Abstract
Introduction
African swine fever (ASF) was officially reported in Vietnam in February 2019 and spread across the whole country, affecting all 63 provinces and cities.
Material and Methods
In this study, ASF virus (ASFV) VN/Pig/HaNam/2019 (VN/Pig/HN/19) strain was isolated in primary porcine alveolar macrophage (PAM) cells from a sample originating from an outbreak farm in Vietnam’s Red River Delta region. The isolate was characterised using the haemadsorption (HAD) test, real-time PCR, and sequencing. The activity of antimicrobial feed products was evaluated via a contaminated ASFV feed assay.
Results
Phylogenetic analysis of the viral p72 and EP402R genes placed VN/Pig/HN/19 in genotype II and serogroup 8 and related it closely to Eastern European and Chinese strains. Infectious titres of the virus propagated in primary PAMs were 106 HAD50/ml. Our study reports the activity against ASFV VN/Pig/HN/19 strain of antimicrobial Sal CURB RM E Liquid, F2 Dry and K2 Liquid. Our feed assay findings suggest that the antimicrobial RM E Liquid has a strong effect against ASFV replication. These results suggest that among the Sal CURB products, the antimicrobial RM E Liquid may have the most potential as a mitigant feed additive for ASFV infection. Therefore, further studies on the use of antimicrobial Sal CURB RM E Liquid in vivo are required.
Conclusions
Our study demonstrates the threat of ASFV and emphasises the need to control and eradicate it in Vietnam by multiple measures.
Keywords: pig, African swine fever virus, virus isolation, Sal CURB, Vietnam
Introduction
African swine fever (ASF) is a pernicious and devastating disease of the world swine industry. It has been demonstrated that ASF is caused by an enveloped DNA virus and this virus replicates in pigs in the cytoplasm of infected cells (6, 8). It is a recognised DNA arbovirus and member of the Asfarviridae family and Asfivirus genus (6, 9, 13). Many previous studies have reported that African swine fever virus (ASFV) infects both domestic pigs and wild boars, and can easily be transmitted by many vectors such as mosquitoes, ticks, or other arthropods. ASFV is a highly contagious virus and pigs’ exposure to it resulted in up to 100% morbidity, the mortality of ASF having been shown to depend on the virulence of the virus, the host, and transmission cycles (13, 21). The first case of ASF was reported in Kenya in 1921 (8) and it was confined to Africa until it spread to Eurasia in the middle and to South America and the Caribbean at the close of the last century (8). In total, 24 genotypes and 8 serotypes of ASFV have been identified (8, 9, 21, 24). The first outbreak of ASF in Vietnam was officially reported in February 2019, and the disease has now spread to 63 out of 63 provinces/cities of Vietnam. Approximately six million pigs have been culled on infected farms, suggesting the risk of spread of this virus. Many active measures have been imposed by the Vietnamese government to inhibit the rapid spread of ASFV. In this study, we isolated the first ASFV in Vietnam from a spleen sample from a pig which died during an ASF outbreak in Ha Nam province, in February 2019. The isolate was characterised using genotyping and the haemadsorption (HAD) test. This investigation also used primary porcine alveolar macrophage (PAM) cell-propagated virus to inoculate commercial swine feed samples after combination with three different Sal CURB-branded antimicrobial feed products at different time points to investigate their activity against ASFV.
Recent research indicated that contaminated feeds can be a potential vector for ASFV transmission to domestic pigs and that it can be easily transmitted through the natural consumption of contaminated feed and liquid (19). As this new information confirmed feed as a risk factor, it became imperative to find solutions to reduce it. A recent study was conducted to evaluate whether liquid and dry antimicrobial products containing formaldehyde and organic acids or organic acids only could lessen the danger of infected feed. The study was based on previous publications indicating that such products containing formaldehyde and organic acids reduced the titre of Salmonella species and/or porcine epidemic diarrhoea virus (PEDV) in feed and pigs (3, 7, 25, 26). Additionally, other experiments indicated that formaldehyde treatment reduced organ infection by highly pathogenic coronavirus in turkey poults, whereas other treatments failed to ameliorate its negative effects (4, 5). In this study, we evaluated three Sal CURB-branded antimicrobial feed products to examine the survivability of ASFV in feed through detection by haemadsorption (HAD) test and real-time PCR. As PEDV and ASFV are both enveloped viruses, it was hypothesised that formaldehyde treatment of ASFV contaminated feed may induce an anti-viral effect, prevent contamination of feed and potentially reduce infectivity in the pig post consumption. This is the first report on antiviral activity against the emerging Vietnamese ASFV of liquid and dry antimicrobial products containing formaldehyde and organic acids.
