Abstract
Cherubism is a rare genetic disorder caused primarily by mutations in SH3BP2 resulting in excessive bone resorption and fibrous tissue overgrowth in the lower portions of the face. Bone marrow derived cell cultures derived from a murine model of cherubism display poor osteogenesis and spontaneous osteoclast formation. To develop a deeper understanding for the potential underlying mechanisms contributing to these phenotypes in mice, we compared global gene expression changes in hematopoietic and mesenchymal cell populations between cherubism and wild type mice. In the hematopoietic population, not surprisingly, upregulated genes were significantly enriched for functions related to osteoclastogenesis. However, these upregulated genes were also significantly enriched for functions associated with inflammation including arachidonic acid/prostaglandin signaling, regulators of coagulation and autoinflammation, extracellular matrix remodeling, and chemokine expression. In the mesenchymal population, we observed down regulation of osteoblast and adventitial reticular cell marker genes. Regulators of BMP and Wnt pathway associated genes showed numerous changes in gene expression, likely implicating the down regulation of BMP signaling and possibly the activation of certain Wnt pathways. Analyses of the cherubism derived mesenchymal population also revealed interesting changes in gene expression related to inflammation including the expression of distinct granzymes, chemokines, and sulfo-transferases. These studies reveal complex changes in gene expression elicited from a cherubic mutation in Sh3bp2 that are informative to the mechanisms responding to inflammatory stimuli and repressing osteogenesis. The outcomes of this work are likely to have relevance not only to cherubism, but other inflammatory conditions impacting the skeleton.
Keywords: Cherubism, Stromal cell, Osteoblast, Osteoclast, Bone, Inflammation
1. Introduction
Cherubism is a rare autosomal dominant craniofacial disorder af-fecting pre-pubertal children. It was first described in 1933 as a multilocular cystic dysplasia of the jaw that led to outward bulging of cheek and upward shift of eyes that gave a ‘cherub’ like appearance to these children [1]. Extensive bone resorption, fibrous tissue overgrowth and immune cell invasion in jawbones further characterize the cherubism phenotype [2]. In most cases, the disease is known to self-regress after puberty over a period of several years [3–5]. However, in more ag-gressive cases, surgical interventions are required to restore severe facial deformation and normal functions like eating and breathing. The majority of cherubism cases (80%) are caused by gain-of-function mutations in Src Homology 3-domain binding protein 2 (SH3BP2), which is located on chromosome 4 (4p16.3 region) in humans [6,7].
SH3BP2 is a cytoplasmic adaptor protein that plays an indispensable role in relaying signals from various cell surface molecules to their downstream effectors by assembling stable enzyme-substrate com-plexes. In humans, it is a 561-aa protein that consists of an N-terminal pleckstrin homology (PH) domain, a proline-rich Src Homology 3 (SH3) binding domain and a C-terminal Src Homology 2 (SH2) domain. SH3BP2 has been shown to take part in various cellular processes by its involvement in key signaling pathways, including the Syk and Src family of kinases [8]. Protein levels of SH3BP2 are itself regulated through tankyrase mediated ADP ribosylation followed by RNF146 mediated ubiquitination and proteasomal degradation [9,10]. There is evidence to suggest that cherubism mutations in SH3BP2 lead to dis-ruption in tankyrase binding resulting in overaccumulation of SH3BP2 protein in cells [9]. Consequently, it is generally assumed that SH3BP2 associated signaling pathways are increased, which ultimately leads to the pathology of cherubism. However, why this mechanistic outcome would contribute to a disease that is largely restricted to the craniofacial region remains a mystery.
To better understand the physiological and molecular pathology of cherubism, a mouse model of the disease was generated by knocking-in one of the most common mutations Pro416Arg into Sh3bp2 (Sh3bp2KI/ KI) [11]. While the generation of Sh3bp2KI/KI mice has not faithfully recapitulated the selective craniofacial aspects of cherubism patients, nonetheless, the animal model has yielded valuable evidence on the dysfunction of Sh3bp2 in certain cell types, such as the heightened activation of inflammatory and RANK signaling pathways that systemically impacts bone resorption [11–13].
One of the unexplained etiologies of cherubism relates to the excessive fibrous tissue overgrowth that occurs, which is analogous to other fibrotic conditions. Consistent with fibrotic tissue, mandibular and maxillary lesions in cherubism patients contain a complex mixture of cell types of hematopoietic and mesenchymal origin, both of which are likely to have roles in perpetuating the fibrotic state. While not of craniofacial origin, adherent bone marrow cultures from long bones contain a rich mixture of hematopoietic and mesenchymal cell types and past work by us has shown that bone marrow cells derived from cherubism mice display poor osteogenesis and spontaneously form osteoclasts [14]. Further, the osteoblasts in these cultures not only fail to mature but typically organize into swirling patterns possibly suggesting the existence of fibrotic mechanisms. Consistent with this thinking, we showed that decreasing TGFβ signaling, a major fibrotic pathway, could decrease osteoclast formation and rescue osteoblast differentiation [14]. Unfortunately, in our hands, anti-TGFβ treatment in cherubism mice did not alleviate bone resorption or overall animal health (data not shown). Therefore, to develop a broader understanding of the mechanisms occurring between hematopoietic and mesenchymal cell lineages, we characterized global gene expression patterns in sorted hematopoietic and mesenchymal cells from day 7 bone marrow cultures derived from femurs using RNA sequencing (RNA-Seq). Here we report our findings from differential expression analyses, which revealed surprising alterations in gene expression both within the hematopoietic and mesenchymal cell lineages that are likely to be relevant not just to cherubism, but to a variety of inflammatory skeletal diseases.
2. Methods
2.1. Reagents
For cell culture, MEM Alpha (12571–063), PBS (10010–023), HEPES 1M (15630–080), 100× Penicillin-Streptomycin (15140122), and Hanks’ balanced salt solution 10× (HBSS) (14185–052) were obtained from Life Technologies. Fetal Bovine Serum (FBS) (S01520) was purchased from Biowest and Accutase (AT104) was purchased from Innovative Cell Technologies.
For cell sorting, FITC conjugated mouse anti-CD45 (130–110-796) and APC conjugated mouse anti-SCA-1 (130–102-833) antibody were bought from Miltenyi Biotec. SUPERase In RNase Inhibitor (20 U/μL) (AM2694) was from ThermoFisher Scientific. UltraPure distilled water (DNase and RNase free) (10977–015) was obtained from Invitrogen by Life Technologies. RNase free BSA (B6917) was obtained from Sigma--Aldrich. RNAprotect cell reagent (76526) was obtained from Qiagen.
For RNA purification, library preparation, and sequencing, Direct-zol RNA Miniprep Plus (R2072) was purchased from Zymo Research. SMARTer Stranded RNA-Seq Kit (634836) and RiboGone-Mammalian (634847) were obtained from Takara Biosciences. Agencourt AMPure XP, 5 mL (A63881) was bought from Beckman Coulter. The sequencing kit, NextSeq® 500/550 High Output Kit v2 (150 cycles) (TG-160–2002), was bought from Illumina. Experion RNA HighSens Analysis Kit (7007105) was purchased from Bio-Rad. High Sensitivity DNA Kit (5067–4626) was obtained from Agilent. Qubit dsDNA HS Assay Kit (Q32851) was bought from ThermoFisher Scientific. NEBNext Library Quant Kit for Illumina (E7630S) was bought from New England Biolabs.
2.2. Animals
Sh3bp2 KI/+ mice were generously provided by Dr. Yasuyoshi Ueki. Col1a1–2.3 EGFP osteoblast reporter mice were generously provided by Dr. David Rowe. Male and female Sh3bp2KI/+ mice were intercrossed with Col1a1–2.3 EGFP mice to generate experimental animals, which were maintained on a C57BL/6 background. Genotyping by PCR for the Sh3bp2 knock-in allele was performed using PCR primers 5′-CCACTA TATGACAATACCTG-3′ and 5′-CATAGTCTTCATCTGAGTCC-3′. Animal studies were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and animal protocols were approved by the Animal Care Committee of the University of Connecticut Health (approved protocols 101060–0518, 101777–0121).
2.3. Primary bone marrow stromal cell cultures
Bone marrow cultures were prepared from 6- to 8-week old mice. Hindlimbs were dissected out and placed in ice cold PBS. Under a cell culture hood, muscle and connective tissues were removed from long bones using a scalpel. Bones were then cut in half to expose the bone marrow cavity and kept wet in cell culture media until all bones were prepared. Long bone halves were vertically inserted with marrow opening facing down in a 1.5 mL microfuge tube containing precut-pipet tips to hold bone pieces. 200 μL of cell culture media was added to each microfuge tube to keep cells wet following centrifugation. Bones were centrifuged at 6000 rpm for 1 min to spin out bone marrow cells. Bone pieces and pipet tips were removed from microfuge tubes under the c ll culture hood with sterile forceps. Cell pellets were resuspended in cell culture media by gentle pipetting. Extra cell culture media (0.5 mL–1 mL) was added to facilitate cell suspension to generate a single cell suspension. Cell suspensions were then filtered through a 70-μm cell strainer. Cells obtained from one mouse were plated in one 10 cm tissue culture dish or one six-well plate with 15 mL of media. On the third day of culture, cells were washed with PBS and re-supple-mented with media.
2.4. Flow cytometry
On day 7 of culture, adherent cells were washed twice with PBS, digested with 5 mL of accutase (Innovative Cell Technologies) for 5 min at 37 °C, gently scraped and pipetted up and down to generate a single cell suspension. 10 mL of PBS was added to dilute accutase. Cells were then centrifuged and resuspended in FACS staining buffer (1× HBSS, 10 mM HEPES, 2% BSA (RNase free), 2 mM EDTA, pH 7.4). Cell surface staining was carried out with anti-Sca1-APC and anti-CD45-FITC according to manufacturer’s recommendations (Miltenyi Biotech). To a small volume of FACS staining buffer, Sytox blue was added to detect and remove dead cells during FACS, and SUPERase In RNase Inhibitor was added to prevent RNA degradation. Sorting was carried out on a Becton-Dickinson FACS-ARIA II (UCH Flow Cytometry Core). Two populations Sca1+CD45− (mesenchymal stromal cells) and Sca1+CD45+ (hematopoietic cells) were collected into RNA protect obtained from Qiagen.
