Abstract
Immunotherapy, designed to exploit the functions of the host immune system against tumors, has shown considerable potential against several malignancies. However, the utility of immunotherapy is heavily limited due to the low response rate and various side effects in the clinical setting. Immune escape of tumor cells may be a critical reason for such low response rates. Noncoding RNAs (ncRNAs) have been identified as key regulatory factors in tumors and the immune system. Consequently, ncRNAs show promise as targets to improve the efficacy of immunotherapy in tumors. However, the relationship between ncRNAs and tumor immune escape (TIE) has not yet been comprehensively summarized. In this review, we provide a detailed account of the current knowledge on ncRNAs associated with TIE and their potential roles in tumor growth and survival mechanisms. This review bridges the gap between ncRNAs and TIE and broadens our understanding of their relationship, providing new insights and strategies to improve immunotherapy response rates by specifically targeting the ncRNAs involved in TIE.
Subject terms: Tumour immunology, Non-coding RNAs
Introduction
According to the immunoediting theory, immune escape is the key to tumor survival.1 There are many mechanisms of tumor immune escape (TIE), including defects in tumor antigen presentation to escape recognition by the immune system, alterations in the tumor death pathways to achieve increased resistance to cytotoxic immune responses, metabolic alterations to promote TIE, and acquisition of stem cell-like phenotypes to escape immune-based recognition and destruction. In addition, some cytokines in the tumor microenvironment (TME), abnormal expression of immune checkpoint molecules on tumor or immune cell surfaces, and some immunosuppressive cells are all involved in TIE. Collectively, these factors may enable TIE, leading to a low response rate to immunotherapy in different malignancies.
Noncoding RNAs (ncRNAs), which cannot be translated into proteins, comprise 98% of the transcriptome. Generally, ncRNAs less than 50 nucleotides in length are defined as small ncRNAs (sncRNAs), including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small interfering RNAs (siRNAs).2 A recent study reported the presence of partial sncRNAs derived from tRNAs, such as tRNA halves (tiRNAs) and tRNA fragments (tRFs).3 ncRNAs with more than 200 nucleotides are defined as long ncRNAs (lncRNAs), including long or large intergenic ncRNAs (lincRNAs), some circular RNAs (circRNAs), and ribosomal RNAs (rRNAs).4 The biological functions of ncRNAs, such as regulating gene expression at the transcriptional and translational levels, guiding DNA synthesis or gene rearrangement, and protecting the genome from foreign nucleic acids, have been gradually elucidated.5 An increasing number of studies indicate that ncRNAs are indispensable in tumorigenesis by regulating the expression of tumor-related genes. Mechanistically, lncRNAs regulate gene expression mainly by acting as transcription factors, regulating chromatin remodeling, or participating in posttranscriptional regulation as ceRNAs.6 circRNAs can regulate gene expression at epigenetic, transcriptional, and posttranscriptional levels (primarily as ceRNAs).7 miRNAs mostly regulate gene expression at the posttranscriptional level through RNA interference by binding to the 3′-untranslated region (3′UTR) (rarely 5′UTR or coding sequence) of protein-coding mRNAs.8–10 In addition, some TRFs and tiRNAs can participate in gene regulation and gene silencing via complementary binding with target genes, and the mechanism is similar to that of miRNA.11
Currently, ncRNAs involved in TIE are gradually emerging and are promising potential targets of antitumor therapy. Several studies have reported that ncRNAs play pivotal roles in TIE.12,13 Therefore, it is essential to systematically elaborate the complex regulatory network of TIE regulated by ncRNAs. In this review, we provide a detailed account of the molecular regulatory mechanisms underlying ncRNA involvement in TIE. We hope this review will broaden our understanding of the relationship between ncRNAs and TIE and provide new insights to target ncRNAs in TIE-associated therapeutic strategies.
Defective antigen presentation and TIE
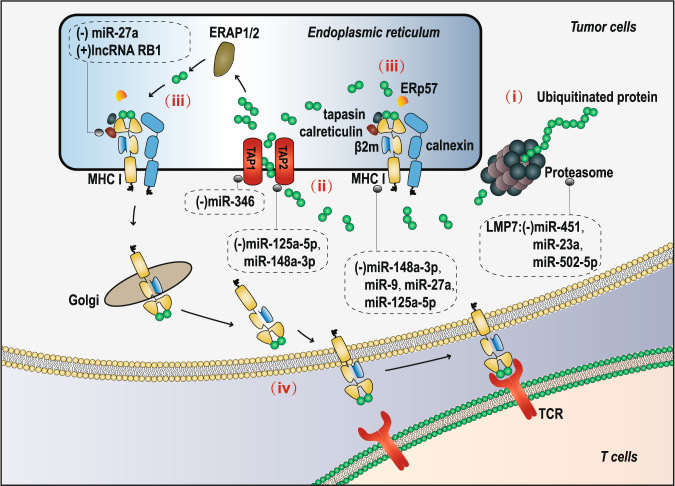
Tumor cells can achieve TIE by inhibiting antigen presentation, which enables T cells and/or natural killer (NK) cells to recognize and destroy target tumor cells. The regulation of the antigen processing and presenting machinery (APM) in tumor cells is dependent on the ubiquitin-protease system and the major histocompatibility complex (MHC) class Ι molecules. The mediation of the APM by MHC class Ι molecules is divided into four steps: (i) peptide generation and modification; (ii) peptide transport; (iii) assembly of the peptide-MHC class I complex; and (iv) antigen presentation (Fig. 1). ncRNAs have been reported to regulate the APM in tumors during these four steps. First, peptide generation and modification can be inhibited by low molecular mass protein 2 (LMP2), LMP7, and LMP10 in tumor cells.14–16 In liver cancer17 and gastric cancer,18 miR-23a and miR-502-5p, respectively, can inhibit the expression of LMP7 by interfering with interferon regulatory factor-1 (IRF-1), which is an essential factor in the INF-γ-mediated increase in LMP7;19 miR-451 has also been found to be able to directly regulate LMP7 in diabetic nephropathy,20 but whether such a regulatory relationship exists in tumors remains to be studied. Next, the expression of transporters associated with antigen processing (TAP) is blocked in tumors, thereby preventing peptide transport to the endoplasmic reticulum.15,21 miR-125a-5p and miR-148a-3p can reduce the levels of TAP2 and MHC molecules by binding to the 3′UTR of TAP2 mRNA in esophageal adenocarcinoma.22 miR-346 has been shown to bind to the 3′UTR of TAP1 mRNA and decrease TAP1 expression.23 Moreover, formation of the peptide-MHC class I complex is prevented via suppression of the expression of chaperone proteins (calnexin, ERp57, calreticulin, and tapasin) and the subsequent loading of peptides on MHC class I molecules.21 lncRNA RB1 can positively regulate calreticulin in multiple tumor cell lines.24 miR-27a has been found to downregulate the expression of calreticulin and MHC class I molecules in colorectal cancer, and the infiltration and cytotoxic activity of CD8+ T cells (the release of perforin) are inhibited.25 Furthermore, defective surface expression of MHC class I molecules, which would repress the APM, has been shown in other tumors.26–28 miR-27a, miR-148a-3p, miR-125a-5p and miR-9 have been found to inhibit the surface expression of MHC class I molecules in colorectal cancer, esophageal adenocarcinoma and nasopharyngeal carcinoma.22,25,29 ncRNAs participating in the inhibition of tumor antigen presentation by repressing key proteins provide direct evidence that these ncRNAs play important roles in the development of TIE. Interfering with the roles of ncRNAs in tumor antigen presentation may provide a new direction for improving the effect of tumor immunotherapy.
Fig. 1.
Regulation of ncRNAs in the four steps of MHC class Ι molecule-mediated APM
Tumor death pathways and TIE
By changing the balance between pro-death signals and anti-death signals, tumor cells can gain resistance to cytotoxic immune responses and thus achieve TIE. First, increased expression of various antiapoptotic proteins (BCL-2, BCL-xL, and MCL-1) in tumor cells can enhance the apoptotic resistance of tumor cells.30–32 Second, inhibition of the expression of apoptosis-related receptors (FAS, DR4, and DR5) and ligands (FASL and TRAIL) can also enable tumor cells to escape apoptotic pathway-induced cell death.33,34 Third, tumor cells can escape cytotoxic T lymphocyte (CTL)- and NK cell-mediated death by blocking the perforin/granzyme pathway in immune cells. For example, tumor cells can escape CTL-mediated cytotoxicity by overexpressing inhibitors of the perforin/granzyme pathway.35 Moreover, one study identified a receptor that acts as a decoy ligand, thereby protecting tumor cells from apoptosis.36
Several studies have confirmed that ncRNAs can assist tumors in achieving TIE by regulating the abovementioned molecules and proteins, which could disrupt the balance between anti-death and pro-death signals (Table 1). For example, miR-195, miR-24-2 and miR-365 can downregulate the expression of Bcl-2 and promote the apoptosis of tumor cells in breast cancer,37 while miR-125b and miR-106a can upregulate the expression of Bcl-2 to inhibit the apoptosis of leukemia cells and breast cancer cells and promote their proliferation and infiltration.38,39 miR-133a targets and inhibits the expression of Bcl-xL and Mcl-1, promoting apoptosis in osteosarcoma cells.40 Similarly, miR-25 inhibits DR4 expression in cholangiocarcinoma cells, thereby enabling these cells to escape apoptosis induced via TNF-related apoptosis-inducing ligand (TRAIL).41 In addition, lncRNA MAGI2-AS3 upregulates the expression of FAS and FASL and promotes apoptosis in breast cancer cells.42 In CTLs extracted from the pleural effusion of lung cancer patients, miR-23a was highly expressed and could inhibit the antitumor ability of CTLs by repressing the expression of granzyme B.43 These ncRNAs regulate the expression of death signal-related molecules and further help tumors achieve immune escape. Targeting these ncRNAs to reduce the interference of death signals may be of great significance to improve the efficiency of antitumor therapy.
Table 1.
ncRNAs influence TIE via regulating tumor cells death signals by targeting death-related genes
| ncRNAs | Target genes and function | Type of cancer | Refs. |
|---|---|---|---|
| miR-195, 24-2 and 365 | Downregulate Bcl-2 and promote apoptosis of tumor cells | Breast cancer | 37 |
| miR-125b, miR-106a | Upregulate Bcl-2 and inhibit apoptosis of tumor cells | Leukemia, Breast cancer | 38,39 |
| miR-133a | Downregulates Bcl-xL and McL-1, and promotes apoptosis of tumor cells | Osteosarcoma | 40 |
| lncRNA HELH | Upregulates Bcl-xL, which mediated by miR-939, and inhibits apoptosis of tumor cells | Colorectal Cancer | 288 |
| lncRNA ASNR | Inhibits the degradation of Bcl-2 by targeting AUF1, and inhibits apoptosis of tumor cells | Stomach cancer, Colon cancer, Liver cancer, Lung cancer | 289 |
| lncRNA OPI5-AS1 | Upregulates Bcl-2, which mediated by miR-448, and inhibits apoptosis of tumor cells | Lung adenocarcinoma | 290 |
| miR-125a-5p, 26a, 193b, 363, 101, 29a, 29b, 106a, 181b, 302b and 320 | Downregulate MCL-1 and promote apoptosis of tumor cells | Colon cancer, Breast cancer, lymphomas, Multiple myeloma, Ovarian cancer, Acute myeloid leukemia, HCC, Cervical cancer | 291–301 |
| miR-205,133b and 218 | upregulate MCL-1 and inhibit apoptosis of tumor cells | Lung cancer | 302,303 |
| miR-25 | Downregulates DR4 and promotes apoptosis of tumor cells | Cholangiocarcinoma | 41 |
| miR-133b | Increases the sensitivity of tumor cells to TRAIL- mediated apoptosis by targeting FAIM | PC-3 and HeLa cell lines | 304 |
| miR-942 | Reduces the sensitivity of tumor cells to TRAIL-mediated apoptosis by targeting ISG12a | Hepatocellular carcinoma, Gastric cancer | 305 |
| miR-221 and 222 | Reduce the sensitivity to TRAIL-mediated apoptosis by targeting p27kip1 | NSCLC | 306 |
| miR-212 | Reduces the sensitivity of tumor cells to TRAIL-induced apoptosis by targeting PED | NSCLC, Liver cancer | 307 |
| miR-130a | Enhances the sensitivity of tumor cells to TRAIL-induced apoptosis by targeting MET | Lung cancer | 308 |
| miR-145, 216, 182 and 96 | Reduce the sensitivity of tumor cells to TRAIL-induced apoptosis by targeting DR4/5, FADD | Breast cancer | 309 |
| miR-200c | Reduces the sensitivity of tumor cells to FAS-mediated apoptosis by targeting FAP-1 | Human kidney clear cell cancer | 310 |
| miR-21 | Downregulates FASL and inhibits apoptosis of tumor cells | Pancreatic cancer | 311 |
| miR-590 and 20a | Downregulate FASL and FAS, and inhibit apoptosis of tumor cells | Osteosarcoma | 312 |
| miR-128a | Downregulates FAS and inhibits apoptosis of tumor cells | Acute T-cell leukemia | 313 |
| lncRNA MAGI2-AS3 | Upregulates FASL and FAS, and inhibits apoptosis of tumor cells | Breast cancer | 42 |
| miR-23a | Downregulates granzyme B and inhibits CTL-mediated death | Lung cancer | 43 |
| miR-27a | Downregulates granzyme B and perforin, inhibits CTL-mediated death | Colorectal cancer cell line sw260 | 314 |
AUF1 ARE/poly (U)-binding/degradation factor 1, FAIM Fas apoptosis inhibitory molecule, ISG12a interferon stimulated gene 12a, PED PED/PEA-15, FADD Fas-associated death domain, FAP-1 Fas-associated phosphatase-1, NSCLC non-small cell lung cancer
Abnormal metabolism and TIE
Aerobic glycolysis and TIE
Owing to mitochondrial dysfunction and despite being in an aerobic environment, tumor cells prefer to produce energy through glycolysis, which is accompanied by the production of a large amount of lactate. Such aerobic glycolysis phenomenon is termed the Warburg effect.44 This particular mode of metabolism provides the energy and macromolecules essential for the rapid growth and invasion of tumor cells. Lactate produced by aerobic glycolysis acidifies the TME, which can lead to the dysfunction of immune cells [cytotoxic T cells, dendritic cells (DCs), NK cells, and macrophages] and inhibit the secretion and function of several antitumor response cytokines. These alterations can subsequently lead to immunosuppression and promote tumor cells to escape destruction by the immune system.45,46 An increasing number of studies have found that ncRNAs can regulate tumor aerobic glycolysis directly (by targeting enzymes related to aerobic glycolysis) or indirectly (by targeting HIF-1α or tricarboxylic acid cycle (TAC)-related enzymes) to help tumors achieve TIE.
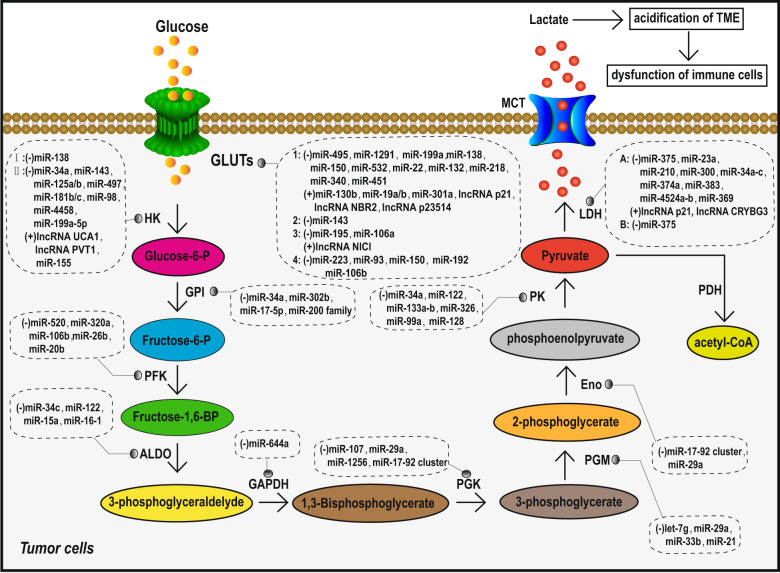
Glucose transporters (GLUTs) are membrane proteins that transport glucose into cells. Abnormal GLUT expression on the tumor cell surface promotes glucose transport into the cell and increases aerobic glycolysis. ncRNAs have been found to be involved in the regulation of GLUTs in human cancers (Table 2, Fig. 2). For example, miR-340, miR-1291, miR-495, miR-22, and miR-132 downregulate GLUT1 expression in various tumors,47–51 whereas miR-130b, miR-301a, miR-19a/b, lncRNA p23154, lncRNA NBR2, and lncRNA p21 promote GLUT1 expression.52–55 miR-150 and miR-195-5p downregulation promotes GLUT4 and GLUT3 expression in pancreatic cancer and bladder cancer, respectively.56,57
Table 2.
ncRNAs influence TIE via regulating abnormal metabolism of tumor by targeting key enzymes
| Target genes | ncRNAs | Function | Refs. |
|---|---|---|---|
| Aerobic glycolysis | |||
| GLUT1 | miR-495, 1291, 199a, 138, 150, 532, 22, 132, 218, 340 and 451 | Downregulate GLUT1 and inhibit aerobic glycolysis | 47–51,315–317 |
| miR-130b, 301a, 19a/b, lncRNA P21, lncRNA NBR2, lncRNA p23514 | Upregulate GLUT1 and promote aerobic glycolysis | 52–55 | |
| GLUT2 | miR-143 | Downregulates GLUT2 and inhibits aerobic glycolysis | 318 |
| GLUT3 | miR-195 and miR-106a | Downregulate GLUT3 and inhibit aerobic glycolysis | 57,319 |
| lncRNA NICI | Upregulates GLUT3 and promotes aerobic glycolysis | 320 | |
| GLUT4 | miR-223, 93, 150, 192 and 106b | Downregulate GLUT4 and inhibit aerobic glycolysis | 321–324 |
| HK1 | miR-138 | Downregulates HK1 and inhibits aerobic glycolysis | 59 |
| HK2 | miR-34a, 143, 125a/b, 497, 181b/c, 98, 4458 and 199a-5p | Downregulate HK2 and inhibit aerobic glycolysis | 62,325–332 |
| miR-155, lncRNA PVT1 and lncRNA UCA1 | Upregulate HK2 and promote aerobic glycolysis | 60,61,327 | |
| GPI | miR-34a, 302b, 17-5p and 200 family | Downregulate GPI and inhibit aerobic glycolysis | 333–335 |
| PFK | miR-520, 320a, 106b, 26b and 20b | Downregulate PFK and inhibit aerobic glycolysis | 336–341 |
| Aldo | miR-34c, 122, 15a and 16-1 | Downregulate Aldo and inhibit aerobic glycolysis | 342–345 |
| GAPDH | miR-644a | Downregulates GAPDH and inhibits aerobic glycolysis | 346 |
| PGK | miR-107, 29a, 1256 and 17-92 cluster | Downregulate PGK and inhibit aerobic glycolysis | 77,347–349 |
| PGM | let-7g, miR-29a, 33b and 21 | Downregulate PGM and inhibit aerobic glycolysis | 340,349–351 |
| Eno | miR-17-92 cluster and miR-29a | Downregulate Eno and inhibit aerobic glycolysis | 348,349 |
| PK | miR-34a, 122, 133a-b, 326, 99a and 128 | Downregulate PK and inhibit aerobic glycolysis | 64,333,352–354 |
| LDHA | miR-375, 23a, 210, 300, 34a-c, 374a, 383, 4524a-b and 369 | Downregulate LDHA and inhibit aerobic glycolysis | 65,66,355–360 |
| lncRNA p21 and lncRNA CRYBG3 | Upregulate LDHA and promote aerobic glycolysis | 67,361 | |
| LDHB | miR-375 | Downregulates LDHB and inhibits aerobic glycolysis | 65 |
| IDH | miR-183 | Downregulates IDH, inhibits TAC and promotes aerobic glycolysis | 70 |
| SDH | miR-210 | Downregulates SDH, inhibits TAC and promotes aerobic glycolysis | 71 |
| Cytochrome c oxidase 1 / 2 | miR-181c and miR-338 | Downregulate Cytochrome c oxidase 1 / 2, inhibit electron transport links and promotes aerobic glycolysis | 72,73 |
| HIF-1α | miR-17-92 cluster, 22, 33a, 107, 128, 138, 155, 186, 195, 516c and circEPHB4 | Downregulate HIF-1α and inhibit aerobic glycolysis | 78–80,269,270,348,354,362–369 |
| lncRNA SNHG1, 00152, DANCR, miR-21 and circRNA PIP5KA | Upregulate HIF-1α and promote aerobic glycolysis | 81,83,370,371 | |
| Arachidonic acid metabolism | |||
| PGE2 | miR-206 | Downregulates PGE2 and inhibits arachidonic acid metabolism of tumor | 88 |
| COX-2 | miR-128, 146a, 101 and 143 | Downregulate COX-2 and inhibit arachidonic acid metabolism of tumor | 89–92 |
| Tryptophan metabolism | |||
| IDO | miR-153-3p | Downregulates IDO and inhibits tryptophan metabolism of tumor | 98 |
| lncRNA SNHG1, lncRNA MALAT1, | Upregulate IDO and promote tryptophan metabolism of tumor | 96,97 | |
Fig. 2.
