Abstract
Research on the formation of mitotic chromosomes from interphase chromatin domains, ongoing for several decades, made significant progress in recent years. It was stimulated by the development of advanced microscopic techniques and implementation of chromatin conformation capture methods that provide new insights into chromosome ultrastructure. This review aims to summarize and compare several models of chromatin fiber folding to form mitotic chromosomes and discusses them in the light of the novel findings. Functional genomics studies in several organisms confirmed condensins and cohesins as the major players in chromosome condensation. Here we compare available data on the role of these proteins across lower and higher eukaryotes and point to differences indicating evolutionary different pathways to shape mitotic chromosomes. Moreover, we discuss a controversial phenomenon of the mitotic chromosome ultrastructure – chromosome cavities – and using our super-resolution microscopy data, we contribute to its elucidation.
Keywords: Chromatin fiber folding, Chromosome cavities, Chromosome condensation, Chromosome conformation capture, Chromosome scaffold, Structural maintenance of chromosomes proteins
1. Introduction
The ability to transmit genetic information to new generations is one of the fundamental features of living organisms. In eukaryotes, this process is ensured at the cellular level by folding long DNA molecules into condensed chromosomes. These consist of pairs of genetically identical chromatids whose separation and movement to opposite poles of the mitotic spindle result in two genetically identical daughter nuclei and cells. Similarly, the condensed state and presence of sister chromatids is important during meiosis, when haploid gametes are formed with new combinations of parental alleles as a consequence of independent assortment and crossing over.
Although the phenomenon of chromosome condensation has been studied for over hundred years, the internal structure of chromosomes and molecular mechanisms underlying the condensation process itself have not yet been fully clarified. Nevertheless, the last decade marked a significant progress and the knowledge of the internal chromosome structure increased considerably. This development has been stimulated mainly by advances in microscopic techniques, such as super-resolution microscopy [1], [2], [3], electron microscopy (EM) [3], [4], and by the analysis of data obtained by chromosome conformation capture techniques such as Hi-C [5], [6]. Continual improvements of sample preparation and probing for electron microscopy enable observing chromosomes in a close-to-native state with a higher resolution and a specific protein/DNA labeling [7], [8], [9], [10].
Most of the knowledge of mitotic chromosome ultrastructure originates from studies on vertebrates and yeasts, but considerable amount of data is also available for model organisms, such as Drosophila and Caenorhabditis. On the other hand, information on plant chromosomes is limited predominantly to observations using EM. The available data suggest that although basic principles of chromatin folding are conserved among eukaryotes, they may result in a diverse mitotic chromosome ultrastructure in different species. This review summarizes the current knowledge of higher-order chromosome structure of condensed mitotic metaphase chromosomes and highlights common features as well as differences in the evolutionary context.
2. Higher-order structure of mitotic chromosomes
Chromatin structure changes dramatically during the cell cycle. In contrast to compact metaphase chromosomes, transcription and DNA synthesis, including chromosome reduplication in interphase nuclei require decondensed chromatin. During interphase, eukaryotic chromosomes occupy distinct spaces within the nucleus, so-called chromosome territories [11]. While the chromatin with active transcription and open structures is located in so-called A compartments, largely inactive regions are separated in B compartments and consist of more condensed chromatin [6]. Within the compartments, chromatin fibers are organized into topologically associated domains (TADs) [12]. In vertebrates, TADs span several hundred kilobases to 1 Mb DNA, while in the less complex genome of Drosophila, they are smaller by one order, ranging from tens to several hundred kilobases [13]. TADs are characterized by preferential intra-domain interactions and only sparse contacts occur across domain borders [14]. TADs in animals were shown to be not only structural, but also functional elements with an important role in the regulation of gene expression [15], [16]. However, mechanisms of their formation and function in plants remain to be investigated [17], [18].
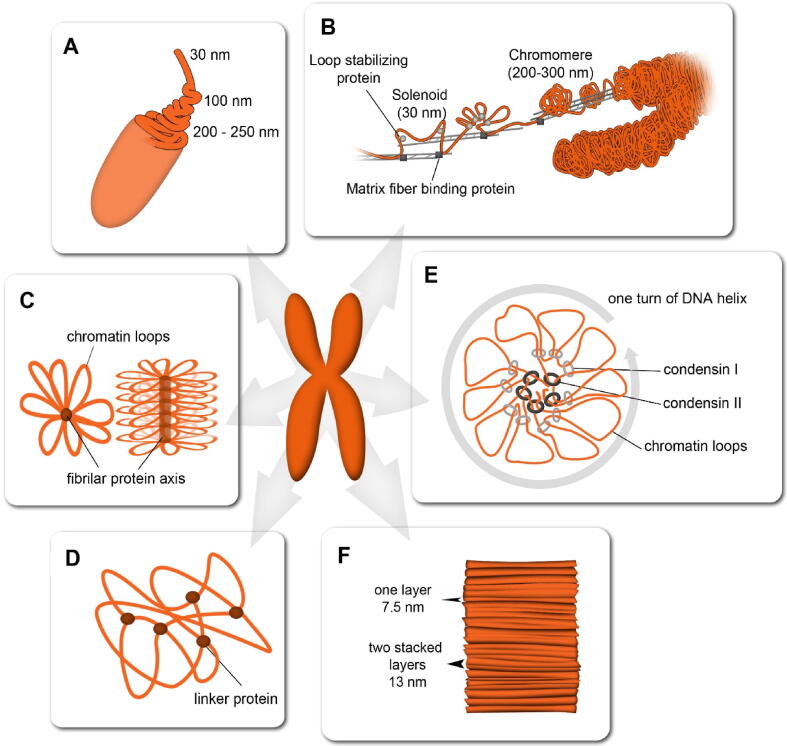
During mitosis, chromatin condenses and forms a uniform structure along the chromosome arms, whereas centromeres and secondary constrictions contain less chromatin. Several models were proposed to address the higher-order structure of the interstitial chromosome space. The classical “hierarchical helical folding” model suggests that the metaphase chromosome shape is established by sequential helical winding of a chromatin fiber from the nucleosome level to the condensed metaphase chromosome [19] (Fig. 1A). This model was based on electron and light microscopy analysis of Drosophila and human chromosomes and postulates that chromatin is a regular 30-nm fiber structure called solenoid, which is formed by consecutive self-coiling or zig-zagging of a 10-nm nucleosome fiber [20], [21]. By further coiling, the solenoid forms 100-nm and subsequently 200–250-nm fibers. The model was supported by many earlier observations by EM of vertebrate and plant chromosomes [22], [23], [24]. However, since the in-vivo formation of the 30-nm fiber was questioned by several recent studies [25], [26], [27], this model is currently not much supported.
