Abstract
Histone three lysine four dimethylation (H3k4me2) in sperm is conserved across species and is linked to transgenerational epigenetic inheritance. To test whether H3K4me2 is a target for transgenerational inheritance of toxicity, a daily gavage bolus exposure of trichloroethylene (TCE) (1000 mg/kg/day) was given to rats for 14 weeks, then epididymal sperm were isolated and native chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) of H3K4me2 was performed. Differential region analysis determined there were 2608 significantly differential H3K4me2 regions after TCE exposure, 477 were significantly increased and 2131 were significantly decreased. Z-score enrichment of differential regions determined there were significantly decreased H3k4me2 in the coding and regulatory regions of genes in the PKA signaling pathway. These changes account for TCE induced spermatozoal toxicity and show H3K4me2 is a target for paternal inheritance of toxicity.
Keywords: Trichloroethylene, dimethylated histone three lysine four, PKA signaling, chromatin, paternal epigenetic inheritance
Dear Editor,
During spermatogenesis, most sperm histones are removed and replaced with protamines. The few histones that remain in the fully formed, mature spermatids have potential epigenetic roles during embryonic development [1]. Histone modifications such as dimethylated histone three lysine four (H3K4me2) and H3K4me3 in sperm critically influence chromatin structure in the embryo [2]. The chromatin structure found in the zygote is more similar to a sperm than an oocyte [3], suggesting that the chromosome patterning is paternally influenced. Altered locations or modification of retained histones in sperm can be deleterious to the offspring. We have previously reported that localization of H3K4me2 to developmental genetic loci is similar in Rattus norvegicus sperm as in mouse and human sperm [4]. Rats are widely used as models of toxicity, and in this letter, we describe how exposure to a toxicant altered localization of H3K4me2 in mature rat sperm measured using native chromatin immunoprecipitation followed by next-generation sequencing (nChIP-seq).
Trichloroethylene (TCE) is used as an industrial solvent and in organic extraction. It is also contained in many consumer products such as rug-cleaning fluids, metal degreasers, and printer ink. TCE has been found to cause sperm effects in rats [5]. Here, we describe TCE effects on H3K4me2 as a potential mechanism of transgenerational paternal epigenetic inheritance in the rat model.
Four vehicle controls and three TCE-exposed adult male Fisher replace with (CDF; Charles River, Wilmington, MA) rats (oral gavage, 1000 mg/kg TCE dissolved in corn oil at 2 mL/kg) were exposed five times per week for 14 weeks, at which time the rats were euthanized and sperm was collected from the cauda of the epididymis. The TCE exposure paradigm is used as a model of disruption and was chosen based on previous results that show significant changes in sperm motility [5]. Sperms were prepared for nChIP-seq as previously described (Abcam ab7766 H3K4me2 antibody) [4]. Matched immunoprecipitated (IP) and genomic control (GC) DNA libraries were made using the Nugen Ovation Ultralow system V2 (cat# 0344 and 0344NB) according to manufacturer’s instructions on each rat sperm sample individually. DNA libraries were randomly multiplexed into two wells and sequenced (Illumina HiSeq2500, single end 1 × 50 bp). Reads were aligned to the rn4 genome using bowtie, where the IP samples averaged 90.9% and GC samples averaged 95.2% mappable reads. The duplicates accounted for 2.4% of GC and 23.4% of IP reads. Diffreps [6] was used to determine the regions with differential enrichment in the exposed rats compared to controls. Duplicates had a negligible effect on differential region analysis (Supplemental Figures S1–S10). The log2-fold-change values from the Diffreps analysis in the gene body, proximal promoter, 1-kb upstream promoter (1 k promoter), and 3-kb upstream promoter (3 k promoter) regions were imported into ingenuity pathway analysis (IPA; Qiagen Germantown, MD, USA) software to find z-score enrichment or depletion in canonical signaling pathways. Principal component (PC) analysis (PCA) and transcription start site (TSS) enrichment analysis were performed using deeptools [7] on normalized IP to GC reads for each sample.
