Abstract
With the recent outbreak of COVID-19, fast diagnostic testing has become one of the major challenges due to the critical shortage of test kit. Pneumonia, a major effect of COVID-19, needs to be urgently diagnosed along with its underlying reasons. In this paper, deep learning aided automated COVID-19 and other pneumonia detection schemes are proposed utilizing a small amount of COVID-19 chest X-rays. A deep convolutional neural network (CNN) based architecture, named as CovXNet, is proposed that utilizes depthwise convolution with varying dilation rates for efficiently extracting diversified features from chest X-rays. Since the chest X-ray images corresponding to COVID-19 caused pneumonia and other traditional pneumonias have significant similarities, at first, a large number of chest X-rays corresponding to normal and (viral/bacterial) pneumonia patients are used to train the proposed CovXNet. Learning of this initial training phase is transferred with some additional fine-tuning layers that are further trained with a smaller number of chest X-rays corresponding to COVID-19 and other pneumonia patients. In the proposed method, different forms of CovXNets are designed and trained with X-ray images of various resolutions and for further optimization of their predictions, a stacking algorithm is employed. Finally, a gradient-based discriminative localization is integrated to distinguish the abnormal regions of X-ray images referring to different types of pneumonia. Extensive experimentations using two different datasets provide very satisfactory detection performance with accuracy of 97.4% for COVID/Normal, 96.9% for COVID/Viral pneumonia, 94.7% for COVID/Bacterial pneumonia, and 90.2% for multiclass COVID/normal/Viral/Bacterial pneumonias. Hence, the proposed schemes can serve as an efficient tool in the current state of COVID-19 pandemic. All the architectures are made publicly available at: https://github.com/Perceptron21/CovXNet.
Keywords: COVID-19 diagnosis, Imaging informatics, Neural network, Pneumonia diagnosis, Transfer learning, X-ray
Highlights
-
•
A novel deep neural network architecture is proposed based on depthwise dilated convolutions.
-
•
Larger database containing non-COVID pneumonia X-rays are used for initial training stage that are effectively transferred for utilizing smaller database containing COVID-19 X-rays.
-
•
Features extracted from different resolutions of X-rays are jointly converged by the proposed stacking algorithm.
-
•
Clinical investigation is carried out by analyzing the gradient based activation mapping.
1. Introduction
Coronavirus disease (COVID-19), caused by SARS-CoV-2, has been declared as a global pandemic by WHO that almost collapsed the healthcare systems in many of the countries [1], [2]. The mortality rate is increasing alarmingly throughout the world demanding an early response to diagnose and prevent the rapid spread of this disease. Because of having no specific drugs and treatments, the situation has become frightening to billions of individuals [3]. Symptoms ranging from dry cough, sore throats, and fever to organ failure, septic shock, severe pneumonia, and Acute Respiratory Distress Syndrome (ARDS) are detected from COVID-19 patients [2]. Reverse transcription-polymerase chain reaction (RT-PCR), the most commonly used diagnostic test of COVID-19, suffers from low sensitivity in early stages with elongated test period assisting further transmission [4]. Furthermore, the extreme scarcity of this expensive test kit [5] exacerbating the situation. Hence, a chest scan such as X-rays and Computer tomography (CT) scans are prescribed to all individuals with potential pneumonia symptoms for faster diagnosis and isolation of the infected individuals. With a serious shortage of experts, while having large similarities of COVID-19 with traditional pneumonia, an artificial intelligence (AI) assisted automated detection scheme can be a significant milestone towards a drastic reduction of testing time.
In [6], [7], CT scans are used with deep learning-based systems for automated COVID-19 pneumonia detection. Though CT scans provide finer details, X-rays are quicker, easier to take, less injurious and more economical alternative. However, due to the scarcity of COVID-19 X-rays, it is extremely difficult to train a very deep network effectively. Hence, transfer-learning can be a viable solution in this circumstance that have been widely adopted in many recently proposed COVID-19 detection schemes [8], [9], [10], [11]. Yet, the traditional scheme of transfer-learning that uses established deep networks pre-trained on the ImageNet database for transferring its initial learning cannot be a good choice as the characteristics of COVID-19 X-rays are solely different from images intended for other applications. To the best of our knowledge, no established method has been yet reported to utilize chest X-rays for the challenging task of separating pneumonia patients with traditional viral/bacterial infection from COVID-19 patients that contains significantly overlapping features.
In this paper, an efficient scheme is proposed utilizing relevant available X-ray images for training an efficient deep neural network so that the trained parameters can be effectively utilized for detecting COVID-19 cases even with very smaller size of available COVID-19 X-rays. At first, instead of using other traditional databases used for disparate applications, a larger database containing X-rays from normal and other non-COVID pneumonia patients are used for transfer learning. A deep neural network is proposed, named as CovXNet, to detect COVID-19 from X-rays, which is built from a basic structural unit utilizing depthwise convolutions with varying dilation rates to incorporate local and global features extracted from diversified receptive fields. Furthermore, a stacking algorithm is developed that utilizes a meta-learner to optimize the predictions of different forms of CovXNet operating with different resolutions of X-rays and thus covering diverse receptive fields. Next, the initially trained convolutional layers are transferred directly with some additional fine-tuning layers to train on the smaller COVID-19 X-rays along with other X-rays. This modified network incorporates all its initial learning on X-rays into further exploration of the COVID-19 X-rays for proper diagnosis. Moreover, a gradient-based localization is integrated for further investigation by circumscribing the significant portions of X-rays that instigated the prediction. Intense experimentations of the proposed methods exhibit significant performance in all traditional evaluation metrics.