Fig. 1.
Models of mitotic chromosome folding. (A) Hierarchical helical folding model [19]; (B) Dynamic matrix model [31]; (C) Radial loop model [33]; (D) Chromatin network model [38]; (E) Consecutive/nested loop model [56]; (F) Stacked layer loop model [60].
In addition to the microscopical techniques, the ultrastructure of mitotic chromosomes has been analyzed by Hi-C [5], [6]. This approach belongs to the group of Chromosome Conformation Capture (3C) methods, which were developed to study the spatial organization of chromatin in interphase nuclei [28]. The 3C-based techniques detect and quantify interactions between genomic loci that arise due to their spatial proximity, regardless of their genomic distance [29]. The principle of the 3C methods lies in the fixation (cross-linking) of chromatin by formaldehyde, which preserves the protein-mediated contacts of nearby chromatin segments. DNA is then digested and proximity-ligated to create a contact library that can be further analyzed by PCR or sequencing to identify DNA regions localized in a close proximity within the nucleus. Amongst these methods, Hi-C represents a whole-genome sequencing-based approach. Importantly, Hi-C analysis on human mitotic chromosomes did not support the classical hierarchical folding model described above since a decrease in the frequency of interactions with increasing genomic distance was much lower than predicted by the model [30]. Another argument against the hierarchical folding is that it would require a molecular machinery to manipulate chromatin at the micrometer and multi-megabase scale [30]. Thus, it is becoming increasingly apparent that the hierarchical helical folding cannot explain all features of the mitotic chromosome ultrastructure.
In 2000, a dynamic matrix model that shares basic principle with the hierarchical model was proposed by Wanner and Formanek, who investigated the ultrastructure of barley mitotic chromosomes by EM [31]. During a controlled decondensation of chromosomes, they observed that solenoid chromatin structures are attached to proteinaceous parallel matrix fibers forming loops. These solenoid loops further condense into 200–300 nm wide knobby structures called chromomeres and were suggested to be stabilized by cross-linker proteins. Moreover, the authors proposed that the matrix fibers move in anti-parallel direction similarly to actin and are responsible for elastic properties of the chromosomes [31], [32] (Fig. 1B). Although the dynamic matrix model still considers the controversial 30-nm fiber, the features like loops and cross-linking elements are common with the most recent models discussed below.
Another model of chromosome structure, named “radial loop model”, was proposed in 1977 (Fig. 1C) and assumes that mitotic metaphase chromosomes comprise a non-histone proteinaceous scaffold with chromatin loops consecutively emanating from its center [33], [34]. Compared to the hierarchical model, the radial loop model is more consistent with Hi-C data that support the consecutive looping and also the dynamic nature of chromatin fibers [30]. Previous observations of histone depleted chromatin indicated the presence of such a scaffold in the center of each chromatid of mammalian mitotic chromosomes [34]. However, more recent fluorescence and super-resolution microscopy observations on human mitotic chromosomes revealed that the chromosome scaffold was not made from a continuous fiber, but rather from discontinuously arranged protein complexes forming either a central double helix axis [35], or an alternating pattern of scaffold protein centers [36], [37].
The latter findings are in agreement with the chromatin network model proposed by Poirier and Marko (2002) based on their experiments with mechanical properties of amphibian mitotic chromosomes [38]. The authors demonstrated that not the proteinaceous scaffold, but chromatin was the only continuous component of chromosomes. Thus, a model was elaborated, where mitotic chromosomes were formed by a chromatin network crosslinked by non-histone linker proteins (Fig. 1D). This model was recently extended as a chromatin-gel model, considering also the role of histone methylation that stiffens the chromosome structure [39]. The chromatin network model relied on proteins suggested to be involved in chromosome condensation [40], [41].
Some of the anticipated proteins have already been confirmed as scaffolding proteins, namely condensins and topoisomerase II (topoII) and appear to be the major players in chromatin condensation [36], [42], [43], [44]. Besides them, other proteins associated with the chromosome scaffold have been found in vertebrates, such as the kinesin family member KIF4 [45] and the zinc-finger protein BAZ1B [46]. All these proteins shape the chromosomes by a coordinated chromatin manipulation.
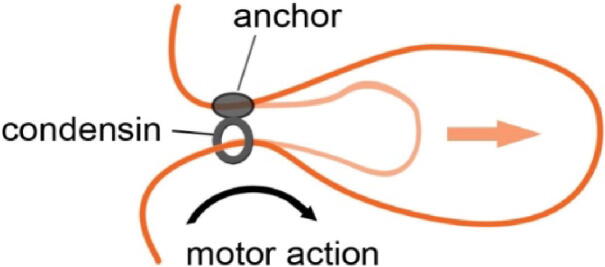
TopoII is a highly conserved enzyme involved in sister chromatid resolution and condensation. It tangles or untangles chromatin fibers by cutting and rejoining DNA fibers [43]. Condensin complexes, namely condensin I and II, together with cohesin belong to the family of structural maintenance of chromosomes (SMC) proteins [47]. Condensins constrain the movement of chromatin fiber by forming a ring structure that holds two distant parts of the fiber together, creating a chromatin loop by loop extrusion [48], [49]. This concept was reinforced by a computational analysis and simulations suggesting that mitotic chromosomes could be formed entirely by loop extrusion where a fiber forms consecutive loop arrays [50]. Recent in vitro experiment with nascent DNA confirmed the concept of loop formation by the extrusion mechanism, where the loop is formed by sliding of the condensin ring complex along a chromatin fiber [51] (Fig. 2) These findings contradict the chromatin network model where constraints are based on crosslinks rather than loop formations [38]. The advantage of the loop extrusion model over simple crosslinking is the ability to avoid crosslinking between both chromatid fibers which would prevent chromatid resolution during cell division [50]. It is not yet clear to what extent this mechanism applies in vivo. Although both condensin I and II share the same principle of action, each of them shows a different spatio-temporal behavior. Condensin II is localized in the nucleus during the entire cell cycle and participates in mitotic chromosome folding starting in prophase. Instead, condensin I is present in cytoplasm during interphase and loads onto chromatin after the nuclear envelope breakdown [52]. Another difference between the two complexes consists in their mode of action. RNA interference and protein depletion experiments in chicken showed that condensin II promotes axial shortening, while condensin I compacts chromosomes laterally [53]. More details on the mechanisms of condensin action and their role in mitotic chromatin folding can be found in dedicated reviews [54], [55].