These experiments are of importance because African swine fever is an endemic disease in the whole of Vietnam and no vaccine, drug nor effective therapy is available.
Material and Methods
Sample collection. The pigs from Ha Nam province in the Red River Delta of Vietnam were selected for the current study based on clinical signs and post-mortem lesions. Tissues, including mesenteric lymph nodes and spleens, were collected according to the guidelines of the OIE (20).
Virus isolation, DNA extraction, and PCR assay. Primary porcine alveolar macrophages (PAMs) were collected from 10–20-day-old SPF pigs that were purchased from the Pig Research Centre of the National Institute of Animal Science, Vietnam, and the cells were maintained in 10% FBS RPMI 1640 medium (Thermo Scientific, Waltham, MA, USA) at 37°C with 5% CO2. The homogenate of the field pig spleen sample was virus-positive by conventional PCR using a p72U/p72D-specific primer (data not shown) (2) and the haemadsorption (HAD) assay recommended by the OIE was used to inoculate PAMs for virus isolation (20). The cell supernatants were collected after four days’ inoculation and the HAD assay for infectious virus particles was performed. The genomic DNA of ASFV was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) from cell supernatants. For molecular analysis, different PCRs were set up on virus isolates: (i) the DNA of ASFV in the supernatant was detected by the real-time PCR for the viral p72 gene recommended by the OIE (20) and was carried out on an AriaMx Real-Time PCR System (Agilent, Santa Clara, CA, USA) according to the OIE-recommended procedure described by King et al. (14); (ii) the C-terminal region of the p72 protein was amplified using p72-U and p72-D primers for genotype analysis (2); and (iii) a fragment of the EP402R gene encoding the CD2v protein was amplified using a CD2-2F/2R-specific primer according to a protocol previously described for serotype analysis (23). The positive amplification control consisted of DNA known to be of ASFV, and the negative amplification control consisted of nuclease-free sterile water.
Sequence analysis of the ASFV isolate. The correctly sized amplicons of the p72 and EP402R partial gene products were electrophoresed on a 1.5% agarose gel against a 100 bp DNA leader marker (Thermo Scientific) and visualised by UV irradiation and ethidium bromide staining (Sigma-Aldrich, St Louis, MO, USA). Amplicons of the correct size were excised from the agarose gel and purified using the QIAQuick gel extraction kit (Qiagen) according to the manufacturer’s specifications. The chromatograms of the amplicons and probe binding site sequence were analysed using the BioEdit and DNAstar programmes (DNASTAR Inc., Madison, WI, USA). The nucleotide identity of the ASFV strain in Vietnam was compared to other sequences using the BLAST tool in the National Center for Biotechnology Information (NCBI) GenBank database and published sequence information. Multiple sequence alignment was performed using Laser gene software (DNASTAR Inc.). Phylogenetic analyses of nucleotide sequences of partial p72 and EP402R genes encoding the ASFV CD2v protein were constructed using the neighbour-joining method with a bootstrap value of 1,000 in the MEGA7 programme (16).
HAD assay. The HAD assay was performed as previously described (18). Primary PAM cells were seeded in 96-well plates and the samples were then added to the plates and titrated in triplicate using 10 × dilutions. The quantity of ASFV was determined by identification of characteristic rosette formations representing haemadsorption of erythrocytes around infected cells, according to the OIE guidelines (20). HAD was observed for four days, and 50% HAD doses (HAD50) were calculated using the method of Reed and Muench (22).