2. Methodology
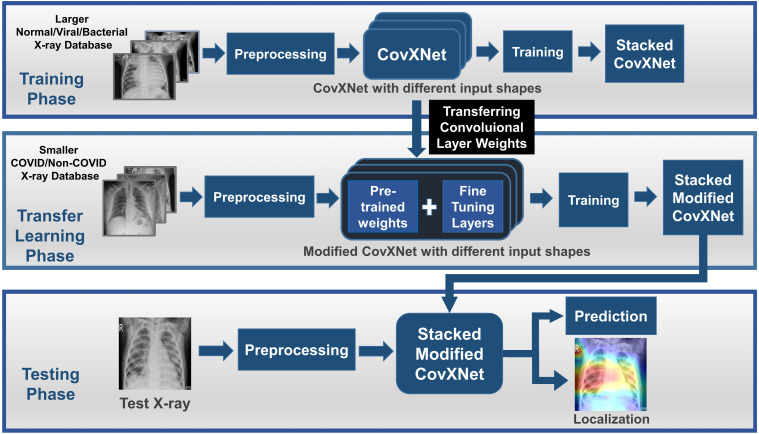
The workflow of the proposed method is schematically shown in Fig. 1. As pneumonia caused by COVID-19 contains a high degree of similarity with traditional pneumonia from both clinical and physiological perspectives [12], [13], transferring knowledge gained from a large number of chest X-rays collected from normal and other traditional pneumonia patients can be an effective way to utilize smaller COVID-19 X-rays for extracting additional features. Therefore, in the initial training phase, a larger database containing X-rays collected from normal and other non-COVID viral/bacterial pneumonia patients are used for training the proposed CovXNet. Here, after pre-processing, different resolutions of input X-rays are deployed to separately train different CovXNet architectures. Afterward, a stacking algorithm is employed to optimize the predictions of all these networks through a meta-learner. As the convolutional layers are optimized to extract significant spatial features from X-rays, weights of these layers are directly transferred in the transfer learning phase. Next, a smaller database containing COVID-19 and other pneumonia patients are used to train the additional fine-tuning layers integrated with the CovXNet. Finally, in the testing phase, this trained, fine-tuned, stacked modified CovXNet is employed to efficiently predict the test X-ray image class. Moreover, a gradient-based localization algorithm is used to visually localize the significant portion of X-ray that mainly contribute to the decision.
Fig. 1.
The complete workflow is represented schematically. In the training phase, a larger non-COVID X-ray database is used to train CovXNet. Predictions from different CovXNet architectures for different resolutions of X-rays are optimized through a stacking algorithm. In the transfer learning phase, a smaller COVID/non-COVID X-ray database is used to train the additional fine-tuning layers of the modified CovXNet. In the testing phase, prediction of class along with localization of significant portion in test X-ray is provided by the stacked modified CovXNet.
2.1. Preprocessing
The collected X-rays pass through minimal preprocessing to make the testing process faster and easier to implement. Images are reshaped to uniform sizes followed by min–max normalization for further processing with the proposed CovXNet.
2.2. Proposed structural units
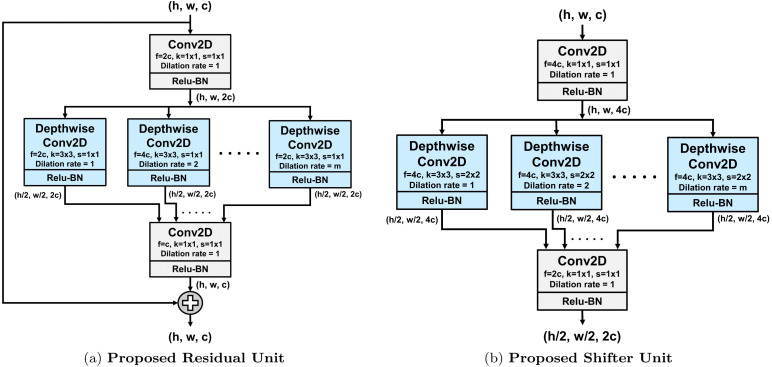
Two structural units are proposed, as shown in Fig. 2, which are the main building blocks of the proposed CovXNet architecture. Depthwise dilated convolutions are efficiently introduced in these units to effectively extract distinctive features from X-rays to identify pneumonia.
Fig. 2.
Proposed structural units. Here, , , and denote the height, width and no. of channels of the feature map, respectively, while ‘’ stands for kernel size, ‘’ for strides and ‘’ for number of filters in the convolution. In depthwise convolution, dilation rate will be varied from to ‘’.
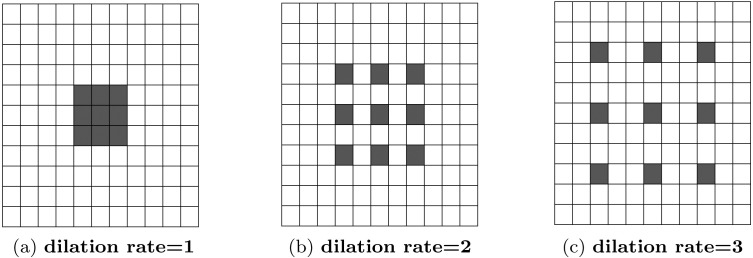
As the features of pneumonia can be very localized (consolidated) or diffusely distributed over a larger area of the X-rays, it is necessary to incorporate features from different levels of observations [12], [14], [15]. In [16], dilated convolution is introduced to broaden the receptive field of the convolution without increasing the total number of parameters of kernels by increasing dilation rates. This process is presented visually in Fig. 3. Various features extracted from different convolutions with varying dilation rates will integrate more diversity in the feature extraction process.
Fig. 3.
Dilated Convolution for different dilation rates with kernel size are encompassing different receptive areas. With increased dilation rate, the receptive area also gets bigger, though kernel size is kept unchanged.
Moreover, traditional convolution can be divided into depthwise convolution followed by a pointwise convolution that makes the process extremely computationally efficient [17]. In depthwise convolution, i.e. a spatial convolution, each input channel is individually filtered by separate filters without combining them. Afterward, a pointwise convolution, i.e. traditional convolution with 1 × 1 windows, is performed for projecting the inter-channel features into a new space.
| (1) |
| (2) |
In the proposed structural units, depthwise dilated convolutions along with pointwise convolutions are introduced efficiently. Firstly, the input feature map undergoes through a pointwise convolution to project the inter-channel information into a broader space. Following that, numerous depthwise convolutions are performed with different spatial kernels with varying dilation rates starting from dilation rate of to a max-dilation rate of . The value of is adjusted according to the shape of the input feature map for covering the necessary receptive area. Hence, these depthwise convolutions are extracting spatial features from various receptive fields ranging from very localized features to broader perspective generalized features. Thereafter, all these variegated features go through another pointwise convolution to merge these inter-channel features into a constricted space.
