Highlights
-
•
Machine learning was used to develop models to predict COVID-19 positive patient.
-
•
Features were extracted from patient data using string matching algorithms.
-
•
Constructed a novel dataset from unstructured hospitalized patient information.
-
•
Used descriptive statistical analysis for frequency calculation of patient symptoms.
-
•
Identified significant symptoms of COVID-19 patients using five different ML models.
Keywords: SARS-Cov-2, COVID-19, Coronavirus, Machine learning, Early stage symptom
Abstract
The recent outbreak of the respiratory ailment COVID-19 caused by novel coronavirus SARS-Cov2 is a severe and urgent global concern. In the absence of effective treatments, the main containment strategy is to reduce the contagion by the isolation of infected individuals; however, isolation of unaffected individuals is highly undesirable. To help make rapid decisions on treatment and isolation needs, it would be useful to determine which features presented by suspected infection cases are the best predictors of a positive diagnosis. This can be done by analyzing patient characteristics, case trajectory, comorbidities, symptoms, diagnosis, and outcomes. We developed a model that employed supervised machine learning algorithms to identify the presentation features predicting COVID-19 disease diagnoses with high accuracy. Features examined included details of the individuals concerned, e.g., age, gender, observation of fever, history of travel, and clinical details such as the severity of cough and incidence of lung infection. We implemented and applied several machine learning algorithms to our collected data and found that the XGBoost algorithm performed with the highest accuracy (>85%) to predict and select features that correctly indicate COVID-19 status for all age groups. Statistical analyses revealed that the most frequent and significant predictive symptoms are fever (41.1%), cough (30.3%), lung infection (13.1%) and runny nose (8.43%). While 54.4% of people examined did not develop any symptoms that could be used for diagnosis, our work indicates that for the remainder, our predictive model could significantly improve the prediction of COVID-19 status, including at early stages of infection.
1. Introduction
There has recently been a rapid spread of the novel SARS-CoV2 coronavirus (Gorbalenya et al., 2020) (designated by the World Health Organization) which gives rise to a respiratory disease COVID-19 (WHO, 2020). The first human coronaviruses, 229E and OC43, were identified during the 1960s from human nasal secretions (Lippi & Plebani, 2020). Other individual virus types classified in this family have been distinguished (such as HCoV NL63 and HKU1) and are thought to arise from zoonotic infections (Huang et al., 2020) as they are endemic in various bat populations. The coronavirus infections known were originally viewed as giving rise to innocuous respiratory human conditions that were not life-threatening. The development incidence of serious and deadly respiratory disorders attributed to beta-coronavirus subfamily members occurred in the last twenty years with the severe acute respiratory syndrome (SARS) and the middle east respiratory syndrome (MERS). The SARS-CoV infections arose first in Foshan, China in 2002 and MERS-CoV in 2012 in Saudi Arabia (Zhavoronkov et al., 2020), both causing international alarm and containment efforts due to their rapid spread and high mortality rates. SARS and MERS were associated with mortality rates of 9.6% and 36%, respectively (Peeri et al., 2020), among those diagnosed patients. These identified coronavirus infections as a significant threat to human health with the potential to cause extreme and lethal respiratory tract infections in people, particularly if person-to-person infection occurs easily (Chan et al., 2020).
The development and spread of the novel coronavirus (Nishiura et al., 2020) causing COVID-19 has vastly outpaced the rate of vaccine and therapeutic development. Nevertheless, within weeks of the first observations of COVID-19 disease, the virus was isolated and characterised. One of the most significant SARS-CoV2 protein targets is a 3C-like protease for which the structure is already known. Much effort has been centred around re-purposing known clinically-tested drugs and virtual screening for possible targets using protein structure data (Zhavoronkov et al., 2020). Priority has been given to the identification of infected individuals in order to isolate and (if necessary) treat them. Central to this is the use of clinical symptoms to optimise identification of infected individuals.
One of the earliest published studies (Tian et al., 2020) showed an analysis of 262 individuals confirmed as COVID-19 infected to determine their clinical and epidemiological characteristics in Beijing, China and found that respiratory and extra respiratory transmission routes may explain the rapid spread of disease.
In February 2020, the noted case fatality rate for COVID-19 in Wuhan, China, was 1.4% (Wu et al., 2020). However, accurate global estimates are far more challenging due to the vastly different response country to country. For example, in Italy during March 2020, it showed a case fatality rate of 7.2% (Onder, Rezza, & Brusaferro, 2020). This may partly reflect the demographic differences between nations, with 23% of the Italian population being over 65. However, even when stratified by age, infection rates remain higher in Italians over 70 years of age compared to China (Onder et al., 2020). This highlights the critical need to have improved screening and prediction methods to stratify those at higher risk of infection in discrete populations in different Track changes is on 39 nations. To this end, machine learning algorithms are ideally suited for improving patient stratification and can be widely and rapidly applied as needed during a pandemic.
In this study, we developed a machine learning methodology to identify the most important and significant clinical symptoms that predict true COVID-19 positive cases. We validated these predictions using COVID-19 patient data from seven provinces in China. The primary features of this machine learning approach are:
-
•
Extraction of features from unstructured raw data (hospitalized patient information in text format) using string matching algorithms and use of this data to construct a processed dataset.
-
•
Identification of the significant symptoms of COVID-19 patients by analyzing their association using five different machine learning approaches.
-
•
Developing a comprehensive predictive model to predict COVID-19 positive patients among suspected and confirmed individuals.
-
•
Analyzing the relationship between patient age and COVID-19 confirmation.
-
•
Identifying patient travel history and measure how it influences disease progression.
-
•
Use statistical analysis to calculate the impact and contribution of particular patient features to COVID-19 diagnosis.
2. Materials & methods
2.1. Data collection
We collected raw hospital data, obtained through GitHub repository (COVID-19-tracker, 2020). A record of their information is made available in anonymised form when a person has presented to hospitals and clinics for diagnosis and treatment. In our datasets, there were data from 6,512 patients from seven different provinces (Anhui, Guangdong, Henan, Jiangsu, Shandong, Shanxi, and Zhejiang) in China. The original dataset was written in Mandarin Chinese, which was translated by Google Translator, and was checked and validated by a native Chinese speaker and researcher (Haoming Xu) to confirm its accuracy.
