Abstract
DISCOVER-Seq (Discovery of In Situ Cas Off-targets and VERification by sequencing) is a broadly applicable approach for unbiased CRISPR-Cas off-target identification in cells and tissues. It leverages the recruitment of DNA repair factors to double-strand breaks (DSBs) after genome editing with CRISPR nucleases. Here we describe a detailed experimental protocol and analysis pipeline to perform DISCOVER-Seq. The principle of this method is to track the precise recruitment of MRE11 to DSBs by chromatin immunoprecipitation followed by next-generation sequencing. A customized open source bioinformatics pipeline, BLENDER (BLunt END findER) (https://github.com/staciawyman/blender), then identifies off-target sequences genome-wide. DISCOVER-Seq is capable of finding and measuring off-targets in primary cells and in situ. The two main advantages of DISCOVER-Seq are (i) low false positive rates because DNA repair enzyme binding is required for the appearance of genome edits, and (ii) its applicability to a wide variety of systems including patient-derived cells and animal models. The whole protocol, including the analysis, can be completed within two weeks.
Keywords: CRISPR-Cas, genome editing, Off-target effects, DNA repair, discover-seq
Introduction
Genome editing fundamentally operates by inducing DNA damage at a targeted locus. This damage is then repaired by the cell to yield an editing outcome. But all known genome editing reagents, from Zinc Finger Nucleases to CRISPR-Cas enzymes, have the potential to affect off-target loci that share sequence similarity with the on-target site1,2. Great strides have been made in developing high-fidelity genome editing enzymes3–6. However, the permanent nature of off-target genome modification can necessitate an approach best characterized as “Doveryai, no proveryai” (“Trust, but verify”). This is particularly the case when developing pre-clinical genome editing protocols in clinically-relevant human cell types, where there is little prospect to counter-select unintended gene marking. Here, we present a detailed workflow for DISCOVER-Seq describing the experimental and computational procedures of the method in detail7. DISCOVER-Seq enables unbiased identification of CRISPR-Cas off-targets in otherwise intractable contexts by monitoring the recruitment of DNA repair factors. We include tips and tricks, as well as further optimization of antibody reagents. Researchers interested in DISCOVER-Seq should have expertise in or access to general molecular biology (for the ChIP-Seq workflow), cell culture, genome editing, next-generation sequencing, and bioinformatics (for processing and analysis of DISCOVER-Seq output).
Development of the Protocol
We developed DISCOVER-Seq as a universally applicable approach to identify and characterize CRISPR-Cas off-targets in any cell type and during editing of living organisms7. DISCOVER-Seq takes advantage of the DNA repair processes inherent to genome editing. All genome editing is the result of DNA repair, and so both on- and off-target loci are processed by DNA repair enzymes. Multiple DNA repair factors localize to a CRISPR-Cas double stranded DNA break, and could theoretically be used to track on- and off-target Cas-induced double-strand breaks (DSBs).
We used chromatin immunoprecipitation followed by sequencing (ChIP-Seq) to identify genomic locations that were bound by DNA repair machinery after inducing a target DSB with CRISPR-Cas9. We selected a panel of proteins involved in repair of DSBs: Ser193-phosphorylated histone 2AX (γH2AX), XRCC6/Ku70, FANCD2 and the three components of the MRN complex: MRE11, RAD50 and NBS1. Our ChIP-Seq data showed that all six DNA repair proteins assembled at the targeted genomic location, but that their binding patterns were distinct7. Both γH2AX and FANCD2 produced a broad peak centered at the target site kilobases (KB) to megabases (MB) in width. In contrast, Ku70 and members of the MRN complex exhibited a defined narrow peak at the cut site, consistent with their known roles in end processing8–11. We found that MRE11 ChIP-Seq is particularly effective in identifying sites of off-target Cas activity, as ~90% of reads in MRE11 ChIP-Seq peaks start or end at the Cas9-induced cut site. This allows one to determine the site of Cas action with single-base precision. In addition, MRE11 has a high-quality chromatin immunoprecipitation (ChIP) antibody that cross-reacts between human and mouse, allowing a single protocol for interrogating both human-derived cells and mouse models7. Choice of repair protein, choice of antibody, optimization and timing of ChIP-Seq, and a specialized bioinformatic pipeline were key to the development of DISCOVER-Seq to successfully identify Cas off-targets on a genome scale.
Alternative methods
Several approaches to discover genome editing off-targets have been described. These broadly fall into in silico, in vitro, in cellula, and in vivo strategies. In silico methods generally rely on sequence similarity of a possible off-target sequence to the on-target sequence, often incorporating differential weighting for the seed or non-seed regions of the protospacer. In silico methods are fast and easy to use, and in some cases can perform well12, but they often result in false positives or leave researchers wondering “what if?”, and in pre-clinical cases are no substitute for experimental data. In vitro methods often isolate genomic DNA from a target cell type, digest it with a recombinant form of the relevant nuclease, and use next generation sequencing (NGS) to detect cleavage events. In vitro approaches are exquisitely sensitive and can interrogate patient-specific genotypes, but they also erase all cellular and tissue context that can influence genome editing, such as cell state and chromatin status, and their sensitivity comes hand-in-hand with hundreds to thousands of false positives13–16. In cellula methods typically determine where an identifying barcode has inserted into a cell’s genome or the location of at least one side of a liberated double strand break17–21. These approaches can be quite sensitive and incorporate chromatin and cellular context, but they are also prone to false positives and thus far only applicable in limited types of cells. A recently published method22 can quantify off-targets in vivo, but requires two entirely separate workflows with initial discovery taking place in vitro followed by extensive testing of putative off-target sites by targeted amplicon sequencing of genomic DNA from mouse tissue. While this approach is highly sensitive, it is laborious and has the same high false-positive rates inherent to in vitro methods. DISCOVER-Seq on the other hand consists of a single workflow that allows off-target discovery directly in the edited tissues.
Applications
DISCOVER-Seq visualizes the process of genome editing itself and does not rely on extra manipulation of living cells, apart from the introduction of editing reagents. Hence, it is broadly applicable to a wide variety of systems where the addition of tracking moieties (e.g. molecular barcodes or handles) is otherwise problematic. For example, we have shown that DISCOVER-Seq yields good data in induced pluripotent stem cells (iPSCs) and during adenoviral in situ editing of the mouse liver7.
We have extensively tested DISCOVER-Seq with both wild-type and high-fidelity S. pyogenes Cas9s, and have optimized the bioinformatic pipeline for these nucleases. ChIP-Seq of MRE11 also performs well with the Cas12a (formerly Cpf1) nuclease23, but a bioinformatic pipeline that can perform genome-wide identification of the overlapping reads stemming from Cas12a is still under development.
For ease of use, we perform DISCOVER-Seq with a commercially available human/mouse cross-reactive anti-MRE11 antibody that allows the same reagents to be used in human and mouse cell lines, primary cells, and during pre-clinical editing of mouse models. DISCOVER-Seq is freely usable without license.
Advantages and Limitations
Because DISCOVER-Seq relies on chromatin immunoprecipitation rather than specific amplification of an edited locus, it has a higher limit of detection and requires more material (>= 5×106 cells) than some in vitro or in cellula off-target identification methods. DISCOVER-seq also requires higher read depth relative to other NGS-based off-target methods such as CIRCLE-Seq14 and GUIDE-Seq17. While CIRCLE-Seq and GUIDE-Seq can be performed on an Illumina MiSeq, DISCOVER-Seq requires instruments capable of greater sequencing depth. We have not exhaustively explored the effect of extremely deep sequencing on DISCOVER-seq data, but generally recommend at least 30 M reads.
We empirically determined that DISCOVER-Seq is capable of finding target sites that result in 0.3 % indels. More sensitive off-target identification methods such as CIRCLE-Seq can find rarer off-targets, but can also have high false-positive rates. So far, we have not found systematic false positives for DISCOVER-Seq. DSBs occurring independent of Cas-induced damage will also be bound by MRE11. However, most of these DSBs will occur at random locations throughout the genome, diluting each DSB signal across millions of cells and resulting in no ChIP-Seq peak. If DSBs occur at genomic fragile sites, these would also be present in the unedited control sample, and would be filtered out by the default BLENDER algorithm (although they could be added back with a simple command line option).
DISCOVER-Seq is agnostic of the repair outcome of the CRISPR-induced double strand break as sites are detected before they are repaired to yield a sequence-based outcome. One must keep this in mind, since it means that DISCOVER-Seq is time sensitive. For example, the timing of DISCOVER-Seq is different when performing RNP editing (where the DNA double-strand break happens almost immediately) as opposed to viral editing (where Cas9 and gRNA must first be expressed from the viral vector).
This must be taken into consideration when planning a DISCOVER-Seq experiment. DISCOVER-Seq’s ability to visualize Cas interventions prior to repair could be used to reveal target sites that are repaired without insertion or deletions or have resulted in large deletions that exceed the PCR amplification range of other methods, but this has not been systematically evaluated.
Finally, it is theoretically possible that not all sites of DNA damage recruit MRE11. While MRE11 is known to play important roles in all DNA repair pathways24–26, we cannot exclude the existence of off-target sites that are missed due to lack of MRE11 at the locus. However, we so far have not found high-frequency off-targets identified by MRE11-independent methods that are missed by DISCOVER-seq.
