Abstract
Engineered microbes are exciting alternatives to current diagnostics and therapeutics. Researchers have developed a wide range of genetic tools and parts to engineer probiotic and commensal microbes. Among these tools and parts, biosensors allow the microbes to sense and record or to sense and respond to chemical and environmental signals in the body, enabling them to report on health conditions of the animal host and/or deliver therapeutics in a controlled manner. In this review, we focus on how biosensing has been applied to engineering “smart” microbes for in vivo diagnostic, therapeutic, and biocontainment goals. We also discuss hurdles that need to be overcome when transitioning from high-throughput in vitro systems to low-throughput in vivo animal models, new technologies that can be implemented to alleviate this experimental gap, and areas where future advancements can be made to maximize the utility of biosensing for medical applications. As technologies for engineering microbes continue to be developed, these engineered organisms will be used to address many medical challenges.
Keywords: Biocontainment, Biosensing, Diagnostic, Probiotic, Therapeutic
1. Introduction
Probiotic and commensal microbes are naturally valuable assets for the host. These microbes can prevent pathogen colonization, reduce the frequency and severity of various ailments, modulate the brain activity through the gut-brain axis, and selectively colonize tumor microenvironments.[1] For example, various strains of Lactobacillus, Lactococcus, Bifidobacterium, and Bacillus inhibit the colonization of many pathogenic bacteria.[2-5] This inhibition occurs through a number of mechanisms, including reduction of the luminal pH, competition for nutritional resources, and excretion of bacteriocin.[6] Some microbes also exhibit tumor-specific colonization that can significantly inhibit the growth of the tumors. This property has been demonstrated and applied using several bacteria, including Clostridium,[7-9] Bifidobacterium,[10] E. coli,[11] and an attenuated version of Salmonella typhimurium (aSt).[12, 13]
Many microbes also alleviate the symptoms or reduce the occurrence of various ailments, including diarrhea,[14] allergy,[15] and gut inflammation.[16] Often, the exact mechanisms of action for these microbes are not well understood. Many of these microbes improve health through interfacing with both host cells and other gut microbes. This communication largely occurs via the production and degradation of various proteins and metabolites that alter the composition of the microbiome, tune the pH of the gut, stimulate the function of the mucosal barrier, and modulate the activity of the immune system.[1]
Microbe-host interactions can also influence the activity of the brain through the gut-brain axis. Some probiotic and commensal microbes can synthesize and degrade brain-modulating neurotransmitters, including catecholamines and serotonin.[17-19] Microbes have also been shown to indirectly tune neurotransmitter levels by interacting with neurotransmitter-producing host epithelial and immune cells and by modulating the composition of other neurotransmitter-regulating microbes in the gut.[20-23] These connections make the gut-brain axis an avenue for microbes to interface with the nervous system to correct neurological malfunctions and help the host cope with stressors.[24]
The natural qualities of probiotic and commensal microbes provide an excellent starting point for engineering microbes with new capabilities. Through synthetic biology, a wide array of new genetic parts may be introduced into these organisms for various applications. A common approach to engineering microbes for health-related goals is to simply express therapeutic proteins from constitutive promoters. These promoters are always active, independent of external stimuli. This approach has been applied for engineering microbes to treat or prevent various diseases and disorders, including hyperammonemia,[25] phenylketonuria,[26] diabetes,[27] AIDS,[28, 29] oral mucositis,[30] inflammatory diseases,[31, 32] obesity,[33] cancers,[34] and bacterial infections.[28]
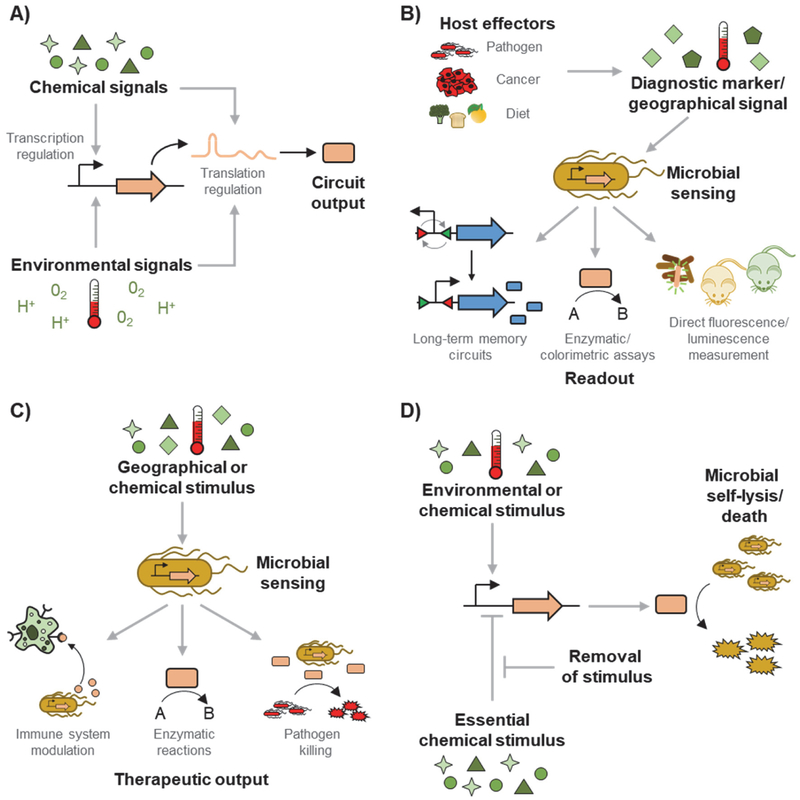
Although constitutively expressing proteins can be effective, a new class of engineered probiotics can be developed with biosensors. In contrast to constitutive protein expression, biosensor-regulated expression provides a means of intelligent control, where chemical cues (including those administered to the host and those naturally present in the body) and environmental cues (including oxygen level, pH, and temperature) determine when and where the probiotic produces the desired proteins (Figure 1A). There are several key advantages to controlling protein expression with biosensors. First, this expression method maximizes the genetic stability of the engineered cell. Heterologous protein expression burdens the cell, increasing the probability of enriching for mutations (e.g., mutations in promoters leading to no expression) that render the cells therapeutically non-functional.[35] Second, limiting protein production to a specified location in the body can minimize potential off-target effects of the proteins, including toxicity and tolerance buildup.[36] Third, the use of biosensors enables microbes to be engineered for diagnostic applications (Figure 1B) as well as therapeutic applications (Figure 1C). Biosensors also allow for the implementation of biocontainment genetic circuits that let the user control the viability of the engineered microbe (Figure 1D).
Figure 1: Chemical and environmental sensors allow microbes to be engineered for diagnostic, therapeutic, and biocontainment applications.
A) Chemical signals (including various sugars, host-produced metabolites, and synthetic compounds) and environmental signals (including oxygen level, pH, and temperature) can regulate the rate of protein production at the transcriptional and translational levels. B) Many host effectors, including pathogenic bacteria, cancers, and diets, affect the levels of various chemical and environmental signals in the host’s gut. Probiotic and commensal microbes can be engineered with sensors that measure and report on the levels of these stimuli. Example reporting methods include uses of memory circuits that maintain their state long-term for measurement outside the body, enzymatic or colorimetric assays that are correlated to the sensed levels, and direct in vivo (e.g., by imaging or electronic sensors) or ex vivo (e.g., using fecal samples) measurements of fluorescence or luminescence. C) Microbes can also be engineered to produce therapeutic outputs only when the microbes sense disease-relevant stimuli. The signals can include those naturally present in the target location, or ones externally administered to the host. Under the induced conditions, the microbe can produce therapeutic proteins for the treatment of diseases and infections. D) Chemical and environmental sensors can also be applied to microbial biocontainment Some circuits control cell viability by inducing cell death with the addition of a chemical or in response to environmental stimulus. With these circuits, the cell grows until the stimulus is applied. Other circuits instead initiate cell death when a chemical is removed. This growth-supporting chemical is added to the cultures when the cell is grown in vitro.
