Abstract
Dorsal root ganglion (DRG) neurons detect sensory inputs and are crucial for pain processing. They are often studied in vitro as dissociated cell cultures with the assumption that this reasonably represents in vivo conditions. However, to our knowledge, no study has directly compared genome-wide transcriptomes of DRG tissue in vivo versus in vitro, or between laboratories and culturing protocols. Comparing RNA sequencing-based transcriptomes of native to cultured (4 days in vitro) human or mouse DRG, we found that the overall expression levels of many ion channels and GPCRs specifically expressed in neurons are markedly lower although still expressed in culture. This suggests that most pharmacological targets expressed in vivo are present under the condition of dissociated cell culture, but with changes in expression levels. The reduced relative expression for neuronal genes in human DRG cultures is likely accounted for by increased expression of genes in fibroblast-like and other proliferating cells, consistent with their mitotic status in these cultures. We found that the expression of a subset of genes typically expressed in neurons increased in human and mouse DRG cultures relative to the intact ganglion, including genes associated with nerve injury or inflammation in preclinical models such as BDNF, MMP9, GAL, and ATF3. We also found a striking upregulation of a number of inflammation-associated genes in DRG cultures, although many were different between mouse and human. Our findings suggest an injury-like phenotype in DRG cultures that has important implications for the use of this model system for pain drug discovery.
Summary
We cataloged gene expression in mouse and human dorsal root ganglion in native and cultured conditions with analysis focused on pain therapeutics discovery and development.
Introduction
Nociceptors within the dorsal root ganglia (DRG) or trigeminal ganglia (TG) are the first neurons in the pain pathway [67]. These neurons are crucial contributors to chronic pain disorders ranging from inflammatory to neuropathic pain [3]. These neurons are frequently studied to gain insight into mechanisms that drive chronic pain and to develop better treatment strategies. Traditionally, investigators have studied rodent nociceptors in vitro as dissociated cell cultures prepared from DRG or TG. More recently, investigators have also started to study DRG nociceptors from human organ donors and surgical patients [16; 43; 51; 53; 54; 62; 76]. This creates a “clinical bridge” for advancing mechanisms or therapeutics from rodents toward the clinic. These models have many advantages; cultures can easily be used for electrophysiology, Ca2+ imaging, biochemical, or other functional studies. These studies have unquestionably advanced the field of pain neurobiology and sensory transduction.
Despite the widespread use of this model system [38], many investigators are skeptical of the degree to which these cells in dissociated culture accurately reflect the status of nociceptors in vivo. Several studies have analyzed the genome-wide RNA profiles of these dissociated cultures [26; 47], but not in the context of changes with respect to the native, acutely dissected ganglia (referred to as “intact” DRG henceforth). A previous study by Thakur et al [57] contrasted RNA sequencing (RNA-seq) profiles of intact DRGs with unsorted, acutely dissociated DRGs. The study found few differences between intact DRG tissue and unsorted, acutely dissociated DRG, suggesting that the process of dissociation does not dramatically alter the molecular phenotype. While some studies have compared expression of a single gene or a handful of genes in these in vitro cultures vs. the intact ganglia [23; 53], we are unaware of any study that has used genome-wide assays to study how gene expression might be altered from native to cultured DRG conditions. We addressed this question by comparing intact versus cultured DRG from human donors and mice using RNA-seq. We designed a series of experiments to study how the transcriptomes of human and mouse native DRG differ under the conditions of dissociated cell cultures relative to native, intact ganglia. Our findings provide a comprehensive, genome-wide evaluation of gene expression changes from native to cultured DRG in both humans and mice. Consistent with previous studies [19; 44], we found that DRG neurons in culture show transcriptional signatures that suggest an injury phenotype [6; 27]. This supports the use of cultured DRG neurons as a model system to study underlying mechanisms of pain. However, our findings point out some shortcomings of using these models to study multiple classes of receptors that show altered expression in culture. Some of these differences do not occur consistently across species, suggesting mouse DRG cultures may not be a good surrogate for human cultures in certain experiments. The data provided in this study will help investigators choose and design appropriate experimental parameters, and can provide an important tool for future experiments in the pain and somatosensory fields.
Methods
Experimental Design
Because genetic variation can be a possible contributor to transcriptome level differences in nervous system samples from human populations [43; 45], we chose a study design wherein we cultured lumbar DRGs from one side in human donors and immediately froze the opposite side from the same donor for RNA sequencing. Although we used an inbred mouse strain (C57BL/6) for parallel mouse studies, we used a similar culturing design where cultures were done in two independent laboratories to look for variability across labs. RNA sequencing was performed at 4 days in vitro (DIV) to stay within the electrophysiologically relevant range of 1 – 7 DIV for human DRG and the biochemical assay range of 4 – 7 DIV for both human and mouse DRG.
Animals
Price Lab: All procedures were approved by the Institutional Animal Care and Use Committee of University of Texas at Dallas and were in strict accordance with the US National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals. Adult C57Bl/6 mice (8–15 weeks of age) were bred in house, and were originally obtained from The Jackson Laboratory. Animals were housed in the University of Texas at Dallas animal facilities on a 12 hour light/dark cycle with access ad libitum to food and water.
Gereau Lab: All procedures were approved by the Animal Care and Use Committee of Washington University and in strict accordance with the US National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals. Adult C57Bl/6 mice (8–15 weeks of age) were bred in house, originally obtained from The Jackson Laboratory. Animals were housed in Washington University School of Medicine animal facilities on a 12 hour light/dark cycle with access ad libitum to food and water.
Intact vs cultured mouse DRG
Price lab: Male and female C57BL/6 mice (4 week-old, ~15–20 g; n=3, for each sex) were anesthetized with isoflurane and killed by decapitation. Male C57BL/6 mice (5 week-old, n=2) were used for RNAscope validation. Mice were not perfused prior to removal of DRGs. Lumbar DRGs (L1–L6) from one side of the spine were frozen in RNAlater (Invitrogen) while DRGs from the other side from the same mouse was cultured and then scraped at 4 DIV into RNAlater. All L1–L6 DRGs were used for RNAscope validation. L1–L6 DRGs for culturing were dissected and placed in chilled HBSS (Invitrogen) until processed. DRGs were then digested in 1 mg/ml collagenase A (Roche) for 25 min at 37°C then subsequently digested in 1 mg/ml collagenase D for 20 min at 37°C. DRGs were then triturated in 1 mg/ml trypsin inhibitor (Roche), then filtered through a 70 μm cell strainer (Corning). Cells were pelleted then resuspended in DMEM/F12 with GlutaMAX (Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 1% penicillin and streptomycin, 5 ng/mL mouse 2.5S NGF (Millipore), and 3 μg/ml 5-fluorouridine with 7 μg/ml uridine. Cells were distributed evenly across 4 wells using a 24-well plate coated with poly-D-lysine (Becton Dickinson). For RNAscope validation cultures, cells were plated as described on an 8-well chamber slide (Nunc Lab-Tek). DRG neurons were maintained in a 37°C incubator containing 5% CO2 with a media change every other day. At 4 DIV, cells were scraped into 500 uL RNAlater and processed for RNA extraction.
Gereau lab: Male and female C57Bl/6 mice (n=3, for each sex) were deeply anesthetized with isoflurane and quickly decapitated. Mice were not perfused prior to removal of DRGs. From one side, L1–6 DRG were extracted, directly placed into 500μL RNAlater, and stored at −80°C. From the other side, L1–6 DRG were extracted and dissociated in freshly made N-methyl-D-glucamine (NMDG) solution (Valtcheva et al 2016). DRG were digested in 15U/mL papain (Worthington Biochemical) for 20min at 37°C, washed, and then further digested in 1.5 mg/mL collagenase type 2 (Sigma) for another 20 min at 37°C. DRG were washed and triturated in DRG media [5% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Corning) in Neurobasal A medium 1x (Gibco) plus Glutamax (Life Technologies) and B27 (Gibco)]. Final solutions of cells were filtered (40 μm, Fisher) and cultured in DRG media on coverslips coated with poly-D-lysine (Sigma) and rat tail collagen (Sigma). Cultures were maintained in an incubator at 37°C containing 5% CO2. On 4 DIV (no media changes), cultured coverslips were scraped in 500 μL RNA later and stored at −80°C.
Intact vs cultured human DRG
Studies involving human DRG were done on de-identified biospecimens and approved by Institutional Review Boards at Washington University in St. Louis and University of Texas at Dallas.
Gereau lab: Human dorsal root ganglia extraction and culturing was performed as described previously (Valtcheva et al 2016), in a similar manner to the mouse culturing protocol. Briefly, in collaboration with Mid-America Transplant Services, L4–L5 DRG were extracted from tissue/organ donors less than 2 hrs after aortic cross clamp. Donor information is presented in Table 1. DRGs were placed in NMDG solution for transport to the lab for fine dissection. From one side, intact L4–5 DRG were directly placed into 500 μL RNAlater, and stored at −80°C. From the other side, L4–5 DRG were minced and cultured. Pieces were dissociated enzymatically with papain and collagenase type 2 for 1hr each, and mechanically with trituration. Final solutions were filtered (100 μm, Fisher) and cultured with DRG media. On 4 DIV, cultured coverslips were scraped in 500μL RNAlater and stored at −80°C.
Table 1.
Human DRG donor characteristics and donor – sample mapping
| Donor id | Age | Sex | Race | Cause of Death | Sample ids |
|---|---|---|---|---|---|
| 1 | 53 | F | White | ICH/Stroke | hDRG-1F, hDRG-1Fre, hDIV4–1F, hDIV4–1Fre |
| 2 | 12 | F | White | Anoxia/OD | hDRG-2F, hDIV4–2F |
| 3 | 26 | M | White | Head trauma/MVA | hDRG-3M, hDIV4–3M |
| 4 | 34 | M | White | Anoxia/OD | hDRG-4M, hDIV4–4M |
| 5 | 18 | F | White | Head trauma/MVA | hDRG-5F, hDIV4–5F |
| 6 | 18 | M | White | Head trauma/GSW | hDRG-6M, hDIV4–6M |
RNA sequencing
Human and mouse DRG tissue/cultured cells were stored in RNAlater and frozen in −80 °C until use. Samples obtained at the Washington University at St Louis were shipped to UT Dallas on dry ice for uniform library preparation. All RNA isolation and sequencing was done in the Price Lab. On the day of use, the frozen tubes were thawed to room temperature. To obtain RNA from tissue samples, the tissue was extracted from RNAlater with ethanol cleaned tweezers and put in 1 mL of QIAzol (QIAGEN Inc.) inside 2 mL tissue homogenizing CKMix tubes (Bertin Instruments). To obtain RNA from cell cultures, cells were spun down to the bottom of the tube by centrifuge at 5000 × g for 10 min. RNAlater was then removed from the tube, and cells were resuspended with 1 mL of QIAzol and transferred to the homogenizing tube. For both tissues and cell cultures, homogenization was performed for 3 × 1 min with Minilys personal homogenizer (Bertin Instruments) at 4 °C. This time course was used to avoid heating during homogenization. RNA extraction was performed with RNeasy Plus Universal Mini Kit (QIAGEN Inc.) with the manufacturer provided protocol. RNA was eluted with 30 μL of RNase free water. Based on the RNA size profile determined by the Fragment Analyzer (Agilent Technologies) with the High Sensitivity Next Generation Sequencing (NGS) fragment analysis kit, we decided to sequence all human samples with total RNA library preparation and all mouse samples with mRNA library preparation. Total RNA was purified and subjected to TruSeq stranded mRNA library preparation for mouse or total RNA Gold library preparation (with ribosomal RNA depletion) for human, according to the manufacturer’s instructions (Illumina). Quality control was performed for RNA extraction and cDNA library preparation steps with Qubit (Invitrogen) and High Sensitivity NGS fragment analysis kit on the Fragment Analyzer (Agilent Technologies). After standardizing the amount of cDNA per sample, the libraries were sequenced on an Illumina NextSeq500 sequencing platform with 75-bp single-end reads in multiplexed sequencing experiments, yielding a median of 22.3 million reads per sample. mRNA library preparation and sequencing was done at the Genome Center in the University of Texas at Dallas Research Core Facilities.
RNAscope-based imaging
RNAscope in situ hybridization (multiplex version 1) [65] assays were conducted based on Advanced Cell Diagnostics (ACD) protocols.
Intact DRG:
Fresh frozen lumbar DRGs were rapidly dissected, frozen in cryomolds with O.C.T (Fisher Scientific; Cat# 23-730-571) over dry ice and sectioned at 20μm onto charged slides. The sections were fixed in cold (4°C) 10% formalin for 15 minutes and then dehydrated in 50% ethanol (5 min), 70% ethanol (5 min) and 100% ethanol (10 min) at room temperature. Slides were briefly air dried and boundaries were then drawn around each section using the hydrophobic ImmEdge PAP pen (Vector Labs). When hydrophobic boundaries had dried, protease IV reagent was used to incubate the sections for 2 minutes and then washed in 1X phosphate buffered saline (PBS). Every slide was placed in a prewarmed humidity control tray (ACD) with dampened filter paper and incubated in a mixture of Channel 1 (Cd68; ACD Cat# 316611), Channel 2 (Calca; ACD Cat#417961), and Channel 3 (P2rx3; ACD Cat# 521611) probes for 2 hours at 40°C. This was performed one slide at a time to avoid liquid evaporation and section drying. Following probe incubation, the slides were washed two times in 1X RNAscope wash buffer, submersed in AMP-1 reagent, and returned to the oven for 30 minutes. Washes and amplification were repeated using AMP-2, AMP-3 and AMP-4B reagents with 15 min, 30 min, and 15 min incubation period, respectively. Slides were then washed two times in 0.1M phosphate buffer (PB, pH7.4) and then submerged in blocking reagent (10% Normal Goat serum and 0.3% Triton-X 100 in 0.1M PB) for 1 hour at room temperature. Slides were incubated in primary antibody (mouse-anti-Neurofilament 200; clone N52; Sigma) at 1:500 in blocking buffer overnight at 4°C. The next day, slides were washed two times in 0.1M PB, and then incubated in secondary antibody (goat-anti-mouse H&L 405; 1:2000) for 1 hour at room temperature. Sections were washed two times in 0.1M PB, air dried, and cover-slipped with Prolong Gold Antifade (Fisher Scientific; Cat# P36930) mounting medium.
Cultured DRG:
At 4 DIV (no media change), media was aspirated from each well and the chambers disassembled from the slide. The slide was washed once in 1X PBS and fixed for 30 minutes at room temperature in 10% formalin. Slides were then washed twice in 1X PBS after which RNAscope in situ hybridization was performed as described above with the noted changes. Protease incubation was 10 minutes at room temperature with protease III reagent (1:30 in 1X PBS). Probe incubation used a Channel 1 (Cd68) probe. Permeabilizing reagent (0.02% Triton-X 100) was added to blocking buffer (10% normal goat serum in 0.1M PB) only for the one hour blocking step. Slides were incubated in primary (rabbit-anti-peripherin; 1:1000; Sigma) and secondary (goat-anti-rabbit H&L 488; 1:2000) antibodies as described. Slides were washed once in 0.1 M PB and incubated in DAPI (1:5000) for 5 minutes at room temperature before washing, mounting and imaging as described.
Imaging:
All images were taken on an Olympus FV3000 confocal microscope using the 20X and 40X objectives. Images were pseudo-colored to show four distinct color frequencies and overlaid, using the CellSens software (Olympus).
Computational analysis
Mapping and TPM quantification:
RNA-seq read files (fastq files) were checked for quality by FastQC (Babraham Bioinformatics, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and read trimming was done based on the Phred score and per-base sequence content (base pairs 13 through 72 were retained). Trimmed Reads were then mapped against the reference genome and transcriptome (Gencode vM16 and GRCm38.p5 for mouse, Gencode v27 and GRCh38.p10 for human [22]) using STAR v2.2.1 [18]. Relative abundances in Transcripts Per Million (TPM) for every gene of every sample was quantified by stringtie v1.3.5 [48]. Downstream analyses were restricted to protein coding genes to make human (total RNA) and mouse (polyA+ RNA) libraries comparable, hence TPMs of only genes annotated as coding genes in the Gencode database were renormalized to sum to a million. Sequencing and mapping statistics reported by STAR are presented in Table 2.
