Abstract
Purpose
Tumors often have different imaging properties and there is no single imaging modality that can visualize all tumors. In CT guided needle placement procedures, image fusion (e.g. with MRI, PET or contrast CT) is often used as image guidance when the tumor is not directly visible in CT. In order to achieve image fusion, interventional CT image needs to be registered to an imaging modality, in which the tumor is visible. However, multi-modality image registration is a very challenging problem. In this work, we develop a deep learning based liver segmentation algorithm and use the segmented surfaces to assist image fusion with the applications in guided needle placement procedures for diagnosing and treating liver tumors.
Methods
The developed segmentation method integrates multi-scale input and multi-scale output features in one single network for context information abstraction. The automatic segmentation results are used to register an interventional CT with a diagnostic image. The registration helps visualize the target and guide the interventional operation.
Results
The segmentation results demonstrated that the developed segmentation method is highly accurate with Dice of 96.1% on 70 CT scans provided by LiTS challenge. The segmentation algorithm is then applied to a set of images acquired for liver tumor intervention for surface-based image fusion. The effectiveness of the proposed methods is demonstrated through a number of clinical cases.
Conclusion
Our study shows that deep learning based image segmentation can obtain useful results to help image fusion for interventional guidance. Such a technique may lead to a number of other potential applications.
Keywords: Image fusion, image segmentation, deep learning, image guided interventions
1. Introduction
Image-guided biopsy and ablation procedures are increasingly used for minimally invasive, local treatment of deep space target tumors in the liver [1]. Despite the increasing availability of alternative imaging techniques for interventional guidance, such as high-quality ultrasound systems and innovative magnetic resonance imaging–compatible guidance systems, computational tomography (CT) remains an important imaging technology for guidance during percutaneous procedures [14]. In fact, the number of CT-guided procedures performed by interventional radiology has increased partly due to the advances in CT imaging technologies [7].
During CT guided procedures, a navigation system uses interventional CT images to show the spatial relationship between devices (e.g. biopsy needle or ablation catheter). However, tumors often have different imaging properties and there is no single imaging modality that can visualize all tumors. When target lesions are not visible in interventional CT images, fusing the interventional CT with different pre-procedural diagnostic images can be very useful to visualize both lesion and interventional devices. In CT guided needle placement procedures, image fusion (e.g. with MRI, PET or contrast CT) is often used as image guidance when the tumor is not directly visible.
In order to achieve image fusion, interventional CT image usually needs to be registered to a pre-procedural image of a different imaging modality, in which the tumor is visible. However, A robust and fast multi-modality image registration is a very challenging problem [9, 19]. Methods for multi-modality image registration, depending on how they exploit the image information, can be divided into intensity based and feature based approaches. The main idea of intensity based registration is to search iteratively for geometric transformation that, when applied to moving imaging modality, optimizes a similarity measure. However, manual and intensity based registration are not robust and can easily fail during the intervention. If these algorithms fail, there is often no time to adjust the parameters and run the algorithms again. Especially, these algorithms often fails due to the large deformation and appearance difference between interventional CT and the imaging modality, in which the tumor is visible. More robust registration algorithm should be used during the intervention to minimize the impact on the clinical workflow. Feature based methods, on the other hand, provide a better solution to focus on local structures [2, 12]. Local representative features are first extracted from images and matched to compute the corresponding transformation. However, matching the feature points itself can be challenging. Using the segmented surfaces of the regions of interest can help robustly register the two modalities. In those methods [2, 12], transform computed from point to point alignment between the surfaces is used to register the corresponding images. Such methods, however, require accurate and fast image segmentation to begin with.
Automatic and robust liver segmentation from CT volumes is a very challenging task due to low intensity contrast between liver and neighboring organs. State-of-the-art medical image segmentation framework are mostly based on deep convolutional neural networks (CNN) [16]. The receptive field increases as convolutional layers are stacked. U-Net, introduced by Ronneberger et al.[21] is the most widely used network architecture for biomedical image segmentation. Incorporating the latest CNN structures into U-Net were the most common changes to the basic U-Net architecture for liver segmentation [3]. For example, Han [8] won ISBI 2017 LiTS Challenge1 by replacing the convolutional layers in U-net with residual blocks from ResNet [11]. For encoder-decoder network architectures like FED-Net [5], the residual connection is integrated into 2D network in which low-level fine appearance information is fused into coarse high-level features through the attention gates between shallow and deep layers. H-DenseUNet [17] proposes to use hybrid features to extract volumetric information. DeepX [24] uses a 29 layer encoder-decoder network for liver segmentation. Most of these works [8, 17, 24] adopt two-step approach to segment the liver, where a coarse step first localize the liver and the other model does the fine segmentation. Specifically, state-of-the-art liver segmentation like the one in [17] require deep 3D neural networks. Although promising liver segmentation can be obtained, the two-step segmentation and 3D convolutions have high demands on the computational environment when transferring to clinical use. Furthermore, the existing liver segmentation methods are mainly developed for segmenting diagnostic CT images, which may not be well suited for interventional CT image segmentation. In order to meet the requirement for interventional use, a method needs to be not only accurate for liver segmentation, but also able to robustly handle various patient positions used for better guidance access in high speed. Multi-scale mechanism, which utilizes contextual information, have shown consistent significant improvement in liver segmentation [6]. Thus, in this work, we propose to use a multi-scale input and multi-scale output feature abstraction network (MIMO-FAN) architecture for 2.5D segmentation of the liver. The network takes three consecutive slices as input and extracts multi-scale appearance features from the beginning. After going through a series of convolutional layers, the multi-scale features are adaptively fused at the end for segmentation. As a result, in our experiments, the developed MIMO-FAN demonstrates a high segmentation accuracy for one-step fast liver segmentation.
