Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected several millions and killed more than quarter of a million worldwide to date. Important questions have remained unanswered: why some patients develop severe disease, while others do not; and what roles do genetic variabilities play in the individual immune response to this viral infection. Here, we discuss the critical role T cells play in the orchestration of the antiviral response underlying the pathogenesis of the disease, COVID-19. We highlight the scientific rationale for comprehensive and longitudinal TCR analyses in COVID-19 and reason that analyzing TCR repertoire in COVID-19 patients would reveal important findings that may explain the outcome disparity observed in these patients. Finally, we provide a framework describing the different strategies, the advantages, and the challenges involved in obtaining useful TCR repertoire data to advance our fight against COVID-19.
Keywords: SARS-CoV-2, COVID-19, T cell receptors, TCR-seq, TCR repertoire
Highlights
The COVID-19 pandemic revealed vulnerability in elderly population and underscored the role of the aging immune system in the fight against emerging infections.
T cells are crucial mediators of antiviral adaptive immunity and delineating the role of the TCR repertoire in the context of SARS-CoV-2 merits research efforts.
The TCR repertoire dynamics, composition, and diversity are influenced by several intrinsic and extrinsic factors such as aging, HLA genetic variabilities, viral infection, etc.
The architecture of the TCR repertoire largely contributes to the performance of the adaptive immune response against SARS-CoV-2.
Advances in sequencing technologies and single-cell immune profiling can be leveraged to monitor adaptive immune responses in COVID-19 patients and guide future SARS-CoV-2 immunotherapy and biomarker development.
The COVID-19 Immune Conundrum
Since its emergence in December of 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (see Glossary) has infected more than 7 million people and killed more than a quarter million people worldwide. In the US, at least 2 million individuals are confirmed positive for the coronavirus disease 2019 (COVID-19) and the death toll exceeds 100 000 people. The clinical manifestations of patients with SARS-CoV-2 infection range from asymptomatic to mild or severe respiratory symptomatic disease with high viral load [1,2]. The current coronavirus infects all ages, yet the severity of the disease is different among the age groups. Approximately 15% of the confirmed cases are severe, of whom the majority are either over 65 years of age or patients with underlying medical conditions [3].
As with any virus, the adaptive immune response plays a central role in clearing SARS-CoV-2 infections and directly influences patients’ clinical outcome. A proper and specific adaptive immune response can successfully eliminate the virus, which likely occurs in the asymptomatic and mild cases, where having an appropriate genetic background [e.g., human leukocyte antigen (HLA)] that elicits specific antiviral immunity, or boosting the immune responses with therapeutic strategies, may support the development of a protective immune response. However, in severe cases, an impaired immune response leads to the viral infection running unchecked and the highly proliferative virus causing destruction of the infected tissues, particularly in the lungs, where the expression of angiotensin-converting enzyme 2 (ACE2), the receptor required by the virus to enter the cells, is high [4,5]. The tissue damage induces innate immune and inflammatory response in the lungs, mediated by proinflammatory macrophages and granulocytes, often causing the life-threatening acute respiratory failure seen in the severe cases [1,6].
Several immune characteristics have emerged from early Wuhan cases and were further confirmed in cases in Italy and in New York hospitals, showing that patients with severe infections exhibit increased serum interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), and c-reactive protein. In addition, patients with severe disease display high levels of neutrophils combined with a significant decrease in total lymphocytes [7]. These proinflammatory immune features were also correlated with disease severity and death [8., 9., 10.]. Recent study has characterized bronchoalveolar lavage fluid immune cells from patients with varying severity of COVID-19 and from healthy people by using single-cell RNA sequencing. The bronchoalveolar lavage fluid from patients with severe COVID-9 exhibited high proinflammatory monocyte-derived macrophages, while moderate cases were characterized by the presence of highly clonally expanded CD8+ T cells [11]. Yet, many important questions remained unanswered: why some patients develop severe disease, while others do not; and how genetic variabilities within the immune system impact individual antiviral immune response.
The Role of T Cells in the Immune Response against SARS-CoV-2
Respiratory viruses have evolved in many ways to evade or neutralize innate immunity; therefore, adaptive immunity, which is comprised of B and T cells, must also become activated for proper response. B cells are derived from the bone marrow and produce antibodies that bind to virus particles in the blood and mucosal surfaces, preventing the spread of the infection (humoral mediated antiviral response). Thus, there is an immense research interest in the identification and the use of the antibodies developed against the SARS-CoV-2 as a biomarker for the development of individual immunity or for therapeutic purposes as in the convalescent plasma therapy [12,13]. There are also substantial ongoing efforts placed to develop serological tests to accurately measure the levels of antiviral antibodies in an individual [14]. This should help the epidemiological assessment of the scale of the viral spread and allow countries to plan for ending lockdowns and the return to normal life. However, current challenges with the sensitivity and specificity for some of these assays are still limiting their wide use.