Overview of the regulation of TIE-associated glycolytic enzymes by ncRNAs in tumor cells
Several enzymes, such as hexokinase (HK), aldose enzyme, glucose phosphate isomerase (GPI), phosphofructokinase (PFK), aldolase (Aldo), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate mutase (PGM), enolase (Eno), pyruvate kinase (PK), pyruvate dehydrogenase (PDH), and lactate dehydrogenase (LDH), are involved in glycolysis reactions. Abnormal expression of ncRNAs has been reported to alter the expression of these enzymes, thus accelerating the process of tumor glycolysis (Table 2, Fig. 2). miR-138 and miR-143 were found to regulate aerobic glycolysis in different types of tumor cells by directly targeting HK1 and HK2, respectively.58,59 miR-155 and lncRNA urothelial cancer associated 1 (lncRNA UCA1) can increase HK2 expression by inhibiting miR-143 in breast cancer and bladder cancer, respectively.60,61 In liver cancer, miR-199a-5p can directly target HK2 and repress HK2 expression to inhibit glycolysis.62 Moreover, miR-122 targets the aldose enzyme in liver cells,63 and miR-326 downregulation is associated with increased PK2 expression in glioblastoma cells,64 both of which can inhibit aerobic glycolysis of tumor cells. PDH is the key enzyme that catalyzes the conversion of pyruvate to acetyl-CoA. Under conditions of aerobic glycolysis in tumor cells, such conversion of pyruvate to acetyl-CoA is inhibited, and pyruvate is converted to lactate by LDH, thus promoting TME acidification and TIE. LDHB is regulated by miR-375 and is increased in esophageal squamous cell carcinoma,65 whereas LDHA, which is also overexpressed in tumor cells, is regulated by miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b in colorectal cancer, leading to more lactate production.66 lncRNA p21 positively regulates LDHA, pyruvate dehydrogenase kinase 4 (PDK4), pyruvate dehydrogenase complex (PDHX), PK2, and GPI simultaneously in different tumor cells to promote aerobic glycolysis and lactate production.67 PDH inhibition can reduce the tricarboxylic acid (TCA) cycle, thus promoting the conversion of pyruvate into lactic acid. miR-23a, miR-375, and miR-138-1* upregulate PDH expression by inhibiting PDK, which is a negative regulator of PDH.68,69
In most tumor cells, the TAC is inhibited, which results in pyruvate being unable to be metabolized through TAC and only converted to lactic acid, exacerbating the acidification of TME. miRNAs can target and inhibit enzymes involved in the TCA cycle and several components of the electron transport chain, thereby inhibiting mitochondrial function and further promoting aerobic glycolysis (Table 2). For example, miR-18370 and miR-21071 can, respectively, target key enzymes isocitrate dehydrogenase (IDH) and succinic acid dehydrogenase (SDH) of the TCA cycle in glioma and lung cancer. miR-181c72 and miR-33873 downregulate cytochrome c oxidase 1/2, respectively, which are components of the electron transport chain.
Recent studies demonstrated that hypoxia-inducible factor-1α (HIF-1α) can promote aerobic glycolysis in tumor cells74 and then promote TME acidification and TIE. First, activated HIF-1α can directly or indirectly increase the expression of all glycolysis-related enzymes and promote glycolysis.75 Second, HIF-1α prevents the conversion of pyruvate to acetyl-CoA, the raw material of the TCA cycle, by inhibiting PDH activity.74,75 Moreover, HIF-1α indirectly promotes glycolysis by inhibiting mitochondrial oxidative phosphorylation.76 ncRNAs have been found to regulate HIF-1α in tumors, which could promote aerobic glycolysis and TIE (Table 2). For example, the miR-17-92 cluster,77 miR-22,78 miR-33a,79 and circRNA EPHB480 have been reported to downregulate HIF-1α expression in different types of tumors, while miR-21 can promote the expression of HIF-1α in prostate cancer cells; lncRNA SNHG1,81 lncRNA 0015282 and circRNA PIP5KA83 function as molecular sponges for miR-18a, miR-138 and miR-600, respectively, to promote HIF-1α expression in different tumors. It has not been reported that ncRNAs and HIF-1α can directly affect the immune escape of tumor cells through complex molecular networks, but this indirect evidence also suggests that this idea merits further exploration.
Arachidonic acid metabolism and TIE
Altered metabolism of arachidonic acid, an unsaturated fatty acid, is also a characteristic of tumors, especially those associated with inflammation, such as colorectal cancer, lung cancer and bladder cancer.84 Most of these tumors show increased expression of prostaglandin E2 (PGE2) and cyclooxygenase-2 (COX-2), two key molecules of the arachidonic acid metabolic pathway. PGE2 and COX-2 overexpression in tumors may be one of the potential mechanisms underlying TIE. First, COX-2 induces DCs to secrete interleukin-10 (IL-10) and transforming growth factor-β (TGF-β), which in turn activate regulatory T cells (Tregs) and promote immunosuppression.85 Second, PGE2 and COX-2 induce the expression of the Treg-specific transcription factor forkhead box P3 (FOXP3), thus increasing the activity of Tregs.86 In addition, PGE2 directly inhibits lymphocyte function by increasing cellular cAMP levels.87 miRNAs have been found to regulate COX-2 and PGE2 in tumors (Table 2). For example, miR-206 inhibits PGE2-induced proliferation and metastasis of colon cancer cells by targeting the transmembrane 4 L six family member 1 (TM4SF1) protein,88 while miR-128,89 miR-146a,90 miR-101,91 and miR-14392 can inhibit tumor progression by decreasing COX-2. The involvement of ncRNAs other than miRNAs in the regulation of COX-2 and PGE2 has not yet been reported. Therefore, the idea that ncRNAs directly regulate the metabolism of arachidonic acid and affect immune escape may be a potential mechanism of TIE.
Tryptophan metabolism and TIE
Indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) are the two key enzymes involved in tryptophan metabolism, and they can be manipulated by tumors to evade immune surveillance. IDO and TDO are highly expressed in tumor cells and can promote the recruitment of Tregs in the TME and induce immunosuppression via increased secretion of the immunosuppressive factors IL-6, IL-10, and TGF-β,93–95 which can help tumors achieve TIE. ncRNAs have been reported to regulate IDO and TDO expression in tumor cells (Table 2). High expression of lncRNA SNHG1 in breast cancer specifically inhibits miR-448, thereby increasing the expression of IDO, promoting Treg differentiation, enhancing immunosuppression, and promoting tumor proliferation and metastasis,96 also providing an advantage for the occurrence of TIE. lncRNA MALAT1 promotes immunosuppression by inducing IDO expression.97 In addition, studies confirmed that miR-153-3p specifically inhibits IDO expression and participates in the development of acute graft-versus-host disease in vitro and in vivo.98 All of the above findings are based on protein level regulation research. Whether ncRNAs can regulate tryptophan metabolism at the functional level and further affect TIE may also become a new potential target research mechanism.
Cancer stem cell-like phenotype and TIE
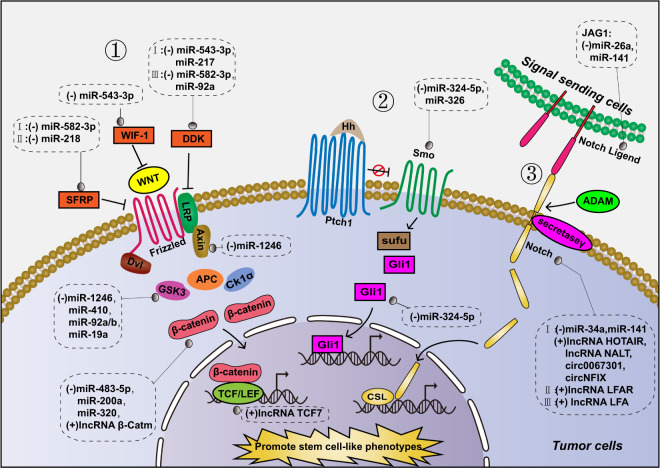
Acquiring a stem cell-like phenotype is considered another strategy adopted by tumors to achieve TIE. This is attributed to the low immunogenicity of cancer stem cells, and their immunoregulatory properties can inhibit the antitumor immune response and help them evade immune recognition.99 The WNT, Notch, and Hedgehog signaling pathways are closely associated with the development of stem cell-like phenotypes in most tumor cells. These three signaling pathways are abnormally activated in tumors to promote the transcription of target genes, thus improving the tumor stem cell-like phenotype (Fig. 3).100 ncRNAs were found to activate these signaling pathways by regulating key proteins involved in these pathways, thereby promoting stem cell-like phenotypes in tumor cells (Table 3, Fig. 3), which is also advantageous for the occurrence of TIE. miR-1246 activates the WNT pathway and promotes tumor stemness by inhibiting two components of the WNT pathway, Axin2 and glycogen synthase kinase 3β (GSK-3β), in liver cancer.101 In colorectal cancer and glioma, circRNA 100290 and circRNA 0000177 upregulate Frizzled receptor expression by acting as molecular sponges for miR-516b and miR-638, respectively, thereby activating the WNT pathway and promoting the stem-cell phenotype of tumor cells.102,103 miR-34a targets Notch1, an important membrane surface receptor of the Notch pathway, thereby inhibiting the stem cell-like phenotype of breast cancer cells.104 miR-324-5p has been found to significantly reduce the activation of the Hedgehog pathway by inhibiting Smoothened (Smo) and Gli zinc‐finger transcription factor-1 (Gli1), thereby inhibiting the stem-cell phenotype of multiple myeloma cells.105
Fig. 3.
Regulation of TIE-associated stem cell signaling pathways by ncRNAs. The pathways include the following: 1. WNT signaling pathway; 2. Hedgehog signaling pathway; and 3. Notch signaling pathway
Table 3.
ncRNAs influence TIE via regulating tumor stem cell-like phenotype by targeting stem cell pathway or stemness genes
| ncRNAs | Target genes | Function | Type of cancer | Refs. |
|---|---|---|---|---|
| WNT signaling pathway | ||||
| miR-543-3p | WIF-1 | Activates WNT pathway by inhibiting WTF-1, and promotes stem cell-like phenotype of tumor cells | Bladder cancer | 372 |
| miR-218 and miR-1301-3p | SFRP | Activate WNT pathway by inhibiting SFPR (an inhibitor of WNT pathway), and promote stem cell-like phenotype of tumor cells | Triple negative breast cancer, Prostate cancer | 373,374 |
| miR-543-3p, miR-217, circRNA 0006427 and circRNA 0000523 | DDK1 | Activate/inhibit the WNT pathway by up-/downregulate the WNT pathway inhibitor DDK1, and regulate stem cell-like phenotype of tumor cells | Bladder cancer, Hepatocellular carcinoma, Lung adenocarcinoma, Colorectal cancer | 372,375–377 |
| circRNA CBFB, 100290, 0000177 and NEK6 | Frizzed receptor3/4/7/8 | Activate Notch pathway by upregulating Frizzed receptor4/7/8, and promote stem cell-like phenotype of tumor cells | Chronic Lymphocytic Leukemia, Colorectal cancer, Glioma, Thyroid cancer | 102,103,378,379 |
| miR-1246, 410, 92a and 19 | GSK-3β | Inhibit the WNT pathway by inhibiting the expression of GSK-3β, and inhibit stem cell-like phenotype of tumor cells | Liver cancer, NSCLC, Colorectal cancer, Lung cancer | 101,380–382 |
| miR-1246 | Axin | Inhibits the WNT pathway by inhibiting the expression of Axin, and inhibits stem cell-like phenotype of tumor cells | Liver cancer | 101 |
| circRNA 0002052 and circRNA 0009361 | APC | Inhibit the WNT pathway by promoting the expression of APC, and inhibit stem cell-like phenotype of tumor cells | Osteosarcoma | 383,384 |
| miR-320, miR-200a and lncRNA β-Catm | β-catenin | Inhibit/activate the WNT pathway by up-/downregulate β-catenin, and regulate stem cell-like phenotype of tumor cells | Prostate cancer, Liver cancer | 385–387 |
| lncRNA TCF7 | TCF7 | Activates the WNT pathway by upregulating the transcription factor TCF7, and promotes stem cell-like phenotype of tumor cells | Colorectal cancer | 388 |
| Hedgehog signaling pathway | ||||
| miR-324-5p | Smo, Gli | Inhibits the Hedgehog pathway by downregulate Smo and Gli, and inhibits stem cell-like phenotype of tumor cells | Multiple myeloma | 105 |
| miR-326 | Smo | Inhibits the Hedgehog pathway by inhibiting Smo, and inhibits stem cell-like phenotype of tumor cells | Chronic myeloid leukemia | 389 |
| Notch signaling pathway | ||||
| miR-26a and miR-141 | JAG1(Notch ligand) | Inhibits the Notch pathway by inhibiting the expression of JAG1, and inhibits stem cell-like phenotype of tumor cells | Osteosarcoma, Glioblastoma | 390,391 |
| lncRNA HOTAIR, lncRNA NALT, circRNA NFIX, circRNA ASH2L | Notch1 | Activates Notch pathway by upregulating the expression of Notch1, and promote stem cell-like phenotype of tumor cells | Intervertebral disc degeneration, Acute lymphoblastic leukemia, Glioma, Pancreatic ductal adenocarcinoma | 392–395 |
| miR-34a | Notch1 | Inhibits Notch pathway by downregulating Notch1, and inhibits stem cell-like phenotype of tumor cells | Breast cancer | 396 |
| lncRNA LFAR | Notch2/3 | Activates the Notch pathway by upregulate Notch2/3, and promote stem cell-like phenotype of tumor cells | Hepatic stellate cells | 397 |
| Stemness related genes | ||||
| miR-34a | SOX2, NANOG and OCT3/4 | Downregulate SOX2, Nanog, and OCT3/4, thereby inhibiting stem cell-like phenotype of tumor cells | Head and neck squamous cell carcinoma | 108 |
| miR-208a | LIN28, SOX2 | Upregulate LIN28, SOX2, thereby inhibiting stem cell-like phenotype of tumor cells | Breast cancer | 398 |
| let- 7, miR-125, 9 and 30 | LIN28 | Downregulate LIN28, thereby inhibiting stem cell-like phenotype of tumor cells | A2780, T47D, MCF7 and HeLa cancer cell lines | 399 |
| miR-21 | OCT4 | Downregulates OCT4, thereby inhibiting stem cell-like phenotype of tumor cells | Liver cancer | 400 |
| lncRNA DYNC2H1-4, SNHG20 and HOTTIP | LIN28, Nanog, SOX2 and OCT4 | Upregulate LIN28, Nanog, SOX2 and OCT4, and promote stem cell-like phenotype of tumor cells | Pancreatic cancer, Oral squamous cell carcinoma, Pancreatic cancer | 401–403 |
| lncRNA FEZF1-AS1, lncRNA ITGB1 and piRNA-823 | Nanog, OCT4 and SOX2 | Upregulate Nanog, SOX2 and OCT4, and promote inhibiting stem cell-like phenotype of tumor cells | Breast cancer, NSCLC, Multiple myeloma | 110,404,405 |
| lncRNA H19 | LIN28 | Enhances upregulate LIN28, and promotes the stem-cell-like phenotype | Breast cancer | 109 |
SFPR secreted frizzled related protein, DDK1 dickkopf-1, APC adenomatous polyposis coli, JAG1 Jagged1
Octamer transcription factor-3/4 (OCT3/4), SRY-box 2 (SOX2), Nanog and LIN28 are genes related to the tumor stem cell-like phenotype that have been proven to be related to TIE.106,107 ncRNAs can also promote the tumor stem cell-like phenotype by directly or indirectly regulating those genes (Table 3). For example, miR-34a targets and inhibits the expression of SOX2, Nanog, and OCT3/4, thereby inhibiting the stem cell-like phenotype of head and neck squamous carcinoma cells.108 lncRNA H19 acts as a molecular sponge for let-7 to upregulate LIN28 and promote the stem cell-like phenotype of breast cancer cells.109 In a study of multiple myeloma, granulocyte-MDCSs increased the expression of SOX2, OCT4, and Nanog in multiple myeloma stem cells by promoting the expression of piRNA-823, which controlled tumor stemness through DNMT3B activation, thereby promoting the tumor stemness phenotype.110 The above research results provide preliminary evidence that these ncRNAs promote the development of TIE by targeting tumor stem cell-like phenotype-related pathways and genes. By inhibiting this process, we may be able to improve resistance to immunotherapy.
Epithelial–mesenchymal transformation (EMT) and TIE
EMT involves molecular changes that transform epithelial cells into mesenchymal cells, and such transformation enables the cells to lose cell-cell adhesion and apical-basal polarity. Therefore, EMT in tumor cells is essential to promote the metastasis of epithelial tumors.111,112 Several studies have reported that EMT may also induce immunosuppression and help tumors achieve TIE. Snail-induced EMT stimulates the production of immunosuppressive factors such as TGF-β and thrombospondin-1 (TSP-1), which could damage DCs, decrease the expression of costimulatory molecules, and increase the expression of IDO, thus indirectly inducing Treg differentiation and promoting immunosuppression.113 A study also proved that Snail-induced EMT in melanoma cells are resistant to CTL lysis.113 Furthermore, compared with breast cancer epithelial cells, mesenchymal cells generated via EMT in breast cancer cells show low expression of MHC class Ι molecules and high expression of programmed death ligand 1 (PD-L1), thereby inducing immune resistance and promoting TIE.114 Therefore, tumor cell EMT can promote immunosuppression in many ways and become one of the potential driving forces of TIE.
