Abstract
Recent applications of mass spectrometry (MS) to study membrane protein complexes are yielding valuable insights into the binding of lipids and their structural and functional roles. To date, most native MS experiments with membrane proteins are based on detergent solubilization. Many insights into the structure and function of membrane proteins have been obtained using detergents, however, these can promote local lipid rearrangement, and can cause fluctuations in the oligomeric state of protein complexes. To overcome these problems, we developed a method that does not use detergents or other chemicals. Here we report a detailed protocol that enables direct ejection of protein complexes from membranes for analysis by native MS. Briefly, lipid vesicles are prepared directly from membranes of different sources and subjected to sonication pulses. The resulting destabilized vesicles are concentrated, introduced into a mass spectrometer and ionized. The mass of the observed protein complexes is determined and this information, in conjunction with ‘omics’-based strategies, is used to determine subunit stoichiometry as well as co-factor and lipid binding. Within this protocol we expand the applications of the method to include peripheral-membrane proteins of the S-layer and amyloid protein export machineries overexpressed in membranes from which the most abundant components have been removed. The described experimental procedure takes approximately 3 days from preparation to MS. The time required for data analysis depends upon the complexity of the protein assemblies embedded in the membrane under investigation.
Introduction
The membrane environment is known to have profound effects in maintaining the delicate interactions and the different conformations of the many proteins embedded within it1–3. However, for most biological studies, it has so far been a necessary to remove the membrane in order to isolate protein complex of interest. While detergent-dependent approaches have been used for decades yielding invaluable information4, the structural information derived when these chemicals are employed is not always entirely accurate – a problem that can be attributed to the difference between the physical properties of the micelles that they form and the physical properties of natural membranes5,6. As such, several methods have been designed either to preserve the membrane environment local to the protein, with the use of styrene maleic acid polymers7, or to emulate it by means of reconstituting the lipid environment within nanodiscs8. While both approaches are extremely powerful, and compatible with electron microscopy9,10 and native mass spectrometry (nMS)7,8, they require the use of acidic reagents or detergents and are not able to mimic the membrane environment as a whole. To circumvent these chemical intervention steps we reported previously an approach that enables ejection of complexes directly from membranes and showed that many protein interactions are maintained for characterization using nMS1.
Here we establish a step-by-step protocol for this process known as Sonication of Lipid Vesicles for Mass Spectrometry (SoLVe-MS). In the first step we examine the morphology of membranes, before and after sonication, and then explore the experimental parameter space for optimal MS. We then apply the method for over expression of outer membrane proteins in Escherichia coli and compare complexes extracted by means of detergent micelles with those ejected directly from membranes. Finally, we consider the applicability of SoLVe-MS to additional biological systems by ejecting complexes directly from bacterial membranes overexpressing recombinant proteins, or stalks layered with membrane associated proteins, without prior chemical disruption of the supporting membrane.
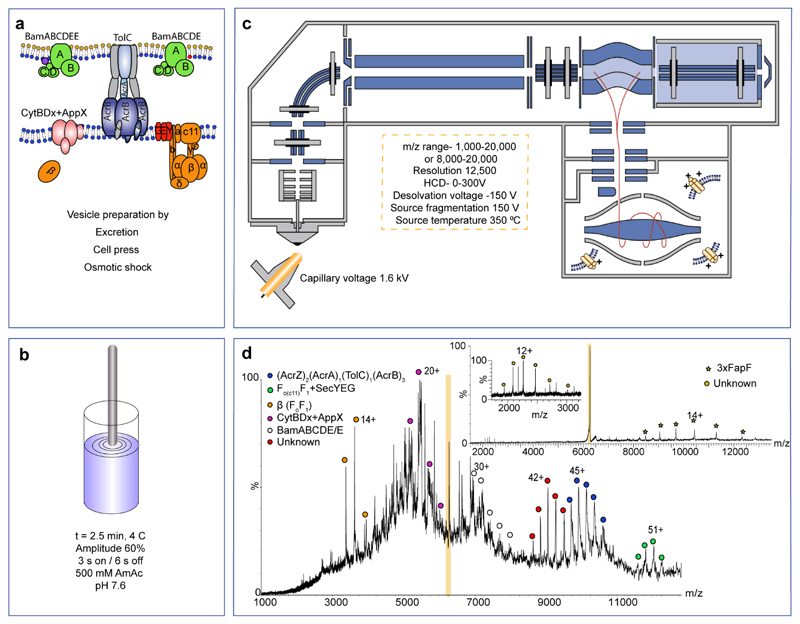
The first step in our method is the formation of vesicles following standard protocols (see procedure-step 1, Fig. 1a). The next step involves recording mass spectra from these vesicles. We first tried injecting the vesicles under standard native MS conditions that use ammonium acetate at neutral pH, but we were unable to deduce any masses from the resulting mass spectra. In order to increase the potential to liberate membrane protein complexes from the vesicles, we also trialed a variety of other MS and solution conditions from which to acquire mass spectra, including exchange into aqueous buffers, addition of low proportions of organic solvents to partially solubilize complexes, solubilization using mild detergents to disrupt the membrane, as well as a high-energy nMS11 to dissociate the lipidic environment from the protein complex. We also employed a desorption electrospray ionization set-up reasoning that this might yield protein complexes from surfaces12 but we were unable to record meaningful mass spectra using any of these approaches. We therefore investigated a physically disruptive process prior to MS, applying short sonication pulses in aqueous buffer (500 mM ammonium acetate pH 7.6). We reasoned that it would be necessary to avoid overheating and therefore maintained the amplitude at 60% (for the specified sonicator) and applied 3 s pulses, separated by 6 s pauses and incubated the vesicle suspension on ice to reduce heating. We optimized the total time for this pulsing, including pauses, and found it to be 2.5 mins (Fig. 1b). For other sonicators, every cycle should dissipate 20 J and 13 W of energy.
Figure 1. Key steps in the process of Sonicated Lipid Vesicle Mass-Spectrometry (SoLVe-MS) performed on E. coli inner and outer membranes.
Vesicles are generated from membranes by standard protocols and collected by ultracentrifugation. (a) Typically 2 mg of total protein in vesicles is then diluted into 20 ml of ammonium acetate and sonicated as described (a and b). Membrane vesicles are then injected into an Q-Exactive UHMR using parameters desgined to record and assign high quality mass spectra (c-d). The parameters listed in panel C represent the starting parameters used to yield an initial spectrum. These parameters can be adapted according to the vesicles under investigations. High energy helps to remove adducts allowing for an initial spectrum, and to abolish many soluble contaminants that may exist. (d) Tandem MS is performed where possible to further validate the identity of the protein complex as well as to increase the resolution, and to infer subunit identity and location; peripheral subunits are expelled first37. SoLVe MS of E. coli mixed membranes expressing FapF reveal multiple complexes, many observed previously, and additional charge states (isolated peak highlighted orange). Following activation the isolated peak yields the FapF trimer bound to an unkonwn subunit with a mass of 27,154±5 Da. The complexes assigned in the spectrum (d) are shown schemtically in (a) with dissociated subunits (grey). Mass spectrometry experiment was performed three times. Spectra were acquired at dessolvation voltage of -200 V, and source fragmentation of 200 V.
