Abstract
Long non-coding RNAs (lncRNAs), associated with various cancers including colorectal cancer (CRC), could be collected from body fluids easily. Our aims were to determine the expression level of HOTTIP lncRNA in plasma samples of healthy individuals and CRC patients as well as their relationship with clinico-pathological characteristics of patients. First, total RNA was extracted from the plasma samples of 100 subjects including 50 patients and 50 age and sex matched healthy persons. Then, gene expression was measured using real-time PCR technique. The sensitivity and specificity of HOTTIP dysregulation in CRC and healthy individual’s plasma was measured by receiver operating characteristic (ROC) analysis. As compared with healthy controls, HOTTIP lncRNA was over expressed in a statistically significant manner in plasma samples of patients (P=0.001). Significant relationship between HOTTIP expression and positive family history of CRC was observed, too (P=0.04). The ROC curve analysis showed an AUC value of 0.775, a specificity of 82%, a sensitivity of 76%, with a cut off value equal to 2.40 (P =0.001). HOTTIP transcript can be proposed as a new biomarker for early diagnosis due to the increased expression in plasma samples of patients with CRC and the relatively high sensitivity and specificity.
Key Words: Biomarker, colorectal neoplasms, long non-coding RNAs, lncRNA HOTTIP
Colorectal cancer (CRC) is ranked third among the most common malignant diseases world-wide (1). Contrary to the many advances in CRC treatment, patients’ survival rate is still changing to a limited extent due to the lack of an effective screening test (2). Age and stage at diagnosis are crucial factors that impress patients survival rate so that the lowest survival is attributed to stage IV of CRC affected patients (3). Therefore, what has attracted the attention of the scientists is finding noninvasive novel biomarkers to diagnose CRC in early stages and predict prognosis to choose an appropriate treatment which is an inevitable need.
Long noncoding RNAs (lncRNAs) are a class of transcribed RNA molecules with a length of greater than 200 nucleotides that are involved in various biological functions such as alternative splicing, chromatin remodeling, and mRNA degradation as well as cell cycle, survival and migration (4, 5). LncRNAs are smaller than mRNAs, contain less but longer exons, and are expressed relatively at lower levels. Most of these molecules represent a specific expression pattern in different tissues so that the aberrant expression of lncRNAs can cause various diseases, including CRC (1, 5-8).
The HOTTIP gene in the homeobox A (HOXA) locus is located at 7p15.2 chromosome and encodes a 4665-bp transcript (9). HOXA locus in mammals contains a cluster containing eleven HOX genes with a series of expression patterns ranging from proximal to distal. In general, HOXA transcript at the distal tip (HOTTIP) lncRNA is able to activate HOX genes by employing histone-modifying enzymes and silencing tumor suppressor genes (10-12). Recently, it was reported that HOTTIP is up regulated in CRC tissue samples compared with healthy adjacent tissues and it has a positive association with T and clinical stages (13). On the other hand, lncRNAs could be collected from body fluids easily (14). So the aims of our study were to determine the expression of HOTTIP in plasma samples of healthy individuals and CRC patients as well as their relationship with clinical and pathological characteristics of patients.
Materials and methods
Patient samples
This case -control study was conducted on 100 plasma samples which fifty of them were obtained from patients suffering from CRC at stages I-III of disease regardless of age and sex with definite clinical and pathological status from Tumor Bank Department of Imam Khomeini Hospital, Tehran, Iran. The other 50 plasma samples were collected from suspected patients who underwent colonoscopy and their biopsy results were reported normal as control samples from the same hospital during 2017 which matched in age and sex with the patient group as much as possible. Following subjects were excluded from study groups: patients who received any treatment, cases with other organ cancers, history of chemotherapy and radiotherapy, systemic diseases such as renal failure, diabetes mellitus, and hypertension. Study protocol was approved by ethical committee of Hamadan University of Medical Sciences (Ethical code: IR.UMSHA.REC.1396.220). Participants signed written informed consent after explanation of the study purpose.
Patients included 22 women and 28 men with an average age of 60.98 ± 13.95 (29-85) and healthy people consisted of 24 women and 26 men with an average age of 59.26 ± 14.38 (22-85).The other clinicopathological information of the patients are shown in Table 1.