EMT is mainly mediated by three transcription factors: zinc-finger E-box-binding 1 (ZEB1), Snail, and Twist1.115 These transcription factors can decrease epithelial cadherin (E-cadherin) and increase neural cadherin (N-cadherin) and vimentin, thereby promoting the occurrence of EMT.112 There is also evidence that these transcription factors are associated with immune escape.113,116 miRNAs such as miR-21, miR-137, miR-34a, and miR-106a/b are known to regulate EMT by targeting these transcription factors (Table 4). In colorectal cancer, miR-21 can downregulate the expression of Snail and E-cadherin to inhibit EMT.117 In ovarian cancer, miR-137 and miR-34a can also downregulate Snail expression to inhibit EMT,118 while miR-106a can upregulate the expression of Snail and promote EMT in glioma cells.119 In hepatocellular carcinoma (HCC), miR-106a/b can inhibit EMT by downregulating Twist1.120 In contrast, miR-23a can upregulate Twist1 expression and promote EMT and cisplatin resistance in tongue squamous cell carcinoma.121 In addition to miRNAs, other ncRNAs have also been found to modulate EMT by targeting these transcription factors. In pancreatic cancer, lncRNA PVT1 promotes EMT by upregulating the expression of ZEB1, Snail, and N-cadherin and downregulating E-cadherin expression.122 In bladder cancer, lncRNA UCA1 can promote EMT by upregulating the expression of N-cadherin, vimentin, and Snail and downregulating the expression of E-cadherin; however, as a competitive endogenous RNA (ceRNA) of miR-145, it also upregulates the expression of ZEB1.123 In melanoma, circRNA 0084043 upregulates Snail expression by acting as a ceRNA of miR-153-3p, thus promoting EMT.124 These dysregulated ncRNAs accelerate EMT by regulating transcription factors in tumors, but their further impact on TIE remains unknown. Whether blocking the mechanism by which these ncRNAs regulate EMT can be conducive to inhibiting TIE and improving the effect of immunotherapy is worth further study.
Table 4.
ncRNAs influence TIE via regulate EMT by targeting EMT- related transcription factors
| Target genes | ncRNAs | Function | Type of cancer | Refs. |
|---|---|---|---|---|
| Twist1 | ||||
| miR-543, 300, 186, 137, 720, 580, 539, 33a, 33b, 520d-5p, 106b, 675,337-3p and 151-5p | Inhibit EMT of tumor cells by targeting and downregulating Twist1, an EMT related transcription factor | Endometrial cancer, Epithelial ovarian cancer, Gastrointestinal stromal tumor, Breast cancer, Osteosarcoma, Melanoma, endometrial carcinoma, HCC, Lung carcinoma | 120,406–417 | |
| miR-23a, lncRNA AK027294, lncRNA ROR and lncRNA AFAP1-AS1 | Promote EMT of tumor cells by targeting and upregulating Twist1, an EMT related transcription factor | Tongue squamous cell carcinoma, Colorectal cancer, Gallbladder cancer | 121,418–420 | |
| Snail | ||||
| miR-21, 137, 34a, 491-5p, 22, 363, 30, 145, 153, 410-3p | Target and downregulate transcription factor Snail, and then inhibit EMT of tumor cells | Colorectal cancer, Ovarian cancer, Gastric cancer, Bladder cancer, Lung cancer, Osteosarcoma, HCC, Breast cancer | 117,118,421–427 | |
| miR-106a, circRNA 0084043, circRNA PRMT5, lncRNA PVT1 and lncRNA UCA1 | Target and upregulate transcription factor Snail, and then promote EMT of tumor cells | Glioma, Melanoma, Bladder carcinoma, Pancreatic cancer, Breast cancer | 119,122,124,428,429 | |
| ZEB1 | ||||
| miR-203, 873, 205-5p, 5702, 126, 186-5p | Target and downregulate transcription factor ZEB1, and then inhibit EMT of tumor cells | Gastric cancer, Breast cancer, Prostate cancer, NSCLC, Cervical cancer, Colorectal cancer | 430–435 | |
| lncRNA MALAT1, ZEB1-AS1, SNHG16, NNT-AS1, HOTTIP, NEAT1, ZNF469-3, TP73-AS1, circRNA TSPAN4 and circRNA PVT1 | Target and upregulate transcription factor ZEB1, and then promote EMT of tumor cells | HCC, NSCLC, Osteosarcoma, Breast cancer, Glioma, Nasopharyngeal carcinoma, Lung adenocarcinoma, Gastric cancer | 436–445 | |
Immunosuppressive cells and TIE
Tregs
Tregs, cells that act as immunosuppressive agents in the body, play important roles in TIE. First, Tregs produce immunosuppressive factors such as IL-10, IL-35, and TGF-β, which inhibit the function of antitumor T cells.125 Second, Tregs inhibit T cell function by expressing coinhibitory factors such as cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed cell death protein 1 (PD-1), and PD-L1.126 In addition to directly affecting T cells, Tregs can also inhibit T cell activation by targeting the maturation and activity of DCs.125 miRNAs play an essential role in Treg maintenance and function. Foxp3-dependent regulation of miR-155 contributes to the proliferative activity and competitive fitness of Tregs.127 miR-146a, highly expressed in Tregs, was found to be able to regulate Treg function, and loss of miR-146a led to increased production of the proinflammatory Th1 cytokine IFN-γ by Foxp3+ Tregs, and transferring purified miR-146a-deficient Tregs together with Foxp3 KO CD4+ effector T cells into lymphopenic recipients failed to repress Th1 responses.128 In tumors, abnormally expressed ncRNAs have been found to regulate Tregs. The miR-17-92 cluster is expressed in many human blood tumors, and studies have shown that the miR-17-92 cluster can regulate the number of Tregs by targeting Bim.129 Some lncRNAs are also involved in Treg regulation. lncRNA HULC, which is highly expressed in HCC, downregulates p18 in liver cirrhosis to affect Treg differentiation.130 In gastric cancer, lncRNA POU3F3 promotes the distribution of Tregs among surrounding T cells by recruiting TGF-β and activating the TGF-β pathway.131 lncRNA SNHG1 regulates Treg differentiation by targeting miR-448/IDO in breast cancer.96
Myeloid-derived suppressor cells (MDSCs)
As a group of heterogeneous cells derived from the bone marrow, MDSCs are precursors of DCs, macrophages, and/or granulocytes. MDSCs can significantly inhibit the cellular immune response and are one of the important driving forces of TIE. First, MDSCs induced by HMGB1 and those with myeloid differentiation potential can mediate TIE by producing high levels of IL-10, inhibiting the activation of antigen-driven CD4+ and CD8+ T cells and the expression of L-selectin in circulating naive T cells.132 Second, tumor cells can also inhibit the function of T cells, NK cells, and DCs by altering the expression of cellular stress sensor C/EBP homologous protein (Chop) and the secretion of IL-6 by MDSCs.133 Moreover, the proliferation of CXCR2+CD11b+Ly6Ghi MDSCs induced by CXCR2 ligands produced by tumor cells inhibits T cell proliferation by L-arginine depletion and exerts local immunosuppressive effects.134 Furthermore, MDSCs can inhibit the host antitumor immune response by inducing Tregs.135 MDSC-derived NO reacts with superoxide to produce peroxynitrite (PNT), which directly inhibits T cells by nitrating the T cell receptors (TCRs) present on the surface of tumor-specific T cells and reducing the reactivity of the associated antigen-MHC complexes.136 ncRNAs have been found to be involved in regulating the immunosuppressive activity of MDSCs. In tumor-bearing mice, lncRNA PVT1 regulates the immunosuppressive activity of MDSCs, and lncRNA PVT1 knockdown significantly inhibits the immunosuppressive activity of MDSCs.137 Similarly, lncRNA HOTAIRM1 negatively regulates the immunosuppressive activity of MDSCs by targeting HOXA1 in lung cancer.138 In addition, some ncRNAs regulate the proliferation, differentiation, and recruitment of MDSCs. miR-34a promotes the proliferation of MDSCs by inhibiting their apoptosis,139 whereas miR-9 regulates the differentiation and function of MDSCs by targeting runt-related transcription factor 1 (Runx1).140 Conversely, the lack of miR-155 in B16-F10 melanoma and Lewis lung carcinoma cell lines leads to the recruitment of MDSCs in the TME and subsequently enhances immunosuppression.141
Tumor-associated macrophages (TAMs)
TAMs have been proven to be the most significant immune cells in the tumor stroma, accounting for more than 50% of the total number of immune cells in the tumor stroma, which are divided into two groups, M1 and M2. M1 macrophages play an important role in the innate immune response to pathogen invasion, whereas M2 macrophages are alternately activated by IL-4, IL-10, IL-13, and glucocorticoids. Studies have found that macrophages in tumor tissues mostly have the phenotype and function of M2 macrophages, which is one of the important driving forces for TIE. First, M2 macrophages produce high levels of the immunosuppressive factor IL-10 in breast tumors, which can inhibit IL-12 expression in tumor DCs and subsequent IL-12-mediated CTL activation, thereby blocking the CTL-dependent antitumor immune response.142 Second, M2 macrophages directly inhibit the activity of CD8+ T cells via the expression of coinhibitory factors such as PD-L1 and B7-H4 but indirectly play an antitumor role by promoting the recruitment of CCL22-mediated Tregs.143 Tumor cells can secrete certain cytokines (CCL2, CSF1, and CXCL12) to promote M2 macrophage recruitment in the TME, thus inducing immunosuppression. In the mouse sarcoma MS-K cell line, miR-342 inhibits the expression of CXCL12 and reduces the recruitment and activation of M2 macrophages.144 lncRNA NIFK-AS1 inhibits the M2 polarization of macrophages by targeting miR-146a, thus reducing immunosuppression and inhibiting the proliferation, migration, and invasion of endometrial cancer.145
In conclusion, immunosuppressive Tregs, MDSCs and M2 macrophages can achieve immunosuppression in many ways, thus reducing the recognition and killing of tumors by the immune system. However, these functions are utilized by ncRNAs to help tumors achieve TIE, which may be effective targets to prevent TIE and improve tumor immunotherapy.
Immune checkpoint molecules and TIE
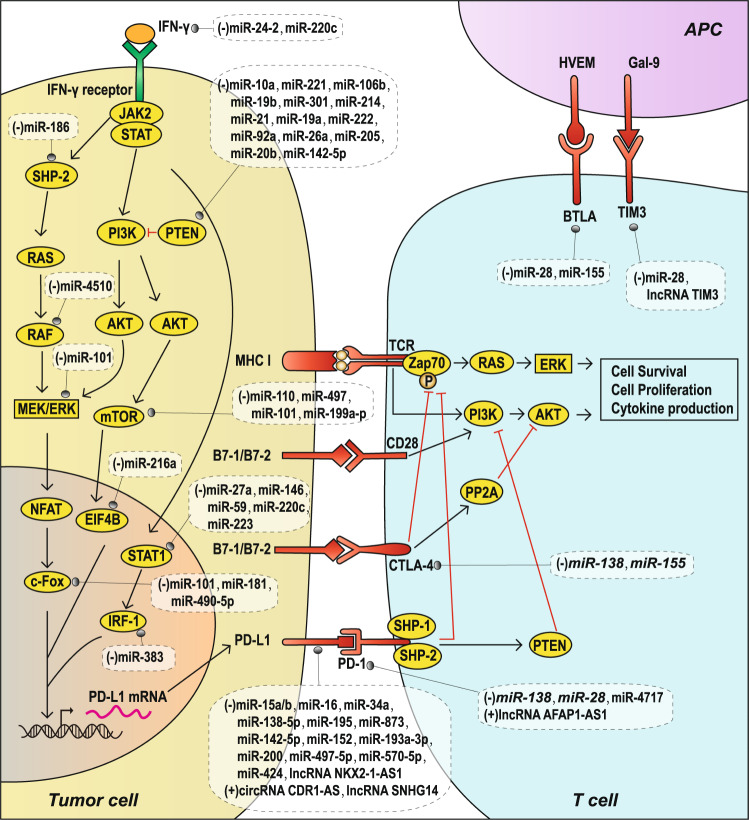
Immune checkpoints refer to a plethora of inhibitory pathways hardwired into the immune system that are crucial for maintaining self-tolerance and modulating the duration and amplitude of physiological immune responses in peripheral tissues to minimize collateral tissue damage. It is now clear that tumors coopt certain immune checkpoint pathways as a major mechanism of tumoral immune resistance, particularly against T cells that are specific for tumor antigens.146 The expression of immune checkpoint proteins can be dysregulated in tumors as an important TIE mechanism.147 Therefore, the study of immune checkpoints is particularly essential to prevent TIE. The currently known TIE-related immune checkpoint molecules are CTLA-4, PD-1, PD-L1, T-cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), and B and T lymphocyte attenuator (BTLA) (Fig. 4). Next, we will summarize the research progress on ncRNAs in the regulation of immune checkpoints (Table 5).
Fig. 4.
TIE and immune checkpoint molecules regulated by ncRNAs. PD-L1 expression induced by the IFN-γ signaling pathway and PTEN/PI3K/AKT/mTOR pathway, as well as the regulation of these pathways by ncRNAs (left).T-cell-activated receptors/ligands (TCR/MHC-I and CD28/B7-1/2) and immune checkpoint molecular receptor-ligands (PD-1/PD-L1, CTLA-4/B7-1/2, BTLA/HVEM and TIM3/Gal-9) regulated by ncRNAs (middle and upper right). The red “T” symbol represents inhibitory modification
Table 5.
ncRNAs influence TIE via regulating immune checkpoint molecules
| Target genes | ncRNAs | Function | Type of cancer | Refs. |
|---|---|---|---|---|
| CTLA-4 | ||||
| miR-138 and miR-155 | Downregulate CTLA-4 on tumor-infiltrating T cells directly, promote the recognition and killing of tumor cells by the immune system | Mouse gliomas, Mouse melanoma | 151,152 | |
| PD-L1 | ||||
| miR-424, 16, 195, 34a, 15a, 15b, 16, 193a-3p, 873, 497-5p, 570, 152, 142-5p, 138-5p and lncRNA NKX2-1-AS1 | Downregulate PD-L1 on tumor cells directly, inhibit immunosuppression and preventing TIE | Ovarian cancer, Prostate cancer, AML, Malignant pleural mesothelioma, Breast cancer, Clear cell renal cell carcinoma, Gastric cancer, Pancreatic cancer, Colorectal cancer, Lung carcinoma | 159–162,164,174,446–450 | |
| circRNA CDR1-AS | Upregulates PD-L1 on tumor cells directly, promotes immunosuppression and TIE | Colon cancer | 173 | |
| lncRNA SNHG14 | Upregulates PD-L1 by SNHG14/miR-5590-3p/ZEB1 positive feedback loop, promotes immunosuppression and TIE | Large B cell lymphoma | 172 | |
| miR-24-2 and miR-200c | Downregulate PD-L1 by inhibiting the IFN-γ in IFN-γ signaling pathway, inhibit immunosuppression and TIE | Cervical cancer | 165 | |
| miR-186 | Downregulates PD-L1 by inhibiting SHP-2 in IFN-γ signaling pathway, inhibits immunosuppression and TIE | Oral squamous cell carcinoma | 166 | |
| miR-4510 | Downregulates PD-L1 by inhibiting RAF1 in IFN-γ signaling pathway, inhibits immunosuppression and TIE | HCC | 451 | |
| miR-101 | Downregulates PD-L1 by inhibiting MEK1 in IFN-γ signaling pathway, inhibits immunosuppression and TIE | Nasopharyngeal carcinoma | 452 | |
| miR-27a, 145, 150, 223 and 200c | Downregulate PD-L1 by inhibiting STAT1 in the IFN-γ signaling pathway, inhibit immunosuppression and TIE | Cervical cancer, Colon cancer, Adult T cell leukemia/lymphoma, Cervical cancer | 165,167,453 | |
| miR-383 | Downregulates PD-L1 by inhibiting IRF1 in the IFN-γ signaling pathway, inhibit immunosuppression and TIE | Testicular embryonal carcinoma | 454 | |
| miR-101, 181b and 490-5p | Downregulate PD-L1 by inhibiting c-FOS in the IFN-γ signaling pathway, inhibit immunosuppression and TIE | Osteosarcoma, Glioma, Bladder cancer | 455–457 | |
| miR-10a, 19a, 19b, 106b, 221, 222, 20b, 21, 130b, 92a, 26a, 205, 214, 301a and 142-5p | Upregulate PD-L1 by inhibiting PTEN in PTEN/PI3K/AKT/mTOR pathway, promote immunosuppression and TIE | NSCLC, Gastric cancer, Colorectal cancer, Lung cancer, Nasopharyngeal carcinoma, Ovarian cancer, Breast cancer, NSCLC | 168,169,458–466 | |
| miR-100, 101, 199a-3p and 497 | Upregulate PD-L1 by downregulating mTOR in PTEN/PI3K/AKT/mTOR pathway, promote immunosuppression and TIE | Bladder cancer, Osteosarcoma cell, Endometrial cancer cell, Ovarian cancer | 170,467–469 | |
| miR-216a | Upregulates PD-L1 by downregulating EIF4B in PTEN/PI3K/AKT/mTOR pathway, promotes immunosuppression and TIE | NSCLC | 171 | |
| PD-1 | ||||
| miR-28, 138 and 4717 | Downregulate PD-1 directly on tumor-infiltrating T cells, promotes the activity and function of T cells and inhibit TIE | Mouse melanoma, Mouse glioma, Chronic HBV | 151,179,470 | |
| ncRNA AFAP1-AS1 | Up- regulates PD-1 expression on tumor-infiltrating lymphocytes, inhibits the activity and function of lymphocytes and promotes TIE | Nasopharyngeal carcinoma | 181 | |
| TIM-3 | ||||
| miR-28 | Reduces T cell exhaustion and increasing TNF-α and IL-2 secretion by downregulating TIM-3 directly, thereby inhibiting TIE | Melanoma | 179 | |
| lncRNA Tim3 | Exacerbates CD8+ T cell exhaustion by specifically binding to TIM-3, thereby promoting TIE | HCC | 184 | |
| BTLA | ||||
| miR-28 and miR-155 | Downregulate BTLA directly and enhance antitumor immune response and inhibit TIE | Melanoma, CD4+ T cell | 179,186 | |
CTLA-4
Activated T cells play vital roles in antitumor immunity, and T cell activation depends on two signals: one involving an interaction between TCR and MHC molecules and another involving the costimulatory signal between CD28 and B7-1/2. To ensure a balance in the function of the immune system, CTLA-4, a homolog of CD28, binds to B7-1/2 and forms an inhibitory signal for T lymphocytes with activated TCRs.148 CTLA-4 binds to B7-1/2 with an affinity higher than that of CD28 and inhibits AKT phosphorylation by activating phosphatase protein phosphatase 2A (PP2A), thereby inhibiting subsequent T cell activation and function (Fig. 4).149 In addition, CTLA-4 inhibits the formation of ζ chain-associated protein kinase 70 (Zap70), thereby affecting TCR signaling and ultimately inhibiting T cell function and promoting T cell apoptosis (Fig. 4).150 Therefore, overexpression of CTLA-4 on the surface of tumor-infiltrating T cells will enhance immune suppression by inhibiting the activation and function of T cells and promoting T cell apoptosis, thus helping tumor cells achieve TIE. Some miRNAs with abnormal expression in tumor cells can regulate the surface expression of CTLA-4 in tumor-infiltrating T cells, thereby inhibiting the antitumor immune response mediated by T cells and promoting TIE (Table 5, Fig. 4). In mice with glioma, miR-138 significantly reduces the expression of CTLA-4 and PD-1 on the surface of tumor-infiltrating T cells.151 In mice with melanoma, miR-155 inhibits CTLA-4 expression on the surface of tumor-infiltrating T cells and promotes the host antitumor immune response.152
PD-L1
