Abstract
Background & Aims
Esophageal adenocarcinomas (EAC) are heterogeneous and often preceded by Barrett’s esophagus (BE). Many genomic changes have been associated with development of BE and EAC, but little is known about epigenetic alterations. We performed epigenetic analyses of BE and EAC tissues, and combined these data with transcriptome and genomic data, to identify mechanisms that control gene expression and genome integrity.
Methods
In a retrospective cohort study, we collected tissue samples and clinical data from 150 BE and 285 EAC cases from the Oesophageal Cancer Classification and Molecular Stratification consortium in the United Kingdom. We analyzed methylation profiles of all BE and EAC tissues and assigned them to subgroups using non-negative matrix factorization with k-means clustering. Data from whole-genome sequencing and transcriptome studies were then incorporated; we performed integrative methylation and RNA-seq analyses to identify genes that were suppressed with increased methylation in promoter regions. Levels of different immune cell types was computed using single-sample gene set enrichment methods. We derived 8 organoids from 8 EAC tissues and tested their sensitivity to different drugs.
Results
BE and EAC samples shared genome-wide methylation features, compared to that with normal tissues (esophageal, gastric, and duodenum; controls) from the same patients and grouped into 4 subtypes. Subtype 1 was characterized by DNA hypermethylation with a high mutation burden and multiple mutations in genes in cell cycle and receptor tyrosine signaling pathways. Subtype 2 was characterized by a gene expression pattern associated with metabolic processes (ATP synthesis and fatty acid oxidation) and lack methylation at specific binding sites for transcription factors; 83% of samples of this subtype were BE and 17% were EAC. The third subtype did not have changes in methylation pattern, compared with control tissue, but had a gene expression pattern that indicated immune cell infiltration; this tumor type was associated with the shortest time of patient survival. The fourth subtype was characterized by DNA hypomethylation associated with structural rearrangements, copy number alterations, with preferential amplification for CCNE1 (cells with this gene amplification have been reported to be sensitive to CDK2 inhibitors). Organoids with reduced levels of MGMT and CHFR expression were sensitive to temozolomide and taxane drugs.
Conclusions
In a comprehensive integrated analysis of methylation, transcriptome, and genome profiles of more than 400 BE and EAC tissues, along with clinical data, we identified 4 subtypes that were associated with patient outcomes and potential responses to therapy.
Keywords: prognostic factor, anti-tumor immune response, response to treatment, gene repression
Esophageal Cancer is the eighth most common cancer type globally1. Esophageal adenocarcinoma (EAC) is the predominant subtype in the western world, particularly amongst white men2; most patients present at an advanced stage and despite some improvements in therapy overall five-year survival rate is under 15%3. Epidemiologically, long-term esophageal exposure to acid and bile reflux appear to be the major risk factors resulting in aberrant differentiation of the cells lining the lower oesophagus to intestinal metaplasia, otherwise known as Barrett’s esophagus4 (BE).
Recent genomic studies have shown that BE harbours a number of point mutations even in cases that never progress to cancer5; however it has a relatively stable genome in terms of copy number alterations and structural variants6, 7. As BE progresses to EAC there is loss of p53 accompanied by an increasingly unstable genome, although the genetic trigger for disease progression has not been established 5, 8. DNA methylation is one of the key epigenetic mechanisms for regulating gene expression and maintaining genome stability9. In a number of different cancer types it has been shown that hypermethylation at CpG islands, including promoter regions, results in gene silencing of tumour suppressor genes, whereas regions undergoing hypomethylation are associated with increased expression of oncogenes and genome instability10.
In EAC, two studies have demonstrated marked variation in the degree of methylation at CpG islands, denoted CpG island methylator phenotype (CIMP) positive and negative respectively11,12. The Cancer Genome Atlas (TCGA) study has shown that methylation profiles of Esophageal Squamous Cell Carcinoma (ESCC) and EAC are distinct and the methylation profile of EAC resembles that of intestinal cancers such as gastric and colon cancer13. However, the detailed landscape of methylation changes across BE and EAC in relation to other genome-wide mutational processes determined from whole genome sequencing (WGS) data remains to be determined.
Here we present methylation data integrated with genomic and transcriptomic information for a large cohort comprising more than 400 cases. The detailed clinical information has enabled us to examine the prognostic significance of the changes and we have used primary organoid models to test the therapeutic relevance of prevalent epigenetically regulated targets.
Methods
Cohort
In this retrospective cohort study, we assessed 150 BE and 285 EAC cases derived from the Biomarker and ICGC study, for which samples are collected through the UK-wide OCCAMS (Oesophageal Cancer Classification and Molecular Stratification) consortium. The procedures for obtaining the samples, quality control processes, extractions and whole genome sequencing are as previously described6. Strict pathology consensus review was observed for these samples with a 70% cellularity requirement before inclusion.
Methylation Profiling and Data Analysis
Methylation profile for all samples were generated using the EPIC array platform. For all samples DNA from fresh frozen material was used. All raw data were processed using minfi14. Samples with less than 96% capture efficiency were not considered in analysis. We filtered probes if they were not significantly detected from background, and are not in CpG context, have known SNPs in the surrounding locus, align to multiple locations in the genome or if they mapped to X and Y chromosomes. Processed methylation data were further normalized using BETA mixture model BMIQ15 implemented in ChAMP package16. Processed data were then corrected for batch effects using limma17.
To identify methylation-dependent subgroups, we performed Non-negative matrix factorization (NMF)18 on 5,000 most variable probes together with k-means clustering. Through NMF we first estimated optimal ranks/metagenes by executing it in combinations of 2–10 metagenes over 200 runs. This analysis identified four optimal metagenes assessed through the cophenetic index. Scores from all four metagenes were further subjected to k-means clustering for identifying the optimal number of subtypes. Using silhouette width as a measure, four optimal subtypes were identified.
Differential analysis on individual probes was performed using linear models implemented in limma17. We selected as differentially methylated only those probes with an absolute difference in β greater than 0.3 and adjusted p-value is less than 0.01. On the other hand, for identifying regions with differential methylation we used the bumphunter19 function implemented in minfi. bumphunter was executed under the following parameter settings: maxGap=500, B=1000, cutoff=0.2 and minProbes=4.
Whole genome sequencing data analysis
WGS data were aligned using BWA-MEM program. We used Strelka20 for calling somatic mutations, ASCAT21 for calling copy number and Manta22 for calling structural variants under similar settings as previously described6. Our methods were benchmarked against various other available methods and have among the best sensitivity and specificity for variant calling (ICGC benchmarking exercise23).
RNA-seq data analysis
Sequencing data were aligned using STAR aligner24. Using ENSEMBL gene annotation, counts of individual genes for all samples were computed using GenomicAlignments25 package from Bioconductor. Based on the counts, sequencing depth of individual samples and gene annotation, Transcripts Per Kilobase Million (TPM) for individual genes was computed across all samples. TPM were further corrected for batch effects using Combat26.