In the proposed residual unit, as shown in Fig. 2(a), this pointwise–depthwise–pointwise convolutional mapping is set to fit a residual mapping by adding the output with the input feature map. This type of residual learning, introduced in [18], is used to capture the identity mapping that helps to produce a very deep network without overfitting. If the proposed residual mapping is denoted by with input tensor such that , the final output mapping can be represented as . These residual units can be stacked in more numbers to produce a deeper network.
In the proposed shifter unit, as presented in Fig. 2(b), the input feature map undergoes through some dimensional transformations. Firstly, the depth of the input feature map is increased by times to introduce more processing for spatial reduction. Later, the spatial dimensions are halved through strided depthwise convolution instead of traditional pooling operation as it loses positional information [5]. Such spatial reduction helps to broaden the receptive field for further processing to introduce more generalization. Finally, the depth of the output feature map is doubled in the final pointwise convolution to increase the filtering operations in later stages.
2.3. Proposed CovXNet architecture
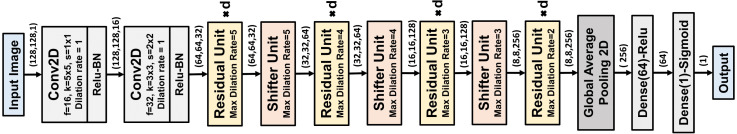
The residual and shifter units are the main building blocks of the proposed CovXNet architecture, as shown in Fig. 4. Firstly, the input image undergoes convolutions with broader kernels to process the information with the larger receptive area. The following convolution introduces some dimensional transformation. Afterward, it passes through a series of residual units. Depth of this stack of residual learning can be increased to produce a deeper network. Shifter units are incorporated in between such stacks to introduce dimensional transformation to generalize the extracted the information further. However, the maximum dilation rate of each residual unit is determined based on the dimension of the input feature map. For processing larger features, is set to be higher to increase the maximum receptive area of the residual unit accordingly to encompass more variations in the extracted features. Finally, the processed feature map passes through global average pooling followed by some densely connected layers before providing final prediction. Moreover, the rectified linear unit (Relu) is instigated after each convolution for non-linear activation with batch normalization to make the convergence faster.
Fig. 4.
Schematic of the Proposed CovXNet architecture optimized for input shape . Each residual unit is replicated for ‘’ times. With decreasing feature map dimension, maximum dilation rate is reduced in both the residual and shifter units to adjust the smaller receptive area.
2.4. Stacking of multiple networks
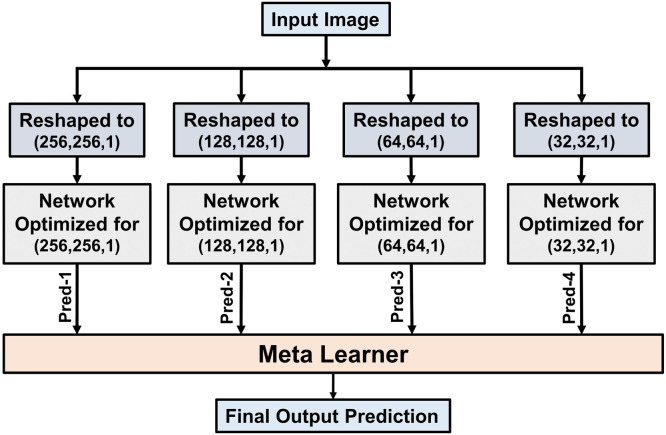
The proposed CovXNet architecture can be optimized for input images with different resolutions by adjusting the number and maximum dilation rates of the structural residual and shifter units. Such introduced architectural variations with changing resolutions of X-rays will force these networks to explore the information content from different levels of observations. Though with the reduction of the resolution, information content of an image decreases, it insists the network on focusing the generalized features by broadening receptive area. In the proposed scheme, a stacking algorithm is incorporated to learn the generalizability of these networks by optimizing their predictions to produce a more accurate final prediction. This step can be considered as a meta-learning process and it is schematically presented in Fig. 5.
Fig. 5.
Individually optimized networks are stacked together by using the meta-learner for obtaining more-optimized predictions. This meta-learner explores the predictions of different individual networks to achieve the most optimized outcome.
Firstly, total training data is divided into two portions: one for training all the individual networks, while other for training the meta-learner. Next, all the individual networks are trained separately with the resized representations of input images. These networks analyze the data from different perspectives for proper prediction. After being properly optimized, these networks are used to generate a prediction on the other portion of data kept for meta learner training. Finally, the meta learner is being optimized by exploring the predictions of all the individual networks to generate the final output. This approach offers the meta learner to optimize the analysis by inspecting diversified viewpoints. As the meta learner deals with the predictions of individually optimized networks, a very small portion of training data is used to train the meta-learner. Hence, shallow neural networks along with other traditional machine learning techniques can be utilized to build the meta-learner. Implementational details of the whole process are given in Algorithm 1.
2.5. Proposed transfer learning method on novel corona virus data using CovXNet
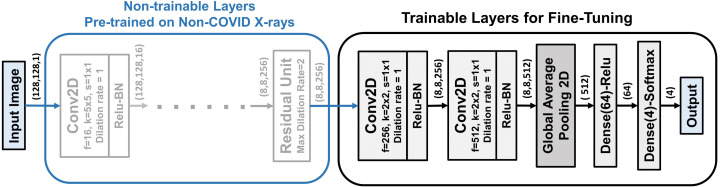
As the CovXNet is optimized for analyzing X-rays using very deep architectures with a large number of convolutional layers, this knowledge can be effectively transferred to learn the representation of novel COVID-19 X-rays. This scheme is presented in Fig. 6. All the convolutional layers including all residual and shifter units that were initially trained on non-COVID X-rays are directly transferred with their pre-trained weights. Additionally, two more convolutional layers are integrated at the bottom for fine-tuning. Afterward, a traditional global pooling layer with a series of densely connected layers are also incorporated for training. As very few images of COVID-19 X-rays are available, it is difficult to train very deep architecture using them. Nevertheless, as most of the pre-trained convolutional layers are directly utilized without further training, very few parameters need to be fine-tuned for the newly integrated layers.
Fig. 6.