PD-L1 (B7-H1) is a transmembrane protein expressed by T cells, B cells, and various tumor cells.153 Binding of PD-L1 with PD-1 on the surface of CTLs can inhibit the proliferation of CTLs and suppress the secretion of CTL cytokines such as IL-2, thereby affecting the function of CTLs and promoting TIE.146 The overexpression of PD-L1 on the surface of tumor cells through different mechanisms can enhance immunosuppression and promote immune escape. Currently, PD-L1 is the main target for tumor immunotherapy, and some PD-L1 monoclonal antibodies, such as durvalumab and atezolizumab, have shown good clinical therapeutic effects.154,155 In addition to factors that directly regulate PD-L1 mRNA, studies have shown that the transcription and expression of PD-L1 in tumor cells are strongly dependent on the interferon-γ (IFN–γ) signaling pathway (Fig. 4).156,157 In addition, PD-L1 is also regulated by the phosphatase and tensin homolog deleted on chromosome ten (PTEN)/phosphatidylinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway in tumor cells (Fig. 4).158 The absence of PTEN promotes AKT and mTOR phosphorylation, which in turn leads to increased PD-L1 translation (Fig. 4).158 miRNAs can affect PD-L1 expression in tumor cells by directly targeting the PD-L1 mRNA or targeting the intermediate links of related signaling pathways (Table 5, Fig. 4). miRNAs such as miR-34a,159 miR-138-5p,160 miR-142-5p,161 miR-152,162 miR-200,163 and miR-424164 directly inhibit PD-L1 by targeting PD-L1 mRNA in different tumor cells. In addition, miRNAs such as miR-24-2,165 miR-186,166 miR-27a,165 and miR-145167 alter PD-L1 expression by regulating the IFN-γ signaling-mediated transcription and expression of PD-L1. Other miRNAs, such as miR-10a,168 miR-221,169 miR-100,170 and miR-216a,171 alter PD-L1 expression by targeting the PTEN/PI3K/AKT/mTOR signaling pathway. In addition to miRNAs, a number of lncRNAs and circRNAs are also involved in regulating the expression of PD-L1 (Table 5, Fig. 4). lncRNA SNHG14 activates PD-L1 expression at the transcriptional level via ZEB1 and miR-5590-3p, thus promoting immune escape of diffuse large B cell lymphoma.172 circRNA CDR1-AS positively regulates PD-L1 levels and leads to poor prognosis in patients with colorectal cancer.173 lncRNA NKX2-1-AS1 inhibits PD-L1 expression in tumor cells at the transcriptional level, thus inhibiting immunosuppression and preventing TIE in lung carcinoma cells.174
PD-1
PD-1 is mainly expressed on the surface of activated T cells, B cells, and DCs.175 The interaction between PD-L1 and PD-1 inhibits TCR-mediated T cell activation (Fig. 4) and is a significant mechanism of TIE. Mechanistically, when PD-L1 and PD-1 interact, intracellular tyrosine of the ligand-bound PD-1 is phosphorylated and thus activated. Src homology 2-containing protein tyrosine phosphatase 1 (SHP-1) and SHP-2 are then recruited to the C-terminal immunoreceptor tyrosine-based switch motif (ITSM) of PD-1, and they inhibit the activation of the RAS/extracellular signal-regulated kinase (ERK) signaling pathway via ZAP70 dephosphorylation (TCR activation signals). This promotes T cell apoptosis and inhibits the proliferation of T cells and secretion of cytokines such as IL-2 (Fig. 4).176,177 In addition, PD-1 can also activate PTEN, indirectly inhibiting the TCR-mediated PI3K/AKT signaling pathway (Fig. 4).178 Multiple miRNAs and lncRNAs have been found to regulate PD-1 expression on the surface of tumor-infiltrating T cells and promote immunosuppression, thus promoting TIE (Table 5, Fig. 4). In melanoma-bearing mice, miR-28 was found to specifically inhibit PD-1 expression on tumor-infiltrating T cells and prevent T cell exhaustion, which can enhance the antitumor immune response, whereas transfection with miR-28 inhibitors increased the number of PD-1-positive exhausted T cells.179miR-138 inhibits the expression of PD-1 and CTLA-4 on tumor-infiltrating T cells, promotes the activity and function of T cells, and inhibits tumor development in mouse GL261 glioma cells.151 miR-4717 was found to inhibit PD-1 expression in the lymphocytes of patients with chronic HBV infection, the leading cause of HCC.180 In addition, lncRNA AFAP1-AS1 positively regulates PD-1 expression on tumor-infiltrating lymphocytes in nasopharyngeal carcinoma.181
TIM-3
TIM-3 is an immune checkpoint molecule expressed on the surface of DCs, NK cells, Tregs, macrophages, and IFN-γ-producing T cells.182 TIM-3 inhibits the function of type 1T helper cells and the secretion of several immune factors, such as IFN-γ and TNF.182 In addition, as an immune checkpoint molecule, TIM-3 can also inhibit antitumor immunity by depleting tumor-infiltrating T cells.183 Therefore, in tumors, regulating the expression of TIM-3 is also one of the mechanisms to achieve TIE, and ncRNAs are significant players in this regulation. For example, in HCC, lncRNA Tim3 can stimulate CD8+ T cell exhaustion and promote the survival of these exhausted CD8+ T cells by specifically binding to TIM-3, thereby inhibiting the T cell-mediated antitumor immune response and promoting TIE.184 In melanoma, miR-28 inhibits the expression of TIM-3, PD-1, and CTLA-4, thereby reducing T cell exhaustion and increasing TNF-α and IL-2 secretion, which could enhance the antitumor immune response and inhibit TIE (Table 5).179
BTLA
Similar to PD-1 and CTLA-4, BTLA is also an inhibitory receptor on the surface of T cells, which could be used by tumors to achieve immune escape. It is expressed on both type 1 and type 2 T helper cells but is not expressed on highly polarized type 2 T helper cells.185 BTLA reduces IL-12 secretion by cross-linking with the antigen receptor, inducing its phosphorylation, and binding to the tyrosine phosphatase SHP-1/2 containing the Src homology 2 (SH2) domain.185 Moreover, BTLA-deficient T cells show increased proliferation.185 As mentioned in the previous section, in melanoma, miR-28 can inhibit the expression of BTLA, PD-1, and CTLA-4 in T cells, which could enhance the antitumor immune response and inhibit TIE.179 In addition, miR-155 can also inhibit BTLA expression, thereby weakening the inhibitory effect of BTLA on T cell activation (Table 5).186
In summary, regulating the expression of immune checkpoints on the surface of tumor cells or immune cells is a significant strategy for tumors to achieve immune escape, and ncRNAs play essential roles in this process. It may be an important strategy to inhibit TIE and improve the efficiency of immunotherapy by inhibiting the expression of these ncRNAs. At present, anti-immune checkpoint molecular targeting drugs have been used in the clinic, and good results have been achieved.154,155 However, targeted therapy for these ncRNAs has not been reported and needs further study.
Cytokines in tme and TIE
TGF-β
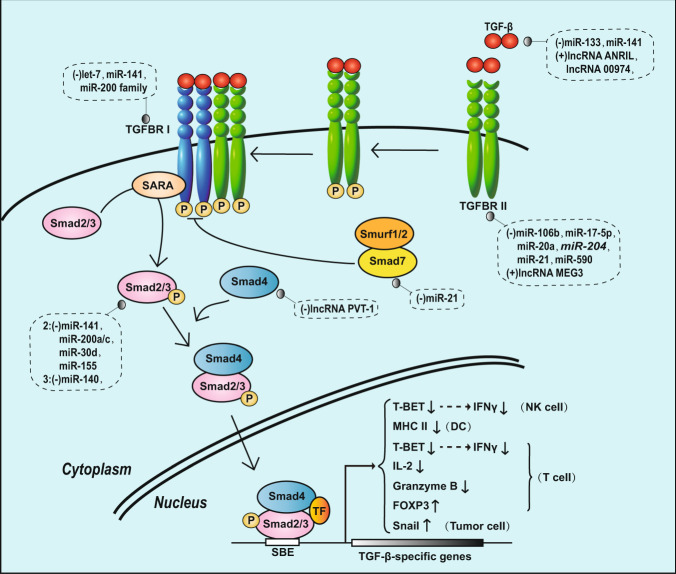
The role of TGF-β in tumorigenesis and development is contradictory. The TGF-β signaling pathway can inhibit tumor growth because the downstream signals of TGF-β family receptors can regulate the expression of DAPK, GADD45β, BIM, SHIP and other apoptosis genes, which can induce apoptosis of tumor cells.187 However, some studies have indicated that TGF-β signaling pathway activation can promote tumor growth and invasion and is critical for TIE.188 The mechanism is as follows: the TGF-β signaling pathway can induce the transcription of relevant target genes to inhibit the activation and/or functions of NK cells, DCs, and T cells and induce the differentiation of Tregs (Fig. 5).187 In NK cells, TGF-β signaling inhibits the expression of the transcription factor TBET, which inhibits IFN-γ expression, thereby inhibiting the function of NK cells.189,190 In DCs, TGF-β signaling inhibits MHC class II gene expression, thus inhibiting its antigen-presenting function.191 In T cells, TGF-β signaling silences the expression of TBET, thereby inhibiting the production of INF-γ.192 TGF-β signaling also inhibits the expression of IL-2 and granzyme B,193,194 thereby preventing the antitumor immune function of T cells. TGF-β signaling can also induce Treg differentiation by inducing the expression of FOXP3, the main transcription factor essential for the Treg phenotype, and then increase immunosuppression.195 Moreover, the TGF-β signaling pathway can also induce the expression of the transcription factor Snail in tumor cells and promote EMT, which may be another mechanism of TGF-β-induced immune escape (Fig. 5).196 Recent research has demonstrated that TGF-β promotes TIE via a mechanism independent of the canonical TGF-β signaling pathway. Tumor progression results in the downregulation of TGFBRII in T cells, enabling TGF-β to directly enter T cells. Once inside the cell, TGF-β molecules bind the Smad protein in mitochondria and disrupt the ATP-coupled respiration of T cells, thereby inhibiting their function and promoting TIE.197
Fig. 5.
TIE and the canonical TGF-β signaling pathway regulated by ncRNAs
TGF-β receptors are serine/threonine kinase receptors divided into type I and type II (TGFBR Ι and TGFBR II) receptors. Binding of the ligand TGF-β to TGFBR II induces the assembly of TGFBRΙ and TGFBR II complexes, and TGFBR II phosphorylates and activates TGFBR I, which then phosphorylates the two C-terminal serine residues of smad2 and Smad3. Phosphorylated Smad2 and Smad3 form heterotrimeric complexes with Smad4; this activated Smad complex enters the nucleus, interacts with other transcription factors, and regulates the expression of the corresponding target genes. Smad7 can form a complex with Smurf1/2 to inhibit the activity of TGFBR I, thereby inhibiting the activity of the pathway (Fig. 5).
ncRNAs in tumors can target components of the TGF-β signaling pathway or directly regulate TGF-β target gene transcription in different tumors, thus inhibiting the antitumor immune response and promoting TIE (Table 6, Fig. 5). For example, miR-133,198 miR-141,199 lncRNA ANRIL,200 and lncRNA 00974201 regulate the expression of TGF-β; Let-7,202 miR-141, and the miR-200 family203 target and regulate TGFBR Ι; miR-106b,204 miR-17-5p,205 miR-204,206 miR-20a,207 miR-21,208 miR-590209 and lncRNA MEG3210 target and regulate TGFBR II; miR-141, miR-200a/c, miR-30d203 and miR-155211 regulate Smad2; miR-140212 and lncRNA PVT1213 regulate the expression of Smad3 and Smad4, respectively; and miR-21 regulates the expression of Smad7 in cervical cancer.214 The abnormal expression of these ncRNAs in tumors can activate the TGF-β signaling pathway in different stages and then help tumors achieve immune escape by inhibiting a variety of immune cells, inducing the differentiation of immune cells and promoting EMT. According to this characteristic of tumors, blocking the activation of the TGF-β signaling pathway by targeting these ncRNAs may become a new direction for tumor immunotherapy.
Table 6.
ncRNAs influence TIE via regulating cytokines
| Target genes | ncRNAs | Function | Type of cancer | Refs. |
|---|---|---|---|---|
| TGF-β | miR-133 and miR-141 | Downregulate TGF-β expression, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Gastric cancer, Myocardial fibrosis | 198,199 |
| lncRNA ANRIL and lncRNA 00974 | Upregulate TGF-β expression, induce immunosuppression and TIE, promote tumor growth and invasion | Esophageal squamous cell carcinoma, HCC | 200,201 | |
| TGFBR Ι | Let-7, miR-141 and miR-200 family | Downregulate the expression of TGFBRΙ, weaken TGF-β signal pathway, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Thyroid carcinomas | 202,203 |
| TGFBR II | miR-106b, 17-5p, 204, 20a, 21, 590 | Downregulate the expression of TGFBR II, weaken TGF-β signal pathway, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Alzheimer’s disease, metastatic cancer, Lung cancer, Leiomyoma | 205–209,471 |
| lncRNA MEG3 | Upregulates the expression of TGFBR II, strengthens TGF-β signal pathway, induce immunosuppression and TIE, promotes tumor growth and invasion | Chondroma | 210 | |
| Smad2 | miR-141, 200a/c, 30d and 155 | Downregulate the expression of Smad2, weaken TGF-β signal pathway, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Thyroid carcinomas | 203,211 |
| Smad3 | miR-140 | Downregulates the expression of Smad3, weakens TGF-β signal pathway, inhibit immunosuppression and TIE, suppresses tumor growth and invasion | 212 | |
| Smad4 | lncRNA PVT1 | Downregulates the expression of Smad4, weakens TGF-β signal pathway, inhibit immunosuppression and TIE, suppresses tumor growth and invasion | Colorectal cancer | 213 |
| Smad7 | miR-21 | Downregulates the expression of Smad4, strengthens TGF-β signal pathway, promotes immunosuppression and TIE | Cervical cancer | 214 |
| IL-6 | miR-33a, 218, 125a, 34a, 217, 26a, 98, and 9 | Downregulate IL-6, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Gallbladder cancer, Lung cancer, Breast cancer, Cardiac myxoma, HCC, Melanoma, HeLa cell line | 220–227 |
| lncRNA HOTTIP, 00460 and UICC | Directly upregulate IL-6, induce immunosuppression and TIE, promote tumor growth and invasion | Ovarian cancer, Nasopharyngeal carcinoma, Cervical cancer | 228–230 | |
| STAT3 | miR-551b-3p, | Upregulates STAT3 expression directly, induces immunosuppression and TIE, promotes tumor growth and invasion | Ovarian cancer | 231 |
| miR-221, 222 and 18a | Downregulate STAT3 expression by negatively regulating the PDLIM2 or PIAS3, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Gastric adenocarcinoma, Colorectal cancer | 232,233 | |
| lncRNA 00518, AB073614 and HOST2 | Activate the JAK2/STAT3 signaling pathway, induce immunosuppression and TIE, promote tumor growth and invasion | Cervical cancer, Colorectal cancer, HCC | 235–237 | |
| IL-10 | miR-98 | Downregulates IL-10, inhibits immunosuppression and TIE, suppresses tumor growth and invasion | HCC | 247 |
| miR-194, miR-193b and lncRNA CCAT1 | Upregulate IL-10, induce immunosuppression and TIE, promote tumor growth and invasion | Laryngeal cancer, Lymphoma, prostate cancer | 248–250 | |
| VEGF | miR-638, 503, 497, 203, 200, 195, 190, 126, 93, 29b and 20 | Downregulate VEGF directly, inhibit immunosuppression and TIE, suppresse tumor growth and invasion | HCC, Cervical cancer, Lung cancer, Oral cancer, Breast cancer | 254–263,265,266 |
| miR-22, 107, 519c, 26a and 145 | Downregulate VEGF indirectly, inhibit immunosuppression and TIE, suppress tumor growth and invasion | Colorectal cancer, HCC | 78,269–272 | |
| lncRNA TDRG1 and lncRNA HOTAIR | Upregulate VEGF directly, induce immunosuppression and TIE, promote tumor growth and invasion | Endometrial carcinoma, Nasopharyngeal carcinoma | 267,268 | |
| lncRNA H19 and lncRNA GAS5 | Upregulate VEGF indirectly, induce immunosuppression and TIE, promote tumor growth and invasion | Mesenchymal stem cells, Colorectal cancer | 273,274 |
PDLIM2 PDZ and LIM domain protein 2, PIAS3 Protein inhibitor of activated signal transducer and activator of transcription
IL‐6
IL-6, commonly secreted by macrophages, DCs, MDSCs, and tumor cells, is a pleiotropic proinflammatory cytokine that is involved in almost all aspects of the immune system, from the infiltration of neutrophils at the site of infection to the generation of T cell responses. IL-6 can be rapidly induced and expressed in large quantities under the conditions of infection and autoimmunity and plays a key role in host defense by stimulating various cell populations (including promoting cytotoxic T cell differentiation, T cell population expansion and activation, and B cell differentiation).215 However, in addition to immune-stimulating effects, IL-6 can also lead to immunosuppression and TIE, most of which are mediated through the IL-6/JAK2/STAT3 signaling pathway. For example, the IL-6/JAK2/STAT3 signaling pathway can make the TME tend towards immunosuppress by attracting and activating MDSCs, TANs, and Tregs.216 In addition, the IL-6/JAK2/STAT3 signaling pathway has also been found to promote the tumor stem cell-like phenotype and EMT.217–219 In tumors, abnormal expression of ncRNAs can promote TIE by regulating the expression of IL-6 or the molecules involved in the IL-6/JAK2/STAT3 signaling pathway (Table 6). miR-33a,220 miR-218,221 miR-125a,222 miR-34a,223 miR-217,224 miR-26a,225 miR-98,226 miR-9,227 lncRNA HOTTIP,228 lncRNA 00460,229 and lncRNA UICC230 directly regulate IL-6 in different cancers. miR-551b-3p directly downregulates STAT3 expression in ovarian cancer cells,231 whereas miR-18a inhibits STAT3 by negatively regulating the expression of the E3 SUMO protein ligase PIAS3 in gastric adenocarcinogenesis.232 miR-221 and miR-222 inhibit STAT3 expression by targeting PDZ and LIM domain protein 2 (PDLIM2) in colorectal cancer.233 Moreover, miR-30 promotes the activation of the JAK/STAT3 pathway by inhibiting the expression of suppressor of cytokine signaling 3 (SOCS3) in glioma stem cells.234 lncRNA 00518, lncRNA AB073614, and lncRNA HOST2 activate the JAK2/STAT3 signaling pathway in cervical cancer, colorectal cancer, and HCC, respectively.235–237 These ncRNAs enhance the immunosuppression of the TME by regulating IL-6 expression and activating the IL-6/JAK2/STAT3 pathway, thus helping tumors achieve immune escape, which may be another target to block TIE and improve the effect of immunotherapy.