Differential analysis of each individual subtype over all other subtypes was performed on counts using the edgeR27 package. Pathway analysis was performed on ranked data from differential analysis using Gene Set Enrichment Analysis (GSEA28). For such analyses, we considered pathways annotated from Gene Ontology, Reactome and other databases.
Enrichment for different immune cell types was computed through gene set variant analysis (GSVA29). Markers for immune cell types were retrieved from publication30.
Identifying epigenetically silenced genes
For assessing which genes undergo transcriptional repression under the influence of gaining methylation in promoter regions, we performed integrative methylation and RNA-seq analysis. For this analysis, we considered samples for which both RNA-seq and methylation were available. For each gene, we identified all probes located 1500 bp both up and downstream from the transcription start site (TSS). We selectively removed all CpG sites that were methylated in normal tissues (mean β-value >0.2). Methylation data was then dichotomised using β-value of ≥0.3 as a threshold (as used in TCGA studies13, 31) for positive DNA methylation, and discarded CpG sites methylated in fewer than 10% of samples. For each probe/gene pair, we then applied the following conditions: 1) categorized samples as either methylated (β ≥0.3) or unmethylated (β <0.3); 2) Compare expression in the methylated and unmethylated groups using the Mann-Whitney test; 3) Compute the correlation between methylation beta and expression TPM. We labelled each individual tumour sample as epigenetically silenced for a specific probe/gene pair selected above if for the probes there is a difference in beta (>0.2) between two groups, difference in distribution of expression of (adjusted p-value < 0.05) and negative correlation between methylation and expression (r < -0.1, adjusted p-value < 0.05). Only genes with multiple probes were considered for this analysis and a sample considered as epigenetically silenced if more than thirty percent of probes for the corresponding gene was also labelled as epigenetically silenced.
Transcription Factor Analysis
We used ELMER32 for understanding which transcription factors are regulated upon perturbations from regulatory regions. Briefly, this method is based on initially identifying differentially methylated distal probes and predicting enriched motifs across them. Methylation levels from motif associated probes are then correlated with expression levels of transcription factor and ranked for any significant associations. We performed supervised analysis where each subtype was compared with others. On doing so we did not find significant results for most of the comparisons except for one, that between Subtype 2 and Subtype 3.
Ethics
The study was registered (UKCRNID 8880), approved by the Institutional Ethics Committees (REC 07/H0305/52 and 10/H0305/1), and all subjects gave individual informed consent.
Data availability
Methylation data is accessible from European Genome-phenome Archive under accession numbers EGAD00010001822, EGAD00010001838 and EGAD00010001834.
Results
To capture comprehensive genome wide methylation changes we used the Illumina MethylationEPIC BeadChip (EPIC, Illumina Inc.). EPIC measures methylation over 850,000 CpG sites covering wide range of regulatory regions of genome (https://emea.illumina.com/products/by-type/microarray-kits/infinium-methylation-epic.html). Compared to its older version Illumina HumanMethylation450 BeadChip (450K, Illumina Inc.) over 90% of 450K probes are included in EPIC along with increased coverage over distal regulatory elements33. In total 435 samples comprising 285 EAC and 150 BE cases along with 100 controls were assayed using the EPIC array. We included control samples from neighbouring tissue types - squamous esophagus (n=39) and gastric cardia (n=38), as well as duodenum (n=23) as a comparison for intestinal differentiation, which is a defining feature of BE and also seen in well-differentiated EAC. Both methylation and RNA-seq specific analysis among the three control tissue types showed that each tissue harbours a unique pattern of methylation (Figure S1J) and RNA expression (Figure S1K). The gene ontology of differentially expressed genes shows enrichment of pathways specific to each individual tissue (Figure S1L). As expected, biological processes related to epidermis development and keratin differentiation are specifically enriched in squamous tissue. Similarly, in gastric tissue we observe upregulation of hormone and gastric acid secretion processes whereas lipid associated metabolic processes are enriched in duodenum. Biological processes such as digestion and ion transport are enriched in both gastric and duodenum tissues in keeping with some common functional roles. For 59% of BE cases and 62% of EAC cases, both WGS, and transcriptomic (RNA-seq) data were available to enable an integrated analysis (Figure S1A,B).
The clinical features of the cohort generated from the UK-wide OCCAMS consortium are in keeping with the expected demographics for this disease (Supplementary Table 1 and 2). Most cases are male (85% EAC, 83% BE) with a median age of 67 years. The most common site of EAC cases is at the gastro-esophageal junction and the majority of patients included are stage 2 or 3 (89%), in keeping with our recruitment in the context of patients entering a curative pathway for whom sample collection is most feasible. Among the premalignant BE cases 57% are non-dysplastic and the remaining 43% are dysplastic. Most of these are taken from patients undergoing surveillance and represent their highest progression grade following multiple years of follow-up. We also included 34 cases with BE adjacent to invasive EAC (see Supplementary Table 2 and Fig. S2C-E for details).
Methylation profiles of BE and EAC reveal four subtypes with independent replication
To elucidate differences between BE and EAC in comparison with controls we carried out principal component analysis on the 5,000 most variable probes selected across all samples. It is apparent that, in keeping with their glandular phenotype, BE and EAC closely resemble gastric cardia and duodenum but are highly distinct from normal squamous esophagus (Figure S1C). Heterogeneous BE profiles overlap more strongly with EAC than with benign gastric and duodenal tissues.