Proposed Transfer learning scheme on CovXNet for fine tuning with small number of images. Pre-trained convolutional layers trained on non-COVID X-rays are directly transferred. Two additional convolutional layers along with the densely connected layers are fine tuned with the smaller database formed with the COVID-19 X-rays.
2.6. Network training and optimization
The network is trained by back propagation algorithm with regularization to minimize the cross entropy loss function , which is given by,
| (3) |
where denotes the weight vector, is the actual level, is the predicted output at node of output for input and is the regularization parameter to reduce overfitting.
The weight of the layer is updated by Adam optimizer [19] at time t with learning rate , which is given by,
| (4) |
| (5) |
| (6) |
Here, is gradient at time , and are the exponential average of gradients and squares of gradients along , respectively, and and are the two hyperparameters.
For multi-class prediction, softmax classifier is used for normalizing the probability vector for any input while sigmoid activation is used for the binary case to normalize the probability prediction and these are given by,
| (7) |
| (8) |
3. Results and discussions
In this section, the performances of the proposed schemes are presented with the visual interpretations of the spatial localization from clinical perspectives. Different cases are analyzed with COVID-19 X-rays to explore the robustness of the method. Finally, some state-of-the-art methods for pneumonia detection along with some traditional networks are also compared.
3.1. Database
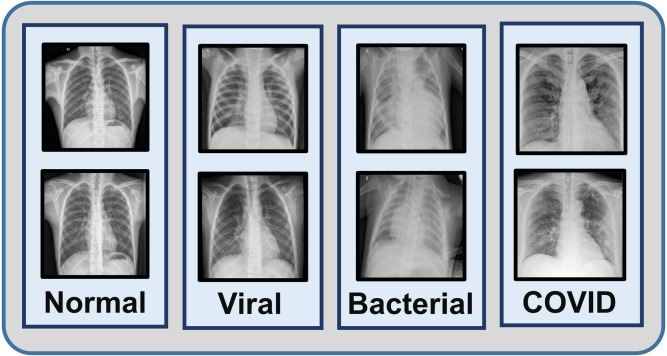
One of the datasets used in this study is a collection of total images consisting normal X-rays, non-COVID viral pneumonia X-rays and bacterial pneumonia X-rays collected in Guangzhou Medical Center, China [20]. Another database containing X-rays of different COVID-19 patients is collected from Sylhet Medical College, Bangladesh which is also verified by expert radiologist panel. Finally, a smaller balanced database is created combining all the COVID-19 X-rays with equal number of normal, viral, bacterial pneumonia X-rays (305 X-rays in each class) that are employed for the transfer learning phase (sample images are shown in Fig. 7). The rest of the X-rays (Normal, viral, bacterial pneumonia) are utilized for the initial training phase. In both these phases, five fold cross validation scheme is employed for the evaluation of the proposed method.
Fig. 7.
Sample X-ray images of normal, viral, bacterial and COVID-19 caused pneumonia patients are shown.
3.2. Experimental setup
Different hyper-parameters of the network are chosen through experimentation for better performance. Intel® Xeon® CPU @2.80 GHz with M Cache and cores along with GB RAM is used for experimentation. For hardware acceleration, x NVIDIA RTX Ti GPU having with CUDA cores running MHz with GB GDDR6 memory is deployed. Numerous traditional metrics of classification tasks are used for evaluating the performance of the proposed architectures, such as accuracy, sensitivity, specificity, area under curve (AUC) score, precision, recall, and F1 score.
3.3. Performance evaluation
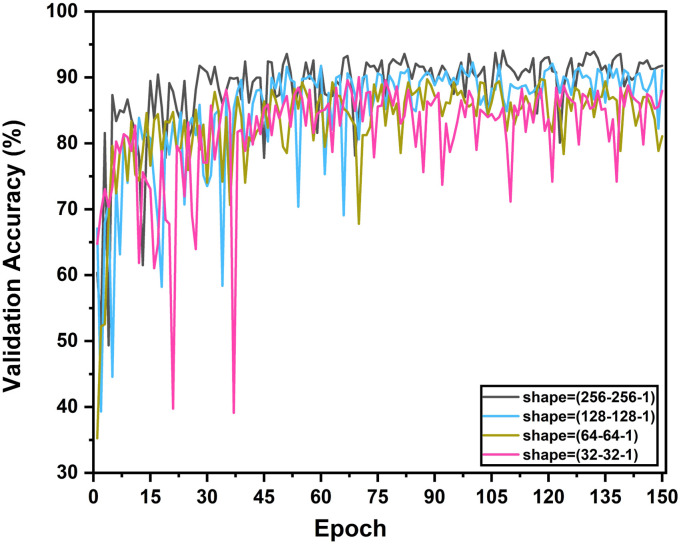
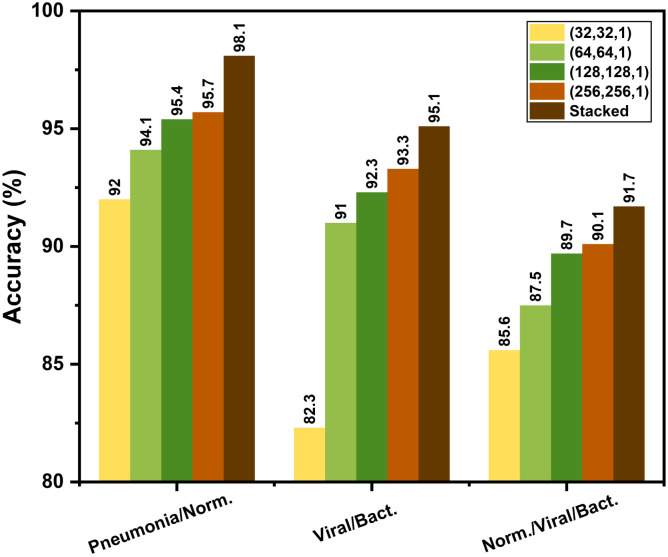
At the initial training phase, the network is optimized for the normal and other non-COVID viral/bacterial pneumonia X-rays. Different combinations of output classes are experimented for analyzing the inter-class relationships. As the CovXNet architecture is highly scalable to adjust the receptive area depending on the input data, performance with different resolutions of images are experimented with targeting different classes of pneumonia. From the multi-class validation accuracy plot for different resolutions over the training epochs, as shown in Fig. 8, it can be observed that the networks with a higher resolution of X-rays lead over smaller ones throughout all the epochs. Nevertheless, the smallest representation still provides comparable performance that indicates the higher generalizability of the proposed CovXNet which can still perform well with very small-scale of information. As a result, utilizing images of different resolutions in the proposed meta-learner, the prediction accuracy is further improved, as shown in Fig. 9. It is clearly observed that the meta-learner optimizes the predictions generated from a different level of data representation and provides a significant rise in accuracy for all types of classifications. As different optimized networks are analyzing the data from diversified perspectives, optimizing all of these predictions through additional meta-learner provides a more generalized decision.
