Abstract
Tuberous sclerosis complex (TSC), a rare genetic disorder caused by a mutation in the TSC1 or TSC2 gene, is characterized by the growth of hamartomas in several organs. This includes the growth of low-grade brain tumors, known as subependymal giant cell astrocytomas (SEGA). Previous studies have shown differential expression of genes related to the extracellular matrix in SEGA. Matrix metalloproteinases (MMPs), and their tissue inhibitors (TIMPs) are responsible for remodeling the extracellular matrix and are associated with tumorigenesis. This study aimed to investigate the MMP/TIMP proteolytic system in SEGA and the regulation of MMPs by microRNAs, which are important post-transcriptional regulators of gene expression. We investigated the expression of MMPs and TIMPs using previously produced RNA-Sequencing data, real-time quantitative PCR and immunohistochemistry in TSC-SEGA samples and controls. We found altered expression of several MMPs and TIMPs in SEGA compared to controls. We identified the lowly expressed miR-320d in SEGA as a potential regulator of MMPs, which can decrease MMP2 expression in human fetal astrocyte cultures. This study provides evidence of a dysregulated MMP/TIMP proteolytic system in SEGA of which MMP2 could be rescued by microRNA-320d. Therefore, further elucidating microRNA-mediated MMP regulation may provide insights into SEGA pathogenesis and identify novel therapeutic targets.
Keywords: Extracellular matrix, Matrix metalloproteinases, MicroRNA, Subependymal giant cell astrocytoma (SEGA), Tuberous sclerosis complex
INTRODUCTION
Tuberous sclerosis complex (TSC) is an autosomal dominantly inherited neurocutaneous disorder affecting approximately 1 million individuals worldwide (1,2). It is caused by inactivating mutations in either the TSC1 or the TSC2 gene (3,4) resulting in constitutive activation of the mammalian target of rapamycin (mTOR) pathway which can affect cell growth and proliferation (5–8). Several brain abnormalities are observed in patients with TSC, including cortical tubers, subependymal nodules and subependymal giant cell astrocytomas (SEGA) (9–11). SEGA are progressive low-grade tumors that develop in the first 2 decades of life in children and adolescents with TSC with a prevalence ranging from 5% to 25% (12–16). They are slow-growing tumors located near the foramen of Monro and extended growth can lead to the obstruction of the cerebrospinal fluid flow and acute hydrocephalus (16,17). Symptoms associated with growing SEGA include headaches, photophobia, diplopia, ataxia, or changes in seizure severity (18). SEGA are believed to develop from subependymal nodules and are characterized by distinctive cytomegalic cells, which display an immature neuroglial phenotype (19–22).
Genetically, there is evidence of second-hit inactivation of TSC1 or TSC2 in SEGA. However, in ∼20% of SEGA these second-hit mutations are not observed, suggesting that additional molecular processes might be involved in SEGA growth (6,23,24). Several studies have performed transcriptional profiling of SEGA identifying differential expression of genes related to the immune system, MAPK family signaling cascades and extracellular matrix (ECM) organization (24–26). Dysregulation of ECM organization has also been seen in TSC cortical tubers with a specific role for matrix metalloproteinases (MMPs) and their endogenous inhibitors (TIMPs), suggesting that the ECM might play an important role in TSC (27–29). MMPs are calcium-dependent zinc-containing endopeptidases that are expressed with a propeptide that needs to be removed for activation (30). TIMPs are “wedge-like”-shaped molecules with 4 residues at N-terminal that form a ridge that can noncovalently bind to the active site of MMPs and thereby inactivate them (31). The MMP/TIMP proteolytic system is known to be involved in the degradation of ECM (32), tissue morphogenesis (33), cell migration (34), angiogenesis (35), blood-brain barrier (BBB) dysfunction (36), wound healing and inflammation (37). Differential expression of MMPs is found in many pathologies including cancer, where they affect proliferation and the metastasis of tumor cells (38–40).
Several MMP inhibitors exist that can regulate MMP overactivity. However, the currently available MMP inhibitors can have a variety of side effects indicating the complexity of MMP regulation (41). MicroRNAs (miRNAs) are short noncoding RNAs which are 20–25 nucleotides long and are able to regulate the expression of protein-coding genes, including MMPs (42,43). They are involved in many physiological processes, such as differentiation, proliferation, and development (44) and have been implicated in neurological disorders (45,46). Previous research showed several miRNAs to be differentially expressed in TSC cortical tubers and SEGA (26,28,29,47,48). In particular, miRNAs have been shown to participate in MMP regulation at the post-transcriptional level in TSC tuber-derived astroglial cultures and glioma cell lines (28,49,50).
In our previous SEGA transcriptome study, we identified differential expression of genes related to several pathways including ECM organization, but did not further study the role of the ECM in SEGA in detail (26). As MMPs and TIMPs may contribute to TSC pathology and cancer, understanding their role in SEGA development could provide new insights into the SEGA biological behavior and potential new therapeutic targets. Therefore, in this study we investigated the expression of MMPs and TIMPs in SEGA. Furthermore, we identified differentially expressed miRNAs in SEGA compared to control tissue that could regulate the expression of MMPs and further investigated the role of these miRNAs in SEGA and their regulation of MMP expression in vitro.
MATERIALS AND METHODS
Subjects
A total of 24 SEGA specimens (male/female: 16/8; age ranging from 1 to 28 years; ethnicity: Caucasian) were collected from the following hospitals: University Medical Centers Amsterdam (location Academic Medical Center, Amsterdam, the Netherlands), the University Medical Center Utrecht (Utrecht, the Netherlands), University Medical Center Groningen (Groningen, the Netherlands), Medical University of Vienna (Vienna, Austria), Children’s Memorial Health Institute (Warsaw, Poland), Anna Meyer Children’s Hospital (Florence, Italy) (Table 1). Twenty-two of the 24 SEGA samples included in this study were obtained from patients who met the clinical diagnostic criteria for TSC, whereas 2 samples were obtained from patients with no other clinical signs of TSC. Histological diagnosis was confirmed following the current WHO classification guidelines (51) by 2 independent neuropathologists. Additional TSC1/TSC2 mutation analysis was performed as part of routine clinical care on blood or tumor sample DNA or was determined using massively parallel sequencing as described previously (23,52), if available (Table 1). Periventricular brain tissue along the lateral ventricle at the level of either the caudothalamic groove or hippocampus plus its associated cortex was obtained from autopsies of patients (postmortem delay less than 24 hours) who did not have TSC, epilepsy or brain tumors and served as control tissue (frozen: n = 8, male/female: 6/2; age ranging from 3 to 87 years; paraffin: n = 4, male/female: 1/3; age ranging from 2 months to 44 years). Specimens were obtained and used in accordance with the Declaration of Helsinki and this study was approved by the Medical Ethics Committee of each institution.
TABLE 1.
Summary of Clinicopathological Features of Patients With SEGA
| SEGA No. | Age (Years) | Gender | Mutation | Location of Tumor | Epilepsy | Age of Epilepsy Onset | Seizure Frequency | AED* | Tumor Size (mm) | Tumor Recurrence/Regrowth | mTOR Inhibitors | Other Clinical Manifestations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 10 | Male | TSC2 | Lateral ventricle | Yes | 3 years | Daily | Yes | 42 | No | No | Unknown |
| 2a | 11 | Female | TSC2 | Lateral ventricle | No | NA | NA | NA | Unknown | No | No | No other signs for TSC |
| 3a,b,c | 8 | Male | TSC2 | Lateral ventricle | Yes | 4 months | None | Yes | 31 | No | No | Cortical tuber, AML |
| 4a | 13 | Female | TSC1 | Lateral ventricle | Yes | 17 months | Monthly | Yes | 40 | No | No | Cortical tuber |
| 5a | 1 | Male | TSC2 | Lateral ventricle | Yes | 1 month | Daily | Yes | 30 | No | Yes (nonresponder) | Multiple SEGAs, cortical tubers, drug resistant epilepsy |
| 6a,b | 13 | Male | TSC2 | Caudate nucleus | Yes | 6 months | Weekly | Yes | 20 | No | Yes (responder) | Tubers, minor psychomotor delay, SEN |
| 7a | 14 | Male | TSC2 | Lateral ventricle | No | NA | NA | NA | 5 | No | No | Renal cysts |
| 8a | 4 | Male | TSC1 | Lateral ventricle | Yes | 3 months | Monthly | Yes | 5 | Yes | No | Cortical tuber |
| 9a,b | 7 | Female | TSC2 | Lateral ventricle | Yes | 5 months | Daily | Yes | 45 | No | No | Unknown |
| 10a,b,c | 17 | Female | TSC2 | Lateral ventricle | No | NA | NA | NA | 27 | No | No | No other signs for TSC |
| 11a | 1 | Female | TSC2 | Lateral ventricle | Yes | 1 month | Daily | Yes | 30 | No | No | Cortical tuber |
| 12a,b,c | 13 | Male | TSC2 | Lateral ventricle | Yes | 4 months | Weekly | Yes | 20 | Yes | No | Cortical tuber |
| 13a,b | 24 | Male | TSC1 | Lateral ventricle | Yes | 6 years | Daily | Yes | 40 | No | No | Unknown |
| 14a,b | 8 | Male | NMI | Caudate nucleus | Yes | 1 year | Weekly | Yes | 30 | No | No | Minor psychomotor delay |
| 15a | 9 | Male | TSC1 | Caudate nucleus | Yes | 2 years | Monthly | Yes | 30 | No | No | None |
| 16a | 28 | Female | TSC2 | Unknown | Yes | Unknown | Unknown | NA | Unknown | Unknown | No | Cortical tuber |
| 17a,b,d | 33 | Male | TSC2 | Basal nuclei | Yes | 6 months | Daily | Yes | 30 | Yes | No | Cortical tubers, autism, drug resistant epilepsy, behavior problems (aggressivity) |
| 18a,b | 1 | Male | TSC2 | Caudate nucleus | Yes | 8 months | Monthly | Yes | 20 | No | No | Minor psychomotor delay, cortical tubers |
| 19a,b | 28 | Male | TSC2 | Lateral ventricle | Yes | 7 years | Daily | Yes | 34 | Yes | No | Unknown |
| 20b,c | 1 | Female | TSC2 | Lateral ventricle | No | NA | NA | NA | 20 | No | No | Cortical tuber |
| 21b,c,d | 4 | Male | TSC2 | Lateral ventricle | Yes | 3 months | Daily | Yes | 18 | No | No | Cortical tuber, AML, autism |
| 22c,d | 16 | Male | TSC1 | Lateral ventricle | No | NA | NA | NA | 37 | No | No | SEN |
| 23c | 10 | Male | TSC2 | 3th ventricle | Yes | 3 months | Seizure free | Yes | 60 | No | No | Cortical tuber, AML, autism |
| 24c | 5 | Female | TSC2 | Lateral ventricle | Yes | 1 month | Daily | Yes | 66 | No | No | Cortical tuber, AML, autism, FCD |
SEGA samples used for RNA-Sequencing (a), RT-qPCR (b), in situ hybridization (c), and immunohistochemistry (d).