Fig. 2.
Model of condensin-mediated loop extrusion. One strand of DNA is anchored by the kleisin and HEAT-repeat subunits of condensin, which extrude the chromatin loop by their motor action through the ring formed by the SMC arms. Adapted from Ganji et al. [51].
In a recent study, Gibcus et al. (2018) analyzed differences in Hi-C data after the depletion of condensin I, condensin II, or both, in chicken DT40 cells [56]. A model inferred from their experiments supported the earlier suggestions that complexes of condensin I and II play a distinct role in mitotic chromosome condensation. Condensin II was proposed to form a basic spiral scaffold, from which consecutive outer chromatin loops emanate. These loops are ~400 kb in size and are further axially compacted by condensin I into clusters of smaller (~80 kb) nested loops that are consequently aggregated by chromatin-to-chromatin attractions [56] (Fig. 1E). The associated loops form a spiral staircase-like structure with one turn of 10–12 Mb, as documented by the increase in long-range contacts with a periodicity of 10–12 Mb, observed in Hi-C data of condensin non-depleted cells [56]. This model was partially reinforced by quantitative super-resolution microscopy analyses on human mitotic cells. While a smaller number of condensin II complexes (~35,000/cell) were observed bound to chromosomal DNA more stably and centrally, the more dynamic condensin I bound peripherally with much higher frequency (~195,000/cell) [37]. Although recent super-resolution microscopy studies do not support the spiral organization of the condensin scaffold [36], [37], the observations are in agreement with the distinct roles of both condensin complexes.
It should be mentioned at this point that the amount of condensins loaded to chromosomes increases as a response to the treatment with anti-microtubule drugs, such as colchicine or nocodazole, that are used to arrest cycling cells in metaphase [36]. Such treatments result in higher degree of condensation and stiffness of mitotic chromosomes in a time-dependent manner [36], [57], [58], [59]. Importantly, Hi-C analyses of human and chicken mitotic chromosomes revealed that nocodazole treatment had only a mild effect on chromatin contact frequency and distribution, if applied for a short period of time (<3 h) [30], [56]. This indicates that short treatments with anti-microtubule drugs do not pose a serious obstacle to obtain a close-to-native picture of chromosome topology.
A different model of mitotic chromosome folding was proposed by Daban et al. (2015) who considered transversal features of mitotic chromosomes, such as G banding pattern, sister chromatid exchange and long-range interaction periodicity [60]. After chromosome analysis by atomic-force microscopy, the authors concluded that human and chicken chromosomes consisted of thin plates of chromatin layers [61]. Cryo-electron tomography and small-angle X-ray scattering (SAXS) methods were employed to show that the chromatids were composed of densely spaced 7.5-nm interdigitated nucleosome layers. An interdigitation of two 7.5-nm layers resulted in stacked layers of 13 nm, which is less than a mere sum of two layers’ thickness [62] (Fig. 1F). This observation is consistent with recent measurements of chromatin fiber parameters in human interphase nuclei and mitotic chromosomes by the ChromEMT technique, which combines electron microscopy tomography (EMT) with the ChromEM labeling method to selectivity enhance the contrast of DNA [9]. The average chromatin fiber width was 14.4 nm, corresponding to two stacked layers.
Although the stacked-layers model is not mutually exclusive with consecutive looping, each model explains in a different way the periodicity in the long-distance interaction frequency that is observed in Hi-C data. According to the consecutive looping model, the periodical increase in contact frequency, with the period of 12 Mb for chicken metaphase chromosomes is a consequence of condensin II-driven winding of the loops [56], while according to the stacked layer model, the long-range contacts reflect the stacking and interdigitating of planar layers already pre-existing in interphase [63]. However, it needs to be clarified to what extent the data for stacked-layers model are affected by the preparation methods.
Taken together, although the stacked layer model explains some features of mitotic chromosomes, it suffers from several shortcomings. First, it does not clarify the mechanism that brings the layers closer to each other during mitosis and separates them during interphase. This model also does not consider the role of architectural proteins such as condensins and topoII, which were shown to be crucial for the chromosome architecture [56], [64]. In contrast, the consecutive looping model of chromosome folding integrates the latest knowledge on the function of condensin I and II, and also data of Hi-C analysis of the mitotic chromosome topology and the features of chromatin fibers [56]. Thus, although the stacked layer model introduced an interesting view of the chromosome structure, the current knowledge is more in favor of the consecutive looping model to explain the compaction of mitotic metaphase chromosomes.
3. Cohesin and condensin play evolutionary distinct roles in mitotic chromosome folding
The current view of higher-order structure of mitotic chromosomes discussed in the previous section is almost exclusively based on findings obtained in vertebrates. Although the general principles of chromosome shaping are considered to be common to all eukaryotic organisms, there are structural and mechanical features that underwent different pathways during the evolution.
