Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly become a global health pandemic. The lack of effective treatments, coupled with its etiology, has resulted in more than 400,000 deaths at the time of writing. The SARS-CoV-2 genome is highly homologous to that of SARS-CoV, the causative agent behind the 2003 SARS outbreak. Based on prior reports, clinicians have pursued the off-label use of several antiviral drugs, while the scientific community has responded by seeking agents against traditional targets, especially viral proteases. However, several avenues remain unexplored, including disrupting E and M protein oligomerization, outcompeting host glycan–virus interactions, interfering with the heparan sulfate proteoglycans–virus interaction, and others. In this review, we highlight some of these opportunities while summarizing the drugs currently in use against coronavirus 2019 (COVID-19).
Teaser
Despite the approval of remdisivir, much remains to be accomplished to address the urgent need for a drug to treat COVID-19. This short review explores several novel avenues for discovering small-molecule antagonists against SARS-CoV-2
Introduction
SARS-CoV-2 is an enveloped positive-sense RNA virus that belongs to the Coronavirinae [1]. SARS-CoV-2 has rapidly become a global pandemic, distinguishing it as the most infectious agent in a century [2]. Coronaviruses, which contribute to nearly one third of common cold infections in humans, have zoonotic origins in a range of mammals and birds [3]. In humans, respiratory infections, such as pneumonia and bronchiolitis, tend to turn fatal, especially in older, pediatric, immunocompromised, or co-morbid patients [4]. The current SARS-CoV-2 pandemic appears to be targeting not only similar populations, but also the young and the healthy. It might also have a gender bias, as was previously shown for SARS-CoV [5].
Four groups, the alpha, beta, gamma, and delta coronaviruses, are categorized based on phylogenetic clustering [1]. The current pandemic causing COVID-19 is of the beta type. The unique feature of all coronaviruses is the ‘club-like’ projection from the surface of the virion, a feature that mimics a crown, or corona, in Latin. The nonsegmented, positive-sense, single-stranded RNA genome of coronavirus is large (∼32 kb) and encodes four structural and 16 nonstructural proteins (NSPs). The NSPs occupy two-thirds (20 kb) of the viral genome, with the remaining third comprising structural and accessory proteins [6]. The NSPs are widely implicated in the modulation of human innate immunity by suppressing interferon (IFN) synthesis and regulating its signaling. The four main structural proteins are Spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, of which S, M, and E are membrane bound, whereas the N protein is located within the virions in complex with the genomic RNA.
Coronavirus attachment and entry into host cells
Overview
The primary pathways for host cell entry of enveloped viruses include either (i) a pH-independent, receptor-mediated pathway, where the viral envelope fuses with the host cell membrane to initiate viral uncoating; or (ii) a pH-dependent, endocytic pathway, where the virus is transported to the endosome (low pH environment) through either clathrin- or caveolin-dependent processes 1, 7. The receptor-mediated pathway is a multistep process that is initiated by virus adherence to the host cell through the receptor-binding domain (RBD) of the S glycoprotein, which forms the prefusion trimeric S-receptor complex [8]. The S glycoprotein participates in two key events during the coronavirus life cycle. First, the S glycoprotein binds to cell surface heparan sulfate (HS) chains of the heparan sulfate proteoglycan (HSPG) receptor and other cellular receptor(s), which initiates internalization. Second, the S glycoprotein promotes fusion between viral and cellular membranes [9]. The accomplishment of these two events drives the release of the viral RNA genome into the host cell and subsequently triggers the viral replication cycle [1].
In addition to mediating viral entry, the S glycoprotein is the principal antigenic determinant and the key target of neutralizing antibodies [10]. Nearly 22 glycosyl chains are present on the S glycoprotein [11], which suggests that the nature of these sugar chains on Asn/Ser/Thr residues determines the specificity of different antibodies and holds the key to developing vaccines [12]. Furthermore, two unexpected O-glycosyl chains have been characterized on the S glycoprotein [13]. Given that glycosylation of influenza virus [14] has posed challenges for vaccine development, similar issues might arise for anti-SARS-CoV-2 vaccine strategies. Thus, the design and/or development of small molecules that limit viral infection is crucial. Interestingly, the membrane fusion domain is highly conserved in the S glycoprotein, which makes it an attractive target to prevent membrane fusion against SARS-CoV outbreaks [15]. Alternatively, panning the S glycoprotein with fusion domains using a small-molecule library is likely to be an attractive approach to develop novel inhibitors of viral entry.
Host cell surface receptors
To date, multiple host cell receptors belonging to different families have been reported to facilitate the attachment and fusion of coronaviruses. For SARS-CoV, the major receptor is ACE2, a zinc metalloprotease and carboxypeptidase expressed as an ectoenzyme in a variety of organs and/or tissues, such as the kidneys, intestinal endothelium, lung, and heart. The ACE2 receptor binds to the RBD of SARS-CoV-2 through an extended set of interactions that span at least eight residues of helix α1 of ACE2 and seven residues of S1 16, 17. Interestingly, superposition of the RBDs of SARS-CoV and SARS-CoV-2 in complex with ACE2 show striking similarity (RMSD = 0.68 Å), despite the large number of substitutions in the interfacial residues.
Other receptors with a role in coronavirus entry include the human aminopeptidase N, a cell-surface metalloprotease expressed on intestine, lung, and kidney epithelial cells. Aminopeptidase N is utilized by human coronavirus-229E [18]. Likewise, dipeptidyl peptidase 4 (DPP4), a serine exopeptidase expressed on the surface of most cell types, is utilized by MERS-CoV [19]. Finally, HCoV-OC43 and HCoV-HKU1 use glycan-based receptors carrying 9-O-acetylated sialic acid residues 20, 21. Coronaviruses of zoonotic origin (i.e., NL63, 229E, HKU1, OC43, and SARS-CoV) have successfully breached the species barrier to become true human pathogens [22]. One reason why SARS-CoV-2 might have transited from bats to humans is the commonality of ACE2 as a receptor [17]. Although not shown as yet for SARS-CoV-2, other coronaviruses exploit DC-SIGN, a receptor highly expressed by macrophages and dendritic cells [23].
