Abstract
In this study, we aimed at determining the impact of naphthalene and different oxygen levels on a biofilm bacterial community originated from a petroleum hydrocarbon–contaminated groundwater. By using cultivation-dependent and cultivation-independent approaches, the enrichment, identification, and isolation of aerobic and oxygen-limited naphthalene degraders was possible. Results indicated that, regardless of the oxygenation conditions, Pseudomonas spp. became the most dominant in the naphthalene-amended selective enrichment cultures. Under low-oxygen conditions, P. veronii/P. extremaustralis lineage affiliating bacteria, and under full aerobic conditions P. laurentiana–related isolates were most probably capable of naphthalene biodegradation. A molecular biological tool has been developed for the detection of naphthalene 1,2-dioxygenase-related 2Fe-2S reductase genes of Gram-negative bacteria. The newly developed COnsensus DEgenerate Hybrid Oligonucleotide Primers (CODEHOP-PCR) technique may be used in the monitoring of the natural attenuation capacity of PAH-contaminated sites. A bacterial strain collection with prolific biofilm-producing and effective naphthalene-degrading organisms was established. The obtained strain collection may be applicable in the future for the development of biofilm-based bioremediation systems for the elimination of PAHs from groundwater (e.g., biofilm-based biobarriers).
Electronic supplementary material
The online version of this article (10.1007/s00253-020-10668-y) contains supplementary material, which is available to authorized users.
Keywords: Biofilm, PAH, Naphthalene, Bioremediation, Oxygen-limited, Biobarrier
Introduction
Polycyclic aromatic hydrocarbons (PAHs), originating either from natural or from anthropogenic sources, are widespread persistent and toxic pollutants of great concern. Many PAHs, consisting of two or more combined benzene rings, are known to be mutagenic and/or carcinogenic (Harvey 1991). The US Environmental Protection Agency (EPA) has listed 16 PAHs (including naphthalene) as priority pollutants (Cerniglia and Heitkamp 1989). PAHs can enter to the environment via several routes including the burning of fossil fuels, processing of gas/coal tar/wood, waste incineration, and fuel leakage (for a review, see Duran and Cravo-Laureau 2016). Apart from acenaphthene, acenaphthylene, and dibenz[a,h]anthracene, which are primarily detected in gasoline and diesel exhaust gas, the majority of the 16 priority pollutant PAHs (13) can be detected in gasoline itself (Zoccolillo et al. 2000; NCBI PubChem 2020).
Microbial degradation is the major mechanism in determining the fate of PAHs in the environment, whether in aquatic (e.g. marine, Cravo-Laureau and Duran 2014) or terrestrial ecosystems (Ghosal et al. 2016). Therefore, microbial resources have been considered for the eco-friendly remediation of PAH-contaminated sites (de Boer and Wagelmans 2016; Bordenave et al. 2004b; Ben Said et al. 2015).
Today, more and more efforts are being made globally on the improvement of sustainable PAH bioremediation strategies. Compared to the physical-chemical approaches, the biological treatment of contaminated sites is a more preferred, efficient, and cost-effective choice (Kuppusamy et al. 2016). Biodegradation of PAHs leads to the complete degradation of the pollutants with greater safety and with reduced environmental disturbance (Habe and Omori 2003). Studies dealing with PAH biodegradation should point in the direction of sustainability from both environmental and financial aspects. Therefore, based on global trends and taking into consideration sustainability, a better understanding of microbial communities responsible for efficient PAH biodegradation under different environmental circumstances—e.g., aquatic versus terrestrial ecosystem, marine versus freshwater environment, aerobic/oxygen-limited/anaerobic conditions—is a must. Related to the oxygenation conditions, our previous studies already pointed out the importance of available oxygen in petroleum hydrocarbon–contaminated environments. As it was found earlier, the presence of enough available oxygen in the contaminated environment could substantially increase the biological degradation rate of petroleum hydrocarbons (crude oil or PAHs). Oxygenation proved to be critical in the process of biodegradation. It was also observed that oxic/anoxic oscillations can be as effective as full aerobic conditions during hydrocarbon biodegradation. However, it has to be added that oxic/anoxic alternations may have an undesired impact upon bacterial community structures, influencing their ability to degrade hydrocarbons and their capacity to reduce hydrocarbon toxicity (Cravo-Laureau et al. 2011; Vitte et al. 2011; Duran et al. 2015; Militon et al. 2015; Terrisse et al. 2017). It was also found previously that, in subsurface hydrocarbon-contaminated groundwater, oxygen availability is one of the main driving factors in the organization of microbial communities (Benedek et al. 2016, 2018).