Using these pulses, we partially disrupt vesicles (Figs. 1b and 3) as evidenced by electron microscopy (EM) imaging of vesicles before and after pulsing. Reproducible differences in many of the vesicles were difficult to detect. For E. coli elongated inner membrane tubes were formed, however, the tips appeared to be partially removed (Figs. 1b and 3), suggesting that membrane vesicles might open, thereby allowing ingress of MS compatible buffer into the vesicle lumen.
Figure 3. The experimental setup required for SoLVe-MS.
(i) Depending on the organism and complexity of the proteins embedded within the sample, as low as 50 μg of protein or as high as 10 mg of proteins embedded in membranes are required per experiment. The mechanism of sonication is unclear, however, before sonication bacterial inner membrane tubes are intact. Post-sonication the tips are removed suggesting sonication allows ingress of MS compatible buffer. Cryo-EM inset scale bar is 0.5 μm. (ii) Sonication is performed with the conditions stated. After sonication, and concentration of the vesicles, this preparation can be injected into an MS instrument. SoLVe-MS has been successfully attempted on a Thermo Q-Exactive UHMR and a Waters Synapt HDMS. Exact conditions are specified in the protocol. The overall suggestion is however to use high energy conditions. Cryo-EM inset scale bar is 0.5 μm. (iii). The entire process should take 2-3 days.
Subjecting these sonicated vesicles to nMS yielded mass spectra of sufficient quality from either a commercially available Synapt HDMS modified for transmission of high mass species, set to high voltages of 200 V cone voltage and 200 V trap voltage as starting conditions, or similarly a Q-Exactive UHMR instrument13 (Parameters are described in Fig. 1c, Equipment Setup, and below). We found that it was necessary to apply higher energy conditions than would normally be employed for nMS of membrane protein complexes14. The capillary voltage was set to 1.6 kV, the capillary inlet heated to 350 °C and applied desolvation (-150 V) and source fragmentation (150 V) as the initial set-up parameters for the SoLVe experiments (Fig. 1c). If complexes are not released, these conditions can then be supplemented with additional voltage, where available, applied to the HCD cell (300 V) throughout the instrument (see Limitations of the Protocol). High energy conditions are required to get proteins ejected from the membrane. These conditions constitute of source desolvation (-150 V), source fragmentation (150 V), variable (up to 300 V) from these 2 parameters as well as the HCD energy (up to additional 300 V). The overall voltage is however optimized according to the properties of the proteins and their complexes and cannot be prescribed in advance.
Limitations of the protocol
It is important to note this protocol was initially implemented on a prototype Q-Exactive UHMR mass spectrometer which allowed voltages in excess of 300 V to be applied in the source region1. The current commercial Q-Exactive UHMR permits a maximum of 300V to be applied in source and additionally up to 300V in the HCD cell. With the transition to the commercial instrument, including software updates and vacuum pump upgrades, applying total combined voltages of 300-400 V using both source and HCD cell, and up to the maximum of 600 V are now available. In our experience this voltage is enough to allow for the analysis of most, if not all, membrane samples. The SoLVe-MS method can also be performed using Q-TOF and ion-mobility mass spectrometers such as the commercially available Waters Synapt HDMS G1 instrument.
It should be noted that although these native membrane vesicles imitate the natural environment of the proteins as closely as possible, it is unlikely that they fully recapitulate all the properties of the native membrane. However, since membrane shape is maintained post sonication1 major perturbation seems unlikely. It is unclear whether lipids might scramble due to sonication, or exchange between membrane leaflets, since standard maintenance by lipid flipases will not occur during vesicle preparation. Moreover, the full lateral forces that exist within intact whole membranes will not be maintained in vesicles due to the change in shape and size of the preparation that is required for the SoLVe-MS approach.
Other limitations are attributed to the amount of protein or membranes required. Depending on the membrane’s level of complexity, whether the origin of the membrane is prokaryotic or eukaryotic, large amounts of material may be required. This may present a hurdle as some membrane vesicles are difficult to purify or produce at high amounts. Synaptic vesicles, membrane protein enriched extracellular vesicles (MPEEVs) and overexpression of proteins in cultured cells may not produce enough protein for analysis. Some membranes such as eukaryotic plasma membranes may be too complicated for analysis, or contain no specifically enriched protein, and might require an additional step. Moreover, as the sample becomes more complex and composed of more proteins, this will invoke the requirement for large amounts of biological material. The anticipated amounts of membranes and proteins are detailed in Fig 2 and 3i. For MPEEVs, two to three T175 flasks seeded at 80% confluency are needed, each flask produces about a 100 μg of protein encapsulated in excreted vesicles. Extended washes are required to remove the vast amount of bovine serum albumin (BSA) existing within the growth medium. For bacteria, a small 5 ml pellet may be enough per 2-3 sonication experiments, each will supply enough processed sample for many MS analyses. We have found that for a single membrane spanning protein a few hundred micrograms are needed, while a membrane containing a large array of enriched protein complexes, whether of natural abundance or overexpressed between 2-10 mg of membranes are typically required.
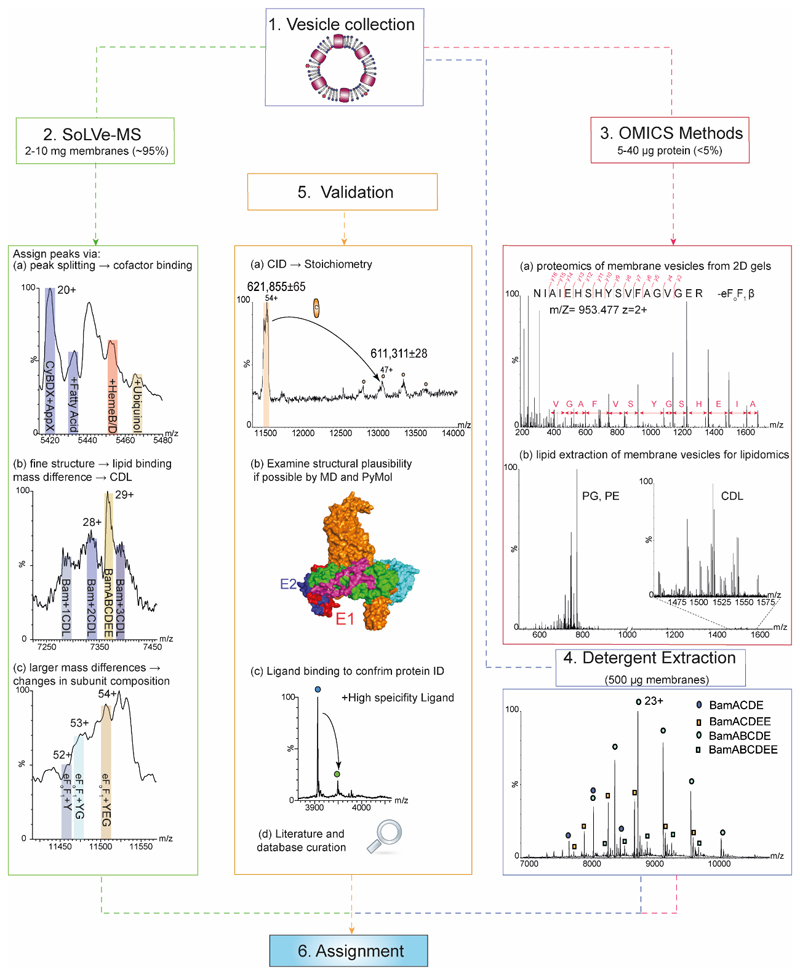
Figure 2. How to approach SoLVe-MS, data analysis and validation.