In summary, we present a deep learning based liver segmentation method for a new workflow of fusion guided intervention. The proposed liver segmentation algorithm segments liver surfaces of interventional CT and thus enables accurate surface based registration of pre-operative diagnostic image and interventional CT. The performance of MIMO-FAN is validated in both public dataset and our own interventional CT images. The application of our developed registration technique is demonstrated through the fusion of various modalities with interventional CT.
2. Methods
This section presents the details of the proposed deep learning segmentation algorithm and the image fusion workflow. Fig. 1 shows an overview of the developed framework, where our proposed MIMI-FAN is used for automatically segmenting an interventional CT and the result is used for surface-based intra-procedural multi-modal image registration.
Fig. 1.
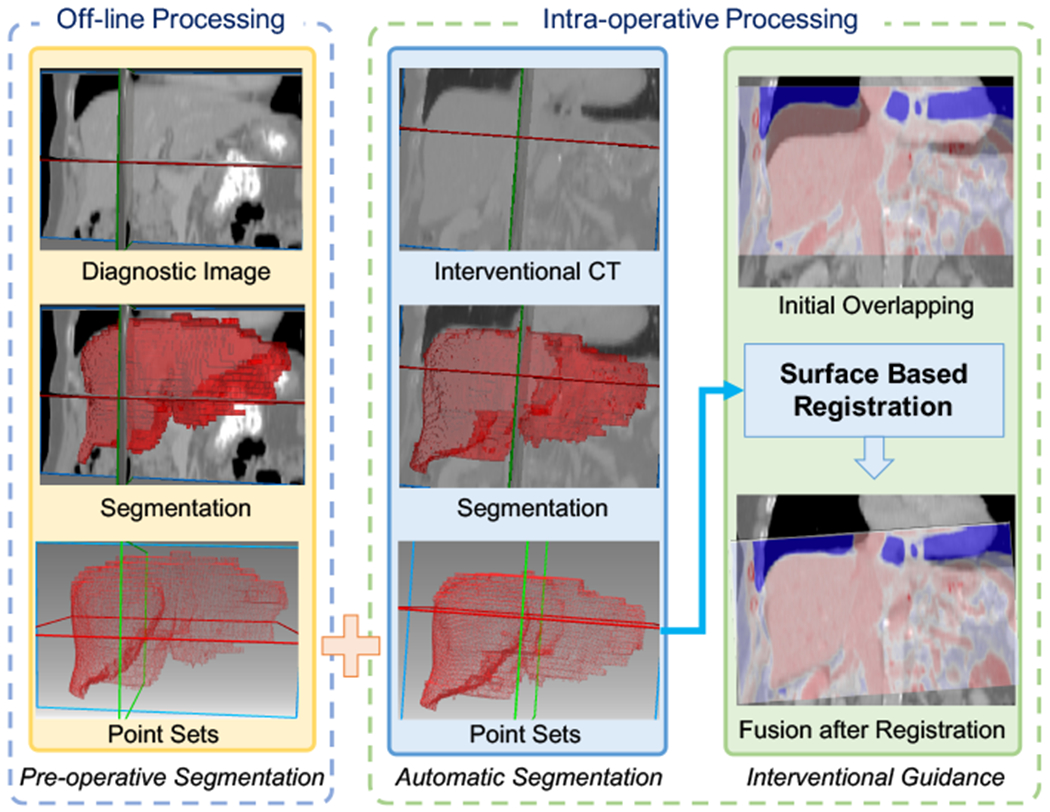
Illustration of the developed image fusion method. Pre-operative diagnostic image is segmented under off-line processing. During intervention, our designed AI model make an automatic and fast segmentation on the interventional CT. The segmentation results are represented in point sets. Then iterative closest point (ICP) algorithm is used to perform surface-based registration to align the pre-operative image and interventional CT in the same coordinate.
2.1. MIMO-FAN for Liver Segmentation
To efficiently exploit the image information for segmentation, in this paper, we design a novel 2.5D deep learning network that uses pyramid input and output architecture to fully abstract multi-scale features. As shown in Fig. 2, the proposed network integrates multi-scale mechanism into a U-shape architecture, which enables the network to extract multi-scale features from the beginning to the end.
Fig. 2.
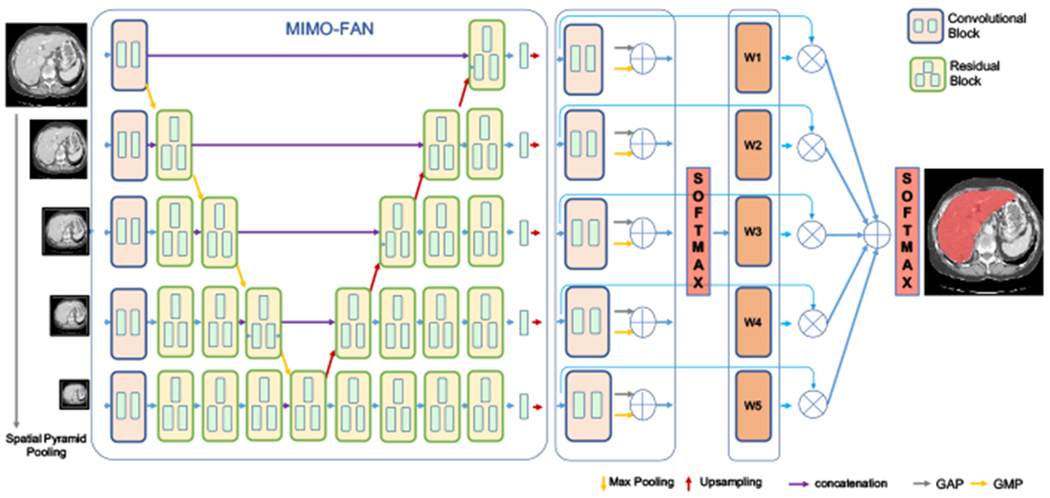
Overview of proposed architecture. Information propagated from multi-scale inputs to hierarchically combination of semantic similar features. Multi-scale segmentation-level features are fused by learnt adaptive weights from a shared convolutional block.
