Abstract
Microalgae and cyanobacteria contribute roughly half of the global photosynthetic carbon assimilation. Faced with limited access to CO2 in aquatic environments, which can vary daily or hourly, these microorganisms have evolved use of an efficient CO2 concentrating mechanism (CCM) to accumulate high internal concentrations of inorganic carbon (Ci) to maintain photosynthetic performance. For eukaryotic algae, a combination of molecular, genetic and physiological studies using the model organism Chlamydomonas reinhardtii, have revealed the function and molecular characteristics of many CCM components, including active Ci uptake systems. Fundamental to eukaryotic Ci uptake systems are Ci transporters/channels located in membranes of various cell compartments, which together facilitate the movement of Ci from the environment into the chloroplast, where primary CO2 assimilation occurs. Two putative plasma membrane Ci transporters, HLA3 and LCI1, are reportedly involved in active Ci uptake. Based on previous studies, HLA3 clearly plays a meaningful role in HCO3− transport, but the function of LCI1 has not yet been thoroughly investigated so remains somewhat obscure. Here we report a crystal structure of the full length LCI1 membrane protein to reveal LCI1 structural characteristics, as well as in vivo physiological studies in an LCI1 loss-of-function mutant to reveal the Ci species preference for LCI1. Together, these new studies demonstrate LCI1 plays an important role in active CO2 uptake and that LCI1 likely functions as a plasma membrane CO2 channel, possibly a gated channel.
Keywords: CO2, Ci, LCI1, CCM, Chlamydomonas, photosynthesis, transporter, channel
Introduction
Incorporation of atmospheric CO2 into organic carbon by photosynthesis is an essential biochemical process that sustains life of almost all living organisms on earth. Yet for many photosynthetic organisms, present-day atmospheric CO2 concentration is a major hindrance to achieve maximum photosynthetic capacity due to the slow catalytic activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), the primary CO2-fixing enzyme in nature. Inefficiency of Rubisco’s catalytic performance in CO2 fixation is characterized by a relatively low affinity for CO2 and a slow carboxylase turnover rate. Furthermore, Rubisco also catalyzes the oxygenation of D-ribulose-1,5-bisphosphate (RuBP), which leads to CO2 loss in the photorespiratory pathway (Spreitzer and Salvucci, 2002). To overcome this limitation, many photosynthetic organisms have evolved a CO2-concentrating mechanism (CCM) that increases local CO2 concentration at the site of Rubisco, thus promoting Rubisco’s carboxylase activity while suppressing its oxygenase activity. Classic examples of CCMs in higher plants are the C4 and Crassulacean acid metabolism (CAM) pathways, in which CO2 is assimilated into a 4-carbon compound followed by its decarboxylation to generate CO2 in the vicinity of Rubisco (Giordano et al., 2005; Edwards, 2019).
In aquatic ecosystems, photosynthetic organisms, such as microalgae and cyanobacteria, often experience a limited CO2 supply, due to slow diffusion of CO2 in water and dramatic changes in the availability of dissolved inorganic carbon (Ci: CO2, HCO3− and CO32−). Thus, the presence of a CCM allows microalgae and cyanobacteria to acclimate to unfavorable environmental conditions. CCMs in cyanobacteria and microalgae are inducible systems that fundamentally rely on active Ci uptake, interconversion of Ci species by carbonic anhydrases and localization of Rubisco in a specific microcompartment. Active Ci uptake is supported by energized HCO3− and CO2 uptake systems that result in the accumulation of an intracellular Ci (Ci pool) significantly higher than the atmospheric CO2 level. This Ci pool, mostly in the form of HCO3−, is subsequently dehydrated to CO2 by the activity of carbonic anhydrase (CA), and the higher concentration of CO2 is fixed by Rubisco.
In CCMs, Ci flux to Rubisco depends on the activity of Ci uptake systems, so much effort has been expended to reveal specific roles of Ci uptake systems. Within the last few decades, significant progress to elucidate eukaryotic Ci uptake systems has been made in the unicellular green alga, Chlamydomonas reinhardtii (hereafter, Chlamydomonas). Through physiological studies, it has been established that Ci flux across membranes in Chlamydomonas is supported via energized Ci uptake of both HCO3− and CO2 (Moroney and Tolbert, 1985; Sültemeyer et al., 1989). However, the components and the molecular mechanisms underlying the operation of HCO3− and CO2 uptake have not been fully characterized. Furthermore, the presence of various cell compartments and the acclimation to multiple CO2 acclimation states add to the complexity of Ci uptake system operation in Chlamydomonas. Genetic and physiological evidence has clearly demonstrated the existence of at least three CO2 acclimation states in Chlamydomonas : a high CO2 (5%–0.5%), a low CO2 (0.4–0.03%) and a very low CO2 (<0.02%) state (Van et al., 2001; Vance and Spalding, 2005; Wang and Spalding, 2006). Interestingly, Ci flux in different CO2 acclimation states, specifically in a low and a very low CO2, apparently is supported by distinct Ci uptake systems (Wang and Spalding, 2014).
In low CO2, active CO2 uptake mediated by LCIB, a novel chloroplast stromal protein, appears to play a critical role in active Ci uptake especially at air-level CO2. LCIB, as part of an LCIB-LCIC complex, apparently functions as a unidirectional CA in capturing CO2 either entering the cells or leaking from the pyrenoid, a microcompartment where Rubisco is sequestered, and hydrating the CO2 to HCO3−, which then enters the bulk stromal Ci pool (Wang and Spalding, 2006; Duanmu et al., 2009a; Wang and Spalding, 2014; Jin et al., 2016). However, the precise mechanism and regulation of LCIB-mediated CO2 uptake requires more exploration.
In very low CO2, both the LCIB-mediated CO2 uptake system and a HCO3− transport system contribute to intracellular Ci accumulation. In this condition, HCO3− uptake is reportedly mediated by a cooperative effort of HLA3, a plasma membrane transporter, and LCIA, a chloroplast envelope protein (Duanmu et al., 2009b; Gao et al., 2015; Yamano et al., 2015). Genetic studies demonstrated that the HLA3-LCIA-mediated HCO3− transport system plays a complementary role with LCIB in very low CO2, since the absence of both is lethal but the absence of only one component has no substantial consequence on growth and photosynthetic activity in very low CO2. Furthermore, physiological studies of LCIA/LCIB double mutants revealed that HLA3-LCIA-mediated HCO3− transport is active in very low CO2 but appears to be inhibited by increasing CO2 concentration in the transition from very low to low CO2 (Wang and Spalding, 2014).
While studies on HLA3, LCIA and LCIB have yielded valuable insights into the mechanisms of Ci uptake systems and their contributions in different CO2 acclimation states, the role of many putative Ci transporters have not yet been clearly defined. At the plasma membrane, in addition to HLA3, LCI1 is reportedly associated with Ci uptake. The Chlamydomonas LCI1 gene encodes a novel, limiting-CO2 induced protein, for which no sequence homologs have been identified even in closely related species, although unrelated, but structurally-similar transporters may exist (Ohnishi et al., 2010). LCI1 is a 21.5 kD, hydrophobic, plasma membrane protein with four transmembrane helices, and it is encoded by nuclear DNA, translated in the cytoplasm, and localized in the plasma membrane. Although predicted to possess four hydrophobic domains, the first, N-terminal domain of LCI1 was presumed to serve as a signal peptide. LCI1 appears to form a complex with HLA3 and LCI1 overexpression reportedly increased Ci uptake in air-level CO2, especially in high pH, suggesting that LCI1 may transport HCO3− (Ohnishi et al., 2010; Mackinder et al., 2017). However, subsequent data indicated that co-expression of LCI1 and LCIA failed to improve Ci affinity at high pH, contradicting the notion that LCI1 may transport HCO3− (Yamano et al., 2015). On the other hand, physiological and genetic studies of an LCI1 insertional mutant presented in the accompanying paper showed that LCI1 has a significant role in supporting photosynthesis in the low CO2 range specifically above air-level CO2 (Kono and Spalding, Accompanying Paper).
Here we demonstrate LCI1 function in active CO2 uptake by elucidating a crystal structure of the full-length LCI1 membrane protein and by further analyzing the in vivo response of an LCI1 loss-of-function mutant.
Results
Overall LCI1 Crystal Structure
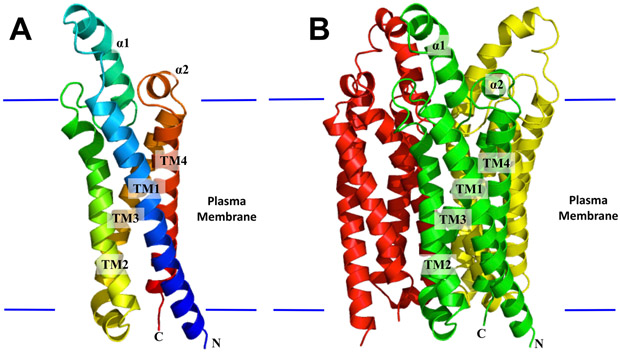
The LCI1 crystal structure was determined to a resolution of 3.19 Å using single isomorphous replacement (Figure 1, Figure S1, Table S1). Three LCI1 molecules, which form a homotrimer, were found in the asymmetric unit, and superimposition of these three protomers gives RMSDs between 0.39 and 0.41 Å (over 165 Cα atoms), suggesting that their conformations are nearly identical to each other. The composition of LCI1 is predominantly hydrophobic, with most amino acids embedded in the transmembrane, and the program TMpred (Hofmann, 1993) predicted its N- and C- terminal ends are located in the cytoplasm, as expected for a plasma membrane protein lacking a signal peptide.
Figure 1.
Structure of the Chlamydomonas reinhardtii LCI1. (A) Ribbon diagram of an LCI1 monomer viewed in the membrane plane. The molecule is colored using a rainbow gradient from the N-terminus (blue) to the C-terminus (red). (B) Ribbon diagram of an LCI1 trimer viewed in the membrane plane. The three protomers are colored green, red and yellow, respectively. The transmembrane segments (TMs) and α-helices (αs) of the front protomer (green) of LCI1 are labeled. The Figure was prepared using PyMOL (http://www.pymol.sourceforge.net).