Step 1- Obtain vesicles-addressed in step 1 of Procedure section. Step 2- SoLVe-MS will require the greatest proportion of the collected vesicles (~95%). Acquire the highest resolution MS data possible and assign peaks to charge states of a protein complex. Careful examination of the spectrum is required to search for peak splitting due to cofactor binding and unique identifiers (a), or diffuse peaks indicating lipid binding and co-factors and other small molecules. (b). Unexpected large mass variation may be observed indicating differences in subunit composition (c). Step 3- Use available omics approaches. This will require μg of protein (Likely less than the remaining 5% of material). Both native and denaturing gels are needed to support interpretation. All gel bands should be excised and subjected to standard proteomics approaches leading to identification of possible candidates and their subunits. Standard proteomics shown here identifies a partial sequence of the β subunit of the eFoF1 (a). The lipid components of the preparation should be examined by lipidomics (b). Step 4- Detergent extraction of proteins directly from the same membrane as a control for stoichiometry and lipid binding, or a recombinant version of a complex, such as the Bam complex purified in C8E4 detergent can be used for comparison. Here both stoichiometries are observed (BamABCDE and a population of BamABCDEE). Step 5- When all data has been acquired, validation is undertaken. This can be done in multiple ways including literature search. A mass table derived from databases or if possible previous literature reports of measured subunit masses should be curated. Examination of structural plausibility utilizing molecular dynamics simulation or examining the structural integrity in PyMOL should be carried out. All these should eventually significantly increase the confidence in the assignment. Representative spectra are of a single repeat from 3-6 biological repeats.
How is SoLVe-MS different from other methods used for nMS?
Our initial experiments, designed to transport membrane proteins from solution to gas phase, were performed using detergent micelles to protect the complex until release once in the mass spectrometer15. Many successful experiments have been reported using this approach11,16–18. It is known however that detergent micelles can both promote and perturb lipid binding19. Concerns about the possible effects of detergent micelles have driven the development of many membrane mimetics including-bicelles, nanodiscs, styrene maleic acid lipid particles (SMALPs) and liposomes (Table 1). These membrane mimetics have provided great insight into the importance of the lipid bilayer, for stability and function, and are widely used for EM studies20,21. It remains challenging however to recapitulate the membrane environment as a whole within a nanodisc for example. The true abundance of lipids22, the force regimes derived from the shape of the membrane and its viscosity, as well as the immense number and diversity of lipids, give rise to properties that are difficult to replicate. The SMALP technology provides one of the closest mimetics for studying membrane proteins23. Attempts to release proteins from these particles for nMS have however proved challenging with only limited resolution reported to date7. Furthermore, these methods in general require overexpression of proteins, which further complicates interpretation of the state of the complex in the native membrane environment.
Table 1. Comparison of different extraction and solubilization methods used for the analysis of membrane proteins by nMS.
| Method | Detergent extraction | Spectral quality | Complexity of spectra | Resemblance to membrane | Protein purification | Protein overexpression | Analysis time |
|---|---|---|---|---|---|---|---|
| Micelles15 | Yes | High | Low | Minimal | Yes | Typically required | Short |
| Bicelles46 | Yes | High | Low | Minimal | Yes | Typically required | Short |
| Amphipols46 | Yes | High | Low | Minimal | Yes | Typically required | Short |
| Nanodiscs8 | Yes | High | High | Medium | Yes | Typically required | Medium |
| SMALPs7 | No | Low | Low | Medium | No | Typically required | Medium |
| SoLVe1 | No | High | High | High | No | Not required, except for low abundance proteins | Variable |
SoLVe-MS by contrast does not employ a membrane mimetic, but uses vesicles formed directly from native membranes. These vesicles can be either naturally secreted by cells24, or created by physical intervention. Importantly however they are not formed through chemical intervention either via detergents, acidic regimes, or hydrophobic polymers. We expect that the membrane proteins and complexes subjected to SoLVe are encapsulated within vesicles as we have not been able to detect membrane “fragments” by EM following sonication, prior to injection. SoLVe-MS therefore allows examination of endogenous membrane proteins, in vesicles derived directly from the original membrane of the host organism. It is important to note that these membrane vesicles are not liposomes. The latter are formed from purified protein inserted into a membrane artificially assembled from selected lipids25. Membranes for SoLVe-MS by contrast are generated directly from their organelle or organism, without chemical intervention, and without the need for overexpression unless specifically required.
How do you define the components of the membrane?
The results derived from the SoLVe protocol were at first unexpected. For example, the bacterial F-type ATP synthase was observed intact while communicating with the core hydrophobic components of the inner membrane insertase SecYEG1,26. This interaction was also recently reported following affinity capture in peptidiscs and analysed by protoemics26. Additionally, two distinct populations of the Bam complex were observed by SoLVe-MS, with and without an additional copy of subunit E. Interestingly, only in the subpopulation of Bam complexes containing a single copy of E, cardiolipin binding was observed (Fig. 2, step 1 and 2).
As described in Fig. 2, to support these observations traditional ‘omics’-based strategies are required (Fig 2. Step 3), including proteomics to identify all protein components, both from denaturing and native gels followed by in-gel digestion and proteomics27. As direct digestion from the vesicles yields relatively low coverage due to large parts of the protein being inaccessible, we have found it is better to perform in-gel digestion. 40 μg of protein per lane is typically used. Moreover, as these membrane proteins are also likely to bind a large array of lipids, lipidomics28,29 must be performed to cover all possible lipid options that might be observed within preparation from native membranes. Because of the prevalence of post translational modifications, and proteoform variations, including a detergent extraction step (Fig. 2, step 4) for membrane proteins may also be required to obtain accurate masses for fitting with intact complexes. Post-acquisition, all information will be needed to understand and interpret the SoLVe mass spectra (Fig. 2, step 3). The ‘omics’ approaches and the SoLVe protocol can be done in any order, and cross-interpreted. The tentative assignments can then be compared and validated against the literature, or incubated with known inhibitors of the expected protein, overexpressed and tested for a known protein, and cross-validated with a variety of supporting methods like molecular dynamics simulations (Fig. 2, Procedure step 5).