The most commonly used AEDs are carbamazepine, vigabatrin, valproate, and topiramate, however each patient had a personalized drug treatment regime.
AED, antiepileptic drugs; AML, angiomyolipoma; FCD, focal cortical dysplasia; NA, not applicable; NMI, no mutation identified; SEGA, subependymal giant cell astrocytomas; SEN, subependymal nodule; TSC, tuberous sclerosis complex.
RNA-Sequencing
RNA-Sequencing (RNA-Seq) data sets produced by our laboratory, were retrieved from the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG (accession number: EGAS00001003787) (26). Data were retrieved for 19 SEGA samples and 8 control and processed as previously described (26). Briefly, sequence reads were trimmed and filtered using FastQC v0.11.5 (Babraham Institute, Babraham, Cambridgeshire, UK) and Trimmomatic v0.36. Paired-end reads were aligned to the human reference genome (GRCh38) with TopHat2 v2.0.13 and default settings (53). For the small RNA-Seq data set no mismatches were allowed between the trimmed reads and the reference genome and small RNA reads were allowed to align a maximum of 10 times. Next, the number of reads that mapped to each gene, based on Gencode v25, was determined using featureCounts from the SubRead package (54). The count matrix was normalized using the R package DESeq2 (55). Genes and small RNAs with a Benjamini-Hochberg adjusted p value < 0.05 were considered differentially expressed. Differentially expressed genes (DEGs) related to the ECM organization were identified by overlapping the ECM organization genelist from the Reactome database with the DEGs produced from the RNA-Seq data (56,57).
To identify miRNAs that could potentially target all 5 MMP genes with a higher expression in SEGA compared to control (MMP2, MMP11, MMP14, MMP15, and MMP19), miRNAs that had lower expression in SEGA (adjusted p value < 0.05) as compared to control and whose expression patterns where inversely correlated with MMP expression were selected. The list of predicted miRNA targets for each of the differentially expressed miRNAs was retrieved from miRWalk2 (58,59). miRNAs that were predicted to target all 5 MMPs (MMP2, MMP11, MMP14, MMP15, and MMP19), were considered potential regulators of MMP expression.
RNA Isolation and Real-Time Quantitative PCR Analysis
For RNA isolation, cell culture or fresh brain material (12 SEGA samples and 8 control samples) was homogenized in Qiazol Lysis Reagent (Qiagen Benelux, Venlo, The Netherlands). RNA was isolated using the miRNeasy Mini kit (Qiagen Benelux, Venlo, The Netherlands) according to manufacturer’s instructions. The concentration and purity of RNA were determined at 260/280 nm using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). mRNA and miRNA expression was evaluated as described previously (60). Briefly, 250 ng of RNA were reverse-transcribed into cDNA using oligo dT primers. Real-time quantitative PCRs (RT-qPCR) were run according to the manufacturer’s guidelines, on a Roche Lightcycler 480 thermocycler (Roche Applied Science, Basel, Switzerland) using LightCycler 480 SYBR Green I Master (Roche Applied Science, Indianapolis, IN) with the following cycling conditions: initial denaturation at 95°C for 5 minutes, followed by 50 cycles of denaturation at 95°C for 10 seconds, 65°C for 10 seconds, and 72°C for 15 seconds, annealing at 95°C for 5 seconds followed by extension on 65°C for 1 minute. PCR primers were designed using Universal Probe library (ProbeFinder version 2.53 for human, Roche Applied Science) (Table 2). Expression of miRNAs was analyzed using TaqMan microRNA assays (Applied Biosystems, Foster City, CA). cDNA was generated from 50 ng of RNA using TaqMan MicroRNA reverse transcription kit (Applied Biosystems). RT-qPCR was performed on a Lightcycler 480 Real-Time PCR System (Roche Applied Science). Quantification of the data was done using the LinRegPCR program in which linear regression on the Log(fluorescence) per cycle number data is applied to determine the amplification efficiency per sample (61). The expression of each specific product was divided by the geometric mean of the concentration of 2 reference genes (mRNA: Elongation factor 1-alpha [EF1α] and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]; miRNA: the U6B small nuclear RNA gene [RNU6B] and RNU44).
TABLE 2.
Primer List
| Gene | Forward (5′->3′) | Reverse (5′->3′) |
|---|---|---|
| MMP2 | ATAACCTGGATGCCGTCGT | AGGCACCCTTGAAGAAGTAGC |
| MMP11 | TCCTGAGGTCAGGTCTTGGT | CAGATTTCCAGGATTGTCAGC |
| MMP14 | GCCTTGGACTGTCAGGAATG | AGGGGTCACTGGAATGCTC |
| MMP15 | CAGGCCCATCAGTGTCTGG | TGGTGCCCTTGTAGAAGTAGG |
| MMP16 | AGGGCATCCAGAAGATATATGG | GGCACTGTCGGTAGAGGTCTT |
| MMP17 | AAGAGGAACCTGTCGTGGAG | CACCTCGTGGAAGTTCAGG |
| MMP19 | ATGCCAGACCCTTGCAGTAG | CCCCCTTGAAAGCATAGGTC |
| MMP25 | TCCTGGGTGGTGGAATCA | GCAACGGAAAAGGTTAACAGC |
| TIMP1 | GGGCTTCACCAAGACCTACA | TGCAGGGGATGGATAAACAG |
| TIMP2 | TGCAGATGTAGTGATCAGGGC | TCTCAGGCCCTTTGAACATC |
| TIMP3 | GCTGGAGGTCAACAAGTACCA | CACAGCCCCGTGTACATCT |
| TIMP4 | TTGGTGCAGAGGGAAAGTCT | GGTACTGTGTAGCAGGTGGTGA |
| EF1α | ATCCACCTTTGGGTCGCTTT | CCGCAACTGTCTGTCTCATATCAC |
| GAPDH | AGGCAACTAGGATGGTGTGG | TTGATTTTGGAGGGATCTCG |
Immunohistochemistry
Sections were deparaffinated in xylene (3x) and rinsed in ethanol (100%, 95%, and 70%). Antigen retrieval was performed by incubating the sections in 0.1 M sodium citrate buffer pH 6.0 at 121°C for 10 minutes using a pressure cooker. Sections were washed with phosphate buffered saline ([PBS], pH 7.4) after which they were blocked with 10% normal goat serum (NGS) for 30 minutes at room temperature. After removal of NGS, primary antibody (1:200 rabbit polyclonal IgG anti-MMP2, Abcam, Cambridge, UK; 1:500 mouse monoclonal IgG 3/κ anti-MMP14, Merck, Darmstadt, Germany) in Normal Antibody Diluent (Immunological, Duiven, The Netherlands) was added to the sections and incubated overnight at 4°C. After incubation with primary antibodies, sections were washed in PBS and stained with a polymer-based peroxidase immunohistochemistry detection kit according to manufacturer’s guidelines (PowerVision Peroxidase system, ImmunoVision, Brisbane, CA). After washing, sections were stained with 3,3′-diaminobenzidine tetrahydrochloride (50 mg diaminobenzidine [DAB], Sigma-Aldrich, Zwijndrecht, the Netherlands) and H2O2 in Tris-HCl (0.015% final concentration). The reaction was stopped by washing with dH2O, the sections were then counterstained with hematoxylin and dehydrated with ethanol (70%, 95%, 100%) and xylene (3x) and coverslipped.
Stainings of MMPs were quantified by measuring the optical density (OD) of SEGA and control tissue using ImageJ. For each case, 2-3 representative photographs of periventricular white matter (wm) (n = 4) and when present nearby gray matter (gm) (n = 3) were taken (20x magnification). Three OD measurements were taken after hematoxylin-DAB color deconvolution for each respective picture. Finally, the mean OD was calculated per case.