The biosensors needed for engineered probiotics can be obtained through part mining of native systems or development of synthetic regulators. Microbes naturally respond to a wide variety of external stimuli, many of which are found in the gut, to control their RNA and protein levels.[37] These natural responses can be leveraged for biosensing in microbes. Alternatively, synthetic protein and RNA sensors can be created through a variety of techniques. For example, novel protein sensors have been developed by fusing the ligand-binding and DNA-binding domains of different proteins[38] and by evolving natural sensors for improved response or altered ligand specificity.[39, 40] Similarly, chemical- or environmental condition-sensing RNA regulators have been created.[41, 42] The topic of biosensor development has been broadly reviewed[43-45], and many approaches are being applied to the development of biosensors in various non-model gut microbes.[46, 47]
The topic of engineered cells for medical applications has been reviewed in the past, with reviews broadly covering engineered microbes for medical applications,[48] focusing on developing engineered live therapeutics with increasing complexity,[49] discussing synthetic biology approaches to developing engineered bacterial and mammalian cells,[50] and focusing on applications of engineered microbes for combating pathogens,[51] treating cancers,[52] and developing biocontainment systems.[53] This review will focus on the application of “biosensing” for the development of smart designer probiotics engineered to sense the status and conditions of the host. Using this information, the probiotics can report on the health of the host (Section 2), respond by generating therapeutics (Section 3), or control their own viability for self-biocontainment (Section 4).
2. Biosensing for reporting on the health and conditions of the host
Many studies have utilized sensing modules in probiotics and commensals to generate living diagnostics (Table 1). One application of smart diagnostic microbes is the detection of gut inflammation. Riglar et al. modified the phage λ CI/Cro bistable switch developed by Kotula et al.[54] to detect tetrathionate, a compound associated with gut inflammation.[55] Specifically, they linked the expression of the cro memory element to the tetrathionate-sensitive, S. typhimurium-native two-component system (TCS) TtrSR and its cognate promoter PttrBCA. They engineered E. coli NGF-1, a strain capable of long-term colonization of the gut, to express a colorimetric enzymatic reporter from the memory switch. This system reliably reported the presence of tetrathionate in IL10−/− mice for up to six months after bacterial administration. Noting that the S. typhimurium PttrBCA promoter requires the oxygen-sensitive global regulator FNR for transcription, Daeffler et al. addressed this unwanted cross-regulation issue by adapting an alternative tetrathionate-responsive TCS (TtrSR-PttrB) from Shewanella baltica that is completely orthogonal to E. coli FNR.[56] They also derived a thiosulfate-sensitive TCS (ThsSR-PphsA) from Shewanella halifaxensis as thiosulfate is another compound associated with gut inflammation. Both TCSs were linked to a fluorescent reporter in E. coli Nissle 1917 (EcN), and they found that only the thiosulfate sensor activated expression of the reporter when exposed to the inflamed mouse gut. Mimee et al. utilized the same thiosulfate sensor in a novel microelectronic-based luminescence detection system, but the thiosulfate sensor was only demonstrated in vitro.[57] As an in vivo demonstration of their technology, they integrated the heme-responsive transcriptional repressor HrtR into EcN to detect bleeding in porcine models. To facilitate heme diffusion into the cell, they also expressed the heme transporter ChuA. The heme-sensing bacteria were loaded into a reservoir in an ingestible capsule with microelectronics capable of detecting the signal from the microbes’ luminescent reporter. This information was wirelessly transmitted outside the body, enabling a novel method of real-time diagnosis.
Table 1:
Bacteria engineered as diagnostics.
| Diagnostic application |
Detected compound or condition |
Sensor type |
Sensor genetic part | Circuit | Reporter | Strain | Ref. |
|---|---|---|---|---|---|---|---|
| Gut inflammation | Tetrathionate | TCS | Salmonella typhimurium TtrSR-PttrBCA | CI/Cro memory switch | β-galactosidase | E. coli NGF-1 | [55] |
| Gut inflammation | Tetrathionate | TCS | Shewanella baltica TtrSR-PttrB | NA | GFP | EcN | [56] |
| Gut inflammation | Thiosulfate | TCS | Shewanella halifaxensis ThsSR-PphsA | NA | GFP | EcN | [56] |
| Gut bleeding | Heme | T-TF | E. coli O157:H7 ChuA-L. lactis HrtR-PL(HrtO) | NA | luxCDABE combined with microelectronics | EcN | [57] |
| Cholera | Cholera autoinducer 1 (CAI-1) | TCS | Engineered chimera† | TetR (inverter) | β-lactamase | L. lactis | [4] |
| Cell division | aTc | TF | TetR-Ptet | CI/Cro memory switch‡ | β-galactosidase | E. coli NGF-1 | [59] |
| Cancer | Tumors colonized by engineered EcN | NA | NA | NA | β-galactosidase converting LuGal into luciferin | EcN | [65] |
| Inflammation | Nitrogen oxides | TF | E. coli NsrR-PYeaR | BxbI or TP901-9 recombinases | GFP | E. coli DH5αZ1 | [64] |
| Diabetes | Glucose | TCS | E. coli CpxAR-PcpxP | BxbI or TP901-9 recombinases | GFP | E. coli DH5αZ1 | [64] |
Abbreviations: AHL, N-acyl homoserine lactone; aTc, anhydrotetracycline; EcN, E. coli Nissle 1917; GFP, green fluorescent protein; TCS, two-component system, consisting of a histidine kinase and a response regulator; TF, transcription factor; T-TF, transporter-transcription factor; LuGal, a soluble conjugate of luciferin and galactose; NA, not applicable. If NA is present in the “Circuit” column, the sensor directly regulates expression of the reporter; otherwise, there is an additional circuit layer.
The TCS is an engineered chimera where the CAI-1-binding domain of V. cholerae CqsQ is fused to the signal transduction domain of Lactococcus lactis NisK. The chimeric histidine kinase interacts with the response regulator NisR.
Switching occurs only in actively dividing engineered E. coli.
Another diagnostic target is host infection. Several efforts have utilized biosensors to detect pathogenic bacteria in vitro,[57, 58] while others have advanced the systems to in vivo diagnostics. For example, Mao et al. engineered Lactococcus lactis to detect cholera infections in vivo.[4] They developed a library of chimeric TCSs with the cholera autoinducer 1 (CAI-1)-binding domain of Vibrio cholerae (Vc) CqsQ fused to the signal transduction domain of L. lactis NisK. The best-performing chimera successfully repressed the cognate promoter PNisR in response to CAI-1. Next, they linked the promoter to a TetR-based inversion module to create an inducible CAI-1-sensing circuit. Using a colorimetric enzymatic reporter, the engineered strain could inform of Vc infection in mice after being isolated from fecal matter. Certain et al. studied the dynamics of microbial infection by employing an inducible CI/Cro memory switch.[54, 59] They demonstrated that the inducer changed the memory state of the switch from OFF to ON only in actively dividing cells. Using this circuit, they sought to interrogate the replication state of memory switch-containing E. coli that had infected mice. Specifically, they exposed the bacterial cells to the inducer and to levofloxacin, an antibiotic that preferentially kills dividing bacteria. They discovered that while levofloxacin treatment reduced bacterial burden at the infection site, the proportion of actively dividing bacteria increased, contrary to the result from in vitro levofloxacin treatment.
Smart diagnostics have also been used to report on other physiological conditions and diseases. Takahashi et al. implemented paper-based platforms using E. coli lysates and toehold switches[60-62] to identify specific species of microbes in the gut microbiome.[63] Fluorescent reporters cis-repressed by toehold hairpin formation were trans-activated by species-specific RNAs from ten different microbes found in human microbiomes. By incorporating the toehold switch into a cell-free paper system, the authors demonstrated a low-cost method of analyzing and quantifying microbiome composition. Courbet et al. utilized whole-cell biosensors to detect clinically relevant biomarkers in urine and plasma samples, including nitrogen oxide for inflammation and glucose for diabetes.[64] Danino et al. relied on EcN’s proclivity to preferentially colonize cancerous masses to detect liver cancer from urine samples.[65] They engineered tumor-colonizing EcN to stably express β-galactosidase (LacZ), and upon tumor colonization, the engineered strain cleaved a luciferin-galactose conjugate using LacZ, releasing luciferin. A luminescence-based assay was then used to detect luciferin in urine.