HSPGs represent an important, but underappreciated, group of host cell surface receptors. They contribute to the adherence and infectivity of multiple viruses 24, 25, 26, 27, 28. Structurally, HSPGs are massive proteoglycans that carry highly sulfated, linear polysaccharide chains, which recruit extracellular signaling ligands, such as growth factors. Nearly all enveloped viruses use their surface glycoproteins to bind to the HS component of the HSPGs to enhance their probability of cell targeting, adherence, and fusion. For coronaviruses, the S glycoprotein has the ability to bind to HS. Enzymatic cleavage of HS or addition of exogenous heparin prevented S glycoprotein of SARS-CoV from binding to the host cell surface and resulted in reduced infectivity [29]. Likewise, a SARS-CoV strain isolated from a severely infected patient was inhibited by ∼50% when treated with 100 mg/ml exogenous heparin [30]. Similarly, avian and murine coronavirus strains also use HSPGs to gain host cell entry 26, 28. Although HSPGs have not been specifically implicated in SARS-CoV-2 entry as yet, several studies using recombinant S1 protein of SARS-CoV-2 have shown tight binding to heparin and/or HS 31, 32, 33. Additionally, this binding appears to induce a conformational change in the RBD, similar to that involved in viral entry (see below), which suggests a role in host cell entry [31].
Molecular players in host cell internalization
Following initial virus adherence to primary host cell receptor (i.e., ACE2 in the case of SARS-CoV), a multistep process ensues, where proteolytic processing of the S glycoprotein and conformational changes are required for high efficiency fusion with the cell membrane [34]. One group of host proteases involved in this process are the type II transmembrane serine proteases TMPRSS2 and TMPRSS11D. There are also several other lung airway serine proteases that support the pH-independent mode of viral entry 35, 36. By contrast, the low pH environment pathway of viral entry involves activation of endosomal proteases, such as cathepsins, which are a family of cysteine proteases. Of interest are cathepsins B and L, which become active in the early and late endosome, respectively, in facilitating fusion with the endosomal membrane to support viral entry [37]. In addition, other host factors could also facilitate virus internalization [38].
A protein with a key role in viral life cycle is actin, which orchestrates rearrangements of the cytoskeleton during endosome formation. Interestingly, previous studies identified imatinib, an inhibitor of Abelson kinase (a tyrosine kinase), as an antiviral agent against SARS-CoV and MERS-CoV [39]. Inhibition of actin cytoskeleton rearrangement has been proposed as the mechanism of action for this agent [40]. However, the efficacy of imatinib in patients with SARS-CoV-2 is yet to be assessed.
Discovering small molecules against SARS-CoV-2
Overview
The number of expressed genes for SARS-CoV-2 (16 NSPs and four structural proteins), identical to that of SARS-CoV, offers multiple avenues for discovering antagonists (Fig. 1 ). More importantly, the two proteomes are ∼95% homologous [41], which implies a high probability of cross-reactive agents. Only six regions of difference in the genome were identified between the two viruses, which could provide additional targets for anti-SARS-CoV-2 agents. Two recently published reviews provide an excellent summary of agents discovered against coronaviruses other than SARS-CoV-2 42, 43. Here, we discuss recent developments in the direction of anti-SARS-CoV-2 agents and highlight opportunities waiting to be explored.
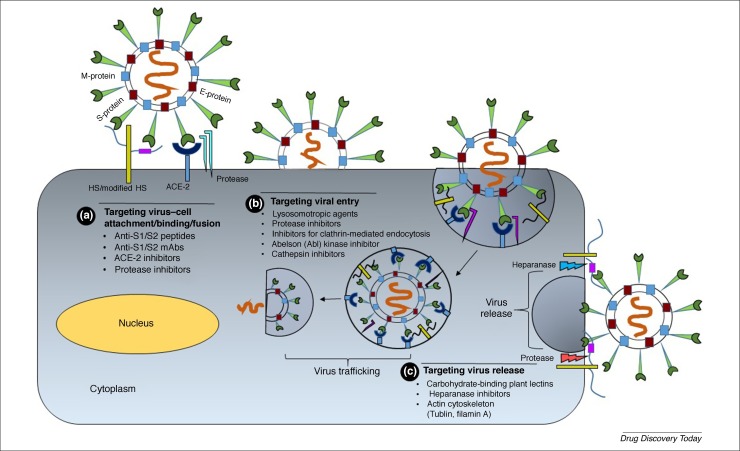
Figure 1.
Developing candidates that interfere with the early stages of virus-receptor attachment and internalization. (a) The early severe acute respiratory syndrome coronavirus (SARS-CoV-2) spike protein–host cell surface heparan sulfate proteoglycan (HSPG) interaction could be targeted by peptide antagonists that mimic S1/S2 subunits, or ACE2 sequence, or are heparin sulfate (HS) mimetics, or anti-Spike glycoprotein (S) antibodies. Likewise, protease inhibitors could prevent fusion with the angiotensin-converting enzyme 2 (ACE2) receptor. (b) The next step of SARS-CoV-2 invasion involving S binding to its fusion receptor (ACE2) offers the possibilities of using soluble ACE2 peptide or anti-ACE2 antibodies. Likewise, inhibitors of endocytosis or cathepsin L could reduce the efficiency of virus–cell fusion. The release of virus from the host cell could also be targeted through inhibitors of proteases or heparanase, which contribute to the process. For example, HS mimetics combined with a cocktail of protease inhibitors might block virus egress. Not shown is the role of other enzymes involved in viral replication, including helicase and RNA-dependent RNA polymerase, which offer a major route to small-molecule discovery. Abbreviation: mAb, monoclonal antibody.
A priori, one of the simplest early approaches would be to target the S glycoprotein–host cell surface HSPG interaction (Fig. 1). Modifiers of S1 and/or S2 subunits could include hydrophobic small molecules, heparin/HS-based oligosaccharides, or HS mimetics. The next step involving ACE2 receptor interactions offers the possibilities of using soluble ACE2 and/or S1 subunit-based peptides or peptidomimetics. The following step involves proteolytic cleavage of the S glycoprotein, which could be prevented by inhibitors of serine or cysteine proteases. Likewise, inhibitors of proteases involved in endocytosis (e.g., cathepsin L) would reduce viral infectivity. Subsequent processes involve several enzymes and nonenzymatic proteins involved in viral replication, such as RNA-dependent RNA polymerase (RdRp) and the E and M proteins, which offer diverse and virus-specific routes to small-molecule discovery. Finally, virus release from the host cell surface involves heparanase and other proteases, which could offer a novel route to effective anti-SARS-CoV-2 agents (Fig. 1). Here, we discuss various aspects of these putative molecular approaches.