What types of membranes are suitable for SoLVe-MS?
The initial developments focused on ejection of overexpressed eukaryotic membrane proteins from MPEEVs, presenting the protein of interest as the sole membrane protein. We then discovered that the approach could be applied to a wide range of protein complexes from native membranes of E. coli and bovine sources. In this protocol we develop the methodology further by demonstrating the ability to eject overexpressed recombinant bacterial operons in suitable bacterial host strains. Additionally, we explore applications to membrane-associated proteins, which were found to be more stable when released from membranes directly. Overall, we have found that SoLVe-MS can be employed to many specialized membranes that contain a high abundance of specific membrane protein complexes.
Materials
Reagents
Stock Ammonium Acetate solution 7.5 M, Sigma, A2706-500 ml
E. coli strain lacking OmpF, OmpA, OmpC and LamB35
B. Taurus heart32. Bovine hearts can be obtained from an abattoir.
BHK-21, clone 1324. Cell line can be obtained from ATCC.
Reagent setup
The ammonium stock solution is made fresh for every experiment, diluted to 500 mM ammonium acetate in milliQ water, and can be stored at 4 °C. for 2-3 days.
Equipment
Glass capillaries, Harvard apparatus, GC120F-10 for UHMR, GC100TF-10 for Synapt.
Amicon Ultra-4 centrifugal filters- 10-100K REF UFC810096
50 ml glass beaker, Pyrex, 10079739
Micro Bio-Spin 6 chromatography columns, MW exclusion limit 6 kDa (Tris buffer, Bio-Rad, cat. no. 732-6221)
Probe sonicator, Sonics and Materials Inc, VC505, 20kHz connected to a large probe CV334, probe tip diameter 12 mm.
Q-Exactive UHMR, Thermo Fisher Scientific, Cat# OPTON 0726090
Optima L-90K Ultracentrifuge, Beckman Coulter, SW32Ti rotor or others and compatible tubes
Thick wall Polycarbonate ultracentrifuge tubes REF355631
Chilled benchtop centrifuge- Hettich Universal 320 R
Synapt HDMS G1 with 32K RF generator (or others), Waters
Equipment Setup
The following are starting parameters for a commercially available Thermo-Fisher Scientific Q-Exactive UHMR and a first-generation Waters Synapt HDMS.
| Q-Exactive UHMR | Synapt G1 HDMS |
|---|---|
| Capillary voltage 1.6 kV | Backing pressure 5.6 mBar |
| Source capillary temperature 350 °C. | Source temperature 90 °C |
| Trapping gas pressure 7.5 | Capillary voltage 1.6 kV |
| Source DC offset 21 | Cone voltage 170 V |
| Source fragmentation 150 V | Trap voltage 100 V |
| In source trapping On | Transfer voltage 5 V |
| Desolvation voltage -150 V | Bias 15 V |
| HCD energy 0 V | *For IMS |
| Detector optimized for high m/z. | Wave speed 250 m/s |
| Noise threshold 3 | Wave Height 7-10 |
| Max inject time 100 | IMS gas 24 ml/min |
| AGC target 1,000,000 | |
| Resolution 12,500 |
Software
Thermo Xcalibur 4.2.47
Unidec 4.1 (Freely available for academic use)33
SUMMIT 1.0 (Free, no longer supported)34
Waters Masslynx 4.2
Procedure (excluding data analysis, O Timing 3-4 days)
Membrane collection- O Timing- 1 day (cell growth may reach one week if cultured eukaryotic cells are needed)
Δ Critical Step The orientation of the proteins within the membrane (inside out or outside in) might be unknown leading to losses of fragile components e.g. F1 soluble complexes from F1Fo ATPases. We advise to have the membrane vesicles imaged by cryo-EM in order to examine how the proteins are oriented. However, it’s possible to avoid this with an additional membrane layer that can be retained by avoiding the osmotic bursting step to protect the proteins as in the case of mitochondrial membranes. Trialing softer sonication conditions such as lower amplitude is also advised.
-
1|
Collect membranes by the method you normally use in your lab, or a method that you think would work well for your experiment. The following options are examples that we have used in our own lab.
| A | MPEEVs | BHK-21 cells |
| B | Inner and outer membranes | E.coli |
| C | Mitochondrial membranes | Bovine heart |
In practice, since membranes are heavily diluted, sonicated and washed in vast excess of ammonium acetate as part of the later steps in this Procedure, it is not generally necessary to change existing membrane preparation protocols. Membrane protein purifications can be modified for SoLVe-MS simply by stopping before the step in which the membrane is dissolved in detergent.
Membranes purified in standard buffers have a lifetime of up to 1-2 weeks at 4 °C or longer at -80 °C. However, it is always recommended to do everything as fresh as possible.
-
A
MPEEVS from mammalian cells
-
(i)
The following is based on the procedure in ref. 24 used to acquire MPEEVs, vesicles excreted naturally to the growth medium from cells overexpressing a protein under a strong promotor. 24 h before transfection, seed 15x106 BHK-21 cells in 40 ml of Glasgow MEM supplemented with 10% fetal calf serum (FCS), 2% tryptose phosphate broth (TPB) and 20 mM HEPES, onto a T175 flask. 2-3 flasks are typically required per experiment depending on expression level and relative abundance, however, sometimes more will be needed. Each flask yields approximately 100 μg of protein.
-
(ii)
On the day of the transfection, change medium to 17 ml growth medium containing 2% FCS.
-
(iii)
Perform transfection according to manufacturer protocol. Use 60 μg of DNA-the protein of interest overexpressed under a strong promotor, and 120 μl of Lipofectamine 2000 transfection reagent, dilute each reagent in 1.5ml of growth medium with 2% FCS to a total mix of 3ml, per flask T175 flask.
-
(iv)
2 h post transfection, wash cells twice with 10 ml of PBS.
-
(v)
Remove PBS completely.
-
(vi)
Supplement the cells with 30 ml of growth media containing 2% FCS, 2% TPB and 20 mM HEPES.
-
(vii)
24 h post transfection, remove growth media, wash cells with at least 100 ml of PBS (20 ml x 5 washes) to remove excess BSA, and replace with 30 ml of growth media that contains 2% TPB, 20 mM HEPES, but does not contain FCS.
Δ Critical Step Insufficient washing will result in BSA contamination of the sample.
-
(viii)
48 h post transfection collect the growth media only and dispose of the flasks with the cells still attached.
-
(ix)
Put collected growth media on ice.
-
(x)
Centrifuge at 1000x g at 4 °C to remove any cells or cell debris for 20 minutes.
