Abstract
Marine animals represent a dynamic and complex habitat for diverse microbial communities. The microbiota associated with bottlenose dolphins (Tursiops truncatus) are believed to influence their health status, but it remains poorly understood. We therefore characterized and compared the bacterial microbiome of bottlenose dolphins from six different anatomical sites that represent four different body systems (respiratory, digestive, reproductive, and integumentary). In this study, a total of 14 free-ranging bottlenose dolphins were sampled during the 2015 Sarasota Bay Dolphin Health Assessment. Bacterial diversity and abundance were assessed by PCR amplification of the hypervariable V3–V4 regions of the bacterial 16S rRNA gene for each sample, followed by sequencing on an Illumina MiSeq platform. Analysis showed that bottlenose dolphins harbor diverse bacterial communities with a unique microbial community at each body system. Additionally, the bottlenose dolphin bacterial microbiome was clearly distinct to the aquatic microbiome from their surrounding habitat. These results are in close agreement with other cetacean microbiome studies, while our study is the first to explore what was found to be a diverse bottlenose dolphin genital microbiome. The core bacterial communities identified in this study in apparently healthy animals might be informative for future health monitoring of bottlenose dolphins.
Keywords: Microbiology, Biological sciences, Veterinary medicine, Health sciences, Bottlenose dolphin, Microbiome, Next-generation sequencing, Health assessment
Microbiology; Biological sciences; Veterinary medicine; Health sciences; Bottlenose dolphin; Microbiome; Next-generation sequencing; Health assessment.
1. Introduction
Marine mammals can serve as sentinels for human and aquatic ecosystem health due to their long lifespans in coastal ecosystems, high trophic feeding level, and propensity for bio-accumulating toxic chemicals in their blubber (Holden, 1970; Ross, 2000; Reddy et al., 2001; Wells et al., 2004; Bossart, 2011). Unfortunately, the health of many marine mammals, including bottlenose dolphins (Tursiops truncatus), is threatened due to marine pollution, habitat degradation, hunting, bycatch, and emerging infectious diseases (Trites et al., 1999; Springer et al., 2003; Read et al., 2006; Estes et al., 2009; Van Bressem et al., 2009; Waltzek et al., 2012). Extensive research has been conducted to evaluate populations of bottlenose dolphins with respect to their behavior, social structure, ecology and conservation needs (Würsig and Würsig, 1979; Shane et al., 1986; Wells, 1991; Segura et al., 2006; Bearzi et al., 2008). However, little is documented about the presence and variation of host associated microbial communities, which play important roles in host health and in a wide range of diseases (Johnson et al., 2009; Bik et al., 2016; Russo et al., 2018; Suzuki et al., 2018).
The host microbiome is composed of all of the microbes in a given environment, including bacteria, viruses, fungi, and protozoa (Blaser and Kirschner, 2007). Host associated microbiota contribute to host physiology (Sommer and Bäckhed, 2013), digestion (Choat and Clements, 1998), vitamin synthesis (LeBlanc et al., 2013), colonization resistance, and host immune health (Hooper et al., 2012; Ottman et al., 2012). Moreover, microbiota are known to establish symbiotic, commensal, or even pathogenic relationships with their hosts, depending upon a wide variety of factors, including host immunocompetence and genetics (Shulzhenko et al., 2011; Sommer and Bäckhed, 2013). Studies have been done to explore the microbial communities of bottlenose dolphins; however, the majority have been based on culture-dependent techniques (Sweeney and Ridgway, 1975; Howard et al., 1983; Buck et al., 2006; Venn-Watson et al., 2008). One major drawback of culture dependent techniques is that a large proportion of bacterial communities are uncultivable (Stewart, 2012). More recently, culture independent approaches using next-generation sequencing (NGS) technologies have gained considerable acceptability to characterize the host associated microbial communities in both humans and animals including aquatic species (Gill et al., 2006; Kim and Isaacson, 2015; Bik et al., 2016; Ahasan et al., 2017).
Although a range of bacterial species have been isolated from moribund bottlenose dolphins using conventional phenotypic identification techniques (Buck et al., 2006; Venn-Watson et al., 2008), the role these bacteria play in disease remains challenging because the same bacteria are often isolated from healthy animals (Buck et al., 2006; Venn-Watson et al., 2008). Importantly, the determination of what comprises “healthy” in wild dolphin health assessments is typically based on limited diagnostic tools applied in one event cycle of an animal's health evaluation. For example, the finding of variations in compromised lung images noted on ultrasound might be correlated with changes in the respiratory microbiome. Unfortunately, microbiome population characteristics, changes and their significance prior, during or after the resolution of these issues cannot be determined with limited sampling.
The distinctive body plans of marine mammals offer many niches for diverse members of bacterial and archaeal kingdoms. These microbial communities may vary by body site due to distinct physiochemical conditions, which exert selective pressures on the microbiota (Costello et al., 2009; Grice et al., 2009). To date, few studies have investigated the microbiota of bottlenose dolphins using NGS platforms. Microbiome studies employing NGS approaches have been limited to the bottlenose dolphin skin (Chiarello et al., 2017; Russo et al., 2018), respiratory tract (Bik et al., 2016) and/or digestive tract (Bik et al., 2016; Soverini et al., 2016). Soverini et al. (2016). Chiarello et al. (2017) sampled managed bottlenose dolphin populations, Russo et al. (2018) sampled free-ranging animals, and Bik et al. (2016) sampled both managed and free-ranging animals. Bik et al. (2016) reported differences in the oral and fecal microbiomes of managed versus free-ranging bottlenose dolphins; however, the two populations differed in geographic location, food sources, and medical care.
The aim of the present study was to explore the body site-specificity patterns of the free-ranging bottlenose dolphin microbiome. We characterized and compared the bacterial communities of four different body systems (respiratory, digestive, reproductive, and integumentary systems) of bottlenose dolphins. Additionally, we evaluated the patterns of bacterial diversity that vary with age, gender, health status, lactation status, and relatedness (mother-calf pairs) in bottlenose dolphins.
2. Materials and methods
2.1. Bottlenose dolphin sample acquisition
The swab and water samples were collected by veterinarians as part of the 2015 Sarasota Bay Dolphin Health Assessment in Sarasota, Florida from May 11th 2015 through May 22nd 2015 following the procedure described by Wells et al. (2004). Additionally, body weight, length, and girth measurements were recorded during the sample collection. An ultrasound evaluation was conducted in adult females for diagnosis of pregnancy. Ultrasound was also used to measure blubber depth and evaluate thoracic and abdominal organs for any abnormality or health concern. The well-being of the animals is a priority, and for that reason, respiratory and behavioral patterns were closely monitored throughout the sampling process. Blood samples were collected by puncturing their fluke, urine samples were taken with a sterile catheter, and milk was collected through a suction system. The age of most of the Sarasota Bay bottlenose dolphins is known through observation and for animals of unknown age, a tooth is extracted under local anesthesia (Hohn et al., 1989). A complete examination and sampling process took up to an hour, and when all the procedures were done, the animal was placed back in the water and released (Wells et al., 2004). Samples were collected under National Marine Fisheries Service Scientific Research Permit No. 15543 and Mote Marine Laboratory IACUC approval.
In the 2015 Sarasota Bay bottlenose dolphin health assessment, sampling included: 14 bottlenose dolphins (9 females and 5 males), including 4 mother-calf pairs and 5 calves (Table S1). Age ranged from 2 to 31 years, as determined by observation of additional newborn calves into the population and tooth extraction from animals of unknown age (Hohn et al., 1989). Health status of the animals was determined through physical examination and ultrasound (Wells et al., 2004). Nine animals were identified with compromised health including seven animals with mild to moderate lung disease (lung changes observed by ultrasonography), including a female with a genital papilloma positive for bottlenose dolphin gammaherpesvirus 1 (confirmed by PCR at the University of Florida's Wildlife and Aquatic Veterinary Disease Laboratory), and two animals with potential kidney disease (renomegaly observed by ultrasonography) (Table S1). Five animals were deemed healthy based on a lack of abnormalities detected by the physical and testing protocols (Table S1). Other than mothers and calves, none of the bottlenose dolphins sampled for this project were closely related, but they are all considered to be long-term members of the resident Sarasota dolphin community (Wells, unpublished data).