Each LCI1 protomer comprises four helical transmembrane segments (TMs) and two extra-membrane α-helices (αs), which are located right above the plasma membrane periplasmic surface. The TMs and αs are designated numerically from the N- to C-termini: TM1 (6–38), α1 (47–62), TM2 (73–95), TM3 (108–133), α2 (136–141) and TM4 (144–169). Of the four TMs, TM1 is relatively long, and part of this helix (residues 34–38) is exposed to solvent. In addition to this top portion of TM1, helices α1 and α2 of this membrane protein constitute a small periplasmic domain. The LCI1 trimer orients in such a way that its pseudo threefold axis is perpendicular to the membrane surface with its threefold axis surrounded by the three closely packed TM3 helices, which seal the central trimer interface against passage.
LCI1 protomer forms a channel
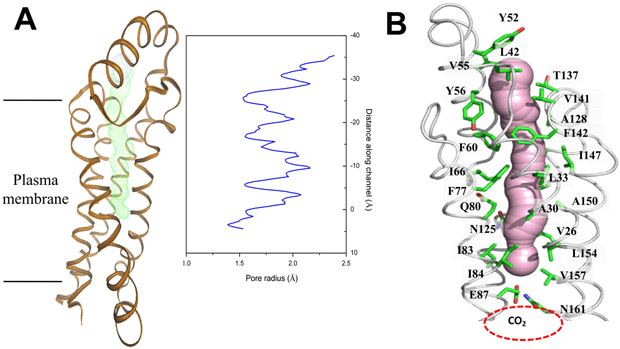
Viewed in parallel to the membrane plane, the LCI1 trimer is about 60 Å tall, 40 Å wide and 40 Å thick. Each LCI1 subunit creates a four antiparallel helix bundle and folds into a cylindrical structure within the transmembrane region. The four antiparallel helix bundles of each LCI1 molecule contribute to create a channel that spans approximately two-thirds of the transmembrane (Figure 2). The two, short α1 and α2 helices, which are exposed to solvent in the periplasm, form the channel entrance, with residues L42, Y52, V55, T137 and V141 found to encircle this entrance. The channel, likely involved in transport or conductance, is hydrophobic in nature, with 19 aromatic and hydrophobic residues (V26, A30, L33, L42, Y52, V55, Y56, F60, I66, F77, I83, I84, A128, V141, F142, I147, A150, L154 and V157) participating to construct this channel, along with three polar residues, Q80, N125 and T137. Apparently, the determined structure captures a closed conformation of the LCI1 channel, where the side chain of a negatively charged residue, E87, within the LCI1 transmembrane region appears to make a hydrogen bond with the side chain of N161 to form a gate that blocks this transmembrane channel (Figure 2).
Figure 2.
(A) Each LCI1 subunit forms a channel (colored green) that spans approximately two-thirds of the transmembrane. The radius of the channel was calculated using the program HOLE (Smart et al., 1993). The channel is closed due to interaction between the sidechains of residues E87 and N161. (B) The LCI1 channel (colored pink) is surrounded by 22 amino acids, including V26, A30, L33, L42, Y52, Y56, F60, I66, F77, Q80, I83, I84, N125, A128, T137, V141, F142, I147, A150, L154 and V157, and containing a potential CO2-binding site located right below residues E87 and N161. This channel representation was generated using the program CAVER (http://loschmidt.chemi.muni.cz/caver). The secondary structural elements of the LCI1 protomer are in gray, and residues involved in forming this channel, along with residues E87 and N161, are represented by green sticks.
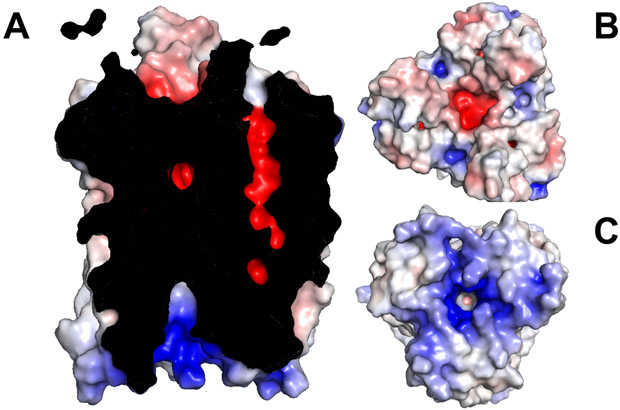
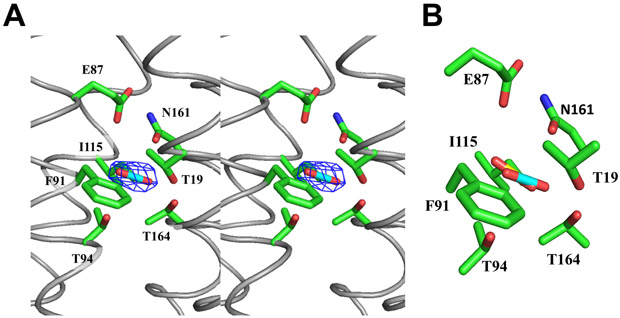
The interior surface of the LCI1 channel is strikingly electronegative (Figure 3), suggesting that it may transport neutral or positively charged substrates. Surprisingly, an extra electron density, having a shape highly consistent with a CO2 molecule (Figure 4), was detected in each LCI1 subunit just to the cytoplasmic side of the putative E87-N161 H-bond gate, revealing a fortuitously bound ligand co-purified and co-crystallized with this membrane protein. At least five amino acids, including T19, T94, F91, I115 and T164, were found within 4.5 Å of and apparently involved in binding of the ligand. Ab initio calculations reveal that all these amino acids have a strong affinity for CO2 (Cundari et al., 2009; Karmakar et al., 2013). In addition, a search for potential CO2-binding sites within the four antiparallel helix bundle of LCI1 using AutoDock Vina (Trott and Olson, 2010) identified only one potential CO2-binding site, and this putative site coincides with the extra electron density (Figure 4). These data are consistent with the fortuitous ligand being CO2 and suggesting that LCI1 may bind and transport CO2 rather than HCO3−.
Figure 3.
Electrostatic surface potentials of LCI1. Surface representations of the inside (A), top view (B) and bottom view (C) of the LCI1 channel colored by charge (red; negative −15 kT/e, blue; positive +15 kT/e).
Figure 4.
Putative CO2 binding site. (A) Stereo view of the Fo - Fc electron density map of bound ligand, presumed to be CO2, in LCI1, with the putative bound CO2 ligand and residues putatively involved in CO2 binding shown as a stick model (cyan, carbon; blue, nitrogen) and as green sticks, respectively. The Fo - Fc map is contoured at 3.0 σ (blue mesh). (B) A composite figure showing the locations of the predicted bound CO2 ligand (yellow) and the putative bound CO2 (cyan) identified in the LCI1 crystal structure.
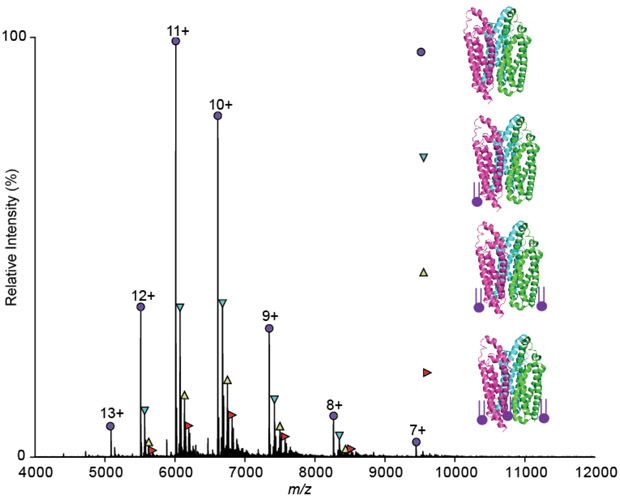
Non-denaturing mass spectrometry (native MS) was used to test the veracity of the predicted trimeric architecture by elucidating the aqueous phase oligomerization state, sample mass, and LCI1 subunit stoichiometry. The native MS spectra showed a well-resolved, charge-state series corresponding to and confirming the trimeric oligomerization of LCI1. It also revealed the presence of bound lipids co-purified with this protein (Figure 5), which were identified, using a lipidomic approach, as phosphatidic acid (16:0/18:1) (PA(16:0/18:1)) and phosphatidic acid (18:1/18:1) (PA(18:1/18:1)) (Figure S2). These lipids are bound within the purified LCI1 in various combinations, with the observed masses of the lipid-LCI1 complexes indicated in Table S2. Co-purified lipids are often specific lipids known to modulate the protein function and oligomeric state in several cases (Testerink et al., 2004; Zhang et al., 2006; Gupta et al., 2017; Bolla et al., 2019). Future studies are required to confirm the importance of PA on LCI1 protein function and fold.
Figure 5.
Mass spectrum of LCI1 obtained under native conditions. The spectra indicate the presence of the LCI1 trimer (purple circles) and its charge state series. The measured mass of this trimer is 66113.01 ± 0.27 Da, which is in good agreement with the theoretical mass of the trimeric LCI1 protein (66115.8 Da) without the first methionine residue on each monomer. The other species (cyan, yellow and red triangles) correspond to the lipid bound trimers, and these are endogenously purified. Lipid analysis on this sample revealed the presence of PA(16:0/18:1) and PA(18:1/18:1) as shown in Figure S2, which are bound in different numbers and combinations. All the molecular masses observed are listed in Table S2.
Molecular Dynamics Simulations
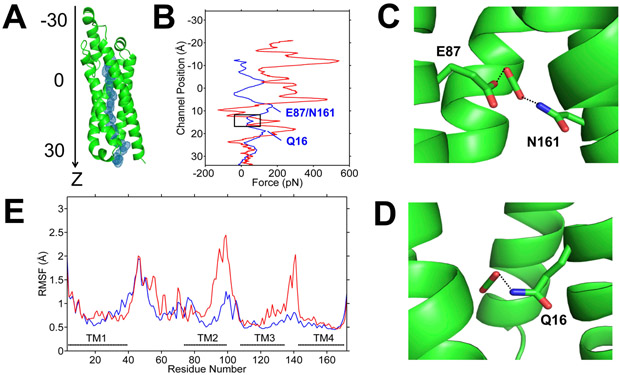
To explore whether LCI1 is capable of transporting CO2 across the membrane, we performed steered molecular dynamics (SMD) simulations on this protein embedded in a palmitoyloleoylphosphatidylethanolamine (POPE) membrane bilayer, measuring the magnitude of applied forces needed to maintain a constant velocity of CO2 through the LCI1 protomer channel. When we pushed CO2 into the LCI1 channel via the opening created by helices α1 and α2, SMD suggested CO2 can be migrated smoothly into the channel. The applied forces needed to maintain a constant velocity of CO2 migration (Figure 6) showed a noticeable reduction in the applied force as CO2 reached the vicinity of the putative CO2-binding site identified by the crystal structure. The SMD calculation suggests that residues lining the wall of this region may have the capacity to push CO2 further into the channel, allowing the CO2 molecule to propagate farther down the channel and then exit to the cytoplasm.