2.5. RNA purification
RNA was isolated from the two sorted cell populations using Zymo Research’s Direct-zol™ RNA Miniprep Plus kit according to manufacturer’s instructions. RNA samples were quantified by NanoDrop 1000 Spectrophotometer (Thermo Scientific) and RNA integrity was assessed by Agilent Bioanalyzer 2100 (Agilent). RNA samples with RNA Integrity Number (RIN) higher than or equal to 8.5 were used for library preparation. Six biological replicates of each group, i.e., WT stromal cells, WT hematopoietic cells, Sh3bp2KI/KI stromal cells, and Sh3bp2KI/KI hematopoietic cells were selected for library preparation.
2.6. Library preparation and validation
RNA-Seq libraries were prepared using SMARTer® Stranded RNA-Seq Kit from Clontech Laboratories, Inc. (currently known as Takara Biosciences) according to manufacturer’s instructions. Briefly, ribo-somal RNA (rRNA) was depleted from 100 ng of RNA sample. rRNA depleted samples were then subjected to SMARTer First-strand cDNA synthesis. cDNA was then purified using magnetic bead pull-down. cDNA samples were amplified for 16 cycles to generate RNA-seq libraries. RNA-Seq libraries were purified using magnetic bead pull-down. Quality check and library validation were performed using Agilent 2100 Bioanalyzer and the High Sensitivity DNA Chip from Agilent’s High Sensitivity DNA Kit. Libraries were quantified through qPCR using the NebNext quant kit specifically designed to quantify Illumina NextSeq libraries.
2.7. Sequencing run
Libraries were pooled in batches of 12 and requantified using pi-cogreen dye and reading the fluorescence through Qubit 3.0 Fluorometer. Pooled libraries were denatured prior to the sequencing run. Sequencing was performed on Illumina NextSeq 500 platform using NextSeq 500/550 High Output v2 kit (150 cycles) with 75 bp paired end reads.
2.8. Quality check and RNA-seq data analyses
Quality of the raw sequenced reads was assessed with the help of FastQC software [15]. MultiQC was used to generate cumulative results from FastQC output files. Sickle was used to trim low quality reads and improve the quality of the reads [16]. Reads were then mapped to mouse reference genome (mm10) using STAR [17]. The output files were then counted for reads mapping to each gene using the htseq_-count function from HTseq [18], a python-based framework designed to process high throughput sequencing data. These count files were then subjected to DESeq2 analysis in R/Bioconductor packages [19]. DESeq2 analysis normalized the reads and provided a list of differentially expressed genes (DEGs). The list of DEGs were examined using a variety of web accessible resources including Ingenuity Pathway Analysis software (Qiagen), Gene Ontology (http://geneontology.org; [20,21]), PathVisio (Pathvisio.org; [22]), and Wikipathways [23,24] to identify significant associations and molecular pathways and make predictions regarding upstream regulators. Raw and processed RNA-Seq data was deposited into the Gene Expression Omnibus (GEO) database and can be found using the accession number GSE139421.
2.9. Quantitative RT-PCR gene expression analyses
RNA purification was carried out on FACS sorted day 7 bone marrow stromal cultures according to the manufacturer’s recommendations (Macherey-Nagel and Zymo Research). cDNA was prepared from 500 ng of RNA/sample using the SuperScript II Reverse Transcriptase kit or Protoscript II kit (Life Technologies, NEB). QPCR was carried out using I-Taq Universal SybrGreen Supermix (Bio-Rad) in a Bio-Rad CFX real-time thermocycler. PCR primer sequences used for gene expression analyses and RNA-Seq validation are listed in Supplementary Table 4.
3. Results
Previously, we reported that cultured bone marrow cells derived from a cherubism mouse model (Sh3bp2KI/KI), in contrast to wild type littermates, displayed defects in osteoblast differentiation while at the same time spontaneously forming osteoclasts [14]. To develop a deeper understanding for the mechanistic differences taking place between cherubism and wild type cells in this culture system, we performed an RNA-Seq study. For this, we decided to separate hematopoietic and mesenchymal cell fractions by FACS on day 7 of culture using the cell surface markers CD45 and SCA1, where CD45+, SCA1+ defined the hematopoietic population and CD45−, Sca1+ defined the mesenchymal population (sFig. 1A) [25–28]. At day 7 of culture, the ratio of hematopoietic cells to mesenchymal cells was consistently 1:1 from both wild type and cherubism mice. RNA from six biological replicates were used for library preparation and sequencing. The high quality of RNA sequencing was confirmed using FASTQC software and outputs were merged with the help of MultiQC software (sFig. 1B) [15,29]. An RNASTAR → HTSeq → DESeq2 work flow was used to align, count, and identify differentially expressed genes [17–19]. Normalization and data analyses were carried out to generate a list of differentially expressed genes (DEGs) comparing cherubism to wild type samples for both hematopoietic and mesenchymal cell populations. Further, we examined the expression of known hematopoietic and mesenchymal cell markers within the wild type sample groups to substantiate the enrichment of cell types by FACS (sFig. 2). DEGs reported here had an additional base mean intensity cut off of ≥50 counts and a log2 fold change ≥ ± 1 with an adjusted p-value > 0.05.
3.1. Differential gene expression in the hematopoietic cell population
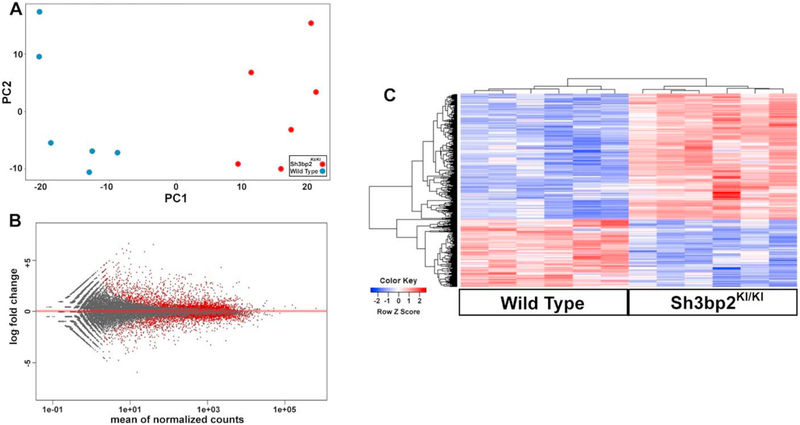
Principal component analysis of the hematopoietic population grouped the wild type (blue) and Sh3bp2KI/KI (red) samples separately indicating differentially expressed gene signatures (Fig. 1A). To ensure no transcriptional bias existed between wild type and Sh3bp2KI/KI samples, an MA plot was also generated (Fig. 1B). The MA plot showed us that the dataset was appropriately normalized as the mean line (red line) passed through 0 on the y-axis. Thus, the PCA and MA plots gave us confidence that the workflow parameters used to align, count, normalize, and identify differentially expressed genes in the dataset were appropriate.
Fig. 1.
Broad comparative analyses of RNAseq outcomes generated from the cherubism and wild type hematopoietic populations. (A) Principal component analysis reveals two distinct groups along the first principle component. (Blue = WT, Red = Cherubism). (B) MA plot indicating proper data normalization. Red dots indicate genes displaying significant differential gene expression. (C) Heatmap view of differential gene expression identifying 299 upregulated genes and 162 downregulated genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To visualize global gene expression differences, a heatmap was generated based on the above mentioned cutoffs that resulted in a total of 461 differentially expressed genes within the hematopoietic population (Fig. 1C). Of the total differentially expressed genes, 299 were upregulated and 162 were downregulated in the Sh3bp2KI/KI hematopoietic population. A list of the differentially expressed genes included in the heatmap and those between Log2 Fold > 1 and < −1 are provided in Supplementary Table 1. To further understand the differentially expressed gene set, we performed gene ontology analyses on both upregulated and downregulated gene sets [30]. Consistent with our previous study, which reported spontaneous osteoclast formation, gene ontology analyses of upregulated genes showed positive regulation of bone resorption and osteoclast differentiation as the highest fold enrichment score (Table 1). Also consistent with other reports detailing an inflammatory phenotype in cherubism [11–13], gene ontology analyses showed enrichment of genes associated with regulation of inflammation and myeloid cell differentiation (Table 1). Gene ontology analyses of downregulated genes indicated that interleukin 2 regulation, lympho-cyte chemotaxis, migration, and negative regulators of cell growth were significantly enriched (Table 2).
Table 1.
GO terms generated from upregulated genes found in the cherubism hematopoietic population.
| GO biological process | Fold enrichment | P–valuea |
|---|---|---|
| Positive regulation of bone resorption (G0:0045780) | 19.51 | 1.42E-02 |
| Positive regulation of bone remodeling (G0:0046852) | 19.51 | 1.42E-02 |
| Osteoclast differentiation (G0:0030316) | 15.96 | 9.80E-04 |
| Positive regulation of tissue remodeling (G0:0034105) | 15.36 | 7.81E-03 |
| Positive regulation of myeloid leukocyte differentiation (GO:0002763) | 13.17 | 6.86E-04 |
| Tissue remodeling (G0:0048771) | 10.47 | 1.10E-05 |
| Positive regulation of myeloid cell differentiation (GO:0045639) | 10.06 | 3.07E-04 |
| Regulation of myeloid leukocyte differentiation (G0:0002761) | 7.2 | 2.22E-02 |
| Positive regulation of leukocyte differentiation (GO: 1902107) | 7.04 | 8.67E-04 |
| Positive regulation of hemopoiesis (GO: 1903708) | 6.49 | 2.48E-04 |
| Regulation of myeloid cell differentiation (G0:0045637) | 5.73 | 7.88E-03 |
| Extracellular matrix organization (G0:0030198) | 4.9 | 4.10E-02 |
| Extracellular structure organization (G0:0043062) | 4.62 | 3.35E-02 |
| Inflammatory response (G0:0006954) | 3.89 | 7.60E-03 |
Generated using a Fisher's Exact test and a Bonferroni correction.
Table 2.