Experimental design
Overview of the workflow
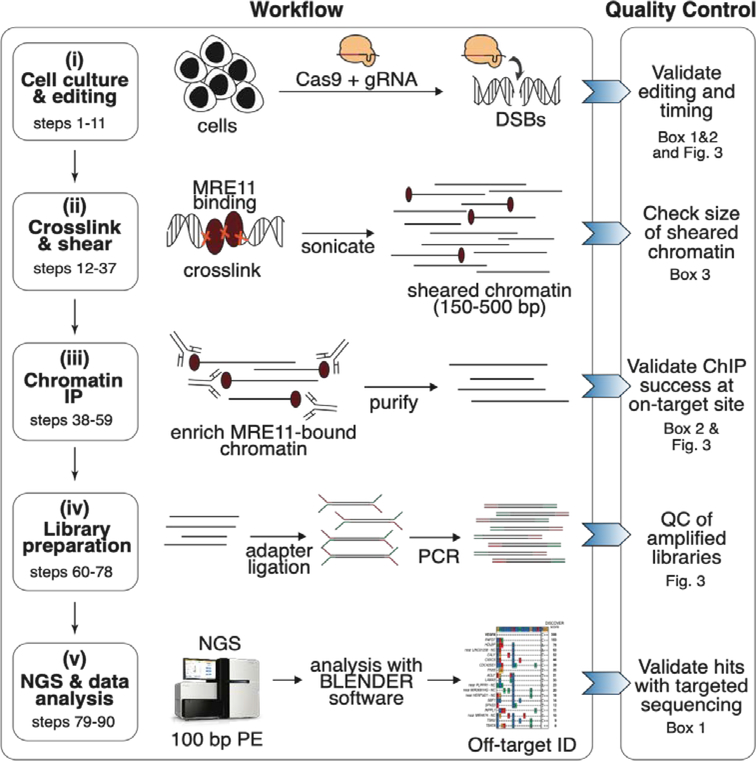
DISCOVER-Seq consists of five major stages (Fig. 1): (i) Cell culture and genome editing of cells, (ii) crosslinking and chromatin shearing, (iii) chromatin immunoprecipitation, (iv) ChIP-Seq library preparation, (v) next-generation sequencing (NGS) and data analysis.
Figure 1: Overview of DISCOVER-Seq workflow and quality controls.
(i) Cells of interest are edited with Cas9 and gRNA to introduce DNA double strand breaks (DSBs) at the target site. Successful genome editing should be confirmed (Box 1 and 2). (ii) Edited cells are crosslinked, lysed and chromatin is sheared to 150–500 bp by sonication. The correct size of sheared chromatin should be confirmed by agarose gel electrophoresis (Box 3). (iii) MRE11-bound chromatin fragments are enriched by immunoprecipitation. Enriched DNA is then purified and reverse crosslinked. Successful MRE11 pulldown should be confirmed by qPCR (Box 2). (iv) If ChIP was successful, purified DNA is prepared for NGS by adapter ligation and PCR amplification of ChIP-Seq libraries. The quality of the amplified libraries should be confirmed by size separation on a fragment analyzer. (v) Libraries are sequenced as 100 bp paired-end (PE) reads on a next generation sequencer. Data is then analyzed using BLENDER software to identify off-target sites. Off-target sites can be validated with targeted amplicon sequencing (Box 1).
Cell numbers required
For DISCOVER-Seq we recommend using approximately 107 edited cells as starting material per chromatin immunoprecipitation. The minimum number of cells to be used as starting material per ChIP is 5×106 cells.
Controls required
As a control one can use cells exposed to a non-targeting gRNA and Cas9, Cas9 only, or unedited cells. The former two options are preferable. The control sample should be treated in exactly the same manner as the sample that was edited with a targeting gRNA and Cas9. The control sample will serve as an indicator for background noise and will help identify ChIP artifacts and blacklisted regions27 in the data analysis step.
Choice of antibody
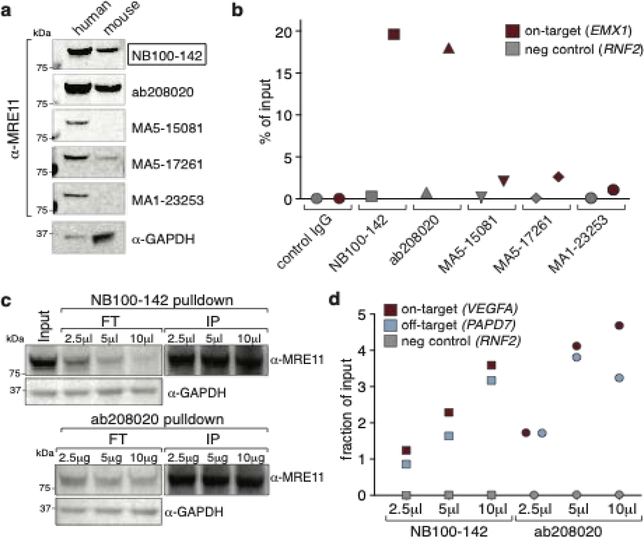
We tested five commercially available antibodies against MRE11 for their cross-reactivity between human and mouse samples and for their suitability for ChIP (Fig. 2 and Supp. Fig. 1).
Figure 2: Comparison of MRE11 antibodies.
(a) Cross-reactivity between human and mouse of five different commercially available MRE11 antibodies as determined by Western Blot. Whole cell extracts (30 μg) from human K562 cells and murine B16-F10 cells were used for the analysis. NB100–142, ab208020 and MA5–17261 are cross-reactive. Black box indicates antibody used in Wienert et al. 20197. (b) Performance of MRE11 antibodies in ChIP. K562 cells were edited with RNPs targeting EMX1. ChIP-qPCR was performed 12 h post nucleofection. Shown is the enrichment of the EMX1 region compared to an unedited negative control region (RNF2). NB100–142 and ab208020 performed best in this assay. (c) Increasing amounts of antibody lead to more MRE11 depletion from cell lysate, as determined by Western Blot. However, increasing pulldown efficiency with larger amounts of antibody is not directly reflected in the levels of eluted MRE11, possibly due to non-linearity of Western blots. FT – flow through, IP - immunoprecipitation. (d) Increasing the amount of antibody increases the on- and off-target signal by qPCR. K562 cells were edited with RNPs targeting VEGFA. ChIP-qPCR was performed 12 h post nucleofection. Shown is the enrichment of the VEGFA region and a known off-target region (PAPD7) compared to an unaffected negative control region (RNF2).
Genome editing
Cells of interest are edited using the CRISPR-Cas9 system. Here we describe the workflow for editing cells by nucleofection using ribonucleoprotein (Cas9-gRNA complexes, RNPs). Theoretically, any type of CRISPR-Cas delivery vehicle (e.g. lipofection, viral transduction) and any form of editing reagent (e.g. RNPs, plasmids, mRNAs) can be used with the DISCOVER-Seq workflow. However, as DISCOVER-Seq measures the amount of DSBs bound by MRE11 at a given time, the timing of the assay may vary between transfection methods (see below). Successful on-target genome editing of samples should be validated before proceeding with the DISCOVER-Seq assay (Box 1).
Box 1: QC checkpoint 1 for editing efficiency.
Before proceeding to cell lysis (Step 26), it is helpful to check if the CRISPR genome editing was successful at the on-target site. This stage requires user-specific primers that are used to amplify a region of 250–300 bp across the on-target site. These amplicons can either (A) be sequenced by Sanger sequencing and edits deconvoluted using web tools such as TIDE (https://tide.deskgen.com/) or ICE (https://ice.synthego.com) or amplicons can be (B) sequenced using next-generation sequencing and analyzed using software such as CRISPResso (http://crispresso.pinellolab.partners.org/) or Cortado (https://github.com/staciawyman/cortado), as described in the Procedure below. The same protocol can be applied to validate edits at off-target sites by using off-target site specific primers.
Collect cells dedicated for checking the editing efficiency of your experiment (from Step 12) in a 10 mL conical tube 72–96 h after nucleofection.
Centrifuge cells at room temperature at 300 g for 5 min.
Wash cells in 1 mL PBS and transfer cell slurry to a 1.5 mL microcentrifuge tube. Spin at 300 g for 5 min. Aspirate supernatant and discard.
-
Extract genomic DNA (gDNA) using a Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s instructions.
PAUSE POINT: Genomic DNA can be stored at −20 °C for up to 6 months.
Adjust gDNA concentration to 100 ng/μl with nuclease-free water.
-
Determine insertion and deletion frequency in your sample by Sanger sequencing (option A) or next generation sequencing (option B).
?Troubleshooting
- Sanger sequencing and ICE analysis
-
Assemble PCR as follows:
Component Volume (μl) Final concentration H2O 9 gDNA (100 ng, 100 ng/μl, from Step 5 of this box 1 4 ng/μl Custom primer F (10 μM) 1.25 0.5 μM Custom primer R (10 μM) 1.25 0.5 μM 2x Q5 Master Mix 12.5 1x Total 25 CRITICAL STEP: Always run a PCR on gDNA from the control sample (unedited) as well as the edited sample. - Perform the PCR using the following thermocycling conditions to amplify on-target locus. Optimize the conditions as required to obtain a single band of the correct size when analyzed by standard 2 % agarose gel electrophoresis.
Step Temperature (C) Time (sec) Cycles Denaturation 98 30 1 Denaturation 98 10 30 Annealing Primer dependent 20 30 Extension 72 20 30 Extension 72 120 1 Hold 4 1 - Sanger sequence the amplicons.
- Determine indel frequencies by uploading files to https://ice.synthego.com.
-
- Next-generation sequencing
- Follow the instructions on https://www.protocols.io/view/preparation-of-pcr-amplicons-from-edited-cells-for-hdsb26e.
If editing was successful and indel frequencies are as expected, proceed to Step 21 in the main procedure.
Note that the formation of indels is not completed at the time that edited cells are prepared for DISCOVER-Seq. Thus, an aliquot of the edited cells is kept in culture for an additional 48–72 h to then be harvested for genomic DNA extraction and validation of off-target sites.
Timing of assay
The ideal time point for DISCOVER-Seq should be determined empirically and depends on the delivery method of editing reagents (e.g. RNP vs. plasmid) and cell type. We summarized the questions that should be addressed prior to performing DISCOVER-Seq with a new cell type in Table 1.