Figure 6.
Steered molecular dynamics (SMD) simulations of the migration of CO2 and HCO3− through LCI1. (A) CO2 trajectory through LCI1 in SMD is shown as blue mesh. LCI1 is oriented with the channel along the z-axis and the position of ligand along the channel is measured as distance from the protein center of mass along the z-axis. (B) Plots of applied forces as a function of ligand positions along the channel (blue, CO2; red, HCO3−), which indicate that pushing HCO3− through the channel requires much larger force, suggesting that CO2 may be the preferred ligand. A black rectangle highlights the region corresponding to the putative CO2-binding site. The two peaks marked in the CO2 plot (blue) indicate local electrostatic interactions between (C) putative bound CO2 and residues E87 and N161; and (D) putative bound CO2 and residue Q16. (E) Root mean square fluctuations (RMSF) of the LCI1 residues during the SMD simulations with CO2 (blue) and HCO3− (red).
SMD simulations performed to push CO2 from the cytoplasmic side indicate that CO2 could only migrate a short distance into the channel before meeting resistance. We observed a significant predicted conformational change between the LCI1 transmembrane helices TMs 1 and 2 as CO2 arrives at the putative CO2-binding site. These two helices are predicted to perform a scissor-liked motion and push CO2 back into the cytosol, suggesting that this channel may be functionally unidirectional.
Similar SMD simulations performed to test the possibility of HCO3− penetration through the channel revealed that HCO3− also can be migrated from the periplasmic to cytoplasmic sides of LCI1 using a similar pathway, but the forces needed to push HCO3− through this channel were at least five times higher than for CO2, suggesting LCI1 favors CO2 over HCO3− (Figure 6). These data are highly consistent with the electronegative nature of the LCI1 channel, which may not easily tolerate negatively charged ions. As with CO2 simulations, SMD calculations using a reverse force to push HCO3− from the cytoplasmic side into the LCI1 channel suggest this ion can only migrate a short distance before encountering resistance.
In Vivo Function: LCI1 Loss-of-function Mutant
To confirm LCI1 Ci species preferences in vivo and to further explore LCI1 roles in low CO2, we carefully examined the impact of an LCI1 loss-of-function mutation on Ci-dependent O2 evolution, exemplified by the “LCI1 uncompensated contribution”, in three different genetic backgrounds: a wild-type, where all other CCM components are present, an lcia mutant, where the LCIA-mediated HCO3− transport system is absent, and an lcib mutant, where the LCIB-mediated CO2 uptake system is absent. To visualize the impact of CO2 and HCO3− species on LCI1 function in vivo, the uncompensated contribution of LCI1 in pH 6, pH 7.3, and pH 9 was simultaneously plotted against calculated CO2 and HCO3− concentrations, and compared to the uncompensated contributions of LCIA (involved in HCO3− transport) and LCIB (involved in active CO2 uptake). The functional relationship between LCI1 and LCIA-associated HCO3− transport or LCIB-mediated CO2 uptake was revealed by comparing the LCI1, LCIA and LCIB uncompensated contribution in the wild-type and mutant backgrounds.
We define the uncompensated contribution of each protein as the magnitude of the decrease in O2 evolution rate in the absence of a specific protein, calculated by subtracting the O2 evolution rate of the added mutant from that of the background strain. For example, the O2 evolution rate of the lci1 mutant was subtracted from that of wild type to estimate the uncompensated contribution of LCI1 in the wild-type background. This value represents only the minimal amount of O2 evolution contributed by LCI1 in wild type, since activity of one or more other components might compensate for the lost LCI1 activity in the absence of LCI1 function — thus representing only the uncompensated LCI1 contribution.
Since the ratio of O2 evolution and CO2 fixation generally is one to one (Stern et al., 1964; Badger, 1985; Shimazaki and Zeiger, 1987), the uncompensated contribution also depicts the amount of CO2 that no longer is able to be fixed by Rubisco during steady state photosynthesis. Thus, the uncompensated contribution represents the exclusive contribution of a specific protein or transporter in Ci uptake to support photosynthetic CO2 assimilation. It is important to note that, due to the complementary function of different Ci uptake systems in the CCM, this calculated uncompensated contribution represents the minimum contribution rather than the actual contribution of a specific protein to Ci uptake, since the actual contribution is likely to be higher than the calculated uncompensated contribution value.
LCI1, LCIA and LCIB Uncompensated Contributions in Wild Type Background
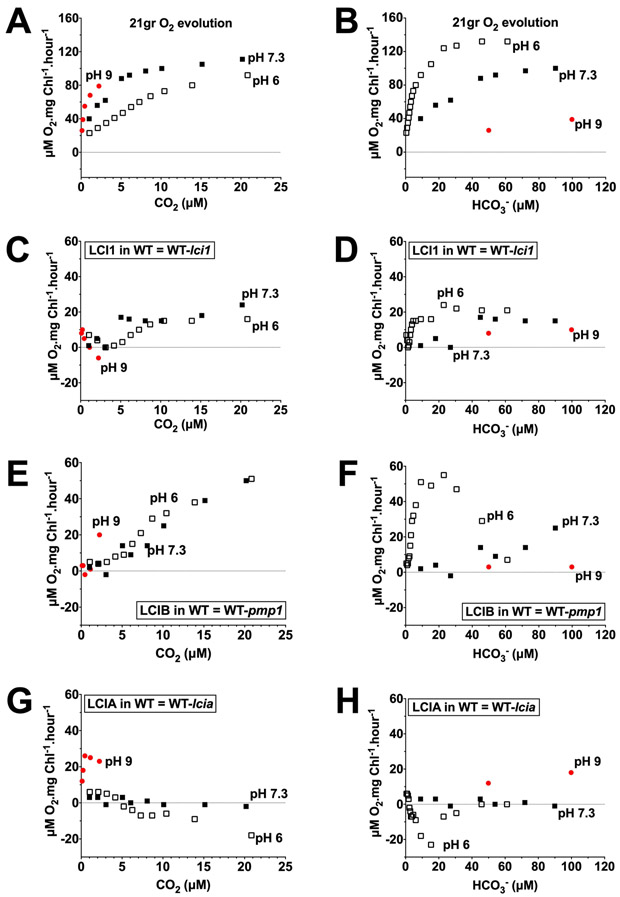
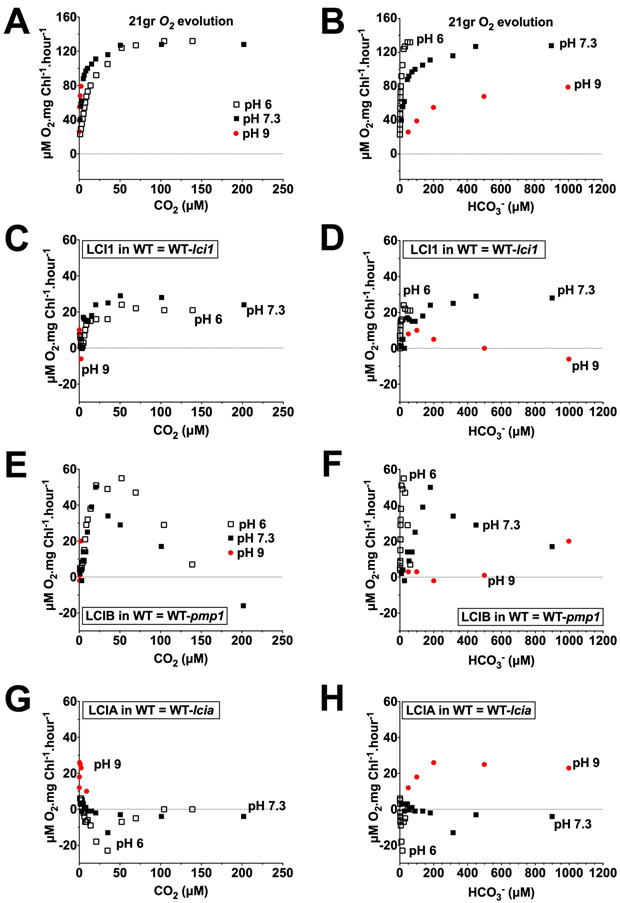
The pattern of LCI1 uncompensated contribution in a wild-type background appears similar at pH 6, pH 7.3 and pH 9 when plotted as a function of calculated CO2 concentration (Figure 7 C and Figure 8 C), but not when plotted as a function of calculated HCO3− concentration (Figure 7 D and Figure 8 D). Because similar patterns for LCI1 uncompensated contributions were observed at acidic and alkaline pH as a function of CO2 but not of HCO3− concentrations, LCI1 appears to respond functionally to CO2 rather than to HCO3−. The LCI1 uncompensated contribution below 5 μM CO2 was negligible at all three pHs and was higher between 5–200 μM CO2 but still very similar in magnitude at pH 6 and pH 7.3. This similarity in the LCI1 uncompensated contribution in both pH 6 and pH 7.3 suggests that the higher HCO3− concentration at pH 7.3 has little or no impact on the LCI1 contribution to wild-type photosynthesis (Figure 8 C). Furthermore, when plotted against calculated HCO3− concentration, LCI1 uncompensated contribution was higher in an acidic pH than in an alkaline pH over the same HCO3− concentration range (Figure 7 D and Figure 8 D). These uncompensated contribution data all argue LCI1 has a definite preference for CO2 rather than HCO3−.
Figure 7.
Uncompensated contribution of LCI1 (C, D), LCIB (E, F), and LCIA (G, H) in a wild-type background at pH 6 (open squares), pH 7.3 (closed squares) and pH 9 (red circles) plotted against calculated CO2 concentration (left) and HCO3− concentration (right). The CO2 and HCO3− concentration ranges covered are 0–25 μM and 0–120 μM, respectively. A and B are the original O2 evolution rates of the wild type. Note that the Y-axis scales on the original O2 evolution plots (A, B) are different than those on the uncompensated contribution plots (C–H). Uncompensated contributions were calculated by subtracting O2 evolution rates of the single mutant from wild type strain (LCI1 in wild type = 21gr-lci1; LCIB in wild type = 21gr-pmp1; LCIA in wild type = 21gr-lcia).
