Abstract
Natural products generally fall into the biologically relevant chemical space and always possess novel biological activities, thus making them a rich source of lead compounds for new drug discovery. With the recent technological advances, natural product-based drug discovery is now reaching a new era. Natural products have also shown promise in epigenetic drug discovery, some of them have advanced into clinical trials or are presently being used in clinic. The histone lysine specific demethylase 1 (LSD1), an important class of histone demethylases, has fundamental roles in the development of various pathological conditions. Targeting LSD1 has been recognized as a promising therapeutic option for cancer treatment. Notably, some natural products with different chemotypes including protoberberine alkaloids, flavones, polyphenols, and cyclic peptides have shown effectiveness against LSD1. These natural products provide novel scaffolds for developing new LSD1 inhibitors. In this review, we mainly discuss the identification of natural LSD1 inhibitors, analysis of the co-crystal structures of LSD1/natural product complex, antitumor activity and their modes of action. We also briefly discuss the challenges faced in this field. We believe this review will provide a landscape of natural LSD1 inhibitors.
KEY WORDS: Epigenetic regulation, Histone demethylase, Natural products, LSD1 inhibitors, Drug discovery, Cancer therapy
Abbreviations: α-MG, α-mangostin; ΔΨm, mitochondrial transmembrane potential; AML, acute myeloid leukemia; CCC, cut countercurrent chromatography; CD11b, integrin alpha M; CD14, cluster of differentiation 14; CD86, cluster of differentiation 86; CoREST, RE1-silencing transcription factor co-repressor; COVID-19, coronavirus disease; EdU, 5-ethynyl-20-deoxyuridine; EMT, epithelial–mesenchymal transition; EVOO, extra virgin olive oil; FAD, flavin adenine dinucleotide; FDA, U.S. Food and Drug Administration; GGA, geranylgeranoic acid; H3K4, histone H3 lysine 4; H3K9, histone H3 lysine 9; HDAC, histone deacetylase; HRP, horseradish peroxidase; iPS, induced pluripotent stem; Kt, competitive inhibition constant; LSD1, lysine-specific histone demethylase 1A; MAO-A, monoamine oxidase A; MHC, myosin heavy chain; MMA, methylmalonic acid; mRNA, messenger RNA; NAD, nicotinamide adenine dinucleotide; NTRK2, neurotrophic receptor tyrosine kinase 2; PDX, patient-derived xenograft; SARs, structure–activity relationship studies; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; siRNA, small interfering RNA; SIRT1, sirtuin 1; SOX2, sex determining region Y-box 2; SPR, surface plasmon resonance; TCP, tranylcypromine; THF, tetrahydrofolate; Tm, melting temperature
Graphical abstract
In this review, the identification of natural LSD1 inhibitors, analysis of the co-crystal structures of LSD1/natural product complex, antitumor activity, their modes of action, and the challenges faced in this field were mainly discussed. This review will provide a landscape of natural LSD1 inhibitors.
1. Introduction
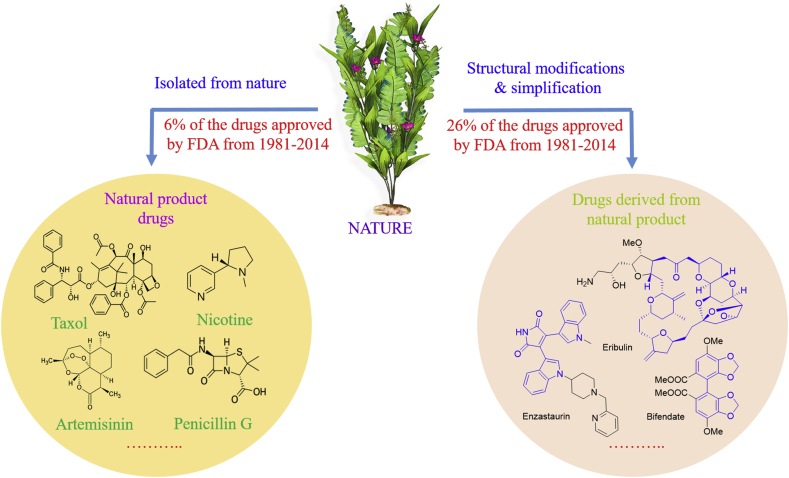
Natural products have long been recognized as a rich source of leads for new drug discovery. Relative to synthetic compounds, natural products possess diverse scaffolds and retain a high degree of stereochemistry1. Natural products are inherently within regions of biologically relevant chemical space and more importantly cover larger space than synthetic small-molecule libraries2. As reported, 48.6% (85) of the 175 small-molecule antitumor agents approved from around the 1940s to 2012 are either natural products or analogs derived natural products (Fig. 1)3,4. William C. Campbell, Satoshi Omura, and Youyou Tu were awarded the 2015 Nobel Prize in Physiology or Medicine for their achievements on the discovery of naturally occurring avermectins and artemisinin, respectively, for treating devastating parasite diseases5. With the recent technological advances, particularly the functional assays and phenotypic screening, many natural products are currently being used as valuable starting points in drug discovery campaigns for novel therapeutics beyond their traditional antimicrobial and anticancer indications6, 7, 8. With the epidemic of coronavirus disease (COVID-19), numerous efforts have been devoted to identifying effective drugs/vaccines for combating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Of note, some natural products have shown effectiveness for treating 2019 novel coronavirus pneumonia9. It is evident that the natural product-based drug discovery is now reaching a new golden age.
Figure 1.
Selected examples of natural product-based drugs.
In last decades, natural products have been used as starting points for epigenetic probe or drug discovery10. Some natural epigenetic modulators have advanced into clinical trials or are currently being used in clinic. For example, romidepsin, a selective class I and II histone deacetylase (HDAC) inhibitor obtained from cultures of Chromobacterium violaceum, was approved by the U.S. Food and Drug Administration (FDA) in 2009 for treating cutaneous T-cell lymphoma11. Mechanistically, the disulfide bridge of this bicyclic compound is reduced by glutathione to give free thiol groups, which then interact with Zn ions within the active site of HDAC enzymes, thus inhibiting the deacetylase activity12.