With the spread of the novel coronavirus, the accumulation of related national epidemiology data, and its availability can be used for ML studies. However, much of this data was in the form of unstructured text information which can be difficult to process. The data used here were collected from a study by a group at Beijing University’s Big Data High-accuracy Center. They collected these datasets from the official channels of the national government websites (COVID-19-tracker, 2020). The detail of the dataset is as follows – basic information regarding gender, age, habitual residence, work and Wuhan/Hubei contact history; trajectory information is time, place, transportation and event up to February 20, 2020. We extracted important features of basic information (age, gender), symptoms (fever, cough, muscle soreness), diagnostic results (lung infection, radiographic imaging), prior disease/symptom history (pneumonia, diarrhea, runny nose) and some trajectory information (isolation treatment status, travel history) that are directly or indirectly related to COVID-19 disease.
2.2. Data preprocessing
The original Chinese datasets did not include information about which patients were suspected positive and which were confirmed for all patients. The definition of a suspected case is the patients who develop symptoms and have communication with confirmed COVID-19 patients but didn’t confirm as COVID-19 after diagnosis. Moreover, confirmed cases defined as, the patients who are confirmed as positive for COVID-19 in the CDC approved test report or the doctors mentioned confirmed cases after diagnosis in the root dataset. The data contain patient symptoms in a text format. For this reason, we find symptoms of every individual patient and some trajectory information applying various string matching algorithms. In detail, we selected some keywords for each feature then we matched those keywords to text data and extract the features individually. Lastly, we generated our final dataset which contained the following features (described in the Table 1 ): gender, age, fever, tussis (cough), rhinorrhoea (runny nose), pneumonia, lung infection, muscle soreness, diarrhea, travel history and isolation treatment status. This dataset consists of 1,572 cases of confirmed COVID-19 and 4,940 suspected cases. All the patients did not develop the same symptoms, although, diarrhea and, muscle soreness occurred only rarely. Then we preprocessed the dataset, firstly cleaned the dataset and eliminated unwanted fields. One of the important issues with missing value is the missing value mechanism. It’s important because it affects how much the missing value biases our results, so we took it into account when choosing a method to deal with the missing value. Our dataset contained 2.1% missing values only in the gender and age fields, and the propensity for the data point to be missing gender and age fields were completely random, i.e., Missing Completely at Random (MCAR) types of missing data. There’s no relationship between whether a data point is missing and any values in the dataset. Thus we imputed the gender field with random values according to the male/female ratio for the total data and impute age with random values within the interquartile range (IQR) values. In our dataset most of the values were binary, but the age field was as an integer value, so feature scaling was done on the age field by using standard scaling methods. Feature scaling is a technique to standardise the re-scaling technique which uses 0 as a mean value and 1 as variance (Gupta, 2019). The new feature value for a feature X is calculated by, . After those two steps, we obtained a structured, clean and preprocessed dataset.
Table 1.
Feature descriptions.
| Feature | Type | Description |
|---|---|---|
| Gender | String | Almost same ratio of male and female patients |
| Age | Integer | The age range is 0–96 years |
| Fever | Boolean | Develops symptoms with a high body temperature of 38 °C or more |
| Cough | Boolean | Develops symptoms with a dry cough or cough with sputum |
| Pneumonia | Boolean | Develops symptom of pneumonia and admitted to hospital |
| Lung Infection | Boolean | Radiographic or CT scan indicates chest imaging changes as lung infection |
| Runny Nose | Boolean | Develops the symptom of runny nose |
| Muscle Soreness | Boolean | Develops symptoms of limb or muscle soreness |
| Diarrhea | Boolean | Develops symptom of diarrhea and admitted to hospital |
| Travel History | Boolean | Patients are marked as suspected for travelling to one or more track |
| Isolation | Boolean | Isolation treatment status at designated hospitals |
2.3. Methods
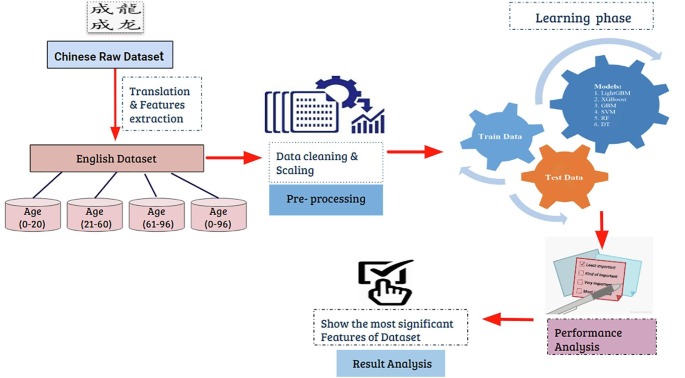
Since identifying the most predictive symptoms is challenging at the early stages of disease, we used ML models to identify them. Our methodology is shown in Fig. 1 . As indicated, using the training datasets we trained five ML algorithms that are described below.
Fig. 1.
Proposed methodology.
2.3.1. Decision tree
Decision Tree algorithms can be utilized to optimize both classification and data regression (Karim & Rahman, 2013). It utilizes tree representation in which each leaf node corresponds to a group of attributes and a branch corresponds to a value. This algorithm is developed in a recursive manner.Consider we have a variable Y whose k potential values have probabilities . The estimations of Y on the observation is known as the entropy. Y is characterised as (Li, Li, & Wang, 2009)
| (1) |
This main idea of Decision Tree algorithms is to build a tree for the entire data and process a unique output at every leaf. According to the target classification, how well a given attribute separates the training set can be measured by a statistical property, known as information gain. An attribute at a node with high information gain can split the training data to achieve improve classification accuracy. We can calculate the information gain IG of an attribute X, relative to a set of training data D, where E is Entropy, as
| (2) |
Here, the set of values of the attribute X is defined as and is the subset of D for which the attribute X has value v.