Specimen collection consisted of six samples per individual (for exceptions see Table S1) from five anatomical regions including: gastric contents, external nares (blowhole wall swabs) and exhaled blow (i.e. blow plate), skin, anal, and genital opening rayon swabs (Puritan Medical Products). The blowhole swab was inserted into the blowhole/nares and contacted the wall of the nares. Exhaled respiratory material was also collected in a sterile petri dish and then the internal surface of the dish was swabbed and placed in a sterile cryovial. Gastric fluid was obtained with an equine stomach tube inserted into the fundic section of the stomach. The fecal swab was obtained by gently inserting the swab into the anal opening. Skin swabs were collected by swabbing the axillary region in an attempt to decrease the contamination by people handling the dolphins. The genital swab was collected by inserting a swab into the vaginal or preputial opening. All swabs tips were placed into sterile Nalgene cryovials and the plastic stems cut to the level of the cryovial top with a pair of scissors to allow closure of the vials. Nalgene cryovials were then immediately placed on dry ice until they could be transferred to a -80 °C freezer for storage.
Seven water samples (500 ml) were collected at each site (Table S1) in a sterile 500 ml glass reagent bottle by submerging the sampling bottle 12 inches below the water surface, removing the cap, and recapping underwater once filled. At the processing boat, an extension set was opened under sterile conditions and placed in the sampling bottle. By using a 3-way stopcock and a 60 ml syringe, aliquots were repeatedly aspirated and pushed through a Sterivex™ filter unit (Millipore catalog# SVGPL10RC). Once this procedure was completed, the filter was sterilely placed in a whirl-pack bag, placed on dry ice, and frozen until being transferred to a -80 °C freezer for storage.
2.2. Bacterial DNA extraction and amplification
Thawed swabs and gastric fluid samples (200 μl per sample) were added to 2 ml lysis bead tubes (0.5 mm high impact zirconium) from Benchmark Scientific, Inc. ATL buffer was added (360 μl for swabs and 180 μl for gastric fluid samples) to the bead tubes and processed in a TissueLyser II (Qiagen) for 1 min at a frequency of 30.0. Bead tubes were then briefly centrifuged before adding Proteinase K (40 μl to tubes with swabs and 20 μl to tubes with gastric fluid), vortexed, briefly centrifuged, and incubated at 56 °C for 15 min. Following the protein digestion, 200 μl of the sample was transferred to a sterile 2 ml microcentrifuge tube and the remainder of the DNA extraction process was automated in a QIAcube® (Qiagen) using the QIAamp Mini body fluid setting. Sterivex™ filters were thawed and DNA extracted using a PowerWater® Sterivex DNA Isolation Kit as specified by the manufacturer (MoBio laboratories). The concentrations of the resulting purified DNA samples were quantified using a Qubit 3.0 fluorometer (Thermo Fischer Scientific) prior to storage in a -80 °C freezer.
2.3. 16S library generation and sequencing
The 16S metagenomic sequencing libraries were prepared for sequencing on a MiSeq sequencer (Illumina) according to the manufacturer's instructions. Briefly, amplicon PCR primers targeting approximately ~460 bp of the variable V3–V4 region of the 16S rRNA gene were employed. The forward primer sequence is 5′- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGG
GNGGCWGCAG - 3′ and the reverse primer sequence is 5′ – GTCTCGTGGGCTCGGAGATGTG
TATAAGAGACAGGACTACHVGGGTATCTAATCC - 3′. The 35 μl PCR mastermix consisted of 0.175 μl of Platinum Taq DNA Polymerase (Invitrogen), 3.5 μl of 10x PCR buffer, 1.4 μl of 50 mM MgCl2, 0.7 μl of 10 mM dNTPs, 1.75 μl of 1 μM forward and reverse primers, 20.5 μl of molecular grade water, and 5.25 μl of DNA at a concentration of 5 ng/μl. The PCR reactions included an initial denaturation of 3 min at 95 °C; followed by 30 cycles of denaturation at 95 °C for 30 s; annealing at 55 °C for 30 s; extension at 72 °C for 30 s; and a final elongation step at 72 °C for 5 min. Products were run on a 1.5% agarose gel to confirm amplification. The PCR amplicons were purified from primers and primer dimers using AMPure XP beads (Beckman) as instructed by the manufacturer.
Each amplicon received dual indices and Illumina sequencing adapters using the Nextera XT Index Kit. The 50 μl PCR mastermix consisted of 0.25 μl of Platinum Taq DNA Polymerase (Invitrogen), 5.0 μl of 10x PCR buffer, 2.0 μl of 50 mM MgCl2, 1.0 μl of 10 mM dNTPs, 5.0 μl of forward and reverse indexing primers, 26.75 μl of molecular grade water, and 5.0 μl of the purified amplicon. The PCR reactions included an initial denaturation of 3 min at 95 °C; followed by 8 cycles of denaturation at 95 °C for 30 s; annealing at 55 °C for 30 s; extension at 72 °C for 30 s; and a final elongation step at 72 °C for 5 min. Following amplification, the indexed amplicons were again purified from primers and primer dimer using AMPure XP beads. The resulting indexed amplicon concentrations were measured using a Qubit 3.0 fluorometer (Thermo Fisher Scientific), normalized, and pooled. The equimolar pool was then sequenced on an Illumina MiSeq platform (San Diego, CA, USA) using a 600-cycle v3 MiSeq Reagent Kit.
2.4. Microbiome data analysis
Reads were classified using the Ribosomal Database Project (RDP) classifier version 2.2 (Wang et al., 2007). Shannon and Inverse Simpson indices were calculated using the diversity function of vegan (Oksanen et al., 2007) and compared using a pairwise t-test. Richness was calculated by rarefying to a sample size representing the sample with the fewest reads.
Raw OTU counts were normalized and log10 transformed using the following formula:
where RC is the read count for a particular OTU in a particular sample, n is the total number of reads in that sample, the sum of x is the total number of reads in all samples and N is the total number of samples (McCafferty et al., 2013).
The Principle Coordinate Analysis (PCoA) was generated from the Bray-Curtis distance (Bray and Curtis, 1957) of the normalized and log10 transformed counts. Analysis of similarities (ANOSIM) was performed comparing each pair of sites, and then the p-values were corrected for multiple hypothesis testing using the method of Benjamini & Hochberg (Benjamini and Hochberg, 1995; Oksanen et al., 2007).
Linear models were used to identify taxa significantly associated with each of our variables. Water samples were not included in these analyses. Taxa present in less than one quarter of the samples were also removed. To identify taxa associated with particular body sites, we used the following model:
| relAbun ~ bodySite + ϵ |
where relAbun is the log10 normalized abundance of a particular taxa and bodySite indicates the site the sample was taken from. For all other variables, the p-values for that variable were taken from the following models:
| relAbun ~ bodySite + animalID + ϵ |
| relAbun ~ bodySite + gender + ϵ |
| relAbun ~ bodySite + age + ϵ |
| relAbun ~ bodySite + healthStatus + ϵ |
| relAbun ~ bodySite + lactationStatus + ϵ |
| relAbun ~ bodySite + motherCalfStatus + ϵ |
where animalID indicates which dolphin was sampled, gender represents male or female, age represents whether the sample is from an adult or calf, and healthStatus represents whether the sample was from a healthy or unhealthy dolphin. lactationStatus represents the lactation status of the dolphin and was divided into lactating animals, non-lactating females, and the rest of the community. For motherCalfStatus (i.e., relatedness), each mother-calf pair was represented by a group, and all other dolphins were grouped together. All models were fit using the lm function in R (R Core, 2013). The p-values were corrected for multiple hypothesis testing using the method of Benjamini & Hochberg (Benjamini and Hochberg, 1995).