Table 1.
Association between lncRNA HOTTIP expression and clinico-pathological data in CRC
| Variables | Number of cases | % of patients | Mean rank | P-value |
|---|---|---|---|---|
| Age (year) | ||||
| >50 <50 |
39 11 |
78 22 |
25.49 25.55 |
0.9** |
| Gender | ||||
| Male Female |
28 22 |
56 44 |
26.89 23.73 |
0.4** |
| Stage | ||||
| I II III |
15 18 16 |
30.6 36.7 32.7 |
22.73 24.28 27.94 |
0.5* |
| Grade | ||||
| I II III |
23 23 4 |
46 46 8 |
25.52 24.39 31.75 |
0.6* |
| Tumor location | ||||
| Rectum Sigmoid Rectosigmoid Colon and cecum |
30 6 7 7 |
60 12 14 14 |
24.57 31.83 29.00 20.57 |
0.3* |
| Histology | ||||
| Adenocarcinoma Mucinous (Colloid) Adenocarcinoma |
44 6 |
88 12 |
25.91 22.50 |
0.5** |
| Tumor Size | ||||
| >5 cm <5 cm |
17 33 |
34 66 |
22.24 27.18 |
0.2** |
| Family history | ||||
| Yes No Unknown |
17 27 6 |
34 54 12 |
32.35 21.07 26 |
0.04* |
| Smoking | ||||
| Non-Smoker Smoker unknown |
32 6 10 |
64 12 20 |
26.16 20.67 21.50 |
0.5* |
| Drinking | ||||
| Alcoholic Non-alcoholic unknown |
3 34 11 |
6 68 22 |
22 25.24 22.91 |
0.8* |
* Kruskal- Wallis test; ** Mann-Whitney test
Relative expression of HOTTIP transcript by quantitative real-time PCR (QRT-PCR)
Total RNA was extracted from the plasma samples using Y TZOL (Yekata Tajhiz Azma Company, Iran) in accordance with the company's protocol. The quality and quantity of the extracted RNA were determined by electrophoresis on gel agarose and NanoDrop spectrophotometer, respectively. Thereafter, cDNA was synthesized by reverse transcription of the extracted RNA using oligo dT primers and the Hyperscript first-strand synthesis kit (Geneall Biotechnology, Korea). Next, QRT-PCR reaction was performed using the Real Q Plus 2x Master Mix Green Kit (Ampliqon A/S, Denmark) by applying specific primers (HOTTIP: Forward sequence 5'AGCTCTTTTCCCCGACA-GTG3', Reverse sequence: 5'CCTTCACCAAGC-TCCCTCTG3', and GAPDH: Forward sequence: 5'CATCAAGAAGGTGGTGAAGCAG3', Reverse sequence: 5'GCGTCAAAGGTGGAGGAGTG3') in duplicate. Primers were designed with Gene runner software and their specificity was evaluated by NCBI primer-BLAST. QRT-PCR and data collection was performed by Step One Plus™ real-time PCR system (Thermo Fisher scientific, USA). The results were normalized against GAPDH and relative expression of HOTTIP was calculated using the 2-∆∆CT method (15).
Statistical analysis
For describing the quantitative variables mean ± SD was used, and qualitative variables were described by percent. Kolmogorov-Smirnov test was used to evaluate normal distribution of the quantitative measures. Mann-Whitney nonpa-rametric test was used to compare lncRNA HOTTIP expression between two groups. Kruskal-wallis and Mann-withney tests were used to investigate the relationship between HOTTIP expression and clinicopathological data of the patients. Statistically, P value <0.05 was supposed significant. The sensitivity and specificity of HOTTIP dysregulation in a CRC and healthy individual’s plasma was measured by receiver operating characteristic (ROC) analysis. Statistical analysis was performed using SPSS software 23.