-
(xi)
Remove supernatant to a different tube.
-
(xii)
Wash ultracentrifuge tubes well with milliQ water and 70% EtOH, and dry before use.
-
(xiii)
Move the supernatant into a thick wall polycarbonate ultracentrifuge tube suitable for a SW32Ti rotor.
-
(xiv)
Using a 5ml syringe with a long needle, insert 5 ml of 20% sucrose in 25 mM HEPES pH=7.4 with 130 mM NaCl, to form a sucrose cushion at the bottom of the tube.
-
(xv)
Centrifuge at 30,000 rpm with a SW32Ti rotor for 2 h at 4 °C.
-
(xvi)
Dispose of the sup patiently.
-
(xvii)
Dry out the tube using kimwipes without touching the pellet.
Δ Critical Step Drying out the tube of the growth media completely is important, as the sample may turn out pink and contaminated with growth media leftovers if not done properly.
-
(xviii)
Add 50-100 μl of 500 mM ammonium acetate to the tube.
-
(xix)
Seal the tube with parafilm and leave at 4 °C overnight for the vesicles to redissolve.
-
(xx)
Collect the sup with the vesicles into an Eppendorf tube the following morning.
<PAUSE POINT> In our experience MPEEVs remain intact for 1-2 weeks at 4 °C.
-
B
Bacterial inner and outer membranes
-
(i)
Follow the procedure described by Baker et al.,35 without change.
-
(ii)
After purification, dilute the vesicles in ammonium acetate, sonicated and treated as described below.
<PAUSE POINT> These can be stored at 1-2 weeks at 4 °C.
-
C
Mitochondrial membranes
-
(i)
To isolate mitochondrial membranes, follow the procedure described in Ref. 3636.
-
(ii)
Stop the procedure after the (last) ultracentrifugation step; and don’t dissolve the membranes in detergent.
<PAUSE POINT> The membranes can be stored for more than a year at -80°C or for 2-3 weeks post-SoLVe at 4 °C.
Sonication- O Timing 1 hour
! Caution- sonication may cause hearing damage and must be performed with appropriate safety precautions. Ideally, a sound proof cabinet should be used. The experimental setup for sonication is described in Fig. 3(i-ii). Details for the sonicator and probe, including catalogue numbers, are in the appropriate equipment section.
-
1|
Prepare an ice bucket.
-
2|
Wash the probe with double distilled water (DDW) filtered through a 0.22 μm filter, followed by a 70% (v/v) Ethanol wash.
-
3|
Dry the probe.
Δ Critical Step We recommend using a 12 mm probe (See Equipment). Do not attempt to sonicate small volumes with a smaller probe, since it is highly likely that membrane vesicle solution will overheat and evaporate.
-
4|
Turn on sonicator. Set pulses to 3 s on /6 s off. Time 2 mins 30 s. Set amplitude to a maximum of 60%. This equates to a total of 20 J and 13 W per 9 s working cycle.
-
5|
Fill a 50 ml glass beaker with chilled 20 ml of 500 mM ammonium acetate at pH 7.6.
-
6|
Place glass beaker firmly into an ice bucket.
-
7|
Place membrane vesicles, collected and formed via standard protocols as described in Step 1 of the procedure, into the ammonium acetate filled beaker.
Δ Critical Step The amount of membrane fraction required is related to the complexity of the system. If a single bacterial protein is of interest, tens of micrograms of proteins embedded within vesicles may be sufficient. For more heterogeneous bacterial inner or outer membranes, a total of 2 mg of the membrane fraction is recommended. For eukaryotic, highly modified proteins, 0.5 mg would be required for a single protein, but for whole membrane fractions up to 10 mg maybe required.
-
8|
Homogenize membrane preparation with a pipette.
-
9|
Place the bucket with the beaker beneath the probe.
-
10|
Slowly raise the beaker until probe is approximately 2-3 mm above the bottom of the beaker. Make sure the probe does not touch the glass.
Δ Critical Step - Do not place the probe too close to the bottom of beaker, the glass may shatter.
Δ Critical Step - Do not place the probe too close to the liquid interface, bubbles might form that induce protein oxidation and deformation.
-
11|
Put on protective hearing equipment.
-
12|
Start sonication.
-
13|
During sonication, constantly examine the preparation to ensure that no bubbles are forming.
Δ Critical Step - The sonication process will heat the preparation, and consequently will melt the ice beneath the beaker, lowering the height of the beaker with respect to the sonication probe. This may cause the probe to create unwanted bubbles or exit the solution.
-
14|
When the sonicator clock reaches 2.5 mins, push the stop button.
Δ Critical Step - Do not perform the procedure for less than 1 min as the process will be ineffective. More than 5 mins will be detrimental to the proteins.
-
15|
Remove the suspension to a labeled 50 ml tube.
-
16|
Clean probe, first with water, then with 70% (v/v) ethanol.
-
17|
Turn off the sonicator.
□ <PAUSE POINT> - Membrane proteins residing within their native membrane in ammonium acetate have an approximate lifetime of 2 weeks at 4 °C.
Protein concentration O Timing 5-6 hours
□ Pause point- Concentrating membranes is time consuming. Concentrator tubes can be stored overnight at 4 °C and concentration can be continued the following day.
-
18|
Select a 10/30/50/100 kDa cut-off 4 ml concentrator tube (Amicon).
Δ Critical Step - The size of the vesicle will not allow it to pass through the pores of any concentrator membrane. However, sometimes a membrane protein complex will contain independent lipoproteins, or soluble components, that might be of interest and may be lost when using a concentrator with a given molecular weight cut-off. Therefore, to maintain these components, a low molecular weight cutoff concentrator may be required. Using a low molecular weight cutoff will result in extended washing times. Moreover, sometimes a soluble contaminant may exist, selecting a cutoff size larger than the mass of this protein may allow its removal. These aspects should be considered when choosing the molecular weight cut-off for the concentrator tube. We recommend starting with a 10 kDa cutoff, and adjusting the choice for subsequent experiments.
-
19|
Equilibrate the concentrator membrane with 2 ml of 500 mM ammonium acetate for 10 mins.
-
20|
Place 2 ml of the sonicated vesicle preparation into the concentrator tube.
Spin at maximum possible speed for a given concentrator tube (7690 x g on a fixed rotor in a Hettich 320 R) for an Amicon Ultra-4 concentrator, at 4 °C.
Δ Critical Step - Make sure centrifuge is balanced at all times.
Δ Critical Step - Do not let the concentrator membrane, containing the preparation, to run dry.
Δ Critical Step - Do not exceed the manufacturer’s suggested centrifugation speed for a particular concentrator. Exceeding this value may induce membrane rupture.
Δ Critical Step - Use of the 4 ml concentrator has been found to be optimal. The physical properties of the membrane in 15 ml concentrators means that the preparation is exposed to too great of a surface area. This can induce losses of material through precipitation or adhesion of the vesicles to the concentrator membranes. Concentration via 0.5 ml concentrators is too time consuming.