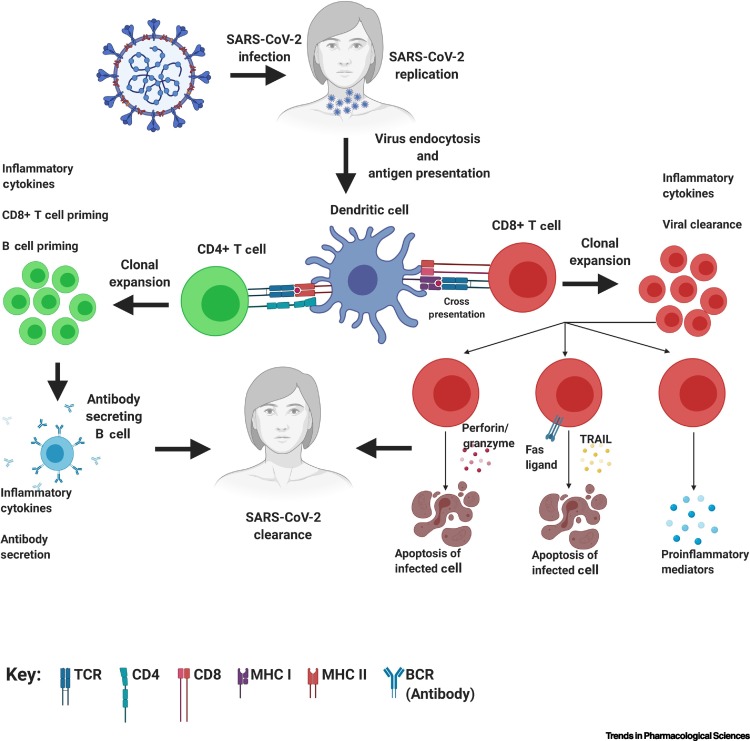
T cells, however, mature in the thymus, and CD8+ T cells kill virus-infected cells (cell-mediated antiviral response) through T cell receptor (TCR)-mediated recognition of viral antigens. Similar to the humoral mediated response, T cells are also essential for antiviral defense. Here, we focus on the role of T cell (cell-mediated) responses and the TCR repertoire, as they are often overlooked in early viral studies. T cells play a crucial role in the immune response to viral infections due to their ability to selectively eliminate virus-infected cells [15]. CD8+ T cells recognize viral antigens through their presentation by major histocompatibility class I (MHC I) molecules on virally infected cells as well as on uninfected dendritic cells and macrophages in the lymph nodes (a process known as cross-presentation) (Figure 1 ) [16]. Once T cells become activated through antigen recognition, they undergo a process termed ‘clonal expansion’, in which the activated T cell rapidly proliferates to generate a large number of T cells with identical TCRs and thus, identical antigen recognition [17]. To eliminate virus-infected cells, expanded CD8+ T cell clones, with TCRs specific for viral antigens, directly lyse them by perforin/granzyme release, induce apoptosis through tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or Fas ligand, and secrete proinflammatory mediators such as interferon-γ (IFN-γ) [18] (Figure 1). T cells are crucial mediators of antiviral adaptive immunity and delineating the role of the TCR in the context of SARS-CoV-2 should garner vast interest.
Figure 1.
T Cell-Mediated Response to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection.
Infection occurs in the patients after respiratory tract exposure. Subsequently, virus entry occurs via binding to angiotensin-converting enzyme 2 (ACE2) on the surface of various cell types and begins viral replication. Antigen-presenting cells such as dendritic cells endocytose the SARS-CoV-2 virus and degrade them through a process called antigen processing. These antigen fragments are then presented by proteins termed ‘MHC class molecules’, on the cell surface and allow recognition by a T cell. If a CD8+ T cell is capable of binding, it will undergo clonal expansion and directly target infected cells through either perforin/granzymes, FAS ligand/tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) pathways, or secretion of proinflammatory mediators. If the CD4+ T cell is capable of binding, it can activate B cells that recognize the antigen by causing them to clonally proliferate and secrete antibodies to target the SARS-CoV-2 virus. Abbreviations: BCR, B cell receptor; MHC, major histocompatibility complex; TCR, T cell receptor.
The Evolution of the TCR Repertoire during Viral Infection
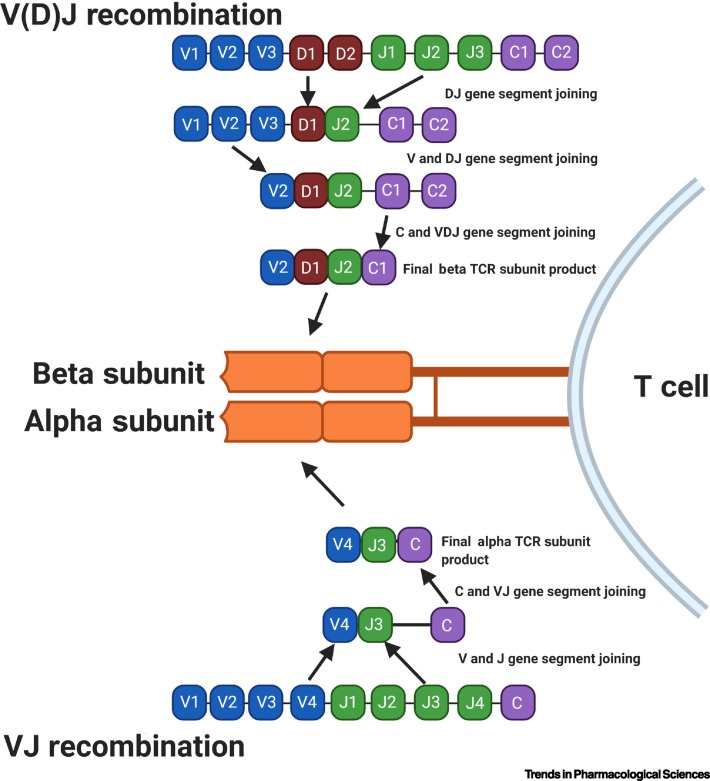
The specificity of T cells toward viral antigens presented by all nucleated cells on MHCs is determined by their TCR [19., 20., 21.]. Roughly 95% of the TCRs on circulating T cells are alpha (α) and beta (β) subunit heterodimers. TCR specificity for any given antigen is shaped through V(D)J recombination (rearrangement) [22]. During recombination, V and J (TCR alpha) or V, D, and J (TCR beta) gene segments are randomly selected from an array of V and J exons [23]. They undergo random addition and deletion of nucleotides to form the final mature TCR gene [24] (Figure 2 ). This process can generate up to 1015 unique TCRs [25,26]. The antigen specificity of each TCR lies in its complementarity-determining region 3 (CDR3), which is located at the V(D)J junction. As a result, each unique T cell clone is identified by its unique CDR3 region and, therefore, unique antigen specificity. The TCR recombination process generates a diverse TCR repertoire capable of recognizing a wide range of antigens and providing potent antiviral immunity [27]. The diversity and the composition of the TCR repertoire changes in response to acute and chronic infection, aging, cancer, and many other external and internal forces, making the repertoire highly dynamic. Exposure to antigen triggers massive expansion of T cells capable of recognizing the particular antigen (antigen-specific T cells), leading to skewing of the TCR repertoire (TCR bias) to favor antigen-specific T cells [28., 29., 30.].