Table 2.
Statistics for RNA-seq experiments
| Sample id | No. of reads sequenced | No. of reads mapped | No. of reads mapped uniquely | No. of coding genes detected |
|---|---|---|---|---|
| mDRG-1Mg | 22,212,434 | 21,048,941 | 16,559,517 | 15,749 |
| mDRG-2Mg | 19,189,967 | 18,300,700 | 14,957,918 | 15,626 |
| mDRG-3Mg | 22,487,076 | 21,509,723 | 17,478,260 | 15,731 |
| mDRG-4Fg | 22,509,372 | 21,402,718 | 17,608,765 | 15,719 |
| mDRG-5Fg | 19,655,816 | 18,756,552 | 15,205,161 | 15,619 |
| mDRG-6Fg | 23,274,760 | 22,290,521 | 18,174,167 | 15,828 |
| mDRG-1Mp | 14,877,799 | 14,315,363 | 11,760,256 | 15,536 |
| mDRG-2Mp | 15,635,533 | 15,082,808 | 12,348,811 | 15,593 |
| mDRG-3Mp | 16,808,435 | 16,186,083 | 13,173,528 | 15,797 |
| mDRG-4Fp | 15,577,724 | 14,993,698 | 12,189,645 | 15,631 |
| mDRG-5Fp | 15,316,097 | 14,756,450 | 11,993,815 | 15,638 |
| mDRG-6Fp | 16,108,903 | 15,531,301 | 12,558,411 | 15,707 |
| mDIV4–1Mg | 19,520,498 | 18,593,009 | 15,426,436 | 15,193 |
| mDIV4–2Mg | 23,861,527 | 22,827,565 | 18,645,619 | 15,402 |
| mDIV4–3Mg | 22,706,726 | 21,755,398 | 17,668,475 | 15,520 |
| mDIV4–4Fg | 14,769,253 | 13,761,459 | 10,464,363 | 15,078 |
| mDIV4–5Fg | 21,735,780 | 20,745,007 | 16,544,691 | 15,362 |
| mDIV4–6Fg | 21,313,704 | 20,495,656 | 16,698,375 | 15,463 |
| mDIV4–1Mp | 15,572,289 | 14,959,958 | 11,966,895 | 14,879 |
| mDIV4–2Mp | 16,189,281 | 15,581,390 | 12,564,211 | 15,105 |
| mDIV4–3Mp | 17,299,306 | 16,653,412 | 13,433,467 | 15,025 |
| mDIV4–4Fp | 15,873,155 | 15,285,290 | 12,296,395 | 14,778 |
| mDIV4–5Fp | 14,300,332 | 13,735,983 | 11,025,628 | 14,767 |
| mDIV4–6Fp | 16,582,457 | 15,955,531 | 12,832,261 | 14,991 |
| hDRG-1F | 37,683,580 | 35,740,910 | 12,227,687 | 15,590 |
| hDRG-1Fre | 60,648,611 | 58,374,576 | 19,659,672 | 16,016 |
| hDRG-2F | 45,504,043 | 43,479,464 | 16,157,986 | 15,881 |
| hDRG-3M | 43,890,090 | 41,819,026 | 14,804,561 | 15,656 |
| hDRG-4M | 83,956,740 | 79,390,702 | 27,625,219 | 16,507 |
| hDRG-5F | 44,290,887 | 42,269,829 | 15,481,526 | 15,594 |
| hDRG-6M | 39,464,882 | 36,862,011 | 12,529,277 | 15,684 |
| hDIV4–1F | 29,815,327 | 28,497,933 | 9,368,106 | 14,420 |
| hDIV4–1Fre | 42,975,599 | 40,614,264 | 15,459,290 | 15,230 |
| hDIV4–2F | 35,923,946 | 34,767,067 | 19,764,235 | 14,886 |
| hDIV4–3M | 37,956,210 | 36,724,983 | 19,909,457 | 14,979 |
| hDIV4–4M | 49,113,664 | 47,509,436 | 26,123,855 | 15,253 |
| hDIV4–5F | 27,378,169 | 26,484,024 | 12,695,780 | 14,388 |
| hDIV4–6M | 36,524,530 | 35,316,646 | 19,947,805 | 14,829 |
Hierarchical clustering:
RNA-seq samples for each species were analyzed for similarity by performing hierarchical clustering. The distance metric used for clustering was (1 – Correlation Coefficient) based on Pearson’s Correlation Coefficient [46], and average linkage was used to generate the dendrogram from the distance matrix. The hierarchical clustering was then used to determine whether there were any transcriptome-wide differences in the RNA profiles based on sex, or based on technical factors that were changing across laboratories (for the mouse samples).
Outlier analysis:
In human cultured DRG samples, we detected an outlier (sample id hDIV4–1F, Figure 1A). To rule out incorrect library construction, we sequenced this sample again using another independently prepared library. However, the new library was still an outlier upon sequencing but very similar to the original library (suggesting low technical variability in our library preparation and sequencing steps). In contrast to the other human DRG cultures, this sample had negligible expression levels for many neuronal markers like CALCA, TRPV1, and SCN10A (Supplementary file 1, sheet 1) suggesting that few neurons survived the culturing process for this sample. Consistent with this, experimental notes regarding cultures from hDIV4–1F indicated very sparse apparent neurons in the cultures (not shown). Thus, this sample and its paired intact DRG sample (sample id hDRG-1F), were excluded from further analysis. A mouse outlier sample (sample id mDIV4–4Fg, Figure 1B) was similarly analyzed, but expression of neuronal marker genes was considered sufficient for retention in the analysis.
Figure 1.
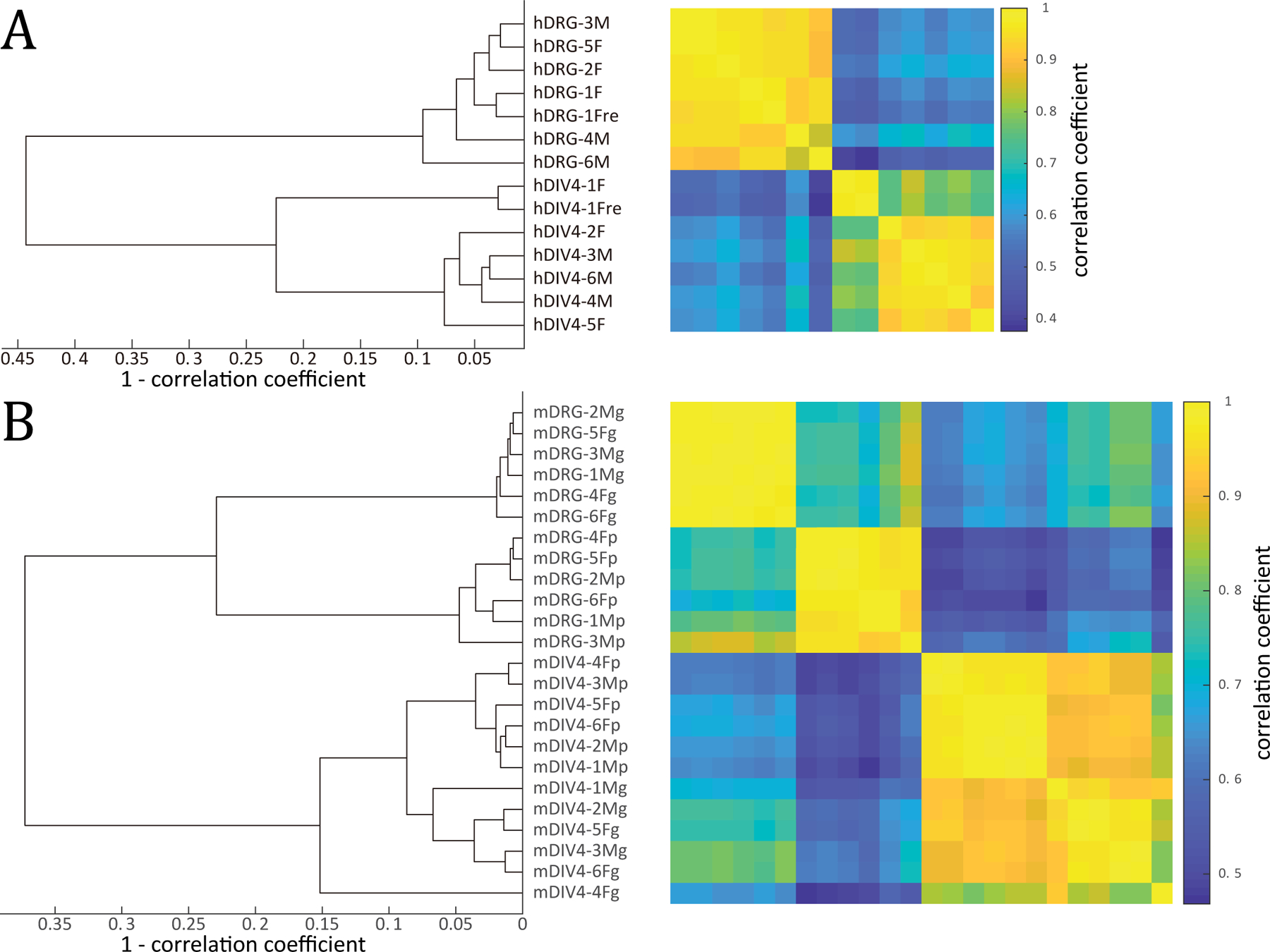
Hierarchical clustering of all human (A) and mouse (B) samples based on TPM-based whole genome gene abundances. A. Cultured and intact human DRG tissue samples are separated into two clusters. The outlier sample hDIV-1F and its paired dissected sample (hDRG-1F) were excluded from further analysis. B. Cultured and intact mouse DRG samples also segregate into separate clusters. Subclusters in the cultured DRG and dissected DRG clusters correspond to sample generated in Gereau and Price laboratories. The outlier sample mDIV4–4Fg shows moderate expression of neuronal genes, and clusters with other Gereau laboratory cultured samples when unrooted clustering is performed for cultured mouse DRG samples. (Sample id nomenclature -- Prefix: h - human; m - mouse; Infix: DRG - intact DRG samples; DIV4 – 4 days in vitro (4 DIV) DRG cultures; Suffix: M - male; F - female; p - Price laboratory; g - Gereau laboratory; re – repeated library preparation and sequencing.
Identification of consistently detectable genes:
Previous studies on whole DRG tissue have found functional responses for GPCRs with < 0.4 TPMs (e.g. GRM2 functionally studied and abundance quantified in the papers [16; 51]). This suggests that the approach of picking an expression threshold (in TPMs) to classify a gene either as “on” or “off” is likely to miss functionally relevant gene products based on traditional thresholds (~ 1 TPM, as in North et al [43]). Instead, we classified consistently detectable genes based on reads being detected in the exonic region in 80% or more of the samples in a particular condition (i.e. in at least 4 of 5 human replicates, or in at least 10 of 12 mouse replicates). Assuming iid probabilities for detecting a read emanating from a particular gene in an RNA-seq experiment, this criterion causes the sensitivity of our approach to be suitable for our purpose, calling consistently detectable genes to be those that have ≥1 read in 7 million coding gene reads in an RNAseq library, as :
Differential expression metrics:
Due to small sample sizes in humans, stringent statistical hypothesis testing using Student’s t test [56] with Benjamini-Hochberg multi-testing correction [4] yield few statistically significant differences.
We therefore decided to use strictly standardized mean difference (SSMD) to discover genes with systematically altered expression levels between experimental conditions. For each human and mouse coding gene, we report fold change and the SSMD across conditions. SSMD is the difference of means controlled by the variance of the sample measurements. We used SSMD as a secondary effect size since it is well suited for small sample sizes as in our human samples [43; 77], while simultaneously taking into account the dispersion of the data points. For determining SSMD thresholds that identify genes that are systematically changing between conditions, we use the notion of the related Bhattacharyya coefficient [7], which is used to calculate the amount of overlap in the area under the curve of the two sample distributions in order to control for false positives in differential expression analysis. For homoskedastic Gaussian distributions, we find that based on the Bhattacharyya coefficient, the less stringent constraint | SSMD | > 2.0 corresponds to a 36.8% overlap in the area under the curve of the two sample distributions being tested, while the more stringent | SSMD | > 3.0 corresponds to a 10.5% overlap. The less stringent criterion was used to select differentially expressed genes in gene sets of pharmacological interest, since genes with a moderate amount (< 36.8%) of overlap in TPM distributions between intact and cultured DRG should likely not be targeted for pharmacological purposes. The more stringent constraint corresponding to little or no overlap in sample distributions (<10.5%) was used to identify differentially expressed genes at the genome wide level.
Since our data are paired, we report several variations of the standard fold change metric. We calculated the ratio of means across conditions to compare cohort level statistics, but also calculate the mean of ratios of paired samples to better control for individual to individual variations in the transcriptome. However, the mean of ratios is more susceptible to outlier values, so we further modified it to calculate the median of ratios. All fold changes are reported as log2 fold changes, for symmetric scaling of fold changes in both directions. Since naïve filtering or ranking by log-fold change can produce incorrect results [49], we constrain differentially expressed genes by SSMD threshold. However, we do additionally constrain that the fold change (ratio of means or median of ratios) be > 1.5, since dosage-based functional effects are unlikely to be manifested as a result of lower fold changes.
To avoid issues in calculations of these metrics for genes with no detectable reads in one or both conditions, a smoothing factor of 0.01 was added to both the numerator and denominator when calculating fold changes, and to the denominator when calculating the SSMD. We also provide uncorrected p values for paired, two sample, two tailed t tests conducted for individual genes.
These cohort and inter-cohort statistics, along with individual sample TPMs, and cohort means, are provided in Supplementary file 1, sheets 1 and 2.
Estimation of density functions:
To estimate the density functions of fold change (ratio of means) and SSMD for human and mouse pharmacologically relevant genes, we used the inbuilt ksdensity function in Matlab, using normal kernel smoothing.
Human – mouse gene orthology mapping and gene expression change comparisons across species:
Orthologous genes with a one-to-one mapping between human and mouse genomes were identified using the Ensembl database [25]. Genes from the relevant gene families (GPCRs, ion channels, kinases) were removed from analysis if one-to-one orthology was not identified between human and mouse genes. Additionally, due to the complicated nature of the orthology map in the olfactory receptor and TAS2R families in mice and human [15; 72], these genes families were also excluded from analysis. For all remaining genes in these families that were consistently detected in human or mouse samples, a trend score was calculated by multiplying the SSMD and log median of paired fold change values. The correlation of the human and mouse trend scores were calculated using Pearson’s R [46]. Genes not consistently detected in samples of either species were left out of the analysis to avoid inflating the correlation based on the trend scores.
Marker gene list compilation:
Gene lists used in the paper (ion channel, GPCR, kinase) were acquired from online databases including the Gene Ontology (AMIGO), HUGO Gene Nomenclature Committee (HGNC) and the Human Kinome database [20; 35; 58].
Marker gene lists for constituent cell types in the DRG were sourced from the literature and validated in a recently published mouse nervous system single cell RNA-seq database published by Zeisel et al [75]. We found that many of the traditional protein-based fluorescence markers for these cell types were not ideal for our analyses. Out of the 49 marker genes we sourced from the literature, we looked for enrichment in the relevant cell subpopulations in the Zeisel et al database. Since PNS macrophages and PNS vascular cells were not profiled in the database, the maximum expression levels in subpopulations of CNS immune cells / microglia and CNS vascular cells profiled in the database were used as surrogates. Genes that had expression levels in the Zeisel et al database that were two-fold (or greater) higher in at least one of the subpopulations of the relevant cell type compared to the other constituent DRG cell types (or their surrogates) were considered to be enriched in the corresponding cell types. Out of the 49 marker genes we sourced from the literature, only 34 were found to be enriched in the relevant cell types, and were subject to statistical hypothesis testing using paired t-tests. Benjamini-Hochberg correction for FDR control was performed on these genes since this gene set was determined pre hoc. A complete list of the 49 genes and their expression levels in cultured and intact DRGs, along with statistical hypothesis testing on the 34 validated marker genes is provided in Table 3.