In Situ Hybridization
Paraffin slides from SEGA and autopsy controls (Table 1) were deparaffinated in xylene (3x) and rinsed in ethanol (2x 100%, 70%) and dH2O. Sections were pretreated using a pressure cooker in 0.1 M sodium citrate buffer, pH 6.0, at 121°C for 10 minutes. The oligonucleotide probe for miR-320d (TCCTCUCAACCCAGCTUUT) contained LNA modification, 2-o-methyl modification and digoxigenin (DIG) label (RiboTask ApS, Odense, Denmark). Sections were incubated with the probe (1:750 dilution) in hybridization mix (600 mM NaCl, 10 mM HEPES, 1 mM EDTA, 5× Denhardts, 50% formamide) for 1 hour at 56°C. Sections were washed with saline-sodium citrate (SSC) 2× for 2 minutes, SSC 0.5× for 2 minutes, and SSC 0.2× for 1 minute (in agitation). After washing with sterile PBS, sections were blocked for 15 minutes with 0.02% Tween 20 and 1% NGS. Hybridization was detected with AP labeled with anti-DIG (Roche Applied Science, Basel, Switzerland). Nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt was used as chromogenic substrate for AP (1:50 diluted in NTM-T buffer [100 mM Tris, pH 9.5; 100 mM NaCl; 50 mM MgCl2]; 0.05% Tween 20). Expression of miR-320d was quantified by measuring the OD of SEGA and control tissue using ImageJ. For each case, 2 and 3 representative photographs of periventricular wm (n = 9) and when present nearby gm (n = 7) were taken (20x magnification). Three OD measurements were taken for each respective picture. Finally, the mean OD was calculated per case.
Cell Culture
As SEGA are thought to originate from neuroglial precursor cells that undergo an altered/impaired development, we have chosen to use a glial cell type in a developmental state; (human fetal astrocyte-enriched cells) to study the role of miR-320d on MMP expression under basal conditions. Primary fetal astrocyte-enriched cell cultures were obtained from fetal brain tissue (n = 3, 14–19 weeks of gestation) collected from medically induced abortions as described previously (47,48), with appropriate maternal written consent for brain autopsy. Briefly, blood vessels were removed and the tissue was cut into smaller fragments. The tissue was enzymatically digested by incubating at 37°C for 30 minutes with trypsin. The enzymatic reaction was stopped by adding twice the amount of astrocyte medium (DMEM/F10, 10% FCS, 5% penicillin/streptomycin, and 5% glutamine) and the cell suspension was passed through a 70-µm cell strainer. After washing 2 times with astrocyte medium, the cells were incubated at 37°C, 5% CO2 for 48 hours after which they were thoroughly washed with PBS. Fresh medium was added twice a week and cultures reached confluency after 2 and 3 weeks. Secondary astrocyte cultures for experimental manipulation were established by trypsinizing confluent cultures and subplating onto poly-l-lysine (PLL; 15 mg/mL, Sigma-Aldrich, St. Louis, MO)-precoated 6- and 12-well plates (Costar; 50 000 cells/well in a 12-well plate for RNA isolation and RT-qPCR).
Transfection of Cell Cultures
Cells on PLL-coated plates were transfected with mimic pre-miRNA for hsa-miR-320d (50 µM; Applied Biosystems, Carlsbad, CA) for 24 hours. Cells treated with lipofectamine without mimic were used as a control. Mimics were delivered to the cells using Lipofectamine 2000 transfection reagent (Life Technologies, Grand Island, NY) in a final concentration of 50 nM.
Statistical Analysis
Statistical analyses were performed with GraphPad Prism version 5 (GraphPad Software, La Jolla, CA) and IBM SPSS Statistics 24 (IBM Corp., Armonk, NY) using the nonparametric Mann-Whitney U test or, for multiple groups, the nonparametric Kruskal-Wallis test followed by Mann-Whitney U test. A p value < 0.05 was assumed to indicate a statistical difference. Correlations were assessed with R or IBM SPSS Statistics 24, using the Spearman’s rank correlation test (p < 0.05 and 0.7 < r > −0.7 was considered statistically significant).
RESULTS
MMP and TIMP Expression in SEGA
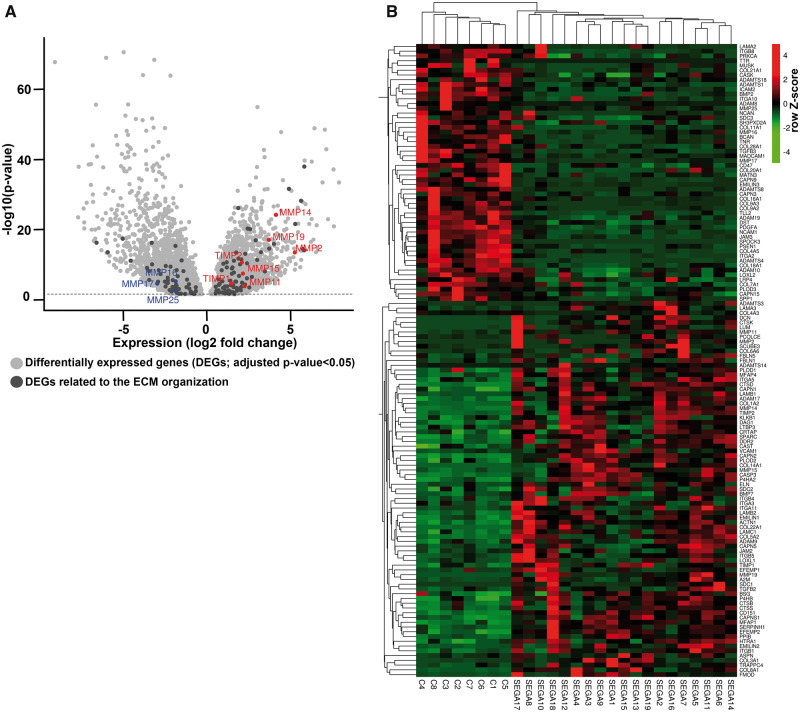
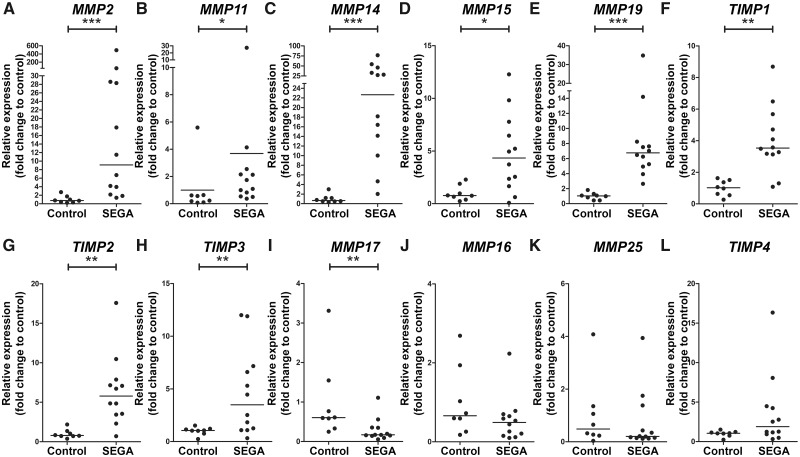
RNA-Seq data previously produced by our laboratory was used to specifically investigate the expression of MMPs and TIMPs in SEGA (26). In total, 19 surgical SEGA samples (obtained from 17 TSC patients and 2 patients without any clinical signs of TSC) and 8 area-matched periventricular controls (autopsy specimens) without a history of other neurological diseases were subjected to RNA-Seq (26) (Table 1). Differential gene expression analysis identified 4621 genes overexpressed and 4779 underexpressed (adjusted p value < 0.05) in SEGA samples compared to control tissue, of which 133 genes belong to the Reactome-based ECM organization pathway (Fig. 1A). Furthermore, the gene expression of these 133 genes could be used to separate the control and SEGA samples (Fig. 1B). Among the 133 genes identified MMP2, MMP11, MMP14, MMP15, MMP16, MMP17, MMP19, MMP25, TIMP1 and TIMP2 were differentially expressed (Fig. 1A, B). Though not present in the ECM organization pathway, the other 2 TIMPs found in the brain, TIMP3 and TIMP4, were also among the 9400 DEGs (Table 3). These MMPs and TIMPs were selected for further validation with RT-qPCR. Higher expression of MMP2, MMP11, MMP14, MMP15, MMP19, TIMP1, TIMP2, and TIMP3 was found in SEGA (n = 12) as compared to control tissue (n = 8; p < 0.05; Fig. 2A–H). The expression of MMP17 (Fig. 2I) was lower in SEGA as compared to control tissue, whereas MMP16, MMP25, and TIMP4 expression did not change (Fig. 2J–L). RNA expression of MMPs and TIMPs did not correlate with the age at epilepsy onset, age at surgery, or size of the tumor (p > 0.05 and −0.7 < r > 0.7 for both RT-qPCR and RNA-Seq data). Furthermore, RNA expression of MMPs and TIMPs was not associated with the clinical features such as gender, TSC mutation, epilepsy, or preoperative seizure frequency (Mann-Whitney U test or, for multiple groups, the nonparametric Kruskal-Wallis test; p > 0.05 for both RT-qPCR and RNA-Seq data). MMP19 expression based on the RT-qPCR was higher in recurrent/regrown SEGA compared to nonrecurrent/regrown SEGA (Supplementary DataFig. S1; p = 0.0485). However, RNA-Seq data did not show differences between recurrent/regrown SEGA and nonrecurrent/regrown SEGA (p = 0.96).