IL-10
IL-10 is an immune cytokine produced by immune cells that plays a dual role in tumorigenesis and development.238 In the early tumor stage, the main role of IL-10 is to activate the immune system to kill tumor cells by stimulating NK cell- and CTL cell-mediated antitumor responses. However, with tumor development, some tumors (melanoma,239 lung cancer240 and bladder cancer241) have been found to be able to utilize the immunosuppressive effects of IL-10 to achieve TIE.242 Specific mechanisms are described below. First, IL-10 can inhibit the expression of MHC class II molecules on antigen-presenting cells and MHC class I molecules in tumor cells to inhibit tumor antigen presentation.243,244 Second, IL-10 can inhibit the activation of the CD28 costimulatory pathway in T cells, resulting in T cell dysfunction.245 Finally, tumor cells express IL-10 and IL-10 receptors by themselves, activate the downstream STAT3/Twist pathway by autocrine signaling, and promote EMT of tumor cells.246 It has been revealed that ncRNAs such as miR-98,247 miR-193b,248 miR-194,249 and lncRNA CCAT1250 can directly or indirectly regulate IL-10 expression in different tumors to help achieve TIE (Table 6). Therefore, IL-10 may be considered another important target for ncRNA-mediated regulation of TIE. Through the abnormal expression of ncRNAs in tumors, the regulation of IL-10 is likely to be a strategy of tumors to achieve TIE. Therefore, we need to further study these mechanisms, which may become effective targets for cancer treatment in the future.
VEGF
Vascular endothelial growth factor (VEGF) is an endothelial cell-specific mitogen that is an angiogenesis inducer in a variety of in vivo models and has important physiological functions.251 In addition, plenty of evidence shows that VEGF is also an important regulator of pathological angiogenesis. In situ hybridization studies have shown that VEGF mRNA is expressed in most human tumors and can promote tumor progression and help tumors achieve TIE.252 Mechanistically, VEGF promotes the proliferation of immunosuppressive cells, inhibits the recruitment of T cells in the TME, and promotes the exhaustion of T cells, thus promoting TIE.252 VEGF can also act as a chemokine to recruit Tregs into the TME, thereby affecting the antitumor immune response of immune cells.253 In many tumors, it has been found that some abnormally expressed ncRNAs can directly or indirectly regulate the expression of VEGF to promote the development of tumors (Table 6). For example, miR-638,254 miR-503,255 miR-497,256 miR-203,257 miR-200,258 miR-195,259 miR-190,260 miR-126,261,262 miR-93,263,264 miR-29b,265 miR-20,266 lncRNA TDRG1267 and lncRNA HOTAIR268 directly up/downregulate VEGF expression in various tumors. In contrast, miR-22,78 miR-107,269 miR-519c,270 miR-26a,271 miR-145,272 lncRNA H19,273 and lncRNA GAS5274 were found indirectly to up/downregulate VEGF expression in different tumors. Therefore, similar to TGF-β, IL-6 and IL-10, tumors are likely to utilize the immunosuppressive function of VEGF through ncRNAs to finally achieve TIE. This is likely to become another target of antitumor immunotherapy.
Tumor exosomes (TEX) and TIE
Exosomes are extracellular vesicles 40–150 nm in diameter that participate in intercellular communication. TEX can transport various ncRNAs and support the associated immunosuppressive functions.12,275 Currently, ncRNAs transported by exosomes from various tumor cells have been reported to be associated with immunosuppression and TIE (Table 7). For example, in HCC, miR-23a in TEX can inhibit the expression of PTEN and upregulate PD-L1 expression in macrophages, decrease antitumor immunity and promote TIE.276 Furthermore, miR-23a derived from hypoxic TEX can also act as an immunosuppressive factor that directly targets CD107 in NK cells, thus inhibiting the killing function of NK cells on tumors.277 Similarly, miR-24-3p, miR-891a, miR-106a-5p, miR-20a-5p, and miR-1980 in nasopharyngeal carcinoma-derived TEX were also found to promote immunosuppression and TIE. These miRNAs can induce the differentiation of Tregs and cause T cell dysfunction, including dysregulation of proliferation, differentiation, and cytokine secretion, by targeting the downregulation of the MAPK1 and JAK/STAT pathways.278 In mouse models of lung cancer and sarcoma, TEX-derived miR-214 inhibits PTEN and the signals downstream of PTEN (to some extent) in T cells, thereby promoting the differentiation of Tregs and enhancing immunosuppression.279 TEX-derived lncRNAs were also found to be associated with immunosuppression and TIE. lncRNA ZFAS1 from gastric cancer-derived TEX can suppress tumor cell apoptosis and promote EMT, which may help gastric cancer to achieve TIE.280 TEX isolated from the bladder cancer cell line 5637 show high lncRNA UCA1 expression, which promotes bladder cancer progression by promoting EMT in tumor cells.281 In add ition, TEX derived from lung cancer show increased lncRNA MALAT1 expression, which promotes tumor growth and metastasis and inhibits tumor cell apoptosis.282 These TEX-derived ncRNAs can promote immunosuppression, inhibit tumor apoptosis and promote tumor EMT through a variety of mechanisms, which are significant driving forces for TIE. This may also be one of the potential causes of immunotherapy tolerance, which needs to be studied in depth.
Table 7.
ncRNAs transferred by TEX participate in TIE
| ncRNAs transferred by TEX | Function | Tumor type | Refs. |
|---|---|---|---|
| miR-24-3p, 891a, 106a-5p, 20a-5p and 1908 | Promote T cell dysfunction by downregulating the MAPK1 pathway, thereby inhibiting the antitumor immunity and promote TIE | Nasopharyngeal carcinoma | 278 |
| miR-214 and miR-214 | Promote the Treg phenotype by inhibiting PTEN in T cells, thereby inhibiting the antitumor immunity and promote TIE | Mouse lung cancer and sarcoma, NSCLC, Multiple myeloma | 279,472,473 |
| miR-23a | Upregulates PD-L1 expression in macrophages and inhibits NK cells via downregulate CD107, thereby inhibiting the antitumor immunity and promote TIE | HCC, CML | 276,277 |
| miR-212-3p | Inhibits antigen presentation process by downregulating MHC class II molecules in immature DC cells, thereby inhibiting the antitumor immunity and promote TIE | Pancreatic cancer | 474 |
| miR-21 and miR-29a | Lead to the M2-type polarization of TAMs and promotes immunosuppression and TIE | NSCLC | 475,476 |
| lncRNA ZFAS1 | Suppresses apoptosis of tumor cells and promotes the process of EMT, thereby promote TIE | Gastric cancer | 280 |
| lncRNA UCA1 | Promotes the process of EMT, thereby promote TIE | Bladder cancer | 281 |
| lncRNA MALAT1 | Promotes tumor growth, metastasis and inhibits tumor cell apoptosis, thereby promote TIE | Lung cancer | 282 |
CML chronic myeloid leukemia
Conclusions and perspectives
In this review, we discussed the different TIE mechanisms and summarized the regulatory roles of ncRNAs involved in these mechanisms. Although ncRNAs have been confirmed to be directly associated with TIE, the precise molecular mechanisms underlying such regulation are yet to be elucidated. Immunotherapy often results in low response rates owing to the multiple TIE mechanisms active in tumor cells, and the role of ncRNAs in TIE may be underestimated. Thus, ncRNAs may be considered potential candidates to therapeutically target such TIE mechanisms and are expected to be the key to overcoming the challenges associated with immunotherapy.
Several recent studies have reported the role of ncRNAs in tumor therapy, demonstrating the therapeutic potential of ncRNAs.283–286 For example, miR-122 expression is low in liver cancer cells, and delivery of miR-122 in liver tumor cells using LNP-DP1, a cationic lipid nanoparticle formulation, can effectively suppress tumor growth by inhibiting target genes and angiogenesis.287 Although the potential of ncRNAs as tumor therapeutic targets has been reported, no study or clinical experiment has reported whether ncRNAs can be considered targets to inhibit TIE. Therefore, considering the key role of ncRNAs in TIE, further research is warranted to explore such therapeutic approaches to improve the efficiency of tumor immunotherapy and reduce the associated side effects. Because ncRNAs are critical in promoting TIE, the potential of ncRNAs as targets for TIE therapy should not be underestimated.
This review summarizes the complex regulatory network of ncRNAs specific to TIE. Some studies provide direct evidence of the involvement of certain ncRNAs in TIE, whereas other studies report observations suggestive of the involvement of certain ncRNAs in the regulation of immune escape-related mechanisms. Although not conclusively investigated, these ncRNAs may be indirectly involved in TIE and can also be explored as potential targets for TIE therapy. Some ncRNAs have both tumorigenic and antitumorigenic properties, and studies involving such ncRNAs may provide new insights into TIE mechanisms and immunotherapy. Further studies verifying the regulatory relationship between ncRNAs and TIE will provide a direction for future studies aimed at developing novel cancer therapeutic approaches. Aside from miRNAs, lncRNAs, and circRNAs, other ncRNAs, such as tRNAs (including their derived tiRNAs and tRFs), rRNAs, and snRNAs, have not been reported to be involved in the regulation of TIE, and further research in this area is essential.
Acknowledgements
This work was supported by funding from the Project Nn10 of Harbin Medical University Cancer Hospital (Grant Number Nn102017-02), National Natural Science Foundation of China (Grant Number 81602323, 81872149), Outstanding Youth Project of Heilongjiang Provincial Natural Science Foundation (Grant Number YQ2019H027), Distinguished Young Scholars of Harbin Medical University Cancer Hospital (Grant Number JCQN2018-03), Yong Elite Training Foundation Grant of Harbin Medical University Cancer Hospital (Grant Number JY2016-02) and Haiyan Fund Project of Harbin Medical University Cancer Hospital (Grant Number JJQN 2018-10).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Lei Liu, Qin Wang
Contributor Information
Da Pang, Email: pangda@ems.hrbmu.edu.cn.
Shouping Xu, Email: Shoupingxu@hrbmu.edu.cn.
References
- 1.Dunn GP, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 2.Shahrouki P, Larsson E. The non-coding oncogene: a case of missing DNA evidence? Front Genet. 2012;3:170. doi: 10.3389/fgene.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green D, Fraser WD, Dalmay T. Transfer RNA-derived small RNAs in the cancer transcriptome. Pflug. Arch. 2016;468:1041–1047. doi: 10.1007/s00424-016-1822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, L., et al. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 7, 3101–3109 (2018). [DOI] [PMC free article] [PubMed]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Huang S, et al. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010;38:7211–7218. doi: 10.1093/nar/gkq564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J. Mol. Med. (Berl.) 2018;96:1167–1176. doi: 10.1007/s00109-018-1693-y. [DOI] [PubMed] [Google Scholar]
- 12.Eichmuller, S. B., Osen, W., Mandelboim, O. & Seliger, B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J. Natl. Cancer Inst. 109, 10.1093/jnci/djx034 (2017). [DOI] [PubMed]
- 13.Ding, L., Lu, S. & Li, Y. Regulation of PD-1/PD-L1 pathway in cancer by noncoding RNAs. Pathol. Oncol Res. 26, 651–663 (2019). [DOI] [PubMed]
- 14.Hoves S, et al. In situ analysis of the antigen-processing machinery in acute myeloid leukaemic blasts by tissue microarray. Leukemia. 2009;23:877–885. doi: 10.1038/leu.2008.391. [DOI] [PubMed] [Google Scholar]
- 15.Meissner M, et al. Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: association with clinical outcome. Clin. Cancer Res. 2005;11:2552–2560. doi: 10.1158/1078-0432.CCR-04-2146. [DOI] [PubMed] [Google Scholar]
- 16.Ogino T, et al. HLA class I antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res. 2006;66:9281–9289. doi: 10.1158/0008-5472.CAN-06-0488. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, et al. MicroRNA-23a downregulates the expression of interferon regulatory factor-1 in hepatocellular carcinoma cells. Oncol. Rep. 2016;36:633–640. doi: 10.3892/or.2016.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, et al. Rs56288038 (C/G) in 3’UTR of IRF-1 regulated by MiR-502-5p promotes gastric cancer development. Cell Physiol. Biochem. 2016;40:391–399. doi: 10.1159/000452554. [DOI] [PubMed] [Google Scholar]
- 19.Namiki S, et al. IRF-1 mediates upregulation of LMP7 by IFN-gamma and concerted expression of immunosubunits of the proteasome. FEBS Lett. 2005;579:2781–2787. doi: 10.1016/j.febslet.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, et al. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell Endocrinol. 2016;433:75–86. doi: 10.1016/j.mce.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Hirata T, et al. Characterization of the immune escape phenotype of human gastric cancers with and without high-frequency microsatellite instability. J. Pathol. 2007;211:516–523. doi: 10.1002/path.2142. [DOI] [PubMed] [Google Scholar]
- 22.Mari L, et al. microRNA 125a regulates MHC-I expression on esophageal adenocarcinoma cells, associated with suppression of antitumor immune response and poor outcomes of patients. Gastroenterology. 2018;155:784–798. doi: 10.1053/j.gastro.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Bartoszewski R, et al. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J. Biol. Chem. 2011;286:41862–41870. doi: 10.1074/jbc.M111.304956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musahl AS, et al. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene. 2015;34:5046–5054. doi: 10.1038/onc.2014.424. [DOI] [PubMed] [Google Scholar]
- 25.Colangelo T, et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016;7:e2120. doi: 10.1038/cddis.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maleno I, et al. Distribution of HLA class I altered phenotypes in colorectal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics. 2004;56:244–253. doi: 10.1007/s00251-004-0692-z. [DOI] [PubMed] [Google Scholar]
- 27.Menon AG, et al. Down-regulation of HLA-A expression correlates with a better prognosis in colorectal cancer patients. Lab Invest. 2002;82:1725–1733. doi: 10.1097/01.lab.0000043124.75633.ed. [DOI] [PubMed] [Google Scholar]
- 28.Browning M, et al. Mechanisms of loss of HLA class I expression on colorectal tumor cells. Tissue Antigens. 1996;47:364–371. doi: 10.1111/j.1399-0039.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 29.Gao F, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem. Biophys. Res Commun. 2013;431:610–616. doi: 10.1016/j.bbrc.2012.12.097. [DOI] [PubMed] [Google Scholar]
- 30.Juin P, et al. Decoding and unlocking the BCL-2 dependency of cancer cells. Nat. Rev. Cancer. 2013;13:455–465. doi: 10.1038/nrc3538. [DOI] [PubMed] [Google Scholar]
- 31.Espana L, et al. Overexpression of Bcl-xL in human breast cancer cells enhances organ-selective lymph node metastasis. Breast Cancer Res. Treat. 2004;87:33–44. doi: 10.1023/B:BREA.0000041579.51902.89. [DOI] [PubMed] [Google Scholar]
- 32.Goncharenko-Khaider N, et al. Ovarian cancer ascites increase Mcl-1 expression in tumor cells through ERK1/2-Elk-1 signaling to attenuate TRAIL-induced apoptosis. Mol. Cancer. 2012;11:84. doi: 10.1186/1476-4598-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Significance of Fas and FasL protein expression in cardiac carcinoma and local lymph node tissues. Int J. Clin. Exp. Pathol. 2015;8:11915–11920. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Twomey JD, et al. Spatial dynamics of TRAIL death receptors in cancer cells. Drug Resist Updat. 2015;19:13–21. doi: 10.1016/j.drup.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Soriano C, et al. Increased proteinase inhibitor-9 (PI-9) and reduced granzyme B in lung cancer: mechanism for immune evasion? Lung Cancer. 2012;77:38–45. doi: 10.1016/j.lungcan.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Zhou XN, et al. Knockdown of decoy receptor 3 impairs growth and invasiveness of hepatocellular carcinoma cell line of HepG2. Chin. Med. J. (Engl.) 2016;129:2623–2629. doi: 10.4103/0366-6999.192775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Saini N. Downregulation of BCL2 by miRNAs augments drug-induced apoptosis–a combined computational and experimental approach. J. Cell Sci. 2012;125:1568–1578. doi: 10.1242/jcs.095976. [DOI] [PubMed] [Google Scholar]
- 38.Liu, J. & Tong, C. Q. Inhibitory effect of miR-125b down-regulation on proliferation of leukemia cell K562. Zhongguo Shi Yan Xue Ye Xue Za Zhi26, 336–340 (2018). [DOI] [PubMed]
- 39.You F, et al. miRNA-106a promotes breast cancer cell proliferation, clonogenicity, migration, and invasion through inhibiting apoptosis and chemosensitivity. DNA Cell Biol. 2019;38:198–207. doi: 10.1089/dna.2018.4282. [DOI] [PubMed] [Google Scholar]
- 40.Ji F, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Razumilava N, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, et al. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum. Cell. 2018;31:232–241. doi: 10.1007/s13577-018-0206-1. [DOI] [PubMed] [Google Scholar]
- 43.Lin R, et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. Invest. 2014;124:5352–5367. doi: 10.1172/JCI76561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi SY, Collins CC, Gout PW, Wang Y. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J. Pathol. 2013;230:350–355. doi: 10.1002/path.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellone M, et al. The acidity of the tumor microenvironment is a mechanism of immune escape that can be overcome by proton pump inhibitors. Oncoimmunology. 2013;2:e22058. doi: 10.4161/onci.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu P, et al. MicroRNA-340 mediates metabolic shift in oral squamous cell carcinoma by targeting glucose transporter-1. J. Oral. Maxillofac. Surg. 2016;74:844–850. doi: 10.1016/j.joms.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 48.Yamasaki T, et al. Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Sci. 2013;104:1411–1419. doi: 10.1111/cas.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie S, et al. miR-495 mediates metabolic shift in glioma cells via targeting Glut1. J. Craniofac. Surg. 2015;26:e155–e158. doi: 10.1097/SCS.0000000000001385. [DOI] [PubMed] [Google Scholar]
- 50.Chen B, et al. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015;356:410–417. doi: 10.1016/j.canlet.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 51.Qu W, et al. miR-132 mediates a metabolic shift in prostate cancer cells by targeting Glut1. FEBS Open Bio. 2016;6:735–741. doi: 10.1002/2211-5463.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi: 10.1016/j.canlet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 54.Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle. 2016;15:3471–3481. doi: 10.1080/15384101.2016.1249545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chow TF, et al. The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J. Urol. 2010;183:743–751. doi: 10.1016/j.juro.2009.09.086. [DOI] [PubMed] [Google Scholar]
- 56.Srivastava SK, et al. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fei X, et al. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586:392–397. doi: 10.1016/j.febslet.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Fang R, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 2012;287:23227–23235. doi: 10.1074/jbc.M112.373084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peschiaroli A, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 60.Jiang S, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–1998. doi: 10.1038/emboj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, et al. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–955. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo W, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 63.Fabani MM, Gait MJ. miR-122 targeting with LNA/2’-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kefas B, et al. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–1112. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kinoshita T, et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int J. Oncol. 2012;40:185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, et al. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget. 2015;6:19456–19468. doi: 10.18632/oncotarget.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hung CL, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl Acad. Sci. USA. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Sun X. MicroRNA-375 inhibits the proliferation, migration and invasion of kidney cancer cells by triggering apoptosis and modulation of PDK1 expression. Environ. Toxicol. Pharm. 2018;62:227–233. doi: 10.1016/j.etap.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, et al. miR-138-1* regulates aflatoxin B1-induced malignant transformation of BEAS-2B cells by targeting PDK1. Arch. Toxicol. 2016;90:1239–1249. doi: 10.1007/s00204-015-1551-4. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka H, et al. MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J. Neurooncol. 2013;111:273–283. doi: 10.1007/s11060-012-1027-9. [DOI] [PubMed] [Google Scholar]
- 71.Puissegur MP, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung KA, Lee S, Kwak MK. NFE2L2/NRF2 activity is linked to mitochondria and AMP-activated protein kinase signaling in cancers through miR-181c/mitochondria-encoded cytochrome c oxidase regulation. Antioxid. Redox Signal. 2017;27:945–961. doi: 10.1089/ars.2016.6797. [DOI] [PubMed] [Google Scholar]
- 73.Aschrafi A, et al. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharm. Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutendra G, et al. Mitochondrial activation by inhibition of PDKII suppresses HIF1a signaling and angiogenesis in cancer. Oncogene. 2013;32:1638–1650. doi: 10.1038/onc.2012.198. [DOI] [PubMed] [Google Scholar]