Table 3.
Differential expression analysis in human and mouse DRG for constituent cell type marker genes.
| HUMAN DATASETS | MOUSE DATASETS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ensembl gene id | ensembl gene name | Cultured DRG mean | Acutely dissected DRG mean | SSMD (smoothe d) | Uncorrect ed paired, 2-tailed t- test p- value | Is B-H FDR correction at α = 0.05 statistically significant? | ensembl gene id | ensembl gene name | Cultured DRG mean | Acutely dissected DRG mean | SSMD (smoothe d) | Uncorrect ed paired, 2-tailed t- test p- value | Is B-H FDR correction at α = 0.05 statistically significant? |
| Literature-based neuronal subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in DRG neurons | |||||||||||||
| ENSG00000167281.18 | RBFOX3 | 0.3 | 24.9 | −3.0 | 0.003 | TRUE | ENSMUSG00000025576.17 | Rbfox3 | 33.0 | 253.7 | −4.6 | 0.000 | TRUE |
| ENSG00000110680.12 | CALCA | 220.5 | 675.2 | −3.7 | 0.001 | TRUE | ENSMUSG00000030669.13 | Calca | 857.3 | 3572.5 | −4.6 | 0.000 | TRUE |
| ENSG00000175868.13 | CALCB | 52.8 | 245.7 | −5.5 | 0.000 | TRUE | ENSMUSG00000030666.11 | Calcb | 42.0 | 308.5 | −4.2 | 0.000 | TRUE |
| ENSG00000142185.16 | TRPM2 | 10.0 | 43.1 | −3.1 | 0.005 | TRUE | ENSMUSG00000009292.18 | Trpm2 | 0.9 | 23.2 | −3.5 | 0.000 | TRUE |
| ENSG00000196689.11 | TRPV1 | 13.0 | 128.1 | −5.6 | 0.000 | TRUE | ENSMUSG00000005952.15 | Trpv1 | 29.9 | 115.2 | −3.5 | 0.000 | TRUE |
| ENSG00000154864.11 | PIEZO2 | 15.8 | 148.6 | −7.0 | 0.000 | TRUE | ENSMUSG00000041482.16 | Piezo2 | 41.8 | 111.6 | −2.1 | 0.000 | TRUE |
| ENSG00000100285.9 | NEFH | 57.4 | 1153.1 | −8.4 | 0.000 | TRUE | ENSMUSG00000020396.8 | Nefh | 109.8 | 1379.3 | −7.4 | 0.000 | TRUE |
| ENSG00000277586.2 | NEFL | 81.5 | 1127.3 | −6.6 | 0.000 | TRUE | ENSMUSG00000022055.7 | Nefl | 612.6 | 3928.3 | −5.5 | 0.000 | TRUE |
| ENSG00000104722.13 | NEFM | 14.7 | 501.1 | −9.3 | 0.000 | TRUE | ENSMUSG00000022054.11 | Nefm | 309.0 | 2524.0 | −4.8 | 0.000 | TRUE |
| ENSG00000135406.13 | PRPH | 333.6 | 2225.8 | −2.1 | 0.018 | TRUE | ENSMUSG00000023484.13 | Prph | 919.2 | 3083.2 | −3.5 | 0.000 | TRUE |
| ENSG00000164220.6 | F2RL2 | 1.4 | 11.4 | −6.5 | 0.000 | TRUE | ENSMUSG00000021675.4 | F2rl2 | 14.2 | 86.0 | −4.1 | 0.000 | TRUE |
| Literature-based Schwann cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in Schwann cells | |||||||||||||
| ENSG00000158887.15 | MPZ | 190.3 | 1984.1 | −5.6 | 0.000 | TRUE | ENSMUSG00000056569.10 | Mpz | 211.5 | 13564.9 | −1.2 | 0.002 | TRUE |
| ENSG00000197971.14 | MBP | 63.5 | 592.0 | −2.9 | 0.004 | TRUE | ENSMUSG00000041607.16 | Mbp | 200.8 | 1186.5 | −1.1 | 0.004 | TRUE |
| Literature-based neural tissue vascular cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in CNS vascular cells | |||||||||||||
| ENSG00000197467.13 | COL13A1 | 15.7 | 1.1 | 2.3 | 0.006 | TRUE | ENSMUSG00000058806.14 | Col13a1 | 0.5 | 1.0 | −0.4 | 0.209 | FALSE |
| ENSG00000204291.10 | COL15A1 | 489.9 | 87.4 | 2.6 | 0.002 | TRUE | ENSMUSG00000028339.17 | Col15a1 | 206.7 | 40.3 | 1.4 | 0.000 | TRUE |
| ENSG00000168542.14 | COL3A1 | 5873.5 | 273.4 | 2.6 | 0.004 | TRUE | ENSMUSG00000026043.18 | Col3a1 | 682.9 | 109.0 | 2.3 | 0.000 | TRUE |
| ENSG00000187498.14 | COL4A1 | 901.3 | 344.0 | 5.2 | 0.000 | TRUE | ENSMUSG00000031502.11 | Col4a1 | 824.2 | 115.7 | 4.0 | 0.000 | TRUE |
| ENSG00000134871.17 | COL4A2 | 661.4 | 413.8 | 2.9 | 0.001 | TRUE | ENSMUSG00000031503.13 | Col4a2 | 648.9 | 89.2 | 3.4 | 0.000 | TRUE |
| ENSG00000188153.12 | COL4A5 | 3.6 | 32.6 | −3.5 | 0.002 | TRUE | ENSMUSG00000031274.16 | Col4a5 | 4.8 | 1.3 | 1.5 | 0.000 | TRUE |
| ENSG00000204262.11 | COL5A2 | 933.5 | 95.6 | 3.8 | 0.001 | TRUE | ENSMUSG00000026042.16 | Col5a2 | 1311.2 | 26.5 | 2.5 | 0.000 | TRUE |
| ENSG00000142156.14 | COL6A1 | 760.6 | 351.8 | 2.4 | 0.003 | TRUE | ENSMUSG00000001119.7 | Col6a1 | 140.6 | 35.9 | 1.4 | 0.000 | TRUE |
| ENSG00000142173.14 | COL6A2 | 1660.9 | 446.3 | 3.5 | 0.000 | TRUE | ENSMUSG00000020241.13 | Col6a2 | 49.8 | 41.2 | 0.5 | 0.014 | TRUE |
| ENSG00000163359.15 | COL6A3 | 676.8 | 65.5 | 5.5 | 0.000 | TRUE | ENSMUSG00000048126.16 | Col6a3 | 119.6 | 10.4 | 2.0 | 0.000 | TRUE |
| ENSG00000115414.18 | FN1 | 5452.9 | 476.4 | 3.3 | 0.002 | TRUE | ENSMUSG00000026193.15 | Fn1 | 1160.4 | 36.9 | 2.1 | 0.000 | TRUE |
| Literature-based neural tissue immune cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in CNS immune cells | |||||||||||||
| ENSG00000129226.13 | CD68 | 91.6 | 41.6 | 0.9 | 0.111 | FALSE | ENSMUSG00000018774.13 | Cd68 | 63.4 | 6.6 | 3.2 | 0.000 | TRUE |
| Literature-based Satellite Glial Cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in proliferating SGCs | |||||||||||||
| ENSG00000117399.13 | CDC20 | 77.4 | 6.9 | 2.5 | 0.005 | TRUE | ENSMUSG00000006398.15 | Cdc20 | 130.0 | 3.3 | 8.7 | 0.000 | TRUE |
| ENSG00000168078.9 | PBK | 23.0 | 0.3 | 1.5 | 0.027 | TRUE | ENSMUSG00000022033.9 | Pbk | 98.0 | 0.7 | 5.3 | 0.000 | TRUE |
| ENSG00000175063.16 | UBE2C | 76.9 | 1.3 | 3.2 | 0.001 | TRUE | ENSMUSG00000001403.13 | Ube2c | 222.4 | 1.9 | 4.8 | 0.000 | TRUE |
| ENSG00000089685.14 | BIRC5 | 23.3 | 2.3 | 1.8 | 0.015 | TRUE | ENSMUSG00000017716.15 | Birc5 | 178.7 | 7.8 | 7.4 | 0.000 | TRUE |
| Literature-based Satellite Glial Cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in proliferating SGCs, also expressed in all SGCs | |||||||||||||
| ENSG00000219435.5 | CATSPERZ | 0.1 | 3.3 | −2.2 | 0.010 | TRUE | ENSMUSG00000050623.4 | Catsperz | 2.8 | 24.4 | −4.7 | 0.000 | TRUE |
| Literature-based Satellite Glial Cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in all SGCs | |||||||||||||
| ENSG00000163520.13 | FBLN2 | 138.4 | 141.1 | 0.0 | 0.919 | FALSE | ENSMUSG00000064080.12 | Fbln2 | 1674.7 | 143.1 | 4.2 | 0.000 | TRUE |
| ENSG00000107165.12 | TYRP1 | 0.1 | 2.8 | −1.9 | 0.015 | TRUE | ENSMUSG00000005994.14 | Tyrp1 | 3.1 | 11.2 | −2.0 | 0.000 | TRUE |
| ENSG00000146250.6 | PRSS35 | 2.0 | 0.3 | 2.1 | 0.013 | TRUE | ENSMUSG00000033491.13 | Prss35 | 41.0 | 28.5 | 0.9 | 0.018 | TRUE |
| Literature-based Satellite Glial Cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in all SGCs, also expressed in Schwann cells | |||||||||||||
| ENSG00000140092.14 | FBLN5 | 145.9 | 293.5 | −2.1 | 0.002 | TRUE | ENSMUSG00000021186.9 | Fbln5 | 91.3 | 51.5 | 0.6 | 0.029 | TRUE |
| Literature-based neuronal subpopulation markers, with undetectable mouse DRG expression | |||||||||||||
| ENSG00000072315.3 | TRPC5 | 0.7 | 3.4 | Not shown | ENSMUSG00000041710.4 | Trpc5 | 0.0 | 0.0 | Not shown | ||||
| Literature-based neuronal subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in other cell types alongside DRG neurons | |||||||||||||
| ENSG00000124813.20 | RUNX2 | 133.3 | 34.1 | Not shown | ENSMUSG00000039153.16 | Runx2 | 20.7 | 2.3 | Not shown | ||||
| Literature-based Schwann cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in other cell types alongside Schwann cells | |||||||||||||
| ENSG00000172020.12 | GAP43 | 105.8 | 60.8 | Not shown | ENSMUSG00000047261.9 | Gap43 | 483.4 | 385.0 | Not shown | ||||
| ENSG00000149294.16 | NCAM1 | 74.2 | 478.5 | Not shown | ENSMUSG00000039542.16 | Ncam1 | 162.6 | 65.0 | Not shown | ||||
| ENSG00000154654.14 | NCAM2 | 3.8 | 33.1 | Not shown | ENSMUSG00000022762.17 | Ncam2 | 1.5 | 2.0 | Not shown | ||||
| Literature-based neural tissue vascular cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in other cell types alongside CNS vascular cells | |||||||||||||
| ENSG00000196739.14 | COL27A1 | 11.3 | 121.6 | Not shown | ENSMUSG00000045672.15 | Col27a1 | 23.1 | 12.5 | Not shown | ||||
| ENSG00000150093.18 | ITGB1 | 1271.5 | 303.9 | Not shown | ENSMUSG00000025809.15 | Itgb1 | 1346.0 | 202.6 | Not shown | ||||
| Literature-based neural tissue immune cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in other cell types alongside CNS immune cells | |||||||||||||
| ENSG00000130203.9 | APOE | 1015.5 | 1152.0 | Not shown | ENSMUSG00000002985.16 | Apoe | 517.6 | 3483.4 | Not shown | ||||
| ENSG00000164434.11 | FABP7 | 457.6 | 596.0 | Not shown | ENSMUSG00000019874.11 | Fabp7 | 603.3 | 590.2 | Not shown | ||||
| ENSG00000155368.16 | DBI | 529.3 | 347.6 | Not shown | ENSMUSG00000026385.16 | Dbi | 1714.6 | 1027.7 | Not shown | ||||
| ENSG00000121594.11 | CD80 | 3.5 | 2.7 | Not shown | ENSMUSG00000075122.4 | Cd80 | 25.4 | 0.2 | Not shown | ||||
| ENSG00000184557.4 | SOCS3 | 5.0 | 2.4 | Not shown | ENSMUSG00000053113.3 | Socs3 | 79.9 | 11.2 | Not shown | ||||
| Literature-based Satellite Glial Cell subpopulation markers, with mouse Zeisel et al scRNA-seq expression enriched in other cell types alongside SGCs | |||||||||||||
| ENSG00000139549.2 | DHH | 3.7 | 13.1 | Not shown | ENSMUSG00000023000.4 | Dhh | 102.6 | 85.5 | Not shown | ||||
| ENSG00000170312.15 | CDK1 | 34.9 | 2.9 | Not shown | ENSMUSG00000019942.13 | Cdk1 | 153.4 | 2.3 | Not shown | ||||
| No human ortholog for Ceacam10 | ENSMUSG00000054169.7 | Ceacam10 | 0.3 | 17.9 | Not shown | ||||||||
Based on our analysis in the Zeisel et al database, the literature-based markers Gap43, Ncam1 and Ncam2 for non-myelinating Schwann cells were also found to be expressed in Satellite Glial Cells (SGCs) and/or neurons. Similarly, SGC markers Dhh, Fbln5, and Ceacam10 are expressed in both Schwann cells and SGCs. Fbnl2, Tyrp1, and Prss35 were found to be comparably enriched in proliferating and non-proliferating SGCs. Microglial / macrophage markers Apoe, Fabp7 and Dbi were also found to be expressed in SGCs and not used as markers. Finally, Trpc5 was found to be absent in mouse sensory neurons.
Code:
Coding was done in Matlab, and data visualization was performed in Matlab and GraphPad Prism V8. Normalized counts and analysis are presented in a companion website: https://bbs.utdallas.edu/painneurosciencelab/sensoryomics/culturetxome/
Results
Hierarchical clustering of human and mouse samples reveal whole transcriptome differences between cultured and intact DRG
We used hierarchical clustering to assess differences between RNA-seq samples analyzed in this study. As shown in Figure 1, the top-level split of the hierarchical clustering for both human and mouse samples was between cultured and intact DRG tissue, showing consistent whole transcriptome changes between the two. We identified broad changes in the transcriptome between intact and cultured DRGs, with 2440 human and 2941 mouse genes having a fold change (ratio of means and median of ratios) > 1.5, and | SSMD | > 3.0 between compared conditions (Supplementary file 1, sheets 1 and 2). The smaller number of changed genes that we detect in human can be attributed to a smaller number of detected genes that increase in abundance in culture in humans compared to mouse. Of the differentially expressed genes, only 443 (18%) of the human genes and 1156 (39%) of the mouse genes have increased abundances in cultured conditions, which suggests that a majority of the differentially expressed genes gain in relative abundance in intact DRGs compared to culture. Controlled laboratory conditions and a similar genome (belonging to the same mouse strain) potentially causes lower within-group variation at the level of individual genes in the mouse samples with respect to the human samples. The smaller number of human genes detected to be increasing in culture can likely be attributed to higher within-group variation in human samples, since genes that show significantly increased expression in cultured conditions have more moderate changes (median across ratio of means in genes satisfying differential expression criterion - human: 2.8 fold, mouse: 3.5 fold) in expression compared to genes that show significantly increased expression in intact DRGs (median in human: 5.4 fold, mouse: 5.1 fold). They are therefore less likely to be detected in a lower signal to noise ratio scenario.