-
21|
Decrease the volume of the preparation as much as possible by centrifugation.
-
22|
Add more of the sonicated preparation to the same concentrator tube.
-
23|
Decrease the volume as much as possible (100-200 μl).
-
24|
Repeat steps 22-23 until the volume of the sonicated preparation is depleted.
-
25|
Decrease volume as much as possible as in step 23.
-
26|
Add 500 mM ammonium acetate to fill to the top tube to equilibrate the lipid vesicles in the mass spectrometry compatible buffer.
-
27|
Repeat steps 25-26 at least 3 times. An approximate 10,000-fold dilution is advised to effectively desalt the vesicles.
Δ Critical Step - Insufficient desalting will likely result in a poor-quality mass spectrum with a low signal to noise ratio. > 10,000-fold buffer dilution factor is recommended.
-
28|
Reduce preparation to a final volume of 100-200 μl.
Δ Critical Step - If centrifugation is too slow, the lipid vesicle preparation can be split into two or more concentrator tubes. This may however result in increased loss of proteins through adsorption to the additional concentrator filter membrane.
□ Pause point- Membranes and proteins at this point can be stored for ~2 weeks in ammonium acetate at 4 °C or years at -80 °C. However, long-term storage specifically in ammonium acetate is not recommended.
Native mass spectrometry analysis O Timing 5-6 hours
Δ Critical- The parameters in fig. 1c and Equipment setup represent starting conditions and can be adapted according to the preparation used. These steps can be adapted according to the advice described in Step 35.
! Caution- Glass capillaries are sharp and may break. Wear gloves and eye protection.
! Caution- High voltage may cause injury.
-
29|
Insert 2-3 μl of fully processed preparation into a gold-coated glass capillary using a gel loading tip37.
-
30|
Ensure that the solution has reached the end of the capillary using a small benchtop centrifuge and a capillary holder for short bursts at maximum speed at room temperature, or manually with a flick of the wrist.
-
31|
Remove capillary tip under a microscope using tweezers. Membrane vesicles can range between 100-500 nm in diameter or length, depending on the membrane of origin. It is therefore sometimes necessary to form a large orifice to allow vesicles to exit the capillary and form a stable spray (See fig. 4).
-
32|
Lock the nano-spray source on the MS instrument.
The starting parameters for both the Waters Synapt HDMS and Thermo Scientific Q-Exactive UHMR can be found in the equipment setup section and in Fig. 1c and Fig. 3(iii).
Δ Critical- These represent starting conditions and can be adapted according to the preparation used.
-
33|
Perform the mass spectrometry experiment, and examine the results.
□ Pause point- Membranes and proteins at this point can be stored for ~2 weeks in ammonium acetate at 4 °C or years at -80 °C. However, long-term storage specifically in ammonium acetate is not recommended.
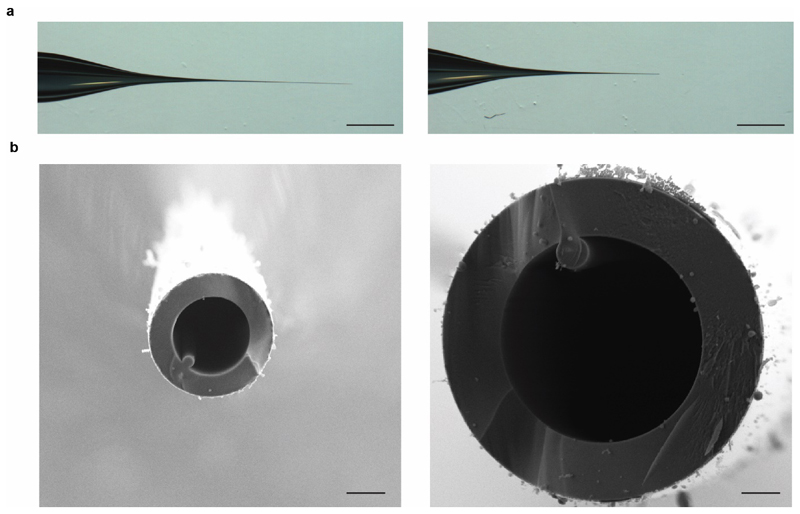
Figure 4. Enhancing the spectra by modifying the capillary tip.
(a) The vesicle diameter varies from 100-500 nm depending on the membrane’s origin, therefore a capillary cut in the standard way (LH panel), may not allow for sufficient exit of vesicles, and may require further opening (RH panel). Scale bar is 1 mm. (b) Scanning electron microscopy of the capillary tips cut in the standard way (LH panel) and further broken (RH panel). Scale bar is 1 μm.
Adapting the parameters for Nano-electrospray of membrane vesicles
-
34|
Modify the capillary tip to enable MS of membrane vesicles Introducing lipid vesicles into a mass spectrometer and optimizing the instrument conditions is not trivial. The vesicles prepared here range from 100 to 500 nm in diameter, therefore sometimes the tip of the capillary will need to be broken such that a large part is removed, facilitating a larger orifice (an increase of ~3x in the diameter) than would typically be required for native MS (Fig. 4a and b).
-
35|
Adjust the MS parameters per sample Optimization of the electrospray is in many ways similar to the procedures used for membranes proteins encapsulated in detergent micelles38,39. In this case high energy is required to remove the membrane as opposed to the micelle. Insufficient energy will yield unresolved spectra while too much energy will induce fragmentation of protein complexes and loss of metabolites or cofactors (Supplementary figure 1). Very likely, MS under a range of conditions will be required. A relatively unresolved spectrum can still be informative as raw data can be deconvolved by software freely available such as UniDec33,40 and iFAMS41 to indicate additional masses. Although not always the case, vesicles typically require more stringent MS conditions than complexes released from detergent micelles. Moreover, higher mass species typically eject at higher energies. Using this high energy does not favor transmission of soluble contaminants from the solution, as these are not membrane protected.
Trouble shooting
Troubleshooting advice can be found in Table 2.