The structure of mitotic chromosomes in small eukaryotic genomes (~12–14 Mb genome size) diverges from the classical vertebrate model in several aspects. Studies conducted in yeast and the marine micro-alga Ostreococcus tauri reported only small differences in the degree of chromatin condensation between interphase and mitosis [65], [66], [67]. The nucleolus remains undissolved during mitosis in budding yeast and ribosomal DNA (rDNA) forms a specific loop around the nucleolus that splits in anaphase [68]. Modelling based on Hi-C analyses predicts that chromatin loops cover only 30–40% of the mitotic chromatin in yeast, which significantly differs from a 100% coverage estimated for animals [69]. This probably reflects different roles and dynamics of architectural proteins in yeast. Unlike in animals and plants, cohesin appears to be important for mitotic chromosome architecture in yeast [69], which may relate to the absence of condensin II in yeast chromatin [70], [71].
In order to investigate how specific proteins contribute to the spatial organization of chromatin, the ChIA-PET method (Chromatin Interaction Analysis by Paired-End Tag Sequencing) was developed [72]. The technique identifies and quantifies contacts between genomic loci that are mediated by a specific protein. ChIA-PET analyses in fission yeast revealed that chromatin domains formed by cohesin are maintained during the whole cell cycle [73], unlike in plants and animals where cohesin is removed from sister chromatid arms at the entry to mitosis [74], [75]. Nevertheless, like in vertebrates, larger domains of 300 kb–1 Mb mediated by condensin I are present in mitotic chromosomes of fission yeast [73], [76]. Yeast condensin is required especially for rDNA, centromere and pericentromeric chromatin loop formation [69], [77], [78]. The observation that condensin-mediated loops in yeast are of the same size or even larger than in vertebrates is consistent with models suggesting a minor role of chromatin loops for the formation of the yeast mitotic chromosome structure. Moreover, the number of small cohesin-mediated loops is reduced during mitosis in fission yeast compared to interphase [73]. Thus, yeasts exhibit a somewhat different topology of mitotic chromosomes than higher eukaryotes, the major difference being the divergent roles of cohesin and condensin.
In animals and plants, cohesin seems not to be involved in mitotic chromosome folding, since it is removed from mitotic chromosome arms after the onset of prophase [74], [75]. On the contrary, the available data across multiple higher-eukaryote species indicate that the importance of condensin II increases dramatically with the increasing genome complexity. While dysfunctional condensin II led to severe mitotic damage in vertebrates [79], the complex seems dispensable for mitotic chromosome condensation in Drosophila [80], [81] and the primitive red alga Cyanidioschyzon merolae [82]. The condensin II complex was also found completely missing in some insects [83]. In Drosophila and C. merolae, condensin I plays a more important role than condensin II [64], [82], which is enriched in the centromere/inner kinetochore region, similar to human [52]. This observation led to a hypothesis that during the evolution, condensin II was originally involved in centromere structure and resolution [42]. Consistently, in the holocentric species Caenorhabditis elegans, condensin II is predominantly present along chromosomes at the kinetochore centers, while condensin I is distributed more internally [84], thus showing a distinct spatial distribution than in vertebrates [37], [56]. Since Drosophila and C. merolae have small chromosomes as compared to the investigated vertebrates, it is possible that condensin I together with topo II are sufficient to maintain the structure of their mitotic chromosomes. Thus, the consecutive loops might not be the predominant feature of the metaphase chromosome structure in the small genomes, just as in yeasts.
Compared to model species and human, functional condensin studies on mitotic chromosome organization in plants are lagging behind. Although condensin subunits were shown to be essential for interphase centromere arrangement and correct plant development [85], mitotic defects caused by mutations of SMC proteins were not lethal in Arabidopsis [86], [87], perhaps due to the presence of partially functional truncated proteins. To study the localization and dynamics of condensin I and II, Fujimoto et al. used GFP-tagged condensin subunits AtCAP-H and AtCAP-H2 in cultured tobacco cells [88]. They found that the localization of these proteins across the cell cycle and on mitotic chromosomes was similar to vertebrates. Studies in plants clearly demonstrated the importance of condensin for chromatin organization and DNA repair in interphase [85], [89], [90]. However, the precise role of condensin for mitotic chromosome formation needs to be determined.
Although currently the most accepted model of chromosome folding assumes the consecutive looping by both condensin complexes (Fig. 1E), several studies indicate that this model cannot be generalized beyond vertebrates [42], [73], [83]. The fact that condensin II appears dispensable across eukaryotes raises the question whether the crucial function of condensin II was replaced by other proteins in some organisms, or whether condensin I is the major player capable to shape mitotic chromosomes. A partial answer was given by an in-vitro experiment where structures resembling mitotic chromatids were reconstituted from Xenopus laevis sperm-chromatin isolates using only six purified components: core histones, three histone chaperones (nucleoplasmin, Nap1 and FACT), topoII and condensin I [91]. Thus, condensin I and topoII seem to be the top players in chromosome shaping. A supporting evidence came from a study conducted in Drosophila revealing that the loop-forming activity of condensin I directs the topoII enzymatic activity, which is essential in chromatid resolution and condensation processes [64], [92]. The hypothesis on the dispensability of condensin II is also reinforced by the evidences of multiple losses of condensin II during insect evolution [83]. Taken together, the available evidence indicates that the dispensability of condensin II for mitotic chromosome formation is restricted to species with small chromosomes. This suggests that the chromosome (genome) size might be an important factor in the evolution of the condensin-mediated chromosome topology.
4. Chromosome cavities: A fact or artifact?
The number of high-resolution and close-to-native-state images of mitotic chromosomes increased considerably during the last decade and contributed to the development of comprehensive models of higher-order chromatin structure. However, one of the phenomena not entirely addressed in any of the current models is the presence of chromosome cavities, observed as a chromatin-free space within condensed chromosomes. They were identified inside chromatids of mitotic chromosomes by several microscopic techniques, such as scanning electron microscopy (SEM), transmission electron microscopy (TEM) and light microscopy [8], [93], [94], [95], [96]. Because this topic has never been systematically addressed, we suggest distinguishing between two types of observations reported in literature.