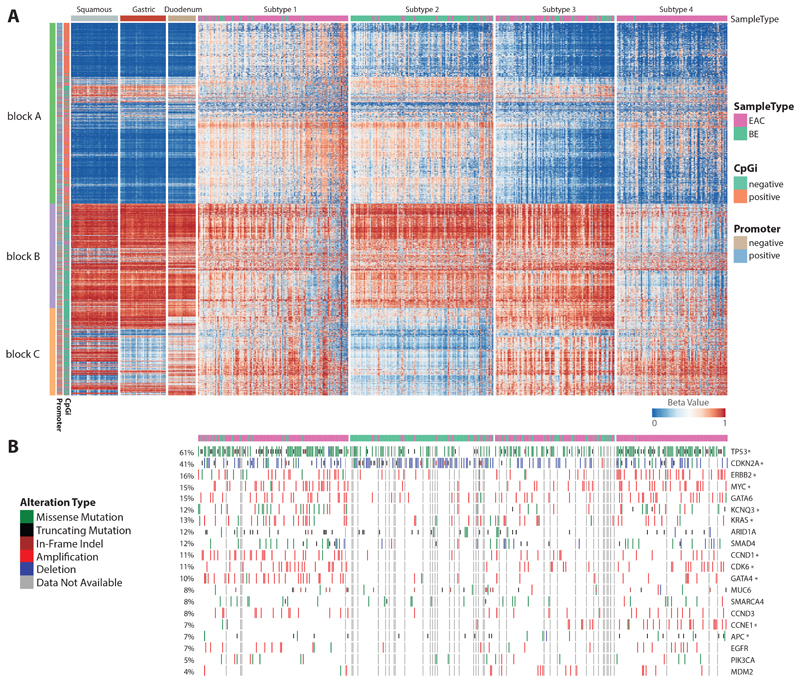
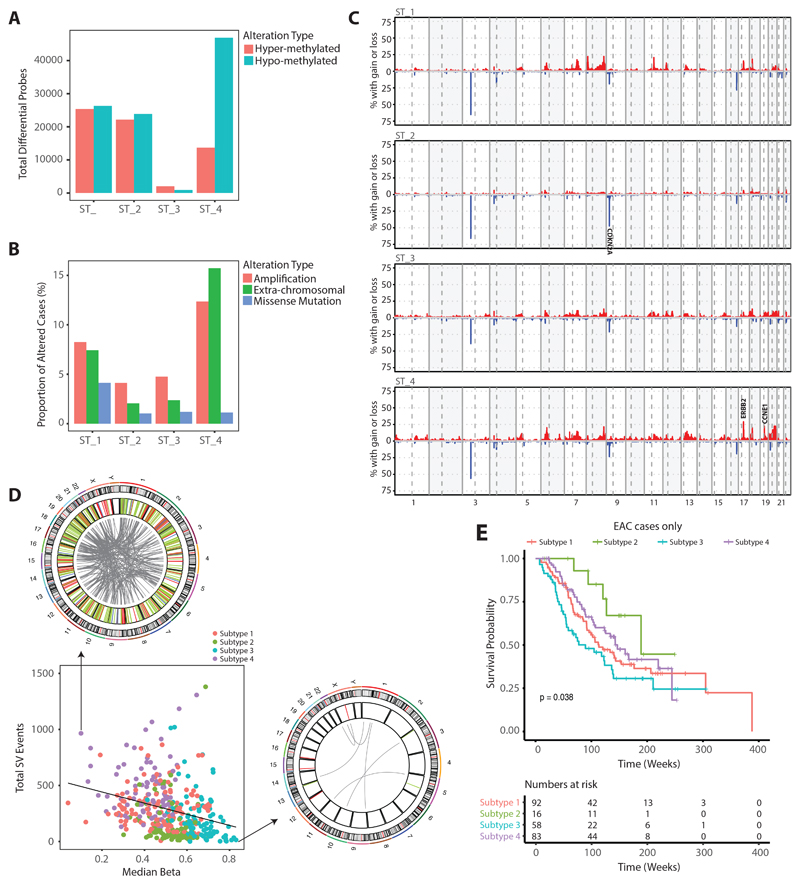
In view of the variability in methylation observed in BE and EAC (Figure S1C) we used Non-Negative Matrix Factorization (NMF) based clustering to identify subtypes. Through this analysis, we were able to identify four optimal metagenes (Figure S1D). Expression measures of these four metagenes were further subjected to k-means clustering, which resulted in four stable subtypes (Figure S1E-F). Figure 1A represents levels of methylation across 5,000 most variables with samples grouped into four identified subtypes. For comparative purposes, levels of methylation from different control samples are also displayed on the left. Interestingly the BE cases are distributed across the four subgroups: 83.2% of the cases in Subtype 2 are BE (n=119; BE=99, EAC=20) with 33.3% (n=99; BE=33, EAC=66) in Subtype 3, 13.6% in Subtype 1 (n=125; BE=17, EAC=108) and a single case in Subtype 4 (n=92; BE=1, EAC=91), (figure 1).
Figure 1. Methylation based BE/EAC subtypes.
a. Heatmap representing methylation levels from top 5000 most variable CpGs across all cases categorised into four subtypes: Subtype 1, 2, 3 and 4 including three different controls (squamous, gastric, duodenum) in extreme left along with annotation of CpG with different colour code. b. Mutation status of driver genes across all cases in same order as displayed in A.
From the heatmap (figure 1A), we can observe that each subtype has a unique methylation pattern. 30.6% of the variable probes are localised within CpGi with the remainder falling in areas designated as shore (2kb outside CpGi boundaries), shelf (2kb outside shore) and open sea. Similarly, in gene centric terms, 42.7% of the most variable probes are localised in promoter regions. For ease of reference we have divided probes into three blocks, A, B and C. In block A, most probes overlap with CpGi (orange) and are located in promoter regions (blue), whereas the majority of probes in block B and C fall within gene bodies and intergenic regions. There is generally a gain in methylation for block A probes in Subtype 1 and 2 when compared to that of controls and the other subgroups. In contrast, probes in block B are relatively hypomethylated in subtype 4 and probes from block C are unmethylated in Subtype 2. For EACs, except for differentiation status we did not find any significant association between subtypes and clinical variables such as tumour location, chemotherapy status, differentiation status (Figure S2A-B). The distribution of BE cases is influenced by the degree of dysplasia, with most of the non-dysplastic BE falling into subtypes 2 and 3 (Figure S2C-E). From here onwards in some figures Subtype 1 is denoted as ST_1, Subtype 2 as ST_2, Subtype 3 as ST_3 and Subtype 4 as ST_4.
To determine whether these subtypes are specific to this cohort or a result of the methodology employed, we examined whether these findings could be replicated in an independent cohort. To do this we examined publicly available methylation data from Australia, comprising 19 BE and 125 EAC cases along with 106 controls (normal esophagus and gastric) profiled using the older 450K array platform11. Remarkably, although the probe overlap between the two platforms was only 55.4% (2,771 of the 5,000 most variable probes), we observed a similar number of metagenes and again four subtypes emerged with very similar methylation profiles to those seen in our cohort (Figure S2F-H).
Methylation profiles in relation to DNA mutation
When integrating the whole genome sequencing data, which were available for the majority of cases (n=391/435), Subtype 1 and 4 are observed to have a significantly higher mutation burden compared to subtypes 2 and Subtype 3 (Figure S1H). The low mutation burden in Subtype 2 is partly explained by the high proportion of premalignant Barrett’s cases but the difference persists in EAC cases 5, 8.
We previously identified 77 genes which, based on their “driver gene” status, are likely to play a critical role in the pathogenesis of EAC7. We mapped the 20 driver genes mutated in at least 4% of EAC cases (Figure 1B, S3). TP53 and CDKN2A are the two most frequently altered genes across the cohort as expected7, wherein TP53 is more preferentially mutated in Subtype 1 (78%) and Subtype 4 (78%) whereas in Subtype 2 and Subtype 3, 37% and 46% are altered. Similarly, CDKN2A is preferentially deleted in Subtype 2, commensurate with the high prevalence of BE (67%, p-value < 0.001). ERBB2 is amplified in both subtype 1 (19%) and subtype 4 (29%). Some genetic events appear to be subtype specific; for example, GATA4 (22%, p-value < 0.001), CCND1 (21%, p-value < 0.001), KCNQ3 (19%, p-value=0.01), MYC (23%, p-value < 0.01), CDK6 (17%, p-value<0.05), and KRAS (18%, p-value < 0.05) are preferentially altered in subtype 1 whereas CCNE1 (21%, p-value < 0.001) and APC (12%, p-value < 0.05) are preferentially altered in subtype 4. Mapping these events to their functional pathways we found that components of the receptor tyrosine kinase (RTK) pathway (GATA4, ERBB2, KRAS) and cell cycle (CCND1, CCNE1, MYC, CDK6) are altered in Subtypes 1 and 4. More specifically, all key drivers of cell cycle aside from CCNE1 are preferentially altered in Subtype 1, whereas components of the Wnt pathway (APC) are dysregulated in Subtype 4. MDM2 is amplified preferentially in subtype 3 (8%, p-value= 0.0643).