Figure 2.
Schematic Diagram of the V(D)J Somatic Recombination to Generate T Cell Receptor (TCR).
The TCR is composed of the alpha (α) and beta (β) subunit. During T cell development, the loci that encode T cell receptor α and β-chains are rearranged. Variable (V), diversity (D), joining (J), and constant (C) gene segments are represented as blue, red, green, and purple rectangles, respectively. Diversity for the β chain is first created via recombination of D and J segments, followed by the V segments, and finally one of two C segments. Diversity for the α chain is created via recombination of V and J segments, followed by the one C segment. This process also involves the deletion and insertion of nucleotides at the V–D, D–J junctions for the β chain and V–J junctions for the α chain, thus adding more potential diversity to the TCR repertoire.
Evidence for TCR bias in antiviral immunity is demonstrated for many viral infections, such as the Epstein-Barr virus (EBV) and influenza. TCR analysis of peripheral blood lymphocytes from either EBV-positive or influenza-positive individuals has revealed several T cell clonotypes with recurrent TCR motifs shared by individuals [31., 32., 33.]. In addition, the expansion and maintenance of TCRs during chronic cytomegalovirus (CMV) infection demonstrates the dynamic and evolving nature of the TCR repertoire in response to viral infection [34]. Therefore, it stands to reason that the presence of a bias exists in the TCR repertoire as a result of the SARS-CoV-2 infection.
Interindividual Variability in the TCR Repertoire Influencing Their Immune Response
The characteristics of the TCR repertoire, such as diversity, composition, and dynamics, are controlled by both genetic and epigenetic factors, leading to interindividual variability. Here we discuss two factors that we believe are relevant to COVID-19 and the observed immune response disparities among patients.
Aging of the TCR Repertoire
It is well known that TCR repertoire diversity declines with aging. This decrease in TCR diversity as we age has also been demonstrated in various studies using high-throughput sequencing [TCR sequencing (TCR-seq)] technology. One study estimated the TCRβ diversity in naïve T cell repertoires was 60–120 million for individuals in the first two decades of life and declined to 8–57 million in individuals over 70 years old [35]. The evidence of aging TCR repertoire is demonstrated in the antiviral response against the human influenza A virus [39,40]. This may be of particular interest in the context of COVID-19, considering that the mortality of elderly patients with COVID-19 is higher than that of young and middle-aged patients [3,36., 37., 38.]. Whether the aged and less diverse TCR repertoire impacts the ability to generate a sufficiently robust T cell response against SARS-CoV-2 in older patients remains to be studied.
Genetic Variabilities in the HLA
Genetic differences in the HLA genes are well known to influence the composition of TCR repertoire by affecting intra- and extrathymic clonal selection, partially accounting for individual variations in the immune response to pathogens [41., 42., 43.]. These genetic differences in the HLA genes directly affect the binding affinity of MHC I/II molecules to antigens and, consequently, influence T cell recognition of pathogen-derived antigens [44., 45., 46., 47.]. One study conducted a comprehensive in silico analysis of viral peptide–MHC class I binding affinity across over a hundred HLA -A, -B, and -C genotypes for all SARS-CoV-2 peptides. It showed that HLA-B*46:01 had the fewest predicted binding peptides for SARS-CoV-2, suggesting higher vulnerability to COVID-19 among individuals with this allele [48]. The same allele was previously associated with susceptibility to SARS [49]. In contrast, HLA-B*15:03 was found to have the greatest capacity to present highly conserved SARS-CoV-2 peptides that are shared among common human coronaviruses, suggesting a potential cross-protective T cell immunity [48]. These observations raise the important question of which alleles confer susceptibility or resistance to the SARS-CoV-2 infection.
Unraveling the TCR Repertoire to Decode the Dynamic of SARS-CoV-2 Immune Response
A protective T cell-driven immune response to unpredicted antigens requires the immune system to generate a diverse TCR repertoire that is ready for any novel pathogen/antigen challenge, such as SARS-CoV-2. Therefore, the size and the diversity of the TCR repertoire are major determinants of the immune response to a given antigen [50]. The TCR repertoire can be further divided into naive and memory. The naive TCR repertoire represents the repertoire of antigen-inexperienced T cells. This population is shaped by thymopoiesis, which introduces new T cell clones and, therefore, new antigen specificities, to diversify the TCR repertoire [51]. Thymopoiesis can generate SAR-CoV-2 reactive T cells where they have not existed previously. The memory TCR repertoire represents the repertoire of antigen-experienced T cells. It is shaped by an individual’s history of antigen exposure, whereby those T cells that have participated in immune responses persist long term. Memory T cells, though they have been selected for by different antigens, such as those generated against prior coronaviruses, may still generate responses against novel viruses such as the SARS-CoV-2 [52]. Indeed, a recent study reported that T cell reactivity to SARS-CoV-2 epitopes is also detected in nonexposed individuals, suggesting crossreactive T cell recognition between circulating ‘common cold’ coronaviruses and SARS-CoV-2 [53].
The development of faster and cheaper sequencing technologies, augmented by the advances in computational tools, support the feasibility of using TCR analyses not only to track SARS-CoV-2-specific T cell expansion post-COVID-19 infection or in the course of treating patients with COVID-19, but also to establish certain features of the TCR repertoire architecture as predictive biomarkers for patients’ clinical outcome. A recent study demonstrated the feasibility and value of these analyses in patients with COVID-19. In ten patients with either early recovery stage or late recovery stage COVID-19 and five healthy donors, TCR-seq analysis showed that T cell expansion was lower in the early recovery group than in the healthy control group. Also, the naïve or central memory T cells showed little clonal expansion, while effector memory T cells, terminal effector CD8+ T cells, and proliferating T cells showed higher expansion levels. The study found that the most highly expanded clone in the early recovered group was TRAV8-6-TRAJ45:TRAV7-8-TRBJ2-1 [54]. Thus, a comprehensive characterization of the dynamics and composition of the TCR repertoires to SARS-CoV-2 infection can largely contribute to the evolving understanding of the functional and mechanistic involvement of the adaptive immune cell response and potentially guide the design of effective treatment. Here we summarize some of the most relevant clinical applications of obtaining comprehensive TCR repertoire data in the setting of COVID-19.