The proposed MIMO-FAN first performs multi-scale analysis to the three consecutive input slices by using spatial pyramid pooling [10] to obtain scene context information. After the first level convolutional blocks with shared kernels, image-level contextual features that interpret the overall scene can be extracted from these inputs in different scales. To fuse features from different scales, a notable feature of MIMO-FAN is that features to be fused at a certain level all go through the same number of convolutional layers, which helps to keep the hierarchical semantic similar feature. Unlike the classical U-net based methods [21], where the scale only reduces when the convolutional depth increases, MIMO-FAN has multi-scale features at each depth and therefore both global and local context information can be fully integrated to augment the extracted features. Furthermore, inspired by the work of deep supervision [23], we further introduce deep pyramid supervision (DPS) to the decoding side for generating and supervising outputs of different scales, which helps to alleviate the gradient vanishing problem and generate good segmentation masks at different scales. DPS also ensures the semantic similar features are learned in the same depth. The training loss is computed by using the output and ground truth segmentation at the same scale. Weighted cross entropy is used as the loss function in our work, which is defined as
| (1) |
where denotes the predicted probability of voxel i belonging to class c (background or liver) in scale s, is the ground truth label in scale s, Ns denotes the number of voxels in the scale s, and is weighting parameter for different classes.
To effectively take advantage of the segmentation-level features from different scales, we design an adaptive weight layer (AWL), which makes use of attention mechanism to learn relative importance of each scale driven by context and fuse the score maps in an automatic and elastic fashion. The score maps are first passed into a shared convolutional block and squeezed into a single channel feature vector. In the convolutional block, the first layer has 2 filters with kernel size 3 × 3 and the second layer has 1 filters with kernel size 1 × 1 to squeeze channel number into one for scale information. To obtain global value for each scale, global average pooling (GAP) and global max pooling (GMP) are applied on the single channel features to extract global feature of each scale. In this work, their sum is applied to extract the global information of each scale. The values from different scales are then concatenated and fed into a softmax layer to get the weights of each scales. The sum of these weight values equals to 1. After resampling to the original image size, the score maps are weighted and summed to be the final score map. Another softmax layer is applied and the threshold value of 0.5 is used to obtain the prediction.
2.2. Surface based Registration for Image Fusion
Surface based registration of CT images and patient’s anatomy in physical space has shown good application to image-guided surgery [12]. It allows physicians determine the position and orientation of surgical tools relative to vertebral anatomy. Different surface based methods can be used for image fusion. In this work, we perform surface-based registration using an independent implementation of iterative closest point (ICP) registration algorithm of Besl and McKay [2]. After segmenting the target organs from interventional CT and preoperative imaging, in which the tumor is visible, point sets will be obtained from the segmentations to compute surface based registration. The standard ICP method and most variants implicitly assume an isotropic noise model [4]. Selected points from these contours were rotated and translated in the x,y and z directions, and zero-mean, normally distributed, isotropic noise was added to the rotated points to simulate a surface acquired in a different imaging modality [12]. We treat the interventional CT as the reference imaging modality and the other as moving imaging modality. The method is a two-step process. Principal component analysis (PCA) alignment is first used to obtain initial guess of correspondences. Then singular value decomposition (SVD) iteratively improves the correspondences. In the iteration, for each transformed source point, the closest target point is assigned as its corresponding point. To evaluate registration error obtained with the proposed method, root mean squared (RMS) distance is used. The optimization stops when a terminal criterion is met. The ICP algorithm always converges to the nearest local minimum with respect to the object function.
3. Experiments
This section presents the details of our experiments and the results. We first present the materials used for training and validating our algorithms. Three clinical cases of image fusion are then demonstrated.
3.1. Materials
For image segmentation, we extensively evaluated our method on the LiTS (Liver tumor segmentation challenge2) dataset. LiTS is the largest liver segmentation dataset that is currently publicly available. The data are composed of 131 training and 70 test datasets. The data were collected from different hospitals and the resolution of the CT scans vary between 0.45mm and 6mm for intra-slice and between 0.6mm and 1.0mm for inter-slices, respectively. To validate the diversity of the data, Li et al. [14] apply model trained on LiTS data on another datasets (3DIRCADb) and obtain state-of-the art liver segmentation performance (dice 0.982) on the dataset.
The clinical datasets used for image fusion are from the Clinical Center at the National Institutes of Health. Pre-procedural diagnostic images were acquired for each patient. In our study, such modalities include Magnetic Resonance Imaging (MRI), Positron Emission Tomography - Computed Tomography (PET/CT), and Contrast-Enhanced CT (CE-CT). The exact modality varies depending on the clinical needs and the tumor characteristics. During the CT-guided procedures, interventional CT scans are obtained to visualize needles or catheters relative to the anatomy. By performing fusion of the interventional CT with the pre-procedural image, we were then able to clearly display the relative spatial relationship between the target regions and the interventional devices. Three different clinical cases are used to demonstrate the effectiveness of the developed techniques.