3. Biosensing for smart expression of therapeutics
3.1. Cancer treatments
One promising application of microbial therapeutics is the treatment of cancer. Engineered cells specifically targeted to tumors can avoid the systemic toxicity of chemotherapeutic agents and enable repeated dosing of a therapeutic at the cancerous site. Many efforts to develop cancer therapies have involved engineering microbes that preferentially colonize hypoxic tumors to deliver constitutively expressed therapeutic proteins.[8, 66-69] Other efforts have focused on engineering the cell to recognize and target the acidic tumor microenvironment[70] or to preferentially bind to cancer cell surfaces.[71] However, these microbes relied on constitutive expression of the therapeutic. Here, we discuss smart therapies that utilize an additional layer of control over the expression of the therapeutic (Table 2A).
Table 2:
Biosensors coupled to therapeutic production.
| Detected compound or condition |
Sensor type |
Sensor genetic part |
Additional circuit component or mechanism of action |
Therapeutic | Therapeutic mechanism of action |
Strain | Ref. |
|---|---|---|---|---|---|---|---|
| A) Cancer: | |||||||
| Low oxygen | TF | E. coli FNR-Vitreoscilla Pvhb | NA | Tumstatin (Tum 5) | Anti-angiogenesis | EcN | [72] |
| Low oxygen | TF | E. coli FNR-Vitreoscilla Pvhb | NA | Tumstatin-p53 fusion | Anti-angiogenesis and cell cycle checkpointing | EcN | [73] |
| Low oxygen | TF | Engineered St FNR-Pfnr | NA | HlyE | Cell lysis | St | [74] |
| Arabinose* | TF | AraC-PBAD | NA | Vibrio vulnifcus FlaB | Immune cell recruitment | St | [75] |
| Cell density (AHL) | TF | LuxR-PluxR | PluxR-luxI, PluxR-φX174E† | HlyE, CCL21, and Bit1-iRGD chimera | Cell lysis and immune cell recruitment | St | [76] |
| B) Metabolic disorder: | |||||||
| Low oxygen | T-TF | PheP-FNR-PfnrS | FNR-PfnrS-pheP | Phenylalanine ammonia-lyase | Phenylalanine removal | EcN | [77] |
| Low oxygen | TF | FNR-PfnrS | NA | Feedback resistant-ArgA | Ammonia removal | EcN | [78] |
| C) Inflammatory bowel disorder/colitis: | |||||||
| Non-permissive condition | NA | NA | Self-lysis‡ | IL-1Ra | IL1-receptor antagonism | Bacillus subtilis | [79] |
| Xylan* | TF | Putative B. ovatus xylanase-inducible promoter | Bacteroides fragilis-derived peptide sequence-mediated secretion | TGF-β1 | Immune suppression | Bacteroides ovatus | [80] |
| D) Infection: | |||||||
| AHL (from PA) | TF | PA LasR-Plas | Plas-Lysin E7** | Pyocin S5 | Cell lysis | E. coli TOP10 | [81] |
| AHL (from PA) | TF | PA LasR-Plas | Plas-Lysin E7** | Pyocin S5 and Dispersin B | Cell lysis and anti-biofilm hydrolase | EcN | [82] |
| AHL (from PA) | TF | PA LasR-Plas | FlgM-mediated secretion | CoPy (Colicin E3-Pyocin S3 chimera) | RNase and cell lysis | E. coli MG1655 | [83] |
| AHL (from PA) | TF | PA LasR-Plas | Plas-cheZ-mediated chemotaxis and YebF-mediated secretion | Microcin S and DNaseI | Biofilm prevention and biofilm destruction | E. coli RP437 ΔcheZ | [84] |
| Cholera autoinducer 1 (CAI-1) | TCS | Vibrio cholerae CqsS-LuxU-LuxO-PtpQrr4 | PtpQrr4-gRNA, Pcon-dCas9, AraC-PBAD, and YebF-mediated secretion†† | Artilysin | Cell lysis | E. coli MG1655 | [85] |
| Pathogenicity and antibiotic resistance | TCS | Vibrio cholerae ToxRS-PompU | Vibrio cholerae SetR-PL-ccdA‡‡ | CcdB | DNA gyrase inhibition | E. coli β3194 | [86] |
| cCF10 | TF | Enterococcus faecalis PgrX-PpgrQ | NA | Enterocin A, Hiracin JM79, and Enterocin P | Cell lysis | L. lactis | [87] |
| Tetrathionate | TCS | Salmonella typhimurium TtRS-PttrBCA | NA | Microcin H47 | ATP synthase inhibition*** | EcN | [88] |
| HPA (from Candida albicans) | T-TF | E. coli HpaX-HpaA-PBC | NA | Burkholderia cenocepacia RpfF-synthesized cis-2-dodecenoic acid | Hypha formation inhibition | E. coli NGF-1 | [89] |
Abbreviations: AHL, N-acyl-homoserine lactone; EcN, E. coli Nissle 1917; HPA, hydroxyphenylacetic acid; PA, Pseudomonas aeruginosa; St, Salmonella typhimurium; TF, transcription factor; T-TF, transporter-transcription factor; NA, not applicable.
Exogenous inducer not directly related to disease state.
PluxR-luxI forms a positive feedback circuit where LuxI synthesizes AHL, promoting a buildup of the therapeutic protein. PluxR-φX174E forms a negative feedback loop, inducing self-lysis of the cell once the population reaches a certain threshold. The lysis enables release of the therapeutic protein. The combined positive-negative feedback results in a bacterial population that completes oscillatory cycles of therapeutic synthesis and lytic release.
Upon reaching the gut, B. subtilis natively senses unfavorable growth conditions and lyses, releasing the therapeutic protein of interest.
Lysin E7 expression induces cellular lysis to more effectively deliver the therapeutic protein to the pathogen.
The CRISPR interference circuit (PtpQrr4-gRNA and Pcon-dCas9) represses the arabinose-inducible promoter that control the expression of artilysin. Under high CAI-1, no gRNA is transcribed from PtpQrr4-gRNA, allowing for arabinose-inducible artilysin production. Localization to the periplasm by YebF causes artilysin to lyse the host cell (E. coli), enabling efficient delivery of artilysin to the pathogen.
In a pathogenic and antibiotic-resistant V. cholerae cell, SetR inhibits the expression of the CcdA antitoxin (by binding to the PL promoter) which prevents CcdB toxin-mediated killing. This therapy is dependent on whole-plasmid conjugation from an E. coli carrier strain to V. cholerae. After conjugation, CcdB toxin-mediated killing occurs only in pathogenic and antibiotic-resistant bacteria that harbor SetR (antibiotic resistance indicator) and ToxR (pathogenicity indicator).
Hypothesized mechanism of action.
The most common strategy to control cancer therapeutic expression is to express the proteins only when the cell is in the hypoxic tumor microenvironment. For example, He et al. controlled expression of an anti-angiogenesis tumstatin gene in EcN using the oxygen-dependent E. coli global regulator FNR and the Vitreoscilla Pvhb promoter.[72] He et al. later improved the therapeutic by fusing a p53 cell cycle checkpointing agent to tumstatin.[73] Ryan et al. placed the cytotoxin HlyE under the control of the S. typhimurium (St) oxygen-sensitive promoter PfnrS in aSt.[74] The cytotoxin was only expressed following colonization of the tumor microenvironment by aSt. Zheng et al. also took advantage of aSt’s ability to selectively colonize the hypoxic regions of tumors by expressing the Vibrio vulnificus FlaB flagellin gene under the control of an arabinose-inducible promoter.[75]. Upon colonization of the hypoxic tumor and supply of exogenous arabinose, the engineered strain significantly increased immune cell recruitment to the tumor site as compared to a non-engineered aSt control. Din et al. also engineered a quorum-sensing circuit in aSt to accumulate and release HlyE in the tumor.[76] Therapeutic protein expression was controlled by the N-acyl homoserine lactone (AHL)-sensitive transcription factor LuxR and its cognate promoter Plux. LuxR-Plux also controlled the expression of the AHL-synthesis protein LuxI in a positive feedback loop and cell lysis protein φX174E in a negative feedback loop. As the aSt cell density in the tumor increased, the cells synthesized increasing amounts of AHL, HlyE, and φX174E. Upon reaching a critical threshold, most cells lysed and released their therapeutic payload at the tumor site, while a few surviving cells began the cycle again. They also created engineered strains that replaced HlyE with a cytokine or apoptotic peptide and determined that a mixture of all three strains was most effective at preventing tumor growth. Finally, they demonstrated that the combination of both chemotherapy and a mixture of all three engineered strains significantly increased the survival of tumor-bearing mice relative to either therapy alone.