Viral enzymes
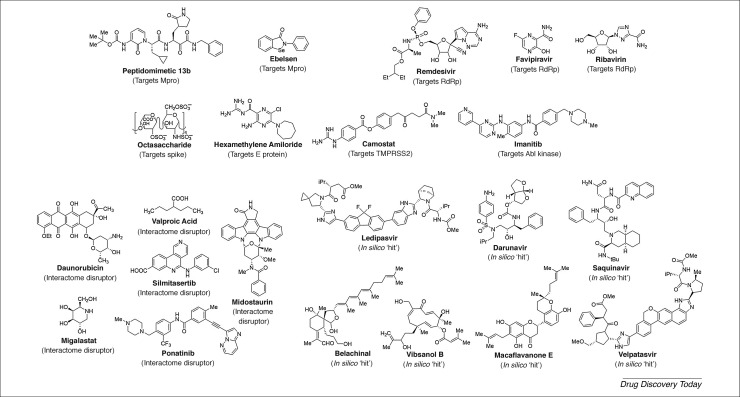
An often-used approach to discover drugs quickly is to identify inhibitors of key enzymes involved viral entry and propagation. For SARS-CoV-2, this group would include members of the NSP family, especially the main or chymotrypsin-like protease (Mpro and CLpro) and the papain-like protease (PLpro). Inhibiting Mpro is likely to be rewarding because it would stop the processing of a large polyprotein, replicase 1ab, which is essential for replication. Additionally, its substrate specificity is unlike that of any human protease. Relying on its 96% sequence identity with Mpro from SARS-CoV, the Hilgenfield group transformed a flexible amide bond of a parent peptide into a structurally restrained, pyridone-containing peptidomimetic 13b (Fig. 2 ), which inhibited SARS-CoV-2 with 670 nM potency and exhibited substantial lung penetration in mice, with no significant adverse effects 44, 45. Peptidomimetic 13b carries an alpha-ketoamide group that irreversibly modifies the active site cysteine of Mpro.
Figure 2.
Structures of in-clinical use and putative anti-severe acute respiratory syndrome coronavirus (SARS-CoV-2) molecules. The putative agents were identified through computational screens, high-throughput interactome analysis, rational design of protease inhibitor analogs, or other approaches. Also shown are agents that were discovered earlier as inhibitors of SARS-CoV, such as hexamethylene amiloride, which are likely to inhibit SARS-CoV-2 owing to their similarity. See main text for details. Abbreviations: Abl, Abelson; E, envelope protein; RdRp, RNA-dependent RNA polymerase; S, Spike protein.
The Mpro has also been the subject of a recent high-throughput screening (HTS) approach [46]. Based on the structure of an earlier Mpro inhibitor, Jin et al. performed in silico drug discovery followed by an high-throughput sequencing (HTS) campaign of 10,000 compounds to identify six diverse molecules (ebselen, disulfiram, tideglusib, carmofur, shikonin, and PX-12) as covalent inhibitors of SARS-CoV-2. Of these, ebselen (Fig. 2) displayed good antiviral potency (4.67 μM). Unfortunately, these agents are likely to be promiscuous. Despite this, Mpro has been the subject of several efforts to identify active site inhibitors through computational and synthetic screening 47, 48, 49.
The PLpro of SARS-CoV is also a replicase-processing enzyme, in which Cys, His, and Asp form the catalytic triad. PLpro has been targeted by both covalent and noncovalent agents 50, 51. The most potent agent identified to date displayed an impressive potency of 150 nM against SARS-CoV, with a good therapeutic index, but with liver microsomal stability of only ∼1 h [52]. Interestingly, despite the high homology (∼95%) of PLpro from the two SARS coronaviruses [41], no inhibitors of the novel coronavirus have been reported as yet.
An enzyme that could be targeted for drug discovery is RdRp (nsp12), which is the target of several agents, including ribavirin, favipiravir, and remdesivir (Fig. 2) 53, 54. All three agents mimic the nucleoside substrate recognized by viral RNA polymerase, leading to inhibition. RdRp inhibition is also a superior approach because, once these substrate mimetics are incorporated, the virus cannot induce ‘repair’, thus permanently blocking replication. All three agents display fairly broad-spectrum antiviral activity because the viral RdRp is substantially conserved across multiple viruses. However, subtle amino acid differences can have profound consequences for the affinity of a particular drug. This is why these drugs exhibit varied in vitro inhibition potencies against different coronaviruses. In fact, early research against a clinical isolate of the SARS-CoV-2 [53] showed that, of the three, only remdesivir displayed good IC 50 (0.77 μM). Extensive clinical trials on all three agents are currently in progress. Nearly 80 Chinese patients displayed promising results with favipiravir, which contributed to its recommendation as a treatment option in China; however, the US Food and Drug Administration (FDA) has guided against its use because of its adverse consequences noted in treatment of patients with influenza A, such as anemia, which raises concerns regarding its use against COVID-19, especially at higher doses.
The first report regarding the use of remdesivir for treating COVID-19 was in a single US patient [55], which was followed by a subsequent report on 53 patients, of whom 36 exhibited clinical improvement [56]. Later, a randomized study on 237 patients reported faster time to clinical improvement with remdesivir than with a placebo, although statistical significance was not obtained [57]. The most extensive randomized, controlled trial thus far, involving 1063 patients, was performed by the US National Institutes of Health, which demonstrated a 31% faster time to recovery [58]. This led the FDA to approve the use of remdesivir in emergency settings, especially for patients with advanced-stage disease in hospitals.
Remdesivir is a good drug but not likely to be the final drug of choice. Major improvements are needed in terms of its efficacy and toxicity. Furthermore, it is a parenteral drug, whereas an oral agent would be more suitable. Hopefully, the recent availability of the cryo-electron microscopy structure of RdRp, the target of remdesivir, should enable the discovery of advanced agents [59].
Another key enzyme is helicase (nsp13), which functions during the postfusion stage. RdRp and helicase belong to the replication-transcription complex that cooperates with other viral factors to rapidly transcribe and propagate in the host cell [60]. Although known to be crucial for replication, the design and/or discovery of inhibitors against helicase and the components of replication-transcription complex, except for RdRp, have been slow over the past decade.