The first type corresponds to large “single cavities” with a diameter of one hundred up to a few hundred nm, which are observed in small numbers, usually 3–5 per chromatid. Single cavities were first observed along the axial region of chromatids in several plant species, being most pronounced during prophase and telophase when chromatin condensation/decondensation takes place [94], [96]. A study in a group of plants indicated that the single cavities are present in species with DNA content per chromatid larger than ~700 Mb [95]. The second type of cavities are “chromatin-free regions” with the size ranging from ~10 nm to ~200 nm, which are present in a higher number and form an interconnected network within chromatids. Such chromatin free-regions were observed in metaphase chromosomes of barley and human prophase/metaphase chromosomes [8], [10], [97].
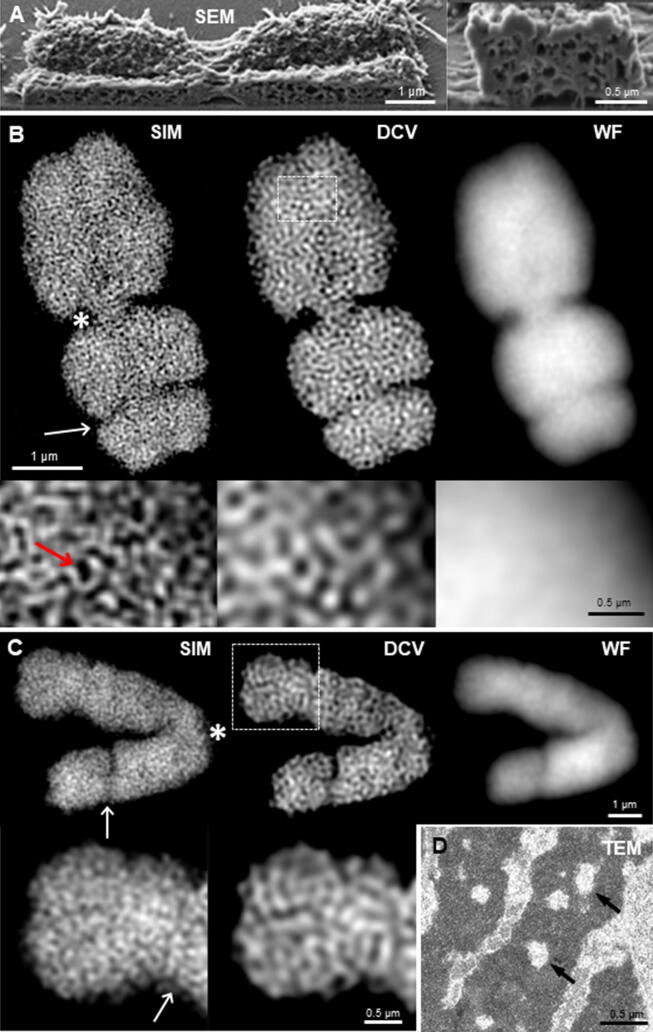
In contrast to an earlier report on large single cavities in plants [95], a detailed SEM study on barley metaphase chromosomes showed that chromosomes contain chromatin-free regions forming a network within the chromatid representing ~19% of the chromosome volume [8] (Fig. 3A). In human prophase chromatids, the chromatin-free regions account to 4.5–7% only [10]. It is noteworthy that the single cavities were observed using TEM, while the chromosome free-regions were observed using SEM. Indeed, a comparison of results obtained by both techniques in barley chromosomes [8], [95] suggests that the cavity type observed depends on the imaging technique used. While TEM displays only a very thin layer and thus the cavities are observed individually, SEM detects a complex structure of chromatin-free regions and their interconnections may be observed. Moreover, several studies pointed out that the occurrence of cavities may be associated with specific sample preparation protocols. Hamano et al. [97] noted that the cavities were not visible in human and barley chromosomes when an ionic liquid method that keeps the chromosomes in a wet state was used. This raised the question whether the cavities are an artifact due to sample drying or whether they indeed exist in vivo but are not visible when filled by the ionic liquid. Chen et al. [10] used serial block-face SEM to establish the whole spatial prophase nuclei structure without changing the nuclear volume, and their results supported the existence of cavities within the human chromosomes.
Fig. 3.
Observation of chromatin-free regions in condensed mitotic chromosomes of barley. The observation of a network of chromatin-free regions (A-C) and large cavities (D) within mitotic barley chromosomes depends on preparation and imaging methods. (A) Focused ion beam SEM of a metaphase chromosome (from Schroeder-Reiter et al. 2009) [8]. (B, C) The meta- (B) and anaphase (C) chromosomes 6H distinguished by a secondary constriction (white arrows) and imaged by SIM show compared to deconvolution (DCV) and wide-field (WF) microscopy the ultrastructure at a resolution of ~100 nm. One representative slice from inside of the chromosome/chromatid per 3D-SIM image stack is shown. Within the chromosome arms and centromeres (asterisks), a network of chromatin-free spaces is present. The size of these spaces may reach up to ~120 × 220 nm (red arrow). The dashed rectangles mark the enlarged regions below. DNA was labeled by DAPI. (D) Anaphase chromosomes showing large cavities (arrows) after imaging by TEM of ultra-thin sections stained with uranyl acetate (from Kuznetsova et al. [95]).
To clarify the presence of cavities in barley meta- and anaphase chromosomes we applied structured illumination microscopy (3D-SIM) to achieve super-resolution at ~100-nm [98]. Root meristems were fixed in ethanol:acetic acid (3:1) and chromosomes were labeled with the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI). In contrast to wide-field and deconvolution microscopy, SIM clearly shows the ultrastructural chromatin organization (Fig. 3B, C). Within the complete 3D-SIM image stacks, both meta- and anaphase chromosomes do not show large cavities as identified via TEM by Kuznetsova et al. [95] (Fig. 3D). Nevertheless, similarly to the observations made with the focused ion-beam SEM by Schroeder-Reiter et al. [8] (Fig. 3A), a network of smaller (up to ~120 × 220 nm) chromatin-free regions was identified (Fig. 3B, C). These results indicate that a chromatin-free network is present within the condensed chromatin of mitotic chromosomes, while the observation of larger cavities may rather be a preparation artifact.