Results
Overall up regulation of HOTTIP in CRC
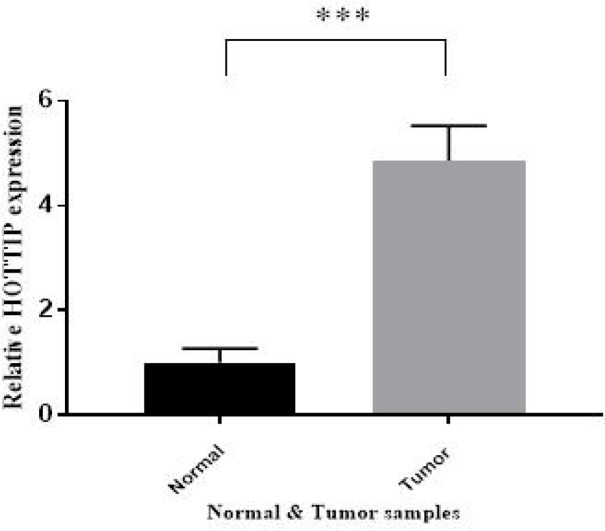
As shown in Figure 1, significant up regulation of HOTTIP was observed in plasma samples of CRC patients in comparison with normal group (P <0.001). According to the 2-∆∆CT method results, data greater and less than 1 was considered up and down regulation, respectively. Higher expression of HOTTIP was obtained in 39 cases of tumor group (78%), individually.
Fig. 1.
Plasma HOTTIP expression. Overall 4.8 fold higher expression of HOTTIP in CRC plasma samples was observed in comparison with of normal plasma samples (***P< 0.001).
Diagnostic value of plasma HOTTIP in CRC patients was assessed by ROC curve analysis. As illustrated in Figure 2, the area under curve (AUC) of the HOTTIP in patients with CRC and control people is equal to 0.775 ± 0.05.
Fig. 2.
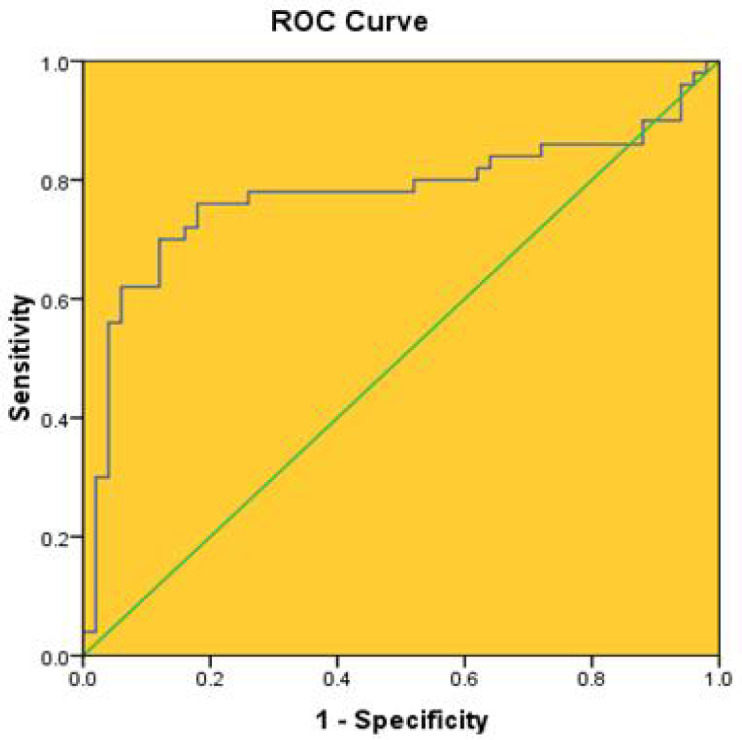
Receiver operating characteristic (ROC) curve for HOTTIP to distinguish CRC disease from healthy persons. The area under the ROC is 0.77
Relationship between HOTTIP expression level and clinico-pathological characteristics of CRC patients
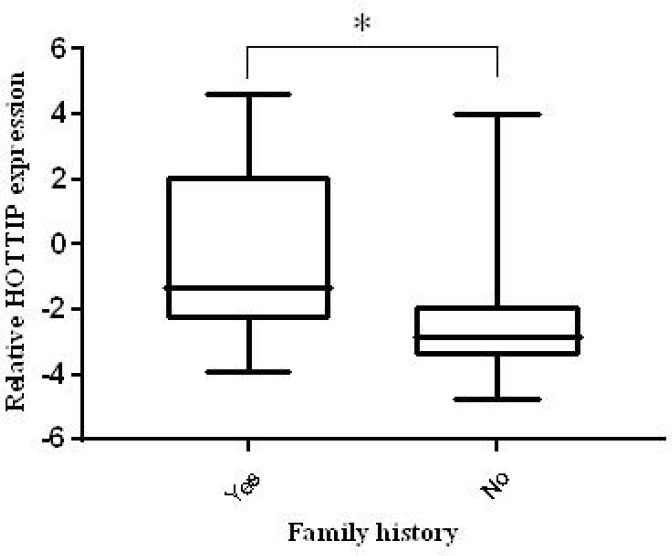
In this study, there was a significant relationship between the HOTTIP expression level and family history of CRC (Figure 3). No significant association was found between HOTTIP dysregulation and other clinico- pathological features such as tumor stage, grade, location and size. The other information are presented in Table1.