Experimental design of feed assay. Samples of 5 g of commercial swine feed (composition in Supplement 1) were treated with Sal CURB RM E Liquid, Sal CURB F2 Dry or Sal CURB K2 Liquid (Kemin Industries, Des Moines, IA, USA; product specifications in Supplement 2) at an application rate of 3.0 kg/tonne and then spiked with 100 μL of minimum essential medium (MEM; Gibco, Thermo Scientific) containing 1 × 105 HAD50 of ASFV. Additional 5 g feed samples were inoculated with phosphate-buffered saline (PBS, Sigma-Aldrich) and used as negative controls in each sampling batch, and feed samples once again of 5 g were inoculated with 1 × 105 HAD50 of ASFV and used as positive controls in 50 mL mini bioreactor tubes (Corning Incorporated, Corning, NY, USA). Treatments of each group or control groups were collected on days 1, 3, and 7 post inoculation. Each sample was resuspended in 15 mL sterile PBS and vortexed for 10 s. The supernatant was aliquoted, centrifuged at 10,000 g for 10 min filtered at 0.45μm pore size (Bio-Rad, Hercules, CA, USA), and subjected to the incubation procedure prior to real-time PCR and virus isolation as described above.
Statistical analysis: Statistical analysis was performed using IBM SPSS software (SPSS 23.0 for Windows; IBM, Armonk, NY, USA). A P < 0.05 value was considered to be statistically significant. Differences among the groups were tested by Duncan’s multiple comparison methods.
Results
Isolation and characterisation of ASFV in vitro. On day 4 post inoculation with the supernatant of the spleen homogenate, primary PAMs showed clear HAD, even in the wells inoculated with 10-fold diluted sample (Fig. 1A). A positive sample was obtained from the real-time PCR (Fig. 1B), and this result together with the clear HAD confirmed that the spleen homogenate contained infectious ASFV. The HAD assay was ASFV-positive for the virus stock and the titre for the first passage stock was 106 HAD50/ml. These results demonstrated that ASFV was successfully isolated from the field outbreak sample. This virus isolate was designated VN/Pig/HaNam/2019, abbreviated to VN/Pig/HN/19.
Fig. 1.
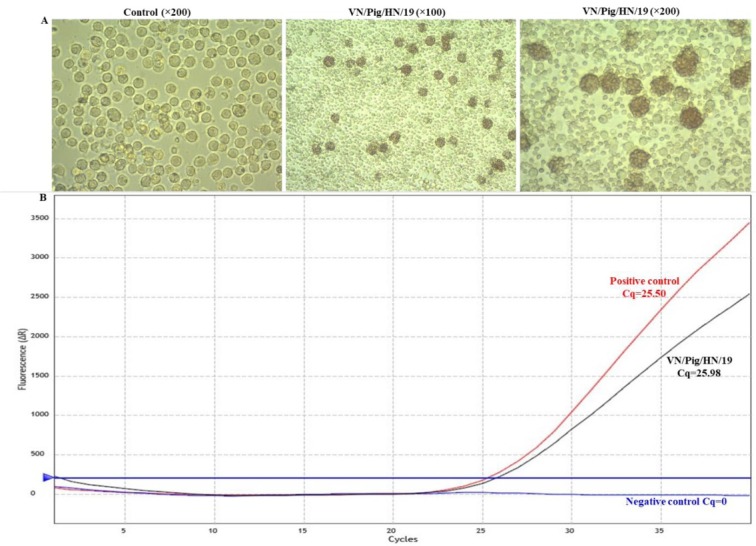
Detection of ASFV in a field sample of spleen tissue. A – HAD assay of the spleen homogenate; B– the real-time PCR amplification of the p72 gene
Sequence analysis of the ASFV isolate. The sequences obtained after amplification of the C-terminal end of p72 suggested that the outbreak VN/Pig/HN/19 strain belongs to genotype II and shares 100% nucleotide identity with the Russia-isolated Krasnodar 2012 and Irkutsk 2017 strains and the Georgia 2007/1, Estonia 2014, and China-isolated ASFV-SY-18 strains as well, based on the p72 gene and this Chinese isolate which caused the first ASF case in Shenyang in that country on 3 August 2018 (Fig. 2A) (17, 27). It was recently reported that the Estonia 2014 strain and ASFV-SY-18 strain in China have an intergenic region (IGR) I variant with additional tandem repeat sequences in the IGR between I73R and I329L (15, 27). We also used the CD2v fragment sequence for phylogenetic analysis of the serogroup. These trees showed that the causative strain (VN/Pig/HN/19) in this study belonged to CD2v serogroup 8 (Fig. 2B). Particularly, the analysis of the amino acid repeats from the VN/Pig/HN/19 strain revealed the presence of three PPPKPC hexamers with identical repeats and the same central interruption as hexamers in the recent outbreaks in China and Russia (data not shown). The results demonstrated that there is a close relationship between these viruses, which suggests that the VN/Pig/HN/19 strain in Vietnam belongs to genotype II, serogroup 8, and the disease was caused by the Eastern European or Chinese strains of ASFV (Fig. 2). The sequences of the p72 and CD2v genes of VN/Pig/HN/19 strain were submitted to the GenBank database with accession numbers MN199633 and MN199634, respectively.