The chromatin-free network was proposed to serve as a “corridor” for functional/signaling molecules in the densely packed chromosomes [8]. Such an organization seems to be essential to facilitate the dynamics of structural proteins and their signaling pathways, and to ensure transcription that occurs also during mitosis, albeit to a much lesser extent than during interphase [99].
5. Conclusions
This review demonstrates that even an evolutionary conserved structure such as the mitotic chromosome displays a certain degree of diversity. This raises the question whether multiple mechanisms of higher-order folding evolved independently to pack the chromatin fiber into the condensed mitotic chromosome. In order to find an answer, comparable data are needed from multiple organisms obtained by a series of techniques, including super-resolution microscopy, 3C-based methods and reverse-genetics approaches to clarify the function of particular proteins involved in shaping the chromosomes. Falling sequencing costs, which enable generating high-resolution Hi-C data even for the most complex genomes, the implementation of new techniques, such as HiChIP [100], Hi-M [101] and super-resolution chromatin tracing [102], as well as advances in computational algorithms used to model chromosome structures promise to clarify one of the biological enigmas in near future.
CRediT authorship contribution statement
Tomáš Beseda: Writing - original draft, Writing - review & editing. Petr Cápal: Visualization, Writing - original draft. Ivona Kubalová: Investigation, Writing - review & editing. Veit Schubert: Funding acquisition, Investigation, Writing - review & editing. Jaroslav Doležel: Funding acquisition, Project administration, Writing - review & editing. Hana Šimková: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work has been supported by the Deutsche Forschungsgemeinschaft (Schu 762/11-1); DFG-GAČR project, award No. 18-14450 and by the ERDF project “Plants as a tool for sustainable global development” (No. CZ.02.1.01/0.0/0.0/16_019/0000827).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.01.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schubert V. Super-resolution microscopy - applications in plant cell research. Front Plant Sci. 2017;8:531. doi: 10.3389/fpls.2017.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walther N., Ellenberg J. Chapter 4 - Quantitative live and super-resolution microscopy of mitotic chromosomes. In: Maiato H., Schuh M., editors. Methods in cell biology. Academic Press; 2018. pp. 65–90. [DOI] [PubMed] [Google Scholar]
- 3.Baroux C., Schubert V. Technical review: microscopy and image processing tools to analyze plant chromatin: practical considerations. In: Bemer M., Baroux C., editors. Plant chromatin dynamics: methods and protocols. Springer; New York: 2018. pp. 537–589. [DOI] [PubMed] [Google Scholar]
- 4.Danev R., Yanagisawa H., Kikkawa M. Cryo-electron microscopy methodology: current aspects and future directions. Trends Biochem Sci. 2019;44:837–848. doi: 10.1016/j.tibs.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Sati S., Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2017;126:33–44. doi: 10.1007/s00412-016-0593-6. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanner G., Schroeder-Reiter E., Ma W., Houben A., Schubert V. The ultrastructure of mono- and holocentric plant centromeres: an immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma. 2015;124:503–517. doi: 10.1007/s00412-015-0521-1. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder-Reiter E., Perez-Willard F., Zeile U., Wanner G. Focused ion beam (FIB) combined with high resolution scanning electron microscopy: a promising tool for 3D analysis of chromosome architecture. J Struct Biol. 2009;165:97–106. doi: 10.1016/j.jsb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O'Shea C.C. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B., Yusuf M., Hashimoto T., Estandarte A.K., Thompson G., Robinson I. Three-dimensional positioning and structure of chromosomes in a human prophase nucleus. Sci Adv. 2017;3:e1602231. doi: 10.1126/sciadv.1602231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cremer T., Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez F., Bhardwaj V., Arrigoni L., Lam K.C., Gruning B.A., Villaveces J. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo Q., Bantignies F., Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon Jesse R., Gorkin David U., Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong P., Tu X., Li H., Zhang J., Grierson D., Li P. Tissue-specific Hi-C analyses of rice, foxtail millet and maize suggest non-canonical function of plant chromatin domains. J Integr Plant Biol. 2019;44 doi: 10.1111/jipb.12809. [DOI] [PubMed] [Google Scholar]
- 18.Stam M., Tark-Dame M., Fransz P. 3D genome organization: a role for phase separation and loop extrusion? Curr Opin Plant Biol. 2019;48:36–46. doi: 10.1016/j.pbi.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Sedat J., Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Finch J.T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodcock C.L., Frado L.L., Rattner J.B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnuki Y. Structure of chromosomes. I. Morphological studies of the spiral structure of human somatic chromosomes. Chromosoma. 1968;25:402–428. doi: 10.1007/BF02327721. [DOI] [PubMed] [Google Scholar]
- 23.Manton I. The spiral structure of chromosomes. Biol Rev Camb Philos Soc. 1950;25:486–508. doi: 10.1111/j.1469-185x.1950.tb00770.x. [DOI] [PubMed] [Google Scholar]
- 24.Rattner J.B., Lin C.C. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42:291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 25.Maeshima K., Ide S., Babokhov M. Dynamic chromatin organization without the 30-nm fiber. Curr Opin Cell Biol. 2019;58:95–104. doi: 10.1016/j.ceb.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Hansen J.C., Connolly M., McDonald C.J., Pan A., Pryamkova A., Ray K. The 10-nm chromatin fiber and its relationship to interphase chromosome organization. Biochem Soc Trans. 2018;46:67–76. doi: 10.1042/BST20170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigoryev S.A., Bascom G., Buckwalter J.M., Schubert M.B., Woodcock C.L., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 29.Davies J.O., Oudelaar A.M., Higgs D.R., Hughes J.R. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14:125–134. doi: 10.1038/nmeth.4146. [DOI] [PubMed] [Google Scholar]