TCR-Based Biomarkers to Evaluate Herd Immunity and Advance the Development of COVID-19 Preventive Vaccines
Recent efforts have focused on identifying shared motifs in core sequences to predict conserved residues driving essential elements of TCR recognition [22,55]. As with any other viral infection, the ability of individuals’ TCR repertoire to effectively clear SARS-CoV-2 infection is highly dependent on its composition. Epitope-specific T cells have been identified in a study using samples obtained from individuals previously infected with SARS-CoV [56]. Similarly, another study demonstrated the presence of epitope-specific public T cells in both healthy and influenza-infected patients [32]. These studies show a correlation between the presence of specific T cell clones and antiviral immunity. Beyond identifying specific clones, TCRβ profiling has demonstrated convergent TCRβ repertoire evolution in large cohorts of unrelated individuals infected with CMV or vaccinated against yellow fever [57,58]. Additional work has demonstrated that information obtained from public TCRs in smallpox vaccinated mice can be used to identify smallpox vaccinated mice with >99% accuracy [59]. Also, it has been demonstrated that influenza A infection induces a polyclonal T cell response in patients, by TCRs predominantly utilizing TRBV19 gene [60].
Collectively, these studies suggest that the TCR repertoire changes in response to infection or vaccination in a potentially predictable manner and can therefore be leveraged to identify clones against SARS-CoV-2. Thus, TCR repertoire profiling in COVID-19 patients after recovery may uncover SARS-CoV-2 reactive clones and/or general TCR repertoire features that differentiate patients with prior infection from those that have never been infected (Figure 3A). TCR repertoire profiling combined with computational strategies may also enable the identification of SARS-CoV-2-specific TCR clones and general TCR repertoire signatures that may serve as a biomarker for herd immunity at the population level (Figure 3B).
Figure 3.
Workflow for Identifying Public T Cell Receptors (TCRs) Associated with Coronavirus Disease 2019 (COVID-19) to Determine Herd Immunity.
(A) Peripheral blood samples collected from COVID-19 infected and healthy uninfected individuals will be the input for the TCR repertoire analyses. Sequences and motif analyses will identify shared TCR clones that are unique to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epitopes. (B) Prediction algorithm generated in (A) should enable the use of TCR sequencing analysis of patient with unknown infection status to determine SARS-CoV-2 exposure history in an individual. This information can be valuable in the fight against COVID-19 as it can be applied to observe the degree of herd immunity in a particular population.
TCR-Based Immunotherapy for COVID-19
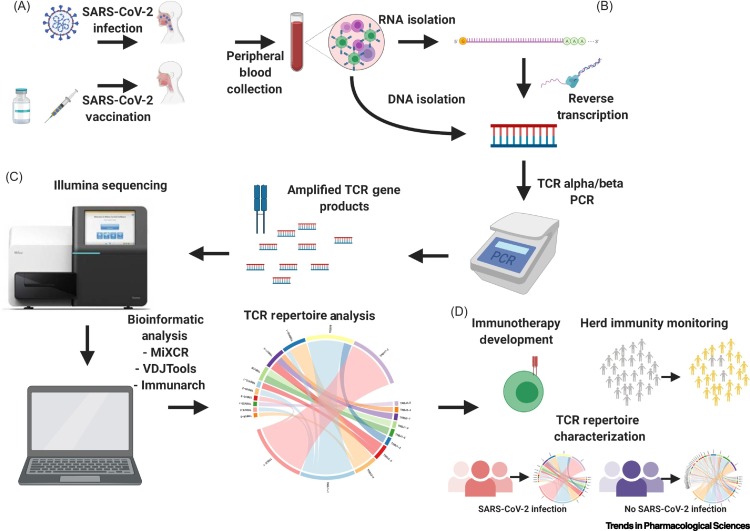
Adoptive transfer of TCR engineered T cell (TCR-T) has been applied to treat viral infections such as hepatitis B and C [61,62]. This was also demonstrated in preclinical models for prior coronaviruses such as SARS; engineered SARS-CoV-specific TCR-T cells from normal individuals have demonstrated antiviral activity that was comparable with that of natural SARS-CoV-specific memory CD8+ T cells [63]. The ability to identify potential TCR clones that are specific to SARS-CoV-2 antigens can be leveraged in the development of highly specific and advanced forms of immunotherapy based on TCR structural and sequence information. Identification of public clones through TCR repertoire profiling in infected and vaccinated individuals may guide the design of future TCR-T immunotherapies against SARS-CoV-2. SARS-CoV-2 reactive T cells can be expanded ex vivo and infused directly into an individual to shorten recovery time and increase the magnitude of response (Figure 4 ).
Figure 4.
T Cell Receptor (TCR) Repertoire Profiling Workflow in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Studies and Applications.
(A) RNA/DNA can be isolated from peripheral blood collected from SARS-CoV-2 infected/vaccinated individuals. (B) Peripheral blood RNA/DNA will be used as an input for TCR sequencing library preparation. (C) TCR sequencing will be done using Illumina or other sequencing platforms and resulting data analyzed using popular bioinformatic tools to characterize the TCR repertoire of SARS-CoV-2 infected/vaccinated individuals. (D) The information obtained can be used to advance immunotherapy development, characterize TCR repertoires in specific populations, and monitor herd immunity.