Figure 8.
Uncompensated contribution of LCI1 (C, D), LCIB (E, F), and LCIA (G, H) in a wild-type background at pH 6 (open squares), pH 7.3 (closed squares) and pH 9 (red circles) plotted against the calculated CO2 concentration (left) and HCO3− concentration (right). The CO2 and HCO3− concentration ranges covered are 0–250 μM and 0–1200 μM, respectively. A and B are the original wild-type O2 evolution rates. Note that the Y-axis scales on the original O2 evolution plots (A, B) are different than those on the uncompensated contribution plots (C–H). Uncompensated contributions in wild type were calculated by subtracting O2 evolution rates of the single mutant from wild type strain (LCI1 in wild type = 21gr-lci1; LCIB in wild type = 21gr-pmp1; LCIA in wild type = 21gr-lcia).
The patterns of LCI1 uncompensated contribution in a wild-type background show more similarity to those for LCIB than to those for LCIA. As was observed for LCI1, LCIB uncompensated contribution is very similar in pattern and in magnitude as a function of calculated CO2 concentration in both pH 6 and pH 7.3 (Figure 7 E and Figure 8 E), even though relative HCO3− abundance is higher at pH 7.3 than at pH 6. And as with LCI1, within the same calculated HCO3− concentration range, LCIB uncompensated contribution was higher in an acidic pH than in an alkaline pH (Figure 7 F, Figure 8 F). These data showing that LCIB responds functionally to CO2 rather than to HCO3− are entirely consistent with its reported function in active CO2 uptake (Wang and Spalding, 2006, 2014).
LCIA uncompensated contribution in the wild-type background, on the other hand, was almost undetectable in pH 6 and pH 7.3, and showed positive values only at pH 9 (Figure 7 G, H and Figure 8 G, H). LCIA uncompensated contribution as a function of the calculated CO2 concentration at pH 9 was higher than that at pH 6 and pH 7.3 (Figure 7 G), in contrast to the data for LCI1 and LCIB. Furthermore, LCIA uncompensated contribution as a function of HCO3− concentration at pH 9 increased as HCO3− concentration increased (Figure 8 H). These data showing that LCIA responds functionally to HCO3− rather than to CO2 are entirely consistent with its reported function in HCO3− transport (Wang and Spalding, 2014; Yamano et al., 2015).
LCI1 and LCIB Uncompensated Contributions in an lcia Mutant Background
Since the function of both LCI1 and LCIB in a wild-type background appears to respond to CO2 rather than HCO3−, we further investigated LCI1 and LCIB function in lcia where the absence of LCIA-dependent HCO3− transport should make photosynthesis in the lcia mutant almost totally dependent on active CO2 uptake. Consistent with data in the wild-type background, the patterns of both LCI1 and LCIB uncompensated contributions in lcia at pH 6, pH 7.3 and pH 9 appeared more similar to each other when plotted as a function of calculated CO2 concentrations (Figure 9 B, C; Figure S3 B, C) than when plotted as a function of calculated HCO3− concentration (Figure 9 E, F, Figure S3 E, F).
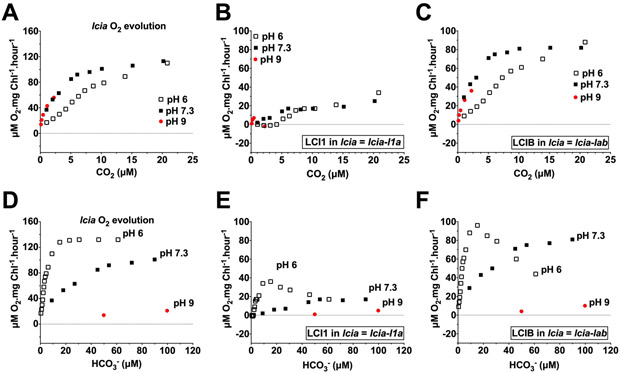
Figure 9.
Uncompensated contributions of LCI1 (B, E) and LCIB (C, F) in an lcia mutant background at pH 6 (open squares), pH 7.3 (closed squares) and pH 9 (red circles) plotted against calculated CO2 concentration (A–C) and HCO3− concentration (D–F). The CO2 and HCO3− concentration ranges covered are 0–25 μM and 0–120 μM, respectively. A and D are the original lcia mutant O2 evolution rates. Note that the Y-axis scales on the original O2 evolution plots (A, D) are different than those on the uncompensated contribution plots (B, C, E, F). Uncompensated contributions were calculated by subtracting O2 evolution rates of the mutant from its background strain: LCI1 in lcia = lcia-l1a; LCIB in lcia = lcia-lab. l1a is the LCI1/LCIA double mutant and lab is the LCIA/LCIB double mutant
The magnitude of LCI1 uncompensated contribution in lcia as a function of CO2 concentration at pH 7.3 was fairly similar to that at pH 6 even though relative abundance of HCO3− was much higher in pH 7.3 (Figure 9 B; Figure S3 B). LCIB uncompensated contributions, on the other hand, were higher at pH 7.3 (< 20 μM CO2 ) and pH 9 (< 2.5 μM CO2) compared to pH 6, presumably because LCIB compensates for the absence of LCIA-mediated HCO3− transport in the lcia mutant (Figure 9 C). Above 20 μM CO2, like LCI1, LCIB uncompensated contribution at pH 6 and pH 7.3 was fairly similar (Figure S3 C). Furthermore, the magnitude of both LCI1 and LCIB uncompensated contributions as a function of HCO3− concentrations were significantly lower in alkaline than in acidic pH (Figure 9 E, F, below 50 μM HCO3−; Figure S3 E, F, between 100–1000 μM HCO3−). Because relative CO2 abundance is higher in acidic pH compared to alkaline pH, the higher LCI1 and LCIB uncompensated contributions in acidic pH can be attributed to their use of CO2 rather than HCO3− species.
Consistent with their profiles in the wild-type background, LCI1 and LCIB uncompensated contributions in the lcia background show that both respond to CO2 rather than to HCO3−, which suggests that, as with the demonstrated CO2 uptake mediator LCIB, LCI1 also appears to function in CO2 uptake, specifically in CO2 uptake across the plasma membrane.
LCI1 and LCIA Uncompensated Contributions in an lcib Mutant Background
We also analyzed and compared the uncompensated contributions of LCI1 and LCIA in pmp1, an lcib mutant in which LCIB is absent. It has been demonstrated that lcib mutants cannot survive in air-level CO2 but exhibit normal growth and nearly normal photosynthesis in very low CO2. The apparent normal growth and photosynthesis in very low CO2 is reportedly supported by LCIA-associated HCO3− transport, which compensates for the absence of LCIB, as reflected by the increasing uncompensated contributions of LCIA in an lcib mutant, ad1, at pH 6 and pH 7.3 specifically in the very low CO2 range (Wang and Spalding, 2014).
In the absence of LCIB, the pattern of LCI1 uncompensated contribution as a function of calculated CO2 concentration appeared complex and slightly different in each pH. In very low CO2 (≤ 5 μM CO2), LCI1 uncompensated contribution was nearly identical in pH 6 and pH 7.3, but it was higher at pH 9 (Figure 10 A). Above 5 μM CO2, LCI1 uncompensated contribution at pH 7.3 is higher than pH 6 but only in certain CO2 concentration (5–10 μM and 35–50 μM CO2), other than that, the LCI1 uncompensated contribution at pH 6 and pH 7.3 is fairly similar (Figure 10 A, Figure S4 A). This peculiar pattern of LCI1 uncompensated contribution observed at pH 7.3 and pH 9 appears to be dictated by the O2 evolution profile of pmp1 since they share an identical pattern (Figure 10 B, Figure S4 B), presumably resulting from the need to compensate for the absence of LCIB.
Figure 10.
Uncompensated contributions of LCI1 (A, C) and LCIA (E, F) in a pmp1 strain (lcib mutant background) at pH 6 (open squares), pH 7.3 (closed squares) and pH 9 (red circles) plotted against the calculated CO2 concentration (A, E) and the HCO3− concentration (C, F). The CO2 and HCO3− concentration ranges covered are 0–25 μM and 0–120 μM, respectively. B and D are the original pmp1 mutant O2 evolution rates. Note that the Y-axis scales on the original O2 evolution plots (B, D) are different than those on the uncompensated contribution plots (A, C, E, F). Uncompensated contributions were calculated by subtracting the O2 evolution rates of the mutant from its background strain: LCI1 in pmp1 = pmp1-l1b; LCIA in pmp1 = pmp1-lab. l1b is the LCI1/LCIB double mutant and lab is the LCIA/LCIB double mutant.
As a function of the calculated HCO3− concentration, the LCI1 uncompensated contribution in pmp1 in acidic pH was always higher than that at alkaline pH within the same HCO3− concentration range and the increasing LCI1 uncompensated contributions in both pH 6 and pH 7.3 occurred in different HCO3− concentrations (Figure 10 C; Figure S4 C). These results are consistent with the profile of LCI1 uncompensated contributions in the wild-type and lcia mutant backgrounds, which argue that CO2 is the preferred Ci species used by LCI1.
Increasing LCIA uncompensated contribution in the pmp1 background as a function of calculated CO2 concentration in all tested pHs was higher at alkaline than at acidic pH (Figure 10 E; Figure S4 E). As a function of calculated HCO3− concentration, the increasing LCIA uncompensated contribution, similar to that of LCI1, occurred in a different HCO3− concentration range at different pHs (Figure 10 F), but it was higher at alkaline pH than at acidic pH within the same HCO3− concentration range (Figure 10 F; Figure S4 F), unlike LCI1, which showed the reverse. This overall LCIA uncompensated contribution profile is consistent with its reported role in HCO3− transport.