No distinct differences at the whole transcriptome level across sexes
In both human and mouse samples, we did not find clear sex differences at the whole transcriptome level though individual sex markers like UTY differ between the sexes (Supplementary file 1, sheets 1 and 2), consistent with previous findings [34]. Thus, male and female samples were grouped together for further analyses.
Small set of differences between cultured mouse DRG transcriptomes across different laboratories
Experiments were performed in 2 laboratories (Gereau laboratory – sample ids with a “g” suffix; and Price laboratory – sample ids with a “p” suffix, Figure 1B) independently for mouse datasets. Although both laboratories used the same strain of mouse, both intact and cultured DRGs had a small but distinct transcriptome difference between the two laboratories, leading us to analyze the magnitude and nature of the laboratory-specific differences.
Changes in intact DRG RNA profiles across laboratories are likely caused by environmental differences between animal facilities. Additionally, while changes in gene expression levels are well known to be different across inbred mouse strains [60], recent research suggests that even for inbred mouse strains separated for over hundreds of generations, mutation profiles diverge and can cause different outcomes in molecular assays, and have been shown to cause changes in immune function related genes [12].
Changes in cultured DRGs across laboratories can additionally be explained by differences in culturing protocol. Among the genes that have a greater than 2-fold change in expression is Ngfr (mean TPM in Price laboratory: 973, in Gereau laboratory: 438), potentially due to the use of NGF in the culturing process in the Price laboratory. Several genes that were detected in one or both laboratories’ cultures had laboratory-specific expression changes with | SSMD | > 2, and are noted in Figure 2. Surprisingly, we saw that inter-laboratory transcriptome differences in cultured mouse DRGs were smaller in cultured samples with respect to intact DRGs (Figure 1B) despite differences in culturing protocols (e.g. without nerve growth factor (NGF) in the Gereau laboratory, and with NGF in the Price laboratory). This is likely due to the fact that neurons have the most plastic molecular profiles, and putatively decline in proportion in cultured DRGs.
Figure 2.
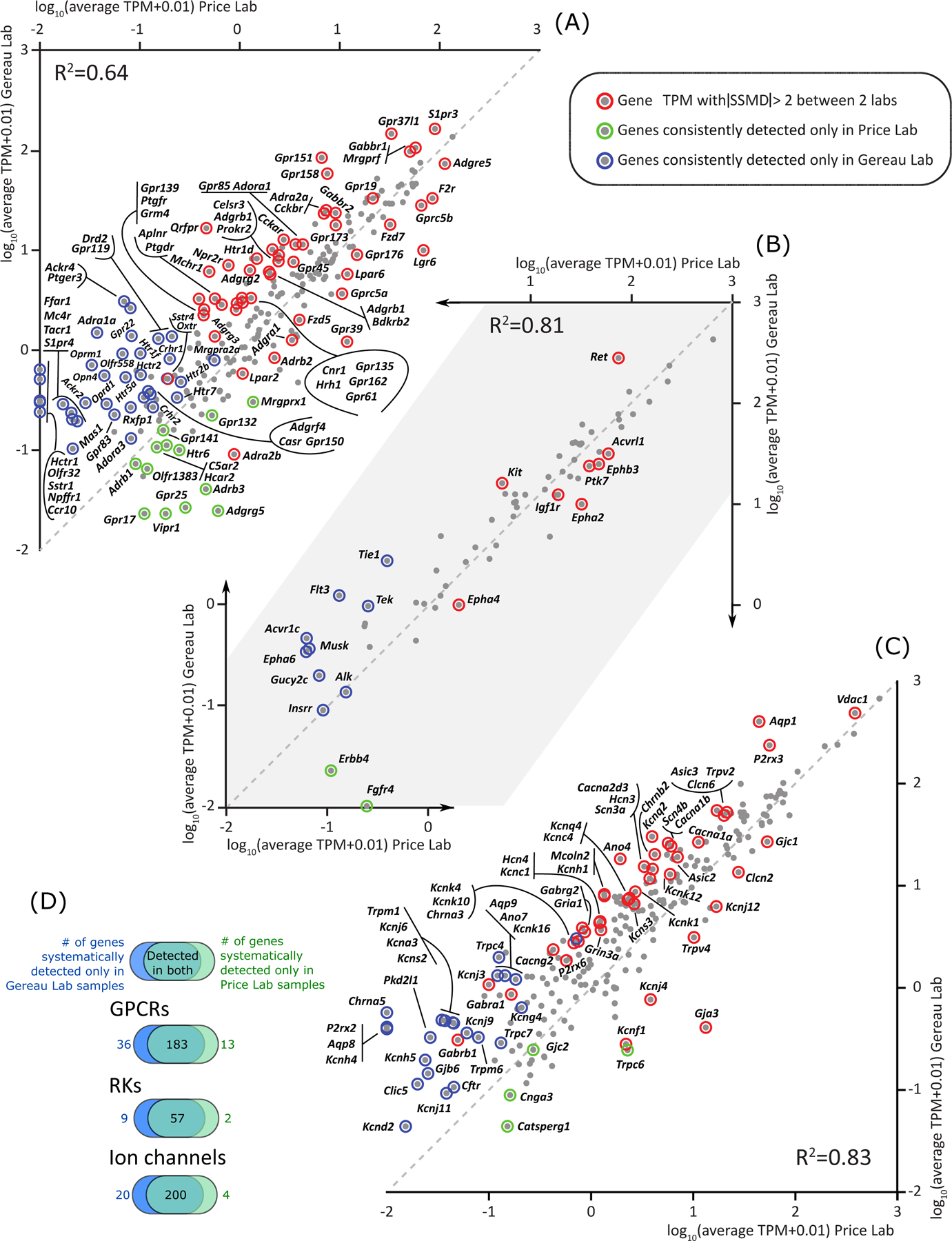
Scatter plot and Venn diagrams showing a small amount of differential expression of GPCR genes (A), RK genes (B), and ion channel genes (C) in culture between the Price and Gereau laboratories. The number of genes consistently detected in RNA-sequencing assays for each laboratory are shown in Venn diagrams separated by gene families in (D). Expression levels of genes in all three families showed consistent correlation between the two laboratories: GPCR genes : Pearson’s R squared: 0.64, p < 0.01, RK genes : Pearson’s R squared: 0.81, p < 0.01, ion channel genes : Pearson’s R squared: 0.83, p < 0.01. Genes like Alk and Insrr are plotted on the diagonal, but marked as consistently detected only in Gereau laboratory samples. This is because they have comparable mean TPMs in samples from both laboratories, but are only consistently detected (in 5 or more samples out of 6) in the Gereau laboratory.
The small amount of changes in DRG culture between the two laboratories can be summarized as follows. A large amount of overlap was found in consistently detected genes for GPCRs (consistently detected in Price laboratory culture: 191, in Gereau laboratory culture: 214, overlap in both labs’ cultures: 183; Figure 2A), RKs (consistently detected in Price laboratory culture: 59, consistently detected in Gereau laboratory culture: 66, overlap in both labs’ cultures: 57; Figure 2B), and ion channels (consistently detected in Price laboratory culture: 204, in Gereau laboratory culture: 217, overlap in both labs’ cultures: 200; Figure 2C and 2D for summary of numbers).
We find that most genes highlighted in Figure 2 have low expression levels (high concentration of genes in the region corresponding to mean TPM < 1.0 for one or more laboratories in scatter plots of Figure 2), or low log fold changes across laboratories (high concentration of genes in the proximal region of the line x = y in scatter plots of Figure 2), or both. This drives the high correlation in RNA profiles between cultured DRGs from both laboratories. Most importantly, for a large majority of the genes that are differentially expressed in DRG cultures between the laboratories, the trend of changes between intact and cultured DRG transcriptome was identical, suggesting that though the degree of change is different for these genes between laboratories, the direction of change is consistent. However, we did identify a small set of pharmacologically relevant genes that had moderate or high gene expression (mean TPM > 1.0) in cultured conditions for at least one laboratory, and a two-fold or greater change in mean TPM between cultures from the two laboratories, but trended in opposite directions between intact and cultured DRG transcriptomes. These genes, which consist of 12 GPCRs (Adgrd1/g3/l2/l4, Adra2a, Aplnr, Bdkrb2, Gpr85/158, Gpr37l1, Mchr1, Prokr2), 1 RK (Kdr), and 10 ion channels (Aqp1, Cacna2d1, Gjb2, Grid1/2, Kcna1/q2, Lrrc8b/8d, P2rx3) show that expression levels for a small set of potential pharmacological targets are influenced by the culturing protocol.
Overall, despite differences in culturing protocol, we find a consistent molecular phenotype in cultured mouse DRGs (Figure 1B) in both laboratories that is further explored in the following sections.
Increases in proliferating SGC and fibroblast markers compensated for by decrease in neuronal and Schwann cell markers in human and mouse cultures
Due to the magnitude of changes, we tested whether the proportion of mRNA sourced from the different constituent cell types of the DRG were different between intact and cultured samples. We profiled the expression levels of neuron, fibroblast-like cell, Schwann cell, SGC, and macrophage marker genes (chosen based on mouse single cell profiles [75]) in both human and mouse cultured and intact DRGs. We found that neuronal markers were broadly downregulated in all cultured samples from mice and humans. Expression levels of neuronal markers in culture were decreased by a median SSMD of 5.55 and 4.20 in human and mouse datasets respectively (Table 3). Conversely, markers for fibroblast-like cells (often of vascular origin) were increased by a median SSMD of 2.90 (human) and 2.03 (mouse) (Table 3) in culture compared to intact samples. We found that myelinating Schwann cell markers (MPZ, MBP) in culture were decreased by a median SSMD of 4.24 (human) and 1.13 (mouse) compared to intact tissues (Table 3) but markers for proliferating SGCs (Table 3) were increased (by a median SSMD of 2.13 and 6.38 in humans and mice respectively). Marker genes for all SGCs show a more mixed set of changes in both species since it is likely that the proportion of proliferating SGCs in culture gain at the expense of other SGC subpopulations (Table 3). The marker gene CD68 for monocyte-derived immune cells also increases in humans (by 0.89, SSMD) and mice (by 3.25, SSMD) (Table 3).
Out of the 49 literature sourced marker genes, 34 marker genes were validated by the Zeisel et al dataset. Of these, 5 SGC marker genes (CATSPERZ, FBLN2, TYRP1, PRSS35, FBLN5) that were found to be expressed in Schwann cell and SGC subpopulations beside proliferating SGCs from the Zeisel et al dataset show a variety of changes between abundances in intact and cultured DRGs. Benjamini-Hochberg corrected p-values (at the level of FDR <= 0.05 based on the pre hoc list of 34 cell type enriched marker genes) in the human and mouse marker genes are consistent in trend and statistically significant for all remaining 29 genes in both species (except for CD68 in humans which shows a consistent trend but higher within-group variability compared to its mouse ortholog; and Col13a1 in mouse and COL4A5 in human that show opposite trends in humans and mice possibly due to evolutionary gene regulatory divergence) (Table 3).
These changes happen broadly (as shown by the density function across pharmacologically relevant gene families, Figure 3) and not just in specific regulatory pathways or gene sets. They indicate that the proportion of mRNA derived from neurons (and possibly Schwann cells) in our RNA-seq libraries decreases in cultured samples. In turn, this suggests that the proportion of neurons (which are post-mitotic) to other cell types decreased in DRG cultures, while the proportion of dividing cells (such as fibroblast-like cells and proliferating SGCs) to other cell types increased. However, Schwann cells, which can be mitotic and proliferate under certain conditions, potentially also decrease in proportion based on our data. This is likely because axonal contact is required for Schwann cell survival [69]. Developmentally established transcription factor expression that define sensory neuronal identity (PRDM12, TLX2, TLX3, POU4F1, DRGX) are all consistently decreased in human and mouse cultures (Supplementary File 1 Sheets 1 and 2), further suggesting that the observed changes are more likely to be caused by changes in relative proportions of cell types rather than molecular plasticity of neurons. These changes were expected, given the different mitotic statuses of these cell types, and were almost certainly the primary factors in distorting the transcriptome from what is seen in vivo. The zero-sum nature of our relative abundance measure (transcripts per million) potentially also amplifies this signal.
Figure 3.
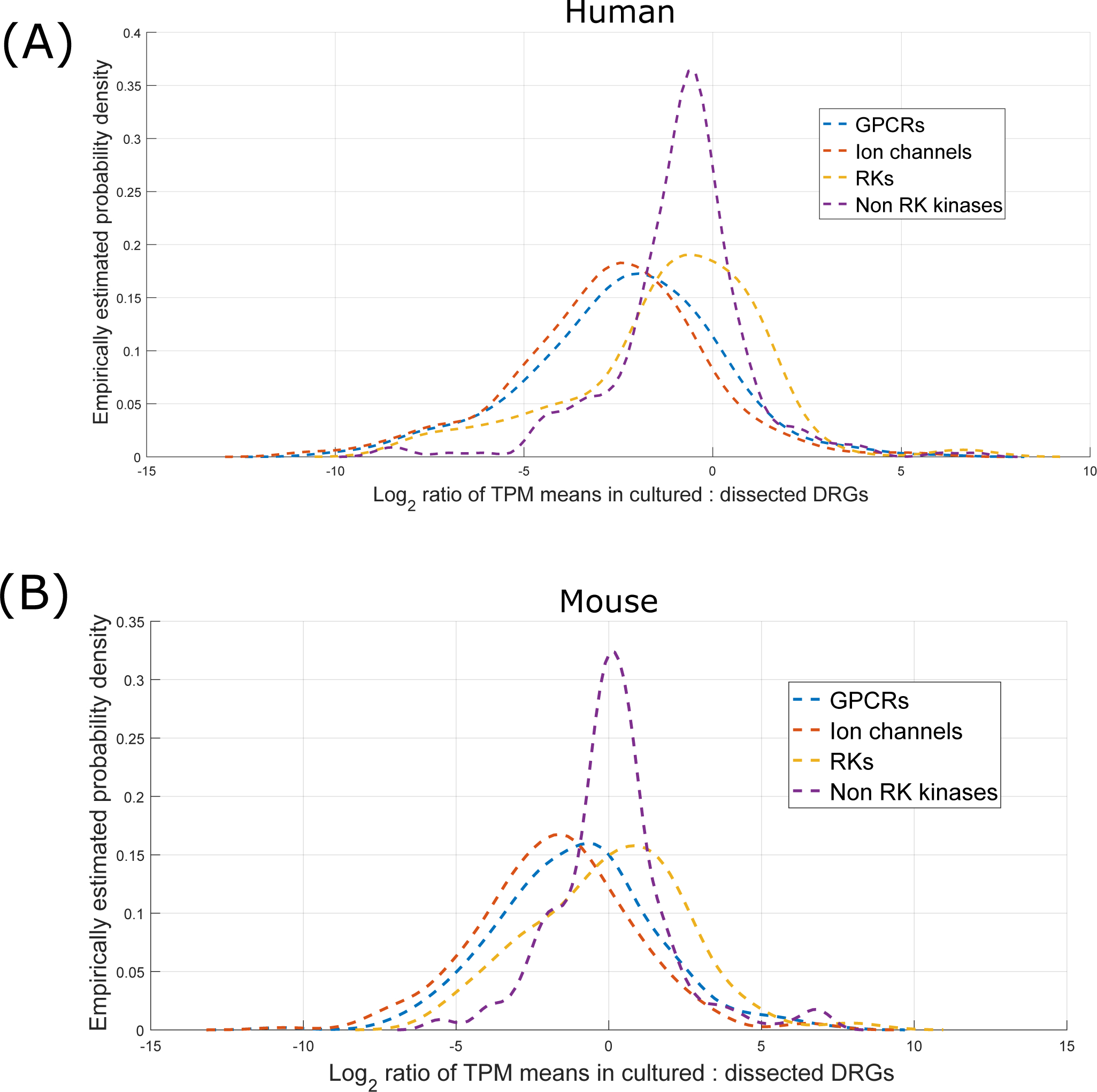
Empirical density distribution of log2 fold changes (ratio of means) for GPCRs, ion channels, RKs, and non-RK kinases in human (A) and mouse (B). RKs and kinases as a group are weakly de-enriched in human and weakly enriched in mouse cultures (in the context of mean expression). However, both GPCRs and ion channels are strongly de-enriched in both human and mouse cultures, likely because of the variety of these genes that are expressed in sensory neurons.