Table 1:
Steps to follow when starting with a new cell type
| Step | Questions to address | Refer to: | ||
|---|---|---|---|---|
| 1. | Genome editing | • What delivery method are you using? • Is genome editing successful at the on-target site? |
Box 1 | |
| 2. | Sonication | • What are the correct sonication settings (intensity, number of cycles) to obtain 200–500 bp fragments of sheared chromatin? → this should be optimized on crosslinked cells |
Box 3 | |
| 3. | Timing | • Using your delivery method, what is the right time point to do DISCOVER-Seq? • Suggested time points: |
Box 2 | |
| RNP electroporation | ∼ 8–16 h | |||
| Adenovirus injection (in vivo) | ∼ 24 h | |||
| RNP lipofection | Determine experimentally | |||
| Plasmid electroporation/lipofection | Determine experimentally | |||
| Lentiviral transduction | Determine experimentally | |||
| AAV delivery (in vitro & in vivo) | Determine experimentally | |||
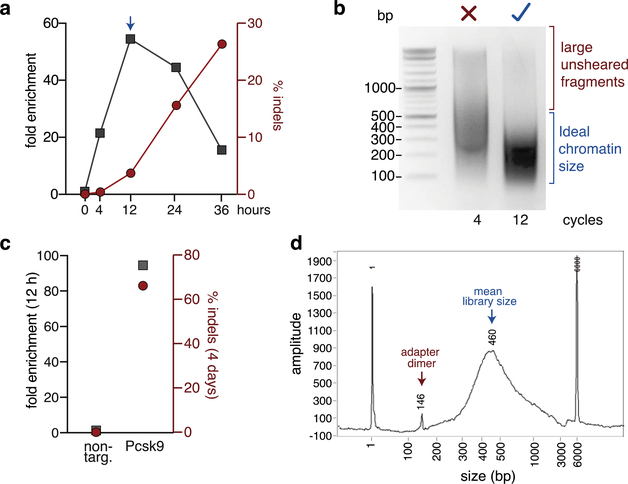
The best signal-to-noise is obtained when edited cells are crosslinked when MRE11 occupancy is maximal. Performing a ChIP-qPCR time course at different times post editing will determine the ideal time point for DISCOVER-Seq in that particular assay and cell system (Fig. 3a and Box 2).
Figure 3: Quality controls of a DISCOVER-Seq experiment.
(a) Example of a ChIP-qPCR time course to determine the ideal time point for DISCOVER-Seq using a Cas9 RNP (Box 2). K562 cells were nucleofected with Cas9 and gRNA targeting the HBB locus. Cells were then crosslinked at indicated time points and MRE11 ChIP performed. Shown is the fold enrichment of the on-target region (HBB) over a negative control region for each sample (left y-axis) and the indel frequencies (right y-axis). For this experiment 12 h post nucleofection was chosen as the ideal time point for DISCOVER-Seq (arrow). (b) Agarose gel electrophoresis of insufficiently (4 cycles) and optimally (12 cycles) sheared chromatin. Sheared chromatin size (150–500 bp) should be checked after sonication before proceeding with the protocol (Box 3). (c) Example of quality control by qPCR for successful isolation of an on-target locus by ChIP before proceeding to ChIP-Seq library preparation. B16-F10 cells were nucleofected with a non-targeting gRNA (non-targ.) or a gRNA targeting Pcsk9. After 12 h, cells were crosslinked and MRE11 ChIP was performed. Shown is the fold enrichment of the on-target site Pcsk9 over a negative control region (left y-axis) and the indel frequencies after 4 days (right y-axis). (d) Example electropherogram of a ChIP-Seq library before sequencing. While the mean library size is optimal, this sample contains adapter dimers. If adapter dimers are present, DNA size selection of the library should be performed (see ?Troubleshooting).
Box 2: QC checkpoint 2 for timing of the assay.
The ideal time point for DISCOVER-Seq analysis to be carried out should be determined experimentally and may vary between cell types and modes of CRISPR-Cas9 delivery. The main procedure describes ideal conditions for K562 and B16-F10 cells using RNP nucleofection. If a different cell type and/or transfection method is used, the ideal time point needs to be determined experimentally by quantifying the enrichment of an on-target locus by quantitative real-time PCR. One needs to design target-specific primers suitable for SYBR-based qPCR assays. The primers should amplify a region within 50–150 bp of the cut site but not span the target site, as the DNA ends at the cut site are still unrepaired and this will result in PCR failure. We design primers to amplify fragments between 60–110 bp in length. Online tools such as NCBI Primer Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) can be useful to design specific primers. To avoid wasting precious ChIP DNA, primers should be first tested on input DNA for linear amplification and specificity. A good primer pair does not show any significant amplification in the no-template control, shows a linear increase in Ct values with decreasing DNA concentrations and shows a single peak in the melting curve profile. If the test run with input DNA was successful, ChIP DNA and input samples should be quantified simultaneously to determine the DNA amounts in each sample as follows.
Transfect nx107 cells of interest (n being the number of time points to be tested) with CRISPR-Cas9 editing reagents (e.g. for 6 time points transfect 6×107 cells).
Harvest cells at different time points post transfection. Choose the time points by taking Cas9/gRNA expression onset and duration into consideration.
Crosslink cells at each time point as described in Steps 12–20 in the main Procedure.
Snap freeze cell pellets at each timepoint. Store pellets at −80 °C until all time points are collected.
-
Isolate nuclei and perform ChIP for all samples following the instructions in Steps 21–59 of the main Procedure.
CRITICAL STEP: Cell pellets from all time points should be processed at the same time to ensure consistency and comparability between samples.
Dilute 1 μL of input DNA from Step 59 in 49 μL of nuclease-free water. Mix well.
Dilute 6 μL of ChIP DNA from Step 59 in 24 μL of nuclease-free water. Mix well. Briefly spin to collect the liquid.
-
Pipette 4.8 μL of diluted input DNA or ChIP DNA into a well of 384-well plate.
CRITICAL STEP: Prepare technical triplicates for each target site.
-
Prepare the following master mixes for each of the qPCR targets:
Reagent Stock Concentration Amount per sample Final concentration PowerUp SYBR green master mix 2x 5 μL 1x Forward Primer 100 μM 0.1 μL 1 μM Reverse Primer 100 μM 0.1 μL 1 μM CRITICAL STEP: Include a standard excess of 10–15 % when making reaction master mixes.
Pipette 5.2 μL of the master mix into the well of the 384-well plate containing the input or ChIP DNA from Step 8 of this box.
-
Seal the plate tightly with a microseal film.
CRITICAL STEP: Ensure that the plate is tightly sealed so liquid does not evaporate during PCR.
Briefly spin the plate to collect liquid at the bottom of the wells.
Carry out a real-time qPCR run with an initial denaturation step at 95 °C for 5 min, then 40 cycles of 95 °C for 3 s followed by 60 °C for 30 s for amplification and detection (or use ‘Fast’ mode with QuantStudio 5).
- To determine the fraction of input for each target calculate the following:
- To determine the enrichment of your target site relative to a negative control region calculate the following:
Determine the point with the highest enrichment (see Fig. 3a for an example) and perform your DISCOVER-Seq experiment at this time point.
Sonication
Chromatin is sheared by sonication to a fragment size of approximately 150–500 bp (Fig. 3b). The exact duration and intensity of sonication may have to be determined empirically for each cell type and instrument. We recommend a Covaris S2 sonicator but checking the size of the sheared chromatin by agarose gel electrophoresis should always be performed before proceeding to chromatin immunoprecipitation when using a new cell type or tissues (Box 3).
Box 3: QC checkpoint 3 for sonication efficiency.
Sonication is a critical step in all ChIP-Seq experiments. Sufficient chromatin shearing is required to obtain ChIP-Seq libraries with good enrichment and resolution. However, one has to be careful not to overheat the sample during the sonication process as this may result in loss of crosslinks, disassembly of protein complexes and protein degradation. We use a Covaris S2 sonicator with a built-in cooling system to address this issue. If using a manual probe sonicator the sample should be kept on ice at all times and one minute breaks should be taken between rounds of active sonication to let the sample cool down. We found that 12 cycles of sonication with the settings described below give us good results on the Covaris S2. However, many variables, such as cell type and number and different sonication systems, can significantly affect the sonication efficiency, and we highly recommend doing a test sonication with your crosslinked cell type of choice before starting the DISCOVER-Seq procedure.
Take out 15 μL of sonicated chromatin into a new microcentrifuge tube.
Add 13 μL of TE buffer.
Add 2 μL of RNase A (1 mg/mL) and incubate at 37 °C for 10 min.
Add 29 μL of elution buffer.
Add 1 μL of Proteinase K (20 mg/mL) and incubate at 65 °C for 1 h.
-
Add 4 μL of 80 % (vol/vol) glycerol to the sample, mix well and analyze 30 μL of it by agarose gel electrophoresis.
CRITICAL STEP: We use glycerol instead of 6x loading dye to avoid interference of the dye with the DNA signal on the gel.
You should expect to see a smear of DNA signal at a size of approximately 150–500 bp (Fig. 3b).
If unsheared fragments are still detected, sonicate the sample for another cycle and repeat Steps 1–6 of this box to check on chromatin size.
Chromatin immunoprecipitation
We describe a ChIP protocol in which all buffers are made in the lab. Alternatively, steps 26–59 can be performed using a commercially available ChIP kit (e.g. from Diagenode cat. no. C01010055). Before sequencing the ChIP samples, the success of the pulldown should be checked by quantitative PCR (qPCR). This can be done by designing primers specific to the on-target site and to a negative control region at an unedited locus within the genome. The enrichment of the on-target region over the negative control region will serve as an indicator for successful pulldown and will be directly correlated to the efficiency of genome editing at that site.
ChIP-Seq library preparation
If the qPCR was successful, samples will be prepared for NGS using a ChIP-Seq library preparation kit. Firstly, DNA is end-repaired, then the 3’ ends are adenylated to allow adapter ligation in the next step. After dual index adapter ligation, DNA is enriched by PCR.