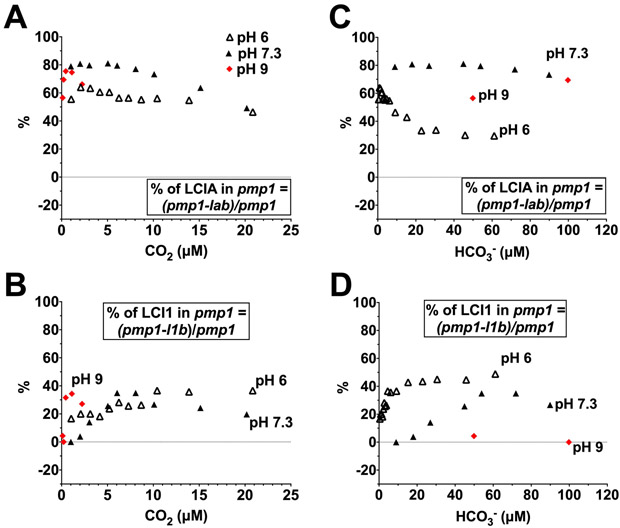
Since the uncompensated contribution patterns of LCI1 and LCIA in the pmp1 background are strongly dictated by the O2 evolution profile of pmp1 itself, we also calculated the percentage of LCI1 or LCIA uncompensated contribution to the pmp1 photosynthesis in order to minimize the influence of the LCIB absence and tease out the responses of LCIA and LCI1 to Ci species. As seen in Figure 11 A, the percentage of LCIA uncompensated contribution in the pmp1 background as a function of CO2 concentration was highest in the two alkaline pHs in very low CO2, but as the CO2 concentration increased, it slowly declined at both pH 6 and pH 7.3, reaching its lowest level at >25 μM CO2 (Figure S5 A). These observations are consistent with photosynthetic O2 evolution of pmp1 (lacking LCIB) being mainly supported by LCIA in the very low CO2 range, but less dependent on LCIA as CO2 concentrations increase (Wang and Spalding, 2014). In contrast, the percentage of LCI1 uncompensated contribution in the pmp1 background in very low CO2 was much lower than that of LCIA, but it increased above 5 μM CO2 (Figure 11 B). Interestingly, LCI1 uncompensated contribution accounted for approximately 50% of total pmp1 O2 evolution at CO2 concentrations >25 μM (Figure S5 B), which was higher than that of LCIA. These data suggest that, in contrast to the apparent dependence of pmp1 photosynthesis on LCIA in very low CO2, at higher than air-level CO2 concentrations, pmp1 photosynthesis depends more on LCI1-mediated Ci uptake.
Figure 11.
Percentage of LCIA (A, C) and LCI1 (B, D) uncompensated contribution to photosynthesis in a pmp1 mutant background at pH 6 (open triangles), pH 7.3 (closed triangles) and pH 9 (red diamonds) plotted as a function of calculated CO2 concentrations of 0–25 μM (A, B) and of calculated HCO3− concentrations of 0–120 μM (C, D).
When plotted as a function of HCO3− concentration in the pmp1 background, the percentage of LCIA uncompensated contribution was significantly higher in alkaline pH than in acidic pH (Figure 11 C), whereas the percentage of LCI1 uncompensated contribution was the reverse, higher in acidic than in alkaline pH (Figure 11 D). These data are consistent with the profile of the total LCI1 uncompensated contribution (Figure 10 and Figure S4) and argue that CO2 is the preferred Ci species used by LCI1. On the other hand, in the pmp1 background LCIA showed a preference for HCO3−, as indicated by a higher total uncompensated contribution, as well as, a higher percentage of LCIA uncompensated contribution in alkaline relative to acidic pH (Figure 10 and Figure 11). Furthermore, at >5 μM CO2, the percentage of LCIA uncompensated contribution declined as CO2 concentration increased, and significant decrease was seen in alkaline versus acidic pH (Figure 11 A; Figure S5 A). As a function of HCO3−, the percentage of LCIA uncompensated contribution significantly decreased in acidic pH where CO2 is more abundant compared to alkaline pH where CO2 is less (Figure 11 C; Figure S5 C). These data are consistent with a report that LCIA-mediated HCO3− uptake is inhibited by increasing CO2 above air-level concentrations at room temperature (Wang and Spalding, 2014).
Complementary Roles of LCI1 and LCIB in Low CO2
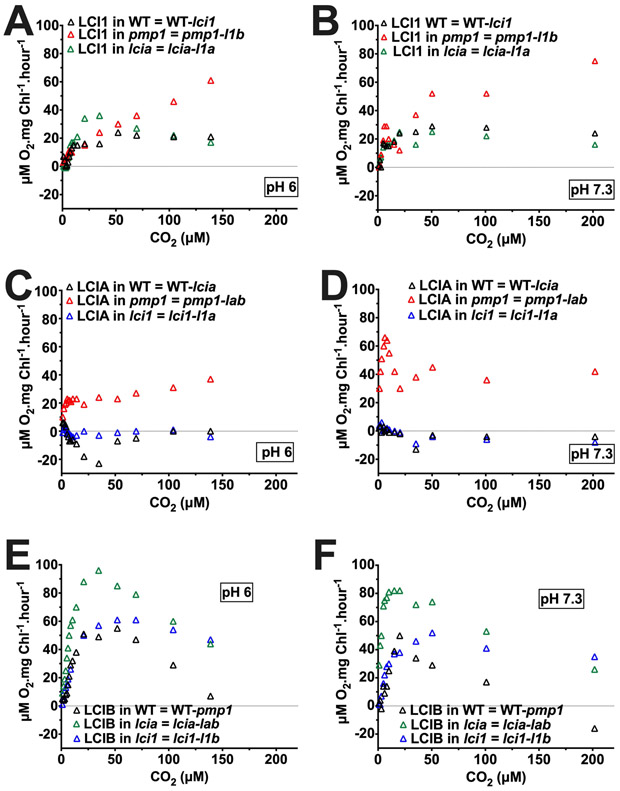
In addition to demonstrating an LCI1 preference for CO2, analysis of LCI1 uncompensated contribution in various genetic backgrounds also revealed relationships between LCI1 and two known Ci uptake pathways: LCIA-mediated HCO3− transport and LCIB-mediated CO2 uptake. In the very low CO2 range in all tested pHs, the uncompensated contributions of LCI1 and LCIA did not change in the absence of each other (Figure S6 A–D, Figure S7 A, B), whereas in all pHs, LCIA uncompensated contribution increased significantly in the absence of LCIB (Figure S6 C, D; Figure S7 B) and LCIB uncompensated contribution increased considerably in the absence LCIA (Figure S6 E, F; Figure S7 C). These results indicate that, in contrast to the reported (and here confirmed) complementarity between LCIA and LCIB, complementary roles may not exist between LCI1 and LCIA in very low CO2. However, since LCI1 uncompensated contribution in pmp1 background (LCIB absent) increased at pH 9, although to a lesser extent than that of LCIA, and LCIB uncompensated contribution also increased in lci1 background (LCI1 absent) (Figure S7 A, C), there may be some complementary relationship between LCI1 and LCIB in very low CO2, at least in this specific condition.
In the low CO2 range above 20 μM CO2, LCI1 uncompensated contribution in pH 6 and pH 7.3 was substantially amplified in the absence of LCIB and was significantly higher than the relatively constant uncompensated contribution of LCIA (Figure 12 A–D). These data indicate that, above air-level CO2, LCI1 plays a more significant role than LCIA in compensating for the absence of LCIB-associated active CO2 uptake. LCIB uncompensated contribution appears to respond to the absence of either LCI1 or LCIA with alterations in its pattern (Figure 12 E, F). The LCIB uncompensated contribution increased substantially in the absence of LCIA only at CO2 concentrations ≤ 35 μM, but, in contrast, increased only at CO2 concentrations ≥35 μM in the absence of LCI1. Interestingly, while LCIB uncompensated contribution in the wild type and lcia backgrounds both declined above 35 μM CO2, in the lci1 background it remained steady both at pH 6 and at pH 7.3. Meanwhile, LCIA uncompensated contribution was not affected by the presence or absence of LCI1 above 35 μM CO2 (Figure 12 C, D). Together, these data argue that above air-level CO2, LCI1 and LCIB (active CO2 uptake) may compensate for each other, while LCI1 and LCIA (HCO3− transport) appear to have no complementary roles.
Figure 12.
Calculated, uncompensated contribution of LCI1 (A, B), LCIA (C, D), and LCIB (E, F) at pH 6 (left) and at pH 7.3 (right) in a wild-type background (black triangles), a pmp1 mutant background (red triangles), an lcia background (green triangles), and an lci1 mutant background (blue triangles). CO2 concentration was calculated from total Ci assuming CO2 and HCO3− are in equilibrium. Uncompensated contributions were calculated by subtracting the O2 evolution rates of the mutant from those of its background strain (LCI1 in wild type = 21gr-lci1; LCI1 in pmp1 = pmp1-l1b; LCI1 in lcia = lcia-l1a; LCIB in wild type = 21gr-pmp1; LCIB in lcia = lcia-lab; LCIB in lci1 = lci1-l1b; LCIA in wild type = 21gr-lcia; LCIA in lci1 = lci1-l1a; LCIA in pmp1 = pmp1-lab). l1b is the LCI1/LCIB double mutant; lab is the LCIA/LCIB double mutant; l1a is the LCI1/LCIA double mutant.
Discussion
In this study combining LCI1 crystal structural data, steered molecular dynamics (SMD) simulations, and in vivo physiological studies of an LCI1 loss-of-function mutant, we determine the Ci species preference of LCI1 and its functional relationship to LCIA-mediated HCO3− transport and LCIB-mediated CO2 uptake, in addition to revealing the molecular structure and hints regarding the mechanism of transport of this novel Ci transport protein.
The determined LCI1 crystal structure revealed that LCI1 forms a homotrimeric functional unit. Although previous predicted protein topology suggested that the hydrophobic domain of LCI1 was too short to form a transmembrane channel (Meyer and Griffiths, 2013), the crystal structure clearly revealed that LCI1 protomer consists of four transmembrane helices that form a putative channel likely representing a Ci transport passageway or channel within each protomer. The interior surface of the channel is extremely electronegative, implying that if LCI1 transports Ci, CO2 is a much more likely transport substrate than HCO3−. The captured LCI1 conformation, although does not contain a full transmembrane channel, appears to represent a “closed” conformation in which a H-bond gate controls flux though the channel, a hypothesis strongly supported by SMD simulations. Furthermore, the detection of a bound ligand with size and shape characteristics of CO2 in a predicted CO2-binding just past the putative H-bond gate site lends further support to the premise that LCI1 transports CO2. SMD simulations also support this premise, since the predicted forces needed to push CO2 through the channel from the periplasm to the cytoplasm were 5-fold lower than those for HCO3−. In addition, the SMD simulations predicted that migration of either CO2 or HCO3− from the cytoplasm to the periplasm would meet strong resistance, suggesting that the channel may be functionally unidirectional, although there is no in vivo supporting evidence. If true, unidirectional CO2 migration would clearly be important to the overall efficiency and energetics of LCI1-mediated CO2 uptake.