Fig. 8.
Multi-class validation accuracy in different training epochs is shown for different resolutions of inputs. With the increase in resolution, trend of validation accuracy becomes higher over the epochs.
Fig. 9.
Effect of using proposed stacking algorithm in the initial training phase: Stacking provides considerable rise of accuracy compared to each individual network operating on different resolutions of X-rays in all the tasks.
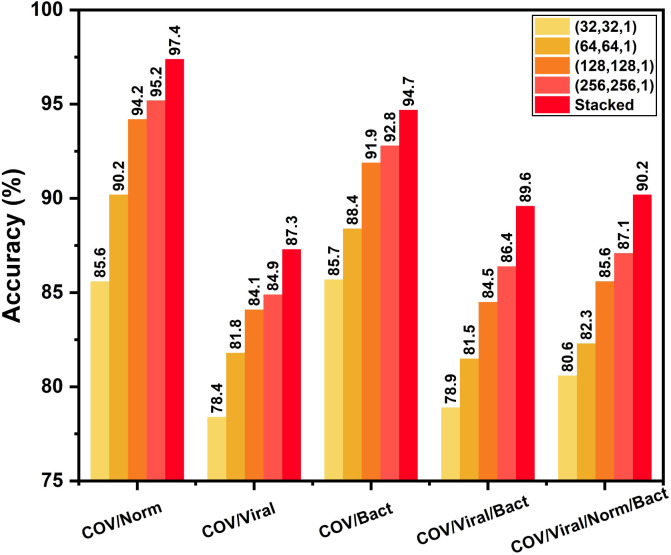
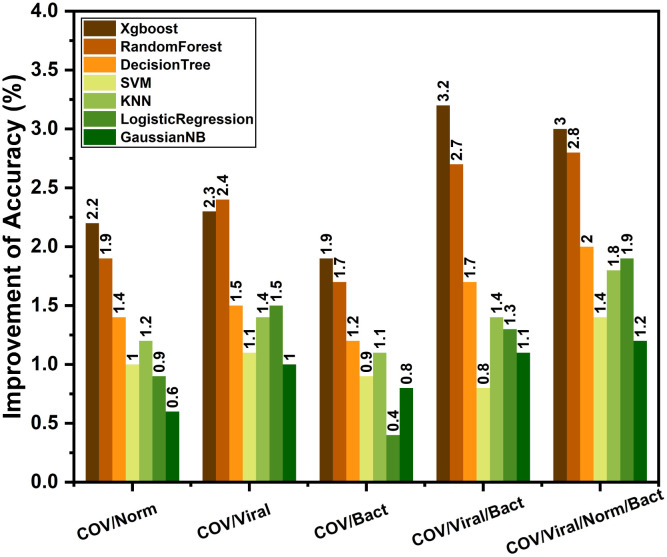
After completing the initial training on non-COVID X-rays, these highly optimized convolutional layers are transferred to train with a smaller database containing COVID-19 X-rays. In this transfer learning phase, COVID-19 X-rays are experimented with different output classes of normal/traditional pneumonia through fine-tuning of the additionally added layers. Similar to the initial training phase, an additional meta-learner is trained to optimize the predictions obtained from different variants of modified CovXNet that are optimized for different resolutions of input X-rays. The performance of these individually trained networks along with the performance obtained after stacking with meta-learner is shown in Fig. 10. As COVID-19 caused pneumonia contains a significant overlap of features with other viral pneumonia [12], [13], it is difficult to isolate these two categories. Hence, comparably smaller accuracy is noticeable for separating COVID-19 and other viral pneumonia X-rays. However, due to significant variations of features between COVID-19 and normal/bacterial pneumonia X-rays [25], [26], higher accuracy is obtained in such cases. Moreover, stacking with meta-learner provides improved performance in all the classification tasks relating to COVID-19. For example, stacking provides 2.2% improvement of accuracy with respect to the best performing individual network in COVID-19/Normal classification. However, this improvement of accuracy may vary depending on the type of supervised classifier to be used in the meta learner phase. For experimentation, different classifiers are tested, such as Xgboost, random forest, decision tree, SVM, KNN, logistic regression and Gaussian naive bias algorithm. Improvement of performance with different meta-learners are shown in Fig. 11 for different classification tasks. Xgboost and RandomForest algorithm provide the best performance as these learners provide prediction after further ensembling of several boosting and bagging algorithms, respectively.
Fig. 10.
Effect of using proposed stacking algorithm in the transfer learning phase: Although moderate accuracy is achieved for COVID-19/Viral pneumonia classification due to overlapping of features, use of the proposed stacking algorithm results in relatively satisfactory performance.
Fig. 11.
Effect of the choice of meta-learner in stacking: Though each one provides some improvement, Xgboost and RandomForest meta-learner provide considerably significant improvement compared to others in most of the tasks.
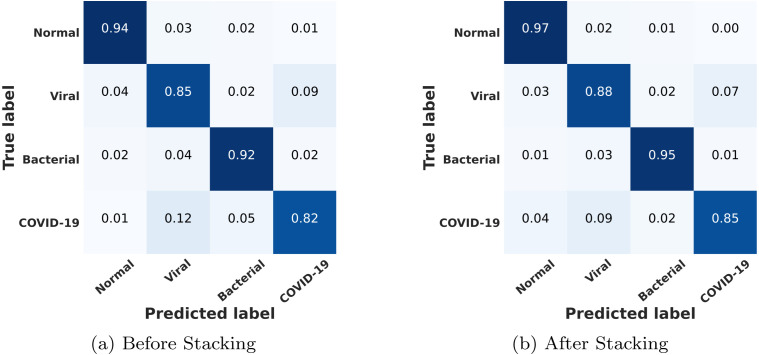
The multi-class confusion matrix is provided in Fig. 12. As expected, due to a high degree of overlapping features, a few COVID-19/viral cases exhibit misclassification. However, very satisfactory performance is obtained for other classification cases. However, recall of all of the classes can be improved further by incorporating the meta-learner through the stacking of different networks.