- 30.Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B.R., Mirny L.A. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wanner G., Formanek H. A new chromosome model. J Struct Biol. 2000;132:147–161. doi: 10.1006/jsbi.2000.4310. [DOI] [PubMed] [Google Scholar]
- 32.Wanner G., Schroeder-Reiter E., Formanek H. 3D Analysis of chromosome architecture: advantages and limitations with SEM. Cytogenet Genome Res. 2005;109:70–78. doi: 10.1159/000082384. [DOI] [PubMed] [Google Scholar]
- 33.Paulson J.R., Laemmli U.K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U.K., Cheng S.M., Adolph K.W., Paulson J.R., Brown J.A., Baumbach W.R. Metaphase chromosome structure: the role of nonhistone proteins. Cold Spring Harb Symp Quant Biol. 1978;42:351–360. doi: 10.1101/sqb.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- 35.Poonperm R., Takata H., Hamano T., Matsuda A., Uchiyama S., Hiraoka Y. Chromosome scaffold is a double-stranded assembly of scaffold proteins. Sci Rep. 2015;5 doi: 10.1038/srep11916. Article 11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun M.X., Biggs R., Hornick J., Marko J.F. Condensin controls mitotic chromosome stiffness and stability without forming a structurally contiguous scaffold. Chromosome Res. 2018;26:277–295. doi: 10.1007/s10577-018-9584-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walther N., Hossain M.J., Politi A.Z., Koch B., Kueblbeck M., Odegard-Fougner O. A quantitative map of human condensins provides new insights into mitotic chromosome architecture. J Cell Biol. 2018;217:2309–2328. doi: 10.1083/jcb.201801048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poirier M.G., Marko J.F. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biggs R., Liu P.Z., Stephens A.D., Marko J.F. Effects of altering histone posttranslational modifications on mitotic chromosome structure and mechanics. Mol Biol Cell. 2019;30:820–827. doi: 10.1091/mbc.E18-09-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura K., Rybenkov V.V., Crisona N.J., Hirano T., Cozzarelli N.R. 13S Condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 41.Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 42.Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 43.Piskadlo E., Oliveira R.A. A Topology-centric view on mitotic chromosome architecture. Int J Mol Sci. 2017;18:E2751. doi: 10.3390/ijms18122751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeshima K., Laemmli U.K. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 45.Poonperm R., Takata H., Uchiyama S., Fukui K. Interdependency and phosphorylation of KIF4 and condensin I are essential for organization of chromosome scaffold. PLoS One. 2017;12:e0183298. doi: 10.1371/journal.pone.0183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohta S., Taniguchi T., Sato N., Hamada M., Taniguchi H., Rappsilber J. Quantitative proteomics of the mitotic chromosome scaffold reveals the association of BAZ1B with chromosomal axes. Mol Cell Proteomics. 2019;18:169–181. doi: 10.1074/mcp.RA118.000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 48.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 49.Alipour E., Marko J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goloborodko A., Marko J.F., Mirny L.A. Chromosome compaction by active loop extrusion. Biophys J. 2016;110:2162–2168. doi: 10.1016/j.bpj.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganji M., Shaltiel I.A., Bisht S., Kim E., Kalichava A., Haering C.H. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ono T., Fang Y., Spector D.L., Hirano T. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green L.C., Kalitsis P., Chang T.M., Cipetic M., Kim J.H., Marshall O. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kakui Y., Uhlmann F. SMC complexes orchestrate the mitotic chromatin interaction landscape. Curr Genet. 2018;4:335–339. doi: 10.1007/s00294-017-0755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi M., Hirota T. Folding the genome into mitotic chromosomes. Curr Opin Cell Biol. 2019;60:19–26. doi: 10.1016/j.ceb.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Gibcus J.H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J. A pathway for mitotic chromosome formation. Science. 2018;359:eaao6135. doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Östergen G. Colchicine mitosis, chromosome contraction, narcosis and protein chain folding. Hereditas. 1944;30:429–467. [Google Scholar]
- 58.Moser S.C., Swedlow J.R. How to be a mitotic chromosome. Chromosome Res. 2011;19:307–319. doi: 10.1007/s10577-011-9198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koyani P.R., Saiyad S.S. Study of effect of colchicine exposure on length of chromosome during mitosis. J. Anat. Soc. India. 2011;60:177–180. [Google Scholar]
- 60.Daban J.R. Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci Rep. 2015;5 doi: 10.1038/srep14891. Article 14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gállego I., Castro-Hartmann P., Caravaca J.M., Caño S., Daban J.-R. Dense chromatin plates in metaphase chromosomes. Eur Biophys J. 2009;38:503–522. doi: 10.1007/s00249-008-0401-1. [DOI] [PubMed] [Google Scholar]
- 62.Chicano A., Crosas E., Oton J., Melero R., Engel B.D., Daban J.R. Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure. Embo J. 2019;38:e99769. doi: 10.15252/embj.201899769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chicano A., Daban J.R. Chromatin plates in the interphase nucleus. FEBS Lett. 2019;593:810–819. doi: 10.1002/1873-3468.13370. [DOI] [PubMed] [Google Scholar]
- 64.Piskadlo E., Tavares A., Oliveira R.A. Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. Elife. 2017;6 doi: 10.7554/eLife.26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C., Lim H.H., Shi J., Tamura S., Maeshima K., Surana U. Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Mol Biol Cell. 2016;27:3357–3368. doi: 10.1091/mbc.E16-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan L., Ladinsky M.S., Jensen G.J. Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma. 2013;122:377–386. doi: 10.1007/s00412-013-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai S., Chen C., Tan Z.Y., Huang Y., Shi J., Gan L. Cryo-ET reveals the macromolecular reorganization of S. pombe mitotic chromosomes in vivo. Proc Natl Acad Sci USA. 2018;115:10977–10982. doi: 10.1073/pnas.1720476115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuchs J., Loidl J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004;12:427–438. doi: 10.1023/B:CHRO.0000034726.05374.db. [DOI] [PubMed] [Google Scholar]