The possibility of LCI1 facilitating the entry of CO2 across the plasma membrane based on crystal structure data is consistent with in vivo photosynthetic O2 evolution profile of LCI1 single, loss-of-function mutant, lci1. Even though lci1 lacks a growth phenotype in any tested CO2 concentrations or pH, its O2 evolution responses are distinctive depending on relative CO2 abundance, normal in pH 9 (less CO2) and decreased at pH 6 and pH 7.3 (more CO2) (Kono and Spalding, Accompanying Paper). This response is similar to that of LCIB single, loss-of-function mutants, which lack the LCIB protein involved in active CO2 uptake (Wang and Spalding, 2014; Kono and Spalding, Accompanying Paper), and substantially different from an LCIA single, loss-of-function mutant, which lacks the LCIA protein involved in HCO3− transport (Duanmu et al., 2009b; Wang and Spalding, 2014; Yamano et al., 2015). To extend the in vivo evaluation of LCI1 Ci species preference, we closely examined the impact of CO2 and HCO3− composition over a range of pH on the LCI1 uncompensated contribution to photosynthesis of wild-type and of lcia and lcib mutants. We also compared the LCI1 uncompensated contribution to the LCIA and LCIB uncompensated contributions.
Consistent with previous reports that any LCIA contribution to Ci uptake is fully compensated by other CCM components at or below neutral pH and with the established role of LCIA in HCO3− transport, the LCIA uncompensated contribution in wild-type, as a function of either CO2 or HCO3−, was near zero or even negative, except at pH 9, where HCO3−/CO2 ratio is extremely high, where Ci uptake is highly dependent on HCO3− uptake, and where LCIA uncompensated contribution was sizable. In the absence of LCIB, the LCIA uncompensated contribution at pH 9, pH 7.3 and pH 6 increased and, over the same CO2 concentration range, was higher at alkaline than at acidic pH. Taken together, the LCIA uncompensated contribution results verify LCIA’s established role in HCO3− transport and clearly confirm LCIA’s preference for HCO3−.
Also consistent with the well-documented function of LCIB in active CO2 uptake, the 20-fold higher HCO3−/CO2 ratio in pH 7.3 did not result in a higher LCIB uncompensated contribution relative to pH 6 in either a wild type or an lcia mutant background above 20 μM CO2, and the apparent response of LCIB to HCO3− below 20 μM CO2 in lcia background probably reflects an increased need to compensate for the absent LCIA-mediated HCO3− uptake under these conditions. Similarly, higher LCIB uncompensated contributions as a function of HCO3− concentration in acidic compared to alkaline pH in either a wild type or an lcia mutant background confirms LCIB’s strong preference for CO2. Taken together, these results show a distinct preference of LCIB for CO2 rather than HCO3− and clearly confirm the previously demonstrated association of LCIB with active CO2 uptake.
The similarity between the LCI1 uncompensated contribution profile and that of LCIB in both wild-type and lcia mutant backgrounds, strongly argues that LCI1, like LCIB, prefers CO2 over HCO3− as substrate. Within the same CO2 concentration range, LCI1 uncompensated contributions are similar in alkaline and acidic pH, displaying little or no functional impact of the higher HCO3− abundance in alkaline pH, and, over the same HCO3− concentration range, LCI1 uncompensated contributions are higher in acidic than alkaline pH, even at higher HCO3− concentration. If LCI1 used HCO3−, LCI1 uncompensated contributions should be, as we saw with LCIA, higher in alkaline pH as a function of CO2 concentration and either higher in alkaline pH or at least similar in both pHs as a function of HCO3− concentration. Taken together, data from wild type and lcia mutant backgrounds argue that LCI1 has a strong preference for CO2 over HCO3−.
Although clearly more complex and superficially similar to the LCIA uncompensated contribution, the LCI1 uncompensated contribution and the percentage of LCI1 uncompensated contribution in the pmp1 mutant background also are consistent with a CO2 preference. The unusual LCI1 uncompensated contribution pattern, i.e., increased over specific CO2 concentration ranges (<5 μM in pH 9 and 5–10 and 25–100 μM in pH 7.3), is somewhat reminiscent of the pattern for LCIA uncompensated contribution but is highly similar to the O2 evolution pattern for pmp1 alone, which suggests that the pattern is imposed by the lack of LCIB rather than by a response of LCI1 uncompensated contribution to available Ci species. Through its unidirectional conversion of CO2 to HCO3−, LCIB is important in active uptake of CO2 into the stromal Ci pool and in maintaining the high Ci pool by recycling pyrenoid CO2 leakage (Duanmu et al., 2009a; Jin et al., 2016). To cope with the absence of LCIB, increased Ci flux into the chloroplast through other Ci uptake systems may be needed to maintain a high stromal Ci pool, which is reflected here as increased uncompensated contributions of LCI1 and LCIA in alkaline pH. This argues that the apparent response of LCI1 or LCIA to HCO3− likely results primarily from the necessity to compensate for the LCIB absence. Importantly, when we analyzed the uncompensated contributions of LCI1 and LCIA in the pmp1 background as a function of the HCO3− concentration, the LCIA uncompensated contribution was significantly higher in alkaline than in acidic pH, as expected given LCIA’s preference for HCO3−, whereas the LCI1 uncompensated contribution was significantly higher in acidic than in alkaline pH, implying LCI1 prefers CO2 and consistent with the results from the wild type and the lcia mutant backgrounds.
In addition to addressing the Ci species specificity of LCI1, analyses of LCI1, LCIA and LCIB uncompensated contributions in various genetic backgrounds also demonstrated and confirmed the complementary function of different Ci uptake systems in the Chlamydomonas CCM within the three defined CO2 acclimation states. Consistent with previous reports, LCIA and LCIB show complementary functions in very low CO2 (Wang and Spalding, 2014), and LCI1 and LCIB exhibit apparent complementary roles in low CO2 specifically above air-level CO2, in which an increasing role of LCI1 was detected (Kono and Spalding, Accompanying Paper). In addition, LCI1 and LCIA did not show complementary roles in low and very low CO2 range, which is consistent with reported LCI1 overexpression studies (Yamano et al., 2015).
In vivo studies analyzing the LCI1 uncompensated contribution profiles in three genetic backgrounds over a wide pH range clearly demonstrate LCI1’s strong preference for CO2 over HCO3−, strongly supporting the crystal-structure-based hypothesis that CO2 is the substrate transported by LCI1, putatively acting as a gated CO2 channel. The LCI1 crystal structure and SMD simulations imply that CO2 can migrate, possibly unidirectionally, from the periplasm, through a channel formed within each LCI1 protomer and exit into the cytoplasm. The small LCI1 periplasmic domain appears likely to be hospitable to both CO2 and HCO3−, although the channel entrance and the interior surface appear more favorable for CO2. The outermost cytoplasmic surface of LCI1, on the other hand, is highly electropositive, and thus more favorable for the negatively charged HCO3−, but also could easily tolerate CO2. Thus, the crystal structure is consistent with entry, transport, and exit of CO2, although an equilibrium mixture of CO2 and HCO3− might be present at either or both channel faces. Because multiple carbonic anhydrase (CA) isoforms may be located at or near the periplasmic and cytoplasmic surfaces of the plasma membrane, for example CAH1 in the periplasm (Coleman et al., 1984; Fukuzawa et al., 1990) and CAH9, recently demonstrated to be localized in the cytosol (Mackinder et al., 2017), transported CO2 could potentially leave the periplasm and/or enter the cytoplasm as an equilibrium mixture of CO2 and HCO3−.
Mackinder et al. (2017), reported that LCI1 and HLA3 physically interact in vitro, and that both proteins also physically interact with ACA4, a P-type ATPase/cation transporter possibly involved in H+ extrusion. ACA4 activity may affect the polarization of plasma membrane thus could change LCI1 conformation and potentially regulate LCI1 activity. Since HLA3 is associated with HCO3− transport and LCI1 with CO2 uptake, the interaction between these two proteins is intriguing, but the implications of this interaction are currently a mystery. Furthermore, the interactions with a putative H+ extrusion protein raise the possibility that either HLA3-mediated HCO3− transport or LCI1-mediated CO2 uptake, or both, may be energized by the proton gradient. Any suggested mechanisms to tie Ci uptake to the proton gradient would be highly speculative, but a combination of physiological studies in various mutant backgrounds with in vitro and in vivo validation of protein complex formation and stability, and with site-directed mutagenesis of the three proteins would provide more insights regarding potential functional interactions among these three plasma membrane proteins.
Although this research provides significant advances in our understanding of the structure and functional role of LCI1 in the Chlamydomonas CCM, many questions remain unanswered regarding how LCI1 functions at the physiological, biochemical and structural levels. The LCI1 structure provides a variety of potentially testable hypotheses regarding its function at the molecular level, including the validation of key amino acids in the LCI1 homotrimeric structure that may be key to its function, including those predicted to bind a CO2 molecule in the putative channel, those involved in the putative H-bond gate.
Materials and Methods
Cloning, expression and purification of Chlamydomonas LCI1
Briefly, the full-length LCI1 membrane protein containing C-terminus 6xHis and GFP tags was overproduced in Pichia pastoris SMD1168H cells possessing the pPICZαΩLCI1 expression vector. Cells were grown at 29 °C and bubbled with pure oxygen (2 l/min) in 2 l of fermentation basal salts medium containing 26.7 ml/l 85%-phosphoric acid, 18.2 g/l K2SO4, 0.93 g/l CaSO4, 18.2 g/l K2SO4, 14.9 g/l MgSO4·7H2O, 4.13 g/l KCl, 20 g/l glycerol and 4.4 ml/l PTM1 trace salts (6.0 g/l CuSO4·5H2O, 0.08 g/l NaI, 3.0 g/l MgSO4·H2O, 0.2 g/l Na2MoO4·2H2O, 0.02 g/l H3BO3, 0.5 g/l CoCl2, 20.0 g/l ZnCl2, zinc chloride, 65.0 g/l FeSO4·7H2O, 0.2 g/l biotin, 5.0 ml/l H2SO4) (Invitrogen). Temperature and pH were automatically maintained at 29 °C and 5.0, respectively, by water-cooling and addition of 30% NH4OH solution. A glycerol fed-batch process was initiated when the glycerol concentration was dropped to zero. 50% glycerol and 12 ml/l PTM1 were fed in a rate of 0.33 ml/min for 8 h. LCI1 expression was induced using 100% methanol and 12 ml/l PTM1 with a rate of 0.06 ml/min, and cells were harvested by centrifugation within 32 h of induction.