3.2. Metabolic and inflammatory disorder treatments
Efforts to treat metabolic disorders with biosensor-augmented engineered probiotics have relied on the detection of the anaerobic gut environment (Table 2B). For example, Isabella et al. constructed a strain of EcN to overexpress the phenylalanine transporter PheP and phenylalanine ammonia lyase (PAL) and tested its ability to reduce phenylalanine levels in an animal model for phenylketonuria.[77] In order to ensure therapeutic expression in the gut, both PheP and PAL were expressed using the E. coli oxygen sensitive promoter PfnrS. Kurtz et al. implemented the same strategy to treat hyperammonemia using EcN.[78] In this therapeutic, PfnrS controlled the expression of the E. coli enzyme N-acetylglutamate synthase, leading to improved consumption of free ammonia.
Gut inflammation is another promising target for smart engineered microbes (Table 2C). Porzio et al. relied on the inherent ability of Bacillus subtilis to lyse in the gut upon sensing the unfavorable growth conditions.[79] They engineered their strain to produce IL-1 receptor antagonist (IL-1Ra), and upon lysis in the gut, free IL-1Ra reduced symptoms of inflammation. Hamady et al. engineered human commensal Bacteroides ovatus to express human transforming growth factor beta 1 (TGF-β1) to treat a murine model of colitis.[80] They utilized the putative B. ovatus xylanase promoter to express TGF-β1 in the gut only in the presence of xylan, a dietary fiber, and incorporated an N-terminal Bacteroides fragilis-derived peptide secretion tag onto TGF-β1 to induce therapeutic release.
3.3. Infection treatments
Microbes can be engineered to sense and kill pathogens by exploiting genetic parts from the pathogen of interest (Table 2D). A commonly used sensor is the Pseudomonas aeruginosa (PA) transcription factor LasR that binds to the PA-specific AHL and induces expression from its cognate promoter, Plas. Saeidi et al. demonstrated that E. coli TOP10 engineered to express the bacteriocin Pyocin S5 from LasR-Plas could selectively kill PA in vitro.[81] They also placed the E7 lysis protein under control of Plas so that the engineered E. coli would lyse and more efficiently deliver its therapeutic payload in response to PA. In a follow-up work, Hwang et al. implemented the same circuit in EcN with the addition of the anti-biofilm enzyme Dispersin B also controlled by Plas.[82] They demonstrated that this engineered strain could work as a prophylactic and therapeutic in C. elegans and mouse infection models. Gupta et al. also targeted PA in vitro using its quorum sensing system to express the chimeric bacteriocin CoPy (Colicin E3-Pyocin S3) in E. coli MG1655.[83] To increase therapeutic efficiency, CoPy was secreted via FlgM. In a separate work, Hwang et al. engineered E. coli RP437 ΔcheZ to move towards and kill PA in vitro.[84] The expression of the chemotaxis protein CheZ, the bacteriocin Microcin S, and the anti-biofilm enzyme DNaseI were all controlled by LasR-Plas to enable movement towards and killing of PA. Microcin S and DNaseI were engineered to be secreted by YebF for extracellular delivery of the pathogen-killing agents.
V. cholerae (Vc) is another common target for pathogen-killing smart microbes. Jayaraman et al. built a CAI-1-responsive circuit in E. coli to kill Vc.[85] Because CAI-1 binding to CqsS represses the promoter PtpQrr4, a CRISPR interference circuit was used to instead induce the expression of the therapeutic lysis protein artilysin in response to CAI-1. Artilysin was also secreted via YebF to enable efficient delivery, inhibiting the growth of Vc in vitro. Mazel et al. designed a system to specifically kill only a pathogenic and antibiotic-resistant Vc cell using plasmid conjugation from E. coli β3194.[86] Upon plasmid conjugation, the TCS ToxRS (associated with pathogenicity) activates transcription of the intein-split toxin CcdB from the cognate promoter PompU. In an antibiotic resistant strain, the resistance-associated transcription factor SetR represses transcription of the CcdA antitoxin from the PL promoter. This application of Boolean AND logic to specifically kill only a pathogenic and antibiotic-resistant Vc cell was demonstrated in a zebrafish model.
Other pathogens have been targeted as well. Borrero et al. engineered L. lactis to detect the Enterococcus faecalis sex pheromone cCF10.[87] They adapted the cCF10-sensitive transcription factor PgrX to drive the expression of three bacteriocins from the promoter PpgrQ, thus killing multi-drug resistant E. faecalis in vitro. Palmer et al. targeted St with a strain of EcN designed to detect tetrathionate, which is associated with St infections in the gut.[88] They used the tetrathionate-responsive St TCS to control the expression of the bacteriocin Microcin H47, which inhibited St growth in vitro. Tscherner et al. utilized the hydroxyphenylacetic acid (HPA) transporter (HpaX), transcription factor (HpaA), and cognate promoter (PBC) to detect HPA, which is produced by the fungus Candida albicans.[89] Upon HPA detection, an enzyme was expressed to synthesize cis-2-dodecenoic acid which inhibits the formation of C. albicans hypha. They demonstrated hypha formation inhibition ex vivo, resulting in reduced filamentation, virulence factor expression, and epithelial damage.
4. Biosensing for biocontainment
While in the host or after released into the environment, both engineered and wild type microbes can evolve and exchange genes with other organisms.[90-93] This adaptation can lead to organisms acquiring competitive growth advantages that disrupt the ecosystem, acquiring metabolic traits that disturb the health of the gut, or developing pathogenic characteristics.[90, 94] These potential adaptations necessitate stringent controls for the biocontainment of engineered organisms used in medical applications. One common indicator for the efficiency of biocontainment methods is the escape frequency, or the ratio of cell counts in the killing condition to the non-killing condition. The NIH guidelines for the escape frequency of engineered organisms is a ratio of less than 10−8.[95] One common approach to achieve this goal with engineered microbes is auxotrophy, the removal of an essential metabolite-producing gene from the genome of the organism.[77, 78, 96] The deficient metabolite can be exogenously provided to maintain growth of the organism. A similar approach is to engineer the cell to be reliant on a nonstandard amino acid.[97, 98] Although these methods can be effective, the engineered strains require additional care or supplementation to grow in vitro and can escape through cross-feeding. A complementary approach includes the use of biosensors, where the organism controls its own viability in response to chemical or environmental signals.
4.1. Chemical sensing to control microbial cell viability
Numerous circuits for directly controlling cell viability with chemical biosensors have been developed for diverse organisms (Table 3A). One common method of controlling cell viability with chemical sensing is to link expression of an essential gene to a chemical-inducible promoter. Kong et al. expressed the essential genes asdA and murA in aSt using an arabinose-inducible promoter, providing a potential Salmonella vaccine strain with biocontainment circuits.[99] Similar circuits have also been developed for Saccharomyces cerevisiae.[100, 101] Agmon et al. used a galactose sensor to control the expression of many essential genes individually and demonstrated an escape frequency of less than 10−7 upon removal of galactose.[100] When Cai et al. added a second layer of control, which involved excising the expression cassette for an essential gene in response to estradiol, the escape frequency was below 10−10.[101]
Table 3:
Genetic circuits applied to the biocontainment of microbes.
| Strain | Controlling inducer | Killed by addition/removal or killing condition |
Log10 escape frequency |
Tested in vivo? |
Generations of stability |
Ref. |
|---|---|---|---|---|---|---|
| A) Chemical sensing: | ||||||
| Saccharomyces cerevisiae | Galactose | Removal | −7.7 | No | ND | [100] |
| Saccharomyces cerevisiae | Estradiol and galactose | Removal of galactose or addition of estradiol | −10 | No | ND | [101] |
| E. coli MC1000 | IPTG | Addition | −4.3 | No | ND | [102] |
| E. coli MG1655 | aTc and IPTG | Removal of aTc or addition of IPTG | −7 | No | ~14 | [103] |
| E. coli MG1655 | Cellobiose, galactose, and IPTG | Survival in only one condition out of the 8 (23) possible inducer combinations† | −8 | No | ~57 | [103] |
| E. coli MG1655 | Arabinose, aTc, and IPTG | Addition | −3 | No | ND | [104] |
| E. coli MG1655 | aTc, IPTG, and biotin (auxotroph) | Removal | −11.9* | No | 110 | [95] |
| E. coli MG1655 | Arabinose | Addition | −7.7 | No | 1700‡ | [105] |
| B) Quorum and environmental sensing: | ||||||
| E. coli TOP10 and MG1655 | AHL | Cell density decrease | NA | No | ND | [106] |
| E. coli NEB10β | Temperature | Temperature decrease | −4 ~ −3 | Yes | ND | [108] |
| E. coli MG1655 | Temperature | Temperature decrease | −5 | Yes | 140** | [109] |
Definitions: escape frequency, the ratio of colony forming units obtained for the strain when grown in the killing condition to the non-killing condition; generations of stability, the maximum number of cell generations before observing a significant increase in escape frequency. Abbreviations: AHL, N-acyl homoserine lactone; aTc, anhydrotetracycline; IPTG, isopropyl-β-D-1-thiogalactoside; NA, value is not applicable to the respective genetic circuit; ND, values were not determined or could not be approximated from the methods of the cited paper.