Integrated analysis of methylation, genomic and expression features in each subgroup
Subtype 1
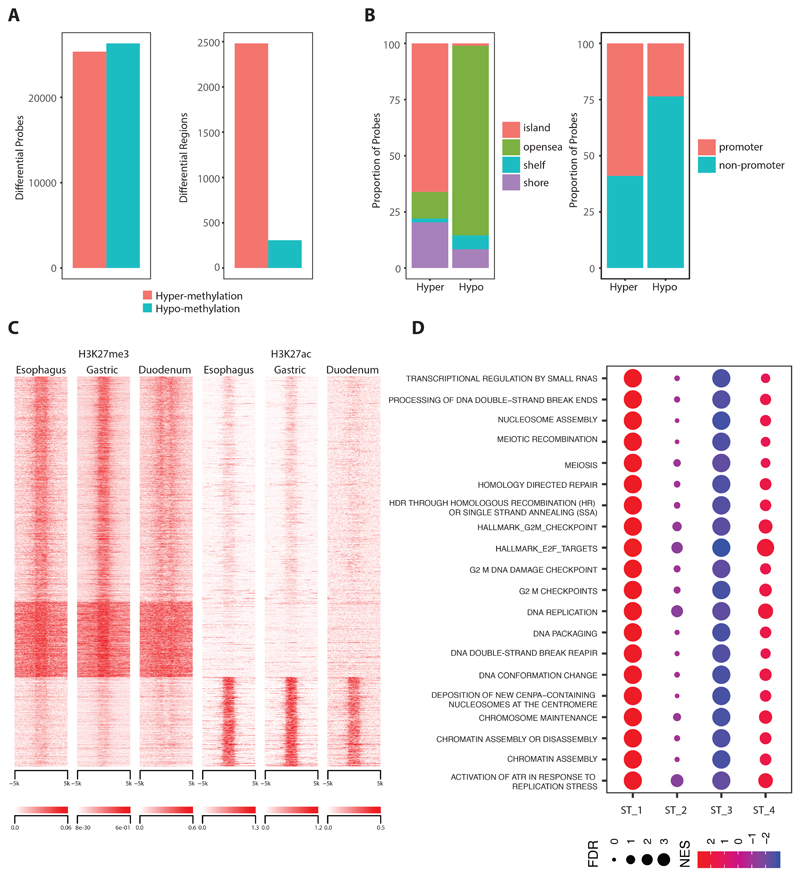
To characterise the highly mutated subtype 1 in more detail we performed a differential analysis in comparison to the controls both at an individual base level and to broad regions for which probes clustered within a distance of 500bp. We found that the proportion of hyper and hypomethylated probes was similar. However, hypomethylation events are spread throughout the genome while hypermethylation is profound in localized regions, mainly promoters rich with CpGi (Figure 2A). Further we observed that 66% of hypermethylated probes and 1% hypomethylated probes overlap with CpG islands and most (59%) occur in promoter regions (Figure 2B), suggesting a CIMP-like phenotype.
Figure 2. Hypermethylation driven Subtype 1.
a. Total number of hyper and hypomethylation events observed in Subtype 1 at individual CpG base level (left) and regions with clustered probes (right). b. Annotation of both of hyper and hypomethylation events with respect to CpG island (left) and gene promoter (right). b. Heatmap quantifying levels of H3K27me3 and H3K27ac in all extend regions undergoing hypermethylation. d. Dot plot with top scored pathways identified in Subtype 1 when compared with other subtypes through gene get enrichment analysis.
Since the state of chromatin can further affect gene regulation we explored markers of closed and open chromatin. To do this we took advantage of histone modification data available from ENCODE34, 35 and the ROADMAP epigenomics consortium36. Using methylation profiles we confirmed tissue specific similarity for normal controls between ENCODE and our dataset (figure S1M-NL). We then compared both repressive Histone 3 methylation at Lysine 27 (H327me3) and activation marks with Histone 3 acetylation at Lysine 27 (H3K27ac) data from squamous, gastric and duodenum tissues available from the ENCODE34, 35 and ROADMAP epigenomics consortium36. This showed that for hypermethylation 77% of regions are marked by H3K27me3 and 23% by H3K27ac (Figure 2C) across all tissues. Hence, the effects of DNA methylation on gene regulation do not appear to be tissue specific.
Transcriptome-based pathway analysis of Subtype 1 in comparison to all other subtypes shows a strong enrichment for pathways related to DNA repair and cell cycle (Figure 2D, supplementary table 6) which is also in line with driver gene alterations (CCND1, CCNE1, MYC, CDK6) described above.
Subtype 2
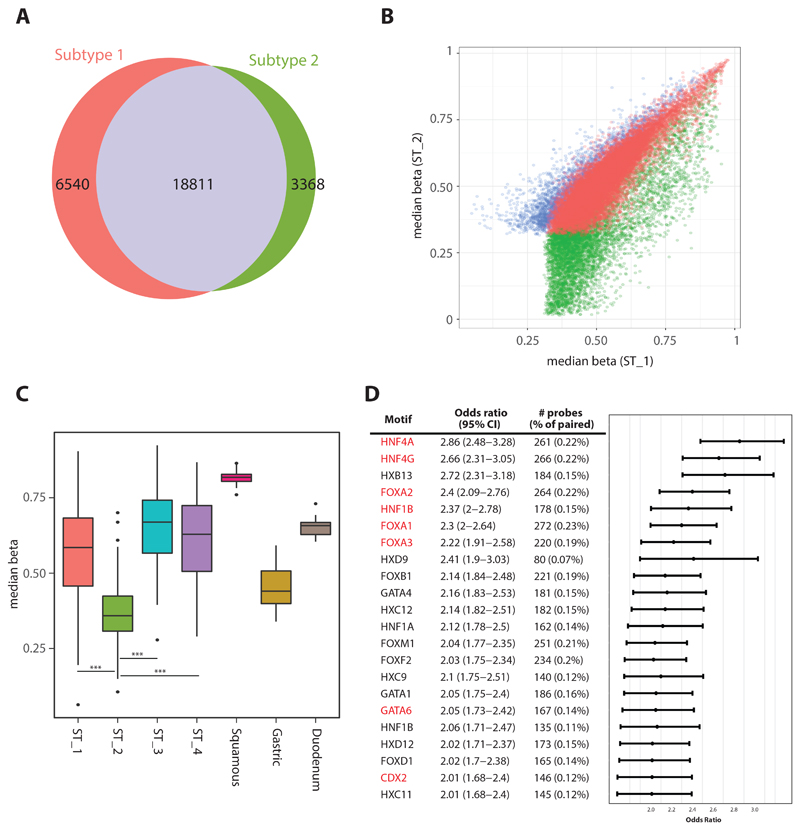
Subtype 2 is dominated by BE cases with hypermethylated CpGi. We were interested to assess whether the hypermethylation changes in this subtype are also seen in EAC, so we compared differentially hypermethylated probes in Subtypes 1 and 2. This showed that the majority (85%) of hypermethylated probes are shared between these subtypes for BE and EAC, suggesting that hypermethylation is an early event (Figure 1A,B).