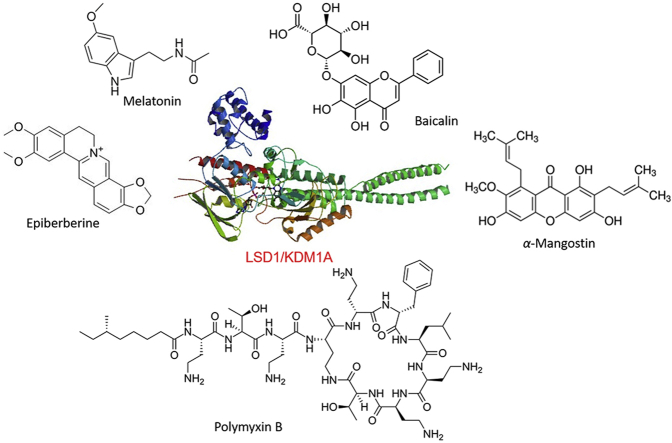
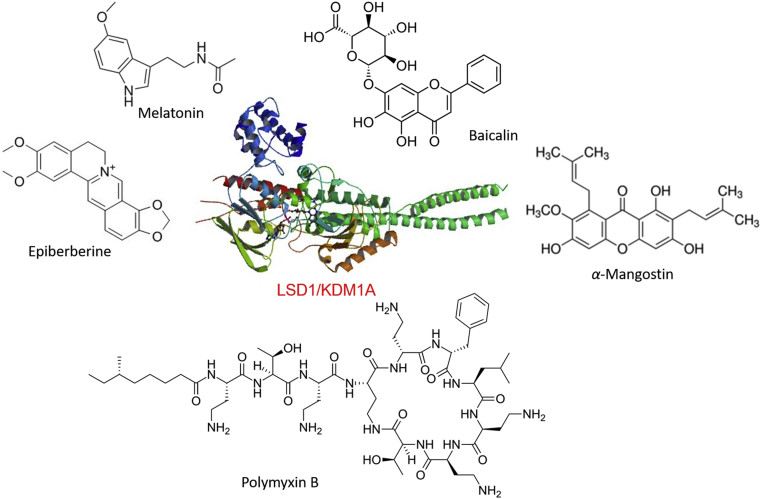
Of the epigenetic targets, the lysine-specific histone demethylase 1A (LSD1, also known as KDM1A) specifically removes methyl marks of histone H3 lysine 4 (H3K4) in flavin adenine dinucleotide (FAD)-dependent manner13. LSD1 has fundamental and diverse roles in physiological processes14 and under pathological conditions, such as cancers15,16, infections17,18, immune modulation19, 20, 21, etc. LSD1 dysregulation has been found in a wide range of cancers and is closely associated with malignant transformation, the epithelial–mesenchymal transition (EMT), stem cell biology, and cell proliferation and differentiation22. LSD1 inhibition by small molecules or genetic knockdown has shown effectiveness in blocking cell growth and migration23,24, inducing differentiation25,26, etc. These findings strongly suggest that targeting LSD1 is a promising option for cancer therapy27, 28, 29, 30. To date, a large number of highly potent and selective LSD1 inhibitors have been reported in last decades31, 32, 33, 34, 35, of which tranylcypromine (TCP), ORY-1001, ORY-2001, GSK-2879552, INCB059872, IMG-7289, and CC-90011 are currently undergoing clinical evaluation at different phases for the treatment of cancers36. Of particular interest is that some natural products could effectively inhibit LSD1 even at nanomolar levels (Fig. 2). Although the significant progress on natural LSD1 inhibitors has been observed, no comprehensive reviews have been published to date. Therefore, it would be necessary and of great interest to give an updated summary of natural product-based LSD1 inhibitors. In this review, an emphasis will be placed on the identification of natural LSD1 inhibitors, analysis of the co-crystal structures of LSD1/natural product complexes, antitumor activity and their modes of action. We will also briefly discuss the opportunities and challenges faced in this field. We believe this review will provide a landscape of natural LSD1 inhibitors and be interesting to the readers in this field.
Figure 2.
Crystal structure of LSD1 (PDB code: 2HKO) and representative natural LSD1 inhibitors (not exhaustive).
2. Natural product-based LSD1 inhibitors
Due to the structural diversity and complexity, natural products generally fall into the biologically relevant chemical space and cover wider space that is not occupied by synthetic compounds, thus always possessing unexpected biological activities. These attributes make natural products a rich source of lead compounds for new drug discovery. To date, some natural products with different chemotypes including protoberberine, flavone, diterpenoid, curcumin, xanthone, stilbene, resveratrol, secoiridoid, indole, phenol, and cyclic peptide, have been reported to be capable of inhibiting LSD1 in vitro and/or in vivo. This section is organized according to the scaffold types of natural LSD1 inhibitors.
2.1. Cyclic peptides
The H3 N-terminal residues bind to an open cleft in LSD1-CoRESt (RE1-silencing transcription factor co-repressor) and form interactions with surrounding negatively charged amino acid residues. Based on the structural features, Speranzini et al.37 screened their in-house focused library containing compounds with positively charged groups by thermal shift assay using ThermoFAD and in vitro anti-LSD1 enzymatic assay. Interestingly, they found that natural cyclic peptides polymyxins B and E (Table 1) could inhibit LSD1 in the thermal shift assay. In structure, polymyxins B and E feature five positively charged amine units (highlighted in blue in Table 1) and possess a linear head group, thus possibly mimicking histone H3 peptide substrates for binding to LSD1. In the thermal shift assay, both compounds increased the melting temperature (ΔTm = 6 °C) of LSD1-CoREST. The enzymatic and fluorescence polarization assays showed that polymyxins B and E competitively bound to LSD1-CoREST with histone H3 peptide and showed favorable affinity with Ki and Kd values of (157–193) and (463–594) nmol/L, respectively (Table 1). In cultured leukemia MV4-11 cells, polymyxin E (1 μmol/L), because of the poor permeability, had no remarkable effects on either H3K4/H3K9 (histone H3 lysine 9) methylation or cell growth.
Table 1.
Chemical structures and biochemical characterization of polymyxins B and E against LSD1.
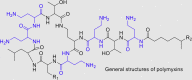 | |||||
|---|---|---|---|---|---|
| Natural product | R1 | R2 | Polymyxins-LSD1 |
||
| ΔTm (°C) | Ki (nmol/L) | Kd (nmol/L) | |||
| Polymyxin B | Ph | Me and Et (mixture) | +6.0 | 157 ± 26 | 463 ± 50 |
| Polymyxin E | i-Propyl | Et | +6.0 | 193 ± 38 | 594 ± 59 |
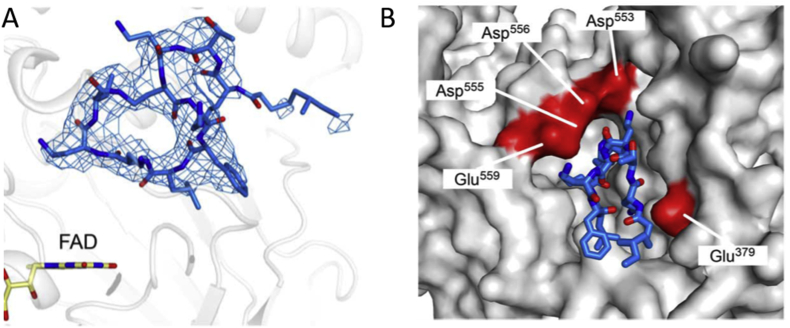
Co-crystal structure (PDB code: 5L3F) of LSD1-CoREST in complex with polymyxin B revealed that this positively charged cyclic peptide bound to LSD1 at the H3 tail-binding region, far away from the flavin (>5 Å) and formed electrostatic interactions with surrounding negatively charged residues (highlighted in red in Fig. 3B). Besides, because of the circular nature and multiple orientations, polymyxin B may adapt other “rotated” orientations with lower occupancies, the evidence supporting this notion is that polymyxin B showed decreased affinity (Kd = 4.7 ± 0.7 μmol/L) by a factor of 10 toward mutant LSD1, in which Glu 379 was substituted with Lys. This substitution from Glu to Lys reversed the charge of the key residue interacting with polymyxin B. This identified highly charged protein surface area is larger than that previously exploited38, and thus particularly attractive for designing new LSD1 inhibitors suppressing the demethylase activity and impairing binding to LSD1-CoREST. Although the serious side effects observed in anti-infection treatments, these findings highlight that the well-known antibiotics polymyxins B and E could be potentially repurposed for regulating bacterial infections and epigenetic processes in the context of leukemia.
Figure 3.
Co-crystal structure (PDB code: 5L3F) of LSD1-CoREST in complex with polymyxin B. (A) No interactions are formed between polymyxin B (shown in blue) and FAD (yellow sticks). (B) Polymyxin B binds to LSD1 at the entrance of the H3 tail-binding cleft and establishes interactions with surrounding negatively charged residues Asp 553, Asp 556, Asp 555, Glu 559, and Glu 379 (depicted in red). Adapted from Ref. 37. Copyright © 2016 American Association for the Advancement of Science.
2.2. Protoberberine alkaloids
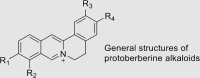
Through screening our in-house compound library containing natural products, we found that the tetracyclic protoberberine alkaloids potently inhibited LSD1 (Table 2)39. Of note, epiberberine showed the strongest inhibitory activity against LSD1 (IC50 = 0.14 μmol/L), while other protoberberine alkaloids showed decreased inhibitory activity, suggesting the crucial roles of substituents on the activity. We also noted that the tetracyclic canadine and tetrahydropalmatine were inactive against LSD1 with the IC50 values of over 100 μmol/L. This strongly indicate that the isoquinoline ring is extremely important for the anti-LSD1 activity. The structure–activity relationship studies (SARs) provide us the essential structural features of protoberberine alkaloids for LSD1 inhibition, we believe the protoberberine alkaloids could be used as starting templates for designing more potent and selective LSD1 inhibitors. Further structural modifications based on the tetracyclic scaffold is currently undergoing in our lab.