GO terms generated from downregulated genes found in the cherubism hematopoietic population.
| GO biological process | Fold enrichment | P–valuea |
|---|---|---|
| Positive regulation of interleukin-2 production (GO:0032743) | 26.19 | 2.19E-02 |
| Lymphocyte chemotaxis (G0:0048247) | 25.79 | 2.25E-03 |
| Lymphocyte migration (G0:0072676) | 18.29 | 1.42E-02 |
| Regulation of interleukin-2 production (G0:0032663) | 17.34 | 1.89E-02 |
| Negative regulation of developmental growth (G0:0048640) | 10.9 | 9.99E-03 |
| Positive regulation of leukocyte proliferation (G0:0070665) | 9.25 | 3.20E-02 |
| Morphogenesis of a branching epithelium (G0:0061138) | 7.86 | 2.80E-02 |
| Morphogenesis of a branching structure (G0:0001763) | 7.47 | 4.16E-02 |
| Regulation of leukocyte proliferation (G0:0070663) | 7.1 | 1.87E-02 |
| Regulation of peptidyl-tyrosine phosphorylation (G0:0050730) | 6.55 | 3.75E-02 |
| Regulation of leukocyte differentiation (GO: 1902105) | 6.47 | 1.37E-02 |
| Positive regulation of response to external stimulus (G0:0032103) | 6.23 | 1.95E-02 |
| Positive regulation of cell adhesion (G0:0045785) | 5.84 | 1.63E-03 |
Generated using a Fisher's Exact test and a Bonferroni correction.
To appreciate the identified gene ontology categories, we examined the dataset for genes associated with osteoclast formation and inflammation. Numerous gene markers of osteoclast differentiation were highly upregulated in the hematopoietic dataset (Table 3). These in-cluded, but were not limited to Acp5, the gene that encodes for the well-known osteoclast secreted acid phosphatase best known as TRAP (tar-trate resistant acid phosphatase) [31]. Atp6v0d2, a gene that encodes for a vacuolar ATPase important for extracellular H+ transport, but also has a role in osteoclast cell fusion [32]. Calcr, a gene that encodes for a receptor that binds calcitonin, a peptide hormone that down regulates bone resorption [33,34]. Proteases cathepsin k (Ctsk) and MMP9, which are highly expressed in osteoclasts and play a role in bone matrix degradation [35–37]. Dcstamp and Ocstamp are genes that encode for transmembrane proteins that participate in osteoclast fusion [38–40]. Integrin β3 (Itgb3) and Oscar are genes that encode for receptors that bind matrix proteins and provide critical signaling cues for osteoclast differentiation [41,42]. Src is a tyrosine kinase whose function is essential for osteoclast activity [43].
Table 3.
Osteoclast upregulated genes found in the cherubism hematopoietic population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Acp5 | Acid phosphatase 5, tartrate resistant (TRAP) | 2.788 | 0.00121 | 5693.97 |
| Atp6v0d2 | ATPase, H + transporting, lysosomal VO subunit D2 | 2.418 | 2.72E-13 | 4674.68 |
| Calcr | Calcitonin receptor | 2.560 | 0.002019 | 103.13 |
| Ctsk | Cathepsin-K | 2.338 | 0.006951 | 10,651.08 |
| Dcstamp | Dendritic cell-specific transmembrane protein | 2.758 | 3.86e-08 | 186.59 |
| Itgb3 | Integrin beta 3 | 2.639 | 1.02e-07 | 877.13 |
| Mmp9 | Matrix metallo-protease 9 | 2.913 | 2.90e-05 | 6047.38 |
| Oscar | Osteoclast-associated receptor | 2.722 | 0.003044 | 67.56 |
| Ocstamp | Osteoclast stimulatory' transmembrane protein | 3.148 | 2.18e-07 | 239.88 |
| Src | Protein tyrosine kinase Src | 2.824 | 1.77e-22 | 877.23 |
Values given in counts.
A variety of genes associated with the regulation of inflammatory processes were also noted in the data set. In particular, genes associated with the production and signaling of certain eicosanoids (sFig. 3). This included Ptges, a gene that encodes for prostaglandin E synthase, which was upregulated Log2 Fold 2.6 in the Sh3bp2KI/KI group while the enzyme capable of breaking down prostaglandins, encoded by Hpdg, was downregulated Log2 Fold −2.9 (Table 4). Cytochrome P450 2s1 (Cyp2s1), which encodes for an enzyme that can metabolize PGG2 and PGH2 into 12-HHT and thromboxane A2 potentially at the expense of making PGE2, was also highly induced (Log2 Fold 3.33) [44–46]. Ephx2, an epoxide hyrolase that is involved in a different arm of ara-chidondic metabolism can convert EETs into DHETs to regulate inflammation was also upregulated (Log2 Fold 2.33) [47,48]. Other genes identified in the dataset that may regulate prostglandin signaling or be induced by prostaglandin signaling include annexin A1 (Log2 Fold 1.03) [49] and Saa3 (Log2 Fold 3.50) [50]. Interestingly, Annexin A1 and SAA ligands have been shown to bind formyl peptide receptors of which Fpr-1 was shown to be upregulated in the dataset [50,51].
Table 4.
Inflammatory upregulated genes found in the cherubism hematopoietic population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Anxal | Annexin A1 | 1.02 | 1.29E-16 | 4996.03 |
| Fprl | Formyl peptide receptor 1 | 1.59 | 0.038477 | 275.73 |
| Saa3 | Serum amyloid A 3 | 3.50 | 1.30E-06 | 183.82 |
| Ptges | Prostaglandin E synthase | Z62 | 3.08E-11 | 249.83 |
| Hpdg | Hydroxyprostaglandin dehydrogenase 15 | −2.90 | 6.21E-09 | 979.01 |
| Ptgfrn | Prostaglandin F2 receptor negative regulator | 2.36 | 1.67E-32 | 819.68 |
| Cyp2sl | Cytochrome P450, family 2, subfamily s, polypeptide 1 | 3.33 | 0.000127 | 221.43 |
| Ephx2 | Epoxide hydrolase 2, cytoplasmic | Z33 | 0.009313 | 81.45 |
| F10 | Coagulation factor X | Z16 | 3.57E-08 | 64.10 |
| F2r | Coagulation factor II (thrombin) receptor | Z53 | 3.97E-05 | 179.81 |
| Pla2g7 | Phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | 1.29 | 1.01E-12 | 6942.66 |
| Mefv | Mediterranean fever | 2.58 | 1.58E-13 | 676.89 |
| NlrplO | NLR family, pyrin domain containing 10 | 1.04 | 1.56E-06 | 62.05 |
| Tnipl | TNFAIP3 interacting protein 1 | 1.05 | 6.46E-15 | 1993.44 |
| C3 | Complement component 3 | 2.09 | 2.75E-13 | 1444.77 |
| Chstl | Carbohydrate sulfotransferase 1 | 1.20 | 0.000243 | 62.16 |
| Ecml | Extracellular matrix protein 1 | 1.06 | 7.00E-05 | 1123.96 |
| Ndstl | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 1.05 | 1.56E-11 | 1377.85 |
| Rasgrpl | RAS guanyl releasing protein 1 | 1.21 | 0.00972 | 103.50 |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 1.49 | 0.000147 | 126.42 |
| Ccl9 | Chemokine (C—C motif) ligand 9 | 1.08 | 0.007658 | 2368.56 |
| Hp | Haptoglobin | 1.57 | 0.001678 | 117.32 |
Values given in counts.
A second group of inflammatory genes were associated with the coagulation cascade. Coagulation factor X (Log2 Fold 2.16) and Coagulation factor II receptor (F2r; Log2 Fold 2.53) also known as Par-1 were both highly upregulated. Additionally, two more genes associated with the coagulation cascade Von Willdebrand Factor and Protein C receptor were upregulated, but just below the Log2Fold cutoff in the dataset. Interestingly, work by others has shown that Activated Protein C can form a complex with Protein C Receptor and F2r to up-regulate target genes Tnfaip3, Birc3, and Sphk1 [52], which were all upregulated in the hematopoietic population, but again just below the Log2 fold cutoff (sFig. 4).
A third group of inflammatory genes have associations with different autoimflammatory disorders (sFig. 4). Mefv encodes for pyrin (Log2 Fold 2.58), an intracellular pattern recognition receptor capable of assembling inflammasome complexes [47]. While known for familial Mediterranean fever disease [53], mutations in Mefv have been associated with juvenile idiopathic arthritis, inflammatory bowel disease, Behcet’s disease, ulcerative colitis, Fibromyalgia, rheumatoid arthritis, and ankylosing spondylitis [54]. TNIP1 (Log2 Fold 1.05) has been implicated by genome wide association studies to be important for psor-iatic arthiritis, systemic sclerosis, systemic lupus erythematosus, and psoriasis [55]. Interestingly, the protein product of Tnip1, whose function is largely anti-inflammatory, can form a complex with Nlrp10 (Log2 Fold 1.04), which is also upregulated in the dataset and can negatively regulate protein levels of Tnip1 upon infection [56]. Nlrp10−/ − mice have also revealed that it is highly important for establishing an adaptive immune response [57]. Studies on Rasgrp1 (Log2 Fold 1.21), a guanine nucleotide exchange factor also implicate this gene in systemic lupus erythematosus and pulmonary alveolar proteinosis [58–60].
Finally, the last group of inflammatory genes has associations with hematopoietic cell migration. This group includes genes that encode for enzymes known to modify the extracellular matrix, Chst1 (Log2 Fold 1.20) and Ndst (Log2 Fold 1.05) thereby indirectly influencing cell migration and cell signaling. Interestingly, Ndst modification of the extracellular matrix has also been shown to negatively regulate BMP signaling [61] and thus, may impact osteoblast differentiation as well. Additional genes upregulated include chemokines Cxcl2 (Log2 Fold 1.49) and Ccl9 (Log2 Fold 1.08). Interesting, Ccl9 also plays a role in osteoclast formation and has been shown to be regulated by TGFβ signaling in myeloid progenitor cells [62,63].