Fig. 3.
Plasma HOTTIP expression according to family history. HOTTIP expression was significantly higher in patients with family history of CRC (* P < 0.05).
Discussion
LncRNAs, a class of RNA molecules with a length of more than 200 nucleotides, are transcribed in the genome and function as regulators in various biological processes. Abnormal expression of this group of molecules participate in various cancers development including CRC (1, 4). LncRNAs may be considered as therapeutic targets and promising biomarker for cancer prognosis (16), and one of well- known lncRNAs with oncogenic role in solid tumors is HOTTIP (17).
In present survey we report that HOTTIP is significantly over expressed in CRC plasma samples. These data are similar to other studies (13, 18). Unlike similar published researches, the value of our study is the evaluation of target gene in matched biological samples rather than tissue samples. Also, all study subjects have not received any treatment prior to sample collection, had no history of chemoradiotherapy due to cancer with other origins, and no systemic disease. In other words, the confounding factors were excluded while the majority of previous surveys have introduced adjacent non-malignant tissues in their study as a control group and have not clarified exclusion criteria, exactly (13, 18-20). Because microscopic adjacent non- malignant cells may have tumoric cell features at molecular level (21), so this is a drawback to this kind of study design.
Our study did not prove any significant statistical relationship between HOTTIP expression and clinical and pathological data, especially tumor stage and size. However, expression level of HOTTIP in grade 2 was higher than the first degree.
Contrary to our results, a positive correlation of higher HOTTIP with T stage and larger tumor size have been reported by Ren et al. study which was conducted on 156 CRC tissues and 21 adjacent non-malignant tissues (13) as well as another investigation which was carried out on 48 paired CRC and adjacent non-tumor colorectal samples (20). However, the later study indicate no relevance of HOTTIP dysregulation to the site and grade of the tumor, lymph nodes metastasis, gender and age (20).
Although, a systematic review and meta-analysis has introduced HOTTIP as a lymph node metastasis indicator in human cancers including CRC, but the authors state that their results may be influenced by ethnic bias because the majority of studies have been conducted on population samples in China (22). Therefore, it is expected to observe the racial differences between Iranian ethnicity and other ethnicities. These inconsistent consequences can be related to low sample size of present and previous studies, different design procedure, and different inclusion and exclusion criteria consideration, too.
In order to discover screening biomarkers for CRC, Zhao et al. evaluated the clinical importance of two HOX transcript antisense RNA (HOTAIR) and colon cancer associated transcript 1 (CCAT1) lncRNAs in plasma samples. They observed increased expression of these two molecules in the plasma of patients. Also, they described that CCAT1 alone, as a diagnostic biomarker for CRC, has 75% sensitivity and 85% specificity (23). Our data illustrated 82% specificity, and 76% sensitivity for plasma HOTTIP. Thus, it is highly suggested to evaluate these two transcripts, CCAT1 and HOTTIP, in a single study because these two transcripts seems to have key properties of novel potential diagnostic biomarkers in combination with each other or one of these transcript alone.