Killing ratio value was obtained when the genetic circuit was paired with auxotrophy.
The authors developed three genetic circuits where the cells survive only in the presence of inducers A and B but not C. Each of the three circuits had a different inducer, IPTG, galactose, or cellobiose, as inducer C.
Long-term stability was determined by applying the CRISPR-based circuit to targeted plasmid degradation rather than cell death caused by targeting the genome.
Long-term stability was tested in vitro.
Many chemical sensing circuits have also been developed for the biocontainment of various E. coli strains. One early biocontainment method for E. coli involved expressing the RelF toxin from an IPTG-inducible promoter.[102] To protect the system from inactivation by random mutations, the authors used two parallel RelF expression cassettes and demonstrated an escape frequency of 5×10−5. Chan et al. developed a genetic circuit, termed “Deadman” using an IPTG-inducible biosensor that paired toxin expression with inducible degradation of an essential protein.[103] In the presence of IPTG, both the EcoRI endonuclease and the mf-Lon protease were expressed. The protease then quickly degraded the tagged essential protein MurC as well as the LacI repressor to further induce the system. Cells with the circuit displayed an escape frequency of less than 10−7, but with poor long-term stability. They also developed an additional set of three-input circuits, termed “Passcode” where biosensors for IPTG, galactose, and cellobiose controlled the same killing mechanisms. Cells with the “Passcode” circuits could survive under only one of the eight possible input conditions. When the authors paired this circuit with an E. coli strain deficient in recombinogenic and mobile elements, they achieved an escape frequency of less than 10−8 with an improved long-term stability.
Riboregulator-based biocontainment has also been developed in E. coli. The first use of riboregulators for biocontainment was a proof-of-concept demonstration of cell lysis, with the expression of two cis-repressed phage genes being regulated by aTc.[104] The expression of the cognate trans-activating RNAs was regulated by arabinose and IPTG. Gallagher et al. used riboregulators to control the expression of two essential genes in parallel, making cell survival dependent on the presence of the IPTG and aTc inducers.[95] Additionally, they combined the riboregulator system with biotin auxotrophy and EcoRI expression. To control the activity of EcoRI, they expressed the EcoRI-inhibiting EcoRI methylase with an aTc sensor. Removal of aTc and IPTG from the culture repressed the expression of the essential genes and EcoRI methylase, allowing EcoRI to cleave the genome. This system achieved a field-best escape frequency of lower than 1.3×10−12.
Caliando and Voigt developed another biocontainment method using CRISPR-based genomic DNA degradation.[105] This system utilized CRISPR-Cas3 and CasABCDE to control the viability of E. coli. They integrated three arabinose-controlled Cas expression cassettes into the genome and constitutively expressed the cognate RNA on a plasmid. Using this system, the authors achieved an escape frequency of 1.9×10−8, nearly meeting the NIH criteria. While they did not demonstrate long-term stability of the genome-targeting system, they showed stable maintenance of a plasmid-cleaving system for 1700 generations.
4.2. Quorum and environmental sensing to control microbial cell viability
Several recent works have implemented biosensing to engineer microbes that control their own viability to prevent survival when released into the environment (Table 3B). To accomplish this goal, Huang et al. utilized the LuxR/LuxI quorum sensing system.[106] When the engineered E. coli cells sense high concentrations of AHL, signifying a high cell density, the cells produce the antibiotic resistance gene allowing them to survive in the presence of the respective antibiotic. However, when the cells leave the general population, the reduced concentration of AHL turns off expression of the antibiotic resistance gene, causing antibiotic-mediated cell death. Although this version of the system cannot be applied in vivo because of the antibiotic-based killing mechanism, it can be modified by applying quorum sensing to an alternative method of viability control, including cell lysis as demonstrated in vivo by Chowdhury et al.[107] Using a quorum-lysis system, Chowdhury et al. also demonstrated localized release of immunotherapeutics and an abscopal effect in a mouse tumor model.
Temperature sensing was also used to control bacterial survival in the environment after excretion from the body.[108, 109] When in the gut, microbes experience a relatively stable temperature near 37°C, but after excretion from the body, the cells usually experience a reduced temperature. Piraner et al. engineered E. coli to sense this temperature downshift using a mutant version of the St-native TlpA temperature sensor.[108] Native TlpA showed half-maximal expression from the PtlpA promoter at ~43.5°C, well above physiologically relevant temperatures, while the optimized mutant demonstrated half-maximal expression at ~36°C. To control cell viability in a temperature-dependent manner, they used the engineered temperature sensor to express the antitoxin CcdA of the CcdB-CcdA toxin-antitoxin system. At high temperatures (>36°C), CcdA is expressed, preventing cell death. At low temperatures, CcdA is no longer expressed, allowing constitutively expressed CcdB to kill the cell. They demonstrated an escape frequency of 10−4~10−3 using this system in mice. Alternatively, Stirling et al. used the E. coli-native PcspA temperature-sensing promoter, which is activated below ~30°C, to differentiate between conditions inside and outside of the gut.[109] In this system, the antitoxin CcdA was constitutively expressed, and the toxin CcdB was expressed by PcspA. They demonstrated that the system can maintain its efficiency for 140 generations in vitro. They also tested the system in vivo, achieving an escape frequency of less than 10−5. These reports have displayed valuable proof-of-concept circuits for environmental biocontainment, with improvements still needed to achieve the NIH recommended escape frequency.
5. Conclusions
Incorporating biosensing into engineered probiotics has provided significant advances in live diagnostics and therapeutics. However, there is still vast potential for improvements and new directions. The synthetic biology technologies to mine and screen for native sensing capabilities of microbes and to design and build novel synthetic sensors will provide a boundless collection of biosensors. Significant advances continue to be made for in vitro biosensing, and many sensors for application-relevant environmental conditions and chemicals have yet to be demonstrated in vivo.[42, 110-114] Importantly, the in vitro development of a sensor does not guarantee successful implementation in the complex gut environment. To be useful in the gut, sensors need to be tuned to respond to physiologically relevant chemical concentrations and conditions and to provide an effective therapeutic level upon activation without imposing a high metabolic burden on the cell.
The throughput of in vitro sensor development is orders of magnitude higher than the throughput of in vivo functionality testing, limiting the rate of applying microbial sensors to health-related applications. To improve the process of selecting strains for in vivo testing, new methods will need to be used that mimic the gut environment. For example, 3-D intestinal scaffolds have been developed that mimic the crypt-villus axis of the small intestine.[115] As a proof-of-principle, this technology was applied to study both the adhesion of common pathogens to the intestine and the efficiency at which different probiotic strains displace said pathogens. In addition, co-culture techniques with intestine-mimicking media can be used to simulate the complex microbial communities of the gut and to improve the relevance of the microbes’ metabolic state.[116] Microfluidic “intestine-on-a-chip” devices have also been developed that foster microenvironments with gut-relevant oxygen gradients, chemical diffusion rates, differential pH regions, microbial communities, and living human intestinal epithelial cells.[117, 118] These technologies can be applied to improve the development of many engineered probiotics.