Fig. 2.
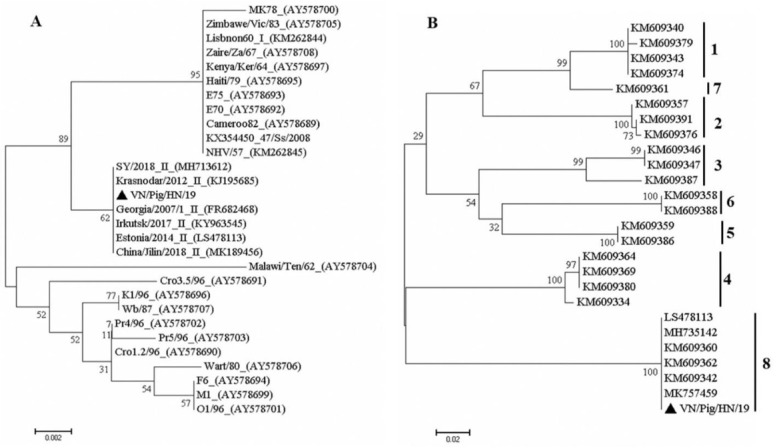
Phylogenetic analysis of VN/Pig/HN/19 based on its partial p72 (A) and EP402R gene encoding the CD2v protein (B) genes. Numbers along branches indicate bootstrap values >70% (1,000 replicates). The black triangle indicates the ASFV isolate from this study. Scale bars indicate nucleotide substitutions per site
Effect of Sal CURB on viral DNA synthesis. The results of the feed sampling are summarised in Fig. 3A with the raw data provided in Fig. 3B. All samples from the negative control groups were real-time PCR-negative and all nine feed samples from the positive control group were positive for the presence of ASFV DNA by real-time PCR, with a mean Cq on day 1 (D1) of 26.38 (range 26.03–27.03), mean Cq on D3 of 26.023 (range 25.51–26.75), and mean Cq on D7 of 27.49 (range 26.96–28.09). The results indicated that ASFV can survive in feed seven days post inoculation. In contrast, the mean Cq value of the feed from the treatment group with Sal CURB RM E Liquid indicated activity against ASFV in feed one, three, and seven days post inoculation. The Cq value on D1 was 29.50 (range 29.39–29.55), on D3 it was 29.27 (range 28.64–29.44), and on D7 it was 33.01 (range 31.55–34.09) (Fig. 3 and Table 1). When analysed by t-test, the difference in the Cq levels between the group treated with Sal CURB RM E Liquid and the positive control group was significant at P < 0.01 for all three days. In addition, the mean Cq value of the feed from the group treated with Sal CURB F2 Dry also showed activity against ASFV in feed after D3 (significant at P < 0.05) and D7 (significant at P < 0.01). Lastly, the Sal CURB K2 Liquid only showed activity against ASFV in the feed on D7 (significant at P < 0.05).
Fig. 3.