The isolation of efficient PAH-degrading bacteria, applicable in the development of innovative, relatively cheap PAH remediation technologies, like biobarriers, small bioreactor platform technology SBPs, enzyme-mediated bioremediation, and mixed cell culture systems—algal/bacterial consortia, etc.—has become also important (Carreghini et al. 2013; Kuppusamy et al. 2016; Menashe and Kurzbaum 2014). As we previously demonstrated, biofilms/microbial mats, developed in hydrocarbon-contaminated environments, represent a microbial resource from which microorganisms with the desired bioremediation potential can be isolated (Bordenave et al. 2004a; Benedek et al. 2016, 2018). In our previous studies, the targeted biofilms (Bugyi site, freshwater environment, Hungary) and microbial mats (Camargue site, marine environment, France) proved to be the habitat of complex networks of chemoorganotrophic and chemolithotrophic bacteria where microorganisms with opposite traits (e.g., aerobic/anaerobic, iron reducers/iron oxidizers) co-existed and interacted on complementary processes (Benedek et al. 2016). The collected samples proved to be the reservoir of hydrocarbonoclastic bacteria capable of aerobic and oxygen-limited (hypoxic) degradation of simple aromatic hydrocarbons (BTEX), as well as low-molecular-weight (LMW < 4 rings) PAHs. Moreover, the investigated biofilm samples also proved to be the source of catabolic genes involved in aerobic (I.2.A and I.2.B catechol 2,3-dioxygenases), microaerobic (I.2.C catechol 2,3-dioxygenases) and anaerobic (bssA) simple aromatic hydrocarbon degradation, as well as in LMW-PAH biodegradation (Bordenave et al. 2004a; Benedek et al. 2016, 2018).
In this study, we aimed at determining the impact of naphthalene (LMW-PAH) on a biofilm bacterial community, which developed initially in hydrocarbon-contaminated groundwater at the Bugyi site (Hungary), where the concentration of oxygen is generally low. Based on our previous studies (Benedek et al. 2016, 2018), we believe that the investigated biofilm, with high phylogenetic and functional diversity, is an accurate and easy-to-handle microbial community to investigate the impact of PAHs (e.g., naphthalene) on bacterial communities under different oxygen levels. It is hypothesized that distinct aerophilic conditions will shape the versatile bacterial community and induce the emergence of bacterial genera adapted to aerobic and oxygen-limited degradation of PAHs. To test the hypothesis, naphthalene will be used as a model PAH compound. We also expect to isolate PAH-degrading and biofilm-producing bacteria, which can be useful for the development of semipermeable biofilm-based biobarriers. Overall, our results provide valuable information regarding PAH biodegradation in hypoxic subsurface freshwater ecosystems.
Materials and Methods
Site description, pollution history, and biofilm sampling
The bacterial biofilm sample was collected in January 2018 in the Central Region of Hungary. The sample was taken from the stainless steel surface of a submersible pump, which belonged to a Pump and Treat system (P&T system) treating gasoline-contaminated groundwater. In 2007, due to an accident, ~ 27 m3 of gasoline leaked into the shallow groundwater leading to aliphatic and aromatic hydrocarbons contamination of the sampling site. Within the contamination plume, the installation of a rapid in situ decontamination P&T system was crucial for ecological and public health reasons. On the date of system startup, the main groundwater pollutants were BTEX—benzene (7 mg l−1), toluene (28.5 mg l−1), ethyl-benzene (1 mg l−1), xylenes (10.4 mg l−1)—and total aliphatic petroleum hydrocarbons (TAPH C5–C40, 10 mg l−1). For more details regarding site characterization and P&T system operation, please see Benedek et al. (2016, 2018).