3.2. Differential gene expression in the mesenchymal cell population
The computational analysis of the mesenchymal population was performed in a manner identical to the hematopoietic population de-tailed above. In brief, principal component analysis of the mesenchymal cell population grouped the wild type (blue) and Sh3bp2KI/KI (red) samples separately indicating differentially expressed gene signatures (Fig. 2A). An MA plot was also generated (Fig. 2B) with the mean line (red line) passed through 0 on the y-axis, indicating that no transcript-tional bias existed between wild-type and Sh3bp2KI/KI samples. Thus, the PCA and MA plots confirmed that the workflow parameters used to align, count, normalize, and identify differentially expressed genes in the dataset were appropriate.
Fig. 2.
Broad comparative analyses of RNAseq outcomes generated from the cherubism and wild type mesenchymal populations. (A) Principal component analysis reveals two distinct groups along the first principle component. (Blue = WT, Red = Cherubism). (B) MA plot indicating proper data normalization. Red dots indicate genes displaying significant differential gene expression. (C) Heatmap view of differential gene expression identifying 77 upregulated genes and 169 downregulated genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To visualize global gene expression differences, a heatmap was generated based on the above mentioned cutoffs that resulted in a total of 246 differentially expressed genes within the mesenchymal cell population (Fig. 2C). Of the total differentially expressed genes, 77 genes were upregulated and 169 genes were downregulated. A list of the differentially expressed genes included in the heatmap and those between Log2 Fold > 1 and < −1 are provided in Supplementary Table 2. To further understand the differentially expressed gene set, we performed gene ontology analyses on both upregulated and downregulated gene sets. Consistent with previous work [14], which reported impaired osteogenesis, gene ontology analyses of the downregulated gene set showed high enrichment scores related to bone mineralization, ossifi-cation, and osteoblast differentiation (Table 5). Additionally, consistent with cherubism mice having an inflammatory phenotype [12,13], gene ontology analyses of upregulated genes showed enrichment of genes associated granzyme mediated apoptotic signaling, proteoglycan synthesis, and chemotaxis (Table 6).
Table 5.
GO terms generated from downregulated genes found in the cherubism mesenchymal population.
| GO biological process | Fold enrichment | P–valuea |
|---|---|---|
| Bone trabecula morphogenesis (G0:0061430) | 25.73 | 3.56E-04 |
| Animal organ maturation (GCh0048799) | 23.47 | 4.70E-05 |
| Positive regulation of bone mineralization (G0:0030501) | 17.15 | 2.80E-06 |
| Endochondral ossification (G0:0001958) | 15.38 | 2.03E-04 |
| Replacement ossification (G0:0036075) | 15.38 | 2.03E-04 |
| Positive regulation of cartilage development (G0:0061036) | 14.86 | 2.28E-04 |
| Bone mineralization (G0:0030282) | 14.54 | 6.64E-06 |
| Negative regulation of endothelial cell apoptotic process (G0:2000352) | 14.38 | 2.56E-04 |
| Positive regulation of biomineral tissue development (G0:0070169) | 14.23 | 7.43E-06 |
| Positive regulation of osteoblast differentiation (G0:0045669) | 11.65 | 4.30E-06 |
| Regulation of bone mineralization (G0:0030500) | 11.43 | 9.95E-07 |
| Branching involved in blood vessel morphogenesis (G0:0001569) | 11.15 | 6.23E-04 |
Generated using a Fisher's Exact test and a Bonferroni correction.
Table 6.
GO terms generated from upregulated genes found in the cherubism mesenchymal population.
| GO biological process | Fold enrichment | P–valuea |
|---|---|---|
| Granzyme-mediated apoptotic signaling pathway (G0:0008626) | 91.1 | 3.08E-07 |
| Proteoglycan biosynthetic process (G0:0030166) | 20.88 | 5.46E-05 |
| Positive regulation of animal organ morphogenesis (G0:0110110) | 12.65 | 5.98E-05 |
| Limb morphogenesis (G0:0035108) | 8.95 | 6.89E-05 |
| Appendage morphogenesis (G0:0035107) | 8.95 | 6.89E-05 |
| Neuron projection guidance (G0:0097485) | 8.51 | 2.28E-05 |
| Chemotaxis (G0:0006935) | 7.03 | 4.99E-08 |
Generated using a Fisher's Exact test and a Bonferroni correction.
While bone marrow cells were cultured for only seven days and grown under non-osteogenic conditions, examination of genes associated with osteoblast differentiation revealed that most, but not all were downregulated (Table 7, sFig. 5). Most early and intermediate markers of osteogenic differentiation including alkaline phosphatase (Alpl; Log2 Fold −1.38), osterix (Sp7; Log2 Fold −1.37), and bone sialoprotein (Ibsp; Log2 Fold −3.64) were downregulated. Additionally genes with known roles in osteoblast differentiation including osteomodulin (Omd; Log2 Fold −1.19), parathyroid hormone receptor (Pth1r; Log2 Fold −1.60), and myocyte enhancer factor 2C (Mef2c; Log2 Fold −1.06) were also downregulated. Interestingly, Tbx3 (Log2 Fold 1.22) and Sox11 (Log2 Fold 1.57), two transcription factors with less defined roles in osteogenesis were up-regulated [64,65].
Table 7.
Osteoblast downregulated genes found in the cherubism mesenchymal population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Alpl | Alkaline phosphatase, liver/bone/kidney | −1.38 | 0.000161 | 5804.69 |
| SP7 | Sp7 transcription factor 7 | −1.37 | 0.000254 | 960.28 |
| Ibsp | Integrin binding sialoprotein | −3.64 | 1.11E−05 | 4817.72 |
| Omd | Osteomodulin | −1.19 | 0.002718 | 422.17 |
| Mef2c | Myocyte enhancer factor 2C | −1.06 | 1.07E−06 | 1167.64 |
| Tbx3 | T−box 3 | 1.22 | 6.83E−15 | 181.12 |
| Soxll | SRY (sex determining region Y)−box 11 | 1.57 | 4.31 E−l 2 | 314.05 |
| Sox6 | SRY (sex determining region Y)−box 6 | −1.16 | 5.62E−05 | 239.61 |
| Pthlr | Parathyroid hormone 1 receptor | −1.60 | 0.000303 | 1945.63 |
| Phex | Phosphate regulating endopeptidase homolog, X−linked | −1.51 | 2.06E−05 | 39.54 |
Values given in counts.
Mesenchymal bone marrow stromal cells, also referred to as adventitial reticular cells, while retaining osteogenic potential also retain a functional importance to maintaining the hematopoietic stem cell niche as well as regulating their differentiation. In doing so, marrow adventitial reticular cells express a variety of genes crucial for this role including Cxcl12 (Log2 Fold −1.71), kit ligand (Kitl) (Log2 Fold −2.19), angiopoietin 1 (Log2 Fold −1.41) and angiopoietin 4 (Log2 Fold −1.06), and leptin receptor (Lepr; Log2Fold −1.15) [66–71]. Interestingly, all of these reticular cell gene markers were downregulated. Work by others has also indicated that many of these adventitial reticular cells serve as important early precursor cells for the osteogenic lineage [72,73]. With this in mind, gremlin 1 (Grem1; Log2 Fold 2.47), a gene that has been reported as a possible marker for skeletal stem cells was highly upregulated (Table 8, sFig. 6) [74]. Grem1, a known BMP inhibitor [75], can also promote angiogenesis through binding Vegfr2 [76].
Table 8.
Changes in marrow stromal cell gene marker expression found in the cherubism mesenchymal population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Greml | Gremlin 1, DAN family BMP antagonist | 2.47 | 1.87E-18 | 1104.66 |
| Angptl | Angiopoietin 1 | −1.41 | 3.47E-13 | 1219.34 |
| Angpt4 | Angiopoietin 4 | −1.06 | 4.55E-09 | 1021.87 |
| Lepr | Leptin receptor | −1.15 | 9.70E-06 | 666.29 |
| Cxdl2 | Chemokine (C-X-C motif) ligand 12 | −1.71 | 1.85E-16 | 9955.47 |
| Kitl | Kit ligand | −2.19 | 2.20E-21 | 1203.63 |
| Cd200 | CD200 Antigen | −1.02 | 0.000718 | 347.42 |
Values given in counts.
In addition to increased expression of Grem 1 implicating changes in BMP signaling, several other genes associated with the BMP pathway were impacted (sFig. 6). The expression of Bmp2 (Log2 Fold −2.74), Bmp4 (Log2 Fold −1.79), Bmp8a (Log2 Fold −2.11), and Gdf6 (Log2 Fold −2.69) were all downregulated in the Sh3bp2KI/KI mesenchymal population. BMPs are well known inducers of osteogenesis [77]. While the significance of GDF6 in joint formation is known, its role in osteogenesis remains less defined. There is evidence to suggest that GDF6 may be anti-osteogenic [78]. However, GDF6 can also heterodimerize with BMPs and therefore may have additional regulatory roles [79]. Genes encoding for secreted BMP inhibitors follistatin (Log2 Fold −1.24) and chordin-like 1 (Log2 Fold −1.91) were downregulated, while receptor Bmp1rb (Log2 Fold 1.26) was upregulated. Taken to-gether, these data suggest that BMP signaling is strongly downregulated (Table 9).
Table 9.
BMP and Wnt pathway gene expression changes in found in the cherubism mesenchymal population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Greml | Gremlin 1, DAN family BMP antagonist | 2.47 | 1.87E-18 | 1104.66 |
| Fst | Follistatin | −1.24 | 3.47E-06 | 833.74 |
| Chrdll | Chordin-like 1 | −1.91 | 1.01E-17 | 238.60 |
| Bmprlb | Bone morphogenetic protein receptor, type IB | 1.26 | 5.11E-05 | 104.62 |
| Bmp4 | Bone morphogenetic protein 4 | −1.79 | 1.45E-08 | 234.74 |
| Bmp2 | Bone morphogenetic protein 2 | −2.74 | 7.80E-16 | 34.50 |
| Bmp8a | Bone morphogenetic protein 8a | −2.11 | 5.15E-05 | 38.59 |
| Gdf6 | Growth differentiation factor 6 | −2.69 | 4.26E-07 | 28.58 |
| Wnt2b | Wingless-type MMTV integration site family, member 2B | 1.49 | 6.46E-07 | 32.72 |
| Sfrpl | Secreted frizzled-related protein 1 | −1.91 | 5.99E-13 | 1725.02 |
| Sfrp2 | Secreted frizzled-related protein 2 | −2.13 | 2.35E-20 | 1981.96 |
| Frzb | Frizzled-related protein | −2.03 | 7.00E-06 | 31.79 |
| Win | Wnt inhibitory factor 1 | −1.83 | 3.15E-05 | 35.95 |
| Lgr5 | Leucine rich repeat containing G protein coupled receptor 5 | −1.65 | 0.000235 | 288.12 |
| Rspo3 | R-spondin 3 | −1.67 | 5.53E-07 | 228.73 |
Values given in counts.