Structural proteins
S glycoprotein
The S glycoprotein of coronaviruses is a distinguishing feature that is the basis not only for its name, but also for its myriad roles in facilitating host cell attachment and entry [61]. The S glycoprotein is a viral membrane-bound protein comprising three S1 subunits arranged in the form of a crown on top of three S2 stalks, each of which connect to a transmembrane segment followed by an intracellular tail. The S1 subunit carries the RBD that is recognized by one or more host cell receptors. The major receptor for SARS-CoV and SARS-CoV-2 is human ACE2 16, 17, 62. The trimeric S glycoprotein is cleaved into S1 and S2 subunits during the process of entry. Interestingly, the S glycoprotein of SARS-CoV-2 differs from that of other SARS-related coronaviruses in that it contains a furin-cleavable site at the S1/S2 boundary [63]. In fact, cleavage following, but not before, host cell receptor engagement enhances viral entry because of the importance of appropriate conformational changes for virus–host cell fusion [64]. Several human airway proteases, such as trypsin, plasmin, TMPRSS2, TMPRSS4, TMPRSS11a, and HAT, or endosomal cathepsin L, induce further cleavage of the S glycoprotein to enhance efficient virus attachment and fusion 65, 66, 67. A recent report showed that SARS-CoV-2 utilizes a fusion and receptor-dependent syncytium formation for host cell entry, which might contribute to rapid virus spread [67].
The S glycoprotein is the primary target for the discovery of antiviral agents, especially antibodies 15, 68, 69. At least three clinical trials (NCT04334980, NCT04283461, and NCT04324606) are in progress to develop vaccines that limit viral spread. Unfortunately, no small-molecule modulator of S glycoprotein function has been identified as yet. A promising idea is to antagonize RBD–ACE2 recognition through the use of soluble peptides. One of the earliest approaches in this direction was the design of a 438Tyr-Lys-Tyr-Arg-Tyr-Leu443 sequence, derived from the RBD of SARS-CoV S glycoprotein, which bound to hACE2 receptor with ∼46 μM affinity [70]. An improved approach was devised recently through the use of the 23-mer ACE2 α1 helix, which forms nearly seven selective interactions when binding to SARS-CoV-2 RBD. The affinity of this peptide was 47 nM [71]. The use of these peptides could limit binding and attachment, thereby limiting SARS-CoV-2 infection.
Another approach is to target the interactions of the S2 subunit during the fusion process, which is mediated by heptad repeat 1 (HR1) and HR2 domains. Crystal structure studies indicated that these interactions were enhanced in SARS-CoV-2 compared with SARS-CoV, which led to the design of a cholesterol-modified 36-mer peptide as a potent inhibitor of virus–cell fusion [72]. Although the in vivo stability of these all-natural sequences is not known, their high affinity makes for an attractive approach to design more stable analogs and/or peptidomimetics as competitive inhibitors.
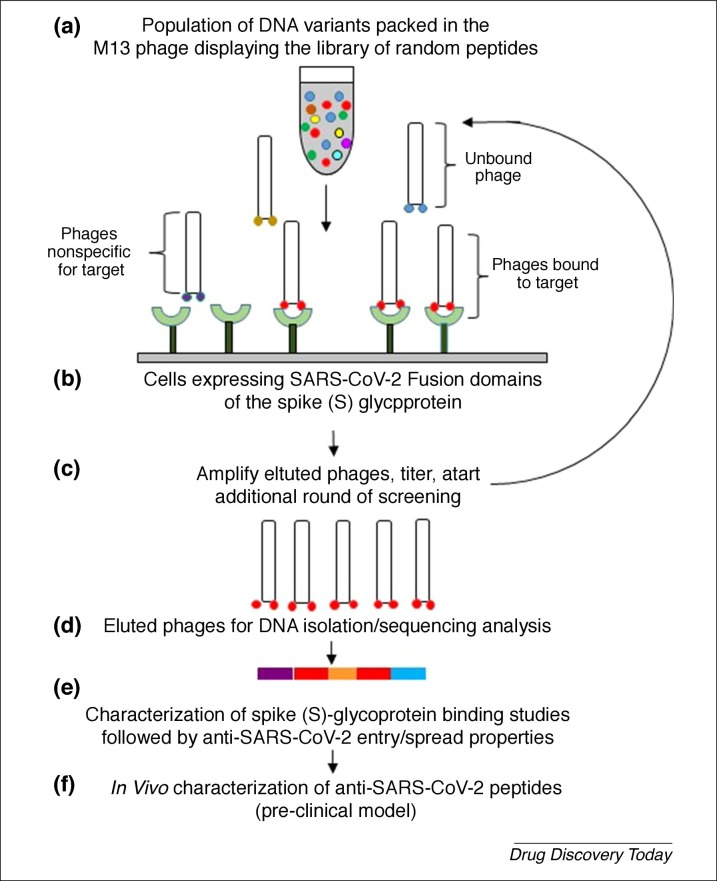
A novel approach that might rapidly identify promising peptidic agents against SARS-CoV-2 is the filamentous bacteriophage surface display technology (Fig. 3 ). Earlier work on herpes simplex virus (HSV) identified multiple candidate peptides that competed with 3-O-sulfated HS and inhibited infection in vivo 73, 74. Likewise, library panning against fusion domains of SARS-CoV-2 viral glycoproteins S1 and S2 could help identify better, smaller peptidic agents. Alternatively, an ACE2 receptor-based panning could isolate antifusogenic peptides that bind the S glycoprotein with high affinity.
Figure 3.
Diagram showing how a panning experiment (a–d) to identify anti-severe acute respiratory syndrome coronavirus (SARS-CoV-2) peptide could be identified using phage display screening of random peptide libraries, such as against fusion domains of the Spike (S) glycoprotein. The technology, implemented earlier for herpes simplex virus (HSV)-binding peptides, is an in vitro selection technique in which a peptide is genetically fused to a coat protein of a nonlytic bacteriophage (M13). This results in the display of the fused protein on the exterior of the phage virion, whereas the DNA encoding the fusion resides within the virion. The physical linkage between the displayed peptide and the DNA encoding it allows screening of more than 1 billion variant peptides against the SARS-CoV-2 S protein. The phages binding to the angiotensin-converting enzyme 2 (ACE2) receptor will have to be sequenced to generate peptides (e,f) for the development and characterization pf anti-S peptides to prevent SARS-CoV-2 infection.