3.2. Implementation Details of Deep Learning
The proposed MIMO-FAN can be considered as a 2.5D segmentation approach, since it takes three consecutive slices as its input to enhance the spatial dependency. The implementation is based on the open-source platform PyTorch [20]. All the convolutional operations are followed by batch normalization and ReLU activation. Weighted cross entropy is used as the loss function in our work. Empirically, we set the weights of 0.2 and 1.2 for the background and the liver, respectively. For network training, we use the RMSprop optimizer. We set the initial learning rate to be 0.002 and the maximum number of training epochs to be 2500. The learning rate decays by 0.01 after every 40 epochs. For the first 2000 epochs, deep supervised losses are applied to focus on MIMO’s feature abstraction ability on each scale. For the remaining 500 epochs, adaptive weighting layer are introduced and only this layer for fusing multi-scale features is trained. We only keep the CT imaging HU values in the range of [−200, 200] to have a good contrast on the liver. For each epoch, we randomly crop a patch with size of 224×224×3 from each volume as input to the network. During testing, four patches are cropped from one slice and segmented, then recombined into one probability map of the slice. All segmented slices are then combined as the segmentation volume. After obtaining the segmentation volume, the connected component analysis was performed to divide all labeled voxels into different connected components, only the largest component is kept as the final segmentation result.
3.3. Segmentation Results
Most of the state-of-the-art methods on liver CT image segmentation have two steps to complete the segmentation, where a coarse segmentation is used to locate the liver followed by fine segmentation step to obtain the final segmentation [8, 17]. However, such two-step methods can be computationally expensive and thus time consuming, which may add delay to clinical procedures. For example during training, the method in [17] takes 21 hours to finetune a pretrained 2D DenseUNet and another 9 hours to finetune the H-DenseUNet with two Titan Xp GPUs. In contrast, our proposed method can be trained on a single Titan Xp GPU in 3 hours. More importantly, when segmenting a CT volume, our method only takes 0.04s for one slice on a single GPU, which is, to the best of our knowledge, the fastest segmentation method compared to other reported methods. In the same time, we are able to obtain the same performance measured by Dice similarity and even better symmetric surface distance (SSD), which computes Euclidean distances from points on the boundary of segmented region to the boundary of the ground truth, and vice versa. The Average SSD, Maximum SSD, Minimum SSD of our algorithm and H-DenseUNet are 1.413, 24.408, 2.421 and 1.450, 27.118, 3.150 respectively. Table 1 shows the performance comparison with other published state-of-the-art methods on LiTS challenge test dataset. Despite its simplicity, our proposed 2D network segments the liver in a single step and can obtain a very competitive performance with less than 0.2% drop in Dice, compared to the top performing method – DeepX [24] on the leader board.
Table 1.
Comparison of segmentation accuracy on the test dataset. Results are from the challenge website (accessed on September 11, 2019).
We further compared our proposed MIMO-FAN against several other classical 2D segmentation networks, including U-Net [21], ResU-Net [8], and DenseU-Net [17], to demonstrate the effectiveness of DPS and AWL. Some example results are shown in Fig. 3. Our MIMO-FAN is based on UNet[21] and ResU-Net[8], so we use them for ablation study. For fair comparison, all these networks are 19-layer networks. The DenseU-Net is the same architecture as 2D DenseU-Net in [17] and the encoder part is Densenet-169 [13]. All these 2D networks are trained from scratch in the same environment. We evaluate the performance of the above networks on LiTS challenge training dataset through five-fold cross validation. We also included one open sourced Nvidia Clara AIAA model ”segment_ct_liver_and_tumor” [18] for comparison on liver segmentation, which is integrated in 3D slicer [15]. Segmented tumor and liver are merged into the whole liver. Some example results are shown in Fig. 4. The five-fold cross validation results are shown in Table 2. The conducted one-tailed t-test on these paired sample shows that MIMO-FAN significantly outperforms Nvidia AIAA Clara, U-Net, ResU-Net and DenseU-Net with p-values of 0.0006, 0.0003, 0.0056, and 0.0004, respectively, all less than 0.01.
Fig. 3.
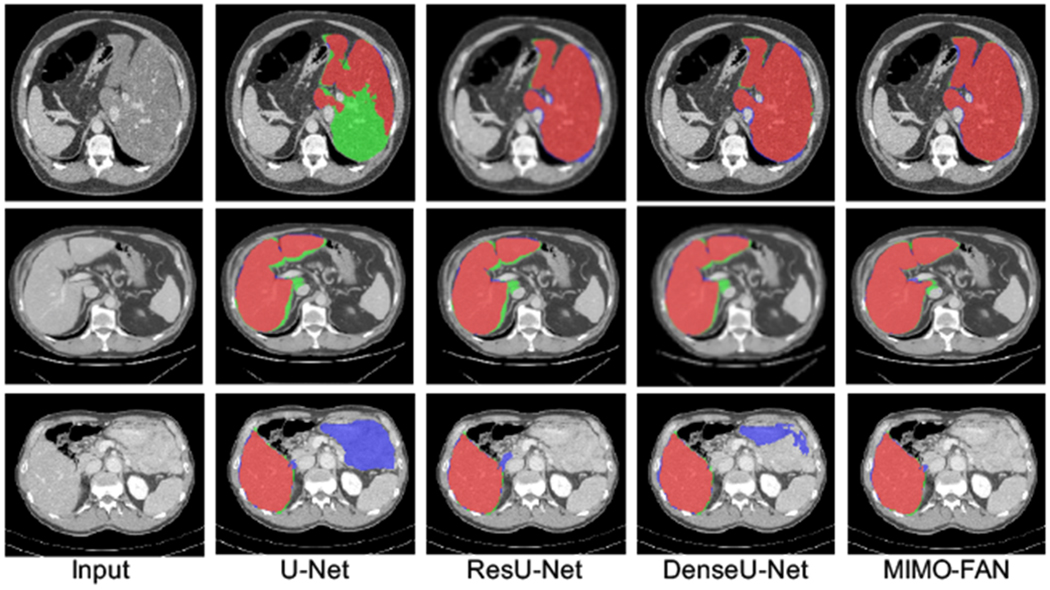
Segmentation examples of different methods. Red color depicts the correctly segmented liver area, blue shows the false positive, and green indicates the false negative.
Fig. 4.