Collected yeast cells were resuspended in low salt buffer (100 mM sodium phosphate (pH 7.2), 10 % glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM phenylmethanesulfonyl fluoride (PMSF)) and disrupted with a French pressure cell. Debris was removed by centrifugation (1500 x g), and the membrane fraction was collected and washed twice with high salt buffer (20 mM sodium phosphate (pH 7.2), 2 M KCl, 10 % glycerol, 1 mM EDTA and 1 mM PMSF), and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM PMSF, as described previously (Long et al., 2010). The membrane protein was solubilized in 2% (w/v) n-dodecyl-β-D-maltoside (DDM) and insoluble material removed by ultracentrifugation (100,000 x g). The extracted protein was purified on a Co2+-affinity column and the purified protein loaded into a PD-10 desalting column (GE Healthcare Bio-Sciences) to remove imidazole, then concentrated to 10 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5) and 0.03% DDM. The C-terminus 6xHis and GFP tags were cleaved by adding 1 unit of thrombin (Fisher Scientific) per 10 mg of purified LCI1 at room temperature for 20 h. A G200 size exclusion column loaded with buffer solution containing 20 mM Na-HEPES (pH 7.5) and 1.1% n-octyl-β-D-glucopyranoside (β-OG) formed the final purification step. LCI1 protein purity (>95%) was judged using Coomassie Brilliant Blue stained 15% SDS-PAGE. Purified protein was concentrated to 10 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5) and 1.1% β-OG.
Crystallization of Chlamydomonas LCI1
Crystals of the LCI1 protein were obtained using hanging-drop vapor diffusion. The LCI1 crystals were grown at room temperature in 24-well plates with the following procedures. A 0.5 μl protein solution containing 10 mg/ml LCI1 in 20 mM Na-HEPES (pH 7.5) and 1.1% (w/v) β-OG was mixed with a 0.5 μl of reservoir solution containing 18% polyethylene glycol (PEG) 2000 MME, 0.1 M sodium citrate (pH 6.5) and 0.01 M NiCl2. The resultant mixture was equilibrated against 500 μl of the reservoir solution at 25°C. LCI1 crystals grew to a full size in the drops within two months, with typical dimensions of 0.1 mm x 0.1 mm x 0.05 mm. These LCI1 crystals diffracted x-rays to >4 Å resolution. To improve the resolution limit, crystals of the mercury derivative were prepared by incubating the LCI1 crystals in solution containing 18% PEG 2000 MME, 0.1 M sodium citrate (pH 6.5), 0.01 M NiCl2 and 1 mM HgCl2 for 15 min at 25°C. Cryoprotection of the crystals was achieved by raising the glycerol concentration stepwise to 25%, with a 5% increment in each step.
Data collection, structural determination and refinement
All diffraction data were collected at 100K at beamline 24ID-C located at the Advanced Photon Source, using a Platus 6M detector (Dectris Ltd., Switzerland). Diffraction data were processed using DENZO and scaled using SCALEPACK (Otwinowski and Minor, 1997). LCI1 crystals belong to space group P3121 (Table S1). Based on the LCI1 molecular weight (21.4 kDa), the asymmetric unit is expected to contain three membrane protein molecules with a solvent content of 64.1%. Three mercury sites were identified using SHELXD (Schneider and Sheldrick, 2002) as implemented in the HKL2MAP package (Pape and Schneider, 2004). These heavy-atom sites were refined by single anomalous dispersion (SAD) at a resolution of 4 Å using the program AutoSol implemented in PHENIX (Adams et al., 2002). Phases were then subjected to density modification, NCS averaging, and phase extension to 3.19 Å-resolution using the program RESOLVE (Terwilliger, 2001). The resulting phases were of excellent quality, which allowed us to trace most of the secondary structural features of the three LCI1 molecules within the asymmetric unit. After tracing the initial model manually using the program Coot (Emsley and Cowtan, 2004), the model was refined using PHENIX (Adams et al., 2002) leaving 5% of reflections in Free-R set. Feature-enhanced maps calculated using PHENIX and B-factor sharpening maps created using CCP4 were employed to ascertain loop-regions and side chains in the structure. Iterations of refinement using PHENIX (Adams et al., 2002) and CNS (Brunger et al., 1998) and model building in Coot (Emsley and Cowtan, 2004) lead to the 3.19 Å-resolution structural model of the LCI1 trimer with excellent geometrical characteristics (Table S1).
Docking of CO2
The program AutoDock Vina (Trott and Olson, 2010) was used to predict the CO2-binding mode. A monomer of the LCI1 structure with the bound CO2 molecule removed was used for docking. A grid of 35 Å x 35 Å x 35 Å with 0.375 Å spacing was calculated around the docking area using AutoGrid. The iterated local search global optimizer algorithm was used to predict the binding free energies for these compounds.
Native mass spectrometry
Purified LCI1 protein was buffer exchanged in 200 mM ammonium acetate (pH 8.0) and 0.05% lauryldimethylamine oxide (LDAO) using a Biospin-6 column (BioRad) prior to mass spectrometry analysis. The protein was directly introduced into the mass spectrometer using gold-coated capillary needles prepared in house (Hernandez and Robinson, 2007). Data was collected on a modified QExactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Berman, Germany) optimized for analyzing high mass complexes (Gault et al., 2016). Optimized instrument parameters were as follows: capillary voltage 1.2kV, S-lens RF potential 100V, quadrupole selection range between 2,000 and 20,000 m/z, collisional activation in the HCD cell 100V, argon pressure in the HCD cell 1.12×10–9 mbar, and resolution of the instrument was acquired at 17,500 with m/z = 200 (transient time = 64 ms).
Lipid analysis
Lipid analysis was carried out by following a previously described protocol (Bechara et al., 2015). Briefly, intact LCI1 protein was digested with trypsin overnight at 37 °C, lyophilized and re-dissolved in 35% acetonitrile. The peptide/lipid mixture was loaded onto a C18 column (Acclaim PepMap 100, C18, 75 μm × 15 cm; Thermo Scientific) and separated with a linear gradient of 35–100% acetonitrile. The column eluent was delivered to a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) coupled to the column. The LTQ-Orbitrap XL was operated in negative ion mode and in data-dependent acquisition set-up to perform five MS/MS scans per MS scan. Survey full-scan MS spectra were acquired in the orbitrap (m/z 350–2000) with a resolution of 60,000.
Steered Molecular Dynamics
A 120 Å x 120 Å palmitoyl-oleyl-phosphatidylethanolamine (POPE) membrane bilayer was constructed using the membrane builder plugin in NAMD (Phillips et al., 2005) with the membrane normal parallel to the z-axis. The protein was embedded in the membrane such that its threefold symmetrical axis was perpendicular to the membrane plane. The system was then solvated by the addition of 5 Å of water layers on both sides of the lipid bilayer. Na+ and Cl− ions were also included to ensure the balance of charges, resulting in a total number of 112,470 atoms (112,472 atoms in the case of HCO3−) in the system. After, the system was equilibrated at 10K and the temperature was slowly increased to 310 K in steps of 1 K per 1 ps each step for 200,000 steps. A harmonic restraint of 1 kcal/mol Å2 was applied to the protein during the process of raising the temperature. A 100 ps equilibration run was then performed at 310K, in which all atoms were allowed to move freely. Langevin dynamics (1 ps−1 damping coefficient) was used to maintain constant temperature. A cutoff distance of 12 Å was used to mimic the effect of van der Waals interactions. A periodic boundary condition was imposed using the particle mesh Ewald method with a 1 Å grid spacing to evaluate long-range full electrostatic interactions. In all simulations, NAMD (Phillips et al., 2005) with CHARMM27 parameter set (Schlenkrich et al., 1996; MacKerell et al., 1998) was used. The topology and parameter files for CO2 were generated using the SwissParam server (MacKerell et al., 1998; Zoete et al., 2011) and those for HCO3− were obtained using the Paratool plugin in VMD (Humphrey et al., 1996).
SMD simulations were carried out to investigate the transport pathway of CO2 and HCO3−. These ligands were placed accordingly at one end of the channel. Constant velocity steered MD (cv-SMD) was applied. Specifically the carbon atom of CO2 or HCO3− was pulled in a direction perpendicular to the membrane plane (Z-direction) using a harmonic constraint at a velocity of 30 Å/ns with a force constant (k) of 5 kcal mol−1Å−2. To prevent overall translational motion of the system, specific residues at the periphery of the periplasmic side (T39, V59, P73, N135 and D143) and cytoplasmic side (V10, V99, M107 and S170) of the protein were harmonically restrained with a force constant of 5 kcal/mol Å2 in the z-direction.
Chlamydomonas Strains and Culture Conditions
Wild-type strain of Chlamydomonas reinhardtii, 21gr mt+ (CC-1690), was obtained from the Chlamydomonas culture center, Duke University, Durham, NC, USA. The LCI1 mutant, lci1 cw15 mt− (LMJ.RY0402.191570; lci1–570 ), and its wild-type progenitor, cw15 mt− (CC-4533), were acquired from the collection of indexed mutants in Chlamydomonas Library Project (Li et al., 2016). The LCIA mutant, lcia-10B mt+, was generated from backcrossing an lcia90 mt+ (CC-5067) (Wang and Spalding, 2014) seven times to wild-type 21gr mt− (CC-5370), with screening for the lcia mutant allele in each generation. The LCIB mutant, pmp1-2137 mt+ (CC-5378), was isolated from backcrossing of the original pmp1 mutant, 16–5k mt− (CC-4676) (Spalding et al., 1983), seven times to wild-type 2137 (CC-3269), with screening for the lcib mutant allele in each generation. The double mutants of LCI1/LCIA (l1a), LCI1/LCIB (l1b) and LCIA /LCIB (lab) were generated as previously described (Kono and Spalding, Accompanying Paper).
Gas conditions used in this study are: high CO2 (5% CO2 [v/v]), low CO2 (normal air ~0.04% CO2 [v/v]) and very low CO2 (<0.01% CO2 [v/v]). The gas conditions were achieved as previously described (Wang and Spalding, 2006). All strains were regularly maintained on solid Tris-acetate-phosphate (TAP) medium (Gorman and Levine, 1965) and grown at room temperature in high CO2 chamber under low light.
Photosynthetic O2 evolution measurement
Photosynthetic O2 evolution rates in each strain were measured as previously described (Kono and Spalding, Accompanying Paper). The O2 evolution rates presented in the accompanying paper were used to calculate the uncompensated contribution of LCI1, LCIA and LCIB in each genetic background and pH. The calculation of the uncompensated contributions was adapted from Wang and Spalding (2014).