Even though we observe strong similarities in hypermethylation patterns between BE and EAC there is also a prominent pattern of unmethylated block C probes (Figure 1A) which are highly specific to BE cases in this subgroup. We suspect that these are unique regions that maintain tissue specificity in BE and in keeping with this, the levels are comparable with gastric but not with squamous or duodenum phenotypes (Figure 3C). It has been observed through functional studies that different sets of key master transcription factors such as ELF3, GATA6, KLF5, TP63 through their self-regulatory networks can play an important role in esophageal cancer progression37, 38. To predict the behaviour of different transcription factors we took advantage of distal probes and observed that key transcription factor motifs including HNF4A/G, FOXA1/2/3, GATA6 and CDX2 are significantly over-represented in probes specific to distal regulatory regions in Subtype 2 (Figure 3D). Correlation between the average DNA methylation levels at probes enriched for individual transcription factors and the relevant expression level across all subtypes is shown in Supplementary Figure S4. This demonstrates that the probes critical for regulation of master transcription factors to maintain the BE phenotype are unmethylated in Subtype 2 with a gain in methylation at these sites and reduced expression in EAC. At the RNA level there is selective enrichment of ATP synthesis, fatty acid metabolism and oxidation related processes in this subtype, especially in BE (Figure S5A, supplementary table 7).
Figure 3. BE specific Subtype 2.
a. Venn diagram showing level of common probes undergoing hypermethylation between Subtype 1 and 2. b. Correlation plot comparing median level of methylation from all probes undergoing hypermethylation in Subtype 1/2. c. Boxplot comparing median level of methylation across four different subtypes including controls for all probes from block C from figure 1A (p-value: * (<0.05), ** (<0.01), *** (<0.001)). d. Plot shows Odds Ratio with 95% confidence interval for set of transcription factors motifs enriched in Subtype 2. Key transcription factors required for maintaining Barrett’s phenotypes are highlighted in red.
Subtype 3
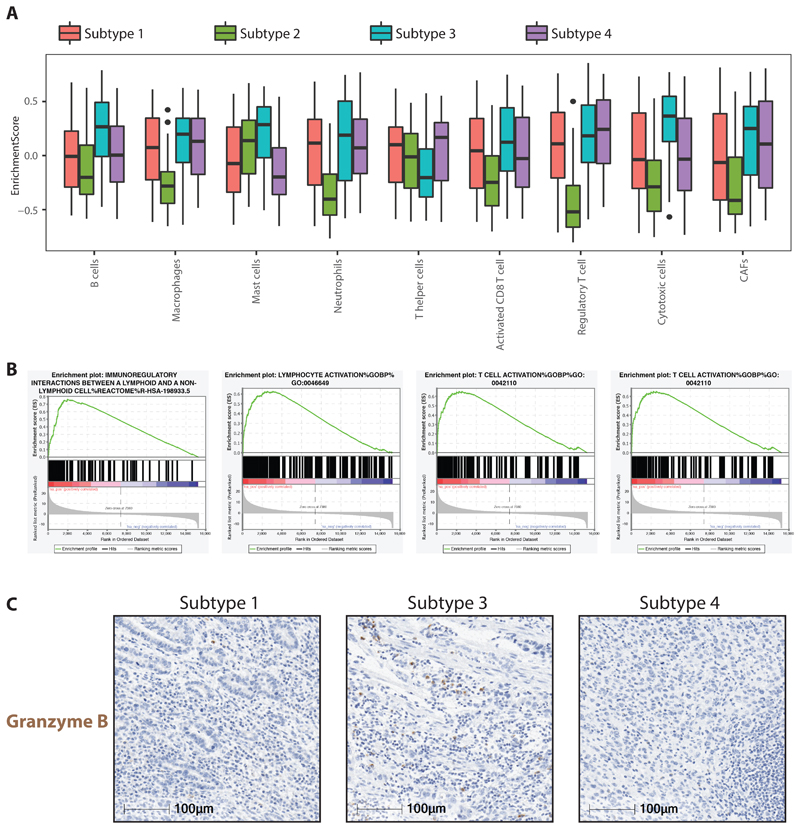
Compared to other subtypes, we did not observe strong changes in methylation in Subtype 3, however from RNA-seq data we observe that subtype 3 has a strong enrichment of both innate and adaptive immune cell types. Particularly we notice strong positive enrichment of cytotoxic cells, B-cells, mast cells and neutrophils along with cancer associated fibroblasts (CAFs) and at the same time we also observe reduced levels of T-helper cells in this subtype (Figure 4A). This contrasts with Subtype 2 which shows no enrichment for immune infiltration (Figure 4A). Consistent with this we observe that all pathways related to immune regulation are strongly enriched (4B, S5B, supplementary table 8). Granzyme B (GZMB), a serine protease protein secreted by cytotoxic and natural killer cells, is well known for its vital role in immune defence mechanisms. Using GZMB as marker of cytotoxic cells we verified their abundance in multiple cases from different subtypes through immuno-histochemical (IHC) staining and confirmed that the relative abundance of GZMB is substantially higher in Subtype 3 as compared to other subtypes (Figure 4C, S5D).
Figure 4. Immune regulated Subtype 3.
a. Boxplot displaying enrichment scores for different immune cell types computed from bulk RNA-seq data across all four different subtypes. b. Gene set enrichment plot for key immune regulated pathways identified on comparing Subtype 3 with all other Subtypes. c. IHC staining for Granzyme B on three different chemo-treated cases representative of Subtype 1,3,4.
The high level of immune infiltration in Subtype 3 also suggests a proportionally lower tumour content (see Figure S1G) as computationally predicted from whole genome sequencing data. To ensure that cellularity is not influencing our subtype classification, we repeated NMF based clustering on samples with computationally predicted cellularity greater than 0.3. On doing so we still retain similar subtypes, suggesting cellularity has no impact on classification.