TCR Repertoire Changes as a Measure of Clinical Outcome
TCR repertoire composition and diversity are major determinants of disease outcomes following viral infection. Multiple studies have provided clear evidence indicating a highly diverse TCR repertoire provides protection against a wide array of antigens [22,64,65]. It is possible that a highly diverse TCR repertoire can allow for the right amount of avidity, affinity, and overall function [66,67]. Similar studies can be performed to track changes in the TCR repertoire in SARS-CoV-2 positive individuals, beginning at hospitalization, to precisely identify changes in the TCR repertoire and composition throughout disease progression. By examining the TCR repertoire composition and diversity in a large patient population, beginning at early stages of infection, we may be able to understand how these two parameters change in association with immune response and outcomes in COVID-19 patients.
Strategies for TCR Repertoire Profiling in Patients with COVID-19
Biological Samples
The selection of proper biological samples and determining an appropriate collection schedule is crucial to prepare the input for TCR analysis. Sputum samples, while feasible, may not offer sufficient depth of the TCR repertoire sequencing, as the number of CD8+ T cells in sputum, relative to other biological materials, is low [68]. Bronchoalveolar lavage and lung tissue samples may be an attractive and informative choice but acquiring such samples is difficult for a variety of reasons, particularly patient willingness and worker safety [69]. In addition, these procedures are invasive to perform on mild cases and otherwise healthy individuals for research purposes. Peripheral blood samples are easy to collect and are the most feasible, particularly to examine the dynamics of the TCR repertoire, as multiple collections could be performed, starting from diagnosis and then weekly till recovery or even afterward to track memory T cell clones. SARS-CoV-2 reactive T cells would transiently appear/persist in blood as they migrate from lymph nodes to tissue sites via peripheral blood circulation and thus could be captured in peripheral blood samples. However, it is possible that their frequency in the blood may be low. It is also possible that the lymphocytopenia observed in COVID-19 patients (particularly in severe cases) could require a larger volume of blood to obtain a sufficient number of T cells for comprehensive TCR repertoire profiling. Another drawback to using peripheral blood samples is that they would not capture SARS-CoV-2-specific T cell expansion as well as samples from infected tissues such as lung samples would, as SARS-CoV-2 T cells would transiently persist in peripheral blood circulation. Ultimately, sample selection and collection schedule will vary based on study objectives and the TCR profiling analysis platform, which would affect sample collection and storage (cells, RNA, or DNA).
Sequencing Platforms
Efforts to characterize the TCR repertoire in health and disease have increased dramatically in recent years due to advances in sequencing technologies. TCR-seq has provided important new insights into the human TCR repertoire, such as the more accurate estimation of the repertoire size and the presence of TCRs that are shared by multiple individuals. TCR-seq is different from standard DNA/RNA-seq in that library preparation is achieved through selective amplification of TCR gene products, allowing the capture of the TCR repertoire at lower overall financial, reagent, and data storage costs. Different methods have been developed to analyze the TCR repertoire via next generation sequencing using genomic DNA (gDNA) and RNA as input. Multiplex PCR-, target enrichment-, and 5′-RACE (RNA only)-based approaches are the most widely used TCR-seq strategies [71]. Multiplex PCR-based methods involve the use of a primer mix that anneals to every V and J exon and the constant region. However, preferential amplification of highly abundant gene products can occur when using multiplex PCR, contributing to inaccuracies in reported clone frequencies. Additionally, intronic sequences and incomplete VDJ recombination products may contribute to data noise when using gDNA samples.
Target enrichment strategies utilize probes to capture TCR gDNA/RNA sequences after initial library preparation using compatible kits (such as the Illumina TruSeq kit or the Agilent SureSelectXT kit) followed by relatively few cycles of multiplex PCR amplification [72,73]. This method reduces the extent of amplification bias seen in standard multiplex PCR but is more labor/time intensive and does not resolve the issues associated with gDNA input. The 5′-RACE method with an RNA input involves the reverse transcription of TCR transcripts followed by the addition of a known sequence at the 5′ end of each cDNA molecule. This 5′ sequence is leveraged along with the highly conserved TCR α and β constant region during nested PCR (separate primers needed to amplify either α or β transcripts) to achieve equally efficient amplification of all TCR transcripts, regardless of abundance [74,75]. While the 5′-RACE approach avoids the PCR bias seen in the other two methods, it requires the use of sequencing platforms capable of long reads, such as the Illumina Miseq, as fragment sizes can exceed 600 bp.
While bulk TCR-seq (TCR-seq of a heterogeneous T cell population) via the earlier-discussed strategies can provide information about the evolution, the dynamic, and the architecture of the TCR repertoire during the course of SARS-CoV-2 infections, it cannot be used to identify the exact TCR α/β pairs that participate in antiviral responses (only draw inferences based on clone frequency changes) [52,57]. Additionally, bulk TCR-seq will likely capture bystander activated and expanded T cells, contributing to data noise. Single-cell TCR-seq in tandem with single-cell flow cytometry sorting is an approach to precisely identify SARS-CoV-2-specific T cells. Single-cell TCR analysis can be limited by their depth and their ability to capture minor clones. Regardless, these approaches generate millions of TCR sequences in one experiment and enable the characterization of the TCR repertoire in COVID-19 patients [73,76].
Computational Tools
Bioinformatic tools have been developed to enable streamlined and standardized TCR-seq data analysis and have been used by our group and others extensively. MiXCRi is a UNIX-based software used to identify and build TCR sequences using raw sequencing data [77]. MiXCR uses a built-in aligner and reference library of V, D, J, and C segment sequences to efficiently and accurately call V, D, J, and C regions from millions of reads and then groups similar reads together to build a final TCR sequence. The assembled TCR sequences along with V, D, and J annotations are written into a text-formatted file that can be used by analysis tools such as VDJToolsii to characterize TCR-seq data and identify changes in the TCR repertoire that may occur during disease progression in COVID-19 patients [78].