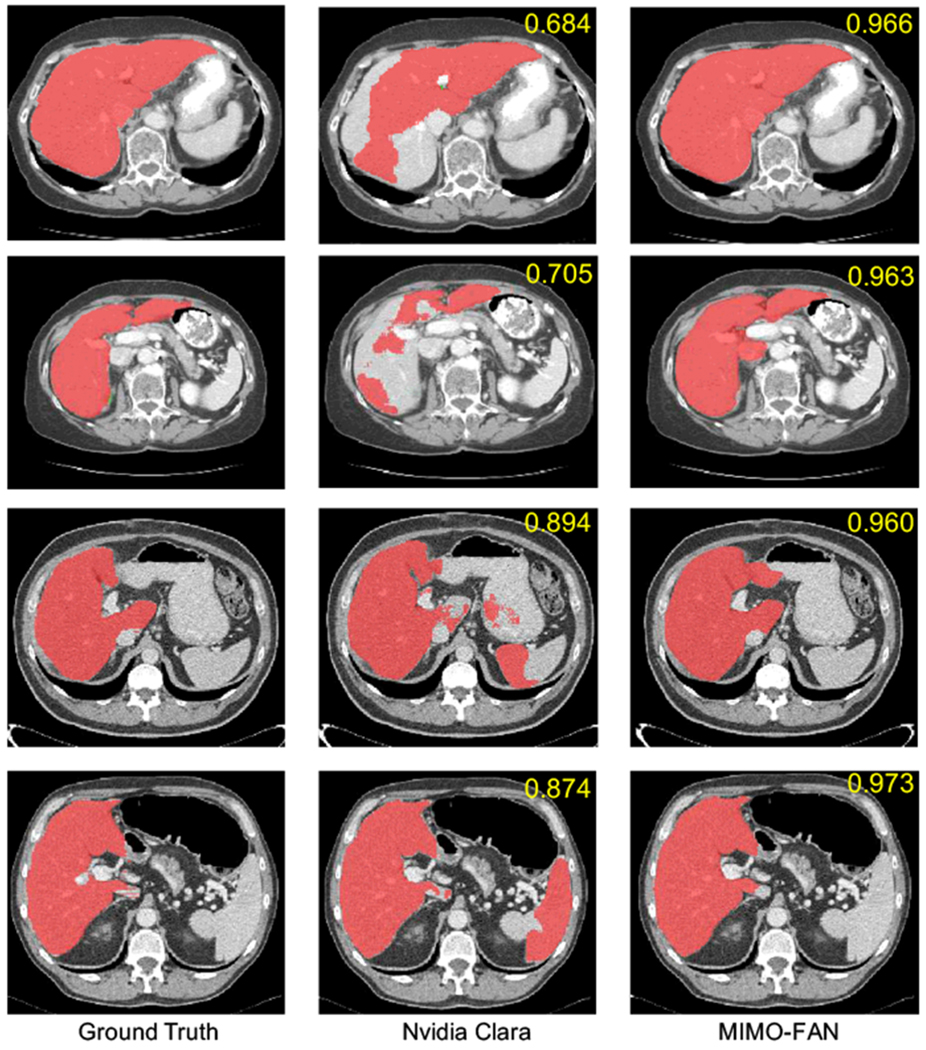
Comparison of our algorithm with open sourced Nvidia AIAA Clara model. From the left to right are ground truth, segmentation examples of Clara model and our MIMO-FAN. Dice accuracy of each volume is labeled in upper right corner.
Table 2.
Network ablation study using five-fold cross validation (Dice %)
| Architecture | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Mean±std |
|---|---|---|---|---|---|---|
| Nvidia Clara | 82.0 | 84.5 | 84.2 | 75.7 | 84.4 | 82.2 ± 3.76 |
| U-Net [21] | 94.5 | 93.8 | 94.1 | 93.0 | 94.1 | 93.9 ± 0.50 |
| ResU-Net [8] | 94.5 | 94.1 | 94.9 | 92.4 | 94.5 | 94.1 ± 0.88 |
| DenseU-Net [17] | 94.1 | 94.2 | 93.9 | 93.6 | 94.5 | 94.1 ± 0.30 |
| MIMO-FAN (DPS) | 95.7 | 95.1 | 95.1 | 94.5 | 96.1 | 95.3 ± 0.62 |
| MIMO-FAN (DPS + AWL) | 96.0 | 95.1 | 95.7 | 95.2 | 96.2 | 95.6 ± 0.48 |
3.4. Clinical Cases of Image Fusion
When the target tumor is not directly visible in interventional CT, image fusion with another pre-operative imaging modality where the tumor is better visualized can be performed to help guide the procedure. The imaging modality to be fused varies depending on clinical application and tumor characteristics. In this paper, we demonstrate the surface registration based fusion through three different clinical scenarios detailed in the sections below.
3.4.1. Fusion of Interventional CT and MRI
Fig. 5 shows the fusion of interventional CT and MRI images. In this case, MRI can provide clear and detailed information about soft tissue as well as tumor that CT imaging cannot give. Through fusion, MRI, as the moving imaging modality, can be mapped to interventional CT to help guide the procedure. MRI can be done before the procedure with manual interaction and the interventional CT is segmented during the procedure. The images are then registered for alignment in the same coordinate system by registering the segmented surfaces. It is worth noting that the patient position in this case is quite different from what is in the LiTS dataset. All the images in the latter are used for diagnostic purpose and thus the patients were in regular supine positions. However, for interventional guidance, patients often have to be positioned for the best access to the target region. Even in this case, our segmentation algorithm performed very well to segment the liver. We contribute this to the use of multi-scale features throughout the network, which enables the superior combination of both high level holistic features and low level image texture details.
Fig. 5.