As the screening rate for engineered microbes improves with in vivo-simulating devices, microbes can be engineered with increasing complexity, stability, and safety. Currently, microbes are being engineered either as diagnostics or as therapeutics (but not as both), often for the same issues such as cancer[65, 74] and inflammation.[55, 56, 79, 80] However, the tools exist to engineer a single microbe with genetic circuits that perform both diagnostic and therapeutic functions simultaneously, to sense the ailment, report on the issue, and solve the problem. Similarly, microbes can be engineered to sense multiple gut-relevant conditions and/or to perform multiple therapeutic functions in parallel, as multiple ailments often accompany each other (e.g., pathogenic infection and gut inflammation).[119] To increase specificity, however, new sensors need to be developed and integrated into multi-input circuits (e.g., AND gates). For example, while oxygen-dependent sensors have been shown to be useful for targeted delivery of therapeutics to both hypoxic tumor cells (Section 3.1) and the anaerobic gut (Section 3.2), better gene circuits would respond to cancer-specific molecules and disease-specific chemicals, respectively, in addition to low oxygen levels. Importantly for each case, biocontainment circuits and technologies should be incorporated into the probiotics to prevent release into the environment.[120] However, increasing the complexity of genetic circuits also increases the burden on the cell. The generation of loss-of-function mutations can enrich for non-functional cells that outcompete the functional microbes. Methods need to be developed and implemented that reduce this burden and increase the genetic stability.[121, 122]
Engineered probiotics have the potential to drastically improve the care of patients with difficult-to-diagnose and -treat disorders and conditions. However, engineering living organisms involves unique potentials and risks as discussed, and many factors must be carefully considered and studied, including (1) in vivo sensor sensitivity, selectivity, and robustness under fluctuating environmental conditions, (2) the effect of many variables such as diet, microbiome composition, and animal host cells on the performance of engineered microbes, (3) mutational stability of engineered circuits in vivo, and (4) biosafety measures. The recent advancements of engineered microbes in clinical trials is providing a valuable precedent for applying synthetic biology to solving health problems.[123] As this progress continues to be made, clinical data can guide the construction of future engineered microbial diagnostics and therapeutics. New problems will continue to be solved with engineered probiotics, and current issues will be tackled with improved efficiency and efficacy.
Acknowledgement
This work was supported by the Office of Naval Research (N00014-17-1-2611 to T.S.M.), the National Institutes of Health (1R01AT009741-01 to T.S.M.), and the National Science Foundation Graduate Research Fellowship Program (DGE-1745038 to M.B.A.).
Abbreviations:
- AHL
N-acyl homoserine lactone
- aSt
attenuated Salmonella typhimurium
- aTc
anhydrotetracycline
- EcN
Escherichia coli Nissle 1917
- HPA
hydroxyphenylacetic acid
- IPTG
Isopropyl-β-D-1-thiogalactoside
- PA
Pseudomonas aeruginosa
- PAL
phenylalanine ammonia lyase
- St
Salmonella typhimurium
- TCS
two component system
- TGF-β1
transforming growth factor beta 1
- Vc
Vibrio cholerae
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
6. References
- [1].Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, et al. , Health benefits of probiotics: a review. ISRN Nutr 2013, 2013, 481651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Munoz-Quezada S, Bermudez-Brito M, Chenoll E, Genoves S, et al. , Competitive inhibition of three novel bacteria isolated from faeces of breast milk-fed infants against selected enteropathogens. Br J Nutr 2013, 109 Suppl 2, S63–69. [DOI] [PubMed] [Google Scholar]
- [3].Kumar M, Dhaka P, Vijay D, Vergis J, et al. , Antimicrobial effects of Lactobacillus plantarum and Lactobacillus acidophilus against multidrug-resistant enteroaggregative Escherichia coli. Int J Antimicrob Agents 2016, 48, 265–270. [DOI] [PubMed] [Google Scholar]
- [4].Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ, Probiotic strains detect and suppress cholera in mice. Sci Transl Med 2018, 10, eaao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Piewngam P, Zheng Y, Nguyen TH, Dickey SW, et al. , Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Collado Mc GMSS, Bioactive Foods in Promoting Health: Probiotics and Prebiotics, Academic Press, Elsevier, London: 2010. [Google Scholar]
- [7].Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B, Combination bacteriolytic therapy for the treatment of experimental tumors. Proc Natl Acad Sci U S A 2001, 98, 15155–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Agrawal N, Bettegowda C, Cheong I, Geschwind JF, et al. , Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc Natl Acad Sci U S A 2004, 101, 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bettegowda C, Dang LH, Abrams R, Huso DL, et al. , Overcoming the hypoxic barrier to radiation therapy with anaerobic bacteria. Proc Natl Acad Sci U S A 2003, 100, 15083–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kimura NT, Taniguchi S, Aoki K, Baba T, Selective localization and growth of Bifidobacterium bifidum in mouse tumors following intravenous administration. Cancer Res 1980, 40, 2061–2068. [PubMed] [Google Scholar]
- [11].Stritzker J, Weibel S, Hill PJ, Oelschlaeger TA, et al. , Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol 2007, 297, 151–162. [DOI] [PubMed] [Google Scholar]
- [12].Low KB, Ittensohn M, Le T, Platt J, et al. , Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol 1999, 17, 37–41. [DOI] [PubMed] [Google Scholar]
- [13].Avogadri F, Martinoli C, Petrovska L, Chiodoni C, et al. , Cancer immunotherapy based on killing of Salmonella-infected tumor cells. Cancer Res 2005, 65, 3920–3927. [DOI] [PubMed] [Google Scholar]
- [14].Allen SJ, Martinez EG, Gregorio GV, Dans LF, Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev 2010, CD003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Berni Canani R, Paparo L, Nocerino R, Di Scala C, et al. , Gut Microbiome as Target for Innovative Strategies Against Food Allergy. Front Immunol 2019, 10, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schultz M, Clinical use of E coli Nissle 1917 in inflammatory bowel disease. Inflammatory bowel diseases 2008, 14, 1012–1018. [DOI] [PubMed] [Google Scholar]
- [17].Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, et al. , Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine. Cell Host & Microbe 2014, 16, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hata T, Asano Y, Yoshihara K, Kimura-Todani T, et al. , Regulation of gut luminal serotonin by commensal microbiota in mice. PLOS ONE 2017, 12, e0180745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Asano Y, Hiramoto T, Nishino R, Aiba Y, et al. , Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. American journal of physiology. Gastrointestinal and liver physiology 2012, 303, G1288–1295. [DOI] [PubMed] [Google Scholar]
- [20].Nzakizwanayo J, Dedi C, Standen G, Macfarlane WM, et al. , Escherichia coli Nissle 1917 enhances bioavailability of serotonin in gut tissues through modulation of synthesis and clearance. Scientific Reports 2015, 5, 17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yano JM, Yu K, Donaldson GP, Shastri GG, et al. , Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Strandwitz P, Kim KH, Terekhova D, Liu JK, et al. , GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 2019, 4, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].El Aidy S, Dinan TG, Cryan JF, Immune modulation of the brain-gut-microbe axis. Front Microbiol 2014, 5, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang H, Braun C, Murphy EF, Enck P, Bifidobacterium longum 1714™ Strain Modulates Brain Activity of Healthy Volunteers During Social Stress. The American Journal of Gastroenterology 2019, 114, 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nicaise C, Prozzi D, Viaene E, Moreno C, et al. , Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology 2008, 48, 1184–1192. [DOI] [PubMed] [Google Scholar]
- [26].Durrer KE, Allen MS, Herbing I. H. v., Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU. PLOS ONE 2017, 12, e0176286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Duan FF, Liu JH, March JC, Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 2015, 64, 1794–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lagenaur LA, Sanders-Beer BE, Brichacek B, Pal R, et al. , Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol 2011, 4, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu X, Lagenaur LA, Simpson DA, Essenmacher KP, et al. , Engineered vaginal lactobacillus strain for mucosal delivery of the human immunodeficiency virus inhibitor cyanovirin-N. Antimicrobial agents and chemotherapy 2006, 50, 3250–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, et al. , AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol 2010, 46, 564–570. [DOI] [PubMed] [Google Scholar]