Table 2.
The inhibitory activity of protoberberine alkaloids against LSD1.
 | |||||
|---|---|---|---|---|---|
| Protoberberine alkaloid | R1 | R2 | R3 | R4 | IC50 (μmol/L) |
| Epiberberine |  |
OMe | OMe | 0.14 ± 0.01 | |
| Columbamine | OMe | OMe | OH | OMe | 0.47 ± 0.01 |
| Jatrorrhizine | OMe | OMe | OMe | OH | 1.55 ± 0.01 |
| Berberine | OMe | OMe |  |
6.97 ± 0.01 | |
| Palmatine | OMe | OMe | OMe | OMe | 12.33 ± 0.01 |
In contrast to LSD1, epiberberine was less potent against monoamine oxidase A/B (MAO-A/B), showing selective inhibition toward LSD1. At 1.0 μmol/L, epiberberine inhibited LSD1 with an inhibitory rate of 89.41%, but had <30% of inhibitory rates against MAO-A/B. Epiberberine reversibly inhibited LSD1 and was non-competitive with LSD1 substrate H3K4me2. In the surface plasmon resonance (SPR) assay, epiberberine bound to LSD1 with a fast association (Ka = 3.55 × 103 L/mol·s) but with a slow dissociation (Kd = 4.32 × 10−2 s−1). In acute myeloid leukemia (AML) cell lines THP-1 and HL-60, epiberberine moderately inhibited cell growth after 8 days’ treatment (IC50 = 12.80 and 23.82 μmol/L, respectively). In THP-1 cells, epiberberine had no effect on LSD1 expression, but induced accumulation of LSD1 substrates H3K4me2 and H3K9me1/2. At 1 μmol/L, epiberberine significantly increased expression of cluster of differentiation 86 (CD86) in THP-1 and HL-60 cells. These data suggest that epiberberine was engaged to LSD1 in THP-1 cells. Consistent with previous report40, epiberberine also induced differentiation related morphological changes and expression of integrin alpha M (CD11b) and cluster of differentiation 14 (CD14), two well-known myelo-monocytic differentiation markers41. However, like the well-known LSD1 inhibitor ORY-1001, epiberberine had no effect on cell cycle distribution and cell apoptosis at low concentrations in AML cells. In nonobese diabetic/severe combined immunodeficiency (NOD/SCID) xenograft model bearing THP-1 cells, epiberberine demonstrated robust efficacy without serious side effects after intravenous administration. Specifically, after treatment with epiberberine (50 mg/kg) for 14 days, the average tumor volume was about 469 mm3, significantly smaller than that of the vehicle group (963 mm3). Relative to the vehicle group, epiberberine significantly prolonged mean survival of the NOD/SCID mice bearing THP-1 cells from 19 (the vehicle group) to 25 days (the group treated with 50 mg/kg).
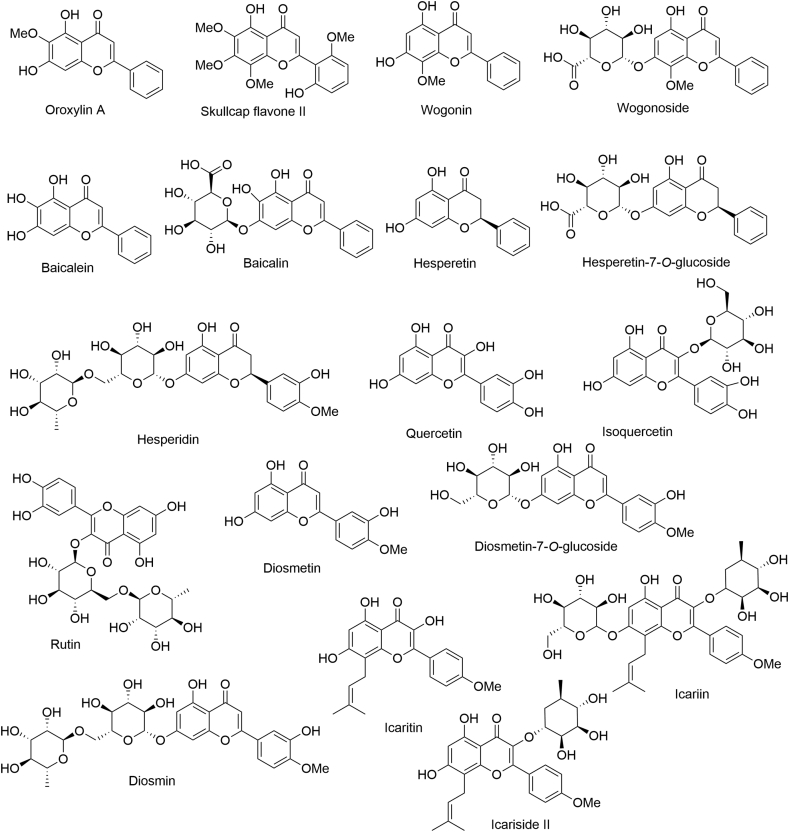
2.3. Flavones
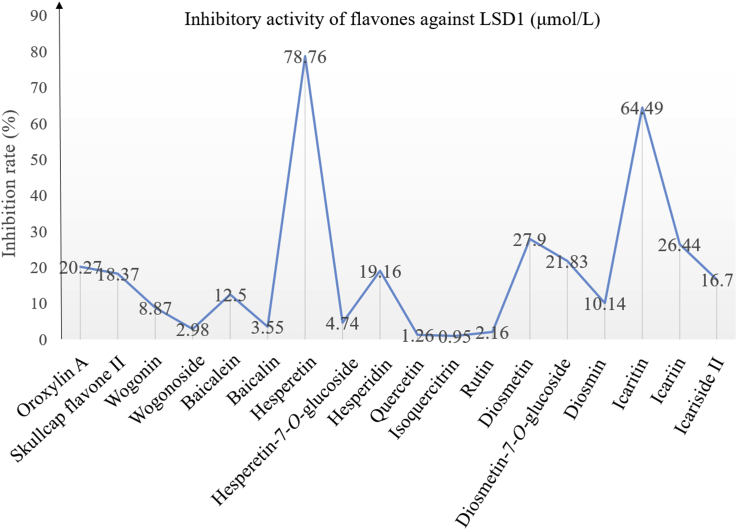
The flavones are a large family of natural products that possess diverse biological activities42,43. In recent years, some flavone-based natural compounds have proven to be effective against LSD1 (Fig. 4)44, 45, 46. In structure, the flavones feature the backbone of 2-phenylchromen-4-one with additional phenolic hydroxy groups and/or sugar moiety. The in vitro inhibitory activity of these flavones against LSD1 is depicted in Fig. 5. Clearly, these flavones showed varied anti-LSD1 inhibitory activity dependent on their substituents attached to the scaffold. Of these compounds, isoquercitrin showed the best potency against LSD1 (IC50 = 0.95 μmol/L). These finding may suggest that the flavone may be a privileged scaffold for designing new LSD1 inhibitors.
Figure 4.
Representative flavone-based natural LSD1 inhibitors.
Figure 5.
In vitro inhibitory activity of flavones against LSD1.
In 2016, our group reported that baicalin could inhibit LSD1 with an IC50 of 3.01 μmol/L (similar to that later reported by Han et al.45) and the HillSlope value of 0.6744. Baicalin is the first reported flavone-based natural LSD1 inhibitor. In MGC-803 cells overexpressing LSD1, baicalin dose-dependently induced accumulation of H3K4me2 and CD86 messenger RNA (mRNA) levels, but had no effect on LSD1 expression, indicating cellular target engagement to LSD1. Baicalin inactivated LSD1 reversibly, dilution could recover the demethylase activity. Besides, baicalin inhibited cell proliferation (IC50 = 8.78 ± 0.49 μmol/L) and migration of MGC-803 cells, accompanying the increase of E-cadherin mRNA levels and decrease of N-cadherin mRNA levels.