Concluding Remarks
Recent advances in TCR-seq have provided evidence for a close relationship between TCR diversity and immune responses against viral antigens. Under homeostatic conditions, the TCR repertoire is largely diverse and polyclonal. However, in the context of viral infection, preferential selection of T cell clones can contribute to the narrowing of an antigen-selected TCR repertoire. To what extent SARS-CoV-2 influences the TCR repertoire remains to be assessed. In addition, what impact the COVID-19 resulting lymphocytopenia has on the TCR repertoire architecture, diversity, and composition, and how comorbidities such as diabetes, obesity, and hypertension which are implicated in COVID-19 severity may influence the TCR repertoire, are questions that warrant attention (see Outstanding Questions).
Outstanding Questions.
How does SARS-CoV-2 influence the TCR repertoire?
How and which characteristics of the TCR repertoire may explain the disproportional risk of COVID-19 in older population?
What impact does the lymphocytopenia in COVID-19 have on the TCR repertoire architecture, diversity, and composition?
How does genetic diversity in the HLAs contribute to the SARS-CoV-2-specific TCR repertoire and, potentially, the immune response?
How might factors such as comorbidities influence the TCR repertoire in COVID-19 patients?
Alt-text: Outstanding Questions
Studying the TCR repertoire may also reveal potential public TCRs, which may allow us to track infection as well as understand immunity against this infection. In line with our opinion, several efforts are on the way to perform TCR analysis in COVID-19 patients. A recent study has reported a longitudinal high-throughput TCR-seq to track changes in the T cell repertoire following two mild cases of COVID-19 infection [79]. In addition, clinical trials (clinical trial numbersiii: NCT04379466 and NCT04362865) have been initiated to examine the immune repertoire in COVID-19 patients. The NCT04362865 trial, sponsored by the National Institute of Health, is particularly interesting as it aims to investigate both the B and T cell repertoire and immune response in patients with acute and resolved COVID-19 infection. We should emphasize that the B cell immunoglobulin repertoire analysis is also a significant and an urgently needed task. Indeed, a recent study has applied a single-cell immune profiling approach that combines single-cell transcriptomic analysis with single-cell TCR and B cell receptor (BCR) sequencing technologies to examine the cellular context of the adaptive immune response and immune repertoires of T and B cells in blood samples of COVID-19 patients [54]. We foresee that other platforms can also provide valuable data, such as CyTOF mass cytometry and the 10×Genomicsiv immune profiling platform that utilizes DNA-barcoded peptide-MHC (pMHC) multimers and antibodies conjugated to barcodes to dissect the immune repertoire in the setting of COVID-19. These analyses should reveal clonality, diversity, antigen specificity, and cellular context; and pair α and β chain TCR sequences from individual T cells. It also allows for pairing heavy and light chain immunoglobulin sequences from individual B cells. These data, combined with sophisticated computational methods, will enable the identification of the paired TCR α and β chain sequences with TCR-pMHC specificity corresponding to SARS-CoV-2. More importantly it will simultaneously measure cell surface protein expression with gene expression and uncover meaningful details related to the mechanisms contributing to the pathology of this infection and the patient’s clinical outcome [57]. Together, these technologies can be leveraged to monitor adaptive immune responses in COVID-19 patients and guide future SARS-CoV-2 immunotherapy development. Which characteristics of the TCR repertoire may explain the disproportional risk of COVID-19 in older population and how genetic diversity in the HLAs contribute to the SARS-CoV-2-specific TCR repertoire and potentially the immune response are some of many outstanding questions to be tackled (see Outstanding Questions).
Acknowledgments
We acknowledge the University of Southern California, School of Pharmacy Seed Fund, and The Norris Cancer Center pilot funding that support HA research effort. All figures were made with BioRender (https://biorender.com/).
Glossary
- Adaptive immunity
immunity that occurs in response to specific antigens. Activation of this immunity is dependent on the presentation of antigens by the innate immune system. It is mediated by immunoglobulins and T cells.
- Angiotensin-converting enzyme 2 (ACE2)
a carboxypeptidase that is mainly expressed in vascular endothelial cells and the renal tubular epithelium. ACE2 binds to SARS-CoV-2 spike protein and promotes internalization of the virus into the cells.
- Antigens
molecule that is capable of stimulating an immune response, specifically activating T cells or B cells.
- CD8+ T cells
T cells that express CD8 on their cell surface are selected in the thymus to recognize and respond to foreign peptides that are presented in the groove of MHC class I molecules. Also known as cytotoxic T cells.
- Clonal expansion
the process in which a small number of precursor cells that recognize a specific antigen proliferate into expanded clones, differentiate, and acquire various effector and memory phenotypes, which promote effective immune responses.
- Clonotypes
term used to describe a unique nucleotide sequence that arises during the gene rearrangement process for the T or B cell receptor.
- Complementarity-determining region 3 (CDR3)
antigen binding region within TCRs that are encoded by the region spanning the V and J for the alpha subunit or the V, D, and J junctions for the beta subunit of the TCR. The diversity of the CDR3 loops is significantly increased because of the addition and loss of nucleotides during the joining process. The CDR3 region determines antigen specificity of TCRs.
- Coronavirus disease (COVID-19)
infectious disease caused by SARS-CoV-2 virus and is the official name given by the World Health Organization to the disease caused by this newly identified coronavirus.
- Cytomegalovirus (CMV)
a betaherpesvirus, represents the major infectious cause of birth defects, as well as an important pathogen for immunocompromised individuals. The viral nucleocapsid containing a linear double-stranded DNA of 230 kb is surrounded by a proteinaceous tegument that is enclosed by a lipid bilayer. Establishes lifelong latent infection and is reactivated by immunosuppression.
- Dendritic cells
professional antigen-presenting cells, uniquely able to induce naïve T cell activation and effector differentiation.
- Epstein-Barr virus (EBV)
a human herpesvirus 4 with a large double-stranded DNA genome that belongs to the γ-herpesviridae subfamily. EBV was the first tumor virus identified in humans. The virus is primarily associated with lymphomas and epithelial cell cancers and is the causative agent of mononucleosis after infection.