FIGURE 1.
The extracellular matrix (ECM) organization in subependymal giant cell astrocytomas (SEGA). (A) Volcano plot showing the differentially expressed genes ([DEGs]; adjusted p value < 0.05; light gray) between SEGA and control tissue. A total of 4621 mRNAs were found to be overexpressed and 4779 underexpressed in SEGA (n = 19) as compared to control tissue (n = 8). A total of 133 DEGs were related to the Reactome-based ECM organization pathway (dark gray). Among the 133 genes MMP2, MMP11, MMP14, MMP15, MMP19, TIMP1, TIMP2 were overexpressed and MMP16, MMP17 and MMP25 were underexpressed. (B) Heatmap with Z-score hierarchical clustering for the RNA expression data of DEGs related to the Reactome-based ECM organization pathway in SEGA (n = 19) and control tissue (n = 8). The color scale means the gene expression standard deviations from the mean, with green for low expression and red for the high expression.
TABLE 3.
Differentially Expressed MMPs and TIMPs in SEGAs as Compared to Control Tissue
| mRNA | MMP2 | MMP11 | MMP14 | MMP15 | MMP16 | MMP17 | MMP19 | MMP25 | TIMP1 | TIMP2 | TIMP3 | TIMP4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BaseMean | 7918.05 | 37.645 | 10816.6 | 498.068 | 402.112 | 129.329 | 407.115 | 32.0523 | 3772.49 | 27817.6 | 7473.16 | 533.5 |
| Log2 FoldChange | 5.26395 | 2.27464 | 4.13891 | 2.16889 | −1.8413 | −2.9409 | 3.71133 | −1.8555 | 1.49335 | 2.10445 | 1.16444 | 1.77748 |
| LfcSE | 0.69286 | 0.59681 | 0.40108 | 0.39237 | 0.40482 | 0.69567 | 0.4314 | 0.62099 | 0.35208 | 0.31444 | 0.39523 | 0.6906 |
| Stat | 7.59742 | 3.81131 | 10.3194 | 5.52764 | −4.5484 | −4.2274 | 8.60299 | −2.9879 | 4.24149 | 6.69264 | 2.94624 | 2.57382 |
| p Value | 3E−14 | 0.00014 | 5.8E−25 | 3.2E−08 | 5.4E−06 | 2.4E−05 | 7.8E−18 | 0.00281 | 2.2E−05 | 2.2E−11 | 0.00322 | 0.01006 |
| Adjusted p value | 8.4E−13 | 0.00057 | 6.5E−23 | 3.2E−07 | 3.2E−05 | 0.00012 | 3.8E−16 | 0.00802 | 0.00011 | 4E−10 | 0.00901 | 0.02403 |
| Control 1 | 182.645 | 0 | 275.352 | 117.612 | 711.21 | 395.731 | 16.6041 | 22.1388 | 599.132 | 8743.45 | 4679.59 | 253.213 |
| Control 2 | 453.634 | 13.9939 | 2239.02 | 141.105 | 928.26 | 165.594 | 45.4801 | 46.6462 | 3266.4 | 6040.69 | 2222.69 | 198.246 |
| Control 3 | 42.8228 | 0 | 1082.38 | 47.2527 | 218.544 | 160.955 | 57.5892 | 270.226 | 3397.76 | 6622.76 | 7322.69 | 243.647 |
| Control 4 | 224.35 | 0 | 213.81 | 213.81 | 2567.23 | 1075.08 | 30.1142 | 76.7911 | 1017.86 | 4381.61 | 3014.43 | 367.393 |
| Control 5 | 252.687 | 19.151 | 588.894 | 120.758 | 808.067 | 170.231 | 32.9823 | 26.0667 | 691.565 | 6271.96 | 3574.86 | 165.976 |
| Control 6 | 61.4008 | 20.4669 | 467.816 | 273.38 | 381.562 | 251.451 | 48.2435 | 58.4769 | 2150.49 | 12322.6 | 4942.76 | 0 |
| Control 7 | 96.9527 | 9.61515 | 1185.87 | 118.587 | 497.584 | 83.3313 | 18.429 | 26.4416 | 1578.49 | 10136 | 4299.57 | 92.9464 |
| Control 8 | 26.5541 | 6.88439 | 70.8108 | 63.9265 | 541.9 | 518.296 | 57.0421 | 21.6366 | 215.383 | 10492.8 | 1072 | 33.4385 |
| SEGA1 | 350.288 | 99.906 | 10408.7 | 1214.91 | 83.8717 | 3.70022 | 145.542 | 0 | 2070.89 | 8752.26 | 29806.5 | 477.329 |
| SEGA2 | 19 637.7 | 35.9809 | 27384.9 | 721.305 | 174.845 | 38.2298 | 278.29 | 12.9307 | 1977.83 | 59003.1 | 4009.06 | 558.267 |
| SEGA3 | 326.138 | 14.7288 | 25176.8 | 1391.87 | 131.507 | 0 | 193.579 | 7.36442 | 5079.34 | 36881 | 25576.6 | 26.3015 |
| SEGA4 | 170.591 | 28.7514 | 12355.4 | 1046.55 | 55.586 | 0.95838 | 208.927 | 17.2508 | 2749.59 | 28491.6 | 8542.03 | 411.144 |
| SEGA5 | 3984.96 | 62.718 | 17916.6 | 324.24 | 427.192 | 77.51 | 1091.06 | 21.8921 | 3878.46 | 51157.2 | 4553.56 | 186.971 |
| SEGA6 | 15 097.6 | 29.4384 | 12479.7 | 817.734 | 711.974 | 7.63219 | 390.332 | 6.54187 | 3086.67 | 41390.4 | 4661.08 | 757.767 |
| SEGA7 | 48 776.7 | 90.7741 | 20215.5 | 380.941 | 216.461 | 33.3614 | 443.785 | 0 | 3766.74 | 55441.3 | 6153.24 | 2215.04 |
| SEGA8 | 924.957 | 28.444 | 6737.01 | 243.354 | 92.7064 | 189.627 | 31.6044 | 4.21393 | 3188.89 | 29829.3 | 6329.32 | 51.6206 |
| SEGA9 | 1171.21 | 8.16435 | 12124.8 | 866.163 | 107.621 | 5.1955 | 235.282 | 27.4619 | 3664.31 | 40081.8 | 6475.81 | 565.567 |
| SEGA10 | 215.259 | 11.5576 | 1533.54 | 18.781 | 85.237 | 4.33408 | 1272.78 | 26.0045 | 9095.07 | 6490.29 | 3561.89 | 0 |
| SEGA11 | 1669.2 | 53.9765 | 14927.1 | 1188.5 | 250.533 | 99.8056 | 1758.82 | 42.7738 | 3814 | 39267.4 | 7084.16 | 89.6214 |
| SEGA12 | 31 142.2 | 17.1046 | 30274.5 | 599.319 | 276.305 | 3.94722 | 809.18 | 7.23657 | 2407.8 | 68810.6 | 1033.51 | 360.513 |
| SEGA13 | 5142.81 | 22.1056 | 7551.02 | 466.819 | 202.852 | 0 | 274.37 | 50.7129 | 6522.46 | 16450.5 | 10090.6 | 3808.67 |
| SEGA14 | 7749.68 | 11.8189 | 19932.6 | 424.3 | 87.4601 | 11.8189 | 579.128 | 0 | 5109.33 | 45514.7 | 9754.17 | 33.093 |
| SEGA15 | 444.297 | 56.1474 | 9724.48 | 961.829 | 69.5739 | 9.76476 | 281.957 | 4.88238 | 6996.45 | 18311.4 | 13725.6 | 86.6623 |
| SEGA16 | 13 231 | 11.3008 | 16272.1 | 386.488 | 479.155 | 75.7155 | 305.122 | 2.26017 | 2356.22 | 48061.3 | 2552.86 | 1237.44 |
| SEGA17 | 44 702.6 | 271.937 | 21 368.3 | 243.825 | 195.06 | 89.4982 | 523.794 | 12.0478 | 5108.28 | 57 122.2 | 16 237 | 1060.78 |
| SEGA18 | 4069.25 | 76.7546 | 12 831.9 | 469.336 | 211.39 | 20.1324 | 1323.7 | 22.6489 | 9297.37 | 13 926.6 | 5554.01 | 54.1057 |
| SEGA19 | 13 639.8 | 14.6952 | 6709.02 | 585.136 | 343.333 | 0 | 538.378 | 50.7652 | 8770.36 | 21 080.9 | 4945.6 | 1068.74 |
MMPs, matrix metalloproteinases; SEGA, subependymal giant cell astrocytomas; TIMPs, tissue-inhibitors of metalloproteinases.
FIGURE 2.
Gene expression MMPs and TIMPs differentially expression in subependymal giant cell astrocytomas (SEGA) compared to controls. (A–L) Gene expression was studied using RT-qPCR analysis of MMP2 (A), MMP11 (B), MMP14 (C), MMP15 (D), MMP19 (E), TIMP1 (F), TIMP2 (G), TIMP3 (H), MMP17 (I), MMP16 (J), MMP25 (K), TIMP4 (L) in SEGA (n = 12) and controls (n = 8). Data are expressed relative to the expression observed in control tissue. *p < 0.05, **p < 0.01, ***p < 0.001, Mann-Whitney U test.