Accession Code
Atomic coordinates and structure factors for the structure of LCI1 have been deposited at the RCSB Protein Data Bank with an accession code 6BHP.
Supplementary Material
Figure S1. Stereo view of the electron density maps of LCI1 at a resolution of 3.19 Å.
Figure S2. Identified bound lipid molecules on the LCI1 membrane protein.
Figure S3. Uncompensated contributions of LCI1 and LCIB in an lcia mutant background at pH 6, pH 7.3 and pH 9 plotted against calculated CO2 and HCO3− concentration of 0–250 μM and of 0–1200 μM, respectively.
Figure S4. Uncompensated contributions of LCI1 and LCIA in a pmp1 strain at pH 6, pH 7.3 and pH 9 plotted against the calculated CO2 concentration of 0–250 μM and the HCO3− concentration of 0–1200 μM.
Figure S5. Percentage of LCI1 and LCIA uncompensated contribution to photosynthesis in a pmp1 mutant background at pH 6, pH 7.3 and pH 9 plotted as a function of calculated CO2 concentrations of 0–250 μM.
Figure S6. Calculated, uncompensated contribution of LCI1, LCIA, and LCIB at pH 6 and pH 7.3 in a wild-type, a pmp1, an lcia, and an lci1 background plotted against calculated CO2 concentration with a focus on the very low CO2 range (0–5 μM CO2).
Figure S7. Calculated, uncompensated contribution of LCI1, LCIA, and LCIB at pH 9 in a wild-type, a pmp1, an lcia, and an lci1 plotted against calculated CO2 concentration with a focus on the very low CO2 range (0–5 μM CO2).
Table S1. Data collection, phasing and structural refinement statistics.
Table S2. Observed masses of the lipid-LCI1 complexes.
Acknowledgements
This work was supported by a U. S. National Science Foundation Grant MCB–0952323 (M.H.S.), a U. S. Department of Energy Grant DEFG02–12ER16335 (M.H.S.), and an NIH Grant R01GM086431 (E.W.Y.). The crystal structure work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by an award GM103403 from the National Institutes of General Medical Sciences. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02–06CH11357.
Footnotes
Competing Interests
The authors declare that there is no conflict of interest associated with the manuscript.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Badger MR (1985). Photosynthetic oxygen exchange. Annual Review of Plant Physiology 36, 27–53. [Google Scholar]
- Bechara C, Noll A, Morgner N, Degiacomi MT, Tampe R, and Robinson CV (2015). A subset of annular lipids is linked to the flippase activity of an ABC transporter. Nat Chem 7, 255–262. [DOI] [PubMed] [Google Scholar]
- Bolla JR, Agasid MT, Mehmood S, and Robinson CV (2019). Membrane protein–lipid interactions probed using mass spectrometry. Annual Review of Biochemistry 88, 85–111. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, and Warren GL (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Coleman JR, Berry JA, Togasaki RK, and Grossman AR (1984). Identification of extracellular carbonic anhydrase of Chlamydomonas reinhardtii. Plant physiology 76, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundari TR, Wilson AK, Drummond ML, Gonzalez HE, Jorgensen KR, Payne S, Braunfeld J, De Jesus M, and Johnson VM (2009). CO2-formatics: how do proteins bind carbon dioxide? Journal of chemical information and modeling 49, 2111–2115. [DOI] [PubMed] [Google Scholar]
- Duanmu D, Wang Y, and Spalding MH (2009a). Thylakoid lumen carbonic anhydrase (CAH3) mutation suppresses air-dier phenotype of LCIB mutant in Chlamydomonas reinhardtii. Plant physiology 149, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Miller AR, Horken KM, Weeks DP, and Spalding MH (2009b). Knockdown of limiting-CO2–induced gene HLA3 decreases HCO3− transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences 106, 5990–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ (2019). Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist 223, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Tachiki A, and Miyachi S (1990). Nucleotide sequences of two genes CAH1 and CAH2 which encode carbonic anhydrase polypeptides in Chlamydomonas reinhardtii. Nucleic acids research 18, 6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang Y, Fei X, Wright DA, and Spalding MH (2015). Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii. The Plant Journal 82, 1–11. [DOI] [PubMed] [Google Scholar]
- Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, Wu B, Kleanthous C, Belov M, Damoc E, Makarov A, and Robinson CV (2016). High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods 13, 333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, and Raven JA (2005). CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131. [DOI] [PubMed] [Google Scholar]
- Gorman DS, and Levine RP (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proceedings of the National Academy of Sciences of the United States of America 54, 1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Donlan JA, Hopper JT, Uzdavinys P, Landreh M, Struwe WB, Drew D, Baldwin AJ, Stansfeld PJ, and Robinson CV (2017). The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, and Robinson CV (2007). Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc 2, 715–726. [DOI] [PubMed] [Google Scholar]
- Hofmann K (1993). TMbase-A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374, 166. [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Jin S, Sun J, Wunder T, Tang D, Cousins AB, Sze SK, Mueller-Cajar O, and Gao Y-G (2016). Structural insights into the LCIB protein family reveals a new group of β-carbonic anhydrases. Proceedings of the National Academy of Sciences, 201616294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar T, Periyasamy G, and Balasubramanian S (2013). CO2 migration pathways in oxalate decarboxylase and clues about its active site. The Journal of Physical Chemistry B 117, 12451–12460. [DOI] [PubMed] [Google Scholar]
- Kono A, and Spalding MH (Accompanying Paper). [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, Grossman AR, and Jonikas MC (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. The Plant Cell 28, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, and Yu EW (2010). Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, and Karplus M (1998). All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Mackinder LC, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, and Jonikas MC (2017). A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147. e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, and Griffiths H (2013). Origins and diversity of eukaryotic CO2-concentrating mechanisms: lessons for the future. Journal of experimental botany 64, 769–786. [DOI] [PubMed] [Google Scholar]
- Moroney JV, and Tolbert NE (1985). Inorganic carbon uptake by Chlamydomonas reinhardtii. Plant Physiology 77, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Mukherjee B, Tsujikawa T, Yanase M, Nakano H, Moroney JV, and Fukuzawa H (2010). Expression of a low CO2–inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. The Plant Cell 22, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). [20] Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pape T, and Schneider TR (2004). HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. Journal of applied crystallography 37, 843–844. [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, and Schulten K (2005). Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenkrich M, Brickmann J, MacKerell AD Jr, and Karplus M (1996). An empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In Biological Membranes (Springer), pp. 31–81. [Google Scholar]
- Schneider TR, and Sheldrick GM (2002). Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Shimazaki K-I, and Zeiger E (1987). Red light-dependent CO2 uptake and oxygen evolution in guard cell protoplasts of Vicia faba L.: evidence for photosynthetic CO2 fixation. Plant Physiology 84, 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Goodfellow JM, and Wallace B (1993). The pore dimensions of gramicidin A. Biophysical journal 65, 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Spreitzer RJ, and Ogren WL (1983). Reduced inorganic carbon transport in a CO2-requiring mutant of Chlamydomonas reinhardii. Plant physiology 73, 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, and Salvucci ME (2002). Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annual review of plant biology 53, 449–475. [DOI] [PubMed] [Google Scholar]
- Stern AI, Schiff JA, and Epstein H (1964). Studies of chloroplast development in Euglena. V. Pigment biosynthesis, photosynthetic oxygen evolution and carbon dioxide fixation during chloroplast development. Plant physiology 39, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sültemeyer DF, Miller AG, Espie GS, Fock HP, and Canvin DT (1989). Active CO2 transport by the green alga Chlamydomonas reinhardtii. Plant Physiology 89, 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2001). Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr D Biol Crystallogr 57, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim Z-Y, Johns MK, Holmes AB, de Koster CG, Ktistakis NT, and Munnik T (2004). Isolation and identification of phosphatidic acid targets from plants. The Plant Journal 39, 527–536. [DOI] [PubMed] [Google Scholar]
- Trott O, and Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K, Wang Y, Nakamura Y, and Spalding MH (2001). Insertional mutants of Chlamydomonas reinhardtii that require elevated CO2 for survival. Plant physiology 127, 607–614. [PMC free article] [PubMed] [Google Scholar]
- Vance P, and Spalding MH (2005). Growth, photosynthesis, and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Canadian journal of botany 83, 796–809. [Google Scholar]
- Wang Y, and Spalding MH (2006). An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences 103, 10110–10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Spalding MH (2014). Acclimation to very low CO2: Contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant physiology 166, 2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Sato E, Iguchi H, Fukuda Y, and Fukuzawa H (2015). Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences 112, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Liu Y, Zhang Q, Wei Q, and Zhang W (2006). Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 224, 545–555. [DOI] [PubMed] [Google Scholar]
- Zoete V, Cuendet MA, Grosdidier A, and Michielin O (2011). SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32, 2359–2368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Stereo view of the electron density maps of LCI1 at a resolution of 3.19 Å.
Figure S2. Identified bound lipid molecules on the LCI1 membrane protein.
Figure S3. Uncompensated contributions of LCI1 and LCIB in an lcia mutant background at pH 6, pH 7.3 and pH 9 plotted against calculated CO2 and HCO3− concentration of 0–250 μM and of 0–1200 μM, respectively.
Figure S4. Uncompensated contributions of LCI1 and LCIA in a pmp1 strain at pH 6, pH 7.3 and pH 9 plotted against the calculated CO2 concentration of 0–250 μM and the HCO3− concentration of 0–1200 μM.
Figure S5. Percentage of LCI1 and LCIA uncompensated contribution to photosynthesis in a pmp1 mutant background at pH 6, pH 7.3 and pH 9 plotted as a function of calculated CO2 concentrations of 0–250 μM.
Figure S6. Calculated, uncompensated contribution of LCI1, LCIA, and LCIB at pH 6 and pH 7.3 in a wild-type, a pmp1, an lcia, and an lci1 background plotted against calculated CO2 concentration with a focus on the very low CO2 range (0–5 μM CO2).
Figure S7. Calculated, uncompensated contribution of LCI1, LCIA, and LCIB at pH 9 in a wild-type, a pmp1, an lcia, and an lci1 plotted against calculated CO2 concentration with a focus on the very low CO2 range (0–5 μM CO2).
Table S1. Data collection, phasing and structural refinement statistics.
Table S2. Observed masses of the lipid-LCI1 complexes.