Based on the bioactivity-guided cut countercurrent chromatography (CCC) strategy, Han et al.45 isolated 6 flavone-based natural LSD1 inhibitors (baicalin, wogonoside, baicalein, wogonin, skullcap flavone II and oroxylin in Fig. 4) from Scutellaria baicalensis Georgi. Wogonoside demonstrated the strongest inhibitory activity against LSD1 (IC50 = 2.98 μmol/L) and reversibly inhibited LSD1. In MDA-MB-231, wogonoside significantly induced accumulation of H3K4me2 and also increased mRNA levels of CD86, a cellular surrogate biomarker for the LSD1 activity. After treatment of MDA-MB-231 cells with wogonoside for 48 h, the cell viability was inhibited dose-dependently with an IC50 value of 14.94 μmol/L. Han et al. also found that wogonoside significantly inhibited migration of MDA-MB-231 cells accompanied by increased E-cadherin mRNA levels and decreased N-cadherin mRNA levels.
Subsequently, Xu et al.46 identified another 12 natural flavone-based LSD1 inhibitors including four aglycones, four monoglycosides, and four diglycosides. The SARs studies revealed that all monoglycosides displayed better potency against LSD1 than their aglycone counterparts independent on the sugar type and position attached. Among 12 flavone-based natural LSD1 inhibitors, isoquercitrin had the strongest inhibitory activity against LSD1 (IC50 = 0.95 μmol/L), 20 times stronger than tranylcypromine (IC50 = 19.11 μmol/L). Besides, isoquercitrin competed with H3K4me2 to inhibit LSD1 with an estimated competitive inhibition constant (Kt) of 1.0 μmol/L. Isoquercitrin dose-dependently increased levels of H3K4me1/me2 and H3K9me2 in MDA-MB-231 cells, indicating its cellular activity. Further mechanistic studies showed that in LSD1 knockdown MDA-MB-231 cells, isoquercitrin dose-dependently inhibited cell proliferation. In the 5-ethynyl-20-deoxyuridine (EdU) incorporation assay, isoquercitrin reduced the number of EdU-positive cells that were treated with LSD1 small interfering RNA (siRNA). Similarly, co-treatment of MDA-MB-231 cells with isoquercitrin and siRNA, chromatin condensation and chromatinorrhexis were observed. For cells treated with isoquercitrin and siRNA, the apoptotic rate was 38.9%, significantly higher than that (10.7%) of the group treated with isoquercitrin alone. They also found that isoquercitrin induced apoptosis of MDA-MB-231 cells via the LSD1-based methylmalonic acid (MMA) pathway. Specifically, relative to the control group or isoquercitrin-treated group, co-treatment with isoquercitrin and siRNA caused lower mitochondrial transmembrane potential (ΔΨm) and higher BAX/BCL-2 protein ratio as well as induced expression of cleaved caspase-3, caspase-7, caspase-9, and PARP.
2.4. Xanthones
Xanthones are an important class of polyphenolic compounds with diverse pharmacological activities, thus making them promising scaffolds for new drug discovery47, 48, 49, 50. Of which α-mangostin (α-MG, Fig. 6), the main constituent of the mangosteen fruit, has been proved to possess diverse pharmacological properties51,52.
Figure 6.
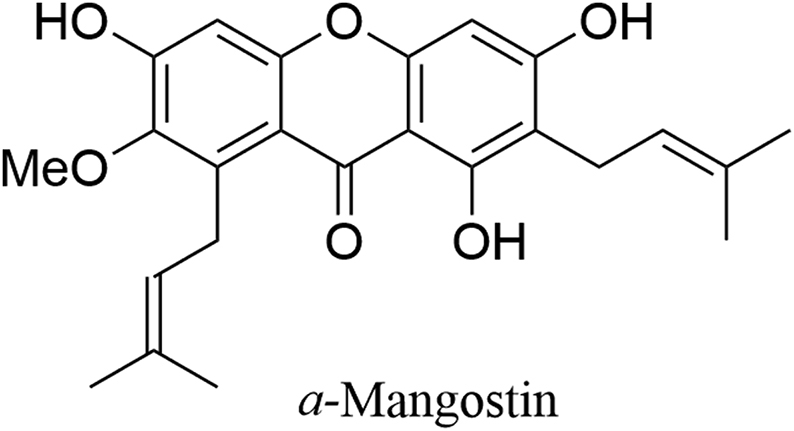
Chemical structure of α-mangostin.
Han et al.53 reported that α-mangostin inhibited LSD1 potently and reversibly (IC50 = 2.81 μmol/L), 7-fold much more potent than tranylcypromine. In MDA-MB-231 cells, α-mangostin dose-dependently induced accumulation of H3K4me2 without affecting LSD1 expression and significantly induced CD86 expression. As a cellularly active LSD1 inhibitor, α-mangostin dose- and time-dependently inhibited viability of MDA-MB-231 cells (IC50 = 12.75 μmol/L) and cell migration. The Western blotting analysis showed that like above flavone-based LSD1 inhibitors, α-mangostin also affected expression of E-cadherin and N-cadherin. Collectively, α-mangostin is the first xanthone-based LSD1 inhibitor, thus making xanthones as novel scaffolds for the search of new LSD1 inhibitors.
2.5. Stilbenes
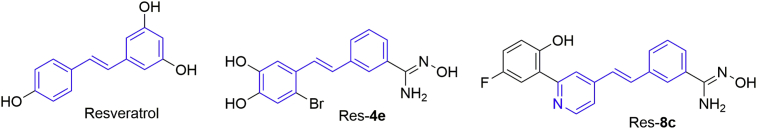
Stilbenes are an important class of biologically active scaffolds structurally featuring a trans-1,2-diphenylethylene nucleus (highlighted in blue in Fig. 7). The stilbene scaffolds are prevalent in natural products including resveratrol (Fig. 7) and biologically active compounds, thus being recognized as privileged scaffolds in drug discovery54, 55, 56, 57. Resveratrol has shown diverse biological activities and is therapeutically potential in preventing or slowing the progression of various pathological conditions, including cancers, cardiovascular diseases, type two diabetes, etc58. Additionally, it has been reported that resveratrol could activate the nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase sirtuin 1 (SIRT1)59, thus potentially achieving epigenetic regulation.
Figure 7.
Chemical structures of resveratrol and analogs.
In 2013, Abdulla et al.60 reported that resveratrol dose-dependently inhibited the enzymatic activity of LSD1 (IC50 = 15 μmol/L) and also inhibited the demethylation process of H3K4me2. The inhibitory activity of resveratrol against LSD1 was independent on its antioxidant properties and was specific, not through SIRT1 activation. For myogenic differentiation of C2C12 fibroblasts, LSD1 is required61. The authors found that in C2C12 cells, resveratrol effectively inhibited myocyte formation and myosin heavy chain (MHC) expression. Finally, they speculated that resveratrol bound to LSD1 to inhibit its demethylase activity. The data may suggest that resveratrol would be a good starting point for designing new LSD1 inhibitors.
Inspired by above work and the prevalence of the amidoxime in LSD1 inhibitors, our group designed new stilbene-based LSD1 inhibitors by introducing the amidoxime to the stilbene scaffold62,63, of which compounds Res-4e and Res-8c showed promising activity against LSD1 and in LSD1 overexpressed cancer cells. Res-4e inhibited LSD1 potently (IC50 = 121 nmol/L) and reversibly62. In MGC-803 cells, Res-4e dose-dependently induced accumulation of H3K4me2 and CD86 mRNA levels, indicating its cellular target engagement to LSD1. Following this work, our group further performed structural modifications to obtain new stilbene derivatives as new LSD1 inhibitors63. Res-8c effectively inhibited LSD1 (IC50 = 283 nmol/L) and showed superior selectivity over MAO-A/B (IC50 > 50 μmol/L). Res-8c inhibited LSD1 reversibly and FAD-competitively. In the SPR assay, Res-8c showed tight binding to LSD1 (KD = 5.49 × 10−6 mol/L) with a fast association and slow dissociation (Ka = 1.82 × 103 L/mol·s, Kd = 1.02 × 10−2 s−1). Res-8c exhibited moderate inhibition against human MOLM-13 and THP-1 cell lines with the IC50 values ranging from 5.76 to 8.34 μmol/L. In THP-1 cells, Res-8c does-dependently inhibited colony formation and increased CD86 expression. Res-8c induced accumulation of H3K4me1 and H3K4me2 dose-dependently after treatment for 5 days, supporting the target engagement of Res-8c in THP-1 cells. Besides, treatment of THP-1 cells with Res-8c dose-dependently caused characteristic morphological changes, including chromatin shrinkage and blurred cell membrane boundaries.