The Wnt pathway was another pathway that showed considerable changes in gene expression. Several inhibitors of the Wnt signaling pathway including Sfrp1 (Log2 Fold −1.91), Sfrp2 (Log2 Fold −2.13), Frzb (Log2 Fold −2.03), and Wif (Log2 Fold −1.83) were downregulated, while Wnt2b (Log2 Fold 1.49) was upregulated. This possibly suggests that the activity of certain downstream Wnt pathways may be heightened. However, Lrg5 (Log2 Fold −1.65) and R-spondin 3 (Log2 Fold −1.67), which have been shown to positively support Wnt signaling were also downregulated [80]. Thus, while our studies clearly detail significant changes in genes associated with Wnt signaling, it will be important to determine how these changes in gene expression impact specific downstream signaling events.
While the majority of genes showing downregulated expression in the Sh3bp2KI/KI mesenchymal population were associated with osteogenesis, most upregulated genes had associations with inflammation and/or supporting osteoclast formation (Table 10, sFig. 5). Gzmc (Log2 Fold 2.47), Gzmd (Log2 Fold 2.2), and Gzme (Log2 Fold 1.78), belonging to the granzyme family of serine proteases, which are notoriously secreted by immune cells, were highly upregulated in the mesenchymal population. Chemokines Cxcl5 (Log2 Fold 2.46) and Cxcl1 (Log2 Fold 1.32), critical for the recruitment of inflammatory cells were also up-regulated. Related to osteoclast formation, there is evidence to suggest Cxcl1 promotes the recruitment of osteoclast precursors while Cxcl5 induces the expression of Rankl (Tnfsf11; Log2 Fold 1.01) [81,82], which was also upregulated in the mesenchymal cell population. Consistent with what we previously reported [14], OPG, a decoy RANK receptor, was downregulated in the data set, but just below the Log2fold cutoff. Facilitating the remodeling of heparan sulfate, several members of the 3-O sulfotransferase family were up regulated including Hs3st1 (Log2 Fold 2.37), Hs3st3a1 (Log2 Fold 1.77), Hs3st3b1 (Log2 Fold 1.69). Through the modification of heparan, these enzymes can regulate processes such as coagulation and epithelial to mesenchymal transition [83,84]. Thrombomodulin (Thbd; Log2 Fold 1.07), which is modestly upregulated has important roles in anti-coagulation and anti-in-flammation through direct binding of thrombin and also through the activation of Protein C [85–87]. F-spondin (Spon1; Log2 Fold 2.38) a gene highly upregulated in the Sh3bp2KI/KI mesenchymal population encodes for a protein containing six thrombospondin domains. Interestingly, similar to thrombospondin-1, f-spondin has also been shown to activate latent TGFβ1 and lead to increased synthesis of PGE2 [88]. Finally, while the expression of most genes associated with inflammation were upregulated, Il1rn (Log2 Fold −2.06), a gene that encodes for Interleukin Receptor 1 Antagonist was strongly downregulated. One of the valued biological effects of bone marrow derived mesenchymal stem cells (MSCs) is that they possess anti-inflammatory and anti-fibrotic properties. There is evidence to suggest that Il1rn expression in MSCs is a major contributor to these anti-inflammatory and anti-fibrotic properties [89]. Thus, strong down regulation of this receptor expression will likely permit stronger inflammatory and fibrotic outcomes.
Table 10.
Inflammation upregulated genes found in the cherubism mesenchymal population.
| Gene | Description | Log2 fold change | P-value | Base mean intensitya |
|---|---|---|---|---|
| Gzmc | Granzyme C | 2.47 | 4.15E-23 | 91.79 |
| Gzmd | Granzyme D | 2.22 | 3.63E-17 | 302.52 |
| Gzme | Granzyme E | 1.78 | 9.30E-11 | 472.76 |
| Cxcl5 | Chemokine (C-X-C motif) ligand 5 | 2.46 | 1.55E-10 | 54.14 |
| Cxcll | Chemokine (C-X-C motif) ligand 1 | 1.32 | 6.95E-06 | 46.89 |
| Hs3stl | Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | 2.37 | 1.57E-11 | 63.17 |
| Hs3st3al | Heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 | 1.77 | 3.60E-10 | 114.28 |
| Hs3st3bl | Heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 | 1.69 | 1.84E-07 | 78.05 |
| Qist8 | Carbohydrate GY-acetylgalactosamine 4-0) sulfotransferase 8 | 1.39 | 0.00223 | 66.41 |
| Sponl | Spondin 1, (f-spondin) extracellular matrix protein | 2.38 | 1.67E-21 | 246.00 |
| Thbd | Thrombomodulin | 1.07 | 2.60E-06 | 223.72 |
| Tnfsfll | Tumor necrosis factor (ligand) superfamily, member 11 | 1.01 | 9.43E-06 | 204.93 |
| nim | Interleukin 1 receptor antagonist | −2.06 | 1.03E-16 | 4361.82 |
Values given in counts.
4. Discussion
Past studies from our lab have shown how bone marrow cultures derived from a cherubism mouse model (Sh3bp2KI/KI mice) displayed defective osteogenesis and spontaneous osteoclast formation [14]. To gain deeper insight into the mechanistic changes that contribute to these alterations in cell differentiation, we performed global gene expression analysis on sorted hematopoietic and mesenchymal cell populations using RNA-Seq. Our findings presented here have yielded some surprising outcomes regarding the identity of specific genes impacted by cherubic mutations in Sh3bp2. Here we discuss the significance of these gene expression changes and speculate on possible links to regulation of inflammation, osteoclast formation, osteoblast differentiation and fibrosis.
It is well established that inflammation remains a core element contributing to cherubism. During the early stages of this disease in patients, the submandibular and cervical lymph nodes are enlarged and within cherubism lesions various immune cell types including giant multinucleated osteoclasts exist [5,90,91]. Studies in mice have gone on to provide evidence that cherubic mutations in Sh3bp2 lead to greater sensitivity to pathogen and damage associated molecular patterns (PAMPS and DAMPS), Rankl stimulation, and homozygous knockin mice systemically have higher levels of TNFα [11,12,92,93]. Consistent with these studies, evaluation of the dataset by Ingenuity Pathway Analysis predicts many of the same inflammatory signaling molecules (Supplementary Table 3). However, examination of the dataset from the hematopoietic population has also drawn our interest to prostaglandin and coagulation signaling pathways, which have known roles in inflammatory processes, but have not been investigated with regard to cherubism. The data set also draws interest towards genes associated with autoinflammatory conditions including MEFV, a gene that encodes for Pyrin, an inflammatory sensing molecule.
Importantly, changes in gene expression associated with inflammation were not exclusive to the hematopoietic population, but also existed within the mesenchymal population. Understanding how the mesenchymal population responds to and participates in inflammatory processes remains a topically understudied area of re-search, yet it is well appreciated that bone marrow derived MSCs retain anti-inflammatory properties [89]. However, given the down regulation of Il1rn, an interleukin 1 receptor antagonist, and hematopoietic stem cell niche regulators Cxcl12 and Kitl and the up regulation in granzymes C, D, and E, it appears that the marrow derived mesenchymal cells derived from cherubism mice have taken on a very different role in regulating immune cell function. The detection of granzyme expression within the mesenchymal population was unexpected. Granzyme expression is generally thought to be restricted to cytotoxic T cells and natural killer cells [94], but certain granzymes genes have been reported to be expressed in non-hematopoietic cell types including marrow stromal cells [95]. In support of our findings, inspection of granzyme C, D, and E expression in the online accessible gene expression portal BioGPS also selectively detects these granzymes in osteoblasts [96].
In cherubism, excessive bone resorption has largely been attributed to the overactivation of osteoclasts. Work by others has shown how myeloid progenitors derived from cherubism mice are more sensitive to both Rankl and TNFα signaling to augment osteoclast formation [11,93]. Our studies have also implicated several additional genes that are likely to enhance the recruitment and development of osteoclasts. Within the hematopoietic population, this includes genes associated with the generation and stability of prostaglandin PGE2. In vitro studies have shown how PGE2 through its receptor EP2 is a potent enhancer of osteoclast formation [97]. Further, Cxcl2 and Ccl9 have also been shown to be involved in the regulation of osteoclast formation [63,98].
Our studies also indicate that gene expression changes within the mesenchymal cell lineage support osteoclast formation. Not only has the expression of Rankl (Tnfsf11) increased, but Cxcl5, a gene shown to positively regulate Rankl expression was also increased [82]. Further, down-regulation of Kitl expression has also been associated with increased osteoclast formation [99] and the gene product of Spon1 has been shown to activate latent TGFβ1 and increase PGE2 synthesis [88], both of which are enhancers of osteoclast formation.
In cherubism, fibrous tissue overgrowth potentially at the expense of osteoblast differentiation is a characteristic of the cherubism phenotype that mechanistically remains poorly understood. Examination of gene expression changes within the mesenchymal population clearly indicated that down-regulation of the BMP signaling pathway was the likely cause for impaired osteoblast differentiation. However, not only was the expression of different BMP ligands downregulated, but Grem1, a known BMP inhibitor was highly upregulated [75]. Interestingly, down-regulation of BMP signaling and up-regulation of Grem1 expression has also been observed in idiopathic pulmonary fibrosis [100]. While mechanistically not understood, evidence in the literature has shown that an inverse relationship exists between BMP and TGFβ signaling pathways. With regard to regulating osteogenesis, BMP active-tion is pro-ostegenic, while TGFβ activation is inhibitory. As men-tioned, the increased expression of Spon1, which can activate latent TGFβ ligands, may be a likely contributor to inhibiting osteogenesis. In support of this thinking, Spon1 deficient mice have increased bone mass with evidence of enhanced BMP signaling [84]. Additionally, the down-regulation of Il1rn may provide a permissive environment for inflammatory cytokines to inhibit osteogenesis and contribute to the profibrotic state [101].