Expression profiles of several pharmacologically relevant gene families show lower expression levels in DRG culture
A primary use of DRG cultures is to examine pharmacological effects of ligands for receptors with the assumption that this type of experiment reflects what occurs in vivo [38]. An underlying assumption of this type of experiment is that the presence or absence of a tested effect is reflected in consistent expression between in vivo and cultured conditions. To give insight into this assumption, we comprehensively cataloged expression of G-protein coupled receptors (GPCRs), ligand gated ion channels and receptor kinases (RKs) in native and cultured human and mouse DRG. To comprehensively characterize the changes in these gene families, we also characterized expression profiles of non-RK soluble kinases (Supplementary file 1, Sheets 3–10). We limited our soluble kinase comparisons to a well-characterized subset with clear mouse to human orthologs [35].
We find that a number of these genes are consistently detected in intact DRGs but not in culture. This was seen in human gene families of GPCRs (detected in intact DRG: 292; in culture: 190; out of which 176 were detected in both), ion channels (in intact DRG: 239, in culture: 179, in both: 172), RKs (in intact DRG: 68; in culture: 60; in both: 59), and non-RK kinases (in intact DRG: 286; in culture: 277; in both: 272); and a similar trend was observed in the mouse gene families as well. Since sensory neurons express a rich diversity of GPCRs and ion channels, the greater decrease in the number of consistently detected GPCRs and ion channels is likely the result of a proportional decrease of neurons in culture and/or decrease of gene expression in cultured neurons. Lists of consistently detected genes in these gene families are presented in Supplementary File 1, Sheets 3–10.
However, it is important to note that over 75% of the human genes in these families (human: 679 out of 885, mouse: 702 out of 824) that are consistently detected in intact DRG are still detectable in culture. This suggests that at single cell resolution, DRG cultures could be used as a surrogate for in vivo models in preclinical research for a majority of pharmacologically relevant molecular assays.
Next, for genes that are consistently detected in at least one condition, we identified the ones in these gene families that have | SSMD | > 2.0 (Tables 4 and 5, for human and mouse genes). Based on the SSMD values, while comparable numbers of GPCRs, ion channels and kinases were found to be decreased in cultured DRGs (GPCRs – human: 85, mouse: 95; ion channels – human: 109, mouse: 122; kinases – human: 106, mouse: 70), more mouse genes were detected to be systematically trending in the opposite direction as compared to their human counterparts (GPCRs – human: 7, mouse: 20; ion channels – human: 7, mouse: 14; kinases – human: 22, mouse: 66). As noted before, within-group variation is likely lower in mice due to controlled laboratory conditions and similar genetic backgrounds, and this enables us to detect more expression changes that have smaller effect sizes (as in the case of genes that are increased in cultured conditions).
Table 4.
Differentially expressed pharmacologically relevant genes (RKs boldfaced) in intact vs cultured human DRGs with | SSMD | > 2, partitioned by mean TPM in condition of higher expression. The number of genes in each column is shown in parentheses. Genes known to be associated with pain from GWAS / functional association databases and the literature are underlined. Typically, smaller TPM brackets have higher technical variance.
| GPCRs | Ion channels | Kinases | |||
|---|---|---|---|---|---|
| Up in native DRG (85) | Up in cultured DRG (7) | Up in native DRG (109) | Up in cultured DRG (7) | Up in native DRG (105) | Up in cultured DRG (22) |
| > 50 TPM | |||||
| ADGRA2, ADGRB3, ADGRL1, ADGRL3, ADGRV1, GABBR1, GPR17, LGR5, LPAR6, TACR2 | GPR33, P2RY11 | ANO5, AQP1, CACNA1A, CACNA1B, CACNA2D1, CACNA2D3, CACNB1, CACNB3, CLCN2, CLCN6, GRIK2, GRIN1, ITPR3, KCNQ2, P2RX3, PKD2L2, RYR1, RYR2, RYR3, SCN10A, SCN11A, SCN1B, SCN4B, SCN8A, SCN9A, TPCN1, TRPV1, ZACN | CLIC1, CLIC4, P2RX4, ANO6 | BCR, BRAF, BRD2, BRSK2, CAMK1D, CAMK2A, CAMK2B, CAMK2G, CAMKK1, CDK10, CDK9, CDKL1, CLK1, CLK2, CLK4, CSNK1G2, DCLK2, ERBB3, FER, FGFR1, KALRN, KSR1, LIMK1, MAP2K5, MAP3K5, MAP3K6, MAPK12, MARK1, MARK2, MAST1, MAST2, MAST3, NEK1, NRBP2, NPR2, NTRK1, NTRK3, PINK1, PRKACA, PRKCA, PRKCZ, RET, ROCK2, SPEG, SRPK2, TAF1, TGFBR2, TLK2, TRRAP, TYRO3, ULK2, ULK3, WNK1 | ABL2, AXL, CDK2, CDK4, DAPK3, ILK, MAPKAPK2, MYLK, NRBP1, PDGFRB, PTK7, TRIO |
| > 25 TPM | |||||
| ADGRB1, ADGRG2, CELSR2, GABBR2, GPR18, GPR183, MRGPRE | ADGRF5, ADGRL4 | ANO3, ANO8, AQP11, CACNA1C, CACNA1D, CHRNA7, CHRNE, CLCN5, GABRA2, GABRB3, GRIA4, GRIK3, GRIK5, HCN2, MCOLN3, SCN1A, TRPM2 | KCNN4 | BMPR2, DCLK1, EGFR, HIPK2, HSPB8, LRRK1, LRRK2, MAPK11, MST1, NTRK2, PAK3, PRKACB, PRKCB, PRKCE, PRKCI, SLK, TSSK3, TSSK4, TTBK2, TTN, ULK1 | DYRK4, FLT1, MAPKAPK3, MELK |
| > 12.5 TPM | |||||
| CCR10, CHRM5, CXCR6, GNRHR, GPR146, GPR171, GPR37L1, GPR62, GPRC5B, GRM7, HTR2B, OR10AD1, OR2A1, OR2A42, OR52B6, P2RY12, P2RY14, SSTR1, TAS1R3, TAS2R40 | CXCR4 | ANO4, ASIC1, CACNB2, CACNB4, CACNG7, CHRNB1, CLCN4, GABRG2, GJA9, GLRB, GRIA2, GRIK1, GRIN3B, KCNC1, KCND1, KCNH2, KCNQ5, KCNT2, LRRC8B, NALCN, P2RX5, P2RX6, SCNN1D | AQP9 | ACVR2A, ADCK1, BRD3, CDKL2, DAPK2, DSTYK, EPHA5, EPHA6, EPHB3, EPHB6, MAK, MAP2K6, MUSK, MYT1, OBSCN, POMK, PRKX, TNK1, TTBK1, WNK2 | IRAK2, PLK1, TRIB3, TTK |
| > 6.3 TPM | |||||
| AVPR2, CELSR3, F2RL2, FZD2, GPER1, GPR158, GPR162, GPR182, GPR82, GPR87, GRM4, LPAR3, OPRM1, OR1K1, OR2A7, OR3A3, OR51I2, OR51J1, OXER1, PTGER4, TAS2R39, TSHR | - | ANO2, CFTR, CHRNG, CLIC3, GJA4, GJB7, GJC3, GRIN2B, HCN1, KCNA2, KCNH7, KCNH8, KCNK12, KCNN2, KCNS1, LRRC8D, SCN3B | - | EPHA10, FLT3, HIPK4, LMTK2, NEK5, PNCK, STKLD1 | NEK2 |
| > 3.1 TPM | |||||
| GPR149, GPR45, GRM3, GRM6, MRGPRF, OR11L1, OR13J1, OR5C1, OR6F1, OR6V1, PITPNM3 | GPR176, F2R | AQP12B, GJB3, GRIN3A, KCNB2, KCNH1, KCNH3, KCNH6, KCNJ3, KCNK1, KCNK15, TRPC5 | - | KSR2, PRKAA2 | DYRK3 |
| <= 3.1 TPM | |||||
| ADRA2C, CCKBR, CXCR3, GPR135, GPR179, GPR26, GRM8, HTR5A, HTR6, OPRL1, OR1Q1, OR2AE1, OR5AS1, SSTR2, TBXA2R | - | GLRA3, HCN3, KCNK10, KCNH5, KCNJ10, KCNJ12, KCNJ5, KCNJ9, KCNQ4, KCNV1, LRRC8E, SCN2B, TRPC3 | KCNJ8 | FRK, PRKCQ | - |
Table 5.
Differentially expressed pharmacologically relevant genes (RKs boldfaced) in intact vs cultured mouse DRGs, with | SSMD | > 2 partitioned by mean TPM in condition of higher expression. The number of genes in each column is shown in parentheses. Typically, smaller TPM brackets have higher technical variance.
| GPCRs | Ion channels | Kinases | |||
|---|---|---|---|---|---|
| Up in native DRG (95) | Up in cultured DRG (20) | Up in native DRG (122) | Up in cultured DRG (14) | Up in native DRG (70) | Up in cultured DRG (66) |
| > 50 TPM | |||||
| Adora1, F2rl2, Gabbr1, Gabbr2, Gpr137, Lpar3, Mrgprd | Adgre5, Mrgprf, Smo | Ano3, Asic1, Asic2, Asic3, Cacna1a, Cacnb1, Cacnb3, Cacng7, Clcn2, Clcn6, Gabrg2, Glrb, Grik1, Grik5, Grin1, Hcn2, Htr3a, Kcna2, Kcnc4, Kcnd1, Kcnk1, Kcns1, Kcns3, Kcnt1, Mcoln1, Scn10a, Scn11a, Scn1b, Scn4b, Trpc3, Trpm4, Trpv1 | Clic1, Clic4, Gja1, Kcnn4, Lrrc8c | Akt3, Brsk1, Brsk2, Camk2a, Camk2b, Camk2g, Cdk10, Cdk5, Cdkl2, Kit, Limk1, Limk2, Map2k4, Mapk11, Mapk12, Mast1, Nek1, Nek7, Npr2, Nrbp2, Ntrk1, Ntrk2, Phkg2, Pim3, Pink1, Prkaca, Prkacb, Prkca, Prkcb, Prkcd, Prkce, Prkcq, Prkcz | Acvr1, Aurka, Aurkb, Axl, Bckdk, Bmpr1a, Cdk4, Ddr1, Erbb3, Ilk, Mapkapk2, Mark3, Pak2, Pbk, Pdgfrb, Plk1, Plk2, Riok3, Rock2, Ryk, Srm |
| > 25 TPM | |||||
| Adgrb1, Cysltr2, Gpr45, Gprc5c, Grm7, Htr1d, P2ry1, Ptgdr, Pth1r | Adgra2, Fzd1, Fzd2 | Ano8, Cacna1h, Cacna2d3, Chrna6, Chrnb2, Gabrb3, Gria4, Hcn3, Kcnb1, Kcnc1, Kcnc3, Kcnj12, Kcnn1, Kcnq4, Tpcn1, Trpa1 | Gjc1, Kcnk5, Pkd2 | Acvr1b, Camk1g, Dclk3, Mark1, Myt1, Ntrk3, Pim2, Pnck, Ttbk1 | Abl1, Bub1, Cdk2, Cdk7, Ddr2, Ephb2, Ephb3, Erbb2, Map3k3, Melk, Met, Nek2, Peak1, Pkd2, Pkn2, Plk4, Prag1, Ripk1, Ripk3, Tec, Tgfbr1, Tgfbr2 |
| > 12.5 TPM | |||||
| Adgrb2, Adgrg2, Adra2c, Agtr1a, Cckar, Cxcr4, Gpr161, Gpr162, Gpr27, Gpr35, Grm4, Mrgpra2a, Mrgpra2b, Mrgprx1, Npy2r, Opn3, Oprl1, Ptger1, Ptgfr | Ackr3, Fzd7, S1pr2 | Cacna2d2, Cacng5, Chrna3, Chrna7, Gabra1, Gabra2, Grik4, Htr3b, Kcnb2, Kcng2, Kcnh1, Kcnh6, Kcnk2, Kcnk3, Kcnk4, Kcnn2, P2rx6, Scn1a, Scn2b, Trpm2 | Clcn5 | Camk1d, Camkk1, Dapk2, Flt3, Lmtk3, Map3k5, Pdk1, Wnk2 | Cdc7, Cdk6, Ephb4, Fer, Haspin, Irak4, Jak2, Lyn, Mastl, Mlkl, Pask, Riok1, Rock1, Scyl3, Trib3, Ttk, Vrk1, Vrk2, Wee1 |
| > 6.3 TPM | |||||
| Adora2a, Chrm4, Cnr1, Gpr135, Gpr139, Gpr146, Gpr149, Gpr157, Gpr37, Gpr61, Gpr75, Grm8, Hcrtr1, Htr1b, Htr7, Lgr5, Lpar5, Prokr1, Ptger3, Ptger4, Sstr2 | C3ar1, C5ar1, Celsr1, Cmklr1, Fzd6, Lpar6 | Aqp11, Cacna1d, Cacng2, Cacng4, Chrnb3, Chrnb4, Gabra3, Gabrg1, Gjb6, Gria1, Grin3a, Hcn4, Kcna4, Kcnc2, Kcng4, Kcnh7, Kcnj11, Kcnj3, Kcnj4, Kcnq3, Kcnq5, Kcnu1, Kcnv1, Nalcn, P2rx2, P2rx5, Ryr2, Trpc6, Trpm8 | - | Amhr2, Cdkl1, Mst1r, Prkaa2, Tie1 | Ikbke, Map3k2 |
| > 3.1 TPM | |||||
| Chrm2, Crhr2, Gpr156, Gpr160, Gpr22, Gpr3, Gpr68, Hcrtr2, Hrh3, Htr1a, Htr4, Nmur1, Ntsr2, Pitpnm3 | Adra1b, F2rl1, P2ry6 | Cacna1i, Gria2, Kcnk18, Scn5a | Gjb5, Lrrc8e | Brdt, Camk4, Epha10, Ksr2, Npr1, Tek | Ror1 |
| <= 3.1 TPM | |||||
| Adrb1, Ccr10, Ccr2, Chrm1, Crhr1, Galr1, Gpr150, Gpr174, Gpr179, Gpr182, Gpr21, Gpr26, Gpr62, Gpr83, Gpr88, Hcar1, Hrh2, Htr5a, Lhcgr, Mas1, Oprd1, Oprk1, Ptafr, Ptger2, Rho | Ccr5, Gpr84 | Aqp4, Best1, Cacna1e, Cacng3, Chrna10, Chrna4, Clic5, Gabrb1, Gabrb2, Gjd2, Kcnd2, Kcng1, Kcng3, Kcnh5, Kcnj13, Kcnj14, Kcnj16, Kcnj2, Kcnj8, Kcnj9, Kcnn3, Kcns2, Scnn1a | Chrna1, Gjb4, Itpr2 | Bmx, Cdkl4, Epha6, Epha8, Ephb1, Insrr, Itk, Nek11, Sbk3 | Frk |
We also characterized the degree of change in expression by estimating the probability density of the fold change (ratio of means) for all the genes in these families. The empirically estimated probability density for the ratio of means (intact DRG: cultured DRG) of the human and mouse pharmacologically relevant genes (Figure 4), shows a clear trend of decreased expression for a majority of the human ion channels and GPCRs.
Figure 4.