Next-generation sequencing
DISCOVER-Seq samples should be sequenced as 100 bp paired-end reads on a next-generation sequencer such as an Illumina HiSeq4000 or similar. We aim for a sequencing depth of at least 30–40 M reads per ChIP sample. In theory, deeper sequencing should be able to identify even more rare MRE11 binding events. But this must be balanced with sequencing budget. It is important to sequence a negative control sample (see Controls required) at the same sequencing depth as the samples to be analyzed to account for the increase in background reads. This sample serves as a control in the data analysis and will be used to eliminate ChIP artifacts and background reads.
DISCOVER-Seq data analysis
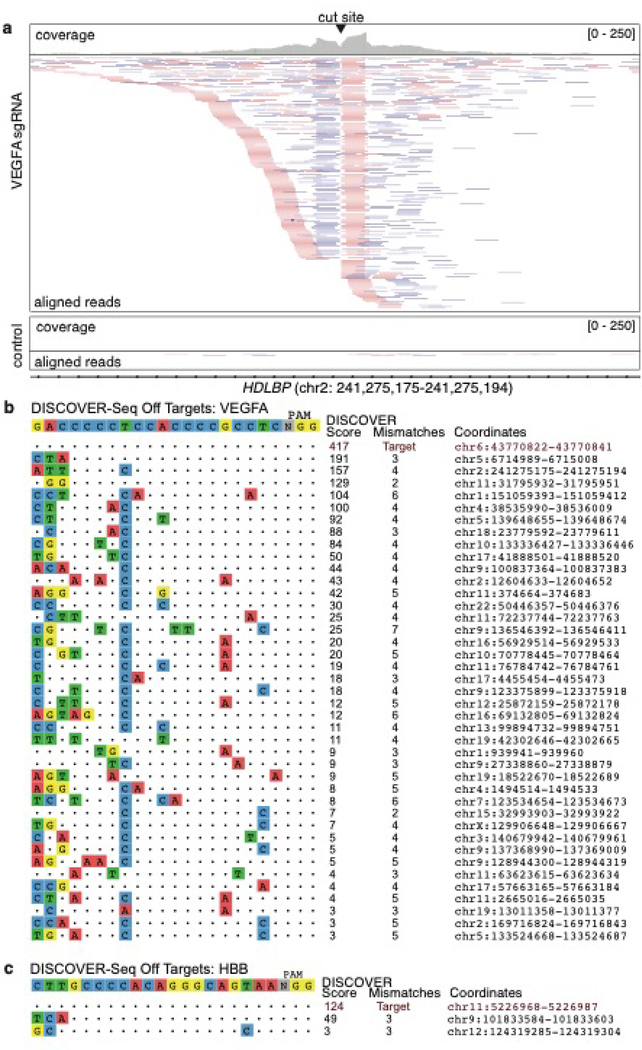
Fastq files from sequencing can be analyzed with our freely available, open source analysis pipeline called BLENDER (BLunt END findER) (https://github.com/staciawyman/blender) available under the GNU Affero General Public License. BLENDER takes a guide sequence and analysis-ready bam files for the ChIP and control samples and traverses the genome identifying start and ends of all reads. These locations are then examined for putative cut sites by identifying characteristic stacks of reads starting and ending at the same nucleotide and then looking for an adjacent PAM (default is NGG,NAG, but user can provide their own). For all loci with a valid PAM, BLENDER calculates a DISCOVER score by summing up the read ends in a 10 bp window around the cut site. Mismatches between putative guide sequence and input protospacer are calculated and then loci are filtered through the following default scoring scheme, which can be adjusted on the command line. If the DISCOVER score is two or more, the mismatches between input protospacer and putative guide can be no higher than 3; for a DISCOVER score of three or more, the mismatches can be as high as 5; and for a DISCOVER score of 4 or more, as many as 7 mismatches are allowed. For all loci passing the scoring scheme, the coordinates and cut site, guide sequence, PAM sequence and orientation plus the DISCOVER score are reported. BLENDER makes use of the ENCODE ChIP-Seq blacklists for human and mouse to filter out loci which have artifactually large numbers of reads. These lists, the control bam file and user-specific PAM sequences and guides are used to filter out false positives. BLENDER runs on mouse mm10 and human hg38 genomes (blacklist coordinates are for these genomes). BLENDER outputs a table of off-target loci as well as an SVG image of the aligned hits (Fig. 4).
Figure 4: Example output of a successful DISCOVER-Seq experiment.
(a) Snapshot of aligned bam files in IGV from K562 cells edited with VEGFA site 2 gRNA (top) and non-targeting gRNA (bottom, control). Shown is one of the off-target sites for the VEGFA site 2 gRNA. The top track for each sample shows the read coverage while the bottom track shows the aligned reads. Forward reads are shown in pink, reverse reads are shown in purple and grey lines connect read pairs. Many reads start and end in the cut site (centered). The control sample displays the level of noise that can be expected from MRE11 pulldown in unedited cells. (b) Example BLENDER output showing the DISCOVER-Seq hits in K562 cells edited with VEGFA site 2 gRNA and Cas9 RNP. The intended target sequence is shown on top and identified sites are ordered from top to bottom by DISCOVER score. Identity to the intended target is shown as dots, mismatches are indicated by colored nucleotides, and the hg38 genomic coordinates of each off-target are listed on the right. (c) Example BLENDER output showing the DISCOVER-Seq hits in K562 cells edited with HBB gRNA and Cas9 RNP.
Materials
Biological materials
K562 cells (ATCC Cat# CCL-243, RRID:CVCL_0004)
B16-F10 cells (ATCC Cat# CRL-6475, RRID:CVCL_0159)
CAUTION: Before use, cells should always be checked for authenticity and for contamination by mycoplasma.
Reagents
Mammalian cell culture
RPMI 1640 Medium GlutaMAX Supplement (Thermo Fisher Scientific, cat. no. 61870127)
DMEM Medium GlutaMAX Supplement (Thermo Fisher Scientific, cat. no. 10566016)
Trypsin-EDTA (0.25 %) (Thermo Fisher Scientific, cat. no. 25200056)
FBS (Thermo Fisher Scientific, cat. no. 10082147)
Penicillin–streptomycin (Thermo Fisher Scientific, cat. no. 15070063)
PBS (Thermo Fisher Scientific, cat. no. 14190250)
Trypan blue (Thermo Fisher Scientific, cat. no. 15250061)
Genome editing of cells
Synthetic gRNA (Synthego) or in vitro transcribed gRNA28
Recombinant Cas9 protein (QB3 Macrolabs, Berkeley)
HEPES (Sigma, cat. no. H4034)
Sodium Chloride (Sigma, cat. no. S7653)
Potassium Chloride (Sigma, cat. no. P9333)
Magnesium Chloride (Sigma, cat. no. M2393)
Ethylenediaminetetraacetic acid (EDTA) (Sigma, cat. no. E6758)
Nuclease-free water (Thermo Fisher Scientific, cat. no. 10977015)
Q5 2x PCR master mix (New England Biolabs, cat. no. M0492)
Agarose (Thermo Fisher Scientific, cat. no. 16500500)
Custom primers (IDT) for amplification of on- and off-target sites in PCR and qPCR
Crosslinking
Formaldehyde solution (37 % (wt/vol) (Sigma, cat. no. 252549)
CAUTION: Formaldehyde is toxic.
Glycine (Sigma, cat. no. G8898)
Cell lysis
Halt™ Protease Inhibitor Cocktail (100X) (Thermo Fisher Scientific, cat. no. 78429)
N-Lauroylsarcosine sodium salt solution 20 % (Sigma, cat. no. L7414)
Glycerol (Sigma, cat. no. G5516)
Igepal CA-630 (Sigma, cat. no. I8896)
Triton X-100 (Sigma, cat. no. T8787)
Tris base (Sigma, cat. no. 93362)
Sodium deoxycholate (Sigma, cat. no. 30970)
CAUTION: Na-Deoxycholate is an irritant which may cause respiratory irritation. Avoid breathing dust and wear a mask when weighing.
KOH solution for pH adjustment (Sigma, cat. no. P4494)
CAUTION: KOH is highly corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
NaOH solution for pH adjustment (Sigma, cat. no. 72068)
CAUTION: NaOH is highly corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
HCl solution for pH adjustment (Sigma, cat. no. 84429)
CAUTION: HCl is highly corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
Chromatin IP
Dynabeads Protein A (Thermo Fisher Scientific, cat. no. 10002D)
MRE11 antibody
Option 1: rabbit polyclonal, cross-reactive with mouse and human MRE11 (Novus Cat# NB 100–142, RRID:AB_350079)
Option 2: recombinant rabbit monoclonal, cross-reactive with mouse and human MRE11 (Abcam, Cat# ab208020, RRID:AB_2814655)
CRITICAL: We tested several antibodies and these two performed the best.
Bovine Serum Albumin (BSA) (Sigma, cat. no. A9418)
Lithium chloride (Sigma, cat. no. L4408)
Sodium dodecyl sulfate (SDS) (Sigma, cat. no. L6026)
CAUTION: SDS is a strong irritant. Wear a mask when weighing SDS and cleaning the area, as it disperses easily.
RNase A, DNase free (Thermo Fisher Scientific, cat. no. EN0531)
Proteinase K (Thermo Fisher Scientific, cat. no. AM2546)
Sodium acetate (NaOAc; 3 M (pH 5.5); Thermo Fisher Scientific, cat. no. AM9740)
MinElute PCR Purification Kit (Qiagen, cat. no. 28004)
PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, A25742)
Custom qPCR primers for amplification of on-target site and a control region (e.g. Integrated DNA Technologies)
ChIP-Seq library preparation
Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, cat. no. Q32854)
AMPure XP SPRI beads (Beckman Coulter, cat. no. A63881)
Reagent Setup
CRITICAL: Caution should be taken to avoid contamination of ChIP samples with PCR amplicons. Use amplicon-free reagents and materials and use filter tips for pipetting.