Fig. 12.
Multi-class confusion matrices are shown before and after stacking. Sensitivity is lower for COVID-19 and viral cases compared to normal and bacterial pneumonia due to overlapping of features. Through stacking, sensitivity can be increased for all the classes.
The performance of the proposed schemes in the initial training phase on non-COVID X-rays is compared with other existing approaches in Table 1. Here, the performance of different traditional architectures [18], [27], [28], developed for other computer vision applications, are compared with our proposed CovXNet. Additionally, performance of some state-of-the-art AI-based pneumonia detection schemes [21], [22], [23] are also compared. Rajraman et al. [21], Kermany et al. [22], and Chouhan et al. [23] utilized conventional transfer learning schemes using pre-trained networks on ImageNet database for traditional pneumonia detection. The proposed schemes outperform most other approaches by a considerable margin.
Table 1.
Performance comparison of the proposed method with other state-of-the-art approaches in non-COVID pneumonia detection.
| Task | Methods | Accuracy (%) | AUC score (%) | Precision (%) | Recall (%) | Specificity (%) | F1 score (%) |
|---|---|---|---|---|---|---|---|
| Normal/ Pneumonia |
Proposed | 98.1 | 99.4 | 98.0 | 98.5 | 97.9 | 98.3 |
| Residual | 91.2 | 96.4 | 90.7 | 95.9 | 84.1 | 93.4 | |
| Inception | 88.7 | 92.6 | 88.9 | 94.1 | 80.2 | 91.1 | |
| VGG-19 | 87.2 | 90.7 | 85.6 | 91.1 | 77.9 | 89.3 | |
| [21] | 95.7 | 99.0 | 95.1 | 98.3 | 91.5 | 96.7 | |
| [22] | 92.8 | 96.8 | – | 93.2 | 90.1 | – | |
| [23] | 96.4 | 99.3 | 93.3 | 99.6 | – | – | |
| Viral/ Bacterial Pneumonia |
Proposed | 95.1 | 97.6 | 94.9 | 96.1 | 94.3 | 95.5 |
| Residual | 89.5 | 92.4 | 88.3 | 96.9 | 78.1 | 92.4 | |
| Inception | 85.8 | 90.6 | 84.5 | 93.8 | 72.1 | 88.9 | |
| VGG-19 | 83.2 | 88.5 | 81.1 | 91.3 | 71.7 | 86.6 | |
| [21] | 93.6 | 96.2 | 92.0 | 98.4 | 86.0 | 95.1 | |
| [22] | 90.7 | 94.0 | – | 88.6 | 90.9 | – | |
| Normal/ Viral/ Bacterial/ Pneumonia |
Proposed | 91.7 | 94.1 | 92.9 | 92.1 | 93.6 | 92.6 |
| Residual | 86.3 | 88.5 | 86.3 | 88.5 | 93.5 | 87.4 | |
| Inception | 81.1 | 84.6 | 75.4 | 84.9 | 86.2 | 78.9 | |
| VGG-19 | 79.8 | 83.1 | 74.5 | 82.9 | 83.4 | 77.9 | |
| [21] | 91.7 | 93.8 | 91.7 | 90.5 | 95.8 | 91.1 | |
In Table 2, the performance of the proposed CovXNet is compared with other traditional networks on COVID-19 and other types of pneumonia detection. It can be observed that the proposed CovXNet architecture provides significantly better performance in different classification tasks handling with COVID-19 X-rays compared to other traditional architectures. Moreover, in Table 3, the proposed method is compared with other existing state-of-the-art approaches for COVID-19 detection from X-rays. As the proposed schemes utilized all the non-COVID X-rays in the initial learning phase, final training and evaluation is carried out on the separated balanced database containing X-rays of COVID patients. Ozturk et al. [24] proposed a deep neural network based approach without applying transfer learning strategies. Whereas, Wang. et al. [8], Ioannis et al. [9], Sethy et al. [10], and Narin et al. [11] used traditional networks with conventional transfer learning scheme from ImageNet database. In most of these cases, the obtained result is biased due to the small amount of COVID-19 X-rays. It should be noticed that the proposed schemes provide consisting performance in different combinations of classification with balanced set of data. Moreover, the larger number of non-COVID X-rays are properly utilized for initial training phase that is effectively transferred for diagnosing COVID-19 and other pneumonias in the final transfer learning phase.
Table 2.
Performance comparison of the proposed method with other traditional networks on COVID-19 and other pneumonia detection.