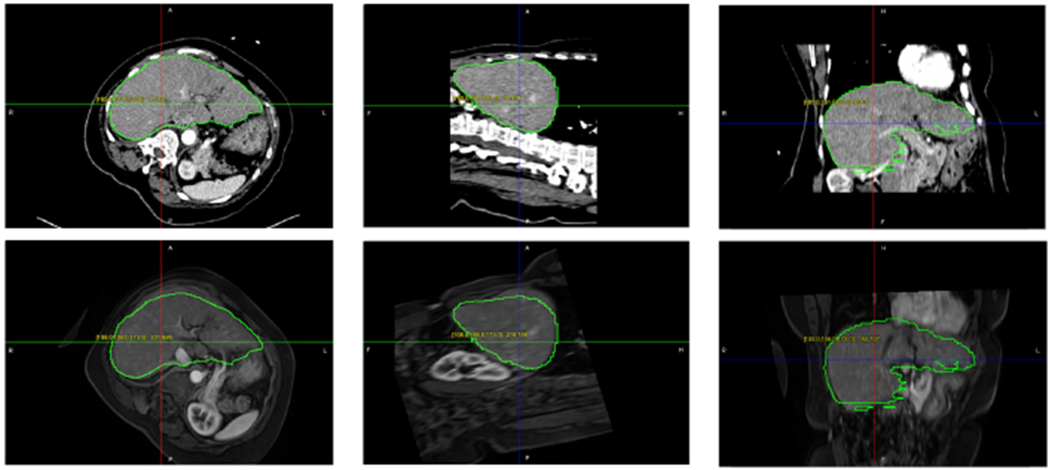
(Top) Starting point of ICP, Segmentation contour of MRI and CT is overlapped on interventional CT; (Middle) Fusion of interventional CT and MRI with segmented contour after ICP; (Bottom) Deformable registration with AI CT segmentation on deformed MRI focusing on liver tumor.
3.4.2. Fusion of Interventional CT and CE-CT
Fig. 6 shows the fusion of interventional CT and CE-CT images. By using contrast enhancing agent, CE-CT can provide good visualization of tumor and vascular structures. Through fusion, CE-CT, as the moving imaging modality in this case, can be mapped to interventional CT for interventional guidance. CE-CT was acquired and segmented before the procedure with manual interaction and the interventional CT is segmented during the procedure. Image registration is then performed by aligning the segmented surfaces.
Fig. 6.
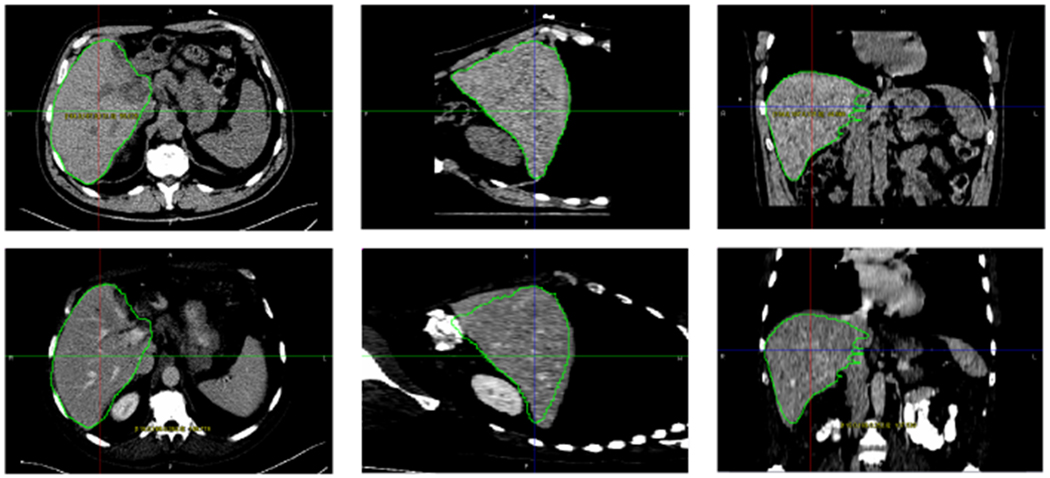
(Top) Intervention CT image and the segmentation result; (Bottom) Fusion of interventional CT and CE-CT with the segmented contour from interventional CT superimposed on the CE-CT image.
3.4.3. Fusion of Interventional CT and PET/CT
Fig. 7 displays a case of fusing interventional CT and PET, through the inherently registered CT component of a PET/CT scan. In this case, functional imaging obtained by PET and intra-procedural guidance imaging performed by interventional CT are combined. PET imaging is low-resolution imaging modality, but can visualize the functional activities of tumor very well. However, due to the lack of structure information, it is hard to directly register PET with interventional CT. Therefore, the CT image component in the PET/CT scan is used as a bridge for registration, which is registered to the interventional CT through aligning the segmented surfaces. By fusing PET image with interventional CT, tumors can be easily observed during a surgical procedure. Fig. 7 shows the three imaging modalities and the fusion result, where the tumor is circled in light gold in the three views.
Fig. 7.