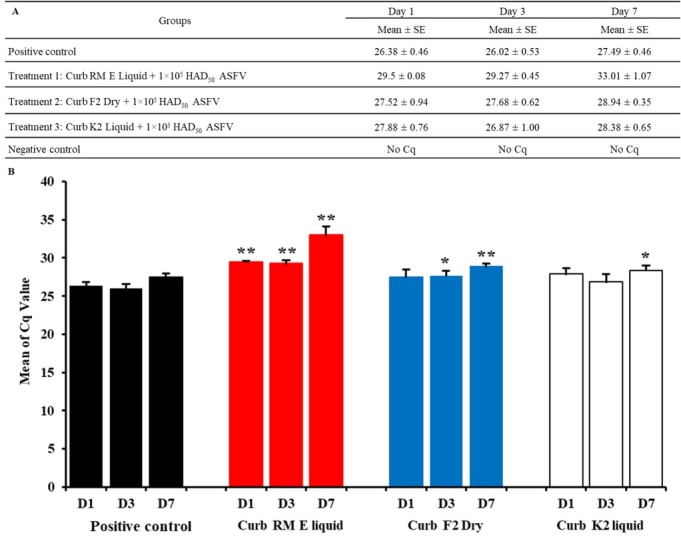
(A, B). The combination of feed with the Sal CURB RM E Liquid, Sal CURB F2 Dry or Sal CURB K2 Liquid and 1×105/mL HAD50 of VN/Pig/HN/19 strain one, three, and seven days post inoculation. Values represent mean and standard deviation results from three independent experiments. Significant differences compared to control are denoted by * for P < 0.05 and ** for P < 0.01
Effect of continuous treatment on ASFV infection. The results of microscopic observation of ASFV-infected cells are shown in Figs 4A and 4B. As shown in the figures, activity was indicated on D3 and D7. The Sal CURB RM E Liquid decreased the viral titre from 4.83 log HAD50/ml to 4.76 log HAD50/ml and 4.16 log HAD50/ml (P < 0.05), respectively. The Sal CURB F2 Dry reduced the viral titre of ASFV in PAM cells from 4.83 log HAD50/ml to 4.57 log HAD50/ml (P < 0.05) on D7 but did not lower the titre on D3. In contrast, the Sal CURB K2 Liquid showed no activity against ASFV replication when it was added at one, three or seven days post inoculation. The viral titre in positive control groups was not changed after one, three or seven days post inoculation and no viral titre in negative control groups was observed (Fig. 4). The results clearly demonstrated that Sal CURB RM E Liquid had cumulative activity against VN/Pig/HN/19 infection when administered over seven days post inoculation.
Fig. 4.
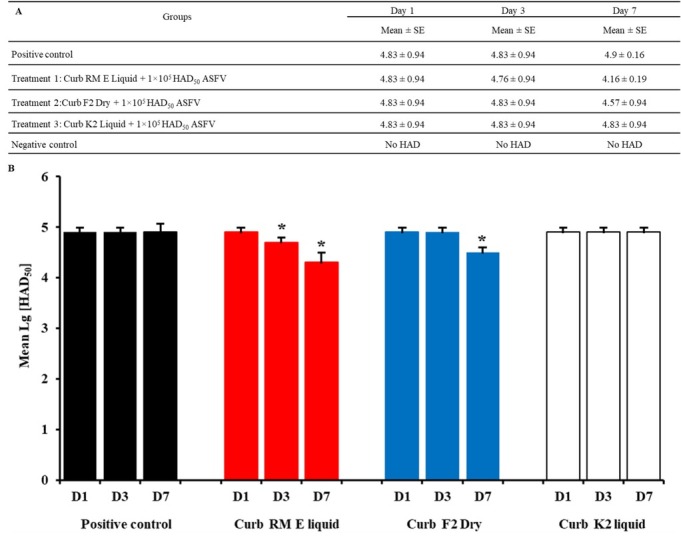
(A, B). The combination of feed with the Sal CURB RM E Liquid, Sal CURB F2 Dry or Sal CURB K2 Liquid and 1×105/mL HAD50 of VN/Pig/HN/19 strain one, three, and seven days post inoculation. Values represent mean and standard deviation results from three independent experiments. Significant differences compared to control are denoted by * for P < 0.05 and ** for P < 0.01
Discussion
In this study, we successfully isolated the first ASFV from field outbreak samples in Vietnam. The isolate, VN/Pig/HN/19, was characterised using the HAD assay, sequencing analysis, and real-time PCR. Our results demonstrated that, based on the p72 and EP402R genes encoding the CD2v protein genes, the VN/Pig/HN/19 strain isolated in Vietnam shared 100% nucleotide identity with virus strains isolated in Georgia, Russia, Estonia, and China such as Georgia 2007/1, Krasnodar 2012, Irkutsk 2017, Estonia 2014, ASFV-SY18 or China/Jilin/2018/boar (15, 17, 27). The results indicated that the ASFV VN/Pig/HN/19 strain belongs to genotype II, serogroup 8, and is genetically close to ASFV prevalent in Eastern Europe and China. It replicated efficiently in primary PAM cells and its titre reached 106 HAD50/ml. The VN/Pig/HN/19 strain of ASFV is highly virulent and causes acute ASF in domestic pigs. Since the ASF outbreak in Ha Nam, subsequent ASF outbreaks occurred in Vietnam from February to November 2019 (outbreaks were officially reported in 63 provinces and cities out of 63), raising concerns that ASFV will be introduced into a total population of 27 million pigs in Vietnam. This continued and far-reaching spread of ASFV in Vietnam demonstrates the threat of the disease’s emergence and deeper spread to Southeast Asia.