| Task | Methods | Accuracy (%) | AUC score (%) | Precision (%) | Recall (%) | Specificity (%) | F1 score (%) |
|---|---|---|---|---|---|---|---|
| COVID/ Normal |
Proposed | 97.4 | 96.9 | 96.3 | 97.8 | 94.7 | 97.1 |
| Residual | 92.1 | 91.2 | 90.4 | 93.4 | 89.2 | 91.9 | |
| Inception | 89.5 | 84.3 | 89.1 | 87.7 | 83.2 | 88.4 | |
| VGG-19 | 85.3 | 82.7 | 86.3 | 83.9 | 79.9 | 85.1 | |
| COVID/ Viral Pneumonia |
Proposed | 87.3 | 92.1 | 88.1 | 87.4 | 85.5 | 87.8 |
| Residual | 80.4 | 78.9 | 81.1 | 79.3 | 77.1 | 80.2 | |
| Inception | 78.2 | 75.5 | 76.8 | 79 | 75.4 | 77.9 | |
| VGG-19 | 72.1 | 67.7 | 70.9 | 74.7 | 69.3 | 72.8 | |
| COVID/ Bacterial Pneumonia |
Proposed | 94.7 | 95.1 | 93.5 | 94.4 | 93.3 | 93.9 |
| Residual | 84.2 | 80.3 | 86.7 | 83.5 | 82.4 | 85.1 | |
| Inception | 83.1 | 79.9 | 82.2 | 85.2 | 83.6 | 83.7 | |
| VGG-19 | 77.2 | 75.5 | 73.3 | 80.3 | 71.4 | 76.8 | |
| COVID/ Viral/ Bacterial Pneumonia |
Proposed | 89.6 | 90.7 | 88.5 | 90.3 | 87.6 | 89.4 |
| Residual | 82.1 | 79.8 | 81.5 | 80.3 | 78.5 | 80.9 | |
| Inception | 84.3 | 83.1 | 81.4 | 85.9 | 80.8 | 83.7 | |
| VGG-19 | 79.1 | 77.5 | 76.5 | 80.7 | 77.2 | 78.6 | |
| COVID/ Normal/ Viral/ Bacterial |
Proposed | 90.2 | 91.1 | 90.8 | 89.9 | 89.1 | 90.4 |
| Residual | 82.3 | 80.7 | 82.7 | 79.5 | 80.7 | 81.1 | |
| Inception | 82.9 | 79.8 | 80.6 | 84.3 | 82.4 | 82.5 | |
| VGG-19 | 80.8 | 78.5 | 77.4 | 81.6 | 78.1 | 79.5 | |
Table 3.
Performance comparison of the proposed scheme with other state-of-the-art approaches on COVID-19 and other pneumonia detection.
| Work | Amount of chest X-rays | Architecture | Accuracy (%) |
|---|---|---|---|
| Ozturk et al. [24] | 125 COVID-19 + 500 No finding | DarkCovidNet | 98.08 |
| 125 COVID-19 + 500 Pneumonia + 500 No finding |
87.02 | ||
| Wang et al. [8] | 53 COVID-19 + 5526 Non-COVID | COVID-Net | 92.4 |
| Ioannis et al. [9] | 224 COVID-19 + 700 Pneumonia + 504 Normal |
VGG-19 | 93.48 |
| Sethy et al. [10] | 25 COVID-19 + 25 Non-COVID | ReNet-50/SVM | 95.38 |
| Narin et al. [11] | 50 COVID-19 + 50 Non-COVID | ResNet-50 | 98 |
| Proposed | 305 COVID-19 + 305 Normal | Stacked Multi-resolution CovXNet |
97.4 |
| 305 COVID-19 + 305 Viral Pneumonia | 87.3 | ||
| 305 COVID-19 + 305 Bacterial pneumonia | 94.7 | ||
| 305 COVID-19 + 305 Viral Pneumonia + 305 Bacterial pneumonia |
89.6 | ||
| 305 COVID-19 + 305 Normal + 305 Viral Pneumonia + 305 Bacterial Pneumonia |
90.3 | ||
3.4. Discriminative localization obtained by proposed CovXNet
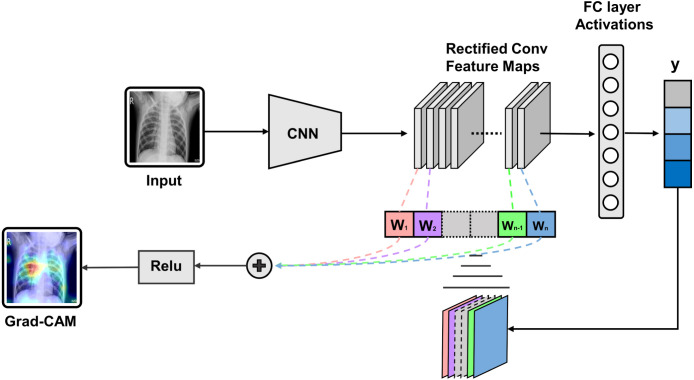
Gradient-based class activation mapping (Grad-CAM) algorithm [29] is integrated with the proposed CovXNet to generate the class activation mapping for localizing the particular portion of the X-rays that mainly instigated the decision, as shown in Fig. 13. By superimposing the heatmap with the input X-rays, such localizations are studied further to interpret the learning of the network from the clinical perspective. In Fig. 14, some of the X-rays with imposed localization are shown. Following findings are summarized:
Fig. 13.
Process of gradient weighted class activation mapping (Grad-CAM): Gradient of the target output class with respect to the final feature map is used to from a coarse localization of significant portions.
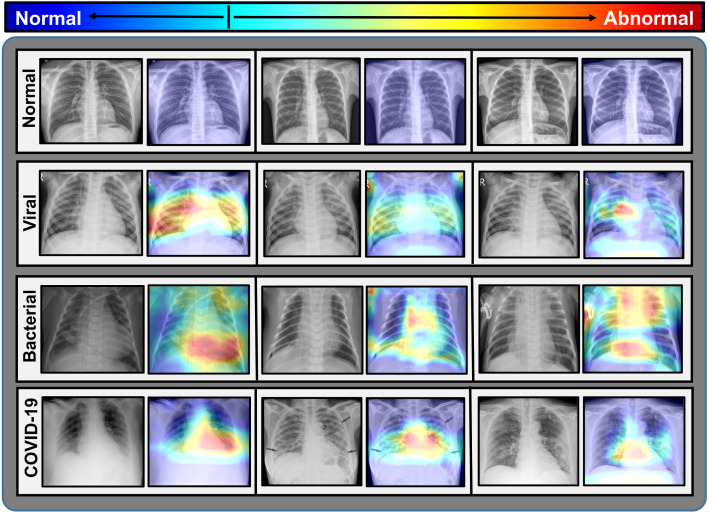
Fig. 14.
Significant portions of the test X-rays that instigate the decision are localized by imposing the activation heatmap obtained from CovXNet.
-
•
In normal X-rays, no kind of opacity is present that isolates the normal patients from all kinds of pneumonia patients having some form of opacities [14], [15], [30]. In Fig. 14, it is observed that no significant region is localized for normal X-rays. As it is more distinguishable, it is easier to isolate from other patients.
-
•
By carefully examining the heatmaps generated for traditional viral pneumonia, it can be observed that our model has localized regions with bilateral multifocal ground-glass opacities (GGO) along with patchy consolidations in some of the cases. Additionally, some localized regions contain diffused GGOs and multilobar infiltrations. These localized features are also commonly approved radiological features of traditional viral pneumonia [4], [12], [14], [30].