- High-throughput sequencing
technology capable of sequencing multiple DNA molecules in parallel to enable millions of DNA molecules to be sequenced at once. This is much quicker and cheaper than the previously used Sanger sequencing and, as such, revolutionized the study of genomics and molecular biology.
- Human leukocyte antigen (HLA)
gene cluster that encodes the major histocompatibility complex proteins, which present antigen peptides to the host immune system. These are among the most polymorphic genes in the human genome.
- Influenza
a virus commonly known as the ‘flu’ that attacks the respiratory system. Influenza viruses are members of the family Orthomyxoviridae. This family represents enveloped viruses, the genome of which consists of segmented negative-sense single-strand RNA segments. There are four genera of this family: types A, B, C, and Thogotovirus, of which, however, only genera A and B are clinically relevant for humans.
- Innate immunity
an immunological subsystem that comprises the cells and mechanisms that provide the first line of defense from infection in a nonspecific manner. It detects and responds to pathogen associated molecular patterns (PAMPS) that are not naturally occurring within the entire organism.
- Interferon-γ (IFN-γ)
cytokine that has since been characterized as a homodimeric glycoprotein with pleiotropic immunologic functions. IFN-γ is primarily secreted by activated T cells and natural killer cells and can promote macrophage activation, mediate antiviral and antibacterial immunity, enhance antigen presentation, orchestrate activation of the innate immune system, coordinate lymphocyte–endothelium interaction, regulate Th1/Th2 balance, and control cellular proliferation and apoptosis.
- Lymphocyte
white blood cells that have important immune functions. The main populations of lymphocytes are B cells, T cells, and natural killer (NK) cells.
- Macrophage
myeloid cells that have important innate immune functions. Macrophages reside in tissues and respond to infection by producing inflammatory mediators and engulfing bacteria.
- Major histocompatibility class I (MHC I)
protein found on the surface of all nucleated cells that functions in antigen presentation to CD8+ T cells.
- Memory T cells
a CD8 T cell that has responded to cognate antigen (Ag) and persists long-term. These cells can populate peripherally and are poised to immediately proliferate, execute cytotoxic functions, and secrete effector cytokines upon Ag re-encounter and exist in different metabolic, transcriptional, and epigenetic states.
- Naïve T cell
a T cell pool that is generally considered to be a fairly quiescent, homogeneous pool of antigen-inexperienced cells. In response to encountering and interacting with cognate antigens in the periphery, naive T cells will proliferate and differentiate into different types of effector and memory T cells, which can migrate to different tissues for local antigen patrol.
- Public T cells
refers to T cells that carry TCR sequences that occur in multiple unrelated individuals, often in response to the same antigen.
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
severe acute respiratory syndrome (SARS)-like coronavirus that belongs to the family Coronaviridae and the subfamily Coronavirinae. It uses the angiotensin-converting enzyme 2 (ACE 2) receptor to enter into the human cells. It is the causative agent of COVID-19.
- T cell
white blood cells that are important for adaptive immunity. They have unique cell surface receptors that are generated by randomly assorting genes. These receptors allow T cells to sense and respond to diverse types of infection.
- T cell receptor (TCR)
protein that recognizes small protein fragments that are displayed by the MHC molecule on the surface of nucleated cells. Each T cell expresses a unique TCR that is generated by randomly recombined genes. This ensures that T cells can respond (at the population level) to almost any infection.
- TCR bias
refers to the over-representation of TCR genes or clones in the TCR repertoire. The bias in TCR usage can be categorized based on the diversity in a specific T cell population. TCR bias can be influenced by many factors, such as thymic selection and antigen-driven selection, which can narrow the available T cell repertoire over time.
- TCR engineered T cell (TCR-T)
T cells engineered by viral vectors to express the TCR gene with defined specificity. These cells can be modified to target cancer cells or bacterial/virus-infected cells.
- TCR repertoire
the sum of all TCRs by the T cells of one individual generated by somatic DNA rearrangements.
- TCR sequencing (TCR-seq)
high-throughput sequencing technologies used in the examination of antigen receptor repertoires at single-nucleotide and, more recently, single-cell resolution.
- Thymopoiesis
process that turns thymocytes into mature T cells according to selection processes. This selection process is vitally important in shaping the population of thymocytes into a peripheral pool of T cells that are able to respond to foreign pathogens but remain tolerant towards the body’s own antigens.
- V(D)J recombination
the process by which T cells randomly assemble different gene segments, known as variable (V), diversity (D), and joining (J) genes, in order to generate unique receptors (known as T-cell receptors) that can collectively recognize many different types of molecule.