For a particular node in the tree, information gain is calculated for all the attributes, and the attribute with the highest information gain is selected as the best attribute that splits the data properly.
2.3.2. Random forest
Random Forest is an ensemble of regression and classification trees, which can train a similar size of training datasets called bootstraps, and at the end combine them for a more accurate result. The bootstraps are created by random re-sampling from the training dataset (Sarica, Cerasa, & Quattrone, 2017). Random Forests perform far better than a single tree. This approach can work with higher dimensional large datasets with comparatively greater accuracy. The model will be built with the following equations (Singh, 2019).
Calculate the constant value and initialise the model
| (3) |
Compute the pseudo-residuals r for
| (4) |
Here, is a model, is a training set and is differentiable loss function.
2.3.3. Gradient Boosting Machine
Gradient Boosting Machine (GBM) is a fixed size decision tree-based learning algorithm that combines many simple predictors (Biau, Cadre, & Rouviére, 2019). It fabricates the model in a phase insightful manner as other boosting strategies do, and it sums them up by permitting enhancement of a self-assertive differentiable loss function. A definitive objective of the GBM is to discover a function , which limits its loss function , through iterative back-fitting as – By definition, a supported predicted model is a weighted straight of the base learners
| (5) |
where is a base learners parameter.
2.3.4. Extreme Gradient Boosting
Extreme Gradient Boosting (XGBoost) is another decision tree-based machine learning algorithm that uses a gradient boosting framework. It is an end to end tree boosting scalable system widely used in data science. XGBoost can solve real-world scale problem utilizing comparatively fewer resources (Chen & Guestrin, 2016). Suppose, a dataset S consists with p examples and q features, where, . So the decision tree model uses m additive functions to forecast the output (Chen & Guestrin, 2016).
| (6) |
Where n indicates to the structure of each tree that maps a guide to the relating leaves nodes and T is the amount of the leafs in the tree. Every relates to an autonomous tree structure n and leaf loads w.
2.3.5. Support Vector Machine
Support Vector Machine (SVM) is one of the most well-known, flexible supervised machine learning algorithms. It is utilized for both regression and classifications tasks. It is typically favoured for medium and little-measured informational collection. The primary target of SVM is to locate the ideal hyper-plane which directly isolates the information focuses on two-part by augmenting the edge. The SVM can guarantee the advancement capacity of the machine model, so it is generally utilized in different fields. The goal of the support vector machine algorithm is to discover a hyper-plane in N-dimensional space (N – the quantity of highlights) that particularly classifies the information focuses (Wei & Hui-Mei, 2014).
2.4. Evaluation criteria:
There are various assessment parameters in our approach, for example, precision, recall, F1-score, Log loss, and area under the ROC curve (AUC). These parameters are used to estimate our prediction accuracy.
-
•Precision: Precision is a legitimate finding of assessment metric when we need to be extremely positive about our prediction. It measures the proportion of anticipated positives that are true positives. So it is dependant on True Positive (TP) and False Positive (FP) values (Agarwal, 2019).
(7) -
•Recall: Recall is another admissible decision of assessment metric when we need to identify the number of positives as could reasonably be expected (Agarwal, 2019). It indicates the ratio of actual Positives correctly classified. True positive (TP) and False negative (FN) values are used to measure recall.
(8) -
•F1 Score: F1 score keeps up a harmony between the precision and recall for your classifier. The F1 score is a number somewhere in the range of 0 and 1 and is the consonant means of precision & recall (Agarwal, 2019).
(9) -
•Area Under the Curve (AUC): AUC is the area under the ROC curve and demonstrates, how well the probabilities from the positive classes are isolated from the negative classes. Where True positive rate or TPR is only the range of trues we are utilizing our calculation (Agarwal, 2019).
(10) -
•Log Loss: Log Loss is the most significant order metric dependent on probabilities. It’s difficult to decipher raw log-loss values, yet log-loss is a decent measurement for looking at models. A lower log-loss value implies better predictions (Kiapour, 2018). The function of log-loss is-
where y is the level of target variable, is the predicted probability of the point for the target value and is the calculated value of log loss.(11)
3. Experimental results analysis
3.1. Statistical analysis
In this study, some statistical analysis was also performed using the Statistical Package for the Social Sciences (SPSS) software version 25.0 (IBM Corp., Armonk, NY). The median age of the individuals studied was 43 years (range 0 years to 96 years), the interquartile range (IQR) was 32 to 55 years for 3,367 males (51.6%). In Table 3 , shows the association of patient COVID-19 confirmation and some selected demographic information including symptoms. We performed Mann–Whitney U test on age field and Chi-square test on the remaining fields and found that age, travel history, isolation treatment is significant as demographic information; and most of the symptoms including fever, cough, runny nose, pneumonia, and lung infection are significant with p-value <0.001. From those studied patients, there was 2,971 (45.6%) patient who displayed some symptoms whereas, among confirmed patient’s 49.3% develops symptoms. It is also seen, 2,675 patients (41.1%) have a fever, which is the most frequent symptom, and their body temperature was equal or above 38-degree centigrade. Some patients had fatigue, dizziness and headache with fever. The cough was the second most common symptom, with 1,975 (30.3%) affected patients. Some of these patients had a dry cough, and some had coughing with sputum. Radio-graphic or pulmonary or chest imaging results showed that 855 patients (13.1%) had a lung infection. Only 26 patients (0.4%), 37 patients (0.57%), had muscle soreness and diarrhea. Travel history is another important issue in COVID-19 infection, 4,239 patients (65.1%) had travelled recently to one or more places in China or abroad. All patients were hospitalized for treatment, but among those 1,413 patients (21.7%) were received treatment in full isolation. The comparison of suspected and confirmed patients according to developing symptoms, we found that more confirmed patient’s 1,466 (93.26%) develops symptoms than 1,505 (30.47%) suspected patients. There are 1,242 (79.01%) fever, 1,188 (75.57%) cough, 502 (31.93%) runny nose, 402 (25.57%) pneumonia, and 786 (50%) lung infection in confirmed patient’s; on the other hand 1,433 (29.01%) fever, 787 (15.93%) cough, 47 (0.95%) runny nose, 85 (1.72%) pneumonia, and 69 (1.4%) lung infection is suspected patients; which is much lower than confirmed.