MMP2 and MMP14 Protein Expression Increased in SEGA
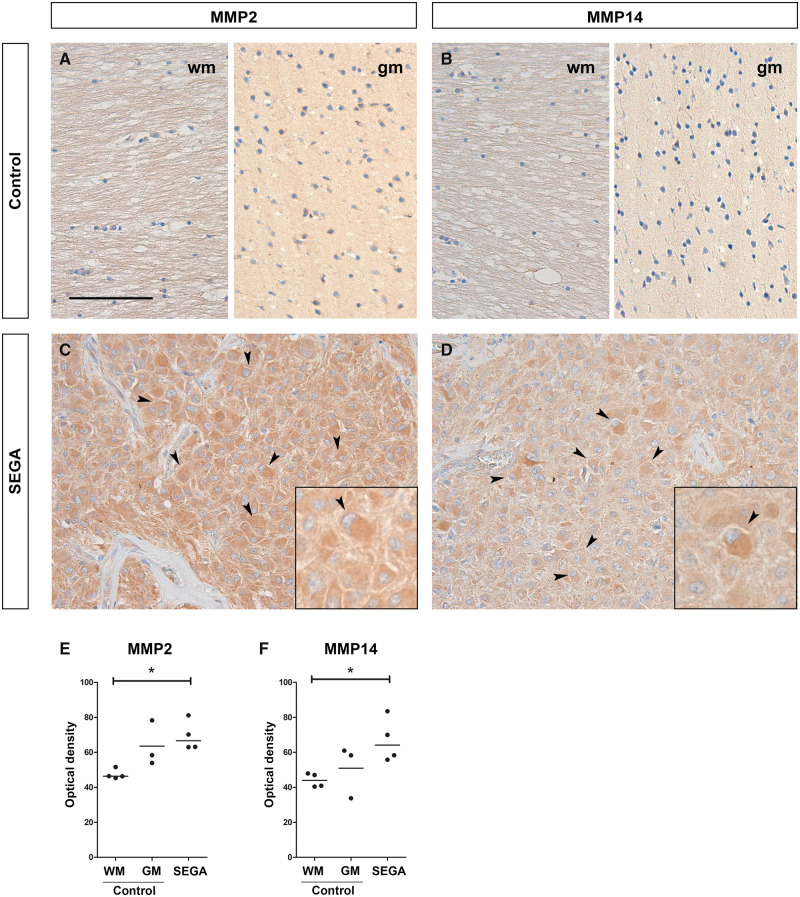
As MMP2 and MMP14 had the most abundant RNA expression in SEGA compared to controls they were selected for further analysis at the protein level. To study cellular localization and distribution, and to quantify protein expression of MMP2 and MMP14 in SEGA, immunohistochemistry was performed on SEGA and periventricular control tissue slides (Fig. 3A–F). In periventricular control tissue both MMP2 and MMP14 were not detected in glial cells in the wm. In nearby cortex tissue, low immunoreactivity of MMP2 and MMP14 was observed in neurons (Fig. 3A, B). In SEGA, moderate to high immunoreactivity of MMP2 and MMP14 was found in giant cells (Fig. 3C, D). The OD of MMP2 and MMP14 was higher in SEGA as compared to periventricular wm of controls (Fig. 3F; p < 0.05).
FIGURE 3.
Protein expression of MMP2, MMP14 in subependymal giant cell astrocytomas (SEGA). (A–D) A moderate to high immunoreactivity of MMP2 (C) and MMP14 (D) was observed in SEGA, while no to low immunoreactivity was seen in periventricular white matter (wm) and gray matter (gm) control tissue, respectively (A, B). Insets show a higher magnification of giant cells (indicated with arrows) in SEGA. Scale bar: 100 µm; insets: 50 µm. (E, F) The optical density per case for the immunoreactivity of MMP2 (E) and MMP14 (F) in periventricular control tissue (n = 3/4) and SEGA (n = 4). *p < 0.05, **p < 0.01, ***p < 0.001, Mann-Whitney U test.
miR-320d as a Predictive Regulator of MMPs Is Expressed at Lower Levels in SEGA Compared to Control
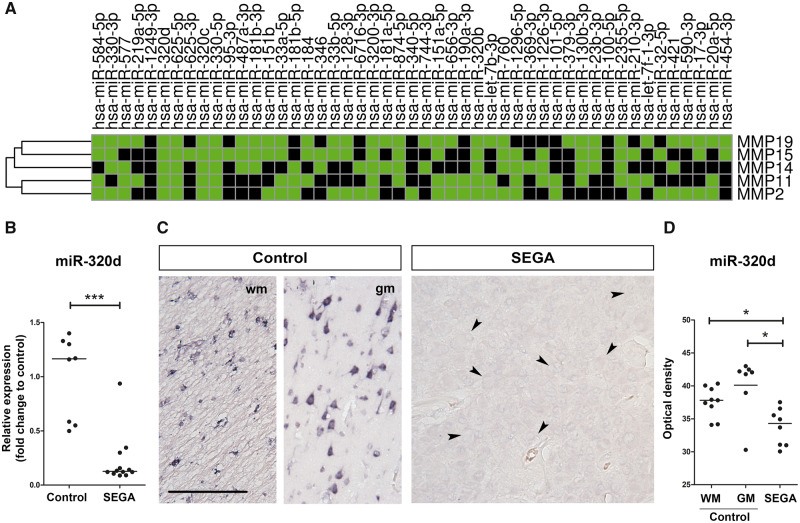
To identify miRNAs that target MMP2, MMP11, MMP14, MMP15, and MMP19, differentially expressed miRNAs with a lower expression in SEGA as compared to controls were evaluated with miRWalk2. Based on small RNA-Seq data, we found 49 miRNAs with lower expression in SEGA as compared to controls. Six miRNAs (miR-320d, miR-320b, miR-320c, miR-625-5p, miR-330-5p, and miR-3200-3p) were predicted to target all 5 MMPs (MMP2, MMP11, MMP14, MMP15, and MMP19) with a higher expression in SEGA as compared to controls (Fig. 4A). Spearman correlations using the RNA-Seq data were calculated between each miRNA and their predicted MMP target (Table 4). miR-320d showed the strongest negative correlation with all 5 MMPs (MMP2: R: −0.484, p = 0.010; MMP11: R: −0.459, p = 0.016; MMP14: R: −0.572, p = 0.002; MMP15: R: −0.471, p = 0.013; MMP19: R: −0.433, p = 0.024) and was therefore selected for further analysis. Using RT-qPCR the lower expression of miR-320d in SEGA as compared to control samples was validated (Fig. 4B). To study the downregulation of miR-320d on a cellular level, in situ hybridization was performed (Fig. 4C). In control tissue, miR-320d is abundant in glial cells in periventricular wm and neuronal cells, whereas in SEGA miR-320d was not detected. The OD of miR-320d was lower in SEGA as compared to controls (Fig. 4D; p < 0.05).
FIGURE 4.
miR-320d is a predictive regulator of MMPs and is lower expressed in subependymal giant cell astrocytomas (SEGA) compared to control. (A) Heatmap showing all underexpressed miRNAs in SEGA as compared to controls (adjusted p value <0.05) predicted to target MMP2, MMP11, MMP14, MMP15, and/or MMP19 (based on miRWalk2). Green boxes indicate MMP19, MMP15, MMP14, MMP11, and/or MMP2 as predicted target of a specific miRNA. (B) Relative expression of miR-320d in SEGA (n = 12) as compared to control tissue (n = 8). Data are expressed relative to the expression observed in control tissue. *p < 0.05, **p < 0.01, ***p < 0.001, Mann-Whitney U test. (C) In situ hybridization of miR-320d in periventricular white matter (wm) and gray matter (gm) control and SEGA tissue. Giant cells are indicated with arrows. Scale bar: 100 µm. (D) The optical density per case for the in situ hybridization signal of miR-320d in periventricular control tissue (n = 7/9) and SEGA (n = 8). *p < 0.05, ***p < 0.001, Mann-Whitney U test.
TABLE 4.