3. Results
3.1. Summary of dolphin and seawater samples used in downstream analyses
Five calves and 9 adults (including one sub-adult) ranging from 2-31 years of age were sampled (Table S1). Four of the five calves were captured as part of mother-calf pairs. These four mothers were lactating as well as another female sampled found without a calf. Bacterial specimens processed for sequencing included 6 body sites (blowhole, blowhole plate, gastric, fecal, skin, and genital) from each of 14 dolphins (9 females and 5 males) and 7 seawater samples from each site where dolphins were captured, sampled, and released. Sixty-six samples were loaded onto the Illumina MiSeq platform resulting in 21,708,338 reads and 17,510,220 reads passing filter (PF) with fairly even coverage across indexes (ranging from 0.4175% to 2.01% PF reads) (Fig. S1). The skin specimen from dolphin TT15022 and blowhole plate specimen from dolphin TT15028 failed to generate significant reads (0.0050% and 0.0099% PF reads, respectively).
3.2. Principal coordinates analysis (PCoA) and ANOSIM
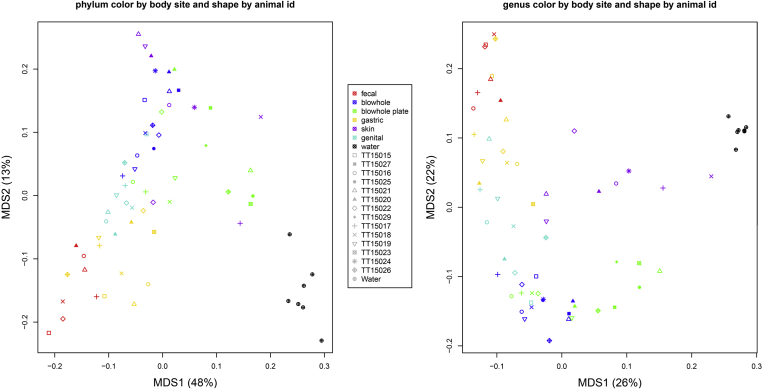
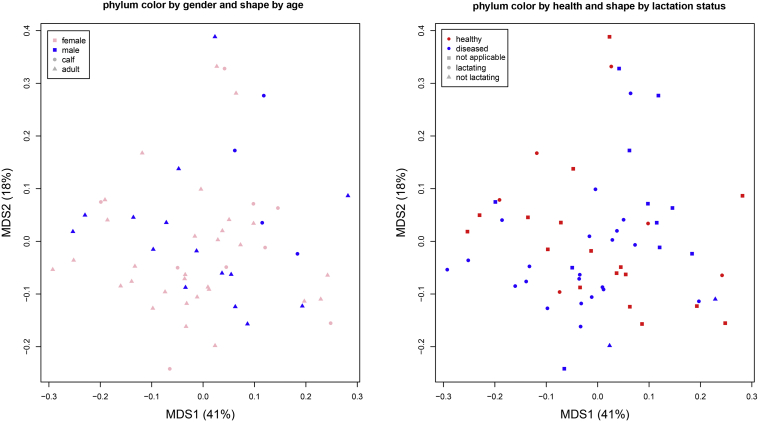
The Principal Coordinates Analysis (PCoA) at all taxonomic levels revealed that specimens from the same body site clustered together (Figure 4). However, seawater samples clustered the most tightly together and were clearly separate from dolphin specimens. At the phylum level, there was no clustering by gender, age, health, lactation status, or relatedness (Figure 5). At the genus level, the greatest similarity was observed between sites of the gastrointestinal tract (gastric and fecal; R = 0.49; p = 0.001 after Benjamini-Hochberg correction for multiple hypothesis testing) and respiratory tract (blowhole and blowhole plate; R = 0.20, p = 0.005). Water samples were the most dissimilar to body sites with significant R values (p ≤ 0.05) varying from 0.70 to 1.0 (Table S2).
Figure 4.
Principal Coordinate Analysis of bacterial communities at the phylum (left) and genus (right) levels from different dolphin body sites and seawater samples. Each color represents a different body site. Shapes represent individual animals.
Figure 5.
Principal Coordinate Analysis of bacterial communities at the phylum level for dolphins from different genders and age classes (left), and health status and lactation status (right). Each data point represents a bacterial community from a specific bottlenose dolphin.
3.3. Diversity boxplots and composition and relative abundance barplots
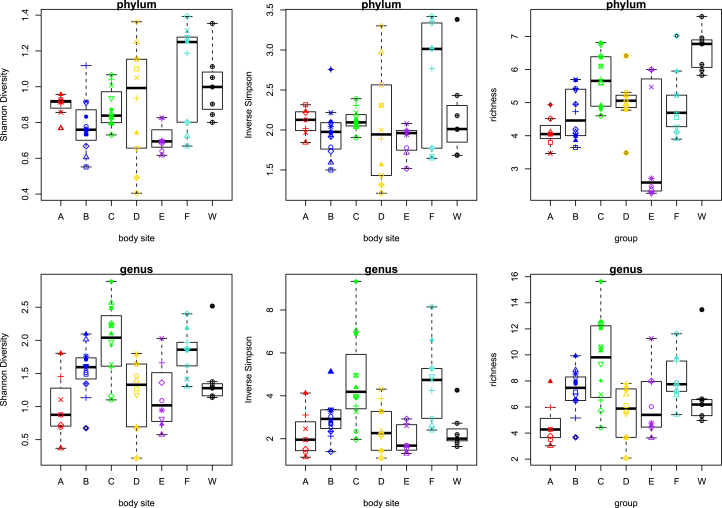
The genital specimens had among the highest microbial diversity at both the phylum and genus levels (Figure 1). Seawater specimens showed the highest bacterial richness at the phylum level (Figure 1). Skin and fecal specimens displayed among the lowest diversity and richness at both the phylum and genus levels (Figure 1). Blowhole plate consistently displayed higher diversity and richness as compared to blowhole specimens at both the phylum and genus levels. Blowhole plate specimens also displayed the highest diversity (Shannon diversity only) and richness at the genus level (Figure 1). The stomach samples consistently displayed moderate microbial diversity and richness as compared to the other specimen types (Figure 1).
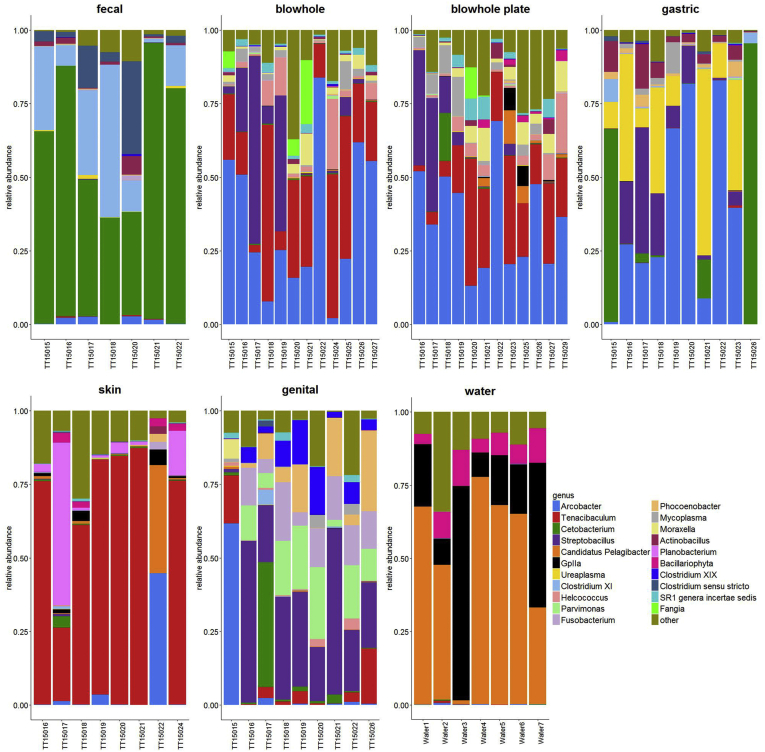
Figure 1.