- 69.Schalbetter S.A., Goloborodko A., Fudenberg G., Belton J.M., Miles C., Yu M. SMC complexes differentially compact mitotic chromosomes according to genomic context. Nat Cell Biol. 2017;19:1071–1080. doi: 10.1038/ncb3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freeman L., Aragon-Alcaide L., Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fullwood M.J., Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanizawa H., Kim K.-D., Iwasaki O., Noma K.-I. Architectural alterations of the fission yeast genome during the cell cycle. Nat Struct Mol Biol. 2017;24:965–976. doi: 10.1038/nsmb.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waizenegger I.C., Hauf S., Meinke A., Peters J.-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 75.Bolaños-Villegas P., De K., Pradillo M., Liu D., Makaroff C.A. In favor of establishment: regulation of chromatid cohesion in plants. Front Plant Sci. 2017;8:846. doi: 10.3389/fpls.2017.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kakui Y., Rabinowitz A., Barry D.J., Uhlmann F. Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast. Nat Genet. 2017;49:1553–1557. doi: 10.1038/ng.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lazar-Stefanita L., Scolari V.F., Mercy G., Muller H., Guerin T.M., Thierry A. Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. 2017;36:2684–2697. doi: 10.15252/embj.201797342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawrimore J., Doshi A., Friedman B., Yeh E., Bloom K. Geometric partitioning of cohesin and condensin is a consequence of chromatin loops. Mol Biol Cell. 2018;29:2737–2750. doi: 10.1091/mbc.E18-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishide K., Hirano T. Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oliveira R.A., Heidmann S., Sunkel C.E. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259–274. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- 81.Herzog S., Nagarkar Jaiswal S., Urban E., Riemer A., Fischer S., Heidmann S.K. Functional dissection of the Drosophila melanogaster condensin subunit Cap-G reveals its exclusive association with condensin I. PLoS Genet. 2013;9:e1003463. doi: 10.1371/journal.pgen.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujiwara T., Tanaka K., Kuroiwa T., Hirano T. Spatiotemporal dynamics of condensins I and II: evolutionary insights from the primitive red alga Cyanidioschyzon merolae. Mol Biol Cell. 2013;24:2515–2527. doi: 10.1091/mbc.E13-04-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.King T.D., Leonard C.J., Cooper J.C., Nguyen S., Joyce E.F., Phadnis N. Recurrent losses and rapid evolution of the condensin II complex in insects. Mol Biol Evol. 2019:2195–2204. doi: 10.1093/molbev/msz140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Csankovszki G., Collette K., Spahl K., Carey J., Snyder M., Petty E. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schubert V., Lermontova I., Schubert I. The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma. 2013;122:517–533. doi: 10.1007/s00412-013-0424-y. [DOI] [PubMed] [Google Scholar]
- 86.Liu C.-M., McElver J., Tzafrir I., Joosen R., Wittich P., Patton D. Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 2002;29:405–415. doi: 10.1046/j.1365-313x.2002.01224.x. [DOI] [PubMed] [Google Scholar]
- 87.Tzafrir I., McElver J.A., Liu C.-M., Yang L.J., Wu J.Q., Martinez A. Diversity of TITAN Functions in Arabidopsis seed development. Plant Physiol. 2002;128:38–51. [PMC free article] [PubMed] [Google Scholar]
- 88.Fujimoto S., Yonemura M., Matsunaga S., Nakagawa T., Uchiyama S., Fukui K. Characterization and dynamic analysis of Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2. Planta. 2005;222:293–300. doi: 10.1007/s00425-005-1546-0. [DOI] [PubMed] [Google Scholar]
- 89.Sakamoto T., Inui Y.T., Uraguchi S., Yoshizumi T., Matsunaga S., Mastui M. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell. 2011;23:3533–3546. doi: 10.1105/tpc.111.086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakamoto T., Sugiyama T., Yamashita T., Matsunaga S. Plant condensin II is required for the correct spatial relationship between centromeres and rDNA arrays. Nucleus. 2019;10:116–125. doi: 10.1080/19491034.2019.1616507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shintomi K., Takahashi T.S., Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17:1014–1023. doi: 10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- 92.Orlandini E., Marenduzzo D., Michieletto D. Synergy of topoisomerase and structural-maintenance-of-chromosomes proteins creates a universal pathway to simplify genome topology. Proc Natl Acad Sci USA. 2019;116:8149–8154. doi: 10.1073/pnas.1815394116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hao S., Jiao M., Huang B. Chromosome organization revealed upon the decondensation of telophase chromosomes in Allium. Chromosoma. 1990;99:371–378. [Google Scholar]
- 94.Hao S., Xing M., Jiao M. Chromatin-free compartments and their contents in anaphase chromosomes of higher plants. Cell Biol Int Rep. 1988;12:627–635. [Google Scholar]
- 95.Kuznetsova M.A., Chaban I.A., Sheval E.V. Visualization of chromosome condensation in plants with large chromosomes. BMC Plant Biol. 2017;17 doi: 10.1186/s12870-017-1102-7. Article 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sparvoli E., Gay H., Kaufmann B.P. Number and pattern of association of chromonemata in chromosomes of Tradescantia. Chromosoma. 1965;16:415–435. doi: 10.1007/BF00343171. [DOI] [PubMed] [Google Scholar]
- 97.Hamano T., Dwiranti A., Kaneyoshi K., Fukuda S., Kometani R., Nakao M. Chromosome interior observation by Focused ion beam/Scanning electron microscopy (FIB/SEM) Using ionic liquid technique. Microsc Microanal. 2014;20:1340–1347. doi: 10.1017/S143192761401280X. [DOI] [PubMed] [Google Scholar]
- 98.Weisshart K., Fuchs J., Schubert V. Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules on flow-sorted arabidopsis nuclei. Bio-protocol. 2016;6:e1725. [Google Scholar]
- 99.Palozola K.C., Liu H., Nicetto D., Zaret K.S. Low-level, global transcription during mitosis and dynamic gene reactivation during mitotic exit. Cold Spring Harb Symp Quant Biol. 2017;82:197–205. doi: 10.1101/sqb.2017.82.034280. [DOI] [PubMed] [Google Scholar]
- 100.Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cardozo Gizzi A.M., Cattoni D.I., Fiche J.-B., Espinola S.M., Gurgo J., Messina O. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell. 2019;74:212–222.e5. doi: 10.1016/j.molcel.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 102.Bintu B., Mateo L.J., Su J.-H., Sinnott-Armstrong N.A., Parker M., Kinrot S. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362:eaau1783. doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.