0.5 % BSA/PBS (wt/vol)
Prepare 50 mL of 0.5 % (wt/vol) BSA/PBS by dissolving 250 mg of BSA in 40 mL of PBS. Adjust the volume to 50 mL with PBS once the powder is dissolved. This solution can be stored at 4 °C for up to a week. For longer storage, the solution needs to be filter sterilized to avoid microbial growth.
2.5 M Glycine
Prepare 100 mL of 2.5 M glycine by dissolving 18.7g of glycine in 80 mL of nuclease-free water. Stir the solution at room temperature to dissolve. Heating might be required. Adjust the volume to 100 mL with nuclease-free water once dissolved. Autoclave or filter sterilize the solution to avoid microbial growth. Once autoclaved, this solution can be stored at room temperature (20–25 °C) for up to a year.
80 % (vol/vol) glycerol
Prepare 100 mL of 80 % (vol/vol) glycerol by diluting 80 mL of glycerol with 20 mL of nuclease free water. Store at room temperature (20–25 °C) for up to a year.
1 M HEPES-KOH
Prepare 500 mL of 1 M HEPES-KOH by dissolving 119.15 g of HEPES in 400 mL of nuclease-free water. Stir the solution to facilitate dissolving. Adjust pH to 7.5 using concentrated KOH and a pH meter. Adjust the volume to 500 mL with nuclease-free water once dissolved. This solution can be stored at room temperature (20–25 °C) for up to a year.
CAUTION: KOH is corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
1 M LiCl
Prepare 500 mL of 1 M LiCl by dissolving 21.2 g of LiCl in 450 mL of nuclease-free water. Stir the solution at room temperature to dissolve. Adjust the volume to 500 mL with nuclease-free water once dissolved. This solution can be stored at room temperature (20–25 °C) for up to a year.
10 % (wt/vol) Na-Deoxycholate
Prepare 50 mL of 10 % (wt/vol) Na-Deoxycholate by dissolving 5 g of Na-Deoxycholate in 40 mL of nuclease-free water. Rotate the solution for 1–2 h at room temperature to facilitate dissolving. Adjust the volume to 50 mL with nuclease-free water once the powder is dissolved. This solution can be stored at room temperature (20–25 °C) for up to a year. Protect from light.
CAUTION: Na-Deoxycholate is an irritant which may cause respiratory irritation. Avoid breathing dust and wear a mask when weighing.
0.5 M EDTA pH 8.0
Prepare 100 mL of 0.5 M EDTA by dissolving 18.61 g of EDTA in 80 mL of nuclease-free water. Stir the solution to facilitate dissolving. Adjust pH to 8.0 using concentrated NaOH and a pH meter. Adjust the volume to 100 mL with nuclease-free water once dissolved. This solution can be stored at room temperature (20–25 °C) for up to a year.
CAUTION: NaOH is highly corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
3 M KCl
Prepare 50 mL of 3 M KCl by dissolving 11.18 g of KCl in 50 mL of nuclease-free water. This solution can be stored at room temperature (20–25 °C) for up to a year.
1 M MgCl2
Prepare 50 mL of 1 M MgCl2 by dissolving 4.76 g of MgCl2 in 50 mL of nuclease-free water. This solution can be stored at room temperature (20–25 °C) for up to a year.
5 M NaCl
Prepare 500 mL of 5 M NaCl by dissolving 146.1 g of NaCl in 450 mL of nuclease-free water. Stir the solution at room temperature or heat to facilitate dissolving. Adjust the volume to 500 mL with nuclease-free water once dissolved. This solution can be stored at room temperature (20–25 °C) for up to a year.
1 M Tris-HCl pH 8.0
Prepare 500 mL of 1 M Tris-HCl by dissolving 60.55 g of Tris in 400 mL of nuclease-free water. Stir the solution to facilitate dissolving. Adjust the pH to 8.0 by adding concentrated HCl using a pH meter. Adjust the volume to 500 mL with nuclease-free water. This solution can be stored at room temperature (20–25 °C) for up to a year.
CAUTION: HCl is highly corrosive when concentrated. Wear appropriate PPE and work in a chemical fume hood.
20 % (vol/vol) Triton X-100
Prepare 50 mL of 20 % (vol/vol) Triton X-100 by diluting 10 mL of Triton X-100 with 40 mL of nuclease free water. Store at room temperature (20–25 °C) for up to a year.
10 % (vol/vol) Igepal CA-630
Prepare 50 mL of 10 % (vol/vol) Igepal CA-630 by diluting 5 mL of Igepal CA-630 with 45 mL of nuclease-free water. Store at room temperature (20–25 °C) for up to a year.
10 % (wt/vol) SDS
Prepare 50 mL of 10 % (wt/vol) SDS by dissolving 5 g of SDS in 40 mL of nuclease-free water. Heat the solution to 60 °C to facilitate dissolving. Adjust the volume to 50 mL with nuclease-free water. This solution can be stored at room temperature for up to 3 months.
CAUTION: SDS is a strong irritant. Wear a mask when weighing SDS and cleaning the area, as it disperses easily.
Cas9 buffer (3x)
For 10 mL of 3x Cas9 buffer prepare the following:
| Volume | Reagent | Stock conc. | 3x conc. | Final conc. |
|---|---|---|---|---|
| 4.12 mL | Nuclease free water | - | - | - |
| 600 μL | HEPES-KOH pH 7.5 | 1 M | 60 mM | 20 mM |
| 1.5 mL | KCl | 3 M | 450 mM | 150 mM |
| 30 μL | MgCl2 | 1 M | 3 mM | 1 mM |
| 3.75 mL | Glycerol | 80 % (vol/vol) | 30 % (vol/vol) | 10 % (vol/vol) |
Filter sterilize the solution through a 0.22 μm filter. Cas9 buffer can be stored at room temperature for several months.
CRITICAL: All reagents have to be RNase free.
Lysis buffer 1 (LB1)
For 200 mL of LB1 combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 146.5 mL | MilliQ water | - | - |
| 10 mL | HEPES-KOH pH 7.5 | 1 M | 50 mM |
| 5.6 mL | NaCl | 5 M | 140 mM |
| 400 μL | EDTA pH 8.0 | 0.5 M | 1 mM |
| 25 mL | Glycerol | 80 % (vol/vol) | 10 % (vol/vol) |
| 10 mL | Igepal CA-630 | 10 % (vol/vol) | 0.5 % (vol/vol) |
| 2.5 mL | Triton X-100 | 20 % (vol/vol) | 0.25 % (vol/vol) |
LB1 can be stored at 4 °C for up to 4 months.
Lysis buffer 2 (LB2)
For 200 mL of LB2 combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 189.6 mL | MilliQ water | - | - |
| 2 mL | Tris-HCl pH 8.0 | 1 M | 10 mM |
| 8 mL | NaCl | 5 M | 200 mM |
| 400 μL | EDTA pH 8.0 | 0.5 M | 1 mM |
LB2 can be stored at 4 °C for up to a year.
Lysis buffer 3 (LB3)
For 250 mL of LB3 combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 233.25 mL | MilliQ water | - | - |
| 2.5 mL | Tris-HCl pH 8.0 | 1 M | 10 mM |
| 5 mL | NaCl | 5 M | 100 mM |
| 500 μL | EDTA pH 8.0 | 0.5 M | 1 mM |
| 2.5 mL | Na-deoxycholate | 10 % (wt/vol) | 0.1 % (vol/vol) |
| 6.25 mL | N-lauroylsarcosine | 20 % | 0.5 % (vol/vol) |
LB3 can be stored at 4 °C for up to 4 months.
RIPA buffer
For 500 mL of RIPA buffer combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 184 mL | MilliQ water | - | - |
| 25 mL | HEPES-KOH pH 7.5 | 1 M | 50 mM |
| 250 mL | LiCl | 1 M | 500 mM |
| 1 mL | EDTA pH 8.0 | 0.5 M | 1 mM |
| 5 mL | Igepal CA-630 | undiluted | 1 % (vol/vol) |
| 35 mL | Na-deoxycholate | 10 % (wt/vol) | 0.7 % (vol/vol) |
RIPA buffer can be stored at 4 °C for up to 4 months.
Tris-EDTA (TE)
For 100 mL TE combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 95 mL | MilliQ water | - | - |
| 1 mL | Tris-HCl pH 8.0 | 1 M | 10 mM |
| 200 μL | EDTA pH 8.0 | 0.5 M | 1 mM |
TE can be stored at room temperature for up to a year.
TBS
For 100 mL of TBS combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 95 mL | MilliQ water | - | - |
| 2 mL | Tris-HCl pH 8.0 | 1 M | 20 mM |
| 3 mL | NaCl | 5 M | 150 mM |
TBS can be stored at 4 °C for up to a year.
Elution buffer
For 50 mL of Elution buffer combine the following:
| Volume | Reagent | Stock conc. | Final conc. |
|---|---|---|---|
| 41.5 mL | MilliQ water | - | - |
| 2.5 mL | Tris-HCl pH 8.0 | 1 M | 50 mM |
| 1 mL | EDTA pH 8.0 | 0.5 M | 10 mM |
| 5 mL | SDS | 10 % (wt/vol) | 1 % (vol/vol) |
Elution buffer can be stored at room temperature for up to a year.