Shannon diversity, Inverse Simpson diversity, and Richness analyses at the phylum and genus levels for bottlenose dolphin body sites versus seawater samples. Each boxplot represents a different body site differentiated by colors and letters. Red is fecal (A), dark blue is blowhole (B), green is blowhole plate (C), yellow is gastric (D), purple is skin (E), light blue is genital (F), and black is water (W). Different shapes represent different individuals.
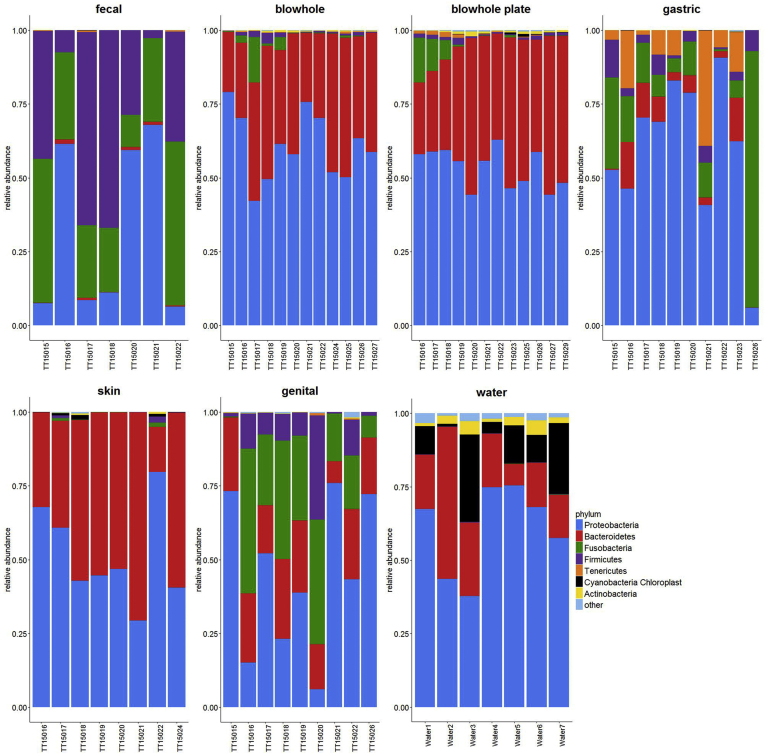
The bacterial communities identified from the sampled bottlenose dolphins and water specimens represent a total of 16 different bacterial phyla and 70 genera with an average relative abundance ≥0.0005 (data not shown). The bacterial phyla with the highest relative abundance identified from the bottlenose dolphins were Proteobacteria, Bacteroidetes, Fusobacteria, Firmicutes, and Tenericutes. The gastric specimens had a high relative abundance of Proteobacteria (54.6%), followed by Fusobacteria (18.5%), Bacteroidetes (17.8%), Tenericutes (4.7%), and Firmicutes (4.1%) (Figure 2). The genera with the highest relative abundance in the gastric specimens were Arcobacter (28.0%), Cetobacterium (21.1%), Tenacibaculum (14.2%), and Ureplasma (10.3%) (Figure 3). The bacterial phyla with the highest relative abundance in the hindgut (fecal) specimens were Firmicutes (35.7%), followed by Proteobacteria (31.8%), and Fusobacteria (31.6%) (Figure 2). Bacteria within the genera Cetobacterium (70.9%), Clostridium XI (15.3%), and Clostridium sensu stricto (4.2%) had the highest relative abundance in the hindgut specimens (Figure 3). Lower abundance genera in the hindgut specimens included Hyrogenimonas, Arcobacter, Clostrdium XIVb, and Actinobacillus. Both blowhole swabs (BS) and blow plate (BP) specimens generated similar results regarding bacterial composition and abundance at both the phylum and genus levels (Figures 2 and 3). Proteobacteria were highest in relative abundance in both the blowhole swab (BS: 57.7%) and blow plate (BP: 46.5%) specimens, followed by Bacteroidetes (BS: 38.4% and BP: 41.8%), with a lower relative abundance of Fusobacteria (BS: 2.3% and BP: 8.4%) and Firmicutes (BS: 1.1% and BP: 2.2%) (Figure 2). At the genus level, Tenacibaculum was the dominant genus in both blowhole swabs (33.6%) and blow plate (27.5%) specimens, followed by Arcobacter (BS: 26.1% and BP: 20.7%) and Streptobacillus (BS: 9.4% and 10.1%) (Figure 3). The respiratory bacterial communities also included genera such as Mycoplasma, Moraxella, and Actinobacillus at a lower relative abundance. Specimens from the blow plate included several bacterial genera observed in seawater specimens (albeit in lower relative abundance – see below). The dominant bacterial phyla within the skin were Proteobacteria (56.8%) and Bacteroidetes (37.9%) (Figure 2). At the genus level, Tenacibaculum (45.2%) was dominant in the skin specimens (Figure 3). The dominant bacterial phyla in the genital specimens were Proteobacteria (48.9%), followed by Bacteroidetes (23.4%), Fusobacteria (18.9%), and Firmicutes (8.1%) (Figure 2). The bacterial genera of high relative abundance in the genital specimens included: Streptobacillus (18.5%), Arcobacter (13.1%), Tenacibaculum (11.4%), Clostridium XIX (8.7%), Parvimonas (7.2%), Cetobacterium (6.9%), and Fusobacterium (5.5%) (Figure 3). The seawater specimens were dominated by bacteria within the phyla Proteobacteria (60.6%), followed by Bacteroidetes (23.4%), and Cyanobacteria/Chloroplast (11.8%) (Figure 2). The bacterial genera with the highest relative abundance in seawater were Candidatus Pelagibacter (51.6%), followed by GpIIa (cyanobacteria) (25.2%), and diatoms of the genus Bacillariophyta (9.8%) (Figure 3).
Figure 2.
Composition and relative abundance of phyla found in bottlenose dolphin body sites versus seawater samples. Each plot represents a different body site with bars representing individual dolphin or water specimens within each plot. The seven colors represent the different bacterial phyla found in the specimens. The average relative abundance of each phyla across specimens was calculated and those with an average relative abundance ≤0.0005 were filtered out.
Figure 3.
Composition and relative abundance of genera found in bottlenose dolphin body sites versus seawater samples. Each plot represents a different body site with bars representing individual dolphin or water specimens within each plot. The 21 colors represent the different bacterial genera found in the specimens. The average relative abundance of each genera across specimens was calculated and those with an average relative abundance ≤0.0005 were filtered out.
3.4. Mixed linear model tests
Linear models were generated to determine statistically significant (adjusted p ≤ 0.05) associations between the relative abundance of bacterial genera, body site, and physiological variables. A total of 94 significant associations were observed involving all body sites and all physiological variables except health status.