Subtype 4
Subtype 4 is dominated by hypomethylation events (figure 5A), which in other studies may be an indication of genome instability39. Widespread hypomethylation has been observed in both early and late stages of many cancer types40–44 including BE and EAC45, 46 causing upregulation of certain coding and non-coding regions. In our analysis when compared to other subtypes, Subtype 4 shows a relatively high number of copy number alterations, which are spread throughout the genome (Figure 5B). For example, focal amplifications of CCNE1, ERBB2 and Chr13 and 20 amplifications are common as compared with other subtypes. Subtype 4 also has more extrachromosomal-like events affecting ERBB2 characterized by more than 10 copies of the gene, whereas in Subtype 1 most events are low level amplifications (Figure 5C). This is consistent with our previous finding that these extrachromosomal-like events are strongly associated with chromosomal rearrangements7. When quantifying the total number of structural variants (SVs), Subtype 4 was found to have significantly more SVs as compared to other subtypes (Suppl. Fig 1I). On a case by case basis, patients in Group 4 with low levels of methylation harbour a high level of SVs (figure 5D), in keeping with the idea that methylation levels may be important for maintaining genome stability.
Figure 5. Hypomethylation driven Subtype 4.
a. Total number of hyper and hypomethylation events observed across all four subtypes. b. Genome wide copy number alteration profile for all cases within individual subtype. c. Proportion of cases harbouring different forms of ERBB2 alternation across all four subtypes. d. Correlation between structural variants and median measure of methylation from probes undergoing hypomethylation from Subtype 4 across all samples from different subtypes. Circos plot (on top and right) representing genome wide structural variants (deletions in red, duplication in light green, inversion in blue and translocations in grey) from individual case undergoing high and low levels of hypomethylation. e. Kaplan-Meier curves for EACs from four different subtypes.
When considering the prognosis of EAC cases according to their methylation profiles (BE cases were removed for this analysis) there are differences in overall survival rates between the subgroups (Figure 5E). The small number of EAC cases in subtype 2 which cluster with the BE cases had the best survival. Surprisingly Subtype 3, which has an immune activation phenotype, a lower mutation burden and fewer oncogenic drivers, has poor survival compared to patients in other subtypes.
Epigenetically silenced genes and relevance to therapy
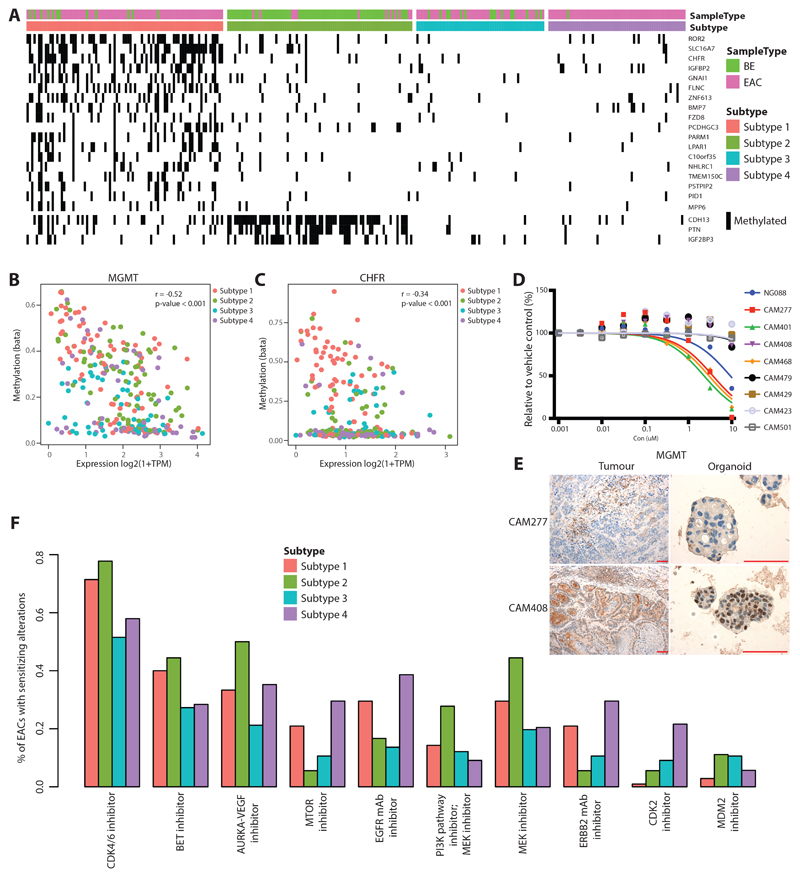
To understand which genes undergo transcriptional repression in association with methylation change, we performed an integrative methylation and transcriptomic analysis. Of the 237 genes with significantly lower expression in relation to increased methylation (Supplementary Table 3), few genes seem to be affected globally across all subtypes, with most silenced genes being more specific to Subtype 1 and 2 (Figure 6A).
Figure 6. Epigenetically Silenced Genes and Clinical Relevance.
a. List of genes which are preferentially silenced either in Subtype 1 or Subtype 2. b. Correlation between methylation and expression for MGMT gene across all cases from four different subtypes. c. Same as B for CHFR gene. d. Growth inhibition responses of eight primary tumour derived organoids and control gastric organoid (NG088) Drug response to Temozolomide. e. MGMT staining in low (CAM277) and stable (CAM408) expressing tumours and derived organoids (scale bar=100uM). f. Drug classes for which sensitivity is indicated by EAC driver genes with data from the Cancer Biomarkers database.
Gene ontology and pathway analysis of silenced genes showed enrichment for biological processes related to transcription and its regulation, along with pathways related to cell cycle (CCND2, RDX, UBE2E2), kinase signalling, stem cell pluripotency, nucleosome assembly, cell adhesion, wnt/β-catenin signalling pathway which has been shown to play a role in the neoplastic transformation of BE47 (Figure S6A-B, Supplementary Table 4-5). We also observe that a few immune regulators (BLNK, CD40, VAV3, IRS2) are also affected by methylation.
Previously we tested different sets of drugs in both EAC cell lines and primary derived organoids and have shown that their response correlates with the specific driver gene alterations7, 48. In view of this, we were interested to identify methylation based drivers and predict their response to known drugs. Previous work has shown that the MGMT gene, a key regulator in DNA repair, is methylated in nearly 50% of glioblastoma cases and these patients benefited from temozolomide chemotherapy more than patients with an unmethylated MGMT promoter49. In our cohort MGMT is strongly regulated by a gain of methylation in promoter regions, affecting 32% cases (Figure 6B, S6C). To examine responses to temozolomide in EAC we took advantage of organoids generated from primary tumours from this cohort48. High sensitivity to Temozolomide was observed in organoids showing low expression of MGMT at both RNA and protein level such as CAM277, in contrast, organoids with stable MGMT expression showing resistance, for example in CAM408 (Figure 6D,E and S6E).
Similarly, CHFR, a cell cycle check point inhibitor, is methylated in many cancer types; in squamous cell carcinoma CHFR methylation sensitizes to taxane chemotherapy50. In our cohort, we observe CHFR to be altered in 18% of cases most of which are preferentially affected in Subtype 1 (Figure 6C, S6D) and in organoid models CHFR expression levels correlate with a differential response to docetaxel (Figure S6D).