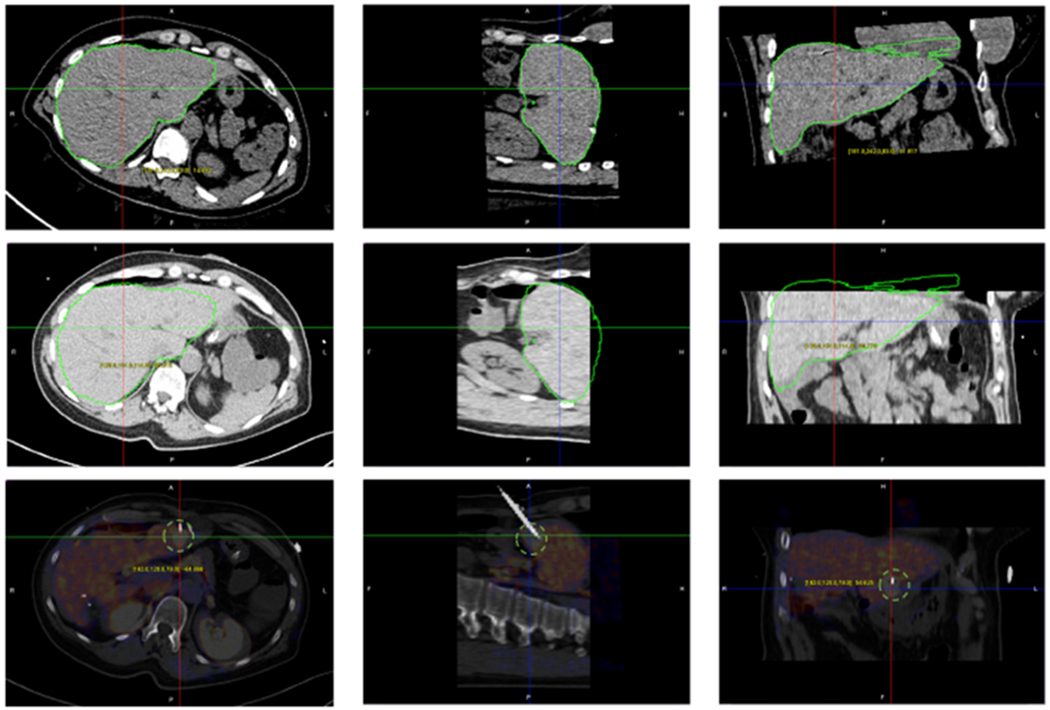
Fusion of PET and interventional CT for guiding biopsy needle placement. (Top) Segmentation of the CT image from a PET/CT scan; (Middle) Superimposed contour from the PET/CT over the interventional CT image after registration; (Bottom) Blended PET and interventional CT images with a biopdy needle reaching a tumor only visible in PET.
4. Discussion
In this section, we analyze some cases that our algorithm fails to segment the liver and then propose some corresponding solutions for improvement. Fig.8 shows three cases. In the first and second case, our algorithm does not discriminate liver from neighboring abdominal organs since these organs have similar HU-value range and distribution. Specifically, these false positive region is near the left lobe of liver and spleen is classified into liver in the second case. A possible solution may be training our algorithm more frequently on patches near the left lobe and spleen region to reduce the false-positive prediction. In the third case, our algorithm classify the liver tumor into background. It may be due to the in-balance between liver tumor and non-tumor liver during training. A possible solution is training our algorithm to segment the liver tumor at the same time. We’d also like to point it out that even in these cases, our algorithm can still be helpful for clinical intervention. In this case, We can see that the boundary of liver is well obtained. After transforming into point sets, these false points can be easily removed from liver surface with manual operation. Segmented images can be then aligned for clearer visualization during intra-operative processing. In this work, we use ICP, a rigid registration method to illustrate the framework. To achieve better image fusion, deformable registration can be implemented to improve current workflow.
Fig. 8.
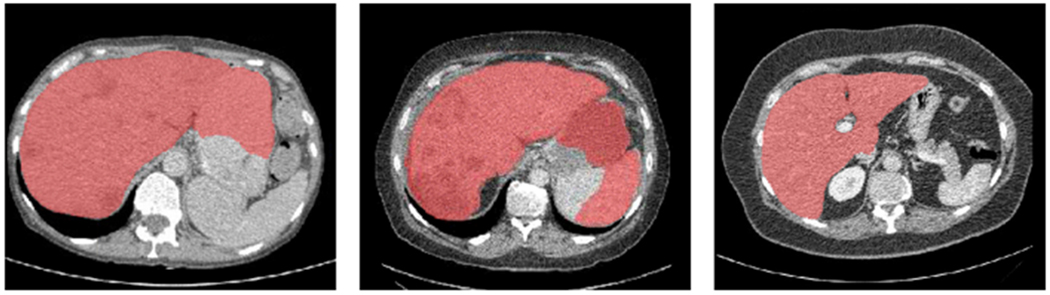
Some results that our algorithm fails to segment the liver. Three cases are shown for illustration.
5. Conclusion
In this paper, we presented a new deep learning based liver CT segmentation algorithm, which can accurately, efficiently, and robustly segment interventional CT images for surface-based image fusion. We then demonstrated the use of this method in three clinical cases, where it facilitates image fusion of interventional CT with diagnostic CE-CT, PET/CT, and MRI, respectively, for image guidance. The developed method may also be used for other applications, including image registration of CT image series for tumor tracking, surface-based deformable image registration between treatment planning, and intra-ablation and post-ablation CT scans for iterative treatment planning and verification.
Acknowledgements
We would also like to thank NVIDIA Corporation for the donation of the Titan Xp GPU used for this research.
This work was supported by National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health (NIH) under awards R21EB028001 and R01EB027898, and through an NIH Bench-to-Bedside award made possible by the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with ethical standards
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee of where the studies were conducted.
Conflict of interest NIH and RPI share intellectual property in the field and one author receive royalties for licensed patents (BW).
Informed consent Informed consent was obtained from all individual participants included in the study.
Contributor Information
Xi Fang, Department of Biomedical Engineering & Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Sheng Xu, National Institutes of Health, Center for Interventional Oncology, Radiology & Imaging Sciences, Bethesda, MD 20892, USA.