Blow plate specimens had the highest relative abundance compared to other body sites for the following genera: Moraxella, Helococcus, SR1_genera_incertae_sedis, Bergeyella, Lutibacter, Mycoplasma, Soonwooa, Tannerella, Bacteriovorax, Arcobacter, Anaerovorax, Flavobacterium, Ornithobacterium, Parcubacteria_genera_incertae_sedis, Saccharibacteria_genera_incertae_sedis, Pseudomonas, Thioreductor, Aureispira, and Saccharofermentans (Table S3). Blowhole specimens had the highest relative abundance for the following genera: Riemerella, Corallibacter, Peptostreptococcus, Aureitalea, Coenonia, Olleya, and Natranaerovirga (Table S3). Genital specimens had the highest relative abundance compared to other body sites for the following genera: Parvimonas, Phocoenobacter, Clostridium XIX, Gemella, Campylobacter, Porphyromonas, Streptobacillus, Fusobacterium, Lachnoanaerobaculum, Treponema, Peptoniphilus, and Guggenheimella (Table S3). Skin specimens had the highest relative abundance compared to other body sites for the following genera: Planobacterium, Tenacibaculum, Enhydrobacter, Bacillariophyta, GpIIa, Ilumatobacter, Psychrobacter, Candidatus Pelagibacter, Lishizhenia, and Pseudoalteromonas (Table S3). Fecal specimens had the highest relative abundance compared to other body sites for the following genera: Anaerobacter, Sporacetigenium, Clostridium XI, Clostridium sensu stricto, Photobacterium, Cetobacterium, and Psychrilyobacter (Table S3). Gastric specimens had the highest relative abundance compared to other body sites for the following genera: Ureaplasma, Paralactobacillus, Helicobacter, Actinobacillus, Sulfurovum, and Lucibacterium (Table S3).
Males displayed a significantly higher relative abundance of Lutibacter and Anaerovorax than females (Table S3). Calves showed a higher relative abundance of Moraxella, Helcococcus, Riemerella, Bergeyella, Tenacibaculum, Lutibacter, Enhydrobacter, Soonwooa, Bacteriovorax, Ornithobacterium, Coenonia, and Pseudoalteromonas than adults (Table S3). Adults showed a higher relative abundance of Clostridium XI and Sulfurovum than calves (Table S3). Mother-calf pairs had a higher relative abundance of Coenonia (TT5015-TT15027) and Fusobacterium (TT5022-TT15029) as compared to weaned animals (Table S3). Lactating females had a higher relative abundance of Anaerobacter and Clostridium XI compared to the non-lactating female and the rest of the community (Table S3). The non-lactating female (TT15023) had a higher abundance of Moraxella, SR1_genera_incertae_sedis, GpIIa, Soonwooa, Tannerella, Flavobacterium, Psychrilyobacter, and Sulfurimonas than the lactating females and the rest of the community (Table S3). The rest of the community had a higher relative abundance of Riemerella, Tenacibaculum, Helococcus, Lutibacter, and Enhydrobacter as compared to the non-lactating and lactating females (Table S3). The relative abundances of the following genera were the highest in the seawater samples: Ilumatobacter (p = 9.53e−13), GpIIa (p = 9.6e−13), Cryptomonadaceae (p = 1.31e−12), Bacillariophyta (p = 6.73e−12), Candidatus Pelagibacter (p = 7.49e−11), Lishizhenia (p = 1.89e−08), Sulfurimonas (p = 2.9e−07), Actibacter (p = 1.94e−06), Desulfosarcina (p = 3.579e−05), Lucibacterium (p = 0.002), and Vibrio (p = 0.005) (data not shown).
4. Discussion
Herein, we reported diverse cutaneous, respiratory, gastrointestinal, and genital microbial communities (16 different bacterial phyla and 70 genera) from a well-studied community of free-ranging bottlenose dolphins and their surrounding aquatic habitat. The PCoA and barplots revealed the uniqueness of the microbial communities identified from the different bottlenose dolphin body sites as has been reported in other mammals, including humans (Peterson et al., 2009) and bottlenose dolphins (Bik et al., 2016). The ANOSIM revealed considerable overlap among specimens collected from the same body system including the respiratory tract (blow and blow plate specimens) as previously reported from samples taken from a managed population of bottlenose dolphins in San Diego Bay, CA (Bik et al., 2016). In agreement with previous studies involving managed (Chiarello et al., 2017) or managed and free-ranging bottlenose dolphins (Bik et al., 2016), the bottlenose dolphin microbiome produced in our study was clearly distinct from the microbiome of their surrounding aquatic habitat.
Proteobacteria, Firmicutes, and Fusobacteria were dominant in the bottlenose dolphin fecal samples as described in previous studies involving bottlenose dolphins (Bik et al., 2016; Soverini et al., 2016). The predominance of Firmicutes has also been reported in the hindgut of fermenting sirenians (Nielsen et al., 2013; Merson et al., 2013). Firmicutes are ubiquitous in the gastrointestinal tracts of mammals due to their ability to harvest energy and nutrients from ingesta (Vos et al., 2011). Cetobacterium, in the phylum Fusobacteria, was found to be in highest relative abundance in the bottlenose dolphin hindgut specimens as previously described (Bik et al., 2016). Cetobacterium spp. are well-known to produce vitamin B12 and butyrate which may contribute to dolphin nutritional health (Bennett and Eley, 1993). Fecal specimens in our study had among the lowest bacterial diversity and richness at the genus level as previously reported (Bik et al., 2016). However, these results may be an artifact of using rectal swabs versus fresh feces to measure microbial communities, as collecting fecal material provides a larger sample mass than swabbing the rectal area (Mealey, 2016).
Proteobacteria, Fusobacteria, Tenericutes, Firmicutes, and Bacteriodetes dominated in the bottlenose dolphin gastric specimens. In contrast, Fusobacteria and Firmicutes were not reported as dominant bacterial phyla in the managed bottlenose dolphin population studied by Bik et al. (2016). Ureaplasma, in the phylum Tenericutes, was in high abundance in the gastric specimens as previously reported by Bik et al. (2016). The genus Arcobacter, within the phylum Proteobacteria, was a dominant group in our gastric specimens but not in the managed bottlenose dolphins studied by Bik et al. (2016). Although arcobacters are primarily animal- and human-associated bacteria, several members of this genus have been shown to be abundantly present in certain environmental niches including sewage, oil, and saline environments (Vandamme et al., 2015).
Our data support previous claims that ‘blow’ samples closely approximate the microbiome of blowhole swabs, thereby eliminating the need for the more invasive and potentially dangerous blowhole swabbing procedures (Lima et al., 2012). Additionally, our study demonstrated that blow plate specimens exhibited slightly higher bacterial diversity and richness than blowhole swabs in accordance with a previous study (Bik et al., 2016). However, we observed a significant number of seawater microbiota contaminants in the blowhole plate specimens including the three dominant genera observed in the seawater specimens: Candidatus Pelagibacter, GpIIa (Cyanobacteria), and diatoms of the genus Bacillariophyta. We hypothesize that seawater on the surface of the dolphin near the blowhole may have been blown on to the blowhole plate during exhalation, altering the observed bacterial communities. Thus, care should be taken when gathering and interpreting the microbiomes of blowhole plate specimens.