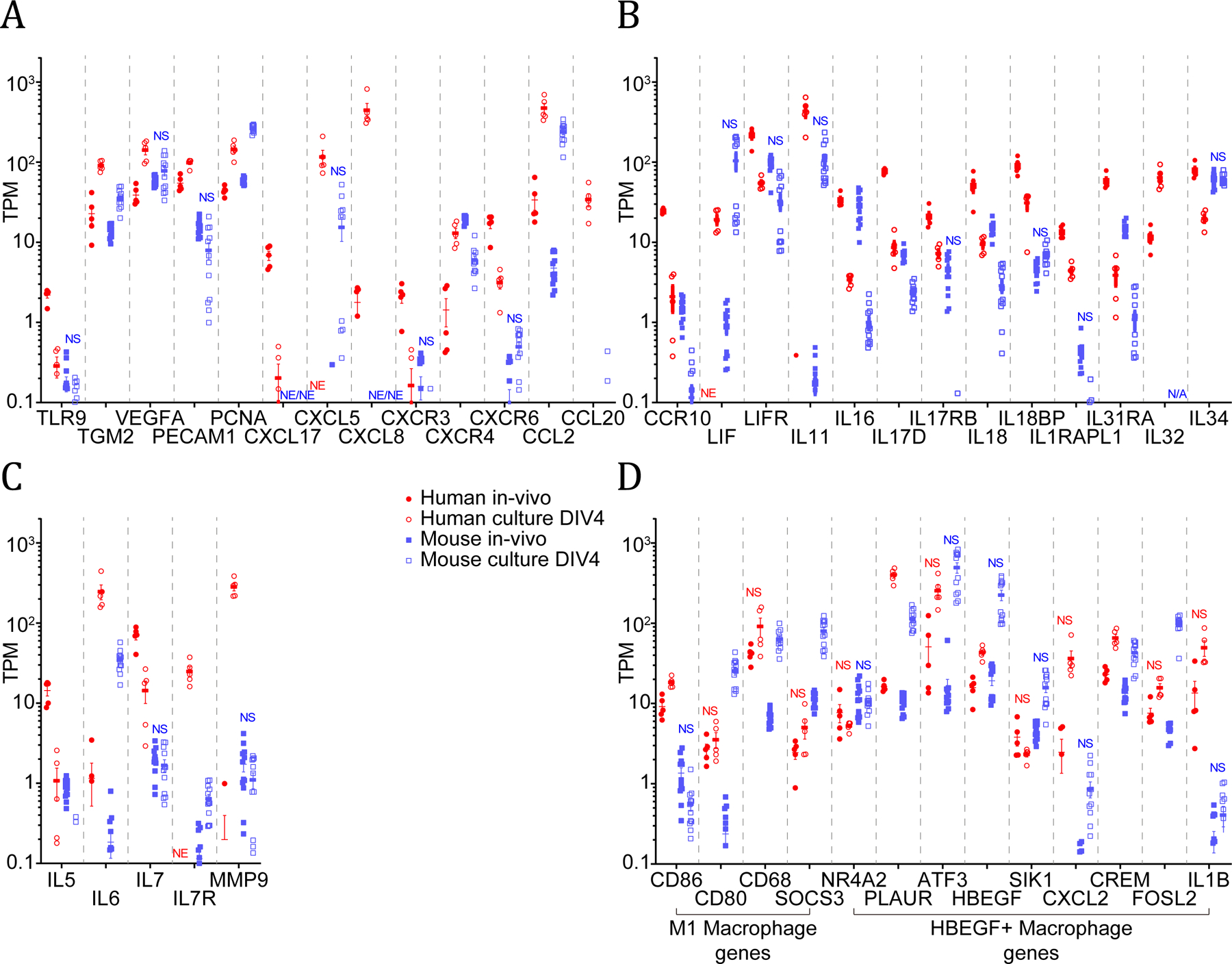
Expression levels in human and mouse intact vs. cultured DRGs. A wide diversity of genes involved in inflammation and proliferation, nerve and neuronal injury and repair, and immune signaling and response are profiled (A, B, and C). Key expressed genes for M1 macrophages and HBEGF+ macrophages are also shown (D). NS: | SSMD | <= 2, NE: not consistently detected for that condition, N/A: not applicable because orthologous gene not identified in that species.
Finally, we analyzed the trends in genes known to be involved in nociception, pain and neuronal plasticity. Genes with | SSMD | > 2.0 between conditions, and known to be associated with pain from the Human Pain Genetics Database, and the Pain – Gene association geneset (from the Comparative Toxicogenomics Database in Harmonizome [17; 52]), as well as from the literature, are underlined in Table 4. They identify pain-associated genes in these pharmacologically relevant families that change in expression between intact and cultured DRGs. Based on changes in consistent detectability between the two conditions, | SSMD | values > 2.0, or ratio of means > 2.0, changes in expression of several genes are discussed below.
Changes in human GPCRs:
Several GPCRs involved in pro-inflammatory pathways, including CCR1, CCRL2, CNR1, CXCR4, F2R, CHRM1 [71; 74] were found to increase in abundance in cultured DRGs. GPCRs found to be decreased in culture included DRD5, HTR5A, HTR6, and some metabotropic glutamate receptors (GRMs) like GRM4 and GRM7, all of which have been shown to be highly neural tissue-enriched in humans (based on neural proportion score > 0.9 in Ray et al [51]). Their mouse orthologs have also been shown to be neuronally expressed in DRG single-cell RNA-seq experiments [61]. Many of these and other GPCRs changing in abundance between intact and cultured DRGs (Table 5, and Supplementary File 1 Sheet 3) have been noted as potential targets for pain treatment [8; 16; 37; 55]. Therefore, our findings suggest that under certain culture conditions false negatives could arise for these targets.
Changes in mouse GPCRs:
Pro-inflammatory mouse GPCRs were also found to be increased in cultured DRGs, including Ccr5, Cxcr6, F2r, and F2rl1 [30; 59; 63]. Several neuronally-expressed mouse GPCRs (based on Usoskin et al [61]), including Chrm2, Htr1a, Htr2c, Htr7, and metabotropic glutamate receptors like Grm4 showed higher expression in intact DRGs (Table 5, and Supplementary File 1 Sheet 4). Many of these genes in the human and mice datasets were from orthologous families of receptors, including cytokine receptors, the protease activated receptor (PAR) family (F2R), 5-HT receptors, and metabotropic glutamate receptors.
Changes in human ion channels:
Among the ion channels increased in abundance in cultured DRGs were the chloride intracellular channels CLIC1 and CLIC4, gap junction protein GJA1, KCNG1 (Kv6.1), KCNJ8 (KIR6.1), KCNN4 (KCa4.2), and P2RX4, TRPV4, and voltage dependent anion channels VDAC1 and VDAC2. Interestingly, many of these ion channels are involved in membrane potential hyperpolarization, suggesting a potential compensatory mechanism to suppress excitability. Neuronally-expressed voltage gated calcium channels such as CACNA1B, CACNA1F, CACNA1I, CACNAG5, CACNAG7 and CACNAG8; glutamate ionotropic receptors GRIA2 and GRIN1; voltage gated potassium channels KCNA1, KCNA2, KCNB2, KCNC3, KCND1, KCND2, KCNH2, KCNH3, KCNH5, KCNJ12, KCNK18, KCNQ2, KCNT1, KCNV1; purinergic receptors P2RX2 and P2RX5; and voltage-gated sodium channels SCN1A, SCN4A, SCNN1A and SCNN1D were found to be increased in intact DRGs. (Table 4, and Supplementary File 1 Sheet 5)
Changes in mouse ion channels:
Changes in mouse ion channel genes were also quantified. (Table 5, and Supplementary File 1 Sheet 6). Genes increased in DRG cultures included several of the same families seen in human, such as chloride intracellular channels Clic1 and Clic4; gap junction proteins Gja1, Gja3, Gjb3, Gjb4, Gjb5 and Gjc1; the glutamate ionotropic receptor Grik3; voltage-gated potassium channels Kcnk5 (K2p5.1) and Kcnn4 (KCa4.2); and purinergic receptors P2rx1, P2rx7. Among ion channels decreased in culture were the chloride intracellular channels Clic3 and Clic5; voltage-gated calcium channels Cacna1i, Cacna1s, Cacng3; cholinergic receptors Chrna10, Chrna6, Chrnb3 and Chrnb4; glutamate ionotropic receptors Grik1, Grin2c; 5-HT receptors Htr3a and Htr3b; voltage-gated potassium channels Kcnd2, Kcng3, Kcng4, Kcnj11, Kcnj13, Kcnn1, Kcnn2 and Kcns1; P2rx2; voltage-gated sodium channels Scn1A and Scn11A; and TRP channels Trpm2 and Trpm8. Most of these genes are well known to be neuronal in expression [61]. Overall, the ion channel subfamilies changing in expression in culture in both species were similar and included primarily voltage-gated calcium/potassium/sodium channels, purinergic receptors and gap junction proteins.
Changes in human RKs and other kinases:
We found that the neuronally-expressed genes from the NTRK family (NTRK1, NTRK2, NTRK3) and the CAMK family (CAMK1D, CAMK1G, CAMK2A, CAMK2B, CAMK2G, and CAMKK1) were decreased in culture in the human DRG. (Table 4, and Supplementary File 1 Sheets 7 and 9)
Changes in mouse RKs and other kinases:
Consistent with what we found in the human cultures, we identify decrease in abundance in neuronally-expressed Ntrk family (Ntrk1, Ntrk2, Ntrk3) and Camk (Camk1g, Camk2a, Camk2b) family genes. The changes in the Ntrk family, responsible for neurotrophin signaling in adult DRG neurons, demonstrates a consistent inter-species trend in culture. Consistent trends in the Camk family genes, which play a vital role in Ca2+-dependent plasticity in the brain [14] and in nociceptors [10; 11; 21], also show conserved patterns in the DRG cultures. (Table 4, and Supplementary File 1 Sheets 8 and 10)
Neuronal injury and inflammation markers were increased in human and mouse DRG cultures
Dissection of the DRG causes an axotomy that may induce an inflammatory phenotype as is seen in vivo after peripheral nerve injury [42]. As shown in Figure 4, many genes associated with inflammation and cell proliferation, neuronal injury and repair, and immune signaling and response, including cytokines [1; 9; 66] and matrix metalloproteases [29] associated with neuropathic pain, were differentially expressed in human and mouse DRG cultures with respect to intact DRG.
Since several of these genes are increased or decreased in cultured samples, we used the mouse DRG single cell RNA-seq profiles [61] to putatively identify cell types of expression among cells constituting the DRG (Supplementary File 1 Sheet 11). Indeed, we find that genes primarily expressed in neurons and Schwann cells decrease in relative abundance, even if they are involved in pro-inflammatory signaling, since it is likely that these cells types are reduced in frequency in DRG cultures. Interestingly, several genes predicted to be primarily expressed in immune cells (TLR9, CXCR3) (Figure 4A) and in vascular cells like IL18BP (Figure 4A), and CXCL17 (Figure 4B) were found to be reduced in relative abundance in cultures, suggesting that potential increase in immune and vascular cell proportions in culture are limited to certain cell subtypes in these categories. As maximal examples of gene expression changes in our datasets, IL6 and MMP9 mRNA expression were increased 100 fold or more in human DRG culture (Figure 4C).
Multiple subtypes of macrophages are involved in inflammatory processes and can be identified with specific markers [24]. In human and mouse, key M1 macrophage genes CD68, CD80, and SOCS3 were all upregulated in culture compared to intact ganglia. As identified in a recent study, HBEGF+ inflammatory macrophages are responsible for fibroblast invasiveness in rheumatoid arthritis patients [31]. We noted that multiple genes expressed in this specific subtype of macrophage (PLAUR, HBEGF, CREM) were increased in human and mouse DRG cultures, suggesting that this particular subtype of macrophage may be present in DRG cultures from both species (Figure 4D).
While specifically identifying the exact subtype of immune cell involved is outside the scope of our bulk RNA-sequencing assay, our findings reveal clearly that many genes involved in neuronal injury, cell proliferation and inflammation, and immune signaling and response are increased in DRG cultures.
Similarities and differences between human and mouse DRG culture transcriptomes in the context of intact DRG transcriptomes
Complicated orthologies and differential evolutionary dynamics between human and mouse gene families [72], and gaps in human to mouse orthology annotation [41] make comparative transcriptomic comparisons difficult between human and mouse transcriptomes. We have previously made similar comparisons between native human and mouse intact DRGs [51], finding overall similarities, but also some changes in gene expression. Since we are analyzing changes in expression at the level of individual genes (such as pharmacologically relevant ones), we limited our analysis to changes in expression in GPCRs, ion channels, and kinases in DRG cultures for tractability.
We calculated trend scores for each GPCR, ion channel, RK, and non-RK kinase, after eliminating genes from the analysis with complicated orthologies between humans and mouse (Supplementary File 1, Sheets 12–15). We find a weak correlation between trend scores of human genes and their mouse orthologs in GPCRs (Pearson’s R: 0.19, one tailed test p value: 0.0008) and ion channels (Pearson’s R: 0.15, one tailed test p value: 0.012). For specific genes, we find consistent increased (eg. F2R, GPRC5A, TRPV4) or decreased (eg. SCN subfamily members, GABAR subfamily members) in cultured samples for both species. This suggests that expression patterns across cell types, which potentially contributes to the trend scores, is likely conserved in these genes across species. However, in several cases, genes may not be consistently detectable in one species but present in one or more conditions in the other (eg. CHRM5 only detectable in human DRGs, CHRNB4 only detectable in mouse DRGs). TRPC5 is expressed at low levels in 3 intact mouse DRG samples, but significantly increased in expression in all human samples. Additionally, several genes are expressed in both species, but have opposing expression trends across intact and cultured DRGs (eg. ACKR4 and CXCR6 decreased in human cultures, but increased in mouse cultures). Such changes are likely due to evolutionary divergence between species in gene expression across cell types, and/or differential transcriptional regulation between species. Both of these involve regulatory evolution.
Supplementary File 1 Sheets 12 – 15 profile members of these gene families, their trend scores, and the number of human and mouse samples where they are detectable. This provides a roadmap for identifying genes changing in cultured versus intact DRGs across species; and creates a resource for the neuroscience community interested in performing molecular assays in cultured DRGs on these genes.
Since several members of the MRGPR family do not have a one-to-one orthology between human and mouse genes, they were not included in the trend score calculation tables (Supplementary File 1 Sheet 12). TPM values from human and mouse cultures for all members of this gene family are presented in their own table because this family of genes plays an important role in sensory neuroscience (Table 6).
Table 6.
MRGPR/Mrgpr family gene expression levels in human and mouse
| Human | Mouse | ||||
|---|---|---|---|---|---|
| Gene name | Mean TPM in intact DRGs | Mean TPM in cultured DRGs | Gene name | Mean TPM in intact DRGs | Mean TPM in cultured DRGs |
| MAS1 | 0.02 | 0.06 | Mas1 | 0.75 | 0.05 |
| No MAS1L ortholog | |||||
| MAS1L | 0.0 | 0.0 | - | - | |
| MRGPRD | 0.85 | 0.21 | Mrgpra1 | 0.77 | 0.04 |
| MRGPRE | 31.41 | 1.53 | Mrgpra2a | 20.91 | 0.67 |
| MRGPRF | 5.45 | 1.84 | Mrgpra2b | 23.85 | 0.96 |
| MRGPRG | 0.00 | 0.00 | Mrgpra3 | 24.50 | 0.02 |
| MRGPRX1 | 4.45 | 0.48 | Mrgpra4 | 0.48 | 0.01 |
| MRGPRX2 | 0.00 | 0.00 | Mrgpra6 | 0.00 | 0.00 |
| MRGPRX3 | 0.32 | 0.08 | Mrgpra9 | 0.98 | 0.01 |
| MRGPRX4 | 0.12 | 0.00 | Mrgprb1 | 0.15 | 0.03 |
| Mrgprb2 | 0.09 | 0.00 | |||
| Mrgprb3 | 0.00 | 0.00 | |||
| Mrgprb4 | 7.33 | 0.00 | |||
| Mrgprb5 | 9.14 | 0.04 | |||
| Mrgprb8 | 0.02 | 0.00 | |||
| Mrgprd | 74.20 | 0.03 | |||
| Mrgpre | 21.22 | 19.68 | |||
| Mrgprf | 7.91 | 73.72 | |||
| Mrgprg | 0.00 | 0.00 | |||
| Mrgprh | 0.23 | 0.04 | |||
| Mrgprx1 | 18.86 | 0.82 | |||
| Mrgprx2 | 0.04 | 0.01 | |||
Cd68 expression profiling using RNAscope
Previous mammalian DRG culture protocols for profiling RNA landscapes in sensory neurons have used mitotic suppressors to inhibit proliferation of mitotic cells [57] in spite of evidence that such inhibitors produce off-target effects on neurons [64]. This lends support to our hypothesis that changes to the cultured DRG transcriptome are at least partly shaped by an increase in proportion of proliferative cells.