Bradford J. Wood, National Institutes of Health, Center for Interventional Oncology, Radiology & Imaging Sciences, Bethesda, MD 20892, USA
Pingkun Yan, Department of Biomedical Engineering & Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
References
- 1.Abi-Jaoudeh N, Kruecker J, Kadoury S, Kobeiter H, Venkatesan AM, Levy E, Wood BJ (2012) Multimodality image fusion-guided procedures: Technique, accuracy, and applications. CardioVascular and Interventional Radiology 35(5):986–998, DOI 10.1007/s00270-012-0446-5, URL 10.1007/s00270-012-0446-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besl PJ, McKay ND (1992) A method for registration of 3-d shapes. IEEE Transactions on Pattern Analysis and Machine Intelligence 14(2):239–256, DOI 10.1109/34.121791 [DOI] [Google Scholar]
- 3.Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, Fu CW, Han X, Heng PA, Hesser J, Kadoury S, Konopczynski T, Le M, Li C, Li X, Lipkovà J, Lowengrub J, Meine H, Moltz JH, Pal C, Piraud M, Qi X, Qi J, Rempfler M, Roth K, Schenk A, Sekuboyina A, Vorontsov E, Zhou P, Hülsemeyer C, Beetz M, Ettlinger F, Gruen F, Kaissis G, Lohöfer F, Braren R, Holch J, Hofmann F, Sommer W, Heinemann V, Jacobs C, Mamani GEH, van Ginneken B, Chartrand G, Tang A, Drozdzal M, BenCohen A, Klang E, Amitai MM, Konen E, Greenspan H, Moreau J, Hostettler A, Soler L, Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L, Menze BH (2019) The liver tumor segmentation benchmark (lits). arXiv preprint arXiv:190104056 [Google Scholar]
- 4.Billings SD, Boctor EM, Taylor RH (2015) Iterative most-likely point registration (imlp): a robust algorithm for computing optimal shape alignment. PloS one 10(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Zhang R, Yan P (2019) Feature fusion encoder decoder network for automatic liver lesion segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp 430–433, DOI 10.1109/ISBI.2019.8759555 [DOI] [Google Scholar]
- 6.Fang X, Du B, Xu S, Wood BJ, Yan P (2019) Unified multi-scale feature abstraction for medical image segmentation. arXiv preprint arXiv:191011456 [Google Scholar]
- 7.Haaga JR (2005) Interventional ct: 30 years’ experience. European Radiology Supplements 15(4):d116–d120 [DOI] [PubMed] [Google Scholar]
- 8.Han X (2017) Automatic Liver Lesion Segmentation Using A Deep Convolutional Neural Network Method. arXiv: 170407239 URL http://arxiv.org/abs/1704.07239 [Google Scholar]
- 9.Haskins G, Kruger U, Yan P (2019) Deep learning in medical image registration: A survey. arXiv preprint arXiv:190302026 [Google Scholar]
- 10.He K, Zhang X, Ren S, Sun J (2015) Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE transactions on pattern analysis and machine intelligence 37(9):1904–1916 [DOI] [PubMed] [Google Scholar]
- 11.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778 [Google Scholar]
- 12.Herring JL, Dawant BM, Maurer CR, Muratore DM, Galloway RL, Fitzpatrick JM (1998) Surface-based registration of ct images to physical space for image-guided surgery of the spine: a sensitivity study. IEEE Transactions on Medical Imaging 17(5):743–752, DOI 10.1109/42.736029 [DOI] [PubMed] [Google Scholar]
- 13.Huang G, Liu Z, van der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: CVPR, pp 2261–2269 [Google Scholar]
- 14.Jones AK, Dixon RG, Collins JD, Walser EM, Nikolic B (2018) Best practice guidelines for ct-guided interventional procedures. Journal of vascular and interventional radiology: JVIR 29(4):518. [DOI] [PubMed] [Google Scholar]
- 15.Kikinis R, Pieper SD, Vosburgh KG (2014) 3d slicer: a platform for subject-specific image analysis, visualization, and clinical support In: Intraoperative imaging and image-guided therapy, Springer, New York, NY, pp 277–289 [Google Scholar]
- 16.Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks In: Pereira F, Burges CJC, Bottou L, Weinberger KQ (eds) Advances in Neural Information Processing Systems 25, Curran Associates, Inc., pp 1097–1105, URL http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf [Google Scholar]
- 17.Li X, Chen H, Qi X, Dou Q, Fu C, Heng P (2018) H-denseunet: Hybrid densely connected unet for liver and tumor segmentation from CT volumes. IEEE Transactions on Medical Imaging 37(12):2663–2674, DOI 10.1109/TMI.2018.2845918 [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Xu D, Zhou SK, Pauly O, Grbic S, Mertelmeier T, Wicklein J, Jerebko A, Cai W, Comaniciu D (2018) 3d anisotropic hybrid network: Transferring convolutional features from 2d images to 3d anisotropic volumes. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 851–858 [Google Scholar]
- 19.Maintz J, Viergever MA (1998) A survey of medical image registration. Medical Image Analysis 2(1):1–36, DOI 10.1016/S1361-8415(01)80026-8, URL http://www.sciencedirect.com/science/article/pii/S1361841501800268 [DOI] [PubMed] [Google Scholar]
- 20.Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, Lerer A (2017) Automatic differentiation in PyTorch. In: NIPS 2017 Workshop Autodiff [Google Scholar]
- 21.Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: MICCAI, pp 234–241 [Google Scholar]
- 22.Vorontsov E, Tang A, Pal C, Kadoury S (2018) Liver lesion segmentation informed by joint liver segmentation. In: ISBI, pp 1332–1335, DOI 10.1109/ISBI.2018.8363817 [DOI] [Google Scholar]
- 23.Wang L, Lee CY, Tu Z, Lazebnik S (2015) Training deeper convolutional networks with deep supervision. arXiv preprint arXiv:150502496 [Google Scholar]
- 24.Yuan Y (2017) Hierarchical convolutional-deconvolutional neural networks for automatic liver and tumor segmentation. arXiv:171004540 [Google Scholar]