There is also evidence from the hematopoietic population that changes in gene expression may contribute to impaired osteogenesis. Work on Saa3, a gene highly increased within the cherubism hematopoietic population, has been shown to inhibit PTH induced osteogenesis [50]. Also, Ndst1, which participates in the synthesis of heparan sulfate chains, has been shown to negatively regulate BMP signaling [61].
In this study, we have thoroughly examined and discussed the gene expression changes that occurred within the hematopoietic and mesenchymal cell populations derived from bone marrow cultures of cherubism mice. In doing so, this work reveals a much broader network of genes involved in the regulation of inflammation, osteoclast formation, and osteoblast differentiation. To the best of our knowledge, few studies have simultaneously examined gene expression changes within the hematopoietic and mesenchymal cell lineages to gain a deeper collective understanding of the complex interplay that exists among these two cell populations. It is important to emphasize that while this study has utilized a cherubism mouse model, what we have learned from this study is likely to have much broader relevance to a variety of inflammatory skeletal diseases. Certain phenotypic characteristics of cherubism resemble other diseases like Noonan syndrome, neurofi-bromatosis type I, craniofacial cutaneous syndrome, and central giant cell granuloma [102]. Thus, mechanisms identified in this study are likely to have relevance for understanding the cross talk between hematopoietic and mesenchymal cell types for a host of diseases involving fibrotic inflammation and focal areas of bone loss.
Supplementary Material
Acknowledgements
This work was supported by grant funding from the National Institute for Craniofacial and Dental Research 5R21DE025933-02 and a stimulus grant from the University of Connecticut.
Footnotes
Disclaimer
None.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bone.2020.115315.
References
- [1].Jones WA, Familial multilocular cystic disease of the jaws, Am. J. Cancer 17 (1933) 946–950, 10.1158/ajc.1933.946. [DOI] [Google Scholar]
- [2].Jones WA G, Cherubism—familial fibrous dysplasia of the jaws, J. Bone Jt. Surg. (32–B) (1950) 334–347. [DOI] [PubMed] [Google Scholar]
- [3].Lima GDMG, Almeida JD, Cabral LAG, Cherubism: clinicoradiographic fea-tures and treatment, J. Oral Maxillofac. Res. 1 (2010) e2, 10.5037/jomr.2010.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Carvalho Silva E, Carvalho Silva GC, Vieira TC, Cherubism: clinicoradiographic features, treatment, and long-term follow-up of 8 cases, J. Oral Maxillofac. Surg.65 (2007) 517–522, 10.1016/j.joms.2006.05.061. [DOI] [PubMed] [Google Scholar]
- [5].Papadaki ME, Lietman SA, Levine MA, Olsen BR, Kaban LB, Reichenberger EJ, Cherubism: best clinical practice, Orphanet J. Rare Dis. 7 (Suppl. 1) (2012) S6, 10.1186/1750-1172-7-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, Ninomiya C, doAmaral C, Peters H, Habal M, Rhee-Morris L, Doss JB, Kreiborg S, Olsen BR, Reichenberger E, Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism, Nat. Genet. 28 (2001) 125–126, 10.1097/00001665-200109000-00022. [DOI] [PubMed] [Google Scholar]
- [7].Tiziani V, Reichenberger E, Buzzo CL, Niazi S, Fukai N, Stiller M, Peters H, Salzano FM, Raposo do Amaral CM, Olsen BR, The gene for cherubism maps to chromosome 4p16, Am. J. Hum. Genet. 65 (1999) 158–166, 10.1086/302456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].GuezGuez A, Prod’homme V, Mouska X, Baudot A, Blin-Wakkach C, Rottapel R, Deckert M, 3BP2 adapter protein is required for receptor activator of NFκB ligand (RANKL)-induced osteoclast differentiation of RAW264.7 cells, J. Biol. Chem. 285 (2010) 20952–20963, 10.1074/jbc.M109.091124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, Cong F, Simoncic PD, Ueki Y, La Rose J, Rottapel R, Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism, Cell 147 (2011) 1324–1339, 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guettler S, Larose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F, Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease, Cell 147 (2011) 1340–1354, 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- [11].Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, Saheki Y, Kawai T, Mukherjee PM, Reichenberger E, Olsen BR, Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice, Cell 128 (2007) 71–83, 10.1016/j.cell.2006.10.047. [DOI] [PubMed] [Google Scholar]
- [12].Yoshitaka T, Mukai T, Kittaka M, Alford LM, Masrani S, Ishida S, Yamaguchi K, Yamada M, Mizuno N, Olsen BR, Reichenberger EJ, Ueki Y, Enhanced TLR-MYD88 signaling stimulates autoinflammation in SH3BP2 cherubism mice and defines the etiology of cherubism, Cell Rep. 8 (2014) 1752–1766, 10.1016/j.celrep.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mukai T, Ishida S, Ishikawa R, Yoshitaka T, Kittaka M, Gallant R, Lin YL, Rottapel R, Brotto M, Reichenberger EJ, Ueki Y, SH3BP2 Cherubism Mutation Potentiates TNF-a – Induced, 29 (2014), pp. 2618–2635, 10.1002/jbmr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu Y, Sharma T, Chen I-P, Reichenberger E, Ueki Y, Arif Y, Parisi D, Maye P, Rescue of a cherubism bone marrow stromal culture phenotype by reducing TGFβ signaling, Bone 111 (2018) 28–35, 10.1016/J.BONE.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].S. A, FastQC: A Quality Control Tool for High Throughput Sequence Data, (2010). [Google Scholar]
- [16].Joshi J, Fass NA, Sickle: A Sliding-window, Adaptive, Quality-based Trimming Tool for FastQ Files, (2011). [Google Scholar]
- [17].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29 (2013) 15–21, 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Anders S, Pyl PT, Huber W, HTSeq—a Python framework to work with high-throughput sequencing data, Bioinformatics 31 (2015) 166–169, 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Love MI, Huber W, Anders S, Moderated estimation of fold change and dis-persion for RNA-seq data with DESeq2, Genome Biol. 15 (2014) 550, 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].The gene ontology resource: 20 years and still GOing strong, Nucleic Acids Res. 47 (2019) D330–D338, 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet. 25 (2000) 25–29, 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT, PathVisio 3: an extendable pathway analysis toolbox, PLoS Comput. Biol. 11 (2015) e1004085,, 10.1371/journal.pcbi.1004085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Mélius J, Waagmeester A, Sinha SR, Miller R, Coort SL, Cirillo E, Smeets B, Evelo CT, Pico AR, WikiPathways: capturing the full diversity of pathway knowledge, Nucleic Acids Res. 44 (2016) D488–D494, 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR, WikiPathways: building research communities on biological pathways, Nucleic Acids Res. 40 (2012) D1301–D1307, 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Short B, Wagey R, Isolation and Culture of Mesenchymal Stem Cells From Mouse Compact Bone, Humana Press, Totowa, NJ, 2013, pp. 335–347, 10.1007/978-1-62703-128-8_21. [DOI] [PubMed] [Google Scholar]
- [26].Short BJ, Brouard N, Simmons PJ, Prospective isolation of mesenchymal stem cells from mouse compact bone, Methods Mol. Biol. 482 (2009) 259–268, 10.1007/978-1-59745-060-7_16. [DOI] [PubMed] [Google Scholar]
- [27].Phinney DG, Isolation of mesenchymal stem cells from murine bone marrow by immunodepletion, Methods Mol. Biol. 449 (2008) 171–186, 10.1007/978-1-60327-169-1_12. [DOI] [PubMed] [Google Scholar]
- [28].Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG, Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection, J. Cell. Biochem. 89 (2003) 1235–1249, 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- [29].Ewels P, Magnusson M, Lundin S, Käller M, MultiQC: summarize analysis results for multiple tools and samples in a single report, Bioinformatics 32 (2016) 3047–3048, 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology, Nat. Genet. 25 (2000) 25–29, 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Minkin C, Bone acid phosphatase: tartrate-resistant acid phosphatase as a marker of osteoclast function, Calcif. Tissue Int. 34 (1982) 285–290. [DOI] [PubMed] [Google Scholar]
- [32].Lee S-H, Rho J, Jeong D, Sul J-Y, Kim T, Kim N, Kang J-S, Miyamoto T, Suda T, Lee S-K, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y, v-ATPase V0 subunit d2–deficient mice exhibit impaired osteoclast fusion and increased bone formation, Nat. Med. 12 (2006) 1403–1409, 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- [33].Chambers TJ, Magnus CJ, Calcitonin alters behaviour of isolated osteoclasts, J. Pathol. 136 (1982) 27–39, 10.1002/path.1711360104. [DOI] [PubMed] [Google Scholar]
- [34].Nicholson GC, Moseley JM, Sexton PM, Mendelsohn FA, Martin TJ, Abundant calcitonin receptors in isolated rat osteoclasts. Biochemical and auto-radiographic characterization, J. Clin. Invest. 78 (1986) 355–360, 10.1172/JCI112584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brömme D, Okamoto K, Wang BB, Biroc S, Human cathepsin O2, a matrix protein-degrading cysteine protease expressed in osteoclasts. Functional expression of human cathepsin O2 in Spodoptera frugiperda and characterization of the enzyme, J. Biol. Chem. 271 (1996) 2126–2132, 10.1074/jbc.271.4.2126. [DOI] [PubMed] [Google Scholar]