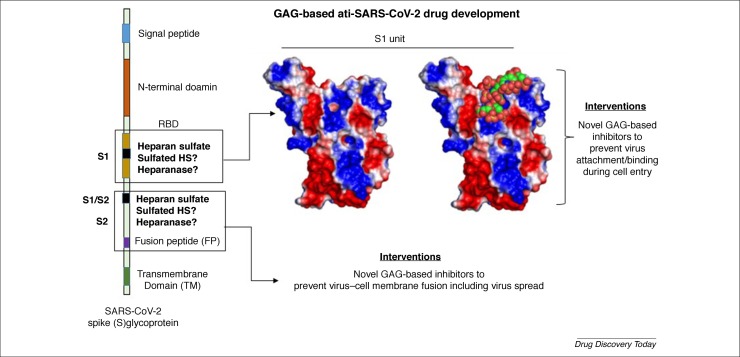
A more recent approach to inhibit coronavirus infection is via competitive inhibition with heparin or HS. Typically, enveloped viruses as distinct as HSV, HIV, cytomegalovirus (CMV), and SARS utilize HSPGs on the host cell surface to facilitate cellular penetration 24, 25, 26, 27, 28, 75, 76. Although much remains to be understood regarding the molecular underpinnings of these processes, the host cell HS–viral glycoprotein interactions might be selective, as exemplified in the case of HSV, in which a sulfated octasaccharide sequence was found to be important for binding to viral glycoprotein D [77]. Recently, the RBD of SARS-CoV-2 was found to interact with pharmaceutical heparin using circular dichroism 31, 32, 33. Whereas the Skidmore lab [31] utilized circular dichroism to show heparin–S glycoprotein interaction, the Linhardt lab [32] showed that heparin is selectively recognized by the S glycoprotein among all the different glycosaminoglycans tested. Furthermore, the Boons lab [33] identified a common octasaccharide sequence (Fig. 2) as the most potent (38 nM) in inhibiting the S–heparin interaction. Interestingly, three possible sites of HS binding on the S glycoprotein, including the RBD, have been predicted [32]. A quick analysis of the electrostatic surface of S1 followed by molecular docking of a small library of HS hexasaccharides based on well-established literature protocols [78] shows high complementarity between the two binding partners (Fig. 4 ). This supports the expectation that heparin-like molecules, such as glycosaminoglycan mimetics, which have been found to potently inhibit HSV and HCMV 79, 80, could also be good inhibitors of SARS-CoV-2.
Figure 4.
Glycosaminoglycan (GAG)-based interventions for targeting severe acute respiratory syndrome coronavirus (SARS-CoV-2). The spike (S) glycoprotein of SARS-CoV-2 contains three heparin sulfate (HS)-binding sites, including the receptor-binding domain (RBD) and S2. Whereas HS engagement of the RBD would directly compete with ACE2 and prevent viral adherence to host cells, HS engagement of the site in S2 would prevent virus–host cell fusion. Also shown is the computerized docking of a HS hexasaccharide binding to the S1 subunit. The model predicts strong interactions between the two. The protein surface is color coded using the electrostatic potential surface (positive in blue and negative in red) calculated through the APBS tool in PyMol. The heparin hexasaccharide sequence (spheres), shown in green, is colored by atom type.
An important area that deserves attention is the glycosylation of proteins involved in virus recognition and entry. All proteins (ACE2, S, etc.) carry multiple glycan chains, especially on Asn residues. In fact, numerous Asn residues on the S glycoprotein are glycosylated with high mannose and complex sugar types, which are predicted to have important roles in bypassing the host defense system [81]. It is possible that soluble glycans or glycan mimetics serve as effective competitors depressing viral attachment. Another possibility is to alter the expression of glycans through modulation of endoplasmic reticulum glucosidases, as exemplified by iminosugar-induced changes in structure of N-glycans of ACE2, which impaired the recognition of SARS-CoV Spike protein [82].
The E and M proteins
The E and M proteins have key roles in virus integrity as well as morphogenesis, ion channel formation, the stabilization of other proteins, and the activation of host inflammatory mediators 83, 84. The E protein is a relatively short protein (76 residues) that forms a symmetric pentamer that yields an ion channel. In the absence of E protein, membrane permeability decreases significantly, which results in inefficient virus maturation. Consistent with this finding, the drug hexamethylene amiloride (Fig. 2) was shown to reduce ion conductance in vitro for several synthetic SARS coronaviruses [85]. The other structural protein, M glycoprotein, is the most abundant protein in coronaviruses. Its transmembrane domain oligomerizes and forms a lattice, which imparts strength for the assembly of the viral membrane and, in turn, the virus. Unfortunately, no small-molecule inhibitors targeting the M protein are known as yet.
A powerful approach that could lead to the discovery of a pan-CoV agent would be to destabilize the E or M proteins. These proteins tend to be highly conserved across many different species and/or strains because of their role in the propagation and completion of the virus life cycle. Thus, small molecules that disrupt the stability of these structural proteins would immediately result in the loss of virus integrity and viral infectivity. Another advantage with these targets is the high number of protein–protein interfaces that could be targeted. The feasibility of such an approach was demonstrated through the computational discovery of a small molecule (MW <500) that disrupts matrix protein 1 of influenza A [86]. To date, no agent has been designed based on this approach for either SARS-CoV-2 or SARS-CoV.
Host proteases
Given that host proteases are required for the rapid internalization of the SARS virus, an effective strategy would be to inhibit these proteases to prevent amplification of infection. In the case of coronavirus entry, the interaction between ACE2 and the S glycoprotein requires priming by serine protease TMPRSS2, which generates S1 and S2 subunits required for successful viral entry. Given that lung epithelial cells express high levels of TMPRSS2 [87], SARS-CoV-2 is likely to use TMPRSS2 for entry. Therefore, the transmembrane protease TMPRSS2 is of major interest as a modulator of viral entry. In fact, the feasibility of this approach was indicated in a 2012 report that demonstrated the use of camostat, a TMPRSS2 inhibitor, and EST, a cathepsin L inhibitor, in efficiently blocking viral entry into human cells [88]. Likewise, studies on SARS-CoV-2 have also shown reasonable support for this approach [89]. This has led to camostat (Fig. 2) being studied in clinical trials (NCT04321096).
Another group of host proteases aiding viral fusion are Abelson (Abl) kinases, which are cytosolic, nonreceptor tyrosine kinases [90]. Previous work identified imatinib (Fig. 2), an anticancer agent, as an inhibitor of Ab1 kinase [91]. Imatinib reduces SARS-CoV virus fusion and syncytia formation, probably by interfering with the dynamics of actin movement during virus–cell fusion [92]. Unfortunately, this area of inhibitor design has not attracted much attention, probably because of the high possibility of adverse consequences that arise from knocking out host proteases.