Table 3.
Accuracy measurement of ML approaches.
| Dataset | Algorithm | Precision | Recall | F1 Score | AUC | Log Loss |
|---|---|---|---|---|---|---|
| Age (0–20) | XGBoost | 0.89 | 0.94 | 0.91 | 0.85 | 4.14 |
| GBM | 0.89 | 0.96 | 0.92 | 0.86 | 3.68 | |
| SVM | 0.92 | 0.98 | 0.95 | 0.91 | 2.30 | |
| Random Forest | 0.90 | 0.92 | 0.91 | 0.86 | 4.14 | |
| Decision Tree | 0.88 | 0.89 | 0.89 | 0.85 | 3.68 | |
| Age (21–60) | XGBoost | 0.95 | 0.92 | 0.93 | 0.87 | 3.57 |
| GBM | 0.98 | 0.86 | 0.91 | 0.89 | 4.35 | |
| SVM | 0.98 | 0.86 | 0.91 | 0.89 | 4.45 | |
| Random Forest | 0.95 | 0.91 | 0.93 | 0.87 | 3.74 | |
| Decision Tree | 0.94 | 0.93 | 0.93 | 0.89 | 3.74 | |
| Age (61–96) | XGBoost | 0.87 | 0.90 | 0.88 | 0.82 | 5.42 |
| GBM | 0.90 | 0.87 | 0.88 | 0.84 | 5.25 | |
| SVM | 0.93 | 0.80 | 0.86 | 0.84 | 5.93 | |
| Random Forest | 0.89 | 0.88 | 0.89 | 0.84 | 5.08 | |
| Decision Tree | 0.88 | 0.89 | 0.88 | 0.83 | 5.25 | |
| Age (0–96) | XGBoost | 0.93 | 0.91 | 0.92 | 0.85 | 4.08 |
| GBM | 0.97 | 0.85 | 0.91 | 0.88 | 4.61 | |
| SVM | 0.97 | 0.84 | 0.90 | 0.88 | 4.93 | |
| Random Forest | 0.92 | 0.89 | 0.91 | 0.83 | 4.74 | |
| Decision Tree | 0.91 | 0.90 | 0.91 | 0.82 | 4.72 | |
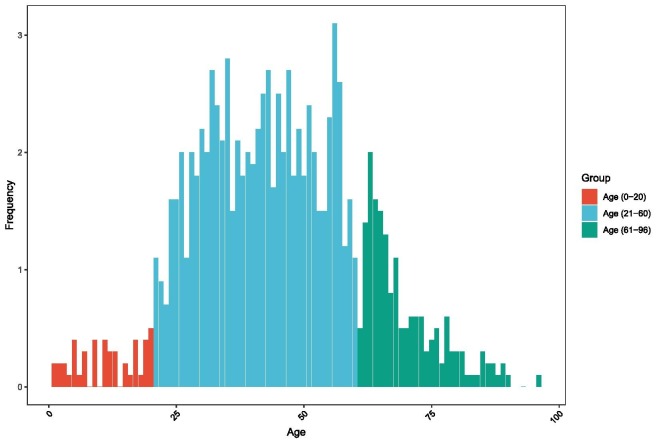
In Fig. 2 is illustrated the age-wise total number of patients. In the age range of 25 years to 65 years, the rate of individuals affected is high. In children and the older adults the affected rate is comparatively low. However, the death rate in older men is high.
Fig. 2.
Impact of age for COVID-19 outbreak.
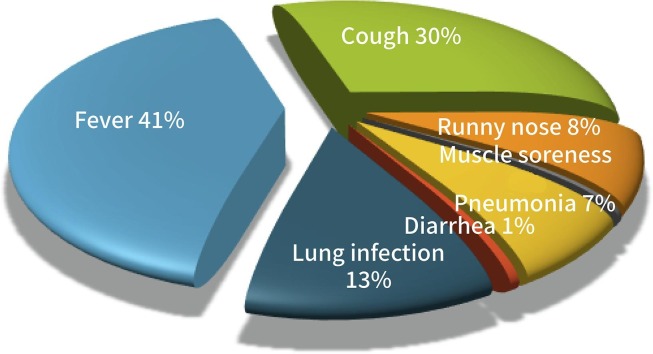
In Fig. 3 , is indicated the frequency of each feature, with most patients displaying fever, cough, lung infection and/or pneumonia. Some patients had a recent travel history; others received treatment in isolation.
Fig. 3.
Illustration of symptoms frequency.
3.2. Machine learning analysis
Firstly we developed a model for our application. In Fig. 1 is shown the pictorial representation of our research. In our workflow, we divided our work into different sections. The first section is data collection, which was described earlier. We prepared our dataset that can be capable to work with different machine learning (ML) approaches.
After preprocessing, we divided our dataset into four criteria (Age 0–20, Age 21–60, Age 61–96 and Age 0–96). We divided our dataset into two parts, one part (70%) for training and another part (30%) for testing. Then we applied the five machine learning algorithms to train our models. The dataset was fitted to ML approaches using the Python programming language (Python 3) (Larose & Larose, 2019). The algorithms used included Decision Tree, Random Forest, XGBoost, Gradient Boosting Machine (GBM) and Support Vector Machine (SVM). Then we analyzed the performances of the algorithms. For each algorithm, we calculated the accuracy of the test dataset. To validate the accuracy, we find confusion matrix, precision, recall, F1-score, AUC and log-loss values. Then we find the feature importance for every algorithm. We calculated the coefficient values for each feature that are significant for COVID-19 patients. Finally, we identified the six most significant features (shown in Table 5) that are strictly related to COVID-19 positive status.
Table 5.