-
•
In the case of bacterial pneumonia, the localized activation heatmaps are mainly involving opacities with consolidation on lower and upper lobes. Additionally, there is also the involvement of both unilateral and bilateral along with peripheral. According to [14], [15], these features mainly represent bacterial pneumonia.
-
•
According to [12], [13], there are lots of similarities between COVID-19 and traditional viral pneumonia both demonstrating bilateral GGOs along with some patchy consolidations. Some more likely features of COVID-19 caused pneumonia are reported in [12], [13], [25], [26], such as peripheral and diffuse distribution, vascular thickening, fine reticular opacity along with the conventional viral-like ground-glass opacities. By carefully examining the generated heatmap from some of the COVID-19 infected X-rays (Fig. 14), it is distinguishable that peripheral and diffuse distribution of such opacities is diagnosed. Moreover, vascular thickening is also localized for some of the cases along with other traditional viral features.
Therefore, the radiological features extracted and localized by the proposed CovXNet provide substantial information about the underlying reasons for pneumonia. This type of localization can assist the clinicians to analyze the prediction obtained from the proposed scheme. All these findings are verified by expert radiologists for detailed investigation from the clinical perspective.
One major challenge is the scarcity of COVID-19 X-ray images which causes few misclassifications as well as some scattering in gradient-based localizations out of the region of interest. Our proposed transfer learning scheme has exploited the few available COVID-19 X-rays effectively. Moreover, this model can be made more accurate and robust through the incorporation of more data. The proposed scheme is highly adaptive and the CovXNet can be more finely tuned in the transfer learning phase with additional COVID-19 X-rays. Further research should be carried out with more diversified data for a thorough investigation of the clinical features of COVID-19. An amalgamation of clinical data and other radiographic findings can substantially improve the accuracy of diagnosis.
4. Conclusion
A deep neural network architecture namely CovXNet is proposed to efficiently detect COVID-19 and other types of pneumonia with distinctive localization from chest X-rays. Instead of using traditional convolution, efficient depthwise convolution is used with varying dilation rates that integrates features from diversified receptive fields to analyze the abnormalities in X-rays from different perspectives. To utilize the small number of COVID-19 X-rays, a larger database is utilized containing X-rays from normal and other traditional pneumonia patients for initially training the deep network. Due to significant overlapping characteristics between COVID-19 and other pneumonia, by transferring the initially trained convolutional layers with some additional fine-tuning layers, a very satisfactory result is obtained with a smaller database containing COVID-19 X-rays. Moreover, it is observed that a stacking algorithm provides additional performance improvement by further optimizing predictions obtained from different variations of CovXNet that are primarily optimized with various resolutions of input X-rays. Furthermore, a generated class activation map provides discriminative localization of the abnormal zones that can assist to diagnose the variations of clinical features of pneumonia on X-rays. The performance of these schemes can be improved further by integrating more sample X-rays of COVID-19 patients for training in the transfer learning phase. Experimental results obtained from extensive simulations suggest that it can a very effective choice for faster diagnosis of COVID-19 and other pneumonia patients. Moreover, the proposed CovXNet is highly scalable with enormous receptive capacity that can also be employed in varieties of other computer vision applications.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to express their sincere gratitude to Dr. Swajal Chandra Das and Dr. Mostaque Ahmed Bhuiyan, department of Radiology and Imaging, Sylhet Medical College and Hospital, for providing necessary assistance in verifying the collected X-rays of local COVID patients and in investigating the activation heatmap localization of the proposed CovXNet from clinical perspectives.
Footnotes
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Contributor Information
Tanvir Mahmud, Email: tanvirmahmud@eee.buet.ac.bd.
Md Awsafur Rahman, Email: mdawsafurrahman@ug.eee.buet.ac.bd.
Shaikh Anowarul Fattah, Email: fattah@eee.buet.ac.bd.
References
- 1.Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World health organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020 doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicro. Ag. 2020:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franquet T. Imaging of pulmonary viral pneumonia. Radiology. 2011;260(1):18–39. doi: 10.1148/radiol.11092149. [DOI] [PubMed] [Google Scholar]
- 5.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 7.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Wong A. 2020. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolopoulos I.D., Mpesiana T.A. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.P.K. Sethy, S.K. Behera, Detection of coronavirus disease (COVID-19) based on deep features, Preprints 2020030300 (2020) 2020.
- 11.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.-B., Wang D.-C., Mei J. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020:200823. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020:200230. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franquet T. Imaging of pneumonia: trends and algorithms. Eur. Respir. J. 2001;18(1):196–208. doi: 10.1183/09031936.01.00213501. [DOI] [PubMed] [Google Scholar]
- 15.Vilar J., Domingo M.L., Soto C., Cogollos J. Radiology of bacterial pneumonia. Eur. J. Radiol. 2004;51(2):102–113. doi: 10.1016/j.ejrad.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Yu F., Koltun V. 2015. Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122. [Google Scholar]
- 17.F. Chollet, Xception: deep learning with depthwise separable convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 1251–1258.
- 18.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.
- 19.Kingma D.P., Ba J. 2014. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- 20.Kermany D., Zhang K., Goldbaum M. Labeled optical coherence tomography (OCT) and chest X-ray images for classification. Mendeley Data. 2018;2 [Google Scholar]
- 21.Rajaraman S., Candemir S., Kim I., Thoma G., Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl. Sci. 2018;8(10):1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C., Damaševičius R., de Albuquerque V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020;10(2):559. [Google Scholar]
- 24.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Lui M.M.-s., Lo C.S.-Y., Leung B., Khong P.-L. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology. 2020;2(1) doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the inception architecture for computer vision, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2818–2826.
- 28.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 29.R.R. Selvaraju, M. Cogswell, A. Das, R. Vedantam, D. Parikh, D. Batra, Grad-CAM: visual explanations from deep networks via gradient-based localization, in: Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 618–626.
- 30.Koo H.J., Lim S., Choe J., Choi S.-H., Sung H., Do K.-H. Radiographic and CT features of viral pneumonia. Radiographics. 2018;38(3):719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]