- [36].Drake FH, Dodds RA, James IE, Connor JR, Debouck C, Richardson S, Lee-Rykaczewski E, Coleman L, Rieman D, Barthlow R, Hastings G, Gowen M, Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts, J. Biol. Chem. 271 (1996) 12511–12516, 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- [37].Sundaram K, Nishimura R, Senn J, Youssef RF, London SD, Reddy SV, RANK ligand signaling modulates the matrix metalloproteinase-9 gene expression during osteoclast differentiation, Exp. Cell Res. 313 (2007) 168–178, 10.1016/J.YEXCR.2006.10.001. [DOI] [PubMed] [Google Scholar]
- [38].Mensah KA, Ritchlin CT, Schwarz EM, RANKL induces heterogeneous DCSTAMP lo and DC-STAMP hi osteoclast precursors of which the DC-STAMP lo precursors are the master fusogens, J. Cell. Physiol. 223 (2009), 10.1002/jcp.22012n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang M, Birnbaum MJ, MacKay CA, Mason-Savas A, Thompson B, Odgren PR, Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation, J. Cell. Physiol. 215 (2008) 497–505, 10.1002/jcp.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T, DCSTAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells, J. Exp. Med. 202 (2005) 345–351, 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ross FP, Chappel J, Alvarez JI, Sander D, Butler WT, Farach-Carson MC, Mintz KA, Robey PG, Teitelbaum SL, Cheresh DA, Interactions between the bone matrix proteins osteopontin and bone sialoprotein and the osteoclast integrin alpha v beta 3 potentiate bone resorption, J. Biol. Chem. 268 (1993) 9901–9907. [PubMed] [Google Scholar]
- [42].Kim N, Takami M, Rho J, Josien R, Choi Y, A novel member of the leukocyte receptor complex regulates osteoclast differentiation, J. Exp. Med. 195 (2002) 201–209, 10.1084/jem.20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne WC, Baron R, Src kinase activity is essential for osteoclast function, J. Biol. Chem. 279 (2004) 17660–17666, 10.1074/jbc.M311032200. [DOI] [PubMed] [Google Scholar]
- [44].Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O, Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids, Drug Metab. Dispos. 39 (2011) 180–190, 10.1124/dmd.110.035121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang C, Li C, Li M, Tong X, Hu X, Yang X, Yan X, He L, Wan C, CYP2S1 depletion enhances colorectal cell proliferation is associated with PGE2-mediated activation of β-catenin signaling, Exp. Cell Res. 331 (2015) 377–386, 10.1016/J.YEXCR.2014.12.008. [DOI] [PubMed] [Google Scholar]
- [46].Frömel T, Kohlstedt K, Popp R, Yin X, Awwad K, Barbosa-Sicard E, Thomas AC, Lieberz R, Mayr M, Fleming I, Cytochrome P4502S1: a novel monocyte/macrophage fatty acid epoxygenase in human atherosclerotic plaques, Basic Res. Cardiol. 108 (2013) 319, 10.1007/s00395-012-0319-8. [DOI] [PubMed] [Google Scholar]
- [47].Heilig R, Broz P, Function and mechanism of the pyrin inflammasome, Eur. J. Immunol. 48 (2018) 230–238, 10.1002/eji.201746947. [DOI] [PubMed] [Google Scholar]
- [48].Luria A, Weldon SM, Kabcenell AK, Ingraham RH, Matera D, Jiang H, Gill R, Morisseau C, Newman JW, Hammock BD, Compensatory mechanism for homeostatic blood pressure regulation in Ephx2 gene-disrupted mice, J. Biol. Chem. 282 (2007) 2891–2898, 10.1074/jbc.M608057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sheikh MH, Solito E, Sheikh MH, Solito E, Annexin A1: uncovering the many talents of an old protein, Int. J. Mol. Sci. 19 (2018) 1045, 10.3390/ijms19041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Choudhary S, Goetjen A, Estus T, Jacome-Galarza CE, Aguila HL, Lorenzo J, Pilbeam C, Serum Amyloid A3 Secreted by Preosteoclasts Inhibits Parathyroid Hormone-stimulated cAMP Signaling in Murine Osteoblasts, (2015), 10.1074/jbc.M115.686576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Trentin PG, Ferreira TPT, Arantes ACS, Ciambarella BT, Cordeiro RSB, Flower RJ, Perretti M, Martins MA, Silva PMR, Annexin A1 mimetic peptide controls the inflammatory and fibrotic effects of silica particles in mice, Br. J. Pharmacol. 172 (2015) 3058–3071, 10.1111/bph.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sarangi PP, Lee H, Kim M, Activated protein C action in inflammation, Br. J. Haematol. 148 (2010) 817–833, 10.1111/j.1365-2141.2009.08020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Manukyan G, Aminov R, Update on pyrin functions and mechanisms of familial Mediterranean fever, Front. Microbiol. 7 (2016) 456, 10.3389/fmicb.2016.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].de Torre-Minguela C, Mesa Del Castillo P, Pelegrín P, The NLRP3 and pyrin inflammasomes: implications in the pathophysiology of autoinflammatory diseases, Front. Immunol. 8 (2017) 43, 10.3389/fimmu.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shamilov R, Aneskievich BJ, TNIP1 in autoimmune diseases: regulation of toll-like receptor signaling, J Immunol Res 2018 (2018) 1–13, 10.1155/2018/3491269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mirza N, Sowa AS, Lautz K, Kufer TA, NLRP10 affects the stability of Abin-1 to control inflammatory responses, J. Immunol. 202 (2019) 218–227, 10.4049/jimmunol.1800334. [DOI] [PubMed] [Google Scholar]
- [57].Eisenbarth SC, Williams A, Colegio OR, Meng H, Strowig T, Rongvaux A, Henao-Mejia J, Thaiss CA, Joly S, Gonzalez DG, Xu L, Zenewicz LA, Haberman AM, Elinav E, Kleinstein SH, Sutterwala FS, Flavell RA, NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells, Nature 484 (2012) 510–513, 10.1038/nature11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sun C, Molineros JE, Looger LL, Zhou X, Kim K, Okada Y, Ma J, Qi Y, Kim-Howard X, Motghare P, Bhattarai K, Adler A, Bang S-Y, Lee H-S, Kim T-H, Kang YM, Suh C-H, Chung WT, Park Y-B, Choe J-Y, Shim SC, Kochi Y, Suzuki A, Kubo M, Sumida T, Yamamoto K, Lee S-S, Kim YJ, Han B-G, Dozmorov M, Kaufman KM, Wren JD, Harley JB, Shen N, Chua KH, Zhang H, Bae S-C, Nath SK, High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry, Nat. Genet. 48 (2016) 323–330, 10.1038/ng.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ferretti A, Fortwendel JR, Gebb SA, Barrington RA, Autoantibody-mediated pulmonary alveolar proteinosis in Rasgrp1-deficient mice, J. Immunol. 197 (2016) 470–479, 10.4049/JIMMUNOL.1502248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mao H, Yang W, Latour S, Yang J, Winter S, Zheng J, Ni K, Lv M, Liu C, Huang H, Chan K-W, Pui-Wah Lee P, Tu W, Fischer A, Lau Y-L, RASGRP1 mutation in autoimmune lymphoproliferative syndrome-like disease, J. Allergy Clin. Immunol. 142 (2018) 595–604.e16, 10.1016/J.JACI.2017.10.026. [DOI] [PubMed] [Google Scholar]
- [61].Hu Zhonghua, Wang Chaochen, Xiao Ying, Sheng Nengyin, Chen Yibin, Xu Ye, Zhang Liang, Mo Wei, Jing Naihe, Hu G, NDST1-dependent heparan sulfate regulates BMP signaling and internalization in lung development, J. Cell Sci. 122 (2008) 1145–1154, 10.1242/jcs.034736. [DOI] [PubMed] [Google Scholar]
- [62].Yan HH, Jiang J, Pang Y, Achyut BR, Lizardo M, Liang X, Hunter K, Khanna C, Hollander C, Yang L, CCL9 induced by TGFβ signaling in myeloid cells enhances tumor cell survival in the premetastatic organ, Cancer Res. 75 (2015) 5283–5298, 10.1158/0008-5472.CAN-15-2282-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lean JM, Murphy C, Fuller K, Chambers TJ, CCL9/MIP-1gamma and its receptor CCR1 are the major chemokine ligand/receptor species expressed by osteoclasts, J. Cell. Biochem. 87 (2002) 386–393, 10.1002/jcb.10319. [DOI] [PubMed] [Google Scholar]
- [64].Govoni KE, Linares GR, Chen S-T, Pourteymoor S, Mohan S, T-box 3 negatively regulates osteoblast differentiation by inhibiting expression of osterix and runx2, J. Cell. Biochem. 106 (2009) 482–490, 10.1002/jcb.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gadi J, Jung S-H, Lee M-J, Jami A, Ruthala K, Kim K-M, Cho N-H, Jung H-S, Kim C-H, Lim S-K, The transcription factor protein Sox11 enhances early osteoblast differentiation by facilitating proliferation and the survival of mesenchymal and osteoblast progenitors, J. Biol. Chem. 288 (2013) 25400–25413, 10.1074/jbc.M112.413377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ding L, Morrison SJ, Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches, Nature 495 (2013) 231–235, 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Greenbaum A, Hsu Y-MS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC, CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance, Nature 495 (2013) 227–230, 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kopp HG, Hooper AT, Avecilla ST, Rafii S, Functional heterogeneity of the bone marrow vascular niche, Ann. N. Y. Acad. Sci. 1176 (2009) 47–54, 10.1111/j.1749-6632.2009.04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, Crystal RG, Besmer P, Lyden D, Moore MAS, Werb Z, Rafii S, Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand, Cell 109 (2002) 625–637, 10.1016/S0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T, Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny, Immunity 19 (2003) 257–267, 10.1016/S1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- [71].Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T, The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche, Immunity 33 (2010) 387–399, 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- [72].Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ, Leptin-receptor-ex-pressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow, Cell Stem Cell 15 (2014) 154–168, 10.1016/J.STEM.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P, Self-renewing os-teoprogenitors in bone marrow sinusoids can organize a hematopoietic micro-environment, Cell 131 (2007) 324–336, 10.1016/J.CELL.2007.08.025. [DOI] [PubMed] [Google Scholar]