Six most significant features for COVID-19 suspected and confirmed patient with algorithms accuracy.
| Dataset | Algorithm | Accuracy | Top Six Features |
|||||
|---|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | 6th | |||
| Age (0–20) | XGBoost | 0.88 | Lung Infection | Cough | Runny Nose | Pneumonia | Isolation | Fever |
| GBM | 0.89 | Fever | Lung Infection | Travel history | Age | Isolation | Cough | |
| SVM | 0.93 | Pneumonia | Cough | Lung Infection | Fever | Muscle Soreness | Isolation | |
| Random Forest | 0.88 | Age | Lung Infection | Fever | Cough | Travel history | Isolation | |
| Decision Tree | 0.89 | Cough | Lung Infection | Age | Isolation | Travel history | Fever | |
| Age (21–60) | XGBoost | 0.90 | Lung Infection | Cough | Pneumonia | Runny Nose | Muscle Soreness | Age |
| GBM | 0.87 | Cough | Lung Infection | Fever | Travel history | Pneumonia | Runny Nose | |
| SVM | 0.87 | Cough | Lung Infection | Pneumonia | Fever | Gender | Travel history | |
| Random Forest | 0.89 | Cough | Age | Lung Infection | Fever | Travel history | Pneumonia | |
| Decision Tree | 0.89 | Cough | Lung Infection | Age | Gender | Pneumonia | Isolation | |
| Age (61–96) | XGBoost | 0.86 | Lung Infection | Cough | Diarrhea | Pneumonia | Isolation | Runny Nose |
| GBM | 0.84 | Lung Infection | Cough | Fever | Pneumonia | Runny nose | Travel history | |
| SVM | 0.83 | Cough | Lung Infection | Pneumonia | Fever | Isolation | Travel history | |
| Random Forest | 0.85 | Lung Infection | Age | Cough | Fever | Pneumonia | Travel history | |
| Decision Tree | 0.85 | Lung Infection | Cough | Age | Gender | Travel history | Isolation | |
| Age (0–96) | XGBoost | 0.88 | Lung Infection | Cough | Pneumonia | Runny Nose | Diarrhea | Muscle Soreness |
| GBM | 0.86 | Lung Infection | Cough | Fever | Runny Nose | Pneumonia | Travel history | |
| SVM | 0.86 | Pneumonia | Cough | Lung Infection | Fever | Isolation | Gender | |
| Random Forest | 0.86 | Age | Lung Infection | Cough | Fever | Pneumonia | Travel history | |
| Decision Tree | 0.86 | Cough | Lung Infection | Age | Pneumonia | Gender | Isolation | |
In our analysis results, we found that every algorithm achieved 88% (0.88) or above accuracy score. The performances of our used algorithms for the different datasets are described below.
-
•
Age (0–20): In Table 3, we showed the accuracy measurement methods and their score for the age range of 0 to 20 years. The precision value for SVM is 0.92, which was the highest, Random Forest achieves 0.90, XGBoost and GBM scored 0.89 and Decision Tree scores 0.88. The SVM provided the highest recall value with 0.98, and the scores of GBM, XGBoost, Random Forest and Decision Tree were 0.96, 0.94, 0.92 and 0.89 respectively. The F1-Score for SVM was the highest is 0.95, GBM, XGBoost, Random Forest and Decision Tree scores 0.92, 0.91, 0.91 and 0.89. The AUC score of SVM is 0.91 is highest, GBM, XGBoost, Random Forest and Decision Tree score 0.86, 0.85, 0.86, and 0.85. Using Log Loss, the SVM algorithm achieved the lowest, 2.30%. GBM, Random Forest, Decision Tree and XGBoost gain gave 3.68, 4.14, 3.68 and 4.14 percent scores, respectively.
Table 2 shows the coefficient values for every feature. We observed that in the age range of 0 to 20 years muscle soreness, diarrhea, runny nose and gender were the least significant features. As Table 4 indicates, we found that lung infection, cough, fever, age, travel history were the most significant features. The SVM algorithm predicts with 93% accuracy result, which is the highest, and the other algorithms, GBM, XGBoost, Random Forest and Decision Tree perform with accuracies of 89%, 88%, 88% and 89%, respectively.
-
•
Age (21–60): For the age range of 21 to 60 years, the accuracy measurement methods and their score are shown in Table 3. After implementing the algorithms, the precision values for GBM & SVM are 0.98; those are the highest. And XGBoost, Random forest & Decision tree algorithms record 0.95, 0.95 and 0.94 values, respectively. The recall value for Decision Tree is highest is 0.93, and the scores of XGBoost, GBM, Random Forest and SVM are 0.92, 0.86, 0.91 and 0.86, respectively. The F1-Scores for XBGoost, Random Forest and Decision Tree are the highest is 0.93; GBM and SVM achieved 0.91 F1-Score. The AUC values of GBM, SVM and Decision Tree are 0.89 is the highest value; XGBoost and Random Forest scored 0.87. XGBoost algorithm achieved score 3.57% Log Loss evaluation criteria, which is the lowest relative to the other algorithms. GBM, SVM, Random Forest and Decision Tree scored 4.35%, 4.45%, 3.74% and 3.74%, respectively.
In Table 2, we present the coefficient values for each feature and in Table 4 is shown the most significant features. For this age range, the most significant features were cough, lung infection, travel history, fever and pneumonia symptoms.
-
•
Age (61–96): In the data presented in Table 3, the age range was restricted to 61 to 96 years and then analyzed as previously described for the five machine learning algorithms. For this data, the precision value was highest for SVM with 0.93, the second highest was GBM which is 0.90. XGBoost, Random Forest, and Decision Tree algorithms scored 0.87, 0.89 and, 0.88, respectively. The recall value of XGBoost, GBM, SVM, Random Forest and Decision tree was 0.90, 0.87, 0.80, 0.88, 0.89, respectively. Random Forest algorithms achieved a 0.89 score, which was the highest value for F1-score; XGBoost, GBM and, Decision Tree gained 0.88 values and SVM gained 0.86. The AUC values for XGBoost, GBM, SVM, Random Forest and Decision Tree are 0.82, 0.84, 0.84, 0.84 and 0.83. The lowest value for Log Loss metrics is 5.08%, which is achieved by the Random Forest algorithm; XGBoost, GBM, SVM, Decision Tree gained 5.42%, 5.25%, 5.93, and 5.25% scores, respectively.