Recent studies indicated that the susceptibility to chemicals and disinfectants depends on viral characteristics in which non-enveloped viruses are more resistant than enveloped ones (12). Additionally, it was demonstrated that some chemicals or extracts from plants such as acacetin, apigenin, genkwanin, rhoifolin, vitexin, and vitexin 2-O-rhamnoside can inhibit or reduce ASFV-specific protein synthesis and viral multiplication in cell culture. ASFV-infected Vero cells continuously treated with apigenin did not display a cytopathic effect (1, 10, 11) but there is limited research investigating the survival of ASFV in feed after treatment with chemicals or antimicrobials. Based on the results of the feed assay, the liquid and dry Sal CURB-branded antimicrobial product-treated feed showed activity against ASFV VN/Pig/HN/19 after one, three, and seven days post inoculation. The positive control feed contained ASFV VN/Pig/HN/19 strain at a scarcely changing level after one, three, or seven days post inoculation and there was no difference in mean Cq and viral titres. Contrastingly, Cq values and the viral titre of ASFV VN/Pig/HN/19 strain in feed treated with Sal CURB RM E Liquid decreased after one, three, or seven days post inoculation and were highly reduced after seven days post inoculation. One interpretation of this observation in conjunction with the swine feed assay data is that the Sal CURB RM E Liquid product had a suppressive effect on viral load and viability, while in its absence the quantity of ASFV remained constant and the virus survived over time. Although it is difficult to be definitive regarding how Sal CURB RM E Liquid shows antimicrobial activity against ASFV infection in feed, our results suggest that Sal CURB RM E Liquid inhibits or reduces ASFV replication in feed. In future studies, we should investigate whether the application of the liquid and dry antimicrobial products may possibly reduce the risk of the introduction of emerging and reemerging ASFV through swine feed. Finally, as feed biosecurity is important for the swine industry, veterinarians, producers, and representatives from the feed industry will need to work together and pursue novel means to implement such a strategy. This aspect is critical as not all countries around the globe have regulations which approve mitigants for use in animal feeds, and many countries have different policies regarding treatment, bio-security, and eradication of ASFV.
In conclusion, in this study an ASFV strain, VN/Pig/HaNam/2019 (VN/Pig/HN/19), was isolated in primary PAMs from a pig sample from an ASF outbreak on a farm in Ha Nam province, Vietnam. The isolate was characterised by the HAD test and real-time PCR. Phylogenetic analysis of the viral p72 and EP402R gene encoding the CD2v protein gene revealed that VN/Pig/HN/19 belongs to genotype II and serogroup 8. Infectious titres of the virus propagated in primary PAMs were as high as 106 HAD50/ml. It is still not known if a single virus crossed into Vietnam and caused all these outbreaks or different ASFV strains are responsible for the Vietnam outbreaks. More viruses need to be isolated and analysed to fully understand the spread of the disease and to develop an effective control strategy. Our study represents an important first report on the activity of antimicrobial Sal CURB RM E Liquid against ASFV. Our feed assay findings suggest that this compound has an inhibitory effect on ASFV replication. These results indicate that the antimicrobial Sal CURB RM E Liquid may be a potential candidate as an antiviral feed or water mitigant for ASFV infection. Therefore, further studies addressing the use of this antimicrobial in vivo are required.