- 76.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 77.Taguchi A, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 78.Yamakuchi M, Yagi S, Ito T, Lowenstein CJ. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS ONE. 2011;6:e20291. doi: 10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, et al. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer Biol. Ther. 2015;16:846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan Y, et al. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1alpha. Mol. Carcinog. 2019;58:875–886. doi: 10.1002/mc.22976. [DOI] [PubMed] [Google Scholar]
- 81.Zhang L, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J. Cell Biochem. 2018;119:5460–5472. doi: 10.1002/jcb.26705. [DOI] [PubMed] [Google Scholar]
- 82.Cai, Q. et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1alpha via miR-138. Open Biol. 7, 160247 (2017). [DOI] [PMC free article] [PubMed] [Retracted]
- 83.Chi Y, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1alpha regulation. J. Cell Biochem. 2019;120:19019–19030. doi: 10.1002/jcb.29225. [DOI] [PubMed] [Google Scholar]
- 84.Sharma S, et al. Tumor cyclooxygenase 2-dependent suppression of dendritic cell function. Clin. Cancer Res. 2003;9:961–968. [PubMed] [Google Scholar]
- 85.Akasaki Y, et al. Induction of a CD4+ T regulatory type 1 response by cyclooxygenase-2-overexpressing glioma. J. Immunol. 2004;173:4352–4359. doi: 10.4049/jimmunol.173.7.4352. [DOI] [PubMed] [Google Scholar]
- 86.Sharma, S. et al. Tumor cyclooxygenase-2/prostaglandin E2-dependent promotion of FOXP3 expression and CD4+ CD25+ T regulatory cell activities in lung cancer. Cancer Res.65, 5211–5220 (2005). [DOI] [PubMed]
- 87.Uotila P. The role of cyclic AMP and oxygen intermediates in the inhibition of cellular immunity in cancer. Cancer Immunol. Immunother. 1996;43:1–9. doi: 10.1007/BF03354243. [DOI] [PubMed] [Google Scholar]
- 88.Park, Y. R. et al. MiRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targetting TM4SF1. Biosci Rep. 38, BSR20180664 (2018). [DOI] [PMC free article] [PubMed]
- 89.Lin Y, Wu Z. MicroRNA-128 inhibits proliferation and invasion of glioma cells by targeting COX-2. Gene. 2018;658:63–69. doi: 10.1016/j.gene.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 90.Cornett AL, Lutz CS. Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA. 2014;20:1419–1430. doi: 10.1261/rna.044149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y, Li H, Zhao C, Jia H. MicroRNA-101 inhibits angiogenesis via COX-2 in endometrial carcinoma. Mol. Cell Biochem. 2018;448:61–69. doi: 10.1007/s11010-018-3313-0. [DOI] [PubMed] [Google Scholar]
- 92.Pham H, et al. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2013;439:6–11. doi: 10.1016/j.bbrc.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wainwright DA, et al. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin. Cancer Res. 2012;18:6110–6121. doi: 10.1158/1078-0432.CCR-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 95.Prendergast GC, Metz R, Muller AJ. Towards a genetic definition of cancer-associated inflammation: role of the IDO pathway. Am. J. Pathol. 2010;176:2082–2087. doi: 10.2353/ajpath.2010.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J. Biol. Macromol. 2018;118:24–30. doi: 10.1016/j.ijbiomac.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 97.Li X, et al. Long non-coding RNA MALAT1 promotes proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing VEGF and IDO. J. Cell Biochem. 2017;118:2780–2791. doi: 10.1002/jcb.25927. [DOI] [PubMed] [Google Scholar]
- 98.Zhao XS, et al. miR-153-3p, a new bio-target, is involved in the pathogenesis of acute graft-versus-host disease via inhibition of indoleamine- 2,3-dioxygenase. Oncotarget. 2016;7:48321–48334. doi: 10.18632/oncotarget.10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maccalli C, Rasul KI, Elawad M, Ferrone S. The role of cancer stem cells in the modulation of anti-tumor immune responses. Semin Cancer Biol. 2018;53:189–200. doi: 10.1016/j.semcancer.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chai S, et al. Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology. 2016;64:2062–2076. doi: 10.1002/hep.28821. [DOI] [PubMed] [Google Scholar]
- 102.Fang G, et al. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2018;504:184–189. doi: 10.1016/j.bbrc.2018.08.152. [DOI] [PubMed] [Google Scholar]
- 103.Chen Z, Duan X. hsa_circ_0000177-miR-638-FZD7-Wnt signaling cascade contributes to the malignant behaviors in glioma. DNA Cell Biol. 2018;37:791–797. doi: 10.1089/dna.2018.4294. [DOI] [PubMed] [Google Scholar]
- 104.Kang L, et al. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci. 2015;106:700–708. doi: 10.1111/cas.12656. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Tang B, et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int J. Cancer. 2018;142:109–120. doi: 10.1002/ijc.31041. [DOI] [PubMed] [Google Scholar]
- 106.Miller TJ, et al. Expression of PD-L1 and SOX2 during rectal tumourigenesis: potential mechanisms for immune escape and tumour cell invasion. Oncol. Lett. 2018;16:5761–5768. doi: 10.3892/ol.2018.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Noh KH, et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J. Clin. Invest. 2012;122:4077–4093. doi: 10.1172/JCI64057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun Z, et al. MicroRNA-34a regulates epithelial-mesenchymal transition and cancer stem cell phenotype of head and neck squamous cell carcinoma in vitro. Int J. Oncol. 2015;47:1339–1350. doi: 10.3892/ijo.2015.3142. [DOI] [PubMed] [Google Scholar]
- 109.Peng F, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ai L, et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer. 2019;18:88. doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann. Oncol. 2010;21(Suppl 7):89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kudo-Saito, C. Shirako, H. Takeuchi, T. & Kawakami, Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell.15, 195–206 (2009). [DOI] [PubMed]
- 114.Dongre A, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanchez-Tillo E, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol. Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Block CJ, et al. A stroma-corrected ZEB1 transcriptional signature is inversely associated with antitumor immune activity in breast cancer. Sci. Rep. 2019;9:17807. doi: 10.1038/s41598-019-54282-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jaca A, Govender P, Locketz M, Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J. Clin. Pathol. 2017;70:331–356. doi: 10.1136/jclinpath-2016-204031. [DOI] [PubMed] [Google Scholar]
- 118.Dong P, et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. 2016;35:132. doi: 10.1186/s13046-016-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Q, et al. The effects and molecular mechanisms of MiR-106a in multidrug resistance reversal in human glioma U87/DDP and U251/G cell lines. PLoS ONE. 2015;10:e0125473. doi: 10.1371/journal.pone.0125473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dong P, et al. MicroRNA-106b modulates epithelial-mesenchymal transition by targeting TWIST1 in invasive endometrial cancer cell lines. Mol. Carcinog. 2014;53:349–359. doi: 10.1002/mc.21983. [DOI] [PubMed] [Google Scholar]
- 121.Peng F, Zhang H, Du Y, Tan P. miR-23a promotes cisplatin chemoresistance and protects against cisplatin-induced apoptosis in tongue squamous cell carcinoma cells through Twist. Oncol. Rep. 2015;33:942–950. doi: 10.3892/or.2014.3664. [DOI] [PubMed] [Google Scholar]
- 122.Wu, B. Q. et al. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res. Treat. 16, 819–827 (2017). [DOI] [PMC free article] [PubMed]
- 123.Xue M, et al. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 2016;107:18–27. doi: 10.1111/cas.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luan W, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem. Biophys. Res. Commun. 2018;502:22–29. doi: 10.1016/j.bbrc.2018.05.114. [DOI] [PubMed] [Google Scholar]
- 125.Vignali, D. A. A., Collison, L. W., & Workman, C. J. How regulatory T cells work. Nat. Rev. Immunol.8, 523–532 (2008). [DOI] [PMC free article] [PubMed]
- 126.Colonna, M. Immunology: an innate regulatory cell. Nature498, 42–43 (2013). [DOI] [PubMed]
- 127.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barron L, et al. Cutting edge: mechanisms of IL-2-dependent maintenance of functional regulatory T cells. J. Immunol. 2010;185:6426–6430. doi: 10.4049/jimmunol.0903940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao J, et al. LncRNA HULC affects the differentiation of Treg in HBV-related liver cirrhosis. Int. Immunopharmacol. 2015;28:901–905. doi: 10.1016/j.intimp.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 131.Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Am. J. Transl. Res. 2015;7:2262–2269. [PMC free article] [PubMed] [Google Scholar]
- 132.Parker KH, et al. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014;74:5723–5733. doi: 10.1158/0008-5472.CAN-13-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thevenot PT, et al. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. 2014;41:389–401. doi: 10.1016/j.immuni.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Highfill SL, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 2014;6:237ra267. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lindau D, et al. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138:105–115. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nagaraj S, et al. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng Y, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol. Cancer. 2019;18:61. doi: 10.1186/s12943-019-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tian X, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol. 2018;9:473. doi: 10.3389/fimmu.2018.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Huang A, et al. miR-34a expands myeloid-derived suppressor cells via apoptosis inhibition. Exp. Cell Res. 2014;326:259–266. doi: 10.1016/j.yexcr.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 140.Tian J, et al. MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J. Immunol. 2015;195:1301–1311. doi: 10.4049/jimmunol.1500209. [DOI] [PubMed] [Google Scholar]
- 141.Wang J, et al. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int. J. Cancer. 2015;136:E602–E613. doi: 10.1002/ijc.29151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ruffell B, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alderton GK. Resistance: turning macrophages on, off and on again. Nat. Rev. Cancer. 2014;14:154–155. doi: 10.1038/nrc3688. [DOI] [PubMed] [Google Scholar]
- 144.Tian Y, et al. MicroRNA-342 inhibits tumor growth via targeting chemokine CXCL12 involved in macrophages recruitment/activation. Genes Cells. 2018;23:1009–1022. doi: 10.1111/gtc.12650. [DOI] [PubMed] [Google Scholar]
- 145.Zhou YX, et al. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018;104:25–33. doi: 10.1016/j.biocel.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 146.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parry RV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell Biol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schneider H, et al. CTLA-4 disrupts ZAP70 microcluster formation with reduced T cell/APC dwell times and calcium mobilization. Eur. J. Immunol. 2008;38:40–47. doi: 10.1002/eji.200737423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wei J, et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 2016;18:639–648. doi: 10.1093/neuonc/nov292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huffaker TB, et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017;292:18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wainwright DA, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin. Cancer Res. 2014;20:5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alvarez-Argote J, Dasanu CA. Durvalumab in cancer medicine: a comprehensive review. Expert Opin. Biol. Ther. 2019;19:927–935. doi: 10.1080/14712598.2019.1635115. [DOI] [PubMed] [Google Scholar]
- 155.Horn L, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N. Engl. J. Med. 2018;379:2220–2229. doi: 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- 156.Eppihimer MJ, et al. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation. 2002;9:133–145. doi: 10.1038/sj/mn/7800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Muhlbauer M, et al. PD-L1 is induced in hepatocytes by viral infection and by interferon-alpha and -gamma and mediates T cell apoptosis. J. Hepatol. 2006;45:520–528. doi: 10.1016/j.jhep.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 158.Parsa AT, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 159.Wang X, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 160.Zhao L, et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jia L, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 162.Wang Y, et al. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget. 2017;8:28125–28134. doi: 10.18632/oncotarget.15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen L, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu S, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang G, et al. Signal transducers and activators of transcription-1 (STAT1) regulates microRNA transcription in interferon gamma-stimulated HeLa cells. PLoS ONE. 2010;5:e11794. doi: 10.1371/journal.pone.0011794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cai Z, Hao XY, Liu FX. MicroRNA-186 serves as a tumor suppressor in oral squamous cell carcinoma by negatively regulating the protein tyrosine phosphatase SHP2 expression. Arch. Oral. Biol. 2018;89:20–25. doi: 10.1016/j.archoralbio.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 167.Gregersen LH, et al. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS ONE. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu T, et al. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239–30250. doi: 10.18632/oncotarget.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chun-Zhi Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu C, et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol. Cancer Ther. 2013;12:207–219. doi: 10.1158/1535-7163.MCT-12-0273. [DOI] [PubMed] [Google Scholar]
- 171.Wang RT, et al. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin. Cancer Res. 2014;20:4705–4716. doi: 10.1158/1078-0432.CCR-14-0517. [DOI] [PubMed] [Google Scholar]
- 172.Zhao L, et al. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10:731. doi: 10.1038/s41419-019-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tanaka E, et al. Expression of circular RNA CDR1AS in colon cancer cells increases cell surface PDL1 protein levels. Oncol. Rep. 2019;42:1459–1466. doi: 10.3892/or.2019.7244. [DOI] [PubMed] [Google Scholar]
- 174.Kathuria H, et al. NKX2-1-AS1 negatively regulates CD274/PD-L1, cell-cell interaction genes, and limits human lung carcinoma cell migration. Sci. Rep. 2018;8:14418. doi: 10.1038/s41598-018-32793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agata Y, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- 176.Patsoukis N, et al. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci. Signal. 2012;5:ra46. doi: 10.1126/scisignal.2002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hofmeyer KA, Jeon H, Zang X. The PD-1/PD-L1 (B7-H1) pathway in chronic infection-induced cytotoxic T lymphocyte exhaustion. J. Biomed. Biotechnol. 2011;2011:451694. doi: 10.1155/2011/451694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Patsoukis N, et al. PD-1 increases PTEN phosphatase activity while decreasing PTEN protein stability by inhibiting casein kinase 2. Mol. Cell Biol. 2013;33:3091–3098. doi: 10.1128/MCB.00319-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li Q, et al. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget. 2016;7:53735–53750. doi: 10.18632/oncotarget.10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang G, et al. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3’ untranslated region: a mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6:18933–18944. doi: 10.18632/oncotarget.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tang Y, et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001–39011. doi: 10.18632/oncotarget.16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol. Rev. 2017;276:97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fourcade J, et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ji J, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018;9:478. doi: 10.1038/s41419-018-0528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Watanabe N, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- 186.Liu Y, et al. B and T lymphocyte attenuator is a target of miR-155 during naive CD4+ T cell activation. Iran. J. Immunol. 2016;13:89–99. [PubMed] [Google Scholar]
- 187.Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li Z, et al. Tumor-derived transforming growth factor-beta is critical for tumor progression and evasion from immune surveillance. Asian Pac. J. Cancer Prev. 2014;15:5181–5186. doi: 10.7314/apjcp.2014.15.13.5181. [DOI] [PubMed] [Google Scholar]
- 189.Yu J, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity. 2006;24:575–590. doi: 10.1016/j.immuni.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 190.Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- 191.Nandan D, Reiner NE. TGF-beta attenuates the class II transactivator and reveals an accessory pathway of IFN-gamma action. J. Immunol. 1997;158:1095–1101. [PubMed] [Google Scholar]
- 192.Gorelik L, Constant S, Flavell RA. Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 2002;195:1499–1505. doi: 10.1084/jem.20012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Brabletz T, et al. Transforming growth factor beta and cyclosporin A inhibit the inducible activity of the interleukin-2 gene in T cells through a noncanonical octamer-binding site. Mol. Cell Biol. 1993;13:1155–1162. doi: 10.1128/mcb.13.2.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 195.Chen W, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dimeloe, S. et al. Tumor-derived TGF-beta inhibits mitochondrial respiration to suppress IFN-gamma production by human CD4(+) T cells. Sci Signal. 12, eaav3334 (2019). [DOI] [PubMed]
- 198.Chen XB, Li W, Chu AX. MicroRNA-133a inhibits gastric cancer cells growth, migration, and epithelial-mesenchymal transition process by targeting presenilin 1. J. Cell Biochem. 2019;120:470–480. doi: 10.1002/jcb.27403. [DOI] [PubMed] [Google Scholar]
- 199.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 200.Chen D, et al. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014;289:91–96. doi: 10.1016/j.cellimm.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 201.Tang J, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tzur G, et al. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS ONE. 2009;4:e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 204.Wang H, et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-beta type II receptor. Brain Res. 2010;1357:166–174. doi: 10.1016/j.brainres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 205.Korpal M, Kang Y. Targeting the transforming growth factor-beta signalling pathway in metastatic cancer. Eur. J. Cancer. 2010;46:1232–1240. doi: 10.1016/j.ejca.2010.02.040. [DOI] [PubMed] [Google Scholar]
- 206.Wang FE, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pan Q, Luo X, Chegini N. microRNA 21: response to hormonal therapies and regulatory function in leiomyoma, transformed leiomyoma and leiomyosarcoma cells. Mol. Hum. Reprod. 2010;16:215–227. doi: 10.1093/molehr/gap093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 209.Shan H, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc. Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- 210.Chen, K., Zhu, H., Zheng, M. Q. & Dong, Q. R. LncRNA MEG3 Inhibits the degradation of the extracellular matrix of chondrocytes in osteoarthritis via targeting miR-93/TGFBR2 axis. Cartilage10.1177/1947603519855759 (2019). [DOI] [PMC free article] [PubMed]
- 211.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J. Biol. Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pais H, et al. Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA. 2010;16:489–494. doi: 10.1261/rna.1701210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Takahashi Y, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br. J. Cancer. 2014;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu, Q., Liu, S. & Wang, D. Overexpression of microRNA-21 decreased the sensitivity of advanced cervical cancer to chemoradiotherapy through SMAD7. Anticancer Drugs 31, 272–281 (2019). [DOI] [PubMed]
- 215.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 216.Liu Q, et al. Targeting interlukin-6 to relieve immunosuppression in tumor microenvironment. Tumour Biol. 2017;39:1010428317712445. doi: 10.1177/1010428317712445. [DOI] [PubMed] [Google Scholar]
- 217.Wang H, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells. 2009;27:2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cheng GZ, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J. Biol. Chem. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang M, et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget. 2016;7:78640–78652. doi: 10.18632/oncotarget.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yang Y, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer. 2017;16:141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Park Y, Kim J. Regulation of IL-6 signaling by miR-125a and let-7e in endothelial cells controls vasculogenic mimicry formation of breast cancer cells. BMB Rep. 2019;52:214–219. doi: 10.5483/BMBRep.2019.52.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Weng YS, et al. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer. 2019;18:42. doi: 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang J, Wang C, Xu H. miR-217 suppresses proliferation and promotes apoptosis in cardiac myxoma by targeting Interleukin-6. Biochem. Biophys. Res. Commun. 2017;490:713–718. doi: 10.1016/j.bbrc.2017.06.106. [DOI] [PubMed] [Google Scholar]