The coefficient values of every feature were consistent in finding the most significant features for this age range were lung infection, cough, fever, travel history, and pneumonia.
-
•
Age (0–96): On the accuracy measurement Table 3, the results for individuals in the age range 0 to 96 years is indicated. We observed that the GBM and SVM algorithms achieved the highest accuracy 0.97 using precision evaluation metrics. XGBoost, Random Forest and Decision Tree showed 0.93, 0.92, & 0.91 accuracy. On the other hand, XGBoost gained the highest 0.91 score using recall evaluation metrics and GBM, SVM, Random Forest and Decision Tree achieved 0.85, 0.84, 0.89, 0.90 scores, respectively. XGBoost scored 0.92 using F1- Score, which is the highest value and GBM, SVM, Random Forst and Decision Tree obtained 0.91, 0.90, 0.91 and 0.91 respectively. The AUC value for XGBoost, GBM, SVM, Random Forest and Decision Tree were 0.85, 0.88, 0.88, 0.83 and 0.82. XGBoost had the lowest value 4.08% from Log Loss metrics and GBM, SVM, Random Forest and Decision Tree gave scores of 4.61, 4.93, 4.74 and 4.72 scores, respectively.
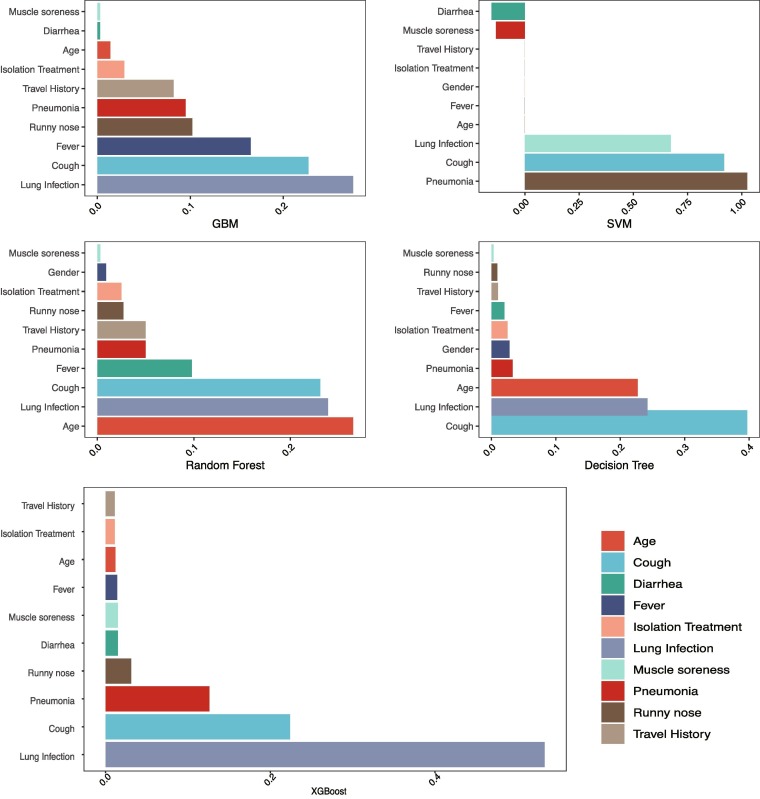
We also analyzed the same parameters using the whole dataset combined (age 0–96 years). We compared combined outcomes with individual outcomes, and we found that there were a few variations in the different age groups, such as lung infection and cough are most significant for all types of age groups. However, in age group 0–20, fever and isolation treatment, in the age group 21–60 and 61–96, fever and pneumonia, in the age group 0–96, age, runny nose and pneumonia were also significant with a lung infection and cough.
Table 2.
Association between patient’s COVID-19 confirmation and selected demographic information including symptoms.
| All patients n = 6,512 | Suspected n = 4,940 | Confirmed n = 1,572 | P value | |
|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | ||
| Demographic information | ||||
| Age, median (IQR), y | 43(32–55) | 43(32–55) | 45(33–57) | <0.001 |
| Gender | ||||
| Male | 3,367(51.70) | 2,564(51.9) | 803(51.08) | – |
| Travel history | 4,239(65.1) | 3,658(74.05) | 581(36.96) | <0.001 |
| Isolation treatment | 1,413(21.7) | 802(16.23) | 611(38.87) | <0.001 |
| Symptoms | ||||
| Fever | 2,675(41.1) | 1,433(29.01) | 1,242(79.01) | <0.001 |
| Cough | 1,975(30.3) | 787(15.93) | 1,188(75.57) | <0.001 |
| Runny nose | 549(8.43) | 47(0.95) | 502(31.93) | <0.001 |
| Muscle soreness | 26(0.4) | 19(0.39) | 7(0.45) | 0.9182 |
| Pneumonia | 487(7.48) | 85(1.72) | 402(25.57) | <0.001 |
| Lung infection | 855(13.1) | 69(1.4) | 786(50) | <0.001 |
| Diarrhea | 37(0.57) | 31(0.63) | 6(0.38) | 0.3488 |
| Have symptoms | 2,971(45.6) | 1,505(30.47) | 1,466(93.26) | – |
Table 4.