Correlations of MMPs and TIMPs With miR-320d, miR-320b, miR-320c, miR-625-5p, miR-330-5p, and miR-3200-3p
| MicroRNA→ Gene↓ | miR-320d (p Value; r) | miR-320b (p Value; r) | miR-320c (p Value; r) | miR-625-5p (p Value; r) | miR-330-5p (p Value; r) | miR-3200-3p (p Value; r) |
|---|---|---|---|---|---|---|
| MMP2 | 0.010; −0.484 | 0.028; −0.423 | 0.052; −0.379 | 0.243; −0.233 | 0.271; −0.22 | 0.493; −0.138 |
| MMP11 | 0.016; −0.459 | 0.024; −0.432 | 0.012; −0.477 | 0.03; −0.419 | 0.072; −0.351 | 0.062; −0.363 |
| MMP14 | 0.002; −0.572 | 0.024; −0.433 | 0.007; −0.503 | 0.019; −0.45 | 0.044; −0.391 | 0.061; −0.366 |
| MMP15 | 0.013; −0.471 | 0.028; −0.422 | 0.011; −0.481 | <0.001; −0.627 | 0.024; −0.433 | <0.001; −0.644 |
| MMP16 | 0.074; 0.349 | 0.315; 0.201 | 0.058; 0.37 | 0.021; 0.443 | 0.06; 0.367 | 0.004; 0.532 |
| MMP17 | 0.040; 0.397 | 0.058; 0.369 | 0.013; 0.47 | 0.002; 0.56 | 0.093; 0.33 | <0.001; 0.633 |
| MMP19 | 0.024; −0.433 | 0.083; −0.340 | 0.031; −0.416 | 0.168; −0.273 | 0.269; −0.22 | 0.518; −0.13 |
| MMP25 | 0.050; 0.380 | 0.125; 0.302 | 0.039; 0.399 | 0.095; 0.328 | 0.085; 0.337 | 0.202; 0.254 |
| TIMP1 | 0.372; −0.179 | 0.634; −0.096 | 0.403; −0.168 | 0.489; −0.139 | 0.565; −0.116 | 0.477; −0.143 |
| TIMP2 | 0.001; −0.590 | 0.014; −0.467 | 0.011; −0.483 | 0.034; −0.41 | 0.014; −0.466 | 0.084; −0.338 |
| TIMP3 | 0.348; −0.188 | 0.475; −0.143 | 0.275; −0.218 | 0.096; −0.327 | 0.073; −0.351 | 0.014; −0.467 |
| TIMP4 | 0.259; −0.225 | 0.102; −0.321 | 0.383; −0.175 | 0.308; −0.204 | 0.251; −0.229 | 0.099; −0.324 |
MMPs, matrix metalloproteinases; TIMPs, tissue-inhibitors of metalloproteinases.
Regulation of MMP by miR-320d Mimic Transfection in Fetal Astrocytes
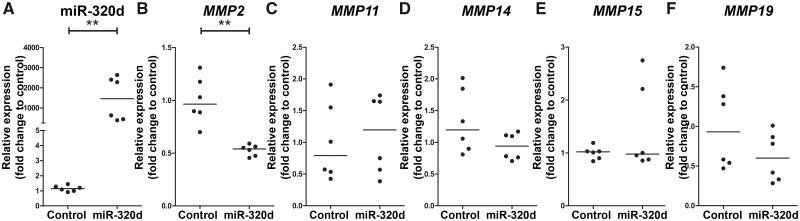
To study regulation of MMP2, MMP11, MMP14, MMP15 and MMP19 by miR-320d, fetal astrocytes were transfected with miR-320d mimic. RT-qPCR revealed that transfection with 50 nM miR-320d mimic led to a higher expression of miR-320d (Fig. 5A; p < 0.05). RNA expression of MMP2, MMP11, MMP14, MMP15 and MMP19 was determined 24 hours after miR-320d transfection using RT-qPCR, and showed a lower expression of MMP2 in miR-320d mimic transfected cells compared to control (Fig. 5B; p = 0.002), whereas the expression of MMP11, MMP14, MMP15 and MMP19 did not differ between conditions (Fig. 5C–F).
FIGURE 5.
MMP expression after transfection with miR-320d in fetal astrocytes. (A–F) TaqMan PCR of miRNA-320d (A) and RT-qPCR of MMP2 (B), MMP11 (C), MMP14 (D), MMP15 (E), and MMP19 (F) in fetal astrocytes transfected with miRNA-320d mimic (miR-320d) for 24 hours (n = 2 biological triplets and 3 technical duplicates). Data are normalized to lipofectamine (control). **p < 0.01, Mann-Whitney U test.
DISCUSSION
This study provides evidence that the MMP/TIMP proteolytic system is dysregulated in SEGA. We showed higher expression of MMP2, MMP11, MMP14, MMP15, MMP19, and their endogenous inhibitors TIMP1, TIMP2 and TIMP3 and lower expression of MMP17 in SEGA as compared to periventricular control tissue. Furthermore, we identified the lowly expressed miR-320d in SEGA compared to controls as a potential MMP regulator that can decrease MMP2 expression in vitro.
MMP and TIMP Expression in SEGA
Dysregulation of ECM organization in TSC cortical tubers and SEGA has been suggested by several transcriptome studies showing differential expression of genes related to the gene ontology term ECM organization, however the ECM has not been studied in detail in SEGA (24,26,27,29). In this study, we identified higher expression of MMP2, MMP11, MMP14, MMP15, MMP16, MMP17, MMP19, MMP25, TIMP1, TIMP2 and TIMP3 in SEGA as compared to controls. This suggests that the dysregulation of the MMP/TIMP proteolytic system is conserved across TSC pathology and might also affect migration during early brain development in TSC. In our previous study, we showed higher expression of MMPs and TIMPs in non-SEGA material of TSC patients. Among others, higher expression of MMP2, MMP14, TIMP1, TIMP2 and TIMP3 was observed in cortical tubers of TSC patients as compared to control cortex. Moreover, increased protein expression of MMP2 and MMP14 was also observed in tubers of TSC patients and the expression of MMPs and TIMPs was associated with neuroinflammation and BBB dysfunction (28). This indicates that the overexpression of MMPs might not be SEGA-specific, but it does underlie its relevance in the dysfunctional microenvironment that is present in TSC-related brain alterations. Multiple studies have shown the presence of neuroinflammation in TSC cortical tubers and SEGA (24,26,27,29,62–64). Previously, we have shown that the immune system is among the most represented enriched pathways in SEGA (26). Furthermore, upregulation of several cytokines, including interleukin 33, TNF Superfamily member 8, C-C Motif Chemokine Ligand 3, and C-C Motif Chemokine Ligand 5 was observed in SEGA compared to controls. Moreover, albumin extravasation, with uptake in astrocytes, has been reported in cortical tubers and SEGA of TSC patients, suggesting that alterations in BBB permeability are associated with the inflammation in TSC brain lesions (62,65). MMPs can contribute to the inflammatory response by cleaving propeptides of cytokines such as TNFα and IL-1β, thereby activating them (66–68). In turn, neuroinflammation can increase the expression and activity of MMPs and infiltrating immune cells such as leukocytes, which are a major source of MMP9 (69,70). The link between the neuroinflammation and MMPs in SEGA deserves further investigation.
MMPs are also known to be involved in cancer pathology in multiple ways, including intravasation and extravasation of tumor cells, migration of tumor cells in the brain and adhesion of metastatic cells to the ECM (38–40). Previous research has shown that higher MMP2 expression can be a predictor for overall survival of patients with astrocytomas (71). In another study, it was found that MMP14 and MMP19 had higher expression in gliomas compared to healthy control tissue, and that coexpression of MMP14 and MMP19 predicted poor survival in human glioma (72,73). Furthermore, a correlation between higher MMP11 and MMP19 expression and increased WHO-grading of malignant tumors has been suggested (74). However, SEGA are slow-growing, WHO grade I tumors that are not associated with poor survival rates, suggesting that in SEGA the higher MMP expression is most likely not linked to overall survival (51). Furthermore, in this study we did not find any correlation with the clinical traits investigated, including preoperative seizure frequency, duration of epilepsy, size of the tumor, and tumor recurrence/regrowth. A previous study found increased expression of TIMP1 and TIMP2 in grade I brain tumors compared to grade II or III tumors (75). Furthermore, it has been shown that TIMP2, is also involved in the activation of pro‐MMP2 together with MMP14 (76,77). We found a higher expression of MMP inhibitors TIMP1, TIMP2 and TIMP3 in SEGA as compared to control tissue. The higher expression of TIMPs may act as a compensatory mechanism for higher MMP expression, which could explain why the higher expression of MMP2, MMP11, MMP14, MMP15, and MMP19 does not lead to metastases and excessive growth in SEGA. Interestingly, we found that MMP19 was highly expressed in recurrent/regrown SEGA as compared to nonrecurrent/regrown SEGA using RT-qPCR analysis. Previous studies found a correlation between MMP19 expression and tumor malignance/invasion (72,74,78). Therefore, MMP19 expression could potentially be utilized as a biomarker for recurrence/regrowth. However, our sample size of recurrent/regrown SEGA was small and this effect was not found in the RNA-Seq data, indicating that further investigation is required.
miR-320d Is a Predictive Regulator of MMPs and Is Expressed at Lower Levels in SEGA Compared to Control
Previous research has shown the importance of miRNAs in TSC and SEGA (26,29) and their role in regulating MMP expression (28,79). In this study, we identified 6 miRNAs (miR-320d, miR-320b, miR-320c, miR-625-5p, miR-330-5p, and miR-3200-3p) with lower expression in SEGA as compared to controls that could potentially target MMP expression. Research from Qin et al (49) has shown that downregulation of miR-320d in gliomas predicted lower survival rates and increased growth and proliferation. However, in our study we did not find a correlation between the expression of miR-320d and the size of tumor or tumor recurrence/regrowth. Furthermore, Qin et al (49) indicated miR-320d as a regulator of MMP2, decreasing MMP2 protein expression in glioma cell lines, inducing cell apoptosis and suppressing cell growth and migration. In accordance with this study, we found that miR-320d can target MMP2 in human fetal astrocytes, but did not affect the expression of MMP11, MMP14, MMP15, and MMP19. Since the interaction of miR-320d was based on prediction tools, it could be that these MMPs are not responsive to this miRNA, however, further investigation is needed. Alternatively, it could be of interest to combine miR-320d with the other 5 miRNAs predicted to target MMP2, MMP11, MMP14, MMP15 and MMP19 (miR-320b, miR-625-5p, miR-320c, miR-330-5p, and miR-3200-3p) to increase the regulatory effect on MMP expression.