The dominant bacterial phyla identified from the respiratory tract specimens were Proteobacteria and Bacteroidetes, which is consistent with the findings of previous studies involving managed and free-ranging bottlenose dolphins using culture dependent (Johnson et al., 2009) and culture independent high-throughput next-generation sequencing approaches (Lima et al., 2012; Bik et al., 2016). However, our study did not find the order Cardibacteriales as an abundant bacterial taxon in contrast to the aforementioned studies (data not shown). At the genus level, Arcobacter, Tenacibaculum, and Helococcus dominated in both the respiratory blowhole and blowhole plate specimens. Arcobacter and Helococcus were previously determined to represent part of the core genera associated with managed common, indo-pacific (Tursiops aduncus), and hybrid bottlenose dolphins (Lima et al., 2012). Using culture dependent methods, Johnson et al. (2009) reported that Tenacibaculum was abundant in the upper respiratory samples of free-ranging bottlenose dolphins from the Indian River Lagoon, FL and Charleston, SC. We also observed a significant abundance of Peptostreptococcus in the blowhole and Mycoplasma in blowhole plate specimens. Members of the genus Peptostreptococcus are commensal bacteria associated with a range of mammalian hosts. However, following traumatic injury these bacteria have been shown to be associated with abscesses and generalized necrotizing soft tissue infections in humans (Könönen et al., 2007). Peptostreptococcus have also been isolated from the peritoneal cavity and “blow” of stranded cetaceans (Bogomolni et al., 2008) and dugongs that succumbed to pneumonia (Nielsen et al., 2013). The significance of Mycoplasma found in the blowhole plate specimens is unclear and deserves further investigation as they have been shown to colonize fish, reptiles, and other aquatic and terrestrial mammals and cause opportunistic respiratory, reproductive, or joint infections (Clippinger et al., 2000; Waltzek et al., 2012; Brown et al., 2015).
The skin associated bacterial community was dominated by Proteobacteria and Bacteroidetes. The high abundance of Proteobacteria (Gammaproteobacteria) was also reported by other studies on managed and wild-captured bottlenose dolphins (Chiarello et al., 2017; Russo et al., 2018). The phylum Bacteroidetes included a high proportion of Flavobacteriaceae, particularly the genus Tenacibaculum. Members of the genus Tenacibaculum have been reported as one of the most dominant skin microbiota of a mysticete, the humpback whale (Megaptera noveangliae) (Apprill et al., 2011, 2014). In this study, we also observed a higher abundance of Planobacterium, Enhydrobacter, Ilumatobacter, and Psychrobacter in skin compared to other body sites. Psychrobacter was previously found to be the most abundant bacterial genus on the skin of managed killer whales (Orca orcinus) and bottlenose dolphins (Chiarello et al., 2017). Moreover, Psychrobacter was found to be one of the 20 most abundant genera recovered from the skin of offshore bottlenose dolphins in California (Russo et al., 2018).
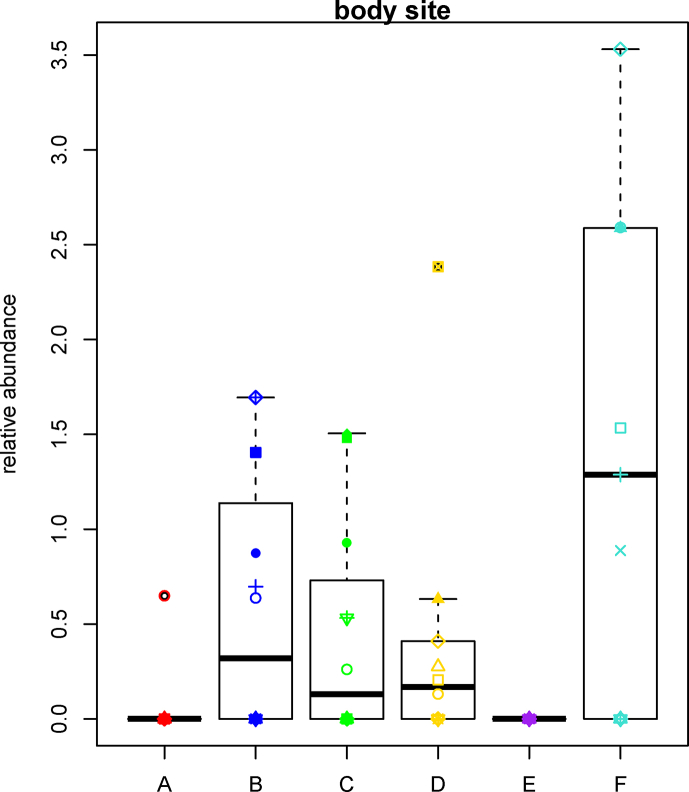
This study is the first to characterize the genital microbiome of bottlenose dolphins which was found to be composed of a diverse bacterial community. The dominant bacterial phyla present in the genital specimens were Proteobacteria, Bacteroidetes, Fusobacteria, and Firmicutes. Bacterial communities at the genus level were unique and were dominated by the following genera: Streptobacillus, Parvimonas, Fusobacterium Phocoenobacter, and Clostridium XIX. The microbiome of a genital specimen from a stranded immature female striped dolphin (Stenella coeruleoalba) revealed Fusobacterium was the most abundant genus, followed by Porphyromonas, Actinobacillus, Parvimonas, Ureaplasma, and Phocoenobacter (Godoy-Vitorino et al., 2017). Phocoenobacter uteri was previously described from the uterus of a harbor porpoise (Phocoena phocoena) that stranded on the Scottish coastline (Foster et al., 2000). Although not among the 20 most abundant genera, Treponema displayed higher relative abundance in bottlenose dolphin genital specimens when compared to other body sites (Figure 6). To our knowledge, Treponema has not previously been reported from marine mammal genital samples by culture-dependent techniques or culture independent high-throughput next-generation approaches. The genus Treponema includes clinically relevant spirochaetes causing syphilis in humans (T. pallidum) (Radolf, 1996), a similar reproductive disease in rabbits (T. paraluis-cuniculi) (Giacani et al., 2004), and dysentery in swine (T. hyodysenteriae) (Weber and Earley, 1991). Further work is needed to determine whether Treponema in dolphins includes one or more novel species and whether any are associated with disease. Our study underscores the need to further explore the reproductive microbiota of bottlenose dolphins and other cetaceans.
Figure 6.
Comparison of the relative abundance of the genus Treponema among all the wild bottlenose dolphin body sites sampled. Each boxplot represents a different body site differentiated by colors and letters. Red is fecal (A), dark blue is blowhole (B), green is blowhole plate (C), yellow is gastric (D), purple is skin (E), and light blue is genital (F). Different shapes represent different individuals. pAdjusted = 0.00476.
The results of our mixed linear model tests indicated some bacterial genera to be significantly different among bottlenose dolphin body sites, relatedness, age class, gender, and lactation status. However, the principal coordinate analyses failed to demonstrate bacterial community clustering with the exception of body sites. Bik et al. (2016) reported body sites defined specific bacterial communities in managed/free-ranging bottlenose dolphins and managed California sea lions (Zalophus californianus). Similiar to our study, they found no other significant patterns associated with age, gender, or animal location.
The main goal of this study was to conduct an in-depth exploration of the bacterial microbiome associated with different body sites of free-ranging bottlenose dolphins and their aquatic ecosystem. We reported a diverse set of bacterial communities present in bottlenose dolphins, which were unique to their surrounding aquatic habitat. Body site was the variable shown to be the major driver of bacterial abundance, diversity, and richness. Our finding supports the recognized notion that different body sites have their own physiochemical conditions which exert selective pressures on the microbiota, and play important roles in shaping the microbiota across different body systems. Our results were in close agreement with other cetacean microbiome studies while this study is the first to explore the bottlenose dolphin genital microbiome. The core bacterial community identified in this study might be informative for future health monitoring of bottlenose dolphins. Finally, we detected several bacterial genera known to be primary or opportunistic pathogens; however, their potential role in disease of bottlenose dolphins remains to be determined.
Declarations
Author contribution statement
María José Robles-Malagamba: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Michael T. Walsh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Mohammad Shamim Ahasan: Analyzed and interpreted the data; Wrote the paper.
Patrick Thompson: Performed the experiments.
Randall S. Wells: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Christian Jobin, Anthony Fodor, Kathryn Winglee: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Thomas B. Waltzek: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Clearwater Marine Aquarium, the University of Florida's Aquatic Animal Health Program, and the National Commission for Science and Technology in Mexico. Dolphin Quest, Inc., funded the sample collection in Sarasota Bay.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to thank the members of the 2015 Sarasota Bay Dolphin Health Assessment team for sample collection and members of the University of Florida's Wildlife and Aquatic Veterinary Disease Laboratory for technical assistance.