In our earlier driver gene analysis, we have shown that more than 50% of EAC (n=551) are predicted to benefit from CDK4/6 inhibitors along with EZH2 and BET inhibitors in a smaller proportion of cases7. In view of this observation we were interested to determine whether the response rate to different inhibitors is also dependent on their methylation profiles. We observe CDK4/6 inhibitors to be effective in EAC, across all subtypes. In contrast, we also observe CDK2 (p-value < 0.001) inhibitors to be more effective in Subtype 4 (Figure 6F). This selective response is due to preferential amplification of CCNE1 in Subtype 4.
Discussion
NMF based clustering demonstrated that both BE/EAC can be broadly classified into four subtypes each with a unique pattern of methylation, mutation (Figure 1) and expression (Figure 2D, S5A-C). Furthermore, these subtypes were shown to be reproducible in an independent cohort from Australia11, even though the data had been generated on a different array platform.
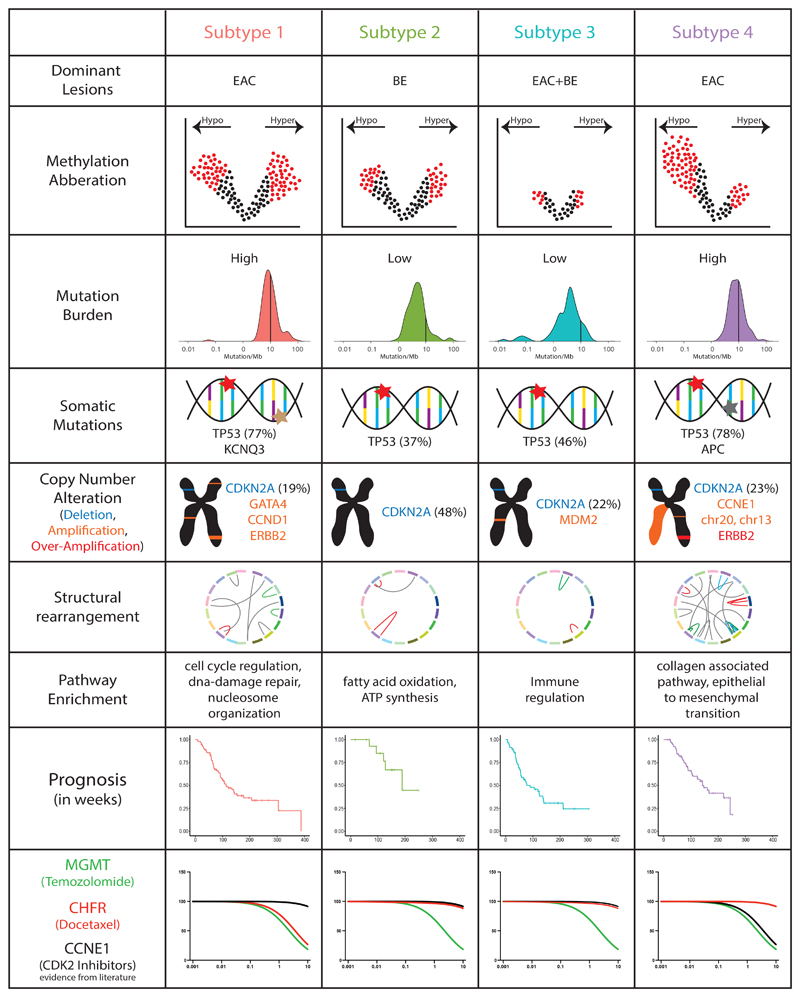
Subtype 1 is dominated by EAC and some BE cases that show a gain in methylation in CpG islands which is representative of a CIMP-like phenotype, with preferential amplification for GATA4, CCND1 and signs of DNA repair. Subtype 2, with a preponderance of BE cases, also shows a gain in CPGi methylation like that of Subtype 1 but with a unique pattern of unmethylation. The transcriptomic profile of this subtype is uniquely enriched for ATP synthesis, fatty acid metabolism and oxidation processes. Methylation levels in Subtype 3 are unremarkable, but show a high-level presence of both myeloid and lymphoid cell lineages. Subtype 4 is characterised by hypomethylation and EAC cases harbouring a high degree of genome stability supported by a high number of copy number alterations and structural variants. Comprehensive molecular and biological features unique to each subtype identified through our analysis are presented in Figure 7.
Figure 7. Overview of different biological features unique to individual subtypes.
We note that although most BE cases cluster together, they are somewhat distributed amongst Subtypes 1 and 3 with the more stable genomes. Out of 108 cases in Subtype 1, 17 cases are BE and detailed inspection revealed that 15/17 cases were dysplastic with high grade dysplasia or intramucosal carcinoma (Figure S2C). On the other hand, some EAC cases (n=20) cluster with the BE Subtype 2. Most of these tumours (11/20) have adjacent Barrett’s oesophagus and are moderately differentiated (Figure S2A), in keeping with better prognosis. This is in keeping with our previous observation that EAC with adjacent BE have a better prognosis51. In future, we would like to compare and study metabolic changes underlying such behaviour.
In terms of prognosis, patients from Subtype 3 with infiltration of immune related cells tend to show a poor prognosis compared to patients in other subtypes. The tumour microenvironment is a complex network of interactions between tumour cells, immune cells and stromal cells. Depending on their composition different immune infiltrates are associated with good or poor prognosis. In general, tumour infiltrating lymphocytes comprising cytotoxic CD8 T-cells, memory T-cells and T-helper cells are associated with a good prognosis, as is evident in many cancer types, such as breast52, ovary53, lung54 whereas regulatory T cells, stromal cells and immune cells of myeloid lineages (such as macrophages, neutrophils, mast cells and others) are indicators of bad prognosis and can promote tumour progression55, 56. In Subtype 3, along with cytotoxic cells we also notice a strong presence of macrophages, neutrophils and CAFs, which could perhaps explain the poor prognosis of cases in this subtype. It is also worth noting that Subtype 3 has a high prevalence of MDM2 amplification (8%), which is associated with resistance to and hyper-progression on immunotherapy57.
In a recent study in EAC has shown that topoisomerase I inhibitors are effective in tumours with high levels of methylation12. Irinotecan is a topoisomerase I inhibitor chemotherapy which is currently used in EAC, however irinotecan treatment has a low monotherapy response rate (~7%). This low response rate could potentially be enhanced if therapy is targeted to methylated tumours. As the TCGA demonstrates EAC to be very similar to CIN gastric cancer, we propose that Subtype 1 representative of CIMP could possibly be sensitive to DNA methyltransferase and topoisomerase I inhibitors.