Taken together, this study provides evidence that both MMPs and TIMPs are highly expressed in SEGA compared to control tissue, indicating that the MMP/TIMP proteolytic system is dysregulated in SEGA as observed in tubers of TSC patients. The MMP-targeting miR-320d is downregulated in SEGA. In human fetal astrocytes, the dysregulation of MMP2 can be normalized by miR-320d. We therefore conclude that targeting MMPs potentially with miR-320d alone or in combination with other MMP targeting miRNAs could be of interest as a therapeutic intervention for TSC patients with SEGA.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all supporters of the TSC brain bank (Laboratory of Molecular and Cellular Neurobiology, International Institute of Molecular and Cell Biology, Warsaw, Poland: J. Jaworski, A. Tempes; The Service d’ Anatomie Pathologique, CHI de Creteil and Inserm U676, Hospital Robert Debre, Paris, France: H. Adle-Biassette; Czech; Department of Neurology and Pathology and Molecular Medicine, Charles University, 2nd Faculty of Medicine, Motol University Hospital, Prague, Czech Republic: P. Krsek, J. Zamecnik; Department of Neuropathology, John Radcliffe Hospital, Oxford, UK: C. Kennard; Department of Anatomic Pathology Sciences, Università Sapienza, Rome, Italy: M. Antonelli, F. Giangaspero; Institute of Neuropathology, Westfälische Wilhelms – Universität Münster, Münster, Germany: W. Paulus, M. Hasselblatt; Department of Neuropathology, University Hospital Erlangen, Erlangen, Germany: R. Coras, I. Blümcke; Bethel Epilepsy Centre, Bielefeld, Germany: T. Polster, C.G. Bien; Laboratory of Neuropathology, Department of Neurology, Hospital de Santa Maria [CHLN], Lisbon, Portugal: J. Pimentel; Department of Pathology, Faculty of Medicine, Hacettepe University, Ankara, Turkey: F. Söylemezoğlu). In this regard we would like to acknowledge all personnel involved in sending us brain tissue. Furthermore, the authors would like to thank Dr Mark Nellist (Department of Clinical Genetics, Erasmus Medical Centre, Rotterdam, The Netherlands) and Dr David J. Kwiatkowski, MD, PhD (Division of Experimental Medicine and Medical Oncology, Brigham and Women’s Hospital, Boston) for performing TSC1/TSC2 mutation analysis. We acknowledge the HIS Mouse Facility of the Amsterdam UMC and the Bloemenhove Clinic (Heemstede, The Netherlands) for providing fetal tissues.
Supplementary Data can be found at academic.oup.com/jnen.
The research leading to these results has received funding from KIKA (Stichting Kinderen Kankervrij; A.B., A.M., A.S., B.S., E.A.); Stichting AMC Foundation (E.A.); Stichting TSC Fonds (E.A.); the Austrian Science Fund (FWF, No. J3499; A.M.); the European Union’s Seventh Framework Programme (FP7/2007–2013) under Grant Agreement No. 602102 (EPITARGET; E.V., E.A.), Grant Agreement No. 602391(EPISTOP; J.S., F.J., T.S., M.F., S.J., A.M., E.A.); the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie Grant Agreement No. 642881 (ECMED; E.A.); the Dutch Epilepsy Foundation, project 16‐05 (D.B., E.V.); the Austrian Epilepsy Society (V.E.G.); the Polish Ministerial funds for science (years 2013–2018) for the implementation of international cofinanced project (K.K., S.J.); and internal research project of the Children’s Memorial Health Institute No. S132/2013 (K.K., S.J.).
The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1. Curatolo P, Bombardieri R, Jozwiak S.. Tuberous sclerosis. Lancet 2008;372:657–68 [DOI] [PubMed] [Google Scholar]
- 2. DiMario FJ., Jr Brain abnormalities in tuberous sclerosis complex. J Child Neurol 2004;19:650–7 [DOI] [PubMed] [Google Scholar]
- 3.European Chromosome 16 Tuberous Sclerosis C. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 1993;75:1305–15 [DOI] [PubMed] [Google Scholar]
- 4. van Slegtenhorst M, de Hoogt R, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 1997;277:805–8 [DOI] [PubMed] [Google Scholar]
- 5. Kwiatkowski DJ. Rhebbing up mTOR: New insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis. Cancer Biol Ther 2003;2:471–6 [DOI] [PubMed] [Google Scholar]
- 6. Chan JA, Zhang H, Roberts PS, et al. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: Biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol 2004;63:1236–42 [DOI] [PubMed] [Google Scholar]
- 7. Inoki K, Li Y, Zhu T, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002;4:648–57 [DOI] [PubMed] [Google Scholar]
- 8. Laplante M, Sabatini DM.. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci 2013;126:1713–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aronica E, Becker AJ, Spreafico R.. Malformations of cortical development. Brain Pathol 2012;22:380–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aronica E, Crino PB.. Epilepsy related to developmental tumors and malformations of cortical development. Neurotherapeutics 2014;11:251–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizuguchi M, Takashima S.. Neuropathology of tuberous sclerosis. Brain Dev 2001;23:508–15 [DOI] [PubMed] [Google Scholar]
- 12. Adriaensen ME, Schaefer-Prokop CM, Stijnen T, et al. Prevalence of subependymal giant cell tumors in patients with tuberous sclerosis and a review of the literature. Eur J Neurol 2009;16:691–6 [DOI] [PubMed] [Google Scholar]
- 13. Kingswood JC, d’Augeres GB, Belousova E, et al. TuberOus SClerosis registry to increase disease Awareness (TOSCA)—Baseline data on 2093 patients. Orphanet J Rare Dis 2017;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kothare SV, Singh K, Chalifoux JR, et al. Severity of manifestations in tuberous sclerosis complex in relation to genotype. Epilepsia 2014;55:1025–9 [DOI] [PubMed] [Google Scholar]
- 15. Goh S, Butler W, Thiele EA.. Subependymal giant cell tumors in tuberous sclerosis complex. Neurology 2004;63:1457–61 [DOI] [PubMed] [Google Scholar]
- 16. Cuccia V, Zuccaro G, Sosa F, et al. Subependymal giant cell astrocytoma in children with tuberous sclerosis. Childs Nerv Syst 2003;19:232–43 [DOI] [PubMed] [Google Scholar]
- 17. Amin S, Carter M, Edwards RJ, et al. The outcome of surgical management of subependymal giant cell astrocytoma in tuberous sclerosis complex. Eur J Paediatr Neurol 2013;17:36–44 [DOI] [PubMed] [Google Scholar]
- 18. Curatolo P, Moavero R, de Vries PJ.. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol 2015;14:733–45 [DOI] [PubMed] [Google Scholar]
- 19. Bonnin JM, Rubinstein LJ, Papasozomenos SC, et al. Subependymal giant cell astrocytoma. Significance and possible cytogenetic implications of an immunohistochemical study. Acta Neuropathol 1984;62:185–93 [DOI] [PubMed] [Google Scholar]
- 20. Buccoliero AM, Franchi A, Castiglione F, et al. Subependymal giant cell astrocytoma (SEGA): Is it an astrocytoma? Morphological, immunohistochemical and ultrastructural study. Neuropathology 2009;29:25–30 [DOI] [PubMed] [Google Scholar]
- 21. Fujiwara S, Takaki T, Hikita T, et al. Subependymal giant-cell astrocytoma associated with tuberous sclerosis. Do subependymal nodules grow? Child's Nerv Syst 1989;5:43–4 [DOI] [PubMed] [Google Scholar]
- 22. Morimoto K, Mogami H.. Sequential CT study of subependymal giant-cell astrocytoma associated with tuberous sclerosis. Case report. J Neurosurg 1986;65:874–7 [DOI] [PubMed] [Google Scholar]
- 23. Bongaarts A, Giannikou K, Reinten RJ, et al. Subependymal giant cell astrocytomas in Tuberous Sclerosis Complex have consistent TSC1/TSC2 biallelic inactivation, and no BRAF mutations. Oncotarget 2017;8:95516–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin KR, Zhou W, Bowman MJ, et al. The genomic landscape of tuberous sclerosis complex. Nat Commun 2017;8:15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tyburczy ME, Kotulska K, Pokarowski P, et al. Novel proteins regulated by mTOR in subependymal giant cell astrocytomas of patients with tuberous sclerosis complex and new therapeutic implications. Am J Pathol 2010;176:1878–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bongaarts A, van Scheppingen J, Korotkov A, et al. The coding and non-coding transcriptional landscape of subependymal giant cell astrocytomas. Brain 2020;143:131–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boer K, Crino PB, Gorter JA, et al. Gene expression analysis of tuberous sclerosis complex cortical tubers reveals increased expression of adhesion and inflammatory factors. Brain Pathol 2009;20:704–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Broekaart DWM, van Scheppingen J, Anink JJ, et al. Increased matrix metalloproteinases expression in tuberous sclerosis complex: Modulation by microRNA 146a and 147b in vitro. Neuropathol Appl Neurobiol 2020;46:142–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mills JD, Iyer AM, van Scheppingen J, et al. Coding and small non-coding transcriptional landscape of tuberous sclerosis complex cortical tubers: Implications for pathophysiology and treatment. Sci Rep 2017;7:8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rivera S, Khrestchatisky M, Kaczmarek L, et al. Metzincin proteases and their inhibitors: Foes or friends in nervous system physiology? J Neurosci 2010;30:15337–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagase H, Visse R, Murphy G.. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res 2006;69:562–73 [DOI] [PubMed] [Google Scholar]