Contributor Information
María José Robles-Malagamba, Email: mj_malagamba@hotmail.com.
Thomas B. Waltzek, Email: tbwaltzek@ufl.edu.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ahasan M.S., Waltzek T.B., Huerlimann R., Ariel E. Fecal bacterial communities of wild-captured and stranded green turtles (Chelonia mydas) on the Great Barrier Reef. FEMS Microbiol. Ecol. 2017;93(12) doi: 10.1093/femsec/fix139. [DOI] [PubMed] [Google Scholar]
- Apprill A., Mooney T.A., Lyman E., Stimpert A.K., Rappé M.S. Humpback whales harbour a combination of specific and variable skin bacteria. Environ. Microbiol. Rep. 2011;3(2):223–232. doi: 10.1111/j.1758-2229.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- Apprill A., Robbins J., Eren A.M., Pack A.A., Reveillaud J., Mattila D., Moore M., Niemeyer M., Moore K.M., Mincer T.J. Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PloS One. 2014;9(3) doi: 10.1371/journal.pone.0090785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzi G., Agazzi S., Bonizzoni S., Costa M., Azzellino A. Dolphins in a bottle: abundance, residency patterns and conservation of bottlenose dolphins Tursiops truncatus in the semi-closed eutrophic Amvrakikos Gulf, Greece. Aquat. Conserv. Mar. Freshw. Ecosyst. 2008;18(2):130–146. [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995:289–300. [Google Scholar]
- Bennett K., Eley A. Fusobacteria: new taxonomy and related diseases. J. Med. Microbiol. 1993;39(4):246–254. doi: 10.1099/00222615-39-4-246. [DOI] [PubMed] [Google Scholar]
- Bik E.M., Costello E.K., Switzer A.D., Callahan B.J., Holmes S.P., Wells R.S., Carlin K.P., Jensen E.D., Venn-Watson S., Relman D.A. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 2016;7:10516. doi: 10.1038/ncomms10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M.J., Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449(7164):843. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- Bogomolni A.L., Gast R.J., Ellis J.C., Dennett M., Pugliares K.R., Lentell B.J., Moore M.J. Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis. Aquat. Org. 2008;81(1):13. doi: 10.3354/dao01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart G. Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 2011;48(3):676–690. doi: 10.1177/0300985810388525. [DOI] [PubMed] [Google Scholar]
- Bray J.R., Curtis T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957;27(4):325–349. [Google Scholar]
- Brown D.R., May M., Bradbury J.M., Balish M.F., Calcutt M.J., Glass J.I., Tasker S., Messick J.B., Johansson K., Neimark H. Mycoplasma. In: Whitman FR W.B., Kämpfer P., Trujillo M., Chun J., DeVos P., Hedlund B., Dedysh S., editors. Bergey's Manual of Systematics of Archaea and Bacteria. 2015. [Google Scholar]
- Buck J.D., Wells R.S., Rhinehart H.L., Hansen L.J. Aerobic microorganisms associated with free-ranging bottlenose dolphins in coastal Gulf of Mexico and Atlantic Ocean waters. J. Wildl. Dis. 2006;42(3):536–544. doi: 10.7589/0090-3558-42.3.536. [DOI] [PubMed] [Google Scholar]
- Chiarello M., Villéger S., Bouvier C., Auguet J., Bouvier T. Captive bottlenose dolphins and killer whales harbor a species-specific skin microbiota that varies among individuals. Sci. Rep. 2017;7(1):15269. doi: 10.1038/s41598-017-15220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choat J., Clements K. Vertebrate herbivores in marine and terrestrial environments: a nutritional ecology perspective. Annu. Rev. Ecol. Systemat. 1998;29(1):375–403. [Google Scholar]
- Clippinger T.L., Avery Bennett R., Johnson C.M., Vliet K.A., Deem S.L., Orós J., Jacobson E.R., Schumacher I.M., Brown D.R., Brown M.B. Morbidity and mortality associated with a new mycoplasma species from captive American alligators (Alligator mississippiensis) J. Zoo Wildl. Med. 2000;31(3):303–314. doi: 10.1638/1042-7260(2000)031[0303:MAMAWA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J., Doak D., Springer A., Williams T. Causes and consequences of marine mammal population declines in southwest Alaska: a food-web perspective. Phil. Trans. Biol. Sci. 2009;364(1524):1647–1658. doi: 10.1098/rstb.2008.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster G., Ross H.M., Malnick H., Willems A., Hutson R.A., Reid R.J., Collins M.D. Phocoenobacter uteri gen. nov., sp. nov., a new member of the family Pasteurellaceae Pohl (1979) 1981 isolated from a harbour porpoise (Phocoena phocoena) Int. J. Syst. Evol. Microbiol. 2000;50(1):135–139. doi: 10.1099/00207713-50-1-135. [DOI] [PubMed] [Google Scholar]
- Giacani L., Sun E.S., Hevner K., Molini B.J., Van Voorhis W.C., Lukehart S.A., Centurion-Lara A. Tpr homologs in Treponema paraluiscuniculi Cuniculi A strain. Infect. Immun. 2004;72(11):6561–6576. doi: 10.1128/IAI.72.11.6561-6576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R., Pop M., DeBoy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Vitorino F. The microbiome of a striped dolphin (Stenella coeruleoalba) stranded in Portugal. Res. Microbiol. 2017;168(1):85–93. doi: 10.1016/j.resmic.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., Bouffard G.G., Blakesley R.W., Murray P.R., Green E.D. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn A.A., Scott M.D., Wells R.S., Sweeney J.C., Irvine A.B. Growth layers in teeth from known-age, free-ranging bottlenose dolphins. Mar. Mamm. Sci. 1989;5(4):315–342. [Google Scholar]
- Holden A. 1970. Monitoring Organochlorine Contamination of the marine Environment by the Analysis of Residues in Seals. [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E.B., Britt J.O.J., Matsumoto G.K., Itahara R., Nagana C.N. Bacterial diseases. In: Howard E.B., editor. Vol. 1. CRC Press; Boca Raton: 1983. pp. 69–118. (Pathobiology of marine Mammal Diseases). [Google Scholar]
- Johnson W.R., Torralba M., Fair P.A., Bossart G.D., Nelson K.E., Morris P.J. Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts. Environ. Microbiol. Rep. 2009;1(6):555–562. doi: 10.1111/j.1758-2229.2009.00080.x. [DOI] [PubMed] [Google Scholar]
- Kim H.B., Isaacson R.E. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015;177(3-4):242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Könönen E., Bryk A., Niemi P., Kanervo-Nordström A. Antimicrobial susceptibilities of Peptostreptococcus anaerobius and the newly described Peptostreptococcus stomatis isolated from various human sources. Antimicrob. Agents Chemother. 2007;51(6):2205–2207. doi: 10.1128/AAC.00056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.G., Milani C., de Giori G.S., Sesma F., Van Sinderen D., Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 2013;24(2):160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Lima N., Rogers T., Acevedo-Whitehouse K., Brown M.V. Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system. Environ. Microbiol. Rep. 2012;4(1):89–96. doi: 10.1111/j.1758-2229.2011.00306.x. [DOI] [PubMed] [Google Scholar]