2.6. Diarylheptanoids
The diarylheptanoids, a class of natural products mainly distributed in the roots, bark and rhizomes of Alpinia, Curcuma, Zingiber and Alnus species, possess the aryl-C7-aryl skeleton and various substituents64. Such compounds have shown diverse biological activities such as anticancer, antimicrobial, and antioxidant activity, thus becoming a promising class of bioactive natural products65. Of particular interest is curcumin (Fig. 8), which is a constituent of turmeric derived from the roots of plant Curcuma longa66. As a natural polyphenol, curcumin has shown numerous biological activities including antioxidant, anti-inflammatory, and anticancer properties and has been one of the focuses for new drug development67,68. Accumulating data have supported the therapeutic potential of curcumin as an epigenetic modulator and warrant further preclinical and clinical studies for exploring its anticancer efficacy69,70.
Figure 8.
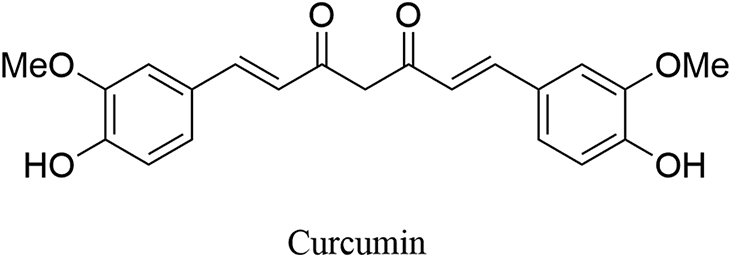
Chemical structure of curcumin.
In 2013, Abdulla et al.60 first reported that curcumin inactivated LSD1 independent on its antioxidant property. Curcumin effectively suppressed myocyte formation of cell differentiation and the expression of both myogenin and MHC. Very recently, Zhao and co-workers71 reported that curcumin inactivated LSD1 (IC50 = 9.6 μmol/L), further structure-based modifications of curcumin gave cinnamamides as new LSD1 inhibitors, of which WB07 demonstrated the best potency (IC50 = 0.8 μmol/L) and was selective over MAO-A/B (IC50 > 50 μmol/L). Also, curcumin suppresses cell growth of A549 cells (IC50 = 9.333 μmol/L). Although curcumin has moderate inhibition toward LSD1, such compound has limitations including low bioavailability, toxicity, etc.72,73.
2.7. Melatonin
Melatonin (Fig. 9), a natural hormone produced primarily by the pineal gland in the brain, is functionally diverse, including anticancer, antioxidant, anti-inflammatory and immune-regulating activity74. Melatonin may have the therapeutic potential against a variety of cancer types75,76, particularly the gastrointestinal cancer77. Interestingly, melatonin could induce histone H3 hyperacetylation78,79 and apoptosis of colorectal cancer cells through histone deacetylase 4 (HDAC4) nuclear import80.
Figure 9.
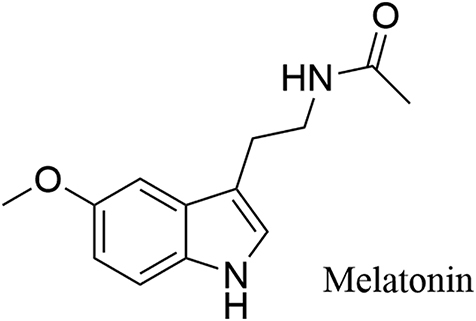
Chemical structure of melatonin.
In 2017, Yang et al.81 revealed that melatonin showed therapeutic potential in patient-derived tumor xenograft (PDTX) models bearing oral cancer cells via suppressing LSD1. After treatment for 42 days, melatonin inhibited tumor growth and weight significantly without observed toxicities. In contrast to the control group, LSD1 expression was significantly lower in the melatonin-treated group. Melatonin induced cell cycle arrest of SAS cells at G0/G1 phase and decreased expression of LSD1 in both SAS and SCC25 cells. In SCC25 cells, melatonin significantly led to elevated levels of P21 and decrease levels of cyclin D1. The data may suggest that melatonin may has therapeutic potential in LSD1-overexpressed oral cancer. However, questions remain exit for the anti-oral cancer effects of melatonin, for example, the authors did not address whether melatonin bound to LSD1 directly or indirectly, the cellular target engagement was not examined as well. More work should be done before carrying out melatonin-based modifications for more potent and selective LSD1 inhibitors.
2.8. Other natural LSD1 inhibitors
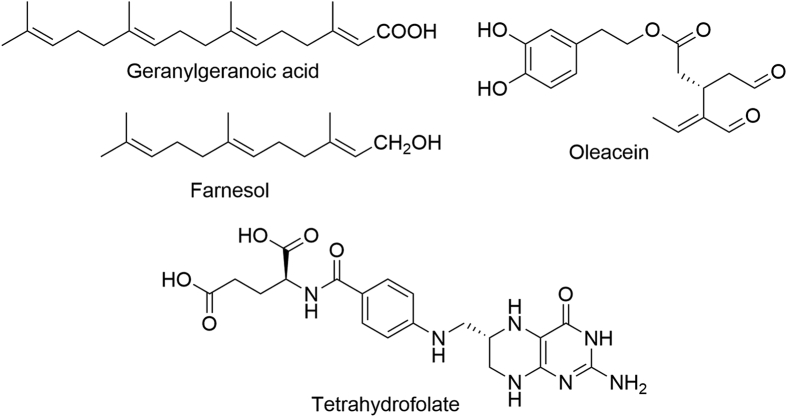
Geranylgeranoic acid (GGA, Fig. 10), a natural diterpenoid, inhibited LSD1 (IC50 = 46.97 μmol/L) in a non-competitive fashion82. GGA induced expression of neurotrophic receptor tyrosine kinase 2 (NTRK2) gene and upregulated H3K4me2 around the P1 and P2 promoter regions of the NTRK2 gene in human neuroblastoma SH-SY5Y cells, thus altering differentiated state of neuroblastoma cells. These findings show promise for GGA to regulate epigenetic modification. Farnesol (Fig. 10), a component of tobacco smoke, also dose-dependently inhibited LSD1 (IC50 ca. 120 μmol/L)83. As a natural phenolic secoiridoid found in extra virgin olive oil (EVOO), oleacein directly inhibited LSD1 (IC50 = 2.5 μmol/L) in the alpha-screen-based in vitro assays. Besides, oleacein completely suppressed the expression of sex determining region Y-box 2 (SOX2) in cancer stem-like and induced pluripotent stem (iPS) cells. Oleacein could be potentially used as a hit compound to design new secoiridoid-based LSD1 inhibitors. It has been reported that the (6R,S)-form of the pentaglutamate form of tetrahydrofolate bound to full-length LSD1 (Kd = 2.8 μmol/L, Fig. 10). Folate is believed to participate in histone demethylation84. Subsequently, Luka et al.85 reported the crystal structure of LSD1‒tetrahydrofolate (THF) complex (PDB code: 4KUM), showing that THF binds to the active center of LSD1 and forms hydrophobic interactions with FAD, Phe 538, Val 333, Ala 809 and Tyr761 in LSD1 through the pterin and p-aminobenzoic rings.
Figure 10.
Chemical structures of geranylgeranoic acid, farnesol, oleacein and tetrahydrofolate.
3. Conclusions and perspectives
Natural products feature diverse and novel molecular scaffolds, and thus are a rich source for identifying new bioactive compounds. Of note, with the recent development of the functional assays and phenotypic screening protocols, more and more natural products have demonstrated novel therapeutics beyond their traditional antimicrobial and anticancer indications. Youyou Tu, Satoshi Omura, and William C. Campbell were awarded the 2015's Nobel Prize in Physiology or Medicine for discovering avermectins and artemisinin for treating parasite diseases. Undoubtedly, natural product-based drug discovery is now reaching a new golden age.