Through our integrated data analysis approach, we have shown how different genes from critical pathways are altered in EAC/BE. We also provide in vitro evidence from organoid models showing how key regulators of DNA repair (MGMT) and cell cycle (CHFR) can be targeted for effective treatment. In an extension of our previous work7, here we have shown other potential inhibitors like CDK2 could be preferentially effective towards subtype 4 cases. Taking all this information together, these results provide wider scope for better stratification and assignment of relevant targeted therapeutics.
It is also worth noting that all observations made in this study are derived from only the CpG sites present on the EPIC array. This is a narrow representation of the whole genome, and may be a limiting factor, as we cannot draw conclusions or understand changes in other parts of the genome and their influence in tumorigenesis. In future, it would be worth studying methylation on a genome-wide scale, perhaps though whole-genome bisulfite sequencing approaches.
In summary, this study elucidates diversity in the methylation landscape across BE and EAC and its influence on gene expression and genome integrity, suggesting a role for DNA methylation alteration in EAC carcinogenesis.
Supplementary Material
Acknowledgement
OCCAMS was funded by a Programme Grant from Cancer Research UK (RG81771/84119), and the laboratory of R.C.F. is funded by a Core Programme Grant from the Medical Research Council (RG84369). We thank the Human Research Tissue Bank, which is supported by the UK National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, from Addenbrooke’s Hospital. Additional infrastructure support was provided from the Cancer Research UK–funded Experimental Cancer Medicine Centre.
We acknowledge Dr. Andrew Beggs and Dr. Celina Whalley from the Institute of Cancer and Genomic Sciences, University of Birmingham, who provided services for profiling methylation on all our samples.
Abbreviations
- BE
Barrett’s Esophagus
- CAF
Cancer Associated Fibroblast
- CpGi
CpG island
- CIMP
CpG Island Methylator Phenotype
- EAC
Esophageal Adenocarcinoma: EAC
- GZMB
Granzyme B
- IHC
Immunohistochemistry
- NMF
Non-Negative Matrix Factorization
Footnotes
Disclosure
The authors declare no competing interests. Simon Tavaré is a consultant for Kallyope Inc. and is a member of the SAB of Ipsen. These are not directly involved in the topic of this paper.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 3.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2006;55:1810–20. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross-Innes CS, Becq J, Warren A, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131–41. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankell AM, Jammula S, Li X, et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat Genet. 2019;51:506–516. doi: 10.1038/s41588-018-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stachler MD, Taylor-Weiner A, Peng S, et al. Paired exome analysis of Barrett's esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–55. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–79. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Krause L, Nones K, Loffler KA, et al. Identification of the CIMP-like subtype and aberrant methylation of members of the chromosomal segregation and spindle assembly pathways in esophageal adenocarcinoma. Carcinogenesis. 2016;37:356–65. doi: 10.1093/carcin/bgw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu M, Maden SK, Stachler M, et al. Subtypes of Barrett's oesophagus and oesophageal adenocarcinoma based on genome-wide methylation analysis. Gut. 2018 doi: 10.1136/gutjnl-2017-314544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortin JP, Triche TJ, Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33:3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunet JP, Tamayo P, Golub TR, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–9. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 21.Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 23.Lee AY, Ewing AD, Ellrott K, et al. Combining accurate tumor genome simulation with crowdsourcing to benchmark somatic structural variant detection. Genome Biol. 2018;19:188. doi: 10.1186/s13059-018-1539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence M, Huber W, Pages H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamborero D, Rubio-Perez C, Muinos F, et al. A Pan-cancer Landscape of Interactions between Solid Tumors and Infiltrating Immune Cell Populations. Clin Cancer Res. 2018;24:3717–3728. doi: 10.1158/1078-0432.CCR-17-3509. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S, Cherniack AD, Dewal N, et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell. 2016;29:723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva TC, Coetzee SG, Gull N, et al. ELMER v.2: an R/Bioconductor package to reconstruct gene regulatory networks from DNA methylation and transcriptome profiles. Bioinformatics. 2019;35:1974–1977. doi: 10.1093/bioinformatics/bty902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis CA, Hitz BC, Sloan CA, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roadmap Epigenomics C. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Huang M, Plummer J, et al. Master transcription factors form interconnected circuitry and orchestrate transcriptional networks in oesophageal adenocarcinoma. Gut. 2019 doi: 10.1136/gutjnl-2019-318325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie JJ, Jiang YY, Jiang Y, et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology. 2018;154:2137–2151 e1. doi: 10.1053/j.gastro.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Sheaffer KL, Elliott EN, Kaestner KH. DNA Hypomethylation Contributes to Genomic Instability and Intestinal Cancer Initiation. Cancer Prev Res (Phila) 2016;9:534–46. doi: 10.1158/1940-6207.CAPR-15-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng P, Schmutte C, Cofer KF, et al. Alterations in DNA methylation are early, but not initial, events in ovarian tumorigenesis. Br J Cancer. 1997;75:396–402. doi: 10.1038/bjc.1997.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–6. [PubMed] [Google Scholar]
- 42.Kim YI, Giuliano A, Hatch KD, et al. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–9. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 43.Feinberg AP, Gehrke CW, Kuo KC, et al. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–61. [PubMed] [Google Scholar]
- 44.Ehrlich M, Jiang G, Fiala E, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez H, Opalinska J, Zhou L, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966 e4. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Cheng Y, Abraham JM, et al. Modeling Wnt signaling by CRISPR-Cas9 genome editing recapitulates neoplasia in human Barrett epithelial organoids. Cancer Lett. 2018;436:109–118. doi: 10.1016/j.canlet.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Francies HE, Secrier M, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05190-9. 2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 50.Yun T, Liu Y, Gao D, et al. Methylation of CHFR sensitizes esophageal squamous cell cancer to docetaxel and paclitaxel. Genes Cancer. 2015;6:38–48. doi: 10.18632/genesandcancer.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawas T, Killcoyne S, Iyer PG, et al. Identification of Prognostic Phenotypes of Esophageal Adenocarcinoma in 2 Independent Cohorts. Gastroenterology. 2018;155:1720–1728 e4. doi: 10.1053/j.gastro.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeschke J, Bizet M, Desmedt C, et al. DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J Clin Invest. 2017;127:3090–3102. doi: 10.1172/JCI91095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leffers N, Gooden MJ, de Jong RA, et al. Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer Immunol Immunother. 2009;58:449–59. doi: 10.1007/s00262-008-0583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruffini E, Asioli S, Filosso PL, et al. Clinical significance of tumor-infiltrating lymphocytes in lung neoplasms. Ann Thorac Surg. 2009;87:365–71. doi: 10.1016/j.athoracsur.2008.10.067. discussion 371-2. [DOI] [PubMed] [Google Scholar]
- 55.Fridman WH, Pages F, Sautes-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 56.Barnes TA, Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117:451–460. doi: 10.1038/bjc.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res. 2017;23:4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Methylation data is accessible from European Genome-phenome Archive under accession numbers EGAD00010001822, EGAD00010001838 and EGAD00010001834.