- 225.Yang X, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 226.Li F, et al. miR-98 suppresses melanoma metastasis through a negative feedback loop with its target gene IL-6. Exp. Mol. Med. 2014;46:e116. doi: 10.1038/emm.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang J, et al. Down-regulation of microRNA-9 leads to activation of IL-6/Jak/STAT3 pathway through directly targeting IL-6 in HeLa cell. Mol. Carcinog. 2016;55:732–742. doi: 10.1002/mc.22317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shang A, et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. J. Exp. Clin. Cancer Res. 2019;38:411. doi: 10.1186/s13046-019-1394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kong YG, et al. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 230.Su K, et al. A novel positive feedback regulation between long noncoding RNA UICC and IL-6/STAT3 signaling promotes cervical cancer progression. Am. J. Cancer Res. 2018;8:1176–1189. [PMC free article] [PubMed] [Google Scholar]
- 231.Chaluvally-Raghavan P, et al. Direct upregulation of STAT3 by MicroRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 2016;15:1493–1504. doi: 10.1016/j.celrep.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wu W, et al. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br. J. Cancer. 2013;108:653–661. doi: 10.1038/bjc.2012.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liu S, et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859 e811. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Che S, et al. miR-30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 2015;36:6805–6811. doi: 10.1007/s13277-015-3400-8. [DOI] [PubMed] [Google Scholar]
- 235.Wang DW, You D, Dong J, Liu TF. Knockdown of long non-coding RNA LINC00518 inhibits cervical cancer proliferation and metastasis by modulating JAK/STAT3 signaling. Eur. Rev. Med. Pharm. Sci. 2019;23:496–506. doi: 10.26355/eurrev_201901_16861. [DOI] [PubMed] [Google Scholar]
- 236.Xue J, et al. LncRNA AB073614 induces epithelial- mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway. Cancer Biomark. 2018;21:849–858. doi: 10.3233/CBM-170780. [DOI] [PubMed] [Google Scholar]
- 237.Wu Y, et al. Long noncoding RNA HOST2 promotes epithelial-mesenchymal transition, proliferation, invasion and migration of hepatocellular carcinoma cells by activating the JAK2-STAT3 Signaling Pathway. Cell Physiol. Biochem. 2018;51:301–314. doi: 10.1159/000495231. [DOI] [PubMed] [Google Scholar]
- 238.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Huang S, Ullrich SE, Bar-Eli M. Regulation of tumor growth and metastasis by interleukin-10: the melanoma experience. J. Interferon Cytokine Res. 1999;19:697–703. doi: 10.1089/107999099313532. [DOI] [PubMed] [Google Scholar]
- 240.Li C, et al. TLR4 signaling pathway in mouse Lewis lung cancer cells promotes the expression of TGF-beta1 and IL-10 and tumor cells migration. Biomed. Mater. Eng. 2014;24:869–875. doi: 10.3233/BME-130879. [DOI] [PubMed] [Google Scholar]
- 241.Wang X, et al. Bladder cancer cells induce immunosuppression of T cells by supporting PD-L1 expression in tumour macrophages partially through interleukin 10. Cell Biol. Int. 2017;41:177–186. doi: 10.1002/cbin.10716. [DOI] [PubMed] [Google Scholar]
- 242.Mannino MH, et al. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367:103–107. doi: 10.1016/j.canlet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 243.Steinbrink K, et al. Interleukin-10-treated human dendritic cells induce a melanoma-antigen-specific anergy in CD8(+) T cells resulting in a failure to lyse tumor cells. Blood. 1999;93:1634–1642. [PubMed] [Google Scholar]
- 244.Adris S, et al. IL-10 expression by CT26 colon carcinoma cells inhibits their malignant phenotype and induces a T cell-mediated tumor rejection in the context of a systemic Th2 response. Gene Ther. 1999;6:1705–1712. doi: 10.1038/sj.gt.3301012. [DOI] [PubMed] [Google Scholar]
- 245.Akdis CA, Blaser K. Mechanisms of interleukin-10-mediated immune suppression. Immunology. 2001;103:131–136. doi: 10.1046/j.1365-2567.2001.01235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lo HW, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li L, et al. MiR-98 suppresses the effects of tumor-associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL-10. Biochimie. 2018;150:23–30. doi: 10.1016/j.biochi.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 248.Zhang S, et al. Primary laryngeal cancer-derived miR-193b induces interleukin-10-expression monocytes. Cancer Invest. 2015;33:29–33. doi: 10.3109/07357907.2014.988344. [DOI] [PubMed] [Google Scholar]
- 249.Harris-Arnold A, et al. Epstein-Barr virus modulates host cell microRNA-194 to promote IL-10 production and B lymphoma cell survival. Am. J. Transpl. 2015;15:2814–2824. doi: 10.1111/ajt.13375. [DOI] [PubMed] [Google Scholar]
- 250.Liu J, et al. Long non-coding RNA CCAT1/miR-148a/PKCzeta prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79:105–112. doi: 10.1002/pros.23716. [DOI] [PubMed] [Google Scholar]
- 251.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr. Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 252.Lapeyre-Prost A, et al. Immunomodulatory Activity of VEGF in Cancer. Int Rev. Cell Mol. Biol. 2017;330:295–342. doi: 10.1016/bs.ircmb.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 253.Hansen W, et al. Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. J. Exp. Med. 2012;209:2001–2016. doi: 10.1084/jem.20111497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cheng J, et al. Downregulation of miRNA-638 promotes angiogenesis and growth of hepatocellular carcinoma by targeting VEGF. Oncotarget. 2016;7:30702–30711. doi: 10.18632/oncotarget.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhou B, et al. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 256.Yan JJ, et al. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6:29527–29542. doi: 10.18632/oncotarget.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zhu X, et al. miR-203 suppresses tumor growth and angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol. Biochem. 2013;32:64–73. doi: 10.1159/000350125. [DOI] [PubMed] [Google Scholar]
- 258.Choi YC, et al. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol. Cells. 2011;32:77–82. doi: 10.1007/s10059-011-1042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang R, et al. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology. 2013;58:642–653. doi: 10.1002/hep.26373. [DOI] [PubMed] [Google Scholar]
- 260.Hao Y, et al. The synergistic regulation of VEGF-mediated angiogenesis through miR-190 and target genes. RNA. 2014;20:1328–1336. doi: 10.1261/rna.044651.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Liu B, et al. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 262.Sasahira T, et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br. J. Cancer. 2012;107:700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li F, et al. Role of microRNA-93 in regulation of angiogenesis. Tumour Biol. 2014;35:10609–10613. doi: 10.1007/s13277-014-2605-6. [DOI] [PubMed] [Google Scholar]
- 264.Long J, et al. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J. Biol. Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chou J, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lei Z, et al. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Chen S, et al. LncRNA TDRG1 enhances tumorigenicity in endometrial carcinoma by binding and targeting VEGF-A protein. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:3013–3021. doi: 10.1016/j.bbadis.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 268.Fu WM, et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7:4712–4723. doi: 10.18632/oncotarget.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yamakuchi M, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl Acad. Sci. USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Cha ST, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675–2685. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 271.Chai ZT, et al. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2alpha/Akt/HIF-1alpha pathway in hepatocellular carcinoma. PLoS ONE. 2013;8:e77957. doi: 10.1371/journal.pone.0077957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yin Y, et al. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta. 2013;1829:239–247. doi: 10.1016/j.bbagrm.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 273.Hou J, et al. Long noncoding RNA H19 upregulates vascular endothelial growth factor A to enhance mesenchymal stem cells survival and angiogenic capacity by inhibiting miR-199a-5p. Stem Cell Res. Ther. 2018;9:109. doi: 10.1186/s13287-018-0861-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Li Y, et al. Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget. 2017;8:13690–13702. doi: 10.18632/oncotarget.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles–diagnostic and therapeutic implications (Review) Int. J. Oncol. 2015;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Liu J, et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. 2019;70:241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Berchem G, et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-beta and miR23a transfer. Oncoimmunology. 2016;5:e1062968. doi: 10.1080/2162402X.2015.1062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ye SB, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Walsh PT, et al. PTEN inhibits IL-2 receptor-mediated expansion of CD4+ CD25+ Tregs. J. Clin. Invest. 2006;116:2521–2531. doi: 10.1172/JCI28057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Pan L, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Xue M, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhang R, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 283.Shah MY, et al. microRNA therapeutics in cancer—an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Smolle MA, Calin HN, Pichler M, Calin GA. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017;284:1952–1966. doi: 10.1111/febs.14030. [DOI] [PubMed] [Google Scholar]
- 285.Barata P, Sood AK, Hong DS. RNA-targeted therapeutics in cancer clinical trials: Current status and future directions. Cancer Treat. Rev. 2016;50:35–47. doi: 10.1016/j.ctrv.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 286.Bakhtiyari S, Haghani K, Basati G, Karimfar MH. siRNA therapeutics in the treatment of diseases. Ther. Deliv. 2013;4:45–57. doi: 10.4155/tde.12.136. [DOI] [PubMed] [Google Scholar]
- 287.Hsu SH, et al. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine. 2013;9:1169–1180. doi: 10.1016/j.nano.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Cui C, et al. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939Mediated transcriptional repression of Bcl-xL. Cancer Res. Treat. 2018;50:992–1008. doi: 10.4143/crt.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chen J, et al. The long noncoding RNA ASNR regulates degradation of Bcl-2 mRNA through its interaction with AUF1. Sci. Rep. 2016;6:32189. doi: 10.1038/srep32189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Deng J, et al. Long non-coding RNA OIP5-AS1 functions as an oncogene in lung adenocarcinoma through targeting miR-448/Bcl-2. Biomed. Pharmacother. 2018;98:102–110. doi: 10.1016/j.biopha.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 291.Gao J, et al. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS ONE. 2013;8:e65138. doi: 10.1371/journal.pone.0065138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Long J, et al. miR-193b modulates resistance to doxorubicin in human breast cancer cells by downregulating MCL-1. Biomed. Res. Int. 2015;2015:373574. doi: 10.1155/2015/373574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang R, et al. MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1 in breast cancer. Med. Oncol. 2014;31:347. doi: 10.1007/s12032-014-0347-3. [DOI] [PubMed] [Google Scholar]
- 294.Liu X, et al. MicroRNA-101 inhibits cell progression and increases paclitaxel sensitivity by suppressing MCL-1 expression in human triple-negative breast cancer. Oncotarget. 2015;6:20070–20083. doi: 10.18632/oncotarget.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Desjobert C, et al. MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood. 2011;117:6627–6637. doi: 10.1182/blood-2010-09-301994. [DOI] [PubMed] [Google Scholar]
- 296.Zhang YK, et al. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem. Biophys. Res. Commun. 2011;414:233–239. doi: 10.1016/j.bbrc.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 297.Rao YM, Shi HR, Ji M, Chen CH. MiR-106a targets Mcl-1 to suppress cisplatin resistance of ovarian cancer A2780 cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013;33:567–572. doi: 10.1007/s11596-013-1160-5. [DOI] [PubMed] [Google Scholar]
- 298.Lu F, et al. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J. Oncol. 2014;45:383–392. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 299.Cai D, et al. MicroRNA-302b enhances the sensitivity of hepatocellular carcinoma cell lines to 5-FU via targeting Mcl-1 and DPYD. Int J. Mol. Sci. 2015;16:23668–23682. doi: 10.3390/ijms161023668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Zhang T, et al. Down-regulation of miR-320 associated with cancer progression and cell apoptosis via targeting Mcl-1 in cervical cancer. Tumour Biol. 2016;37:8931–8940. doi: 10.1007/s13277-015-4771-6. [DOI] [PubMed] [Google Scholar]
- 301.Tong Z, et al. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed. Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 302.Zarogoulidis P, et al. MiR-205 and miR-218 expression is associated with carboplatin chemoresistance and regulation of apoptosis via Mcl-1 and Survivin in lung cancer cells. Cell Signal. 2015;27:1576–1588. doi: 10.1016/j.cellsig.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 303.Crawford M, et al. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 2009;388:483–489. doi: 10.1016/j.bbrc.2009.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Patron JP, et al. MiR-133b targets antiapoptotic genes and enhances death receptor-induced apoptosis. PLoS ONE. 2012;7:e35345. doi: 10.1371/journal.pone.0035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Liu N, et al. miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget. 2014;5:4959–4971. doi: 10.18632/oncotarget.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Garofalo M, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 307.Incoronato M, et al. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70:3638–3646. doi: 10.1158/0008-5472.CAN-09-3341. [DOI] [PubMed] [Google Scholar]
- 308.Acunzo M, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ovcharenko D, et al. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- 310.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol. Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Wang P, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Huang G, et al. miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res. 2012;72:908–916. doi: 10.1158/0008-5472.CAN-11-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Yamada N, et al. Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell leukaemia jurkat cells by modulating expression of fas-associated protein with death domain (FADD) Biochim. Biophys. Acta. 2014;1843:590–602. doi: 10.1016/j.bbamcr.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 314.Kim TD, et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118:5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.King BC, et al. CD46 activation regulates miR-150-mediated control of GLUT1 expression and cytokine secretion in human CD4+ T cells. J. Immunol. 2016;196:1636–1645. doi: 10.4049/jimmunol.1500516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Li P, et al. MicroRNA-218 increases the sensitivity of bladder cancer to cisplatin by targeting Glut1. Cell Physiol. Biochem. 2017;41:921–932. doi: 10.1159/000460505. [DOI] [PubMed] [Google Scholar]
- 317.Guo H, et al. miRNA-451 inhibits glioma cell proliferation and invasion by downregulating glucose transporter 1. Tumour Biol. 2016;37:13751–13761. doi: 10.1007/s13277-016-5219-3. [DOI] [PubMed] [Google Scholar]
- 318.Trakooljul N, Hicks JA, Liu HC. Identification of target genes and pathways associated with chicken microRNA miR-143. Anim. Genet. 2010;41:357–364. doi: 10.1111/j.1365-2052.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 319.Dai DW, et al. Decreased miR-106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer. 2013;13:478. doi: 10.1186/1471-2407-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Lauer, V. et al. Hypoxia drives glucose transporter 3 expression through HIF-mediated induction of the long non-coding RNA NICI. J. Biol. Chem. 295, 4065–4078 (2019). [DOI] [PMC free article] [PubMed]
- 321.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 2010;86:410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 322.Karolina DS, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE. 2011;6:e22839. doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Chen YH, et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278–2286. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Zhou T, et al. Regulation of insulin resistance by multiple MiRNAs via targeting the GLUT4 signalling pathway. Cell Physiol. Biochem. 2016;38:2063–2078. doi: 10.1159/000445565. [DOI] [PubMed] [Google Scholar]
- 325.Zhao S, et al. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013;333:253–260. doi: 10.1016/j.canlet.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 326.Jiang JX, et al. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Mol. Med. Rep. 2014;10:995–1002. doi: 10.3892/mmr.2014.2271. [DOI] [PubMed] [Google Scholar]
- 327.Song J, et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 2017;490:217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 328.Li W, et al. Astragalin reduces hexokinase 2 through Increasing miR-125b to Inhibit the proliferation of hepatocellular carcinoma cells in vitro and in vivo. J. Agric. Food Chem. 2017;65:5961–5972. doi: 10.1021/acs.jafc.7b02120. [DOI] [PubMed] [Google Scholar]
- 329.Tao T, et al. Involvement of EZH2 in aerobic glycolysis of prostate cancer through miR-181b/HK2 axis. Oncol. Rep. 2017;37:1430–1436. doi: 10.3892/or.2017.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Zhu W, et al. MicroRNA-98 suppress warburg effect by targeting HK2 in colon cancer cells. Dig. Dis. Sci. 2017;62:660–668. doi: 10.1007/s10620-016-4418-5. [DOI] [PubMed] [Google Scholar]
- 331.Qin Y, Cheng C, Lu H, Wang Y. miR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cells. Biochem. Biophys. Res. Commun. 2016;469:37–43. doi: 10.1016/j.bbrc.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 332.Lan, H. et al. [miR-181c inhibits glycolysis by targeting hexokinase 2 in cancer-associated fibroblasts]. Nan Fang Yi Ke Da Xue Xue Bao35, 1619–1623 (2015). [PubMed]
- 333.Kim HR, et al. p53 regulates glucose metabolism by miR-34a. Biochem. Biophys. Res. Commun. 2013;437:225–231. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 334.Rengaraj D, et al. Regulation of glucose phosphate isomerase by the 3'UTR-specific miRNAs miR-302b and miR-17-5p in chicken primordial germ cells. Biol. Reprod. 2013;89:33. doi: 10.1095/biolreprod.112.105692. [DOI] [PubMed] [Google Scholar]
- 335.Ahmad A, et al. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71:3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Park YY, et al. Tat-activating regulatory DNA-binding protein regulates glycolysis in hepatocellular carcinoma by regulating the platelet isoform of phosphofructokinase through microRNA 520. Hepatology. 2013;58:182–191. doi: 10.1002/hep.26310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Tang H, et al. Oxidative stress-responsive microRNA-320 regulates glycolysis in diverse biological systems. FASEB J. 2012;26:4710–4721. doi: 10.1096/fj.11-197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.White NM, et al. Dysregulation of kallikrein-related peptidases in renal cell carcinoma: potential targets of miRNAs. Biol. Chem. 2010;391:411–423. doi: 10.1515/BC.2010.041. [DOI] [PubMed] [Google Scholar]
- 339.Du JY, Wang LF, Wang Q, Yu LD. miR-26b inhibits proliferation, migration, invasion and apoptosis induction via the downregulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 driven glycolysis in osteosarcoma cells. Oncol. Rep. 2015;33:1890–1898. doi: 10.3892/or.2015.3797. [DOI] [PubMed] [Google Scholar]
- 340.Seliger B, et al. Linkage of microRNA and proteome-based profiling data sets: a perspective for the priorization of candidate biomarkers in renal cell carcinoma? J. Proteome Res. 2011;10:191–199. doi: 10.1021/pr1011137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Ge X, et al. Overexpression of miR-206 suppresses glycolysis, proliferation and migration in breast cancer cells via PFKFB3 targeting. Biochem. Biophys. Res. Commun. 2015;463:1115–1121. doi: 10.1016/j.bbrc.2015.06.068. [DOI] [PubMed] [Google Scholar]
- 342.Savarimuthu Francis SM, et al. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genomics. 2014;15:88. doi: 10.1186/1471-2164-15-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Boesch-Saadatmandi C, Wagner AE, Wolffram S, Rimbach G. Effect of quercetin on inflammatory gene expression in mice liver in vivo—role of redox factor 1, miRNA-122 and miRNA-125b. Pharm. Res. 2012;65:523–530. doi: 10.1016/j.phrs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 345.Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15:445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sikand K, Singh J, Ebron JS, Shukla GC. Housekeeping gene selection advisory: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin are targets of miR-644a. PLoS ONE. 2012;7:e47510. doi: 10.1371/journal.pone.0047510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.White NM, et al. Identification and validation of dysregulated metabolic pathways in metastatic renal cell carcinoma. Tumour Biol. 2014;35:1833–1846. doi: 10.1007/s13277-013-1245-6. [DOI] [PubMed] [Google Scholar]
- 348.Li Y, et al. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics. 2012;7:940–949. doi: 10.4161/epi.21236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Muniyappa MK, et al. MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur. J. Cancer. 2009;45:3104–3118. doi: 10.1016/j.ejca.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 350.Leder A, et al. Micron-sized iron oxide-containing particles for microRNA-targeted manipulation and MRI-based tracking of transplanted cells. Biomaterials. 2015;51:129–137. [Google Scholar]