When the H3K4me2 localization in sperm from TCE-exposed rats was compared to that of control rats, there were 2608 significantly differential regions after TCE exposure (Supplemental Table S1), 477 were significantly increased and 2131 were significantly decreased. Normalized read counts were scaled relative to all genes in the rn4 genome. These scaled, normalized read counts determined the peak of H3K4me2 enrichment occurred immediately after the TSS, in all samples regardless of TCE exposure (Figure 1A). About, 75% of the differential regions occurred at intergenic loci, 13% occurred in gene bodies, 6% occurred in the 1 k promoter, 4% occurred in the 3 k promoter, and 2% occurred in the proximal promoters. PCA of normalized read counts in the differential regions shows PC1 separates samples based on treatment and accounts for 64.5% of variance (Figure 1B). Browser tracks of 10 differential regions support the findings of differential abundance of DNA fragments at these loci (Supplemental Figures S1–S10). Of the differential regions in gene bodies and promoters, protein kinase A (Pka) signaling was the only pathway that had significantly reduced H3K4me2 modifications across 3 of 4 categories involving multiple genes in this signaling pathway (Figure 1C). Several Pka signaling genes (the catalytic subunit of protein kinase a, phospholipase C, phosphodiesterase 10a, and G-protein subunit beta 3, Map 2 k6, Map 2 k10, and protein tyrosine phosphatase receptor type c) had fewer “activating” H3K4me2 modifications, while inositol triphosphate receptor 1 and Mek1/2 had more H3K4me2 (Figure 1C). Protein tyrosine phosphatase (Ptp) had significantly less H3K4me2 in exposed samples within the gene body, and significantly more H3K4me2 in its promoters.
Figure 1.
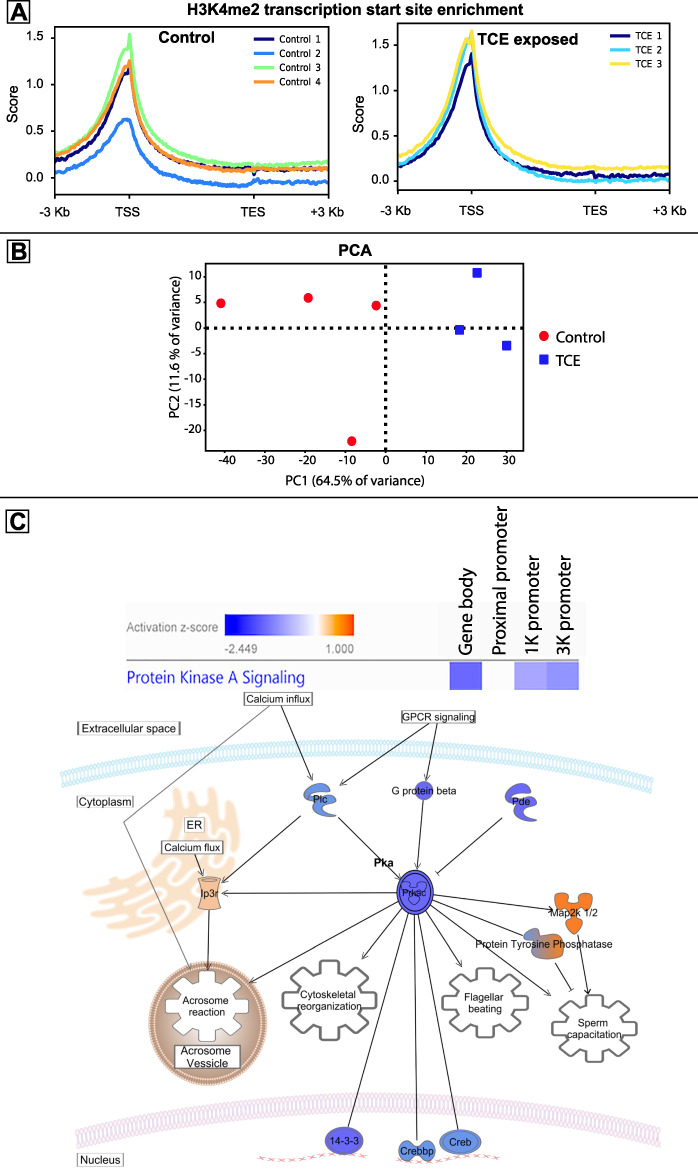
Altered demethylated H3K4me2 localization in rat sperm after exposure to TCE. (A) Enrichment of H3K4me2 as measured by normalized read counts scaled to each gene in the rn4 rat genome, their TSS, TES, and 3000 nucleotides upstream and downstream of the genes. (B) PCA shows how the first two PCs account for most of the variation in the samples. (C) IPA software found protein kinase A (Pka) signaling genes had significant decrease in number of H3K4me2 peaks in the gene body, 1-k promoter, and 3-k promoter regions. IPA pathway builder illustrates the biological relation of genes that are epigenetically altered after TCE exposure. Blue shows where fewer H3K4me2 were found near the genetic loci for the respective gene in TCE compared to controls and orange shows where more H3K4me2 were found. The blue–orange gradient denotes opposite findings in the different gene regions. Two known sperm stimuli, calcium influx, and Zona pellucida binding to g-protein coupled receptors (GPCRs) feed into the PKa signaling pathway. The PKa signaling pathway stimulates four known sperm functions, including: the acrosome reaction, cytoskeletal reorganization, flagellar beating, and sperm capacitation.