Coefficient values for each features for each ML approaches.
| Dataset | Algorithm | Features |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Age | Fever | Cough | Runny Nose | Muscle Soreness | Pneumonia | Diarrhea | Lung Infection | Travel | Isolation | ||
| Age (0–20) | XGBoost | 0.086 | 0.017 | 0.043 | 0.216 | 0.111 | 0 | 0.106 | 0 | 0.402 | 0.031 | 0.052 |
| GBM | 0.034 | 0.132 | 0.256 | 0.063 | 0 | 0.05 | 0.05 | 0.002 | 0.184 | 0.144 | 0.08 | |
| SVM | 0 | 0 | 0 | 0.858 | −1.027 | 0 | 0.98 | −0.164 | 0.629 | 0 | 0 | |
| Random Forest | 0.044 | 0.207 | 0.152 | 0.149 | 0.016 | 0 | 0.028 | 0.005 | 0.191 | 0.111 | 0.096 | |
| Decision Tree | 0.017 | 0.186 | 0.039 | 0.333 | 0.002 | 0 | 0.023 | 0 | 0.208 | 0.088 | 0.1 | |
| Age (21–60) | XGBoost | 0.004 | 0.013 | 0.013 | 0.26 | 0.026 | 0.017 | 0.139 | 0.009 | 0.494 | 0.013 | 0.01 |
| GBM | 0.002 | 0.015 | 0.146 | 0.261 | 0.08 | 0.005 | 0.087 | 0.001 | 0.243 | 0.134 | 0.025 | |
| SVM | 0 | 0 | 0 | 0.913 | −0.547 | −0.146 | 0.63 | −0.136 | 0.662 | 0 | 0 | |
| Random Forest | 0.011 | 0.237 | 0.103 | 0.251 | 0.027 | 0.005 | 0.043 | 0.002 | 0.233 | 0.065 | 0.025 | |
| Decision Tree | 0.031 | 0.198 | 0.018 | 0.422 | 0.008 | 0.006 | 0.03 | 0.002 | 0.246 | 0.018 | 0.021 | |
| Age (61–96) | XGBoost | 0.016 | 0.013 | 0.022 | 0.149 | 0.022 | 0 | 0.114 | 0.114 | 0.475 | 0.021 | 0.023 |
| GBM | 0.021 | 0.065 | 0.116 | 0.19 | 0.093 | 0.003 | 0.101 | 0.012 | 0.278 | 0.073 | 0.046 | |
| SVM | 0 | 0 | 0 | 0.966 | −0.589 | −0.099 | 0.557 | −0.441 | 0.717 | 0 | 0 | |
| Random Forest | 0.036 | 0.234 | 0.086 | 0.194 | 0.031 | 0.001 | 0.057 | 0.009 | 0.247 | 0.055 | 0.045 | |
| Decision Tree | 0.055 | 0.174 | 0.026 | 0.248 | 0.011 | 0.002 | 0.025 | 0.009 | 0.38 | 0.041 | 0.029 | |
| Age (0–96) | XGBoost | 0.005 | 0.012 | 0.014 | 0.224 | 0.031 | 0.015 | 0.126 | 0.015 | 0.533 | 0.011 | 0.011 |
| GBM | 0.001 | 0.014 | 0.165 | 0.227 | 0.102 | 0.003 | 0.095 | 0.003 | 0.275 | 0.082 | 0.029 | |
| SVM | 0 | 0 | 0 | 0.918 | −0.556 | −0.133 | 1.025 | −0.154 | 0.673 | 0 | 0 | |
| Random Forest | 0.009 | 0.265 | 0.098 | 0.231 | 0.027 | 0.003 | 0.05 | 0.003 | 0.239 | 0.05 | 0.025 | |
| Decision Tree | 0.028 | 0.227 | 0.02 | 0.397 | 0.009 | 0.003 | 0.033 | 0.003 | 0.242 | 0.01 | 0.025 | |
Fig. 4 shows the feature ranking according to coefficient values for each applied algorithm. Every algorithm found almost the same sequence of features for all the age groups.
Fig. 4.
Feature importance for COVID-19 patients.
From the above analysis, we also found that among those who displayed a fever, they had body temperatures equal to or above 38-degree centigrade. A small number of individuals also presented with chest tightness. Some patients had a cough with sputum or dry cough, nasal congestion, fatigue, discomfort, pharyngeal discomfort, respiratory symptoms, shortness of breath, headache, dizziness, weakness, nausea, among other symptoms.
4. Discussion and conclusion
The development of the COVID-19 pandemic currently represents a dangerous threat to global health. The key to stopping this spread is the development of methods to identify infected individuals as early as possible. This can be challenging given the delay in symptom presentation; however, machine learning algorithms provide a promising approach to address this problem that can be rapidly and cheaply applied in a pandemic situation. In our study, we developed and tested a range of machine learning approaches and found the most significant clinical COVID-19 predictive features were (in descending order): lung infection, cough, pneumonia, runny nose, travel history, fever, isolation, age, muscle soreness, diarrhea, and gender. Our models were able to predict the stage of COVID-19 based on basic patient information (age and gender), travel and isolation, and clinical symptoms (including fever, cough and runny nose and pneumonia). The accuracy of our algorithms was highest for the age range 0–20 years, with the SVM algorithm with 93% accuracy, but it was notable that the other algorithms performed almost as well with greater than 85% accuracy. In the age range 21 to 60 years the situation was similar, with the highest accuracy of 90% of XGBoost, and others(e.g. SVM, Random Forest and GBM and Decision Tree algorithms) also performed well. In the age range of 61 to 96 years, again XGBoost achieved 86% accuracy but the others gave above 80% accuracy. As might be expected given similar results across different ages (indicating that the symptoms develop similarly in individuals of any age) this pattern is also seen when the whole range of 0 to 96 years was studied and also get above 85% accuracy of prediction. Accordingly, we were able to rank the features that are of importance to the disease prediction.