Acknowledgments
The authors would like to thank Kemin Industries for providing the funding and in-kind resources necessary to complete this project. Thanks to Dr Pham Minh Hang from the Department of Epidemiology and Pathology of NIVR for support in performing the real-time PCR.
Footnotes
Conflict of Interest
Conflict of Interest Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This work was supported by the Ministry of Science and Technology in a disbursement to Hoang Vu Dang (Project code: ĐTĐL.CN-75/19).
Supplements comprise a separate pdf file viewable online at http://content.sciendo.com/view/journals/jvetres/jvetres-overview.xml and doi:10.2478/jvetres-2020-0041.
Animal Rights Statement: The study was conducted in compliance with the institutional rules for the care and use of laboratory animals and using a protocol approved by the Ministry of Agriculture and Rural Development (MARD) Vietnam (TCVN 8402:2010).
References
- 1.Abba Y., Hassim H., Hamzah H., Noordin M.M.. Antiviral activity of resveratrol against human and animal viruses. Adv Virol. 2015;2015:184241. doi: 10.1155/2015/184241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastos A.D., Penrith M.L., Cruciere C., Edrich J.L., Hutchings G., Roger F., Couacy-Hymann E.. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch Virol. 2003;148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- 3.Berge A.C., Wierup M.. Nutritional strategies to combat Salmonella in mono-gastric food animal production. Animal. 2012;6:557–564. doi: 10.1017/S1751731111002217. [DOI] [PubMed] [Google Scholar]
- 4.Brown T.P., Garcia A., Kelley L.. Spiking mortality of turkey poults: 1. Experimental reproduction in isolation facilities. Avian Dis. 1997;41:604–609. [PubMed] [Google Scholar]
- 5.Brown T.P., Garcia A., Kelly L.. Spiking mortality of turkey poults: 2. Effect of six different in vitro disinfection techniques on organ homogenates capable of reproducing SMT. Avian Dis. 1997;41:906–909. [PubMed] [Google Scholar]
- 6.Cubillos C., Gomez-Sebastian S., Moreno N., Nunez M.C., Mulumba-Mfumu L.K., Quembo C.J., Heath L., Etter E.M., Jori F., Escribano J.M., Blanco E.. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. 2013;173:159–167. doi: 10.1016/j.virusres.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Dee S., Neill C., Clement T., Christopher-Hennings J., Nelson E.. An evaluation of a liquid antimicrobial (Sal CURB(R)) for reducing the risk of porcine epidemic diarrhea virus infection of naive pigs during consumption of contaminated feed. BMC Vet Res. 2014;10:220. doi: 10.1186/s12917-014-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galindo I., Alonso C.. African swine fever virus: a review. Viruses. 2017;10:9. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallardo C., Nieto R., Soler A., Pelayo V., Fernandez-Pinero J., Markowska-Daniel I., Pridotkas G., Nurmoja I., Granta R., Simon A., Perez C., Martin E., Fernandez-Pacheco P., Arias M.. Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in Eastern European Union countries: how to improve surveillance and control programs. J Clin Microbiol. 2015;53:2555–2565. doi: 10.1128/JCM.00857-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakobyan A., Arabyan E., Avetisyan A., Abroyan L., Hakobyan L., Zakaryan H.. Apigenin inhibits African swine fever virus infection in vitro. Arch Virol. 2016;161:3445–3453. doi: 10.1007/s00705-016-3061-y. [DOI] [PubMed] [Google Scholar]