The reduced H3K4me2-mediated activation of Pka genes reflected the effects of TCE on sperm, including reduced motility [5]. Pka inhibitors are known to decrease motility and prevent capacitation and the acrosome reaction, while the use of cAMP analogues stimulates these sperm functions [8]. When sperm bind the zona pellucida, G-protein coupled receptors (GCPR) activate signaling through phospholipase C (Plc) and Gβ to mediate fertilization, both of which have fewer H3K4me2 marks after TCE exposure (Figure 1C). Calcium ions influx into sperm, triggering Pka signaling mechanisms for motility, the acrosome reaction, capacitation of sperm, and oocyte fusion [8]. These results indicate a relationship between these H3K4me2 modifications near Pka genes and TCE-induced toxicity to sperm.
Histone modifications in sperm have been shown to transmit metabolic disease in Drosophila [9]. Overexpression of Kdm1a and subsequent loss of H3K4me2 in sperm at many important developmental regions resulted in both intergenerational and transgenerational embryonic developmental defects [10]. Our results show that H3K4me2 is decreased near Pka signaling genes after TCE exposure in rats. Pka signaling is a critical repressive mechanism for Hedgehog (Hh) signaling. Altering this pathway would be expected to alter embryonic notochord development, body axis patterning, and limb formation. Although sperm toxicities of TCE are well documented, how the exposure of the father to an environmental toxicant can effect offspring has not been explored.
Our findings show that altered sperm histone modifications occur in rats. Future studies will explore the role of epigenetic inheritance through altered histone modifications in offspring.
Conflict of interest
The authors have declared that no conflict of interest exists.
Supplementary Material
References
- 1. Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aoshima K., Inoue E., Sawa H., and Okada Y.. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep 2015; 16(7):803–812. doi: 10.15252/embr.201439700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, Chen H, Du Z et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 2017; 170:367–381. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 4. Wilson S., Dere E., Klein D., Schorl C., Hall S., and Boekelheide K.. Localization of dimethylated histone three lysine four in the Rattus norvegicus sperm genome. Biol Reprod 2018; 99(2):266–268. doi: 10.1093/biolre/ioy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stermer AR, Klein D, Wilson SK, Dalaijamts C, Bai CY, Hall SJ, Madnick S, Bianchi E, Chiu WA, Boekelheide K. Differential toxicity of water vs gavage exposure to trichloroethylene in rats. Environ Toxicol Pharmacol 2019; 68:1–3. doi: 10.1016/j.etap.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen L., Shao N.Y., Liu X., Maze I., Feng J., and Nestler E.J.. diffReps: Detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 2013; 8(6):e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016; gkw257. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Visconti PE, Moore GD, Bailey JL, Leclerc P, Connors SA, Pan D, Olds-Clarke P, and Kopf G. S. Capacitation of mouse spermatozoa II: Protein tyrosine phosporylation and capacitation are regulated by a cAMP-dependent pathway. Development 1995; 121(4):1139–1150. [DOI] [PubMed] [Google Scholar]
- 9. Öst A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PV, Stoeckius M, Ruf M, Rajewsky N et al. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 2014; 159:1352–1364. doi: 10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 10. Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur J, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters JHM, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015; 350(6261):651. doi: 10.1126/science.aab2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