Equipment
General laboratory consumables
Filtered sterile pipette tips (Rainin, cat. no. 17005860)
Microcentrifuge tubes (1.5 mL; Axygen, cat. no. 31104051)
Conical tubes (15 mL; Fisher, cat. no. 339651)
Conical tubes (50 mL; Fisher, cat. no. 339653)
Serological pipettes (5 mL; Fisher, cat. no. 170355)
Serological pipettes (10 mL; Fisher, cat. no. 12567603)
Serological pipettes (25 mL; Fisher, cat. no. 12567604)
384-well quantitative PCR plates (Thermo Fisher Scientific, cat. no. 4326270)
Microseals adhesives (Bio-Rad, cat. no. MSB1001)
Screw Cap Micro Tubes (Fisher-Scientific, cat. no. 21403196)
Covaris milliTUBE 1 mL AFA Fiber (Covaris, cat. no. 520130)
Nucleofection cuvettes (Lonza, cat. no. V4XC-2024)
96-well 0.3 mL PCR plate (VWR, cat. no. 10011–228)
LoBind DNA microcentrifuge tubes (1.5 mL; Sigma, cat. no. Z666505)
DNeasy Blood & Tissue Kit (Qiagen, cat. no. 69504)
Qubit assay tubes (Thermo Fisher Scientific, cat. no. Q32856)
TruSeq ChIP Library Preparation Kit (Illumina, cat. no. IP-202–1012)
General laboratory equipment
Cell culture CO2 incubator (Thermo Fisher Scientific, model no. Heracell VIOS 160i)
Benchtop microcentrifuge (Eppendorf, cat. no. 05400002)
Eppendorf ThermoMixer (Eppendorf, cat. no. 5384000020)
Pipettes (Rainin, cat. no. 17014391)
Inverted microscope
Pipet filler (Thermo Fisher Scientific, cat no. 9531)
Hybridization oven
pH meter
qPCR machine (Thermo Fisher Scientific, cat. no. A28140)
Sonicator (Covaris, model S2)
Qubit 4 Fluorometer (Thermo Fisher Scientific)
Magnetic stand (Thermo Fisher Scientific, cat. no. 12321D)
Lonza 4D nucleofector, X unit
Agilent 5300 Fragment Analyzer System
Software
SAMtools: http://www.htslib.org/download
Burrows–Wheeler Aligner (BWA): https://github.com/lh3/bwa
IGV for visualization: https://software.broadinstitute.org/software/igv
Python v3.7, svgwrite package https://www.python.org/downloads/
Procedure
Mammalian cell culture
Timing: 3–5 days
-
1Any human or mouse cells can be used for DISCOVER-Seq. Here, we describe culture of K562 cells (option A), B16-F10 cells (option B) or any cells of interest (option C).
- K562 maintenance culture
- Culture K562 cells in 30 mL RPMI1640-Glutamax supplemented with 10 % (vol/vol) FBS and 1x penicillin–streptomycin at 37 °C and 5 % CO2 in 150-mm tissue culture plates.
- To passage, transfer 3 mL of 90 % confluent cell suspension into a new 150-mm tissue culture plate containing 27 mL of fresh pre-warmed media.
- Harvest cells by transferring the cell suspension into a 50 mL conical tube.
- B16-F10 maintenance culture
- Culture the cells in DMEM-Glutamax medium supplemented with 10 % (vol/vol) FBS and 1x penicillin–streptomycin at 37 °C and 5 % CO2 in 150-mm tissue culture plates.
- To passage, remove the medium and rinse once with 10 mL of PBS. Add 3 mL of pre-warmed 0.25 % (vol/vol) trypsin-EDTA and incubate at 37 °C for 5 min. Add 12 mL of pre-warmed medium to inactivate the trypsin and gently pipette up and down to make single-cell suspension. Add 3 mL of the cell suspension into 27 mL of fresh pre-warmed DMEM medium in a new 150-mm tissue culture plate.
- Harvest cells by removing medium and rinse the cells with 10 mL PBS. Add 3 mL of pre-warmed 0.25 % (vol/vol) trypsin-EDTA and incubate at 37 °C for 5 min. Add 12 mL of pre-warmed medium to inactivate the trypsin and gently pipette up and down to make single-cell suspension. Transfer cell suspension into a 50 mL conical tube.
- Culture cells of interest culture
- Maintain and passage your cells of interest according to pre-established protocols and procedures.
CRITICAL STEP: Avoid letting cells become fully confluent by passaging every 2–3 days at a split ratio of 1:8–1:10.
Genome editing of cells
Timing: 1h
-
2Prepare Cas9-gRNA ribonucleoprotein (RNP) complexes by combining the following (per 107 cells):
Volume Reagent Stock conc Total amount 5 μL Cas9 buffer 3x 1x 7.5 μL Cas9 protein 40 μM 300 pmol 3 μL gRNA 100 μM 300 pmol CRITICAL STEP: Include a negative control in your experiments. This control can be cells that were nucleofected with Cas9 and a non-targeting gRNA, Cas9 only or untransfected cells.
-
3
Incubate at room temperature until ready to nucleofect (10–30 mins).
-
4
Take a 10 μL aliquot of the cell suspension from step 1 and mix 1:1 with Trypan Blue. Count cells using a hemocytometer.
-
5
Transfer an aliquot of 107 cells per ChIP into a conical falcon tube.
-
6
Centrifuge cells at room temperature at 300 g for 5 min, discard the supernatant.
-
7
Wash cells with 1 mL of PBS and transfer to eppendorf tube. Spin cells at room temperature at 300 g for 5 min. Aspirate the supernatant and discard.
CRITICAL STEP: FBS in the growth media may contain nucleases. Always wash cells with PBS before adding editing reagents.
-
8
Add 65 μL of Lonza SF nucleofection buffer including supplements to the RNP complex from Step 3 and slowly pipette up and down avoiding bubble formation. Transfer the whole volume into Eppendorf tube with cell pellet from Step 7 and resuspend cells in SF buffer/RNPs by gently pipetting up and down.
CRITICAL STEP: The type of nucleofection buffer used depends on the cell type. Check Lonza guidelines for the correct buffer for your cell type.
CRITICAL STEP: The amount of nucleofection buffer added depends on the size of the cell pellet. We found that 107 pelleted K562 and B16-F10 cells have a volume of approximately 35 μL. Adding 15ul RNP solution and 65 μL nucleofection buffer makes a total volume of 115 μL which is suitable for a Lonza 100 μL nucleofection cuvette. At least 50 % of the total volume should be nucleofection buffer. If the cell pellet is too big, it can be split up into two nucleofections.
-
9
Transfer cell suspension (115 μL) into Lonza 4D XL nucleofection cuvette and electroporate with a Lonza 4D X nucleofector using settings FF-120 for K562 and DJ-110 for B16-F10 cells.
CRITICAL STEP: Check the Lonza guidelines for ideal nucleofection settings for your cell type of interest.
-
10
After nucleofection, let the cells recover in the nucleofection cuvette at room temperature for 5–10 mins.
-
11
Transfer nucleofected cells into a 150-mm cell culture plate with 30 mL of prewarmed complete media and incubate at 37 °C and 5 % CO2 until ready to harvest.
Crosslinking
Timing: 45 min
-
12
-
Harvest K562 cells
-
iAfter 8–12 h of culture, transfer 500 μL of edited K562 cells into a 100-mm tissue culture plate with 9.5 mL of prewarmed complete RPMI medium. Prepare genomic DNA from these cells after 72–96 h to check for genome editing efficiency (see Box 1 for details).
?Troubleshooting-
iiTransfer the remaining K562 cells into a 50 mL conical tube and immediately proceed to Step 13.
-
i
-
Harvest B16-F10 cells
- After 8–12 h of culture, aspirate medium from edited B16-F10 cells, wash cells with 10 mL of PBS, add 3 mL of pre-warmed trypsin-EDTA and incubate at 37 °C for 5 min. Add 12 mL of pre-warmed medium to inactivate the trypsin and gently pipette up and down to make single-cell suspension. Transfer 250 μL of B16-F10 cells into a 100-mm tissue culture plate with 9.75 mL of prewarmed complete DMEM medium. Prepare genomic DNA from these cells after 72–96h to check for genome editing efficiency (see Box 1 for details).
?Troubleshooting- Transfer the remaining B16-F10 cells into a 50 mL conical tube and immediately proceed to Step 13.
- Harvest cells of interest
- After 8–12h in culture, harvest the cells of interest according to pre-established protocols and procedures. Keep approximately 2×105 cells of edited cells in culture for genomic DNA extraction after 72–96h.
-
Transfer the remaining cells into a 50 mL conical tube and immediately proceed to Step 13.
-
-
13
Centrifuge cells at room temperature at 300 g for 5 min, aspirate the supernatant and discard.
-
14
Resuspend cell pellets in 20 mL of room temperature medium without any supplements (RPMI or DMEM, respectively).
CRITICAL STEP: It is important to obtain a single-cell suspension to ensure even crosslinking in all cells. To achieve this, adherent B16-F10 cells can be passed through a syringe or cell strainer.
-
15
Add 37 % formaldehyde to a final concentration of 1 % (i.e. 540 μL). Invert tube multiple times to ensure equal distribution of formaldehyde in the solution.
CAUTION: Formaldehyde is toxic. Work in a chemical fume hood.
-
16
Incubate tube slowly rotating at room temperature for 15 min.
-
17
Add 1 mL of ice cold 2.5 M glycine (final concentration 125 mM) to quench formaldehyde. Incubate for 3 min on ice.
-
18
Centrifuge cells at 4 °C at 500 g for 5 min, discard the supernatant.
CAUTION: Formaldehyde is toxic. Discard the 1 % formaldehyde waste appropriately.
-
19
Wash cells twice with 10 mL ice cold PBS (500 g, 3 min, 4 C). Transfer cells to a 15 mL conical tube before last spin. Aspirate supernatant and discard.
-
20
Proceed with lysis (Step 26), or snap freeze cells in liquid nitrogen or dry ice-ethanol bath.
PAUSE POINT: Crosslinked cell pellets can be stored at −80 °C for 2–3 months.