Molecular mechanism of action of HOTTIP in CRC development has been little known, and studies on this lncRNA are in the primary stages. In recent years, some studies have tried to elucidate the real role of HOTTIP in tumor behaviors including: a survey that suppressed HOTTIP expression by siRNA and elucidated that tumor cell migration and invasion in vitro are inhibited. This study implies that HOTTIP is somehow involved in tumor metastasis through down regulation of dickkopf WNT signaling pathway inhibitor 1 (DKK1) tumor suppressor gene (18). Also, apoptosis was induced and cell proliferation was suppressed by knockdown of HOTTIP in HCT-116 and SW620 cell lines through targeting serum- and glucocorticoid-inducible kinase 1 (SGK1) gene. Besides, it inhibits GSK3β, β-catenin, cellular myelocytomatosis oncogene (C-MYC), vimentin and matrix metalloproteinase 7 (MMP-7) expression, and results in up-regulation of E-cadherin (19). In addition, HOTTIP participates in CRC cell growth via silencing p21, a cyclin-dependent kinase inhibitor (20, 24).
As mentioned earlier, there is still no comprehensive information available about HOTTIP expression which confirms its application as a biological marker in the diagnosis and treatment of CRC. Thus, functional consequences of altered expression of this lncRNA can be resulted from more extensive examinations.
Our results suggest that higher expression level of HOTTIP has the potential to be a diagnostic marker for CRC patients.
Acknowledgment
The vice Chancellery for Research and Technology, Hamadan University of Medical Sciences supported this research [Grant number: 9603231784].
Conflict of interest
There was no conflict of interest.
References
- 1.Li H, Ma SQ, Huang J, et al. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8:39859–76. doi: 10.18632/oncotarget.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 3.Crooke H, Kobayashi M, Mitchell B, et al. Estimating 1-and 5-year relative survival trends in colorectal cancer (CRC) in the United States: 2004 to 2014. J Clin Oncol . 2018;36(4_suppl):587. [Google Scholar]
- 4.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 8.Han D, Wang M, Ma N, et al. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Fan Y, Yan T, Chai Y, et al. Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin Chim Acta. 2018;482:224–30. doi: 10.1016/j.cca.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 10.Lian Y, Cai Z, Gong H, et al. HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol Biosyst. 2016;12:3247–53. doi: 10.1039/c6mb00475j. [DOI] [PubMed] [Google Scholar]
- 11.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess DJ. Non-coding RNA: HOTTIP goes the distance. Nat Rev Genet. 2011;12:300. doi: 10.1038/nrg2992. [DOI] [PubMed] [Google Scholar]
- 13.Ren YK, Xiao Y, Wan XB, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8:11458–63. [PMC free article] [PubMed] [Google Scholar]
- 14.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–65. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 16.Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–13. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Li N, Kang X, et al. Prognostic value of the long noncoding RNA HOTTIP in human cancers. Oncotarget. 2017;8:59563–9. doi: 10.18632/oncotarget.19166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rui Y, Hu M, Wang P, et al. LncRNA HOTTIP mediated DKK1 downregulation confers metastasis and invasion in colorectal cancer cells. Histol Histopathol. 2019;34:619–30. doi: 10.14670/HH-18-043. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Yu T, Hu H, et al. Knockdown of the long non-coding RNA HOTTIP inhibits colorectal cancer cell proliferation and migration and induces apoptosis by targeting SGK1. Biomed Pharmacother. 2018;98:286–96. doi: 10.1016/j.biopha.2017.12.064. [DOI] [PubMed] [Google Scholar]
- 20.Lian Y, Ding J, Zhang Z, et al. The long noncoding RNA HOXA transcript at the distal tip promotes colorectal cancer growth partially via silencing of p21 expression. Tumour Biol. 2016;37:7431–40. doi: 10.1007/s13277-015-4617-2. [DOI] [PubMed] [Google Scholar]
- 21.Seyedabadi S, Saidijam M, Najafi R, et al. Assessment of CEP55, PLK1 and FOXM1 expression in patients with bladder cancer in comparison with healthy individuals. Cancer Invest. 2018;36:407–14. doi: 10.1080/07357907.2018.1514504. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, He A, Wang D, et al. -Long noncoding RNA HOTTIP as a novel predictor of lymph node metastasis and survival in human cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:14126–32. doi: 10.18632/oncotarget.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W, Song M, Zhang J, et al. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:14131–40. [PMC free article] [PubMed] [Google Scholar]
- 24.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–5. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]