singh A. V.; Sitti M. Bioengineered and biohybrid bacteria-based systems for drug delivery. Adv. Drug Delivery Rev. 2016, 106, 27–44. 10.1016/j.addr.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Singh A. V.; Ansari M. H. D.; Mahajan M.; Srivastava S.; Kashyap S.; Dwivedi P.; Pandit V.; Katha U. Sperm Cell Driven Microrobots—Emerging Opportunities and Challenges for Biologically Inspired Robotic Design. Micromachines 2020, 11 (4), 448. 10.3390/mi11040448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch D. L.; Subramanian G. Collective Hydrodynamics of Swimming Microorganisms: Living Fluids. Annu. Rev. Fluid Mech. 2011, 43 (1), 637–659. 10.1146/annurev-fluid-121108-145434. [DOI] [Google Scholar]
- Wensink H.; Löwen H. Emergent states in dense systems of active rods: from swarming to turbulence. J. Phys.: Condens. Matter 2012, 24 (46), 464130. 10.1088/0953-8984/24/46/464130. [DOI] [PubMed] [Google Scholar]
- Pessot G.; Löwen H.; Menzel A. M. Binary pusher-puller mixtures of active microswimmers and their collective behaviour. Mol. Phys. 2018, 116 (21–22), 3401–3408. 10.1080/00268976.2018.1496291. [DOI] [Google Scholar]
- Hoell C.; Löwen H.; Menzel A. M. Multi-species dynamical density functional theory for microswimmers: Derivation, orientational ordering, trapping potentials, and shear cells. J. Chem. Phys. 2019, 151 (6), 064902. 10.1063/1.5099554. [DOI] [Google Scholar]
- Thomases B.; Guy R. D. Mechanisms of elastic enhancement and hindrance for finite-length undulatory swimmers in viscoelastic fluids. Phys. Rev. Lett. 2014, 113 (9), 098102. 10.1103/PhysRevLett.113.098102. [DOI] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9 (7), 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. V.; Dad Ansari M. H.; Dayan C. B.; Giltinan J.; Wang S.; Yu Y.; Kishore V.; Laux P.; Luch A.; Sitti M. Multifunctional magnetic hairbot for untethered osteogenesis, ultrasound contrast imaging and drug delivery. Biomaterials 2019, 219, 119394. 10.1016/j.biomaterials.2019.119394. [DOI] [PubMed] [Google Scholar]
- Tseng Q.; Duchemin-Pelletier E.; Deshiere A.; Balland M.; Guillou H.; Filhol O.; Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (5), 1506. 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T.; Harris E. H.; Coleman A. W. Portrait of a species: Chlamydomonas reinhardtii. Genetics 2005, 170 (4), 1601–1610. 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sretenovic S.; Stojković B.; Dogsa I.; Kostanjšek R.; Poberaj I.; Stopar D. An early mechanical coupling of planktonic bacteria in dilute suspensions. Nat. Commun. 2017, 8 (1), 213. 10.1038/s41467-017-00295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga E.; Powers T. R. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 2009, 72 (9), 096601. 10.1088/0034-4885/72/9/096601. [DOI] [Google Scholar]
- Chen C.; Liu S.; Shi X.-q.; Chaté H.; Wu Y. Weak synchronization and large-scale collective oscillation in dense bacterial suspensions. Nature 2017, 542 (7640), 210. 10.1038/nature20817. [DOI] [PubMed] [Google Scholar]
- Wilson M. A.; Baljon A. R. C. Microstructural Origins of Nonlinear Response in Associating Polymers under Oscillatory Shear. Polymers 2017, 9 (11), 556. 10.3390/polym9110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram Singh A.; Laux P.; Luch A.; Balkrishnan S.; Prasad Dakua S. Bottom-UP assembly of nanorobots: extending synthetic biology to complex material design. Front. Nanosci. Nanotechnol. 2019, 5, 1–2. 10.15761/FNN.1000S2005. [DOI] [Google Scholar]
- Charlton S. G. V.; White M. A.; Jana S.; Eland L. E.; Jayathilake P. G.; Burgess J. G.; Chen J.; Wipat A.; Curtis T. P. Regulating, measuring and modelling the viscoelasticity of bacterial biofilms. J. Bacteriol. 2019, 201 (18), e00101-19. 10.1128/JB.00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez de Aldecoa A. L.; Zafra O.; González-Pastor J. E. Mechanisms and Regulation of Extracellular DNA Release and Its Biological Roles in Microbial Communities. Front. Microbiol. 2017, 8, 1390–1390. 10.3389/fmicb.2017.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetz G. V.; Artemenko N. K.; Tetz V. V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 2009, 53 (3), 1204–1209. 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffe J. F.Rheological Methods in Food Process Engineering; Freeman Press: 1996. [Google Scholar]
- Wolgemuth C. W. Collective swimming and the dynamics of bacterial turbulence. Biophys. J. 2008, 95 (4), 1564–1574. 10.1529/biophysj.107.118257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga E.; Powers T. R. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 2009, 72 (9), 096601. 10.1088/0034-4885/72/9/096601. [DOI] [Google Scholar]
- Saintillan D.; Shelley M. J. Instabilities, pattern formation, and mixing in active suspensions. Phys. Fluids 2008, 20 (12), 123304. 10.1063/1.3041776. [DOI] [PubMed] [Google Scholar]
- Udekwu K. I.; Parrish N.; Ankomah P.; Baquero F.; Levin B. R. Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 2009, 63 (4), 745–757. 10.1093/jac/dkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen R. W.; Sitti M. Bio-hybrid cell-based actuators for microsystems. Small 2014, 10 (19), 3831–3851. 10.1002/smll.201400384. [DOI] [PubMed] [Google Scholar]
- Vikram Singh A.; Sitti M. Targeted drug delivery and imaging using mobile milli/microrobots: A promising future towards theranostic pharmaceutical design. Curr. Pharm. Des. 2016, 22 (11), 1418–1428. 10.2174/1381612822666151210124326. [DOI] [PubMed] [Google Scholar]
- Zhuang J.; Wright Carlsen R.; Sitti M. pH-Taxis of Biohybrid Microsystems. Sci. Rep. 2015, 5 (1), 11403. 10.1038/srep11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.