- [31].Steidler L, Hans W, Schotte L, Neirynck S, et al. , Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. [DOI] [PubMed] [Google Scholar]
- [32].Marinho FA, Pacifico LG, Miyoshi A, Azevedo V, et al. , An intranasal administration of Lactococcus lactis strains expressing recombinant interleukin-10 modulates acute allergic airway inflammation in a murine model. Clin Exp Allergy 2010, 40, 1541–1551. [DOI] [PubMed] [Google Scholar]
- [33].Chen Z, Guo L, Zhang Y, Walzem RL, et al. , Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest 2014, 124, 3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xiang S, Fruehauf J, Li CJ, Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology 2006, 24, 697–702. [DOI] [PubMed] [Google Scholar]
- [35].Lynch M, Marinov GK, The bioenergetic costs of a gene. Proc Natl Acad Sci U S A 2015, 112, 15690–15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ryan RM, Green J, Lewis CE, Use of bacteria in anti-cancer therapies. Bioessays 2006, 28, 84–94. [DOI] [PubMed] [Google Scholar]
- [37].Naydich AD, Nangle SN, Bues JJ, Trivedi D, et al. , Synthetic Gene Circuits Enable Systems-Level Biosensor Trigger Discovery at the Host-Microbe Interface. mSystems 2019, 4, e00125–00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Oakes BL, Nadler DC, Flamholz A, Fellmann C, et al. , Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol 2016, 34, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, Voigt CA, Escherichia coli "Marionette" strains with 12 highly optimized small-molecule sensors. Nat Chem Biol 2019, 15, 196–204. [DOI] [PubMed] [Google Scholar]
- [40].Ellefson JW, Ledbetter MP, Ellington AD, Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nature chemical biology 2018, 14, 361–367. [DOI] [PubMed] [Google Scholar]
- [41].Espah Borujeni A, Mishler DM, Wang J, Huso W, Salis HM, Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res 2016, 44, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hoynes-O'Connor A, Hinman K, Kirchner L, Moon TS, De novo design of heat-repressible RNA thermosensors in E. coli. Nucleic Acids Res 2015, 43, 6166–6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang J, Jensen MK, Keasling JD, Development of biosensors and their application in metabolic engineering. Curr Opin Chem Biol 2015, 28, 1–8. [DOI] [PubMed] [Google Scholar]
- [44].Dekker L, Polizzi KM, Sense and sensitivity in bioprocessing-detecting cellular metabolites with biosensors. Curr Opin Chem Biol 2017, 40, 31–36. [DOI] [PubMed] [Google Scholar]
- [45].De Paepe B, Peters G, Coussement P, Maertens J, De Mey M, Tailor-made transcriptional biosensors for optimizing microbial cell factories. J Ind Microbiol Biotechnol 2017, 44, 623–645. [DOI] [PubMed] [Google Scholar]
- [46].Mimee M, Tucker AC, Voigt CA, Lu TK, Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Systems 2015, 1, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lim B, Zimmermann M, Barry NA, Goodman AL, Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 2017, 169, 547–558 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Riglar DT, Silver PA, Engineering bacteria for diagnostic and therapeutic applications. Nature Reviews Microbiology 2018, 16, 214–225. [DOI] [PubMed] [Google Scholar]
- [49].Ozdemir T, Fedorec AJH, Danino T, Barnes CP, Synthetic Biology and Engineered Live Biotherapeutics: Toward Increasing System Complexity. Cell Systems 2018, 7, 5–16. [DOI] [PubMed] [Google Scholar]
- [50].Sedlmayer F, Aubel D, Fussenegger M, Synthetic gene circuits for the detection, elimination and prevention of disease. Nature Biomedical Engineering 2018, 2, 399–415. [DOI] [PubMed] [Google Scholar]
- [51].Hwang IY, Lee HL, Huang JG, Lim YY, et al. , Engineering microbes for targeted strikes against human pathogens. Cell. Mol. Life Sci 2018, 75, 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wu MR, Jusiak B, Lu TK, Engineering advanced cancer therapies with synthetic biology. Nat Rev Cancer 2019, 19, 187–195. [DOI] [PubMed] [Google Scholar]
- [53].Lee JW, Chan CTY, Slomovic S, Collins JJ, Next-generation biocontainment systems for engineered organisms. Nature Chemical Biology 2018, 14, 530–537. [DOI] [PubMed] [Google Scholar]
- [54].Kotula JW, Kerns SJ, Shaket LA, Siraj L, et al. , Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A 2014, 111, 4838–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Riglar DT, Giessen TW, Baym M, Kerns SJ, et al. , Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nature Biotechnology 2017, 35, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Daeffler KNM, Galley JD, Sheth RU, Ortiz - Velez LC, et al. , Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Molecular Systems Biology 2017, 13, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mimee M, Nadeau P, Hayward A, Carim S, et al. , An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 2018, 360, 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lubkowicz D, Ho CL, Hwang IY, Yew WS, et al. , Reprogramming Probiotic Lactobacillus reuteri as a Biosensor for Staphylococcus aureus Derived AIP-I Detection. ACS Synth. Biol 2018, 7, 1229–1237. [DOI] [PubMed] [Google Scholar]
- [59].Certain LK, Way JC, Pezone MJ, Collins JJ, Using Engineered Bacteria to Characterize Infection Dynamics and Antibiotic Effects In Vivo. Cell Host Microbe 2017, 22, 263–268 e264. [DOI] [PubMed] [Google Scholar]
- [60].Green AA, Silver PA, Collins JJ, Yin P, Toehold switches: de-novo-designed regulators of gene expression. Cell 2014, 159, 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Green AA, Kim J, Ma D, Silver PA, et al. , Complex cellular logic computation using ribocomputing devices. Nature 2017, 548, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kim S-J, Leong M, Amrofell MB, Lee YJ, Moon TS, Modulating Responses of Toehold Switches by an Inhibitory Hairpin. ACS Synthetic Biology 2019, 8, 601–605. [DOI] [PubMed] [Google Scholar]
- [63].Takahashi MK, Tan X, Dy AJ, Braff D, et al. , A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun 2018, 9, 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Courbet A, Endy D, Renard E, Molina F, Bonnet J, Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci Transl Med 2015, 7, 289ra283. [DOI] [PubMed] [Google Scholar]
- [65].Danino T, Prindle A, Kwong GA, Skalak M, et al. , Programmable probiotics for detection of cancer in urine. Science Translational Medicine 2015, 7, 289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Theys J, Pennington O, Dubois L, Anlezark G, et al. , Repeated cycles of Clostridium-directed enzyme prodrug therapy result in sustained antitumour effects in vivo. Br J Cancer 2006, 95, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fu W, Lan H, Liang S, Gao T, Ren D, Suicide gene/prodrug therapy using salmonella-mediated delivery of Escherichia coli purine nucleoside phosphorylase gene and 6-methoxypurine 2'-deoxyriboside in murine mammary carcinoma 4T1 model. Cancer Sci 2008, 99, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sasaki T, Fujimori M, Hamaji Y, Hama Y, et al. , Genetically engineered Bifidobacterium longum for tumor-targeting enzyme-prodrug therapy of autochthonous mammary tumors in rats. Cancer Sci 2006, 97, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu YZ, Youming Z, Liqiu X, Xiangli Z, et al. , Escherichia coli Nissle 1917 Targets and Restrains Mouse B16 Melanoma and 4T1 Breast Tumors through Expression of Azurin Protein. Appl Environ Microbiol 2012, 78, 7603–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang Y, Ji W, He L, Chen Y, et al. , E. coli Nissle 1917-Derived Minicells for Targeted Delivery of Chemotherapeutic Drug to Hypoxic Regions for Cancer Therapy. Theranostics 2018, 8, 1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ho CL, Tan HQ, Chua KJ, Kang A, et al. , Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nature Biomedical Engineering 2018, 2, 27. [DOI] [PubMed] [Google Scholar]
- [72].He L, Yang H, Liu F, Chen Y, et al. , Escherichia coli Nissle 1917 engineered to express Tum-5 can restrain murine melanoma growth, Oncotarget 2017, pp. 85772–85782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].He L, Yang H, Tang J, Liu Z, et al. , Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum-5 to solid tumors for cancer therapy. Journal of Biological Engineering 2019, 13, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ryan RM, Green J, Williams PJ, Tazzyman S, et al. , Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther 2009, 16, 329–339. [DOI] [PubMed] [Google Scholar]
- [75].Zheng JH, Nguyen VH, Jiang SN, Park SH, et al. , Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med 2017, 9, eaak9537. [DOI] [PubMed] [Google Scholar]