- 32. Sternlicht MD, Werb Z.. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 2001;17:463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi YB, Fu L, Hasebe T, et al. Regulation of extracellular matrix remodeling and cell fate determination by matrix metalloproteinase stromelysin-3 during thyroid hormone-dependent post-embryonic development. Pharmacol Ther 2007;116:391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang L, Zhang ZG, Zhang RL, et al. Matrix metalloproteinase 2 (MMP2) and MMP9 secreted by erythropoietin-activated endothelial cells promote neural progenitor cell migration. J Neurosci 2006;26:5996–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rundhaug JE. Matrix metalloproteinases and angiogenesis. J Cell Mol Med 2005;9:267–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosenberg GA, Yang Y.. Vasogenic edema due to tight junction disruption by matrix metalloproteinases in cerebral ischemia. Neurosurg Focus 2007;22:1–9 [DOI] [PubMed] [Google Scholar]
- 37. Van Lint P, Libert C.. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol 2007;82:1375–81 [DOI] [PubMed] [Google Scholar]
- 38. Nakamura H, Suenaga N, Taniwaki K, et al. Constitutive and induced CD44 shedding by ADAM-like proteases and membrane-type 1 matrix metalloproteinase. Cancer Res 2004;64:876–82 [DOI] [PubMed] [Google Scholar]
- 39. Lee KY, Kim YJ, Yoo H, et al. Human brain endothelial cell-derived COX-2 facilitates extravasation of breast cancer cells across the blood-brain barrier. Anticancer Res 2011;31:4307–13 [PubMed] [Google Scholar]
- 40. Belien AT, Paganetti PA, Schwab ME.. Membrane-type 1 matrix metalloprotease (MT1-MMP) enables invasive migration of glioma cells in central nervous system white matter. J Cell Biol 1999;144:373–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vandenbroucke RE, Libert C.. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov 2014;13:904–27 [DOI] [PubMed] [Google Scholar]
- 42. Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2008;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97 [DOI] [PubMed] [Google Scholar]
- 44. Ha TY. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune Netw 2011;11:135–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cao DD, Li L, Chan WY.. MicroRNAs: Key regulators in the central nervous system and their implication in neurological diseases. Int J Mol Sci 2016;17:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reschke CR, Henshall DC.. MicroRNA and epilepsy. Adv Exp Med Biol 2015;888:41–70 [DOI] [PubMed] [Google Scholar]
- 47. van Scheppingen J, Iyer AM, Prabowo AS, et al. Expression of microRNAs miR21, miR146a, and miR155 in tuberous sclerosis complex cortical tubers and their regulation in human astrocytes and SEGA-derived cell cultures. Glia 2016;64:1066–82 [DOI] [PubMed] [Google Scholar]
- 48. van Scheppingen J, Mills JD, Zimmer TS, et al. miR147b: A novel key regulator of interleukin 1 beta-mediated inflammation in human astrocytes. Glia 2018;66:1082–97 [DOI] [PubMed] [Google Scholar]
- 49. Qin CZ, Lv QL, Yang YT, et al. Downregulation of microRNA-320d predicts poor overall survival and promotes the growth and invasive abilities in glioma. Chem Biol Drug Des 2017;89:806–14 [DOI] [PubMed] [Google Scholar]
- 50. Xia H, Qi Y, Ng SS, et al. MicroRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res 2009;1269:158–65 [DOI] [PubMed] [Google Scholar]
- 51. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol 2016;131:803–20 [DOI] [PubMed] [Google Scholar]
- 52. Northrup H, Krueger DA, Northrup H, et al. International Tuberous Sclerosis Complex Consensus G. Tuberous sclerosis complex diagnostic criteria update: Recommendations of the 2012 International Tuberous Sclerosis Complex Consensus Conference. Pediatr Neurol 2013;49:243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim D, Pertea G, Trapnell C, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liao Y, Smyth GK, Shi W.. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30 [DOI] [PubMed] [Google Scholar]
- 55. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Croft D, O'Kelly G, Wu G, et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res 2011;39:D691–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fabregat A, Jupe S, Matthews L, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2018;46:D649–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dweep H, Gretz N, Sticht C.. miRWalk database for miRNA-target interactions. Methods Mol Biol 2014;1182:289–305 [DOI] [PubMed] [Google Scholar]
- 59. Dweep H, Sticht C, Pandey P, et al. miRWalk—Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 2011;44:839–47 [DOI] [PubMed] [Google Scholar]
- 60. Bongaarts A, Prabowo AS, Arena A, et al. MicroRNA519d and microRNA4758 can identify gangliogliomas from dysembryoplastic neuroepithelial tumours and astrocytomas. Oncotarget 2018;9:28103–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ruijter JM, Ramakers C, Hoogaars WM, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009;37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boer K, Jansen F, Nellist M, et al. Inflammatory processes in cortical tubers and subependymal giant cell tumors of tuberous sclerosis complex. Epilepsy Res 2008;78:7–21 [DOI] [PubMed] [Google Scholar]
- 63. Prabowo AS, Anink JJ, Lammens M, et al. Fetal brain lesions in tuberous sclerosis complex: TORC1 activation and inflammation. Brain Pathol 2013;23:45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang B, Zou J, Rensing NR, et al. Inflammatory mechanisms contribute to the neurological manifestations of tuberous sclerosis complex. Neurobiol Dis 2015;80:70–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Vliet EA, Aronica E, Gorter JA.. Blood-brain barrier dysfunction, seizures and epilepsy. Semin Cell Dev Biol 2015;38:26–34 [DOI] [PubMed] [Google Scholar]
- 66. Van den Steen PE, Proost P, Wuyts A, et al. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood 2000;96:2673–81 [PubMed] [Google Scholar]
- 67. Schonbeck U, Mach F, Libby P.. Generation of biologically active IL-1 beta by matrix metalloproteinases: A novel caspase-1-independent pathway of IL-1 beta processing. J Immunol 1998;161:3340–6 [PubMed] [Google Scholar]
- 68. Mohan MJ, Seaton T, Mitchell J, et al. The tumor necrosis factor-alpha converting enzyme (TACE): A unique metalloproteinase with highly defined substrate selectivity. Biochemistry 2002;41:9462–9 [DOI] [PubMed] [Google Scholar]
- 69. Gidday JM, Gasche YG, Copin JC, et al. Leukocyte-derived matrix metalloproteinase-9 mediates blood-brain barrier breakdown and is proinflammatory after transient focal cerebral ischemia. Am J Physiol Heart Circ Physiol 2005;289:H558–68 [DOI] [PubMed] [Google Scholar]
- 70. Justicia C, Panes J, Sole S, et al. Neutrophil infiltration increases matrix metalloproteinase-9 in the ischemic brain after occlusion/reperfusion of the middle cerebral artery in rats. J Cereb Blood Flow Metab 2003;23:1430–40 [DOI] [PubMed] [Google Scholar]
- 71. Ramachandran RK, Sorensen MD, Aaberg-Jessen C, et al. Expression and prognostic impact of matrix metalloproteinase-2 (MMP-2) in astrocytomas. PLoS One 2017;12:e0172234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang L, Yuan J, Tu Y, et al. Co-expression of MMP-14 and MMP-19 predicts poor survival in human glioma. Clin Transl Oncol 2013;15:139–45 [DOI] [PubMed] [Google Scholar]
- 73. Thorns V, Walter GF, Thorns C.. Expression of MMP-2, MMP-7, MMP-9, MMP-10 and MMP-11 in human astrocytic and oligodendroglial gliomas. Anticancer Res 2003;23:3937–44 [PubMed] [Google Scholar]
- 74. Stojic J, Hagemann C, Haas S, et al. Expression of matrix metalloproteinases MMP-1, MMP-11 and MMP-19 is correlated with the WHO-grading of human malignant gliomas. Neurosci Res 2008;60:40–9 [DOI] [PubMed] [Google Scholar]
- 75. Kachra Z, Beaulieu E, Delbecchi L, et al. Expression of matrix metalloproteinases and their inhibitors in human brain tumors. Clin Exp Metastasis 1999;17:555–66 [DOI] [PubMed] [Google Scholar]
- 76. Atkinson SJ, Crabbe T, Cowell S, et al. Intermolecular autolytic cleavage can contribute to the activation of progelatinase A by cell membranes. J Biol Chem 1995;270:30479–85 [DOI] [PubMed] [Google Scholar]
- 77. Itoh Y, Takamura A, Ito N, et al. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J 2001;20:4782–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lettau I, Hattermann K, Held-Feindt J, et al. Matrix metalloproteinase-19 is highly expressed in astroglial tumors and promotes invasion of glioma cells. J Neuropathol Exp Neurol 2010;69:215–23 [DOI] [PubMed] [Google Scholar]
- 79. Korotkov A, Broekaart DWM, van Scheppingen J, et al. Increased expression of matrix metalloproteinase 3 can be attenuated by inhibition of microRNA-155 in cultured human astrocytes. J Neuroinflamm 2018;15:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.