In the field of epigenetics, natural products have also shown promise in epigenetic drug discovery. To date, numerous natural products have been reported to be able to modulate epigenetic processes, some of them have advanced into clinical trials or are presently being used in clinic. LSD1 has fundamental roles in the development of various pathological conditions, including cancers. Pharmaceutical inhibition of LSD1 has been a promising therapeutic option for cancer treatment30. Some TCP-based LSD1 inhibitors are currently being evaluated for cancer therapy. Notably, some natural products of different types have also shown effectiveness in inhibiting LSD1. Generally, these natural products can be mainly divided into four categories, including cyclic peptides, protoberberine alkaloids, polyphenols, and unsaturated carbonyl compounds (e.g., GGA). These natural products indeed provide novel scaffolds for developing new reversible LSD1 inhibitors. However, most of them show poor to moderate inhibitory activity against LSD1, much efforts should be devoted to identifying new LSD1 inhibitors with novel scaffolds from nature. The “2 + 1” model previously proposed by us33 has been successfully used to design new LSD1 inhibitors in our lab86, 87, 88, 89, 90 and would also be useful to guide the selection of potential hit compounds from natural product library for further screening. We have to admit that biological characterization of natural LSD1 inhibitors is at very early stage, most of them are not fully characterized in vitro and in vivo. Before further structural modifications and/or in-depth mechanistic studies, any hit compound inhibiting LSD1 should be carefully cross-validated to make sure that it is a genuine LSD1 inhibitor. Particular attention should be paid to natural polyphenols, which always consume H2O2 (produced in the screening assay), and then give a false positive reading. Besides, some compounds with aromatic character appear to be LSD1 inhibitors, but are actually horseradish peroxidase (HRP, a component used in the screening assay) inhibitors. This kind of compounds also give a false positive reading and should be excluded. Collectively, herein we provide an updated overview of natural LSD1 inhibitors reported to date and our own perspectives on future development of novel LSD1 inhibitors based on natural products. The scaffolds derived from natural products could be used for designing new LSD1 inhibitors.
Acknowledgments
We are grateful for the financial support from the National Natural Science Foundation of China (Nos. 81703326, 81973177, 81773580 and 81802130), China Postdoctoral Science Foundation (Nos. 2018M630840 and 2019T120641), the Open Project of State Key Laboratory of Natural Medicines (No. SKLNMKF202005, China), and Guangdong Key Laboratory for Translational Cancer Research of Chinese Medicine (No. 2018B030322011, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Qingchun Mu, Email: muqcns@163.com.
Guochao Liao, Email: liao@gzucm.edu.cn.
Bin Yu, Email: zzuyubin@hotmail.com.
Author contributions
Yuan Fang and Chao Yang wrote the draft manuscript; Zhiqiang Yu and Xiaochuan Li polished the language of this manuscript; Qingchun Mu and Guochao Liao gave insightful comments/suggestions on the outline of this manuscript; Bin Yu revised and submitted this manuscript on behalf of other authors.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Barnes E.C., Kumar R., Davis R.A. The use of isolated natural products as scaffolds for the generation of chemically diverse screening libraries for drug discovery. Nat Prod Rep. 2016;33:372–381. doi: 10.1039/c5np00121h. [DOI] [PubMed] [Google Scholar]
- 2.Harvey A.L., Edrada-Ebel R., Quinn R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 3.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2014;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 4.Wang S., Dong G., Sheng C. Structural simplification: an efficient strategy in lead optimization. Acta Pharm Sin B. 2019;9:880–901. doi: 10.1016/j.apsb.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeCorte B.L. Underexplored opportunities for natural products in drug discovery. J Med Chem. 2016;59:9295–9304. doi: 10.1021/acs.jmedchem.6b00473. [DOI] [PubMed] [Google Scholar]
- 7.Harvey A.L. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Li J.W.H., Vederas J.C. Drug discovery and natural products: end of an era or an endless frontier?. Science. 2009;325:161. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 9.Du H.Z., Hou X.Y., Miao Y.H., Huang B.S., Liu D.H. Traditional Chinese medicine: an effective treatment for 2019 novel coronavirus pneumonia (NCP) Chin J Nat Med. 2020;18:206–210. doi: 10.1016/S1875-5364(20)30022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherblanc F.L., Davidson R.W.M., Di Fruscia P., Srimongkolpithak N., Fuchter M.J. Perspectives on natural product epigenetic modulators in chemical biology and medicine. Nat Prod Rep. 2013;30:605–624. doi: 10.1039/c3np20097c. [DOI] [PubMed] [Google Scholar]
- 11.VanderMolen K.M., McCulloch W., Pearce C.J., Oberlies N.H. Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J Antibiot. 2011;64:525–531. doi: 10.1038/ja.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J., Guo J., Kang D., Li Z., Wang G., Wu J. New techniques and strategies in drug discovery. Chin Chem Lett. 2020;31:1695–1708. [Google Scholar]
- 13.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Kozub M., Carr R., Lomberk G., Fernandez-Zapico M. LSD1, a double-edged sword, confers dynamic chromatin regulation but commonly promotes aberrant cell growth. F1000Res. 2017;6:2016. doi: 10.12688/f1000research.12169.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amente S., Lania L., Majello B. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta. 2013;1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Majello B., Gorini F., Saccà D.C., Amente S. Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers. 2019;11:324. doi: 10.3390/cancers11030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill J.M., Quenelle D.C., Cardin R.D., Vogel J.L., Clement C., Bravo F.J. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwergel C., Stazi G., Mai A., Valente S. Trends of LSD1 inhibitors in viral infections. Future Med Chem. 2018;10:1133–1136. doi: 10.4155/fmc-2018-0065. [DOI] [PubMed] [Google Scholar]
- 19.Sheng W., LaFleur M.W., Nguyen T.H., Chen S., Chakravarthy A., Conway J.R. LSD1 Ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;174:549–563. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin Y., Vasilatos S.N., Chen L., Wu H., Cao Z., Fu Y. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene. 2019;38:390–405. doi: 10.1038/s41388-018-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzi K., Geng H., Doane A.S., Meydan C., LaRiviere R., Cardenas M. Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nat Immunol. 2019;20:86–96. doi: 10.1038/s41590-018-0273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdel-Magid A.F. Lysine-specific demethylase 1 (LSD1) inhibitors as potential treatment for different types of cancers. ACS Med Chem Lett. 2017;8:1134–1135. doi: 10.1021/acsmedchemlett.7b00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu L., Wang J., Kong W., Huang J., Dong B., Huang Y. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharm Sin B. 2019;9:324–334. doi: 10.1016/j.apsb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callegari K., Maegawa S., Bravo-Alegria J., Gopalakrishnan V. Pharmacological inhibition of LSD1 activity blocks REST-dependent medulloblastoma cell migration. Cell Commun Signal. 2018;16:60. doi: 10.1186/s12964-018-0275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Z., Yao Y., Zhou C., Chen F., Wu F., Wei L. Pharmacological inhibition of LSD1 for the treatment of MLL-rearranged leukemia. J Hematol Oncol. 2016;9:24. doi: 10.1186/s13045-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barth J., Scheder A.M., Mohr S., Schulz-Fincke J., Schmitt M., Walter A. Lsd1 inhibition induces differentiation and decreases leukemic stem cell frequency in Hoxa9/Meis1-driven AML. Exp Hematol. 2017;53:S125. [Google Scholar]