We chose to profile Cd68 by RNAscope. The gene product for Cd68 is well known as a marker for myeloid lineage immune cells like monocytes and macrophages, including tissue-residential and circulating myeloid cells (microglia /macrophage) in the mouse Central Nervous System [75] and DRG [32].
RNAscope assays were conducted in intact mouse DRGs (Figure 5A). Gene expression for Cd68, and Calca (marker for most nociceptive neuronal subpopulations) and P2rx3 (marker for most non-peptidergic nociceptive neuronal subpopulations) was detected using the RNAscope probes. Additionally, immunostaining of Nf200 (gene product of Nefh, marker for neurofilament cell bodies and afferents) was performed. We also performed RNAscope assays in cultured mouse DRGs (Figure 5B) using only the Cd68 probe identifying Cd68 gene expression, additionally immunostaining Peripherin (gene product of Prph – a pan-sensory neuronal gene marker).
Figure 5.
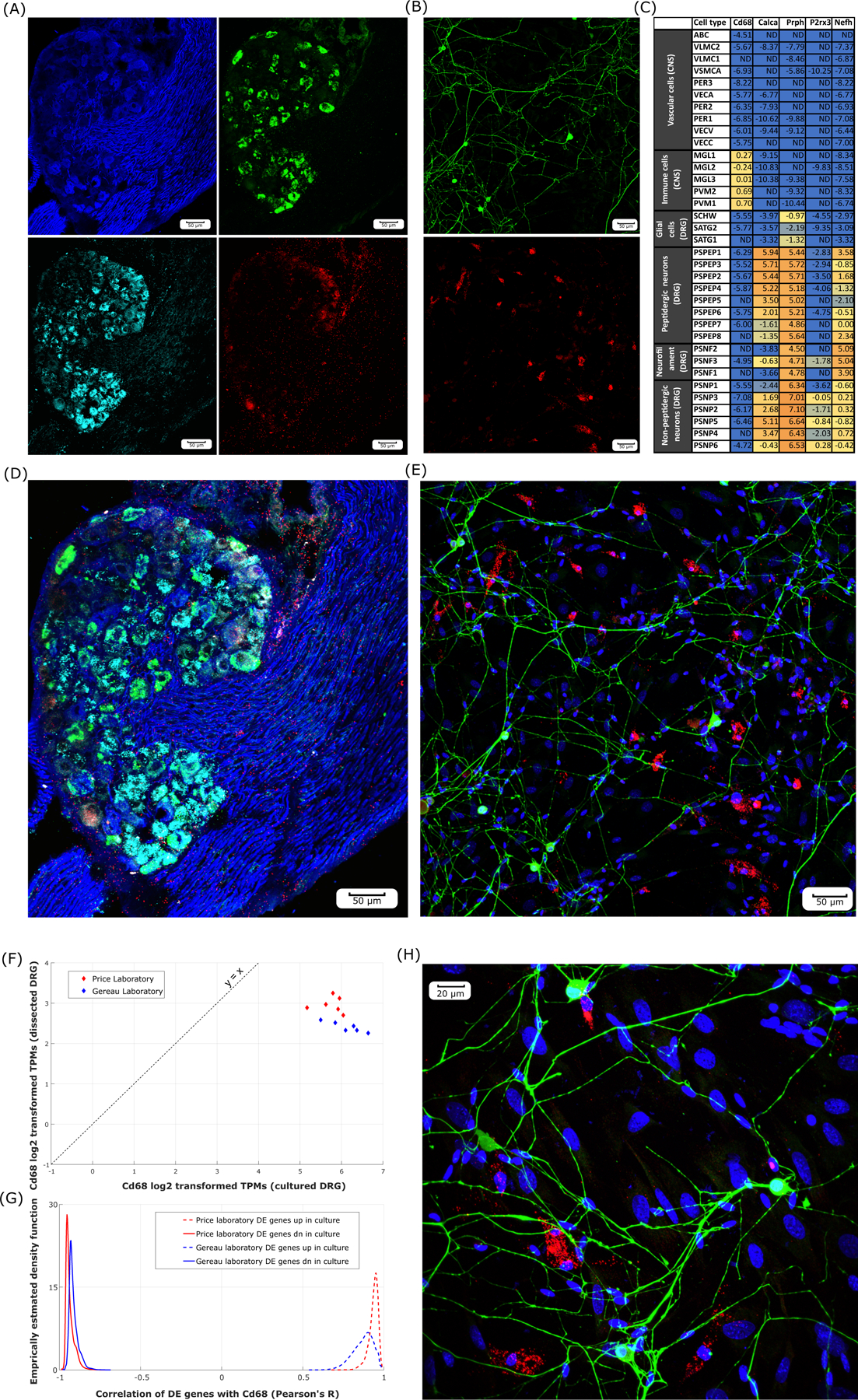
Cd68 expression in intact vs. cultured mouse DRGs. RNAscope in situ hybridization imaging (20X) in pseudo-color for Cd68 (red) in combination with various neuronal markers including Calca (green), P2rx3 (cyan), and immunostained Nf200 (blue) in intact mouse DRG (A). RNAscope in situ hybridization imaging (20X) in pseudo-color for Cd68 (red) and immunostained Peripherin (green) and DAPI staining (blue) in cultured mouse DRG (B). log2 transformed expression levels of Cd68 and neuronal markers from mousebrain.org (C) in mouse DRG neuron and glial subtypes, and nervous system vascular and immune cells show Cd68 is detected only in macrophage-like cells (ND: Not Detectable). Overlay of images show that Cd68 mRNA is not expressed in neurons in either intact (D) or cultured (E) mouse DRG neurons. Expression levels (in log2-transformed TPMs) of Cd68 in intact versus cultured mouse DRGs are plotted (F) to show the consistent increase of Cd68 expression in cultures. Differentially expressed gene TPMs show strong correlation (or anti-correlation) to Cd68 abundance (in TPMs) (G), suggesting a consistent phenotype across samples and laboratories. Overlay of RNAscope in situ hybridization imaging (40X) for cultured mouse DRG (H) suggests Cd68+ cells have consistent shape and size with respect to DRG macrophages. Scale bar for 20X images equal to 50μm and 40x equal to 20μm.
We then queried the mousebrain.org [75] database to identify putative cell type of expression (Figure 5C) of Cd68. The database contained gene expression levels for neurofilament (PSNF1–3), peptidergic (PSPEP1–8) and non-peptidergic (PSNP1–6) sensory neuronal subpopulations as well as Schwann Cell (SCHW) and Satellite Glial Cell (SATG1–2) subpopulations. Since the database did not profile DRG vascular or immune cells, we used CNS vascular and immune cell population gene expression profiles as surrogate. CNS vascular cells profiled include venous/capillary/arterial endothelial cells (VECA/C/V), vascular leptomeningeal cells (ABC, VLMC1–2), pericytes (PER1–2), and arterial smooth muscle cells (VSMCA). CNS immune cells profiled include microglia / tissue-resident macrophages (MGL1–3) and perivascular macrophages (PVM1–2). We thus find (using an external dataset) that Cd68 expression in mouse DRG is non-neuronal, and that it is only expressed in macrophage-like cell populations, in agreement with DRG immune cell studies [32].
Overlays of fluorescence imaging of the intact DRG (Figure 5D) and cultured DRG (Figure 5E, additionally overlaid with DAPI stain to identify nuclear DNA) on the 20X objective show co-fluorescence among the cell type markers. Little to no overlap is seen between Cd68-driven fluorescence and the neuronal markers in intact DRGs, clearly showing that Cd68 expression is non-neuronal in origin. In the cultured DRGs, we find no overlap of Cd68-driven fluorescence and Peripherin staining.
It is well documented that macrophage accumulation occurs in the DRG in mouse models of neuroinflammation [33], and macrophages are also known to upregulate Cd68 expression in response to inflammatory stimuli [13]. Looking at the RNA-seq libraries we generated, we find that Cd68 expression is consistently increased in both Price and Gereau laboratory mouse cultured DRG datasets (Figure 5F). In conjunction with the RNAscope data, this suggests that either Cd68+ macrophages increase in proportion in culture, or gene expression of Cd68 increases in macrophages in culture, or a combination of both occurs.
We then chose all the genes that were identified as differentially expressed between intact and cultured mouse DRGs based on our differential expression criteria (Supplementary File 1, Sheet 2, Column AR), which include Cd68. For both the Price and Gereau Laboratory datasets, expression levels of these genes were tested for correlation with Cd68 expression levels (Figure 5G) showing strong correlation of Cd68 with genes that increased in relative abundance in culture, and strong anticorrelation with genes that decreased in relative abundance in culture. This is also in agreement with our hypothesis of increase in cell type proportions of mitotic cells at the expense of sensory neurons (and possibly Schwann cells).The increased spread of correlation coefficients of genes increasing in Gereau laboratory mouse cultures (with respect to Price laboratory datasets) are in agreement with greater variability in Cd68 expression (in TPM) in Gereau laboratory cultures (std dev = 19.3) compared to Price laboratory cultures (std dev = 10.9).
Using the 40X objective (Figure 5H), we further find that the size and shape of Cd68+ cells are commensurate with previous studies of DRG macrophages [73]. The evidence, in its entirety, points to changes in constituent cell type proportions in DRG cultures, with potential increase of frequency of macrophage-like cells in DRG cultures.
Discussion
We are unaware of any previous studies that have used genome-wide technologies to characterize transcriptomes between intact and cultured DRGs. While in vivo cross-species comparisons have previously been made [51], in vitro transcriptome comparisons between mice and human DRGs have not been performed, despite the obvious need for such knowledge given the reliance on the mouse model for both target and drug discovery work in the pain area [16; 38; 50; 62; 76]. Certain perturbation studies [47] like gene expression knockdowns, DNA editing or optogenetic optimization [39], especially in the context of human research, cannot be performed in vivo, causing DRG cultures to be essential to human, clinical translational research. Our work gives fundamental new insight into some of the most commonly used model systems in the pain field with important implications for future work.
We reach two major conclusions. First, while many pharmacologically meaningful features of the DRG are well-conserved from mouse to human, there are some important differences that need to be considered in future experimental design. Moreover, there are a small but potentially important number of human receptors that simply cannot be studied in culture systems that may be good targets for drug discovery. Second, mouse and human DRG cultures take on an inflammatory-like transcriptomic phenotype that shares some qualities with transcriptomic changes in neuropathic pain [1; 29; 34; 43]. Therefore, the cultured DRG system may reflect certain clinical features that would be advantageous for neuropathic pain mechanism and/or drug discovery, especially in humans where these samples are not readily available except under very unique circumstances [43]. In further support of this conclusion we find a subset of genes that are upregulated in cultures from mouse and human DRGs that are consistent with a transcriptional reprogramming of sensory neurons after axonal injury that is associated with some aspects of neuropathic pain [40].
A critique of using primary neuronal cultures to test pharmacological targets is that cultures are not an accurate representation of native tissue. While there were some specific genes that did not appear in culture when compared to native tissue and vice versa, the main difference between the two conditions was in the expression level of each gene, which our data strongly suggests is due to change in the proportion of cell types. Specifically, the proportion of neurons in culture was decreased when compared to macrophages, fibroblast-like cells and SGCs. To this end, when specific pharmacological targets are being tested in either mouse or human cultures it is important to check that these targets remain expressed and our work provides a comprehensive resource to do this in both species (Supplementary File 1). Because pharmacology is the most common use of cultured DRG, we focused our analysis on pharmacologically relevant targets. Interestingly, both mouse and human cultures displayed an increase in M1 and HBEGF+ macrophage [31] markers when compared to native tissue. This change suggests an increase in the inflammatory macrophage population in culture. We predict that this shift is due to phenotypic shifts and / or increased cell type proportion of tissue resident macrophages caused during the dissociation and culturing process, potentially replicating a nerve injury phenotype [1; 9; 28; 29; 66]. The presence of these cell types in culture could be employed to further study how macrophages and sensory neurons interact, and should be very relevant to the pain community.
Families of genes remained consistently expressed in both species following dissociation and culturing protocols, but individual genes of the same family varied in whether they were present in either mouse or human. For example, Kcna1 was consistently detected only in mouse DRG cultures. Therefore, while most ion channel types are likely to be equally represented in both human and mouse DRG neurons, there is a substantial chance that the specific subtypes of channels that make up those conductances will be different between species, and such changes may be present both in vivo and in culture. In fact, studies focusing on exactly this question for voltage gated sodium channels in DRG between rat and human have found qualitative similarities but key differences that are almost certainly due to differences in expression between species [76]. This is a critical distinction for pharmacology because a primary goal in therapeutic development is ion channel subtype specific targeting [50]. It is vital to understand these similarities and differences when choosing a model system to study a particular target and, critically, we provide a resource to do this. From a discovery perspective, studies performed in vitro in mouse neuronal cultures likely remain a valid and reliable option for researchers as the families of ion channels, GPCRs, and RKs are well conserved from mice to humans (Supplementary File 1, sheets 12–15).
Our study has several limitations to acknowledge. The first is the choice of time point for the cultured DRG RNA-seq studies. We chose 4 DIV for our studies. Given the literature on biochemical and Ca2+ imaging studies (which is too extensive to cite) we think that our findings will provide a substantial resource for studies of this nature as most of them are done between 3 and 7 DIV. This can also be said for many electrophysiological studies on human DRG neurons as most investigators do experiments on these neurons over many days, with 4 DIV falling in the middle of the experimental spectrum for this small, but growing, body of work. The exception is mouse DRG electrophysiology where the vast majority of this very large literature has been done at 24 hrs after culturing. It is possible that some of the changes we observe at 4 DIV are not present at less than 1 DIV and/or that other differences are observed at this early time point. Another limitation is that changes in mRNA expression in culture may not represent differences in functional protein because some of these proteins may have long half-lives. In such a scenario, a down-regulation of mRNA would not lead to any difference in functional protein over the time course of our experiment (4 DIV). This can only be addressed with proteomic or physiological [53; 76] methods, which we have not done. Finally, we have relied on bulk RNA sequencing in the work described here. We acknowledge that single cell sequencing would yield additional insights that will be useful for the field. This will be a goal of future work.
We have focused on using DRGs from uninjured mice. Many studies have demonstrated that cultured DRG neurons from mice with neuropathic pain retain some neuropathic qualities in vitro, in particular spontaneous activity in a sub-population of nociceptors [2; 36; 43; 68; 70]. This also occurs in human DRG neurons taken from people with neuropathic pain [43]. Our transcriptomic studies suggest that cultured DRG neurons from normal mice and human organ donors show some transcriptomic changes consistent with a neuropathic phenotype, but these neurons likely do not generate spontaneous activity. Some transcriptomic changes were found in mice but not conserved in humans, especially relevant for translational pharmacological studies in cultured mouse DRGs. An example is caspase 6 (Casp6) which has been implicated in many neuropathic pain models in mice [5; 6; 36]. This gene was over 3 fold increased in mouse DRG culture but unchanged in human DRG cultures.
An interesting observation emerging from our work is that some macrophages are apparently present in DRG cultures and this macrophage phenotype is inflammatory in nature. An emerging literature describes DRG resident macrophages as key players in development of many chronic pain states, including neuropathic pain [24; 28; 43; 54; 66]. In future studies it may be possible to manipulate these macrophages to interact with DRG neurons in culture to push this neuropathic pain phenotype further toward the generation of spontaneous activity. Such studies could allow for the generation of a neuropathic pain model in vitro. Such an advance would be particularly useful for the human organ donor DRG model, especially considering that neuropathic pain in patients is associated with a macrophage transcriptomic signature, at least in males [43].