Repurposing drugs, clinical candidates, or natural products
Each protein, nucleic, and glycan component of the SARS-CoV-2 could be targeted for discovery of small molecules that modulate their interaction with human biomolecules. This ‘human–virus biomolecular interactome’ is likely to be massive and challenging to decipher. However, curating the interactome might yield the discovery of novel agents that alter key pathways for infection. A recent study of the likely human–virus biomolecular interactome for SARS-CoV-2 offers a peek into navigating this process and deriving new uses for approved drugs, agents in clinical trials, and preclinical agents.
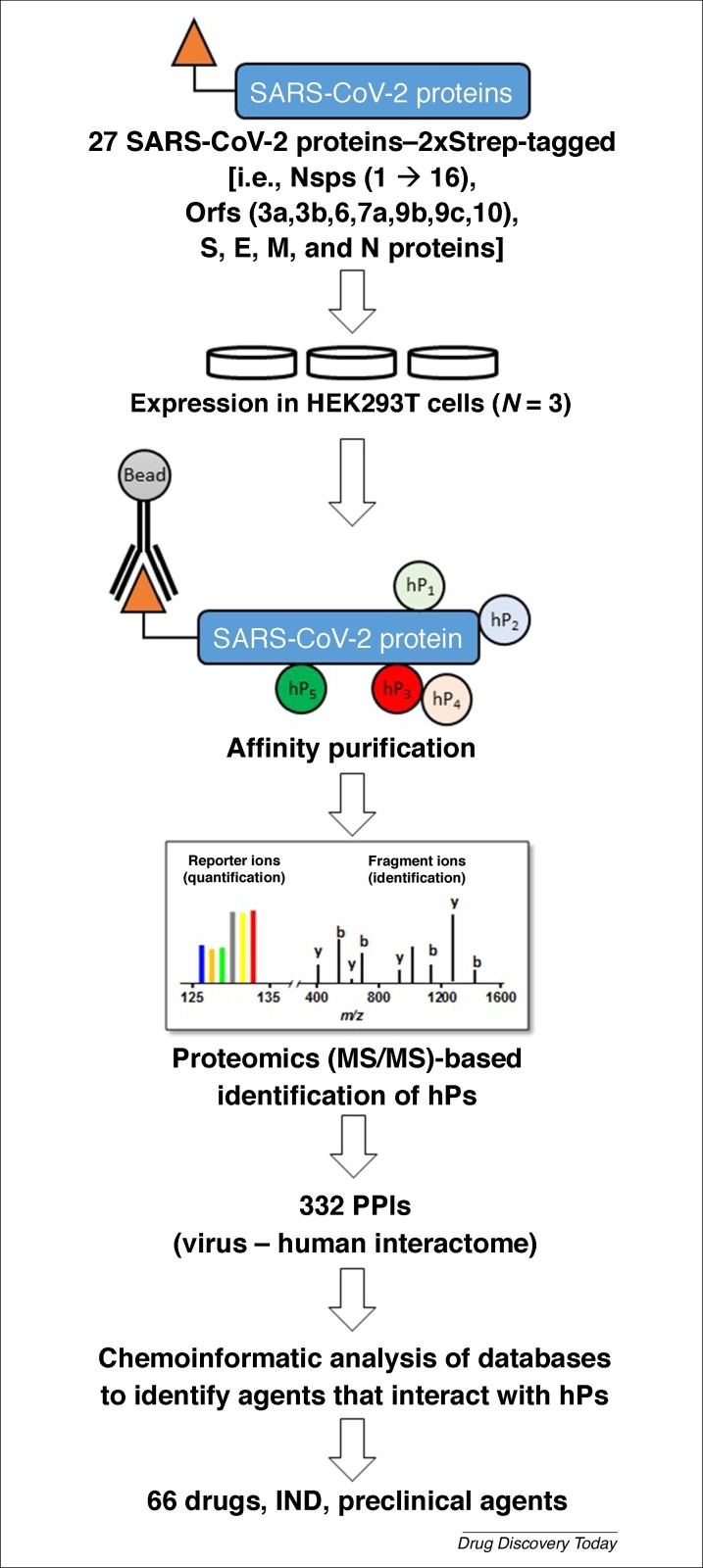
A multilaboratory team assembled by Nevan Krogan of University of California–San Francisco identified using affinity purification–mass spectrometry 332 human protein binding partners to 27 of the 28 SARS-CoV-2 proteins (Fig. 5 ) [93]. The interactions spanned a range of functions, such as gene replication and expression, trafficking between endoplasmic reticulum and Golgi, palmitoylation, interferon signaling, stress response modulation, and ubiquitinylation. The SARS-CoV-2 proteins that invoke these human pathways include ten NSPs (i.e., NSP1, 4, 5, 6, 7, 8, 9, 10, 13, and NSP15), two structural proteins (S and E), and six open-reading frames (i.e., Orf3a, 6, 8, 9b, 9c, and Orf10). Interestingly, Nsp6 of SARS-CoV-2 was found to interact with Sigma receptor, which is known to be targeted by chloroquine, the drug suggested during the early phase of pandemic for treating patients with COVID-19. Likewise, NSP8 binds to human mitochondrial ribosomes, which are known to be off-target partners of azithromycin [93], another drug used extensively to date.
Figure 5.
An affinity purification–mass spectrometry (MS) approach used to identify potential inhibitors of severe acute respiratory syndrome coronavirus (SARS-CoV-2) [93]. By affinity tagging (Strep) every gene of SARS-CoV-2, expressing the constructs in human HEK293T cells, affinity purifying on immobilized antibody beads, and identifying all the human proteins (hPs) that are co-isolated, a multidisciplinary team deduced 332 protein–protein interactions worth targeting. Using chemoinformatics, the team then identified 66 known drugs, or investigational new drug (IND) agents or preclinical promising molecules as plausible inhibitors of SARS-CoV-2. The structures of some of the agents are shown in Figure 2.