Prebinding of antibody
Timing: 30 min
-
21
Add 100 μL Protein A magnetic beads per ChIP (Invitrogen, Dynabeads) to a 1.5 mL microcentrifuge tube. Add 1 mL block solution (0.5 % BSA (w/v) in PBS). When performing multiple ChIPs scale up accordingly. e.g.: for 3 ChIPs: use 300 μL of beads complexes.
-
22
Collect the beads using a magnetic stand. Remove supernatant with pipette and discard. Add 1 mL of block solution to the tube. Keep the tubes on the magnetic stand, but quickly turn them 180º to allow the beads to travel through the solution and facilitate washing.
CRITICAL STEP: Try to avoid aspirating and dispensing magnetic beads with a pipette as much as you can. Magnetic beads tend to stick to the plastic and you will lose precious bead-bound antibody and sample.
-
23
Remove the block solution and repeat blocking in Step 22.
-
24
Remove block solution and resuspend beads in 250 μL block solution and add 10 μL of NB100–142 or 10 μg of ab208020 MRE11 antibody per ChIP.
-
25
Incubate for 1–3 hours on a rotating platform at 4 C.
Cell lysis
Timing: 30 min
-
26
Prepare working amounts of LB1, LB2 and LB3 with 1x protease inhibitors. Keep buffers on ice.
-
27
Resuspend cell pellets from Step 20 in 10 mL of LB1 (+protease inhibitors). Incubate on ice for 10 min.
-
28
Centrifuge at 1000 g for 3 min at 4 °C. Discard supernatant.
-
29
Resuspend pellet in 10 mL of LB2 (+protease inhibitors). Incubate on ice for 5 min.
-
30
Centrifuge at 1000 g for 3 min at 4 °C. Discard supernatant.
-
31
Resuspend nuclei pellet in 1 mL of LB3 (+protease inhibitors).
Sonication
Timing: 30–45 min
-
1
Transfer 1 mL of nuclei suspension into a pre-chilled Covaris milliTube. Keep on ice.
-
2Sonicate lysate using a Covaris S2 with the following settings:
Duty Cycle 5 % Intensity 5 Cycles per burst 200 Time (sec) 60 Number of cycles 12 ?Troubleshooting
CRITICAL STEP: The ideal sonication settings may differ between cell types and sonicators and should be determined empirically. The correct chromatin size is important as it determines the resolution of the ChIP-Seq experiment. See Box 3 for details on sonication optimization and quality control.
-
3
Transfer sonicated solution into a microcentrifuge tube. Spin at 20,000g for 10 min at 4 °C to pellet debris.
-
4
Transfer supernatant to a 15 mL conical tube.
-
5
Add 1.85 mL of LB3 and 150 μL of 20 % (vol/vol) Triton-X (1 % (vol/vol) final concentration) to make it a total volume of 3 mL.
-
6
Transfer 50 μL of cell lysate as input DNA into a screw cap microcentrifuge tube. Store at −20 °C until the next day. Keep the remaining cell lysate on ice until used in Step 42.
Chromatin Immunoprecipitation
Timing: 16–24 h
-
7
Retrieve pre-bound antibody-beads from Step 25.
-
8
Collect beads using a magnetic stand.
-
9
Wash magnetic beads twice with 1 mL of block solution by turning the tubes quickly on the magnetic stand.
-
10
Remove supernatant and resuspend beads in 100 μL block solution.
-
11
Transfer 100 μL antibody/magnetic bead mix to the remaining 2.95 mL cell lysate (from Step 37).
-
12
Gently rotate at 4 °C for immunoprecipitation for at least 6h.
PAUSE POINT: Samples can be incubated rotating at 4 °C overnight.
-
13
Collect beads using a magnetic stand. Remove flowthrough by pipetting.
-
14
Add 1 mL of ice cold RIPA buffer to each tube. Turn tubes to wash beads.
-
15
Collect beads using a magnetic stand. Remove supernatant by pipetting.
-
16
Repeat this wash 5 more times.
-
17
Wash once with 1 mL TBS. Remove supernatant completely by pipetting.
-
18
Add 200 μL of elution buffer. Transfer bead slurry to a 1.5 mL screw cap microcentrifuge tube.
-
19
Thaw 50 μL of the input lysate from Step 37 and add 150 μL of elution buffer.
-
20
Reverse crosslinks and elute complexes from beads in a hybridization oven rotating at 65 °C for at least 6 h or overnight.
PAUSE POINT: Samples can be incubated rotating at 65 °C overnight.
Purify DNA
Timing: 2.5 h
-
21
Briefly spin tubes from Step 51 to collect liquid from lids. Collect beads with magnetic stand.
-
22
Remove the supernatant by pipetting and transfer it to a new microcentrifuge tube.
-
23
Add 100 μL of TE buffer to each tube to dilute the SDS in the elution buffer.
-
24
Add 8 μL of 1 mg/mLl RNaseA. Mix and incubate at 37 °C for 30 min.
-
25
Add 4 μL of 20 mg/mL proteinase K. Mix and incubate at 55 °C for 1 h.
-
26
Purify DNA using a Qiagen MinElute PCR Purification Kit, following the manufacturer’s instructions.
CRITICAL STEP: Check the pH of your reaction by using the pH indicator in buffer PB provided with the Qiagen MinElute PCR Purification Kit. If necessary, add 5 μL of 3 M sodium acetate, pH 5.2 directly to the sample. The color indicator should change to yellow.
-
27
Elute DNA in 41 μL of purified nuclease-free water (will result in ~40 μL eluate).
PAUSE POINT: ChIP samples can be frozen at −20 °C for up to 6 months.
-
28
Before proceeding to ChIP-Seq library preparation (Step 60), check that MRE11 pulldown was successful by quantifying ChIP DNA by qPCR (Fig. 3c). To do so, follow the instructions in Box 2 starting at step VI.
?Troubleshooting
Library preparation and NGS
Timing: 3–4 days
-
29
Transfer all of remaining sample (i.e. 34 μL) from Step 59 into a 96-well 0.3 mL PCR plate
CRITICAL STEP: Often ChIP DNA concentration is too low to be measured accurately by Nanodrop or Qubit. We recommend using the whole sample volume that is available after qPCR for library preparation.
-
30
Adjust the volume to a total of 50 μL using nuclease-free purified water.
-
31
Prepare the libraries using an Illumina TruSeq ChIP library preparation kit by following the manufacturer’s instructions.
-
32
Once individual libraries are amplified and purified, determine the concentration of libraries using a Qubit fluorometer and Qubit dsDNA HS Assay Kit.
-
33
Pool libraries into one LoBind DNA microcentrifuge tube using equal amounts of individual libraries.
-
34
Clean the adapter dimers from the pooled library by adding AMPure XP beads using a ratio of 0.8x beads to sample (e.g. to 50 μL sample add 40 μL AMPure XP beads).
-
35
Gently pipette the entire volume up and down 10 times to mix thoroughly.
-
36
Incubate the tube at room temperature for 15 min.
-
37
Place the tube on a magnetic stand at room temperature for 5 min or until the liquid is clear.
-
38
Remove and discard supernatant.
CRITICAL STEP: Leave the tube on the magnetic stand while performing the following 80 % EtOH wash steps.
-
39
With the tube on the magnetic stand, add 200 μL freshly prepared 80 % EtOH without disturbing the beads.
-
40
Incubate the tube at room temperature for 30 seconds, then remove and discard all of the supernatant.
-
41
Repeat the two previous steps for a total of two 80 % EtOH washes.
-
42
Let the sample air-dry at room temperature for 10 minutes, and then remove the tube from the magnetic stand.
-
43
Resuspend the dried pellet in 15–20 μL of nuclease-free purified water.
-
44
Determine the concentration of libraries using a Qubit fluorometer and Qubit dsDNA HS Assay Kit.
-
45
Run an aliquot of the pooled library on a fragment analyzer to check for successful library enrichment and correct library size (Fig. 3d for an example). If adapter dimers are present, perform additional size selection of the pooled library.
?Troubleshooting
CRITICAL STEP: We recommend to eliminate adapter dimers before sequencing to increase library diversity and sequencing quality.
-
46
Sequence your samples as 100 bp paired-end reads on a next-generation sequencer such as an Illumina HiSeq4000, NovaSeq, or similar following the manufacturer’s instructions.
CRITICAL STEP: Aim for sequencing depth of around 30–40 M reads per ChIP sample. We found that increasing sequencing depth can increase the sensitivity of the assay.
-
47
After sequencing, copy the demultiplexed fastq files to a location accessible to the DISCOVER-Seq analysis pipeline.
DISCOVER-Seq data analysis
Timing: 4–48 h, depending on sequencing depth, optional arguments, and computational speed
-
48Install BLENDER from GitHub on a Linux computer:
$ git clone https://github.com/staciawyman/blender.git $ cd blender
-
49Make sure samtools and BWA are installed on your computer and available in your path. Test this with the “which” command that tells you if a program is in your path by listing where it is or saying not found (for bash shell):
$ which samtools /usr/local/bin/samtools
-
50Make sure the reference genome for your organism has been downloaded. Here, we use hg38 as an example. The reference genome can be downloaded using wget, then create a BWA index from the reference genome fasta file:
$ wget \ http://hgdownload.cse.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz $ gunzip hg38.fa.gz $ bwa index hg38.fa
CRITICAL STEP: Creating a BWA index can take several hours.
-
51Create an output directory with subdirectories for different file types as follows. It works best to run blender from inside the repository and by creating an output directory inside that with directories for the fastq files, bam files and blender output.
$ mkdir -p output/bwa $ mkdir output/blender
-
52BWA runs in three steps. First, use bwa aln on the read one and read two files individually:
$ bwa aln -q 20 -o 1 -t <#threads> \ /path/to/reference/hg38.fa \ /path/to/fastq/edited.R1.fastq.gz > \ output/bwa/edited.R1.sai $ bwa aln -q 20 -o 1 -t <#threads> \ /path/to/reference/hg38.fa \ /path/to/fastq/edited.R2.fastq.gz > \ output/bwa/edited.R2.sai
CRITICAL STEP: The more threads you are able to allocate to bwa, the faster it will run (we typically use a couple fewer threads than the number available on the system).