We have comprehensively characterized transcriptomic changes between native and cultured mouse and human DRG. These tissues are similar between the two species, suggesting that discovery work that is largely done in mice faithfully models many physiological characteristics of human DRG neurons. There are, however, important differences between species and between native and cultured conditions, with minimal impact of the type of culturing protocol used. Our resource brings these differences to light. A priori knowledge of gene expression levels of a potential pharmacological or perturbation assay target in mouse and human cultured and excised DRGs can provide a convenient framework for the pain biologist to decide appropriate model system choice and delineate pharmacological divergences. Additional whole transcriptome assays at varying timepoints in culture, from different DRG culture protocols, in additional species, and subject to various perturbations (including sorted cell type specific cell-pools) can be integrated into our database to get a more comprehensive picture of the RNA landscape of mammalian DRG cultures.
Supplementary Material
Funding:
NIH grants T32DA007261 (LM); NS113457 (CP); NS065926 and NS102161 (TJP); NS106953 and NS042595 (RWG).
Footnotes
The authors declare no conflicts of interest.
References Cited
- [1].Abbadie C, Bhangoo S, De Koninck Y, Malcangio M, Melik-Parsadaniantz S, White FA. Chemokines and pain mechanisms. Brain research reviews 2009;60(1):125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Amir R, Kocsis JD, Devor M. Multiple interacting sites of ectopic spike electrogenesis in primary sensory neurons. J Neurosci 2005;25(10):2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009;139(2):267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Benjamini H, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society: series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- [5].Berta T, Park CK, Xu ZZ, Xie RG, Liu T, Lu N, Liu YC, Ji RR. Extracellular caspase-6 drives murine inflammatory pain via microglial TNF-alpha secretion. J Clin Invest 2014;124(3):1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berta T, Perrin FE, Pertin M, Tonello R, Liu YC, Chamessian A, Kato AC, Ji RR, Decosterd I. Gene Expression Profiling of Cutaneous Injured and Non-Injured Nociceptors in SNI Animal Model of Neuropathic Pain. Sci Rep 2017;7(1):9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bhattacharjee A On a measure of divergence between two statistical populations defined by their probability distributions. Bulletin of the Calcutta Mathametical Society 1943;35:99–109. [Google Scholar]
- [8].Bhave G, Karim F, Carlton SM, Gereau RW. Peripheral group I metabotropic glutamate receptors modulate nociception in mice. Nat Neurosci 2001;4(4):417–423. [DOI] [PubMed] [Google Scholar]
- [9].Campbell JN, Meyer RA. Mechanisms of neuropathic pain. Neuron 2006;52(1):77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carlton SM. Localization of CaMKIIalpha in rat primary sensory neurons: increase in inflammation. Brain Res 2002;947(2):252–259. [DOI] [PubMed] [Google Scholar]
- [11].Chen Y, Yang C, Wang ZJ. Ca2+/calmodulin-dependent protein kinase II alpha is required for the initiation and maintenance of opioid-induced hyperalgesia. The Journal of neuroscience : the official journal of the Society for Neuroscience 2010;30(1):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chisolm DA, Cheng W, Colburn SA, Silva-Sanchez A, Meza-Perez S, Randall TD, Weinmann AS. Defining Genetic Variation in Widely Used Congenic and Backcrossed Mouse Models Reveals Varied Regulation of Genes Important for Immune Responses. Immunity 2019;51(1):155–168 e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev YV. CD68/macrosialin: not just a histochemical marker. Lab Invest 2017;97(1):4–13. [DOI] [PubMed] [Google Scholar]
- [14].Colbran RJ, Brown AM. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr Opin Neurobiol 2004;14(3):318–327. [DOI] [PubMed] [Google Scholar]
- [15].Conte C, Ebeling M, Marcuz A, Nef P, Andres-Barquin PJ. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol Genomics 2003;14(1):73–82. [DOI] [PubMed] [Google Scholar]
- [16].Davidson S, Golden JP, Copits BA, Ray PR, Vogt SK, Valtcheva MV, Schmidt RE, Ghetti A, Price TJ, Gereau RW. Group II mGluRs suppress hyperexcitability in mouse and human nociceptors. Pain 2016;157(9):2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res 2019;47(D1):D948–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dussor GO, Price TJ, Flores CM. Activating transcription factor 3 mRNA is upregulated in primary cultures of trigeminal ganglion neurons. Brain Res Mol Brain Res 2003;118(1–2):156–159. [DOI] [PubMed] [Google Scholar]
- [20].Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res 2006;34(Database issue):D319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ferrari LF, Bogen O, Levine JD. Role of nociceptor alphaCaMKII in transition from acute to chronic pain (hyperalgesic priming) in male and female rats. J Neurosci 2013;33(27):11002–11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, Garcia Giron C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martinez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigo R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47(D1):D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goswami SC, Thierry-Mieg D, Thierry-Mieg J, Mishra S, Hoon MA, Mannes AJ, Iadarola MJ. Itch-associated peptides: RNA-Seq and bioinformatic analysis of natriuretic precursor peptide B and gastrin releasing peptide in dorsal root and trigeminal ganglia, and the spinal cord. Mol Pain 2014;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hamidzadeh K, Christensen SM, Dalby E, Chandrasekaran P, Mosser DM. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu Rev Physiol 2017;79:567–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SM, Amode R, Brent S, Spooner W, Kulesha E, Yates A, Flicek P. Ensembl comparative genomics resources. Database (Oxford) 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hirai T, Mulpuri Y, Cheng Y, Xia Z, Li W, Ruangsri S, Spigelman I, Nishimura I. Aberrant plasticity of peripheral sensory axons in a painful neuropathy. Sci Rep 2017;7(1):3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, Fan G. Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci Rep 2016;6:31851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ji RR, Chamessian A, Zhang YQ. Pain regulation by non-neuronal cells and inflammation. Science 2016;354(6312):572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ji RR, Xu ZZ, Wang X, Lo EH. Matrix metalloprotease regulation of neuropathic pain. Trends Pharmacol Sci 2009;30(7):336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kiguchi N, Maeda T, Kobayashi Y, Fukazawa Y, Kishioka S. Macrophage inflammatory protein-1alpha mediates the development of neuropathic pain following peripheral nerve injury through interleukin-1beta up-regulation. Pain 2010;149(2):305–315. [DOI] [PubMed] [Google Scholar]
- [31].Kuo D, Ding J, Cohn IS, Zhang F, Wei K, Rao DA, Rozo C, Sokhi UK, Shanaj S, Oliver DJ, Echeverria AP, DiCarlo EF, Brenner MB, Bykerk VP, Goodman SM, Raychaudhuri S, Ratsch G, Ivashkiv LB, Donlin LT. HBEGF(+) macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci Transl Med 2019;11(491). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liang Z, Hore Z, Harley P, Stanley FU, Michrowska A, Dahiya M, La Russa F, Jager SE, Villa-Hernandez S, Denk F. A transcriptional toolbox for exploring peripheral neuro-immune interactions. bioRxiv 2019:813980. [DOI] [PubMed] [Google Scholar]
- [33].Lindborg JA, Niemi JP, Howarth MA, Liu KW, Moore CZ, Mahajan D, Zigmond RE. Molecular and cellular identification of the immune response in peripheral ganglia following nerve injury. J Neuroinflammation 2018;15(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lopes DM, Malek N, Edye M, Jager SB, McMurray S, McMahon SB, Denk F. Sex differences in peripheral not central immune responses to pain-inducing injury. Sci Rep 2017;7(1):16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002;298(5600):1912–1934. [DOI] [PubMed] [Google Scholar]
- [36].Megat S, Ray PR, Moy JK, Lou TF, Barragan-Iglesias P, Li Y, Pradhan G, Wanghzou A, Ahmad A, Burton MD, North RY, Dougherty PM, Khoutorsky A, Sonenberg N, Webster KR, Dussor G, Campbell ZT, Price TJ. Nociceptor Translational Profiling Reveals the Ragulator-Rag GTPase Complex as a Critical Generator of Neuropathic Pain. J Neurosci 2019;39(3):393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Megat S, Shiers S, Moy JK, Barragan-Iglesias P, Pradhan G, Seal RP, Dussor G, Price TJ. A Critical Role for Dopamine D5 Receptors in Pain Chronicity in Male Mice. J Neurosci 2018;38(2):379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Melli G, Hoke A. Dorsal Root Ganglia Sensory Neuronal Cultures: a tool for drug discovery for peripheral neuropathies. Expert Opin Drug Discov 2009;4(10):1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mickle AD, Gereau RW. A bright future? Optogenetics in the periphery for pain research and therapy. Pain 2018;159 Suppl 1:S65–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nguyen MQ, Le Pichon CE, Ryba N. Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nichio BTL, Marchaukoski JN, Raittz RT. New Tools in Orthology Analysis: A Brief Review of Promising Perspectives. Front Genet 2017;8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Niemi JP, DeFrancesco-Lisowitz A, Roldan-Hernandez L, Lindborg JA, Mandell D, Zigmond RE. A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J Neurosci 2013;33(41):16236–16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, Dussor G, Kim TH, Price TJ, Dougherty PM. Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 2019;142(5):1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ono K, Xu S, Hitomi S, Inenaga K. Comparison of the electrophysiological and immunohistochemical properties of acutely dissociated and 1-day cultured rat trigeminal ganglion neurons. Neurosci Lett 2012;523(2):162–166. [DOI] [PubMed] [Google Scholar]
- [45].Parisien M, Khoury S, Chabot-Dore AJ, Sotocinal SG, Slade GD, Smith SB, Fillingim RB, Ohrbach R, Greenspan JD, Maixner W, Mogil JS, Belfer I, Diatchenko L. Effect of Human Genetic Variability on Gene Expression in Dorsal Root Ganglia and Association with Pain Phenotypes. Cell Rep 2017;19(9):1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pearson K Notes on regression and inheritance in the case of two parents. Proceedings of the Royal Society of London 1895;58:240–242. [Google Scholar]
- [47].Perry RB, Hezroni H, Goldrich MJ, Ulitsky I. Regulation of Neuroregeneration by Long Noncoding RNAs. Mol Cell 2018;72(3):553–567 e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology 2015;33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 2017;14(7):687–690. [DOI] [PubMed] [Google Scholar]
- [50].Price TJ, Gold MS. From Mechanism to Cure: Renewing the Goal to Eliminate the Disease of Pain. Pain medicine 2018;19(8):1525–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018;159(7):1325–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma’ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sheahan TD, Valtcheva MV, McIlvried LA, Pullen MY, Baranger DAA, Gereau RW. Metabotropic Glutamate Receptor 2/3 (mGluR2/3) Activation Suppresses TRPV1 Sensitization in Mouse, But Not Human, Sensory Neurons. eNeuro 2018;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shepherd AJ, Copits BA, Mickle AD, Karlsson P, Kadunganattil S, Haroutounian S, Tadinada SM, de Kloet AD, Valtcheva MV, McIlvried LA, Sheahan TD, Jain S, Ray PR, Usachev YM, Dussor G, Krause EG, Price TJ, Gereau RW, Mohapatra DP. Angiotensin II Triggers Peripheral Macrophage-to-Sensory Neuron Redox Crosstalk to Elicit Pain. J Neurosci 2018;38(32):7032–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stone LS, Molliver DC. In search of analgesia: emerging roles of GPCRs in pain. Mol Interv 2009;9(5):234–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Student. The probable error of a mean. Biometirka 1908:1–25. [Google Scholar]
- [57].Thakur M, Crow M, Richards N, Davey GI, Levine E, Kelleher JH, Agley CC, Denk F, Harridge SD, McMahon SB. Defining the nociceptor transcriptome. Front Mol Neurosci 2014;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].The Gene Ontology C The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47(D1):D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tillu DV, Hassler SN, Burgos-Vega CC, Quinn TL, Sorge RE, Dussor G, Boitano S, Vagner J, Price TJ. Protease-activated receptor 2 activation is sufficient to induce the transition to a chronic pain state. Pain 2015;156(5):859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Turk R, t Hoen PA, Sterrenburg E, de Menezes RX, de Meijer EJ, Boer JM, van Ommen GJ, den Dunnen JT. Gene expression variation between mouse inbred strains. BMC Genomics 2004;5(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18(1):145–153. [DOI] [PubMed] [Google Scholar]
- [62].Valtcheva MV, Copits BA, Davidson S, Sheahan TD, Pullen MY, McCall JG, Dikranian K, Gereau RW. Surgical extraction of human dorsal root ganglia from organ donors and preparation of primary sensory neuron cultures. Nat Protoc 2016;11(10):1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Vergnolle N, Bunnett NW, Sharkey KA, Brussee V, Compton SJ, Grady EF, Cirino G, Gerard N, Basbaum AI, Andrade-Gordon P, Hollenberg MD, Wallace JL. Proteinase-activated receptor-2 and hyperalgesia: A novel pain pathway. Nat Med 2001;7(7):821–826. [DOI] [PubMed] [Google Scholar]
- [64].Wallace TL, Johnson EM, Jr. Cytosine arabinoside kills postmitotic neurons: evidence that deoxycytidine may have a role in neuronal survival that is independent of DNA synthesis. J Neurosci 1989;9(1):115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Watkins LR, Maier SF. Beyond neurons: evidence that immune and glial cells contribute to pathological pain states. Physiol Rev 2002;82(4):981–1011. [DOI] [PubMed] [Google Scholar]
- [67].Woolf CJ, Ma Q. Nociceptors--noxious stimulus detectors. Neuron 2007;55(3):353–364. [DOI] [PubMed] [Google Scholar]
- [68].Wu Z, Li L, Xie F, Du J, Zuo Y, Frost JA, Carlton SM, Walters ET, Yang Q. Activation of KCNQ Channels Suppresses Spontaneous Activity in Dorsal Root Ganglion Neurons and Reduces Chronic Pain after Spinal Cord Injury. Journal of neurotrauma 2017;34(6):1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang DP, Zhang DP, Mak KS, Bonder DE, Pomeroy SL, Kim HA. Schwann cell proliferation during Wallerian degeneration is not necessary for regeneration and remyelination of the peripheral nerves: axon-dependent removal of newly generated Schwann cells by apoptosis. Mol Cell Neurosci 2008;38(1):80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yang Q, Wu Z, Hadden JK, Odem MA, Zuo Y, Crook RJ, Frost JA, Walters ET. Persistent pain after spinal cord injury is maintained by primary afferent activity. J Neurosci 2014;34(32):10765–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yarwood RE, Imlach WL, Lieu T, Veldhuis NA, Jensen DD, Klein Herenbrink C, Aurelio L, Cai Z, Christie MJ, Poole DP, Porter CJH, McLean P, Hicks GA, Geppetti P, Halls ML, Canals M, Bunnett NW. Endosomal signaling of the receptor for calcitonin gene-related peptide mediates pain transmission. Proc Natl Acad Sci U S A 2017;114(46):12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet 2002;11(5):535–546. [DOI] [PubMed] [Google Scholar]
- [73].Yu X, Liu H, Hamel KA, Morvan MG, Yu S, Leff J, Guan Z, Braz JM, Basbaum AI. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat Commun 2020;11(1):264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yu Y, Huang X, Di Y, Qu L, Fan N. Effect of CXCL12/CXCR4 signaling on neuropathic pain after chronic compression of dorsal root ganglion. Sci Rep 2017;7(1):5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S. Molecular Architecture of the Mouse Nervous System. Cell 2018;174(4):999–1014.e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhang X, Priest BT, Belfer I, Gold MS. Voltage-gated Na(+) currents in human dorsal root ganglion neurons. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhang XD, Marine SD, Ferrer M. Error rates and powers in genome-scale RNAi screens. J Biomol Screen 2009;14(3):230–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