- 27.Magliulo D., Bernardi R., Messina S. Lysine-specific demethylase 1A as a promising target in acute myeloid leukemia. Front Oncol. 2018;8:255. doi: 10.3389/fonc.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G.J., Lei P.M., Wong S.Y., Ma D.L., Leung C.H. Pharmacological inhibition of LSD1 for cancer treatment. Molecules. 2018;23:3194. doi: 10.3390/molecules23123194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macheleidt I.F., Dalvi P.S., Lim S.Y., Meemboor S., Meder L., Käsgen O. Preclinical studies reveal that LSD1 inhibition results in tumor growth arrest in lung adenocarcinoma independently of driver mutations. Mol Oncol. 2018;12:1965–1979. doi: 10.1002/1878-0261.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y., Liao G., Yu B. Targeting histone lysine demethylase LSD1/KDM1A as a new avenue for cancer therapy. Curr Top Med Chem. 2019;19:889–891. doi: 10.2174/156802661911190725094910. [DOI] [PubMed] [Google Scholar]
- 31.Mould D.P., McGonagle A.E., Wiseman D.H., Williams E.L., Jordan A.M. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: clinical significance and progress to date. Med Res Rev. 2015;35:586–618. doi: 10.1002/med.21334. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y.C., Ma J., Wang Z., Li J., Jiang B., Zhou W. A systematic review of histone lysine-specific demethylase 1 and its inhibitors. Med Res Rev. 2015;35:1032–1071. doi: 10.1002/med.21350. [DOI] [PubMed] [Google Scholar]
- 33.Fu X., Zhang P., Yu B. Advances toward LSD1 inhibitors for cancer therapy. Future Med Chem. 2017;9:1227–1242. doi: 10.4155/fmc-2017-0068. [DOI] [PubMed] [Google Scholar]
- 34.Pandey M.R., Wang E.S. What potential is there for LSD1 inhibitors to reach approval for AML?. Expet Opin Emerg Drugs. 2019;24:205–212. doi: 10.1080/14728214.2019.1694001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Huang B., Suzuki T., Liu X., Zhan P. Medicinal chemistry insights in the discovery of novel LSD1 inhibitors. Epigenomics. 2015;7:1379–1396. doi: 10.2217/epi.15.86. [DOI] [PubMed] [Google Scholar]
- 36.Fang Y., Liao G., Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol. 2019;12:129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speranzini V., Rotili D., Ciossani G., Pilotto S., Marrocco B., Forgione M. Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features. Sci Adv. 2016;2 doi: 10.1126/sciadv.1601017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Lu F., Ren Q., Sun H., Xu Z., Lan R. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z.R., Suo F.Z., Guo Y.J., Cheng H.F., Niu S.H., Shen D.D. Natural protoberberine alkaloids, identified as potent selective LSD1 inhibitors, induce AML cell differentiation. Bioorg Chem. 2020;97:103648. doi: 10.1016/j.bioorg.2020.103648. [DOI] [PubMed] [Google Scholar]
- 40.Fiskus W., Sharma S., Shah B., Portier B.P., Devaraj S.G.T., Liu K. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 2014;28:2155–2164. doi: 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schenk T., Chen W.C., Göllner S., Howell L., Jin L., Hebestreit K. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma A.K., Pratap R. The biological potential of flavones. Nat Prod Rep. 2010;27:1571–1593. doi: 10.1039/c004698c. [DOI] [PubMed] [Google Scholar]
- 43.Singh M., Kaur M., Silakari O. Flavones: an important scaffold for medicinal chemistry. Eur J Med Chem. 2014;84:206–239. doi: 10.1016/j.ejmech.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y.C., Shen D.D., Ren M., Liu X.Q., Wang Z.R., Liu Y. Baicalin, a natural LSD1 inhibitor. Bioorg Chem. 2016;69:129–131. doi: 10.1016/j.bioorg.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Han C., Wang S., Li Z., Chen C., Hou J., Xu D. Bioactivity-guided cut countercurrent chromatography for isolation of lysine-specific demethylase 1 inhibitors from Scutellaria baicalensis Georgi. Anal Chim Acta. 2018;1016:59–68. doi: 10.1016/j.aca.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Xu X., Peng W., Liu C., Li S., Lei J., Wang Z. Flavone-based natural product agents as new lysine-specific demethylase 1 inhibitors exhibiting cytotoxicity against breast cancer cells in vitro. Bioorg Med Chem. 2019;27:370–374. doi: 10.1016/j.bmc.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 47.Cruz M.I., Cidade H., Pinto M. Dual/multitargeted xanthone derivatives for Alzheimer's disease: where do we stand?. Future Med Chem. 2017;9:1611–1630. doi: 10.4155/fmc-2017-0086. [DOI] [PubMed] [Google Scholar]
- 48.Klein-Júnior L.C., Campos A., Niero R., Corrêa R., Vander Heyden Y., Filho V.C. Xanthones and cancer: from natural sources to mechanisms of action. Chem Biodivers. 2020;17 doi: 10.1002/cbdv.201900499. [DOI] [PubMed] [Google Scholar]
- 49.Panda S.S., Chand M., Sakhuja R., Jain S.C. Xanthones as potential antioxidants. Curr Med Chem. 2013;20:4481–4507. doi: 10.2174/09298673113209990144. [DOI] [PubMed] [Google Scholar]
- 50.Feng Z., Lu X., Gan L., Zhang Q., Lin L. Xanthones, a promising anti-inflammatory scaffold: structure, activity, and drug likeness analysis. Molecules. 2020;25:598. doi: 10.3390/molecules25030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen G., Li Y., Wang W., Deng L. Bioactivity and pharmacological properties of α-mangostin from the mangosteen fruit: a review. Expert Opin Ther Pat. 2018;28:415–427. doi: 10.1080/13543776.2018.1455829. [DOI] [PubMed] [Google Scholar]
- 52.Tsai S.Y., Chung P.C., Owaga E.E., Tsai I.J., Wang P.Y., Tsai J.I. Alpha-mangostin from mangosteen (Garcinia mangostana Linn.) pericarp extract reduces high fat-diet induced hepatic steatosis in rats by regulating mitochondria function and apoptosis. Nutr Metab. 2016;13:88. doi: 10.1186/s12986-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han C., Li Z., Hou J., Wang Z., Xu D., Xue G. Bioactivity evaluation of natural product α-mangostin as a novel xanthone-based lysine-specific demethylase 1 inhibitor to against tumor metastasis. Bioorg Chem. 2018;76:415–419. doi: 10.1016/j.bioorg.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 54.De Filippis B., Ammazzalorso A., Amoroso R., Giampietro L. Stilbene derivatives as new perspective in antifungal medicinal chemistry. Drug Dev Res. 2019;80:285–293. doi: 10.1002/ddr.21525. [DOI] [PubMed] [Google Scholar]
- 55.Elisa G., Sebastiano R., Laura G., Maurizio R., Marinella R. The use of stilbene scaffold in medicinal chemistry and multi-target drug design. Curr Med Chem. 2016;23:2439–2489. doi: 10.2174/0929867323666160517121629. [DOI] [PubMed] [Google Scholar]
- 56.Lizard G., Latruffe N., Vervandier-Fasseur D. Aza- and azo-stilbenes: bio-isosteric analogs of resveratrol. Molecules. 2020;25:605. doi: 10.3390/molecules25030605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan Z.A., Iqbal A., Shahzad S.A. Synthetic approaches toward stilbenes and their related structures. Mol Divers. 2017;21:483–509. doi: 10.1007/s11030-017-9736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu W., Fu Y.C., Wang W. Cellular and molecular effects of resveratrol in health and disease. J Cell Biochem. 2012;113:752–759. doi: 10.1002/jcb.23431. [DOI] [PubMed] [Google Scholar]