The team then identified 62 agents, either approved drugs, clinical, or preclinical molecules that interact with the human proteins as putative disruptors of the human–SARS-CoV-2 interactome. A number of these agents are anticancer drugs (e.g., ponatinib, silmitasertib, midostaurin and daunorubicin) (Fig. 2), whereas others are very simple agents, such as valproic acid and miglastat [93]. Despite these successes, the S1–ACE2 interaction was not picked up in this study. Likewise, the SARS-CoV-2 proteins involved in host cell glycan recognition (e.g., sialic acids and/or HS) would not be picked up because of its focus on protein–protein interactions. Finally, the computational screening used in the approach was directed toward repurposing drugs against human proteins, and not toward against SARS-CoV-2 proteins. Thus, the knowledge garnered in developing the interactome appears to have been only partially utilized thus far.
Another approach to repurpose approved drugs for SARS-CoV-2 was reported by Zhou et al. [94]. In this approach, a ‘systems pharmacology-based network medicine platform’ was developed for quantifying the projected interactions between SARS-CoV-2 proteins and drugs through calculation of proximity of known drug targets to the coronavirus–human interactome. The logic of this approach was to first identify human proteins that are known to interact with coronavirus proteins and then to sort the proximity of these proteins to known targets of clinically used drugs, which could then be expected to antagonize the interactions of SARS-CoV-2 with host proteins. Based on this network analysis, the authors identified several drugs and drug combinations as potential inhibitors of SARS-CoV-2, including melatonin, mercaptopurine, sirolimus, toremifene, emodin, and others [94].
A good strategy to rapidly discover antivirals against SARS-CoV-2 in a nontargeted manner would be through the phenotypic screening of clinically approved drugs. Examples of this strategy have been in the use of chloroquine/hydroxychloroquine, lopinavir/ritonavir, and remdesivir/fravipiravir in patients with COVID-19. In fact, several clinical trials have begun with other broad-spectrum agents, including darunavir 54, 95. Since the COVID-19 outbreak, several groups have used computational tools to repurpose approved drugs against SARS-CoV-2 proteins, including Mpro 47, 96, 97. Interestingly, these studies identified several different drugs, including remdesivir, saquinavir, darunavir, ledipasvir, and velpatasvir (Fig. 2).
Finally, a group with considerable promise in terms of preventing or treating viral infection is through the use of natural products. A large number of libraries containing natural products are and could be tested for their ability to limit SARS-CoV-2 infection. Following the SARS-CoV outbreak of 2000/2003, several groups pursued natural products as inhibitors of Mpro and realized modest levels of activities 98, 99. Recently, computational modeling using a library of natural products led to the identification of belachinal, macaflavanone E, and vibsanol B (Fig. 2) as possible modulators of SARS-CoV-2 protein E [100]. Yet, the use of these natural compound libraries in target-agnostic phenotypic HTS studies against SARS-CoV-2 has not been implemented so far.
Concluding remarks
The global footprint and high fatality rate of SARS-CoV-2 is a clear and urgent call for the development of novel antiviral interventions. An ideal candidate will be an agent that blocks the early events of viral attachment and cell entry, thereby preventing viral spread. This would be advantageous for preventing human-to-human transmission, especially from asymptomatic individuals. In this regard, the most direct approach will arguably be to target inhibitors of key proteases, such as Mpro, PLpro, TMPRSSs, cathepsin L, RdRp, and helicase, involved in viral infection and spread. Targeting the attachment and fusion processes, involving structural glycoproteins, including S and M, would also stop the virus in its tracks. Unfortunately, research on small-molecule modulators of coronaviruses, especially SARS-CoV, had not progressed enough since the 2003 outbreak to yield a bounty of candidates to screen against SARS-CoV-2.
Yet, several clinically approved drugs have been rapidly evaluated in patients with COVID-19 based on reports from SARS-CoV studies. The pandemic nature of SARS-CoV-2 led to rapid off-label use of chloroquine/hydrochloroquine, lopinavir/ritonavir, ribavirin/favipiravir, and others. However, caution should be exercised regarding the interpretation of the results using these agents. Most off-label studies have been with a limited number of patients without the benefit of rigorous control cohorts. Additionally, the known adverse effects of these drugs introduce additional challenges in infected patients.
The scientific community has quickly responded to the COVID-19 crisis by developing a range of possible therapeutics. These agents include the pyridone-containing mimetic against Mpro, the 23-mer α1 peptide against the RBD, and the cholesterol-containing 36-mer sequence from the S2 subunit. This survey also reveals some major avenues of antiviral drug discovery not being pursued with vigor, including targeting the HSPG–virus interaction, disrupting the oligomerization of E and M proteins, outcompeting host glycan–virus interactions, and the comprehensive screening of all nonstructural and structural proteins of SARS-CoV-2. A glaring omission appears to be high-throughput phenotypic cellular entry-based screening against SARS-CoV-2, which is likely to yield better results because of its target-agnostic nature. Likewise, artificial intelligence (AI)-based drug discovery [101] has not yet been implemented for identifying potent binders of all 20 SARS-CoV-2 proteins.
There is also a possibility that several other promising targets are being sidestepped completely. One example is human heparanase, which is known in some enveloped viruses, such as HSV [102], to contribute to the removal of HS chains from cell surfaces, thereby facilitating virus release from host cells. It is likely that heparanase has a similar role for coronaviruses. Thus, heparanase inhibitors, such as PI-88 and SST0001, which are currently in clinical trials as anticancer agents (NCT00268593 and NCT01764880), might also be effective against SARS-CoV-2.
Overall, the unprecedented COVD-19 pandemic raises the expectations that the scientific community should be ever-ready to rapidly discover and develop efficacious and quality agents to avoid the risk of a pandemic. Hopefully, this experience will ensure that emphasis on understanding the fundamentals of biology and chemistry of these infectious agents remains high for years to come.
Author contributions
V.T. prepared and finalized the manuscript; J.C.B. searched literature and contributed to writing, N.V.S. performed molecular modeling and prepared figures; M.S.M. reviewed and revised the manuscript; and U.R.D. wrote sections and finalized the manuscript.
Conflict of interest
None declared.
Acknowledgments
We thank Rama Gunta and Adam Hawkridge for preparing the structure figure and generic MS image used in Fig. 5, respectively. We also thank the computational resources made available to us through a grant from the National Center for Research Resources (S10 RR027411) to V.C.U. This work was supported, in part, by grants from the NIH including HL107152, HL090586 and CA241951 (U.R.D.).