According to the statistics, the median age was 43 years with IQR 32–55, composed of approximately half males and half females. Most of the patients presented with fever, cough and radio-graphic chest imaging results that indicated that around 50% of confirmed patients had one or both lungs affected by the infection. In suspected patients, 29.01% were affected with fever, whereas 79.01% confirmed patients have fever & 75.57% have a cough. Travel history was notable for being one of the major associated features to COVID-19 infection, as would be expected with 65.1% of patients having recently travelled a long distance. Some other symptoms were also related to COVID-19 status but were less commonly seen, including muscle soreness and diarrhea; these features, particularly diarrhea, were much more prominent in the earlier SARS epidemic. However, it is striking that 6.74% of the confirmed COVID-19 positive and 69.53% of the suspected patients did not develop any type of symptoms. As these patients cannot be detected or predicted by symptoms alone, our machine learning approach is of no use for assessing these people, although it is possible that they may have other factors that may lend themselves to detection in this way. However, the importance of particular social factors are likely to vary over time; notably, foreign travel may come to be less critical as local community transmission becomes the most common form of infection. Contact with infected individuals would be and remains an excellent predictor, but this relies on rigorous contact tracing and social network analysis. Mann–Whitney U test and chi-square tests indicated that all the features were impacted except muscle soreness and diarrhea. These significant symptoms matched with findings from our machine learning analysis.
We implemented machine learning algorithms on different clinical features of patients with COVID-19 infections in a new dataset from mainland China and utilized different classifiers to examine information criterion and assess performance. Our ability to predict the probability and course of COVID-19 infection will improve the capacity of doctors to identify infected patients at an early stage by utilizing predictor clinical features. Some of the classifiers did not, however, show reliable outcomes, presumably because while they demonstrated exactitude, they created one-sided results for these datasets. However, the size of the COVID-19 dataset was probably not extensive enough to give enough statistical power to resolve these issues. In future studies, using much larger datasets, we will have improved capacity to circumvent these limitations and further improve our predictive accuracy.
CRediT authorship contribution statement
Md. Martuza Ahamad: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Sakifa Aktar: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Md. Rashed-Al-Mahfuz: Investigation, Validation, Writing – original draft. Shahadat Uddin: Formal analysis, Validation, Writing – original draft, Writing – review & editing. Pietro Liò: Validation, Writing – original draft. Haoming Xu: Data curation, Supervision, Visualization , Writing – original draft, Writing – review & editing. Matthew A. Summers: Validation, Writing – original draft, Writing – review & editing. Julian M.W. Quinn: Validation, Writing – original draft, Writing – review & editing. Mohammad Ali Moni: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Agarwal, R. (2019). The 5 Classification Evaluation metrics every Data Scientist must know.https://towardsdatascience.com/the-5-classification-evaluation-metrics-you-must-know-aa97784ff226 Accessed 18 April 2020.https://www.aljazeera.com/news/2020/01/countries-confirmed-cases-coronavirus-200125070959786.html Accessed 18 April 2020.
- BDBC-KG-NLP/COVID-19-tracker. GitHub. (2020).https://github.com/BDBC-KG-NLP/COVID-19-tracker Accessed 20 February 2020.
- Biau G., Cadre B., Rouviére L. Accelerated gradient boosting. Machine Learning. 2019;108:971–992. doi: 10.1007/s10994-019-05787-1. [DOI] [Google Scholar]
- Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020;395:514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining – KDD. 2016. XGBoost: A scalable tree boosting system; p. 16. [DOI] [Google Scholar]
- Gorbalenya, A. E., Gulyaeva, G. A., Lauber, C., Sidorov, I. A., Leontovich, A. M., Penzar, D., et al. (2020). The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5, 536–544.https://doi.org/10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed]
- Gupta, M. (2019). ML: Feature Scaling – Part 2. GeeksforGeeks.https://www.geeksforgeeks.org/ml-feature-scaling-part-2/ Accessed 18 April 2020.
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M., Rahman R.M. Decision tree and Naïve Bayes algorithm for classification and generation of actionable knowledge for direct marketing. Journal of Software Engineering and Applications. 2013;06:196–206. doi: 10.4236/jsea.2013.64025. [DOI] [Google Scholar]
- Kiapour A. Bayes, E-Bayes and robust Bayes premium estimation and prediction under the squared log error loss function. Journal of the Iranian Statistical Society. 2018;17:33–47. doi: 10.29252/jirss.17.1.33. [DOI] [Google Scholar]
- Larose C.D., Larose D.T. Wiley; Hoboken: 2019. Data science using Python and R. (Chapter 6) [Google Scholar]
- Li F., Li Y.-Y., Wang C. Uncertain data decision tree classification algorithm. Journal of Computer Applications. 2009;29:3092–3095. doi: 10.3724/sp.j.1087.2009.03092. [DOI] [Google Scholar]
- Lippi G., Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clinica Chimica Acta. 2020;505:190–191. doi: 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naming the coronavirus disease (COVID-19) and the virus that causes it. World Health Organization. (2020).http://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Accessed 18 April 2020.
- Nishiura H., Jung S.-M., Linton N.M., Kinoshita R., Yang Y., Hayashi K. The extent of transmission of novel coronavirus in Wuhan, China, 2020. Journal of Clinical Medicine. 2020;9:330. doi: 10.3390/jcm9020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. Jama. 2020 doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? International Journal of Epidemiology. 2020 doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarica A., Cerasa A., Quattrone A. Random forest algorithm for the classification of neuroimaging data in Alzheimers disease: A systematic review. Frontiers in Aging Neuroscience. 2017;9 doi: 10.3389/fnagi.2017.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., (2019). Mathematics behind Random forest and XGBoost.https://medium.com/analytics-vidhya/mathematics-behind-random-forest-and-xgboost-ea8596657275 Accessed 18 April 2020.
- Tian S., Hu N., Lou J., Chen K., Kang X., Xiang Z. Characteristics of COVID-19 infection in Beijing. Journal of Infection. 2020;80:401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C., & Hui-Mei, Y. (2014). An improved GA-SVM algorithm. In 2014 9th IEEE Conference on Industrial Electronics and Applications.https://doi.org/10.1109/iciea.2014.6931525.
- Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., Salazar P.M.D. Nature Medicine; China: 2020. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A., Aladinskiy V., Zhebrak A., Zagribelnyy B., Terentiev V., Bezrukov D.S. Potential 2019-nCoV 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches. BioRxiv. 2020 doi: 10.26434/chemrxiv.11829102.v1. [DOI] [Google Scholar]