- 59.Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 60.Abdulla A., Zhao X., Yang F. Natural Polyphenols inhibit lysine-specific demethylase-1 in vitro. J Biochem Pharmacol Res. 2013;1:56–63. [PMC free article] [PubMed] [Google Scholar]
- 61.Choi J., Jang H., Kim H., Kim S.T., Cho E.J., Youn H.D. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun. 2010;401:327–332. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Duan Y.C., Guan Y.Y., Zhai X.Y., Ding L.N., Qin W.P., Shen D.D. Discovery of resveratrol derivatives as novel LSD1 inhibitors: design, synthesis and their biological evaluation. Eur J Med Chem. 2017;126:246–258. doi: 10.1016/j.ejmech.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 63.Duan Y., Qin W., Suo F., Zhai X., Guan Y., Wang X. Design, synthesis and in vitro evaluation of stilbene derivatives as novel LSD1 inhibitors for AML therapy. Bioorg Med Chem. 2018;26:6000–6014. doi: 10.1016/j.bmc.2018.10.037. [DOI] [PubMed] [Google Scholar]
- 64.Lv H., She G. Naturally occurring diarylheptanoids. Nat Prod Commun. 2010;5:1687–1708. [PubMed] [Google Scholar]
- 65.Alberti Á., Riethmüller E., Béni S. Characterization of diarylheptanoids: an emerging class of bioactive natural products. J Pharmaceut Biomed Anal. 2018;147:13–34. doi: 10.1016/j.jpba.2017.08.051. [DOI] [PubMed] [Google Scholar]
- 66.Banik U., Parasuraman S., Adhikary A.K., Othman N.H. Curcumin: the spicy modulator of breast carcinogenesis. J Exp Clin Canc Res. 2017;36:98. doi: 10.1186/s13046-017-0566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson K.M., Dahlin J.L., Bisson J., Graham J., Pauli G.F., Walters M.A. The essential medicinal chemistry of curcumin. J Med Chem. 2017;60:1620–1637. doi: 10.1021/acs.jmedchem.6b00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dona S., Jaydip B., Bokyung S., Bharat B.A., Anupam B. Chemopreventive and chemotherapeutic potential of curcumin in breast cancer. Curr Drug Targets. 2012;13:1799–1819. doi: 10.2174/138945012804545632. [DOI] [PubMed] [Google Scholar]
- 69.Fu S., Kurzrock R. Development of curcumin as an epigenetic agent. Cancer. 2010;116:4670–4676. doi: 10.1002/cncr.25414. [DOI] [PubMed] [Google Scholar]
- 70.Hassan F.U., Rehman M.S.U., Khan M.S., Ali M.A., Javed A., Nawaz A. Curcumin as an alternative epigenetic modulator: mechanism of action and potential effects. Front Genet. 2019;10:514. doi: 10.3389/fgene.2019.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J., Zhang X., Yan J., Li W., Jiang Q., Wang X. Design, synthesis and biological evaluation of curcumin analogues as novel LSD1 inhibitors. Bioorg Med Chem Lett. 2019;29:126683. doi: 10.1016/j.bmcl.2019.126683. [DOI] [PubMed] [Google Scholar]
- 72.Kunnumakkara A.B., Harsha C., Banik K., Vikkurthi R., Sailo B.L., Bordoloi D. Is curcumin bioavailability a problem in humans: lessons from clinical trials. Expet Opin Drug Metabol Toxicol. 2019;15:705–733. doi: 10.1080/17425255.2019.1650914. [DOI] [PubMed] [Google Scholar]
- 73.Salehi B., Stojanović-Radić Z., Matejić J., Sharifi-Rad M., Anil Kumar N.V., Martins N. The therapeutic potential of curcumin: a review of clinical trials. Eur J Med Chem. 2019;163:527–545. doi: 10.1016/j.ejmech.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 74.Manchester L.C., Coto-Montes A., Boga J.A., Andersen L.P.H., Zhou Z., Galano A. Melatonin: an ancient molecule that makes oxygen metabolically tolerable. J Pineal Res. 2015;59:403–419. doi: 10.1111/jpi.12267. [DOI] [PubMed] [Google Scholar]
- 75.Bhattacharya S., Patel K.K., Dehari D., Agrawal A.K., Singh S. Melatonin and its ubiquitous anticancer effects. Mol Cell Biochem. 2019;462:133–155. doi: 10.1007/s11010-019-03617-5. [DOI] [PubMed] [Google Scholar]
- 76.Capote-Moreno A., Ramos E., Egea J., López-Muñoz F., Gil-Martín E., Romero A. Potential of melatonin as adjuvant therapy of oral cancer in the era of epigenomics. Cancers. 2019;11:1712. doi: 10.3390/cancers11111712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xin Z., Jiang S., Jiang P., Yan X., Fan C., Di S. Melatonin as a treatment for gastrointestinal cancer: a review. J Pineal Res. 2015;58:375–387. doi: 10.1111/jpi.12227. [DOI] [PubMed] [Google Scholar]
- 78.Niles L.P., Pan Y., Kang S., Lacoul A. Melatonin induces histone hyperacetylation in the rat brain. Neurosci Lett. 2013;541:49–53. doi: 10.1016/j.neulet.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 79.Sharma R., Ottenhof T., Rzeczkowska P.A., Niles L.P. Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene expression in C17.2 neural stem cells. J Pineal Res. 2008;45:277–284. doi: 10.1111/j.1600-079X.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- 80.Wei J.Y., Li W.M., Zhou L.L., Lu Q.N., He W. Melatonin induces apoptosis of colorectal cancer cells through HDAC4 nuclear import mediated by CaMKII inactivation. J Pineal Res. 2015;58:429–438. doi: 10.1111/jpi.12226. [DOI] [PubMed] [Google Scholar]
- 81.Yang C.Y., Lin C.K., Tsao C.H., Hsieh C.C., Lin G.J., Ma K.H. Melatonin exerts anti-oral cancer effect via suppressing LSD1 in patient-derived tumor xenograft models. Oncotarget. 2017;8:33756–33769. doi: 10.18632/oncotarget.16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakane C., Okitsu T., Wada A., Sagami H., Shidoji Y. Inhibition of lysine-specific demethylase 1 by the acyclic diterpenoid geranylgeranoic acid and its derivatives. Biochem Biophys Res Commun. 2014;444:24–29. doi: 10.1016/j.bbrc.2013.12.144. [DOI] [PubMed] [Google Scholar]
- 83.Cuyàs E., Gumuzio J., Lozano-Sánchez J., Carreras D., Verdura S., Llorach-Parés L. Extra virgin olive oil contains a phenolic inhibitor of the histone demethylase LSD1/KDM1A. Nutrients. 2019;11:1656. doi: 10.3390/nu11071656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luka Z., Moss F., Loukachevitch L.V., Bornhop D.J., Wagner C. Histone demethylase LSD1 is a folate-binding protein. Biochemistry. 2011;50:4750–4756. doi: 10.1021/bi200247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luka Z., Pakhomova S., Loukachevitch L.V., Calcutt M.W., Newcomer M.E., Wagner C. Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate. Protein Sci. 2014;23:993–998. doi: 10.1002/pro.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Z., Ding L., Li Z., Wang Z., Suo F., Shen D. Development of the triazole-fused pyrimidine derivatives as highly potent and reversible inhibitors of histone lysine specific demethylase 1 (LSD1/KDM1A) Acta Pharm Sin B. 2019;9:794–808. doi: 10.1016/j.apsb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S., Li Z.R., Suo F.Z., Yuan X.H., Yu B., Liu H.M. Synthesis, structure−activity relationship studies and biological characterization of new [1,2,4]triazolo[1,5-a]pyrimidine-based LSD1/KDM1A inhibitors. Eur J Med Chem. 2019;167:388–401. doi: 10.1016/j.ejmech.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 88.Wang S., Zhao L.J., Zheng Y.C., Shen D.D., Miao E.F., Qiao X.P. Design, synthesis and biological evaluation of [1,2,4] triazolo [1,5-a] pyrimidines as potent lysine specific demethylase 1 (LSD1/KDM1A) inhibitors. Eur J Med Chem. 2017;125:940–951. doi: 10.1016/j.ejmech.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 89.Li Z.R., Wang S., Yang L., Yuan X.H., Suo F.Z., Yu B. Experience-based discovery (EBD) of aryl hydrazines as new scaffolds for the development of LSD1/KDM1A inhibitors. Eur J Med Chem. 2019;166:432–444. doi: 10.1016/j.ejmech.2019.01.075. [DOI] [PubMed] [Google Scholar]
- 90.Li Z.R., Suo F.Z., Hu B., Guo Y.J., Fu D.J., Yu B. Identification of osimertinib (AZD9291) as a lysine specific demethylase 1 inhibitor. Bioorg Chem. 2019;84:164–169. doi: 10.1016/j.bioorg.2018.11.018. [DOI] [PubMed] [Google Scholar]