References
- 1.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verity R. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belouzard S. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses-Basel. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchdoerfer R.N. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belouzard S. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe Y. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020 doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virus Dis. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shajahan A. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020 doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zost S.J. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. USA. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan M. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan R. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang J. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeager C.L. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Doremalen N. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler N. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology. 2006;347:410–421. doi: 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z.W. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regan A.D., Whittaker G.R. Utilization of DC-SIGN for entry of feline coronaviruses into host cells. J. Virol. 2008;82:11992–11996. doi: 10.1128/JVI.01094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shieh M.T. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell. Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilgard P., Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology. 2000;32:1069–1077. doi: 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- 26.de Haan C.A. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005;79:14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalia M. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009;83:12714–12724. doi: 10.1128/JVI.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milewska A. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang J. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE. 2011;6:e23710. doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicenzi E. Coronaviridae and SARS-associated coronavirus strain HSR1. Emerg. Infect. Dis. 2004;10:413–418. doi: 10.3201/eid1003.030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mycroft-West C. The 2019 coronavirus (SARS-CoV-2) surface protein (spike) S1 receptor binding domain undergoes conformational change upon heparin binding. BioRxiv. 2020 doi: 10.1101/2020.02.29.971093. [DOI] [Google Scholar]
- 32.Kim S. Glycosaminoglycan binding motif at S1/S2 proteolytic cleavage site on spike glycoprotein may facilitate novel coronavirus (SARS-CoV-2) host cell entry. BioRxiv. 2020 doi: 10.1101/2020.04.14.041459. [DOI] [Google Scholar]
- 33.Liu L. SARS-CoV-2 spike protein binds heparan sulfate in a length and sequence-dependent manner. BioRxiv. 2020 doi: 10.1101/2020.05.10.087288. [DOI] [Google Scholar]
- 34.Hulswit R.J.G. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. U. S.A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata-Yoshikawa N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93:e01815. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zumla A. Infectious diseases epidemic threats and mass gatherings: refocusing global attention on the continuing spread of the Middle East Respiratory syndrome coronavirus (MERS-CoV) BMC Med. 2016;14:132. doi: 10.1186/s12916-016-0686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman C.M. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley W.D., Koleske A.J. Regulation of cytoskeletal dynamics and cell morphogenesis by Abl family kinases: emerging mechanisms and physiological contexts. J. Cell Sci. 2009;122:3441–3454. doi: 10.1242/jcs.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:E244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillaiyar T. Recent discovery and development of inhibitors targeting coronaviruses. Drug Disc. Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prajapat M. Drug targets for corona virus: a systematic review. Ind. J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verschueren K.H. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 2008;15:597–606. doi: 10.1016/j.chembiol.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin Z. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 47.Khan S.A. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020;13:2020. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 48.Ton A.T. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020;11:2020. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem. 2017;137:450–461. doi: 10.1016/j.ejmech.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Báez-Santos Y.M. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh A.K. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Báez-Santos Y.M. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong L. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 55.Holshue M.L. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grein J. Compassionate use of remdesivir for patients with severe COVID-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y. Remdesivir in adults with severe COVID-19: a randomized, double-blind, placebo-controlled, multicenter trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NIH . NIH; 2020. NIH Clinical Trial Shows Remdesivir Accelerates Recovery from Advanced COVID-19. [Google Scholar]
- 59.Gao Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subissi L. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li F. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 63.Walls A.C. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kam Y.W. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bosch B.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe R. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 2008;82:11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ou X. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian X. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sui J. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Struck A.W. A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 2012;94:288–296. doi: 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang G. The first-in-class peptide binder to the SARS-CoV-2 spike protein. BioRxiv. 2020 doi: 10.1101/2020.03.19.999318. [DOI] [Google Scholar]
- 72.Xia S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tiwari V. Anti-heparan sulfate peptides that block herpes simplex virus infection in vivo. J. Biol. Chem. 2011;286:25406–25415. doi: 10.1074/jbc.M110.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali M.M. A 3-O-sulfated heparan sulfate binding peptide preferentially targets herpes simplex virus 2-infected cells. J. Virol. 2012;86:6434–6443. doi: 10.1128/JVI.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song B.H. Human cytomegalovirus binding to heparan sulfate proteoglycans on the cell surface and/or entry stimulates the expression of human leukocyte antigen class I. J. Gen. Virol. 2001;82:2405–2413. doi: 10.1099/0022-1317-82-10-2405. [DOI] [PubMed] [Google Scholar]
- 76.Baldwin J. A role for 3-O-sulfated heparan sulfate in promoting human cytomegalovirus infection in human iris cells. J. Virol. 2015;89:5185–5192. doi: 10.1128/JVI.00109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Copeland R. Using a 3-O-sulfated heparin octasaccharide to inhibit the entry of herpes simplex virus type 1. Biochemistry. 2008;47:5774–5783. doi: 10.1021/bi800205t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sankaranarayanan N.V. So you think computational approaches to understanding glycosaminoglycan-protein interactions are too dry and too rigid? Think again! Curr. Opin. Struct. Biol. 2018;50:91–100. doi: 10.1016/j.sbi.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Majmudar H. A synthetic glycosaminoglycan mimetic blocks HSV-1 infection in human iris stromal cells. Antiviral Res. 2019;161:154–162. doi: 10.1016/j.antiviral.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 80.Elste J. Inhibition of human cytomegalovirus entry into host cells through a pleiotropic small molecule. Int. J. Mol. Sci. 2020;21:1676. doi: 10.3390/ijms21051676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao X. Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrob. Agents Chemother. 2015;59:206–216. doi: 10.1128/AAC.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siu Y.L. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pervushin K. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mosier P.D. Broad spectrum anti-influenza agents by inhibiting self-association of matrix protein 1. Sci. Rep. 2016;6:32340. doi: 10.1038/srep32340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lukassen S. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kawase M. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rogers E.M. Abelson kinase acts as a robust, multifunctional scaffold in regulating embryonic morphogenesis. Mol. Biol. Cell. 2016;27:2613–2631. doi: 10.1091/mbc.E16-05-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buchdunger E. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 92.Sisk J.M. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol. 2018;99:619–630. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gordon D.E. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Y. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Disc. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Disc. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 96.Chen Y.W. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu C. Analysis of therapeutic targets of SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pillaiyar T. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar V. Anti-SARS coronavirus agents: a patent review (2008–present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 100.Gupta M.K. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557:S55. doi: 10.1038/d41586-018-05267-x. [DOI] [PubMed] [Google Scholar]
- 102.Hadigal S.R. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat. Commun. 2015;6:6985. doi: 10.1038/ncomms7985. [DOI] [PMC free article] [PubMed] [Google Scholar]