- 351.Ramirez CM, et al. MicroRNA 33 regulates glucose metabolism. Mol. Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wong TS, et al. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J. Cancer. 2008;123:251–257. doi: 10.1002/ijc.23583. [DOI] [PubMed] [Google Scholar]
- 353.Li W, et al. Insulin promotes glucose consumption via regulation of miR-99a/mTOR/PKM2 pathway. PLoS ONE. 2013;8:e64924. doi: 10.1371/journal.pone.0064924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Tao T, et al. Loss of SNAIL inhibits cellular growth and metabolism through the miR-128-mediated RPS6KB1/HIF-1alpha/PKM2 signaling pathway in prostate cancer cells. Tumour Biol. 2014;35:8543–8550. doi: 10.1007/s13277-014-2057-z. [DOI] [PubMed] [Google Scholar]
- 355.Saumet A, et al. Estrogen and retinoic acid antagonistically regulate several microRNA genes to control aerobic glycolysis in breast cancer cells. Mol. Biosyst. 2012;8:3242–3253. doi: 10.1039/c2mb25298h. [DOI] [PubMed] [Google Scholar]
- 356.Kaller M, et al. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol. Cell Proteom. 2011;10:M111 010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Zhang, Y., Liu, G. & Gao, X. Attenuation of miR-34a protects cardiomyocytes against hypoxic stress through maintenance of glycolysis. Biosci Rep. 37, BSR20170925 (2017). [DOI] [PMC free article] [PubMed]
- 358.Li L, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 359.Han RL, et al. miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64:244–252. doi: 10.4149/neo_2017_211. [DOI] [PubMed] [Google Scholar]
- 360.Zhang R, et al. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am. J. Cancer Res. 2016;6:312–320. [PMC free article] [PubMed] [Google Scholar]
- 361.Chen H, et al. Long non-coding RNA CRYBG3 regulates glycolysis of lung cancer cells by interacting with lactate dehydrogenase A. J. Cancer. 2018;9:2580–2588. doi: 10.7150/jca.24896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Meng S, et al. MicroRNA 107 partly inhibits endothelial progenitor cells differentiation via HIF-1beta. PLoS ONE. 2012;7:e40323. doi: 10.1371/journal.pone.0040323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Song T, et al. MiR-138 suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in clear cell renal cell carcinoma 786-O cells. Asian Pac. J. Cancer Prev. 2011;12:1307–1311. [PubMed] [Google Scholar]
- 365.Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J. Cancer. 2013;133:867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 366.Bertozzi D, et al. The natural inhibitor of DNA topoisomerase I, camptothecin, modulates HIF-1alpha activity by changing miR expression patterns in human cancer cells. Mol. Cancer Ther. 2014;13:239–248. doi: 10.1158/1535-7163.MCT-13-0729. [DOI] [PubMed] [Google Scholar]
- 367.Hua Z, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Hebert C, et al. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Bai R, et al. MicroRNA-195 induced apoptosis in hypoxic chondrocytes by targeting hypoxia-inducible factor 1 alpha. Eur. Rev. Med Pharm. Sci. 2015;19:545–551. [PubMed] [Google Scholar]
- 370.Liu LZ, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS ONE. 2011;6:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wen X, et al. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8:5676–5689. doi: 10.7150/thno.28538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Asadzadeh Z, et al. microRNAs in cancer stem cells: Biology, pathways, and therapeutic opportunities. J. Cell Physiol. 2019;234:10002–10017. doi: 10.1002/jcp.27885. [DOI] [PubMed] [Google Scholar]
- 373.Taipaleenmaki H, et al. Antagonizing miR-218-5p attenuates Wnt signaling and reduces metastatic bone disease of triple negative breast cancer cells. Oncotarget. 2016;7:79032–79046. doi: 10.18632/oncotarget.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Song XL, et al. miR-1301-3p promotes prostate cancer stem cell expansion by targeting SFRP1 and GSK3beta. Biomed. Pharmacother. 2018;99:369–374. doi: 10.1016/j.biopha.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 375.Jiang C, et al. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol. Rep. 2017;38:2351–2359. doi: 10.3892/or.2017.5924. [DOI] [PubMed] [Google Scholar]
- 376.Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019;508:37–45. doi: 10.1016/j.bbrc.2018.11.079. [DOI] [PubMed] [Google Scholar]
- 377.Jin Y, et al. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz. J. Med. Biol. Res. 2018;51:e7811. doi: 10.1590/1414-431X20187811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Xia L, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/beta-catenin pathway. Biochem. Biophys. Res. Commun. 2018;503:385–390. doi: 10.1016/j.bbrc.2018.06.045. [DOI] [PubMed] [Google Scholar]
- 379.Chen F, et al. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol. Ther. 2018;19:1139–1152. doi: 10.1080/15384047.2018.1480888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Ke X, et al. MiR-410 induces stemness by inhibiting Gsk3beta but upregulating beta-catenin in non-small cells lung cancer. Oncotarget. 2017;8:11356–11371. doi: 10.18632/oncotarget.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zhang GJ, et al. MiR-92a promotes stem cell-like properties by activating Wnt/beta-catenin signaling in colorectal cancer. Oncotarget. 2017;8:101760–101770. doi: 10.18632/oncotarget.21667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zhu J, et al. miR-19 targeting of GSK3beta mediates sulforaphane suppression of lung cancer stem cells. J. Nutr. Biochem. 2017;44:80–91. doi: 10.1016/j.jnutbio.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 383.Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 2018;502:465–471. doi: 10.1016/j.bbrc.2018.05.184. [DOI] [PubMed] [Google Scholar]
- 384.Geng Y, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin. Sci. (Lond.) 2019;133:1197–1213. doi: 10.1042/CS20190286. [DOI] [PubMed] [Google Scholar]
- 385.Hsieh IS, et al. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 386.Su J, et al. MicroRNA-200a suppresses the Wnt/beta-catenin signaling pathway by interacting with beta-catenin. Int J. Oncol. 2012;40:1162–1170. doi: 10.3892/ijo.2011.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zhu P, et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 388.Li T, et al. Long non-coding RNA lncTCF7 activates the Wnt/beta-catenin pathway to promote metastasis and invasion in colorectal cancer. Oncol. Lett. 2017;14:7384–7390. doi: 10.3892/ol.2017.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Babashah S, et al. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J. Cancer. 2013;133:579–589. doi: 10.1002/ijc.28043. [DOI] [PubMed] [Google Scholar]
- 390.Lu J, et al. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231–241. doi: 10.1038/onc.2016.194. [DOI] [PubMed] [Google Scholar]
- 391.Gao X, Zhu X, Sun Y, Liu J. MicroRNA141 inhibits the selfrenewal of glioblastoma stem cells via Jagged1. Mol. Med. Rep. 2017;16:167–173. doi: 10.3892/mmr.2017.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Shao T, et al. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene. 2019;715:144029. doi: 10.1016/j.gene.2019.144029. [DOI] [PubMed] [Google Scholar]
- 393.Wang Y, et al. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci. Rep. 2015;5:13749. doi: 10.1038/srep13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Xu H, et al. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol. Neurosci. 2018;11:225. doi: 10.3389/fnmol.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Chen Y, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2019;38:466. doi: 10.1186/s13046-019-1436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Lan L, et al. Rhamnetin induces apoptosis in human breast cancer cells via the miR-34a/Notch-1 signaling pathway. Oncol. Lett. 2019;17:676–682. doi: 10.3892/ol.2018.9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Zhang K, et al. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFbeta and Notch pathways. Nat. Commun. 2017;8:144. doi: 10.1038/s41467-017-00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Sun X, et al. MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget. 2015;6:32944–32954. doi: 10.18632/oncotarget.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Zhong X, et al. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010;285:41961–41971. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Jiang J, et al. Overexpression of microRNA-21 strengthens stem cell-like characteristics in a hepatocellular carcinoma cell line. World J. Surg. Oncol. 2016;14:278. doi: 10.1186/s12957-016-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Gao Y, et al. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis. 2017;8:e2924. doi: 10.1038/cddis.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wu J, et al. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J. Cell Mol. Med. 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Fu Z, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 404.Zhang Z, et al. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J. Cell Physiol. 2018;233:8630–8638. doi: 10.1002/jcp.26611. [DOI] [PubMed] [Google Scholar]
- 405.Guo L, et al. Knockdown of long non-coding RNA linc-ITGB1 inhibits cancer stemness and epithelial-mesenchymal transition by reducing the expression of Snail in non-small cell lung cancer. Thorac. Cancer. 2019;10:128–136. doi: 10.1111/1759-7714.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yu J, et al. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol. Cancer. 2014;13:121. doi: 10.1186/1476-4598-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Bing L, Hong C, Li-Xin S, Wei G. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch. Gynecol. Obstet. 2014;290:533–541. doi: 10.1007/s00404-014-3219-3. [DOI] [PubMed] [Google Scholar]
- 408.Liu S, et al. MiR-137 regulates epithelial-mesenchymal transition in gastrointestinal stromal tumor. Tumour Biol. 2014;35:9131–9138. doi: 10.1007/s13277-014-2177-5. [DOI] [PubMed] [Google Scholar]
- 409.Zhu X, et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–332. doi: 10.1038/onc.2015.84. [DOI] [PubMed] [Google Scholar]
- 410.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J. Biol. Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Nairismagi ML, et al. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;31:4960–4966. doi: 10.1038/onc.2011.650. [DOI] [PubMed] [Google Scholar]
- 412.Li LZ, et al. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35:469–478. doi: 10.1093/carcin/bgt330. [DOI] [PubMed] [Google Scholar]
- 413.Zhou Y, et al. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J. Exp. Clin. Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zhang P, et al. Cordycepin (3'-deoxyadenosine) suppressed HMGA2, Twist1 and ZEB1-dependent melanoma invasion and metastasis by targeting miR-33b. Oncotarget. 2015;6:9834–9853. doi: 10.18632/oncotarget.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Tsukerman P, et al. MiR-520d-5p directly targets TWIST1 and downregulates the metastamiR miR-10b. Oncotarget. 2014;5:12141–12150. doi: 10.18632/oncotarget.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Hernandez JM, et al. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 2013;20(Suppl 3):S625–S635. doi: 10.1245/s10434-013-3106-3. [DOI] [PubMed] [Google Scholar]
- 417.Nairismagi ML, et al. The proto-oncogene TWIST1 is regulated by microRNAs. PLoS ONE. 2013;8:e66070. doi: 10.1371/journal.pone.0066070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Niu H, et al. Long non-coding RNA AK027294 involves in the process of proliferation, migration, and apoptosis of colorectal cancer cells. Tumour Biol. 2016;37:10097–10105. doi: 10.1007/s13277-015-4350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Ma F, et al. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed. Pharmacother. 2016;84:1249–1255. doi: 10.1016/j.biopha.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 420.Wang SH, et al. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour Biol. 2016;37:12867–12875. doi: 10.1007/s13277-016-5210-z. [DOI] [PubMed] [Google Scholar]
- 421.Yu T, et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109:1393–1403. doi: 10.1111/cas.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Xu M, et al. MiR-22 suppresses epithelial-mesenchymal transition in bladder cancer by inhibiting Snail and MAPK1/Slug/vimentin feedback loop. Cell Death Dis. 2018;9:209. doi: 10.1038/s41419-017-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Cao L, Wan Q, Li F, Tang CE. MiR-363 inhibits cisplatin chemoresistance of epithelial ovarian cancer by regulating snail-induced epithelial-mesenchymal transition. BMB Rep. 2018;51:456–461. doi: 10.5483/BMBRep.2018.51.9.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Fan MJ, et al. MiR-30 suppresses lung cancer cell 95D epithelial mesenchymal transition and invasion through targeted regulating Snail. Eur. Rev. Med Pharm. Sci. 2017;21:2642–2649. [PubMed] [Google Scholar]
- 425.Zhang Z, Zhang M, Chen Q, Zhang Q. Downregulation of microRNA-145 promotes epithelial-mesenchymal transition via regulating Snail in osteosarcoma. Cancer Gene Ther. 2017;24:83–88. doi: 10.1038/cgt.2017.1. [DOI] [PubMed] [Google Scholar]
- 426.Xia W, et al. miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting Snail. Oncol. Rep. 2015;34:655–662. doi: 10.3892/or.2015.4008. [DOI] [PubMed] [Google Scholar]
- 427.Zhang YF, et al. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol. Rep. 2016;36:480–486. doi: 10.3892/or.2016.4828. [DOI] [PubMed] [Google Scholar]
- 428.Chen X, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 429.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharm. Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 430.Jiang Y, et al. MiR-203 acts as a radiosensitizer of gastric cancer cells by directly targeting ZEB1. Onco Targets Ther. 2019;12:6093–6104. doi: 10.2147/OTT.S197539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wang G, et al. Loss of miR-873 contributes to gemcitabine resistance in triple-negative breast cancer via targeting ZEB1. Oncol. Lett. 2019;18:3837–3844. doi: 10.3892/ol.2019.10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Li L, Li S. miR-205-5p inhibits cell migration and invasion in prostatic carcinoma by targeting ZEB1. Oncol. Lett. 2018;16:1715–1721. doi: 10.3892/ol.2018.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Zhang, C., Xue, Q., Xu, Z. & Lu, C. MiR-5702 suppresses proliferation and invasion in non-small-cell lung cancer cells via posttranscriptional suppression of ZEB1. J. Biochem. Mol. Toxicol. e22163 10.1002/jbt.22163 (2018). [DOI] [PubMed]
- 434.Xu J, et al. The inhibition of miR-126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas. 2019;156:11. doi: 10.1186/s41065-019-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Li J, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch. Biochem. Biophys. 2018;640:53–60. doi: 10.1016/j.abb.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 436.Chen L, Yao H, Wang K, Liu X, Long Non-Coding RNA. MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J. Cell Biochem. 2017;118:4836–4843. doi: 10.1002/jcb.26158. [DOI] [PubMed] [Google Scholar]
- 437.Qu R, Chen X, Zhang C. LncRNA ZEB1-AS1/miR-409-3p/ZEB1 feedback loop is involved in the progression of non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018;507:450–456. doi: 10.1016/j.bbrc.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 438.Zhu C, et al. Long noncoding RNA SNHG16 promotes cell proliferation by sponging MicroRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol. Biochem. 2018;51:429–440. doi: 10.1159/000495239. [DOI] [PubMed] [Google Scholar]
- 439.Li Y, et al. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed. Pharmacother. 2018;103:939–946. doi: 10.1016/j.biopha.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 440.Zhang S, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed. Pharmacother. 2017;95:711–720. doi: 10.1016/j.biopha.2017.08.133. [DOI] [PubMed] [Google Scholar]
- 441.Lu Y, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 442.Wang PS, et al. A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene. 2018;37:4662–4678. doi: 10.1038/s41388-018-0293-1. [DOI] [PubMed] [Google Scholar]
- 443.Zou Q, et al. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J. Cell Biochem. 2018;119:2189–2199. doi: 10.1002/jcb.26380. [DOI] [PubMed] [Google Scholar]
- 444.Ying X, Zhu J, Zhang Y. Circular RNA circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR-665. Mol. Genet. Genom. Med. 2019;7:e991. doi: 10.1002/mgg3.991. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 445.Liu, Y. Y., Zhang, L. Y. & Du, W. Z. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through regulates ZEB1 expression by sponging miR-124-3p. Biosci Rep. 39, BSR20193045 (2019). [DOI] [PMC free article] [PubMed]
- 446.Tao Z, et al. MiR-195/-16 family enhances radiotherapy via T cell activation in the tumor microenvironment by blocking the PD-L1 immune checkpoint. Cell Physiol. Biochem. 2018;48:801–814. doi: 10.1159/000491909. [DOI] [PubMed] [Google Scholar]
- 447.Kao SC, et al. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J. Thorac. Oncol. 2017;12:1421–1433. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 448.Gao L, et al. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Qu F, et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J. Drug Target. 2019;27:67–74. doi: 10.1080/1061186X.2018.1479755. [DOI] [PubMed] [Google Scholar]
- 450.Wang W, et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 451.Ghousein, A. et al. miR-4510 blocks hepatocellular carcinoma development through RAF1 targeting and RAS/RAF/MEK/ERK signalling inactivation. Liver Int. 40, 240–251 (2020). [DOI] [PubMed]
- 452.Wu RS, et al. MiR-101 promotes nasopharyngeal carcinoma cell apoptosis through inhibiting Ras/Raf/MEK/ERK signaling pathway. Eur. Rev. Med. Pharm. Sci. 2018;22:150–157. doi: 10.26355/eurrev_201801_14112. [DOI] [PubMed] [Google Scholar]
- 453.Moles R, Bellon M, Nicot C. STAT1: a novel target of miR-150 and miR-223 Is involved in the proliferation of HTLV-I-transformed and ATL cells. Neoplasia. 2015;17:449–462. doi: 10.1016/j.neo.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Lian J, et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010;1:e94. doi: 10.1038/cddis.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Wang Z, et al. MicroRNA-101 has a suppressive role in osteosarcoma cells through the targeting of c-FOS. Exp. Ther. Med. 2016;11:1293–1299. doi: 10.3892/etm.2016.3085. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 456.Tao T, et al. Involvement of FOS-mediated miR-181b/miR-21 signalling in the progression of malignant gliomas. Eur. J. Cancer. 2013;49:3055–3063. doi: 10.1016/j.ejca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 457.Li S, et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem. Biophys. Res. Commun. 2013;441:976–981. doi: 10.1016/j.bbrc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 458.Wang F, et al. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem. Biophys. Res. Commun. 2013;434:688–694. doi: 10.1016/j.bbrc.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 459.Yang TS, et al. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014;588:2162–2169. doi: 10.1016/j.febslet.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 460.Zhu J, et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 461.Zhang G, et al. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig. Dis. Sci. 2014;59:98–107. doi: 10.1007/s10620-013-2858-8. [DOI] [PubMed] [Google Scholar]
- 462.Liu B, et al. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim. Biophys. Acta. 2012;1822:1692–1704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 463.Qu C, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Yang H, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 465.Ma F, et al. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/beta-catenin signaling. Gene. 2014;535:191–197. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 466.Wan J, Ling X, Peng B, Ding G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol. Rep. 2018;40:272–282. doi: 10.3892/or.2018.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Lin S, et al. Effect of microRNA-101 on proliferation and apoptosis of human osteosarcoma cells by targeting mTOR. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014;34:889–895. doi: 10.1007/s11596-014-1369-y. [DOI] [PubMed] [Google Scholar]
- 468.Wu D, Huang HJ, He CN, Wang KY. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR) Int J. Gynecol. Cancer. 2013;23:1191–1197. doi: 10.1097/IGC.0b013e31829ea779. [DOI] [PubMed] [Google Scholar]
- 469.Xu S, et al. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget. 2015;6:26457–26471. doi: 10.18632/oncotarget.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Zhang G, et al. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3' untranslated region: a mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6:18933–18944. doi: 10.18632/oncotarget.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Wang H, et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer's disease targets TGF-beta type II receptor. Brain Res. 2010;1357:166–174. doi: 10.1016/j.brainres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 472.Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018;507:457–464. doi: 10.1016/j.bbrc.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 473.Frassanito MA, et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J. Pathol. 2019;247:241–253. doi: 10.1002/path.5187. [DOI] [PubMed] [Google Scholar]
- 474.Ding G, et al. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget. 2015;6:29877–29888. doi: 10.18632/oncotarget.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Szebeni GJ, Vizler C, Kitajka K, Puskas LG. Inflammation and cancer: extra- and intracellular determinants of tumor-associated macrophages as tumor promoters. Mediators Inflamm. 2017;2017:9294018. doi: 10.1155/2017/9294018. [DOI] [PMC free article] [PubMed] [Google Scholar]