- McCafferty J., Mühlbauer M., Gharaibeh R.Z., Arthur J.C., Perez-Chanona E., Sha W., Jobin C., Fodor A.A. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7(11):2116. doi: 10.1038/ismej.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey R.H. Elsevier Health Sciences; 2016. New Perspectives in Infectious Diseases, an Issue of Veterinary Clinics of North America: Equine Practice, E-Book. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merson S.D., Ouwerkerk D., Gulino L.M., Klieve A., Bonde R.K., Burgess E.A., Lanyon J.M. Variation in the hindgut microbial communities of the Florida manatee, Trichechus manatus latirostris over winter in Crystal River, Florida. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Ecol. 2013;87(3):601–615. doi: 10.1111/1574-6941.12248. [DOI] [PubMed] [Google Scholar]
- Nielsen K.A., Owen H.C., Mills P.C., Flint M., Gibson J.S. Bacteria isolated from dugongs (Dugong dugon) sub-mitted for postmortem examination in Queensland, Australia, 2000–2011. J. Zoo Wildl. Med. 2013;44(1):35–41. doi: 10.1638/1042-7260-44.1.35. [DOI] [PubMed] [Google Scholar]
- Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M.H.H., Oksanen M.J., Suggests M. The vegan package. Community Ecol. Package. 2007;10:631–637. [Google Scholar]
- Ottman N., Smidt H., De Vos W.M., Belzer C. The function of our microbiota: who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J., Garges S., Giovanni M., McInnes P., Wang L., Schloss J.A., Bonazzi V., McEwen J.E., Wetterstrand K.A., Deal C., Baker C.C., Di Francesco V., Howcroft T.K., Karp R.W., Lunsford R.D., Wellington C.R., Belachew T., Wright M., Giblin C., David H., Mills M., Salomon R., Mullins C., Akolkar B., Begg L., Davis C., Grandison L., Humble M., Khalsa J., Little A.R., Peavy H., Pontzer C., Portnoy M., Sayre M.H., Starke-Reed P., Zakhari S., Read J., Watson B., Guyer M. The NIH human microbiome project. Genome. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. URL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core T. R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. URL http://www. R-project. org/.(3.3. 1) Software Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Radolf J.D. Medical Microbiology. University of Texas Medical Branch at Galveston; 1996. Treponema. [Google Scholar]
- Read A.J., Drinker P., Northridge S. Bycatch of marine mammals in US and global fisheries. Conserv. Biol. 2006;20(1):163–169. doi: 10.1111/j.1523-1739.2006.00338.x. [DOI] [PubMed] [Google Scholar]
- Reddy L., Dierauf L.A., Gulland F.M. Marine mammals as sentinels of ocean health. CRC Handbook Mar. Mamm. Med.: Health Dis. Rehabil. 2001:3–13. [Google Scholar]
- Ross P.S. Marine mammals as sentinels in ecological risk assessment. Hum. Ecol. Risk Assess. 2000;6(1):29–46. [Google Scholar]
- Russo C.D., Weller D.W., Nelson K.E., Chivers S.J., Torralba M., Grimes D.J. Bacterial species identified on the skin of bottlenose dolphins off southern California via next generation sequencing techniques. Microb. Ecol. 2018;75(2):303–309. doi: 10.1007/s00248-017-1071-2. [DOI] [PubMed] [Google Scholar]
- Segura I., Rocha-Olivares A., Flores-Ramírez S., Rojas-Bracho L. Conservation implications of the genetic and ecological distinction of Tursiops truncatus ecotypes in the Gulf of California. Biol. Conserv. 2006;133(3):336–346. [Google Scholar]
- Shane S.H., Wells R.S., Würsig B. Ecology, behavior and social organization of the bottlenose dolphin: a review. Mar. Mamm. Sci. 1986;2(1):34–63. [Google Scholar]
- Shulzhenko N., Morgun A., Hsiao W., Battle M., Yao M., Gavrilova O., Orandle M., Mayer L., Macpherson A.J., McCoy K.D. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 2011;17(12):1585. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Bäckhed F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013;11(4):227. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Soverini M., Quercia S., Biancani B., Furlati S., Turroni S., Biagi E., Consolandi C., Peano C., Severgnini M., Rampelli S. The bottlenose dolphin (Tursiops truncatus) faecal microbiota. FEMS Microbiol. Ecol. 2016;92(4) doi: 10.1093/femsec/fiw055. [DOI] [PubMed] [Google Scholar]
- Springer A.M., Estes J., Van Vliet G.B., Williams T., Doak D., Danner E., Forney K., Pfister B. Sequential megafaunal collapse in the North Pacific Ocean: an ongoing legacy of industrial whaling? Proc. Natl. Acad. Sci. Unit. States Am. 2003;100(21):12223–12228. doi: 10.1073/pnas.1635156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E.J. Growing unculturable bacteria. J. Bacteriol. 2012;194(16):4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Segawa T., Sawa S., Nishitani C., Ueda K., Itou T., Asahina K., Suzuki M. Comparison of the gut microbiota of captive common bottlenose dolphins, Tursiops truncatus in three aquaria. J. Appl. Microbiol. 2018 doi: 10.1111/jam.14109. [DOI] [PubMed] [Google Scholar]
- Sweeney J., Ridgway S. Common diseases of small cetaceans. J. Am. Vet. Med. Assoc. 1975;167(7):533–540. [PubMed] [Google Scholar]
- Trites A.W., Livingston P.A., Mackinson S., Vasconcellos M., Springer A.M., Pauly D. 1999. Ecosystem Change and the Decline of marine Mammals in the Eastern Bering Sea: Testing the Ecosystem Shift and Commercial Whaling Hypotheses. [Google Scholar]
- Van Bressem M.-F., Raga J.A., Di Guardo G., Jepson P.D., Duignan P.J., Siebert U., Barrett T., de Oliveira Santos M.C., Moreno I.B., Siciliano S. Emerging infectious diseases in cetaceans worldwide and the possible role of environmental stressors. Dis. Aquat. Org. 2009;(2) doi: 10.3354/dao02101. [DOI] [PubMed] [Google Scholar]
- Vandamme P., Dewhirst F.E., Paster B.J., On S.L. 2015. Arcobacter. Bergey's Manual of Systematics of Archaea and Bacteria. [Google Scholar]
- Venn-Watson S., Smith C.R., Jensen E.D. Primary bacterial pathogens in bottlenose dolphins Tursiops truncatus: needles in haystacks of commensal and environmental microbes. Dis. Aquat. Org. 2008;79(2):87–93. doi: 10.3354/dao01895. [DOI] [PubMed] [Google Scholar]
- Vos P., Garrity G., Jones D., Krieg N.R., Ludwig W., Rainey F.A., Schleifer K.-H., Whitman W.B. Vol. 3. Springer Science & Business Media; 2011. Bergey's Manual of Systematic Bacteriology. (The Firmicutes). [Google Scholar]
- Waltzek T., Cortés-Hinojosa G., Wellehan J., Jr., Gray G.C. Marine mammal zoonoses: a review of disease manifestations. Zoonoses Public Health. 2012;59(8):521–535. doi: 10.1111/j.1863-2378.2012.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Earley D. Novel method for measuring growth of Treponema hyodysenteriae and its application for monitoring susceptibility of clinical isolates to antimicrobial agents. Antimicrob. Agents Chemother. 1991;35(10):2012–2015. doi: 10.1128/aac.35.10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R.S. The role of long-term study in understanding the social structure of a bottlenose dolphin community. Dolphin Soc.: Discov. Puzzles. 1991:199–225. [Google Scholar]
- Wells R.S., Rhinehart H.L., Hansen L.J., Sweeney J.C., Townsend F.I., Stone R., Casper D.R., Scott M.D., Hohn A.A., Rowles T.K. Bottlenose dolphins as marine ecosystem sentinels: developing a health monitoring system. EcoHealth. 2004;1(3):246–254. [Google Scholar]
- Würsig B., Würsig M. Behavior and ecology of the bottlenose dolphin, Tursiops truncatus, in the South Atlantic. Fish. Bull. 1979;(2) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.