CRITICAL STEP: Be sure there are no spaces after the “\” when you run the commands below. Commands may be joined to a single line without the “\”‘s.
-
53Give BWA the sai files and fastq files and create a sam file with bwa sampe. :
$ bwa sampe \ /path/to/reference/hg38.fa \ output/bwa/edited.R1.sai \ output/bwa/edited.R2.sai \ /path/to/fastq/edited.R1.fastq.gz \ /path/to/fastq/edited.R2.fastq.gz > \ output/bwa/edited.sam
-
54Use samtools to convert sam to bam format, sort, and remove duplicates to create an analysis-ready bam file:
$ samtools view -S -b output/bwa/edited.sam | \ samtools sort > output/bwa/edited.sorted.bam $ samtools rmdup output/bwa/edited.sorted.bam \ output/bwa/edited.bam $ samtools index output/bwa/edited.bam
CRITICAL STEP: Removing duplicates eliminates false positives resulting from PCR bias.
-
55
Repeat preceding three steps for the control sample fastq files (substituting “control” for “edited” in the commands above).
-
56You should now have analysis-ready bam files for analysis with BLENDER. To verify the bam files, use the samtools flagstat command to examine summary statistics as follows. It will give you the number of reads aligned in your bam file, or an error if there was an error in the preceding steps resulting in a truncated bam file.
$ samtools flagstat output/bwa/edited.bam $ samtools flagstat output/bwa/control.bam
Example output without errors:$ samtools flagstat BW43 _MRE11_K562_VEGFA_5XL_16h.bam 59097633 + 0 in total (QC-passed reads + QC-failed reads) 0 + 0 secondary 0 + 0 supplementary 0 + 0 duplicates 39316383 + 0 mapped (66.53% : N/A) 59097633 + 0 paired in sequencing 29548839 + 0 read1 29548794 + 0 read2 38098574 + 0 properly paired (64.47% : N/A) 38443007 + 0 with itself and mate mapped 873376 + 0 singletons (1.48% : N/A) 192692 + 0 with mate mapped to a different chr 107671 + 0 with mate mapped to a different chr (mapQ>=5)
-
57Once it is clear the bam files are not truncated, remove the intermediate files:
$ rm output/bwa/edited.sam output/bwa/control.sam $ rm output/bwa/edited.R1.sai output/bwa/edited.R2.sai $ rm output/bwa/control.R1.sai output/bwa/control.R2.sai $ rm output/bwa/edited.sorted.bam output/bwa/edited.control.bam
-
58Run blender by calling the shell script run_blender.sh as follows and giving the editing and control sample bam files, guide sequence, and reference genome for your organism as arguments. This runs blender with the default options (at least 3 stacked reads at the cut site).
$ sh run_blender.sh /path/to/reference/hg38.fa \ output/bwa/edited.bam output/bwa/control.bam \ <guide without PAM> <output directory>
OPTIONAL: It is recommended that you initially run BLENDER with the default parameters, and then again with a more sensitive cutoff to determine if a large number of additional sites are discovered (“-c 2” given at the end of the command to call blender):$ sh run_blender.sh /path/to/reference/hg38.fa \ output/bwa/edited.bam output/bwa/control.bam \ <guide without PAM> <output directory> “-c 2”
Blender outputs three files into the “output/blender” directory.CRITICAL STEP: Using a cutoff of two stacked reads at the cut site will possibly allow for some false positives but also possibly capture some rarer events. For details of different filters see the GitHub website for BLENDER.
-
59
Examine hit locations by looking at the raw reads in both the control and edited bam files using IGV (Fig. 4a).
Troubleshooting
Troubleshooting advice can be found in Table 2
Table 2.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 12 & Box 1 | Editing efficiency is low | (i) low efficiency gRNA (ii) transfection efficiency is low |
(i) rethink the design and choice of your gRNA (ii) optimize the transfection reagents and procedure for your cell type |
| 33 | Presence of unsheared DNA in sonicated samples | (i) Suboptimal sonication settings (ii) nuclei clumps present during sonication |
(i) Optimize the settings by adding more cycles of sonication (ii) Make sure to obtain a single nuclei suspension before sonication (especially when working with crosslinked tissues) |
| 59 & Box 2 | Enrichment at on-target site is low | (i) Editing efficiency is low (ii) Low starting cell numbers |
(i) see troubleshooting above (ii) Repeat experiment using recommended numbers of cells |
| 76 | Poor enrichment of the ChIP-Seq library and adapter dimer peak in fragment analyzer trace | (i) Input concentration of ChIP DNA into library prep too low (ii) presence of adapter dimers before library enrichment that compete with adapter-ligated DNA during the PCR |
(i) Repeat experiment using recommended numbers of cells. If necessary pool ChIP DNA from multiple experiments. (ii) Perform DNA size selection of the adapter-ligated DNA fragments using SPRI beads, PippinPrep or separation by gel electrophoresis |
Timing
Step 1, cell culture: 3–5 d
Steps 2–11, genome editing: 1 h
Steps 12–20, crosslinking: 45 min
Steps 21–37, lysis and sonication: 2 h
Steps 38–51, chromatin immunoprecipitation: 16–24 h
Steps 52–59, DNA purification: 2.5 h
Steps 60–78, Library preparation and NGS, 3–4 d
Steps 79–90, Data analysis with Blender, 2 d
Anticipated results
Before performing the full DISCOVER-Seq protocol, editing efficiency at the intended target site should be tested in the cell type of interest (Box 1). While different gRNAs, transfection methods and cell types can result in a range of editing outcomes, we usually expect between 60 and 90% indel rates at the on-target site when working with an efficient gRNA. It is essential to confirm successful on-target editing in the same sample that will be used for DISCOVER-Seq. This can be done by PCR and Sanger sequencing or PCR and amplicon NGS (Box 1).
If editing was successful and the sample is processed for DISCOVER-Seq, sheared chromatin after sonication should have a median size of 200–500 bp (Fig. 3b). If sheared chromatin size is suboptimal, sonicator settings, length and cycles may have to be optimized. After chromatin immunoprecipitation with MRE11, the success of the pulldown at the on-target site should be tested by qPCR and compared to a negative control region elsewhere in the genome. The fold enrichment at the on-target site is directly correlated with the efficiency of genome editing (% indels). For gRNAs that produce high indel rates (60–90%), we usually expect 80- to 200-fold enrichment of the on-target site over the negative control region by qPCR (Fig. 3c).
Before proceeding to sequencing, the ChIP-Seq libraries should be analyzed with a fragment analyzer to determine the mean library size and the presence of adapter dimers. We expect the mean size of the library to be 300–500 bp (Fig. 3d). Adaptor dimers can be removed by size selecting the sample with SPRI beads, PippinPrep or gel electrophoresis.
After a successful DISCOVER-Seq experiment, the sequencing reads can be visualized in IGV and one should expect to see large numbers of reads at the on-target site (Fig. 4a) and very sparse read coverage in the negative control sample. For most targets, the number of on-target DISCOVER-Seq reads should be >200, using ~30 M paired-end sequencing. BLENDER also outputs a visualization of the off-target sites with respect to gRNA sequence. For a gRNA with many off-target sites (e.g. VEGFA site 2, Fig. 4b) one can expect many (>30) off-target sites, while for a more specific gRNA (e.g. HBB, Fig. 4c) should yield many fewer (1–10) hits. Off-target hits can then be further validated individually by amplicon NGS. We have found that DISCOVER score from BLENDER correlates very well with eventual indel frequencies at both on- and off-target sites7.
Supplementary Material
Acknowledgements
J.E.C. and C.D.Y. are supported by the NOMIS Foundation, the Lotte and Adolf Hotz-Sprenger Stiftung, the Swiss National Science Foundation (310030_188858), the Fanconi Anemia Research Foundation, and the Bill and Melinda Gates Foundation. B.W. is supported by an NHMRC Early Career Fellowship. B.R.C and B.W. received support from the Gladstone Institutes, and NIH grants R01-EY028249, R01-HL130533 and R01-HL13535801. S.K.W. is supported by the Li Ka Shing Foundation. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, which is supported by NIH Instrumentation grant S10 OD018174.
Footnotes
Competing interests
J.E.C. is a cofounder of Spotlight Therapeutics. J.E.C. has received sponsored research support from AstraZeneca and Pfizer. B.R.C is a founder of Tenaya Therapeutics.
Data availability statement
Data that support the findings of this study have been deposited in SRA BioProject accession PRJNA509652.
Code Availability Statement
Our open source analysis pipeline called BLENDER (BLunt END findER) (https://github.com/staciawyman/blender) is freely available under the GNU Affero General Public License. Code in this manuscript has been peer reviewed.
References
- 1.Cheng Y & Tsai SQ Illuminating the genome-wide activity of genome editors for safe and effective therapeutics. Genome Biol. 19, 226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D Collateral damage: benchmarking off-target effects in genome editing. Genome Biol. 20, 114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vakulskas CA et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JS et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, Judge LM, Conklin BR, Mareca M, & Corn JE Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stracker TH & Petrini JHJ The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RS, Williams JS & Tainer JA Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85, 509–520 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi J Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res 45, 473–478 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Richardson CD et al. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Haeussler M et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzarotto CR et al. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc 13, 2615–2642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai SQ et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron P et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Kim D et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–43, 1 p following 243 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannoukos G et al. UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics 19, 212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frock RL et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosetto N et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan WX et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akcakaya P et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rass E et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16, 819–824 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Xie A, Kwok A & Scully R Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16, 814–818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinkelmann M et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 16, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wienert B, Shin J, Zelin E, Pestal K & Corn JE In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol. 16, e2005840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.