- [76].Din MO, Danino T, Prindle A, Skalak M, et al. , Synchronized cycles of bacterial lysis for in vivo delivery. Nature 2016, 536, 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, et al. , Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nature Biotechnology 2018, 36, 857–864. [DOI] [PubMed] [Google Scholar]
- [78].Kurtz CB, Millet YA, Puurunen MK, Perreault M, et al. , An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Science Translational Medicine 2019, 11, eaau7975. [DOI] [PubMed] [Google Scholar]
- [79].Porzio S, Bossu P, Ruggiero P, Boraschi D, Tagliabue A, Mucosal delivery of anti-inflammatory IL-1Ra by sporulating recombinant bacteria. BMC Biotechnol 2004, 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hamady ZZ, Scott N, Farrar MD, Wadhwa M, et al. , Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-beta1 under the control of dietary xylan 1. Inflammatory bowel diseases 2011, 17, 1925–1935. [DOI] [PubMed] [Google Scholar]
- [81].Saeidi N, Wong CK, Lo T-M, Nguyen HX, et al. , Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology 2011, 7, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hwang IY, Koh E, Wong A, March JC, et al. , Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nature communications 2017, 8, 15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gupta S, Bram EE, Weiss R, Genetically Programmable Pathogen Sense and Destroy. ACS Synth. Biol 2013, 2, 715–723. [DOI] [PubMed] [Google Scholar]
- [84].Hwang IY, Tan MH, Koh E, Ho CL, et al. , Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol 2014, 3, 228–237. [DOI] [PubMed] [Google Scholar]
- [85].Jayaraman P, Holowko MB, Yeoh JW, Lim S, Poh CL, Repurposing a Two-Component System-Based Biosensor for the Killing of Vibrio cholerae. ACS Synth Biol 2017, 6, 1403–1415. [DOI] [PubMed] [Google Scholar]
- [86].Mazel RL-I, Joaquín B-B, Alfonso R-P, Jean-Marc G, Didier, Engineered toxin–intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nature Biotechnology 2019, 37, 755–760. [DOI] [PubMed] [Google Scholar]
- [87].Borrero J, Chen Y, Dunny GM, Kaznessis YN, Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth Biol 2015, 4, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Palmer JD, Piattelli E, McCormick BA, Silby MW, et al. , Engineered Probiotic for the Inhibition of Salmonella via Tetrathionate-Induced Production of Microcin H47. ACS Infectious Diseases 2018, 4, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Tscherner M, Giessen TW, Markey L, Kumamoto CA, Silver PA, A Synthetic System That Senses Candida albicans and Inhibits Virulence Factors. ACS Synth Biol 2019, 8, 434–444. [DOI] [PubMed] [Google Scholar]
- [90].Crook N, Ferreiro A, Gasparrini AJ, Pesesky MW, et al. , Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut. Cell Host & Microbe 2019, 25, 499–512.e498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang CM, Ravi US, Daniel EF, Harris H, Real-time capture of horizontal gene transfers from gut microbiota by engineered CRISPR-Cas acquisition. bioRxiv 2018. [Google Scholar]
- [92].Zhao S, Lieberman TD, Poyet M, Kauffman KM, et al. , Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host Microbe 2019, 25, 656–667 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lieberman TD, Flett KB, Yelin I, Martin TR, et al. , Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nature genetics 2014, 46, 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Miskinyte M, Sousa A, Ramiro RS, de Sousa JA, et al. , The genetic basis of Escherichia coli pathoadaptation to macrophages. PLoS Pathog 2013, 9, e1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gallagher RR, Patel JR, Interiano AL, Rovner AJ, Isaacs FJ, Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res 2015, 43, 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Steidler L, Neirynck S, Huyghebaert N, Snoeck V, et al. , Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol 2003, 21, 785–789. [DOI] [PubMed] [Google Scholar]
- [97].Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, et al. , Biocontainment of genetically modified organisms by synthetic protein design. Nature 2015, 518, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rovner AJ, Haimovich AD, Katz SR, Li Z, et al. , Recoded organisms engineered to depend on synthetic amino acids. Nature 2015, 518, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kong W, Wanda SY, Zhang X, Bollen W, et al. , Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc Natl Acad Sci U S A 2008, 105, 9361–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Agmon N, Tang Z, Yang K, Sutter B, et al. , Low escape-rate genome safeguards with minimal molecular perturbation of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2017, 114, E1470–E1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Cai Y, Agmon N, Choi WJ, Ubide A, et al. , Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proc Natl Acad Sci U S A 2015, 112, 1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Knudsen SM, Karlstrom OH, Development of efficient suicide mechanisms for biological containment of bacteria. Appl Environ Microbiol 1991, 57, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ, 'Deadman' and 'Passcode' microbial kill switches for bacterial containment. Nature Chemical Biology 2016, 12, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ, Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci U S A 2010, 107, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Caliando BJ, Voigt CA, Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun 2015, 6, 6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Huang S, Lee AJ, Tsoi R, Wu F, et al. , Coupling spatial segregation with synthetic circuits to control bacterial survival. Molecular Systems Biology 2016, 12, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Chowdhury S, Castro S, Coker C, Hinchliffe TE, et al. , Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med 2019, 25, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A, Shapiro MG, Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nature Chemical Biology 2017, 13, 75–80. [DOI] [PubMed] [Google Scholar]
- [109].Stirling F, Bitzan L, O’Keefe S, Redfield E, et al. , Rational Design of Evolutionarily Stable Microbial Kill Switches. Molecular cell 2017, 68, 686–697.e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ogasawara H, Hasegawa A, Kanda E, Miki T, et al. , Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol 2007, 189, 4791–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bina XR, Howard MF, Taylor-Mulneix DL, Ante VM, et al. , The Vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog 2018, 14, e1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lee SK, Keasling JD, A propionate-inducible expression system for enteric bacteria. Appl Environ Microbiol 2005, 71, 6856–6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sberro H, Fremin BJ, Zlitni S, Edfors F, et al. , Large-Scale Analyses of Human Microbiomes Reveal Thousands of Small, Novel Genes. Cell 2019, 178, 1245–1259 e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].DeLorenzo DM, Moon TS, Construction of Genetic Logic Gates Based on the T7 RNA Polymerase Expression System in Rhodococcus opacus PD630. ACS Synth Biol 2019, 8, 1921–1930. [DOI] [PubMed] [Google Scholar]
- [115].Costello CM, Sorna RM, Goh YL, Cengic I, et al. , 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol Pharm 2014, 11, 2030–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Villageliu DN, Rasmussen S, Lyte M, A microbial endocrinology-based simulated small intestinal medium for the evaluation of neurochemical production by gut microbiota. FEMS Microbiol Ecol 2018, 94. [DOI] [PubMed] [Google Scholar]
- [117].Walsh CL, Babin BM, Kasinskas RW, Foster JA, et al. , A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip 2009, 9, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, et al. , A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature Biomedical Engineering 2019, 3, 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hoynes - O'Connor A, Shopera T, Hinman K, Creamer JP, Moon TS, Enabling complex genetic circuits to respond to extrinsic environmental signals. Biotechnology and Bioengineering 2017, 114, 1626–1631. [DOI] [PubMed] [Google Scholar]
- [120].Cao Z, Cheng S, Wang X, Pang Y, Liu J, Camouflaging bacteria by wrapping with cell membranes. Nat Commun 2019, 10, 3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Deatherage DE, Leon D, Rodriguez AE, Omar SK, Barrick JE, Directed evolution of Escherichia coli with lower-than-natural plasmid mutation rates. Nucleic Acids Res 2018, 46, 9236–9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ceroni F, Algar R, Stan GB, Ellis T, Quantifying cellular capacity identifies gene expression designs with reduced burden. Nature methods 2015, 12, 415–418. [DOI] [PubMed] [Google Scholar]
- [123].Vargason AM, Anselmo AC, Clinical translation of microbe-based therapies: Current clinical landscape and preclinical outlook. Bioeng Transl Med 2018, 3, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]