Table 2. Troubleshooting.
| Step No. | Problem | Possible Reason | Possible Solutions |
|---|---|---|---|
| 4 | Decreased spectra quality at higher m/z. | For UHMR effective resolution at higher m/z is lower. | Change m/z window to encompass only high m/z values i.e. from 10,000 to 20,000 m/z. Set detector to high m/z. Increase HCD (UHMR) or backing (Synapt) pressure to stabilize high mass species. |
| 1,4 | Absence of signal. | Insufficient protein Vesicles do not exit capillary |
Collect additional vesicles. Expand capillary orifice. |
| 3,4 | Broad peaks throughout. | Insufficient desalting, too much sonication, use of non-compatible compounds for native MS. | Perform additional washes of preparation. Reduce sonication amplitude. Avoid having DTT, TCEP, and other small molecules that might complicate the analysis. |
| 4 | Collision induced dissociation not possible. | Not enough energy used throughout. Total energy used is insufficient. | Raise voltage throughout where possible. |
| 5 | Soluble contaminants. | Vesicles not washed well enough. Some maybe intrinsic components of the system. |
Additional wash stages can be presented into the protocol by repeating ultracentrifugation multiple times. Utilization of a concentrator tube with a molecular weight cutoff larger than the contaminant’s mass. Higher energy releases more membranes proteins, making them of higher representation in the spectra when compared to contaminants. |
Anticipated results
To demonstrate SoLVe-MS, and to highlight comparison with detergent solubilized preparations, we considered proteins overexpressed by the Fap operon42 which has been shown to form an amyloid-like structure on the surface of bacterial membranes. While it is possible to perform SoLVe with unseparated wild-type E. coli membranes expressing target protein complex, in the case of the FapF, the spectra are complicated by large amounts of endogenous proteins (Fig. 1d) from both inner and outer membranes. We therefore used membranes derived from a strain of E. coli lacking the four most abundant outer membrane proteins - OmpF, OmpC, LamB and OmpA43. After overexpressing the Fap operon containing FapA-F proteins in this E. coli strain lacking outer membrane proteins, native gel analysis showed almost no traces of any complexes in the sample, but after n-Dodecyl β-D-maltoside (DDM) solubilizationconfirmed the presence FapF by proteomics (Supplementary figure 2a). Initially to check for expression and for comparison with SoLVe we solubilized and purified the assembly in C8E4 detergent micelles. Under these conditions, a trimer of FapF, which forms the membrane spanning pore, as well as a very low population of (FapF)3(FapD)1could be observed (Fig 5a and b, Supplementary figure 3). When ejected directly from membranes (Fig 5a and c), however, additional species were observed with high intensity corresponding to (FapF)3(FapD)1(FapC)1 and (FapF)3(FapD)1(FapC)1(FapE)1. These results are consistent with formation of complexes that correspond to initial stages of amyloid fibril formation on membranes. Furthermore, the results imply that SoLVe-MS is capable of preserving assemblies that form not only within membranes, but also on their surfaces.
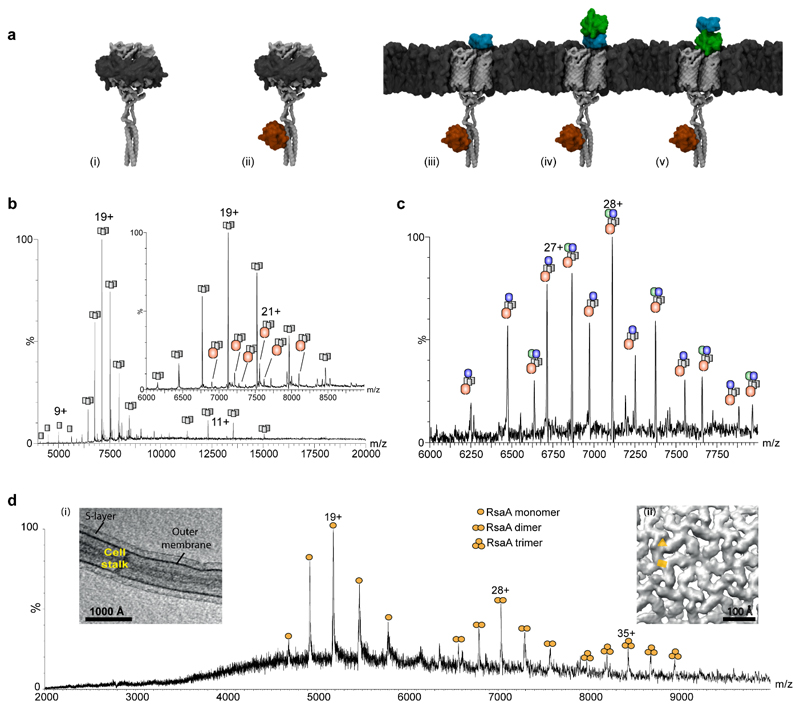
Figure 5. Comparison of SoLVe-MS with low pH and detergent extracted membrane protein complexes and associated proteins.
(a) Models of Fap protein complexes observed by detergent extraction (i)-(ii) and by SoLVe-MS (iii) – (v). The interaction of FapF with FapD is modeled based on BS3 cross-linking and data reported previously45, and models for FapC and E are homology based. (b) Mass spectrum of the Fap operon purified in C8E4 shows predominantly FapF3 (i) (grey) alone, and with a very low population containing a single FapD (ii) (red), or a single misprocessed FapD (red). (c) SoLVe-MS of E. coli membranes lacking major outer membrane components, overexpressing the Fap operon, show association of FapF3 with FapD and FapC (blue) (iii) or FapD, FapC and FapE (green) (iv) or (v). It is not known whether or not FapE (green) displaces FapC (blue), so both models are presented. Spectra were acquired at 400 V (Desolvation -200V, Source fragmentation 200V), and are a representative from 2 biological repeats and multiple MS experiments. (d) SoLVe-MS enables release of RsaA from the surface of the membrane, from the cell stalk. Inset (i) Tomographic slice through a C. crescentus stalk. Inset (ii) Sub-tomogram averaging of the purified cell stalks shows the presence of two-fold and a pseudo-three-fold symmetry axes in the S-layer lattice on native membranes, marked with a yellow rectangle and triangle respectively. Spectrum was recorded at a Desolvation -150 V, source fragmentation 100 V and HCD 150 V.
To examine further the versatility of SoLVe to probe surface associated proteins we attempted to release the peripheral membrane bacterial S-layer protein RsaA directly from the surface of the Caulobacter crescentus cell stalks44. Comparing the SoLVe experiment (Fig 5d) with a preparation in which the standard low pH extraction was used (Supplementary fig. 4), shows similar amounts and masses of RsaA are measured for both. Measured and expected masses for all spectra appear in Supplementary table 1. These comparisons demonstrate the importance of the membrane for preserving the stability of proteins and add to the applications of SoLVe-MS for membrane associated assemblies.
Supplementary Material
Acknowledgements
The authors would like to thank all CVR group members for helpful discussions, Dr. Linna Zhou for her help in photographing the capillaries, Siyun Chen for her help in photographing the experimental setup, Raman Dhaliwal and Dr. Errin Johnson from the Oxford Dunn School EM facility for SEM imaging, Prof. Tzviya Zeev-Ben-Mordehai for her technical assistance with MPEEVs and Dr. Harris D. Bernstein for providing the plasmid pJH114 for expressing the Bam complex. D.S.C., H.T. and C.V.R. are grateful for the support of an ERC grant (695511-ENABLE) and J. G. and C.V.R. for support by a Wellcome Trust Investigator Award (104633/Z/14/Z). J.R.B. is supported by a Medical Research Council grant (MR/N020413/1). T.A.M.B. is supported by the Wellcome Trust and the Royal Society (Grant No. 202231/Z/16/Z). A.V.K., T.A.M.B. and C.V.R. thank the Vallee Foundation for support. S.L.R and S.J.M are supported by a Wellcome Trust (Senior Investigator Award 100280). J.G. is a Junior Research Fellow at the Queen’s College Oxford. L.A.B. was supported by a Human Frontier Science Program Long Term Fellowship and a Canadian Institutes for Health Research Postdoctoral Fellowship. K.G. was supported by a Wellcome Trust Senior Research Fellowship (090895/Z/09/Z) and a core award (090532/Z/09/Z).