Resources
ihttps://mixcr.readthedocs.io/en/master/iihttps://vdjtools-doc.readthedocs.io/en/master/iiihttps://clinicaltrials.gov/ivwww.10xgenomics.com/products/single-cell-immune-profiling/References
- 1.Shi Y. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai W. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao L. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin C. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. Published online March 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao M. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature Medicine. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 12.Shen C. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan K. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Q.X. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nature Medicine. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 15.Ranasinghe S. Antiviral CD8(+) T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity. 2016;45:917–930. doi: 10.1016/j.immuni.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang-Huau T.L. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H. Select sequencing of clonally expanded CD8(+) T cells reveals limits to clonal expansion. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8995–9001. doi: 10.1073/pnas.1902649116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braciale T.J. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 2012;12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song I. Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8(+) T cell epitope. Nat. Struct. Mol. Biol. 2017;24:395–406. doi: 10.1038/nsmb.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen T.H., Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 21.Stumptner-Cuvelette P. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12144–12149. doi: 10.1073/pnas.221256498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dash P. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassing C.H. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 24.Komori T. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 25.Nikolich-Zugich J. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 26.Laydon D.J. Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen J.L. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 28.Chen G. Sequence and structural analyses reveal distinct and highly diverse human CD8(+) TCR repertoires to immunodominant viral antigens. Cell Rep. 2017;19:569–583. doi: 10.1016/j.celrep.2017.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trautmann L. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 30.Pantaleo G. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 31.Callan M.F. T cell selection during the evolution of CD8+ T cell memory in vivo. Eur. J. Immunol. 1998;28:4382–4390. doi: 10.1002/(SICI)1521-4141(199812)28:12<4382::AID-IMMU4382>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 32.Sant S. Single-cell approach to influenza-specific CD8(+) T cell receptor repertoires across different age groups, tissues, and following influenza virus infection. Front. Immunol. 2018;9:1453. doi: 10.3389/fimmu.2018.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gil A. Epstein-Barr virus epitope-major histocompatibility complex interaction combined with convergent recombination drives selection of diverse T cell receptor alpha and beta repertoires. mBio. 2020;11 doi: 10.1128/mBio.00250-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schober K. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat. Immunol. 2020;21:434–441. doi: 10.1038/s41590-020-0628-2. [DOI] [PubMed] [Google Scholar]
- 35.Britanova O.V. Dynamics of individual T cell repertoires: from cord blood to centenarians. J. Immunol. 2016;196:5005–5013. doi: 10.4049/jimmunol.1600005. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. Published online February 24, 2020. [DOI] [PubMed] [Google Scholar]
- 37.Zhou F. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan Q. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen T.H.O. Perturbed CD8(+) T cell immunity across universal influenza epitopes in the elderly. J. Leukoc. Biol. 2018;103:321–339. doi: 10.1189/jlb.5MA0517-207R. [DOI] [PubMed] [Google Scholar]
- 40.Gil A. Narrowing of human influenza A virus-specific T cell receptor alpha and beta repertoires with increasing age. J. Virol. 2015;89:4102–4116. doi: 10.1128/JVI.03020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharon E. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akolkar P.N. Influence of HLA genes on T cell receptor V segment frequencies and expression levels in peripheral blood lymphocytes. J. Immunol. 1993;150:2761–2773. [PubMed] [Google Scholar]
- 43.Reed E.F. Influence of HLA class I and class II antigens on the peripheral T-cell receptor repertoire. Hum. Immunol. 1994;40:111–122. doi: 10.1016/0198-8859(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 44.Kekik C. Relationship between HLA tissue type, CMV infection, and acute graft-vs-host disease after allogeneic hematopoietic stem cell transplantation: single-center experience. Transplant. Proc. 2009;41:3859–3862. doi: 10.1016/j.transproceed.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Quinones-Parra S. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1049–1054. doi: 10.1073/pnas.1322229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falfan-Valencia R. An increased frequency in HLA class I alleles and haplotypes suggests genetic susceptibility to influenza A (H1N1) 2009 pandemic: a case-control study. J Immunol Res. 2018;2018 doi: 10.1155/2018/3174868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hertz T. HLA targeting efficiency correlates with human T-cell response magnitude and with mortality from influenza A infection. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13492–13497. doi: 10.1073/pnas.1221555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen A. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00510-20. Published online April 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin M. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4 doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritmahan W. Revealing factors determining immunodominant responses against dominant epitopes. Immunogenetics. 2020;72:109–118. doi: 10.1007/s00251-019-01134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar B.V. Human T cell development, localization, and function throughout life. Immunity. 2018;48:202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riley T.P. T cell receptor cross-reactivity expanded by dramatic peptide-MHC adaptability. Nat. Chem. Biol. 2018;14:934–942. doi: 10.1038/s41589-018-0130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grifoni A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen W. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glanville J. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emerson R.O. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat. Genet. 2017;49:659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 58.Pogorelyy M.V. Precise tracking of vaccine-responding T cell clones reveals convergent and personalized response in identical twins. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12704–12709. doi: 10.1073/pnas.1809642115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf K. Identifying and tracking low-frequency virus-specific TCR clonotypes using high-throughput sequencing. Cell Rep. 2018;25:2369–2378. doi: 10.1016/j.celrep.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yassai M.B. CDR3 motif generation and selection in the BV19-utilizing subset of the human CD8 T cell repertoire. Mol. Immunol. 2016;72:57–64. doi: 10.1016/j.molimm.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kah J. Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J. Clin. Invest. 2017;127:3177–3188. doi: 10.1172/JCI93024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balasiddaiah A. Hepatitis C virus-specific T cell receptor mRNA-engineered human T cells: impact of antigen specificity on functional properties. J. Virol. 2017;91 doi: 10.1128/JVI.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh H.L.J. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price D.A. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Miconnet I. Large TCR diversity of virus-specific CD8 T cells provides the mechanistic basis for massive TCR renewal after antigen exposure. J. Immunol. 2011;186:7039–7049. doi: 10.4049/jimmunol.1003309. [DOI] [PubMed] [Google Scholar]
- 66.Zehn D. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price D.A. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freeman C.M. Design of a multi-center immunophenotyping analysis of peripheral blood, sputum and bronchoalveolar lavage fluid in the Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS) J. Transl. Med. 2015;13:19. doi: 10.1186/s12967-014-0374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charlton C.L. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosati E. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017;17:61. doi: 10.1186/s12896-017-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okino S.T. Evaluation of bias associated with high-multiplex, target-specific pre-amplification. Biomol. Detect. Quantif. 2016;6:13–21. doi: 10.1016/j.bdq.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Linnemann C. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 74.Mamedov I.Z. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Front. Immunol. 2013;4:456. doi: 10.3389/fimmu.2013.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rapid amplification of 5′ complementary DNA ends (5′ RACE) Nat. Methods. 2005;2:629–630. doi: 10.1038/nmeth0805-629. [DOI] [PubMed] [Google Scholar]
- 76.Woodsworth D.J. Sequence analysis of T-cell repertoires in health and disease. Genome Med. 2013;5:98. doi: 10.1186/gm502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bolotin D.A. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 78.Shugay M. VDJtools: unifying post-analysis of T cell receptor repertoires. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minervina A.A. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T cell memory formation after mild COVID-19 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.100545. Published online May 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]