- 11.Hakobyan A., Arabyan E., Kotsinyan A., Karalyan Z., Sahakyan H., Arakelov V., Nazaryan K., Ferreira F., Zakaryan H.. Inhibition of African swine fever virus infection by genkwanin. Antiviral Res. 2019;167:78–82. doi: 10.1016/j.antiviral.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Juszkiewicz M., Walczak M., Woźniakowski G.. Characteristics of selected active substances used in disinfectants and their virucidal activity against ASFV. J Vet Res. 2019;63:17–25. doi: 10.2478/jvetres-2019-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor R., Sharma B., Kanwar S.S.. Antiviral phytochemicals: an overview. Biochem Physiol. 2017;6:220. doi: 10.4172/2168-9652.1000220. [DOI] [Google Scholar]
- 14.King D.P., Reid S.M., Hutchings G.H., Grierson S.S., Wilkinson P.J., Dixon L.K., Bastos A.D., Drew T.W.. Development of a taqman PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods. 2003;107:53–61. doi: 10.1016/s0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 15.Kolbasov D., Titov I., Tsybanov S., Gogin A., Malogolovkin A.. African swine fever virus, Siberia, Russia, 2017. Emerg Infect Dis. 2018;24:796–798. doi: 10.3201/eid2404.171238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S., Stecher G., Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Wang Q., Ge S., Liu Y., Liu C., Liu F., Hu Y., Li J., Bao J., Ren W., Zhang Y., Xu T., Sun C., Li L., Wang S., Fan X., Huang B., Wu X., Wang Z.. Infection of African swine fever in wild boar, China, 2018. Transbound Emerg Dis. 2019;66:1395–1398. doi: 10.1111/tbed.13114. [DOI] [PubMed] [Google Scholar]
- 18.Malmquist W.A., Hay D.. Hemadsorption and cytopathic effect produced by African swine fever virus in swine bone marrow and buffy coat cultures. Am J Vet Res. 1960;21:104–108. [PubMed] [Google Scholar]
- 19.Niederwerder M.C., Stoian A.M.M., Rowland R.R.R., Dritz S.S., Petrovan V., Constance L.A., Gebhardt J.T., Olcha M., Jones C.K., Woodworth J.C., Fang Y., Liang J., Hefley T.J.. Infectious dose of African swine fever virus when consumed naturally in liquid or feed. Emerg Infect Dis. 2019;25:891–897. doi: 10.3201/eid2505.181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2012, Chapter 2.8.1 African swine fever. World Organisation for Animal Health (OIE); Paris: 2012. [Google Scholar]
- 21.Quembo C.J., Jori F., Vosloo W., Heath L.. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound Emerg Dis. 2018;65:420–431. doi: 10.1111/tbed.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed L.J., Muench H.. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–497. [Google Scholar]
- 23.Sanna G., Dei Giudici S., Bacciu D., Angioi P.P., Giammarioli M., De Mia G.M., Oggiano A.. Improved strategy for molecular characterization of African swine fever viruses from Sardinia, based on analysis of p30, CD2V, and I73R/I329L variable regions. Transbound Emerg Dis. 2017;64:1280–1286. doi: 10.1111/tbed.12504. [DOI] [PubMed] [Google Scholar]
- 24.Simulundu E., Sinkala Y., Chambaro H.M., Chinyemba A., Banda F., Mooya L.E., Ndebe J., Chitanga S., Makungu C., Munthali G., Fandamu P., Takada A., Mweene A.S.. Genetic characterisation of African swine fever virus from 2017 outbreaks in Zambia: identification of p72 genotype II variants in domestic pigs. Onderstepoort J Vet Res. 2018;85:e1–e5. doi: 10.4102/ojvr.v85i1.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wales A.D., Allen V.M., Davies R.H.. Chemical treatment of animal feed and water for the control of Salmonella. Foodborne Pathog Dis. 2010;7:3–15. doi: 10.1089/fpd.2009.0373. [DOI] [PubMed] [Google Scholar]
- 26.Wales A.D., McLaren I., Rabie A., Gosling R.J., Martelli F., Sayers R., Davies R.. Assessment of the anti-Salmonella activity of commercial formulations of organic acid products. Avian Pathol. 2013;42:268–275. doi: 10.1080/03079457.2013.782097. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X., Li N., Luo Y., Liu Y., Miao F., Chen T., Zhang S., Cao P., Li X., Tian K., Qiu H.J., Hu R.. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. 2018;65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]