Footnotes
Data availability
Raw data presented in this manuscript is either deposited in https://doi.org/10.6084/m9.figshare.11376015 or available upon request.
Code availability
No code has been written for this manuscript. Software needed is stated and referenced in text.
Author Contributions
D.S.C. performed the mass spectrometry experiments. H.T. performed the mass spectrometry experiments for RsaA. J.G. optimized the Orbitrap UHMR platform for the transmission and detection of membrane proteins and to enable high-energy regimes. J.R.B. purified and analyzed the recombinant BAM complex. D.W. established and applied the lipidomics platform. S.L.R. and S.J.M. purified Fap proteins and membranes, and modelled the Fap complexes. A.V.K. and T.A.M.B. purified s-layer proteins and membranes, and performed cryo-EM of cell stalks. L.A.B. and K.G. performed the cryo-EM for inner membrane tubes. D.S.C. and C.V.R. supervised the research and wrote the manuscript with contributions from all authors.
Competing interests
The authors declare no competing interests.
References
- 1.Chorev DS, et al. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science. 2018;362:829–834. doi: 10.1126/science.aau0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeev-Ben-Mordehai T, et al. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc Natl Acad Sci U S A. 2016;113:4176–4181. doi: 10.1073/pnas.1523234113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson H. Biological membranes. Essays Biochem. 2015;59:43–69. doi: 10.1042/bse0590043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J, et al. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 2019;364:1068–1075. doi: 10.1126/science.aaw4852. [DOI] [PubMed] [Google Scholar]
- 5.Blum TB, Hahn A, Meier T, Davies KM, Kuhlbrandt W. Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1816556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maki-Yonekura S, et al. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. Elife. 2018;7 doi: 10.7554/eLife.35419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellwig N, et al. Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs. Chem Commun (Camb) 2018;54:13702–13705. doi: 10.1039/c8cc06284f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keener JE, et al. Chemical Additives Enable Native Mass Spectrometry Measurement of Membrane Protein Oligomeric State within Intact Nanodiscs. J Am Chem Soc. 2019;141:1054–1061. doi: 10.1021/jacs.8b11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celia H, et al. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun Biol. 2019;2:358. doi: 10.1038/s42003-019-0604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postis V, et al. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim Biophys Acta. 2015;1848:496–501. doi: 10.1016/j.bbamem.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambrose S, et al. Native Desorption Electrospray Ionization Liberates Soluble and Membrane Protein Complexes from Surfaces. Angew Chem Int Ed Engl. 2017;56:14463–14468. doi: 10.1002/anie.201704849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Waterbeemd M, et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat Methods. 2017;14:283–286. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 14.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 16.Yen HY, et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature. 2018;559:423–427. doi: 10.1038/s41586-018-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habeck M, Kapri-Pardes E, Sharon M, Karlish SJ. Specific phospholipid binding to Na,K-ATPase at two distinct sites. Proc Natl Acad Sci U S A. 2017;114:2904–2909. doi: 10.1073/pnas.1620799114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landreh M, Costeira-Paulo J, Gault J, Marklund EG, Robinson CV. Effects of Detergent Micelles on Lipid Binding to Proteins in Electrospray Ionization Mass Spectrometry. Anal Chem. 2017;89:7425–7430. doi: 10.1021/acs.analchem.7b00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers JB, et al. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature. 2018;564:372–377. doi: 10.1038/s41586-018-0786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao DY, et al. Cryo-EM structure of the native rhodopsin dimer in nanodiscs. J Biol Chem. 2019 doi: 10.1074/jbc.RA119.010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parmar M, et al. Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim Biophys Acta Biomembr. 2018;1860:378–383. doi: 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeev-Ben-Mordehai T, Vasishtan D, Siebert CA, Whittle C, Grunewald K. Extracellular vesicles: a platform for the structure determination of membrane proteins by Cryo-EM. Structure. 2014;22:1687–1692. doi: 10.1016/j.str.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akbarzadeh A, et al. Liposome: classification, preparation, and applications. Nanoscale Res Lett. 2013;8:102. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson ML, et al. Profiling the E. coli membrane interactome captured in peptidisc libraries. Elife. 2019;8 doi: 10.7554/eLife.46615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 28.Bechara C, et al. A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem. 2015;7:255–262. doi: 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- 29.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. Journal of bacteriology. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Molecular microbiology. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 32.Urbani A, et al. Purified F-ATP synthase forms a Ca(2+)-dependent high-conductance channel matching the mitochondrial permeability transition pore. Nature communications. 2019;10 doi: 10.1038/s41467-019-12331-1. 4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marty MT, et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverner T, et al. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc Chem Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 35.Baker LA, et al. Magic-angle-spinning solid-state NMR of membrane proteins. Methods Enzymol. 2015;557:307–328. doi: 10.1016/bs.mie.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 36.Maeda S, et al. Two-dimensional crystallization of intact F-ATP synthase isolated from bovine heart mitochondria. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:1368–1370. doi: 10.1107/S1744309113029072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 38.Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8:639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta K, et al. Identifying key membrane protein lipid interactions using mass spectrometry. Nat Protoc. 2018;13:1106–1120. doi: 10.1038/nprot.2018.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid DJ, et al. MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J Am Soc Mass Spectrom. 2019;30:118–127. doi: 10.1007/s13361-018-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleary SP, Prell JS. Liberating Native Mass Spectrometry from Dependence on Volatile Salt Buffers by Use of Gabor Transform. Chemphyschem. 2019;20:519–523. doi: 10.1002/cphc.201900022. [DOI] [PubMed] [Google Scholar]
- 42.Rouse SL, et al. A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis. Nature communications. 2017;8 doi: 10.1038/s41467-017-00361-6. 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meuskens I, Michalik M, Chauhan N, Linke D, Leo JC. A New Strain Collection for Improved Expression of Outer Membrane Proteins. Front Cell Infect Microbiol. 2017;7:464. doi: 10.3389/fcimb.2017.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharat TAM, et al. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol. 2017;2:17059. doi: 10.1038/nmicrobiol.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bobeica SC, et al. Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease. Elife. 2019;8 doi: 10.7554/eLife.42305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopper JT, et al. Detergent-free mass spectrometry of membrane protein complexes. Nat Methods. 2013;10:1206–1208. doi: 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.