- [74].Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y, Gross S, Renz BW, Setlik W, Martinez AN, Chen X, Nizami S, Lee HG, Kang HP, Caldwell J-M, Asfaha S, Westphalen CB, Graham T, Jin G, Nagar K, Wang H, Kheirbek MA, Kolhe A, Carpenter J, Glaire M, Nair A, Renders S, Manieri N, Muthupalani S, Fox JG, Reichert M, Giraud AS, Schwabe RF, Pradere J-P, Walton K, Prakash A, Gumucio D, Rustgi AK, Stappenbeck TS, Friedman RA, Gershon MD, Sims P, Grikscheit T, Lee FY, Karsenty G, Mukherjee S, Wang TC, Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential, Cell 160 (2015) 269–284, 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM, The xenopus dorsalizing factor gremlin identifies a novel family of secreted proteins that an-tagonize BMP activities, Mol. Cell 1 (1998) 673–683, 10.1016/S1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- [76].Mitola S, Ravelli C, Moroni E, Salvi V, Leali D, Ballmer-Hofer K, Zammataro L, Presta M, Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2, Blood 116 (2010) 3677–3680, 10.1182/blood-2010-06-291930. [DOI] [PubMed] [Google Scholar]
- [77].Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA, Novel regulators of bone formation: molecular clones and activities, Science 242 (1988) 1528–1534, 10.1126/SCIENCE.3201241. [DOI] [PubMed] [Google Scholar]
- [78].Clendenning DE, Mortlock DP, The BMP ligand Gdf6 prevents differentiation of coronal suture mesenchyme in early cranial development, PLoS One 7 (2012) e36789,, 10.1371/journal.pone.0036789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chang C, Hemmati-Brivanlou A, Xenopus GDF6, a new antagonist of noggin and a partner of BMPs, Development 126 (1999). [DOI] [PubMed] [Google Scholar]
- [80].de Lau WB, Snel B, Clevers HC, The R-spondin protein family, Genome Biol. 13 (2012) 242, 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Onan D, Allan EH, Quinn JMW, Gooi JH, Pompolo S, Sims NA, Gillespie MT, Martin TJ, The chemokine Cxcl1 is a novel target gene of parathyroid hormone (PTH)/PTH-related protein in committed osteoblasts, Endocrinology 150 (2009) 2244–2253, 10.1210/en.2008-1597. [DOI] [PubMed] [Google Scholar]
- [82].Sundaram K, Rao DS, Ries WL, Reddy SV, CXCL5 stimulation of RANK ligand expression in Paget’s disease of bone, Lab. Investig. 93 (2013) 472–479, 10.1038/labinvest.2013.5. [DOI] [PubMed] [Google Scholar]
- [83].Smits NC, Kobayashi T, Srivastava PK, Skopelja S, Ivy JA, Elwood DJ, Stan RV, Tsongalis GJ, Sellke FW, Gross PL, Cole MD, DeVries JT, Kaplan AV, Robb JF, Williams SM, Shworak NW, HS3ST1 genotype regulates antithrombin’s inflammomodulatory tone and associates with atherosclerosis, Matrix Biol. 63 (2017) 69–90, 10.1016/J.MATBIO.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhang Z, Jiang H, Wang Y, Shi M, Heparan sulfate D-glucosamine 3-O-sulfo-transferase 3B1 is a novel regulator of transforming growth factor-beta-mediated epithelial-to-mesenchymal transition and regulated by miR-218 in nonsmall cell lung cancer, J. Cancer Res. Ther. 14 (2018) 24, 10.4103/jcrt.JCRT_659_17. [DOI] [PubMed] [Google Scholar]
- [85].Okamoto T, Tanigami H, Suzuki K, Shimaoka M, Thrombomodulin: a bifunc-tional modulator of inflammation and coagulation in sepsis, Crit. Care Res. Prac. 2012 (2012) 614545, 10.1155/2012/614545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Foley JH, Conway EM, Cross Talk Pathways Between Coagulation and Inflammation, (2016), 10.1161/CIRCRESAHA.116.306853. [DOI] [PubMed] [Google Scholar]
- [87].Esmon CT, The regulation of natural anticoagulant pathways, Science 235 (1987) 1348–1352, 10.1126/SCIENCE.3029867. [DOI] [PubMed] [Google Scholar]
- [88].Attur MG, Palmer GD, Al-Mussawir HE, Dave M, Teixeira CC, Rifkin DB, Appleton CTG, Beier F, Abramson SB, F-spondin, a neuroregulatory protein, is up-regulated in osteoarthritis and regulates cartilage metabolism via TGF-beta activation, FASEB J. 23 (2009) 79–89, 10.1096/fj.08-114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ortiz LA, Dutreil M, Fattman C, Pandey AC, Torres G, Go K, Phinney DG, Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury, Proc. Natl. Acad. Sci. U. S. A. 104 (2007) 11002–11007, 10.1073/pnas.0704421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kadlub N, Sessiecq Q, Mandavit M, L’Hermine AC, Badoual C, Galmiche L, Berdal A, Descroix V, Picard A, Coudert AE, Molecular and cellular char-acterizations of human cherubism: disease aggressiveness depends on osteoclast differentiation, Orphanet J. Rare Dis. 13 (2018) 166, 10.1186/s13023-018-0907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ranjan Misra S, Mishra L, Mohanty N, Mohanty S, Cherubism With Multiple Dental Abnormalities: A Rare Presentation. 10.1136/bcr-2014, (n. d.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Prod’Homme V, Boyer L, Dubois N, Mallavialle A, Munro P, Mouska X, Coste I, Rottapel R, Tartare-Deckert S, Deckert M, Cherubism allele heterozygosity am-plifies microbe-induced inflammatory responses in murine macrophages, J. Clin. Invest. 125 (2015) 1396–1400, 10.1172/JCI71081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Levaot N, Simoncic PD, Dimitriou ID, Scotter A, La Rose J, Ng AHM, Willett TL, Wang CJ, Janmohamed S, Grynpas M, Reichenberger E, Rottapel R, Deckert M, Rottapel R, Cicchetti P, Mayer B, Thiel G, Baltimore D, Soriano P, Montgomery C, Geske R, Bradley A, Zou W, Faccio R, Jones W, Gerrie J, Pritchard J, Ueki Y, Ueki Y, Chen G, Lowe C, Yoneda T, Boyce B, Chen H, Mundy G, Soriano P, Wang Z, Ovitt C, Grigoriadis A, MohleSteinlein U, Ruther U, Wagner E, Dumas A, Brigitte M, Moreau M, Chretien F, Basle M, Chappard D, Mao D, Epple H, Uthgenannt B, Novack D, Faccio R, Shukla U, Hatani T, Nakashima K, Ogi K, Sada K, Yagi M, Lee S, Kim K, Lee S, Kim JH, Choi Y, Kim N, McHugh K, Nakamura I, Sato M, Yamamoto M, Crippes B, Sanjay A, Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne W, Baron R, Arias-Salgado E, Lizano S, Sarkar S, Brugge J, Ginsberg M, Shattil S, Maeno K, Foucault I, Le Bras S, Charvet C, Moon C, Altman A, Deckert M, Woodside D, Deckert M, Tartare-Deckert S, Hernandez J, Rottapel R, Altman A, Zakaria S, Wu X, Xu Y, Malladi P, Zhou D, Longaker M, Ducy P, Zhang R, Geoffroy V, Ridall A, Karsenty G, Ren R, Mayer B, Cicchetti P, Baltimore D, Li B, Quarles L, Yohay D, Lever L, Caton R, Wenstrup R, Nagar B, Miyoshi-Akiyama T, Aleman L, Smith J, Adler C, Mayer B, Barila D, Master Z, Tanis K, Veach D, Duewel H, Bornmann W, Koleske A, Brasher B, Van Etten R, Mayer B, Majidi M, Hubbs A, Lichy J, Seeliger M, Barker S, Kassel D, Weigl D, Huang X, Luther M, Knight W, Jin H, Wang J, GuezGuez A, Horne W, Neff L, Chatterjee D, Lomri A, Levy J, Baron R, Boyce B, Yoneda T, Lowe C, Soriano P, Mundy G, Reeve J, Zou W, Liu Y, Maltzman J, Ross F, Teitelbaum S, Mocsai A, Zou W, Reeve J, Liu Y, Teitelbaum S, Ross F, Lanier L, Corliss B, Wu J, Leong C, Phillips J, Insogna K, Shi Y, Alin K, Goff S, Fitter S, Dewar A, Parfitt A, Mousny M, Cao J, Venton L, Sakata T, Halloran B, 3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions, J. Clin. Invest. 121 (2011) 3244–3257, 10.1172/JCI45843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wensink AC, Hack CE, Bovenschen N, Granzymes regulate proinflammatory cytokine responses, J. Immunol. 194 (2015) 491–497, 10.4049/jimmunol.1401214. [DOI] [PubMed] [Google Scholar]
- [95].Yokoi A, Kina T, Minato N, Selective expression and function of granzyme D in lymphohematopoietic stromal cells, Biochem. Biophys. Res. Commun. 264 (1999) 768–773, 10.1006/BBRC.1999.1581. [DOI] [PubMed] [Google Scholar]
- [96].Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, Su AI, BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources, Genome Biol. 10 (2009) R130, 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Li X, Okada Y, Pilbeam CC, Lorenzo JA, Kennedy CRJ, Breyer RM, Raisz LG, Knockout of the Murine Prostaglandin EP 2 Receptor Impairs Osteoclastogenesis In Vitro, (2000). [DOI] [PubMed] [Google Scholar]
- [98].Ha J, Choi H-S, Lee Y, Kwon H-J, Song YW, Kim H-H, CXC chemokine ligand 2 induced by receptor activator of NF-kappa B ligand enhances osteoclastogenesis, J. Immunol. 184 (2010) 4717–4724, 10.4049/jimmunol.0902444. [DOI] [PubMed] [Google Scholar]
- [99].Lotinun S, Krishnamra N, Disruption of c-kit signaling in kit W-sh/W-sh growing mice increases bone turnover, Sci. Rep. 6 (2016) 31515,, 10.1038/srep31515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Koli K, Myllärniemi M, Vuorinen K, Salmenkivi K, Ryynänen MJ, Kinnula VL, Keski-Oja J, Bone morphogenetic protein-4 inhibitor gremlin is overexpressed in idiopathic pulmonary fibrosis, Am. J. Pathol. 169 (2006) 61–71, 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Stashenko P, Dewhirst FE, Rooney ML, Desjardins LA, Heeley JD, Interleukin-1β is a potent inhibitor of bone formation in vitro, J. Bone Miner. Res.2 (2009) 559–565, 10.1002/jbmr.5650020612. [DOI] [PubMed] [Google Scholar]
- [102].Flanagan AM, Speight PM, Giant cell lesions of the craniofacial bones, Head Neck Pathol. 8 (2014) 445, 10.1007/S12105-014-0589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.