Although the concentration of PAHs has not been determined separately, based on literature data, their presence in gasoline-contaminated sites is highly expected. PAHs are a class of chemicals that occur naturally in gasoline (Candeli et al. 1975; Zoccolillo et al. 2000). Marr et al. (1999) by measuring the PAH content of five different gasoline samples (regular, premium, and midgrade quality) found that on average, naphthalene contributes 97 ± 1% of the total concentration of the PAHs measured. PAH concentrations in the fuels ranged from undetectable (< 0.1 mg l−1) for several of the higher molecular weight PAHs to 2600 mg l−1 for naphthalene. Gasoline’s naphthalene content varies by brand and grade, and “premium” gasoline tends to have higher concentrations than “regular” gasoline (Jia and Batterman 2011). Naphthalene content of gasoline has been expressed as 1.04 mg g−1 (Schauer et al. 2002), 69 to 2600 mg l−1 (Marr et al. 1999), and 0.15 to 0.18% (w/w) (Harley et al. 2000).
The temperature of slightly alkaline (pH 7.3–7.5) groundwater is between 10 and 14 °C. The oxygen concentration of the groundwater is in the hypoxic range (≤ 2 mg l−1). The treatment capacity of the system is 30 m3 of contaminated groundwater per day. Owing to the P&T system, the groundwater has been gradually decontaminated. By the time of sampling, in the case of the biofilm sampling well BUT18, the concentration of the abovementioned hydrocarbons (BTEX and TAPH) had decreased significantly below the threshold values defined by the Hungarian Government, Regulation No. 6/2009. (IV.14) KvVM-EÜM-FVM.
In the aforementioned P&T system, on the surface of submersible pumps located in the extraction wells and inside the pipelines, frequent bacterial biofilm formations were observed. For our studies, the biofilm sample was collected from the extraction well BUT18 and placed into sterile 50-ml Falcon conical centrifuge tubes. Samples were stored in dry ice and were processed immediately upon arrival at the laboratory.
Enrichment of naphthalene-degrading biofilm bacteria
Naphthalene-degrading enrichment cultures were similarly initiated as described previously in the case of BTEX-degrading biofilm enrichments (Benedek et al. 2018).
In 100-ml crimp-sealed serum bottles, in mineral salt solution (49 ml) supplemented with vitamins (Fahy et al. 2006), full aerobic (~ 8 mg l−1 O2) and oxygen-limited (≤ 0.5 mg l−1 O2) enrichment microcosms were set up in duplicates. One milliliter of biofilm suspension (1 g of biofilm suspended in 9 ml mineral salt solution) was added to each microcosm as an inoculant. Naphthalene crystals were added to the microcosms in a final concentration of 100 mg l−1 as the only source of carbon and energy (Acros Organics, Belgium). After 1 week of incubation (28 °C at 150 rpm), the naphthalene crystals, which were initially clearly visible to the naked eye, totally disappeared from the aerobic enrichments. Consequently, 1 ml of the aerobic enrichment was transferred to freshly prepared enrichment medium amended again with naphthalene crystals. Six consecutive transfers were conducted. In the case of oxygen-limited enrichments, the transfers were done every 2 weeks, three transfers in total. For the oxygen-limited bacterial community, 2 weeks were needed for the complete degradation of the naphthalene crystals. Before initiating the oxygen-limited enrichments, microcosms were sparged aseptically with N2/CO2 (80:20, v/v) for 10 min. The desired oxygen concentration (0.5 mg l−1) was set by sterile air injection (0.2-μm-pore-size-filtered) through the butyl-rubber septa. Oxygen concentration in the liquid phase was measured non-invasively by using a Fibox 3 trace v3 fiber optic oxygen meter with PSt3 sensor spots (PreSens, Regensburg, Germany). In the case of oxygen depletion, supplementation was performed.
Indirect evidence, namely naphthalene biodegradation screening tests of isolates prior to GC-MS measurements (data not shown), suggest that during the enrichment, naphthalene crystals’ disappearance from the bottles was predominantly due to biodegradation and not to volatilization.
Isolation of naphthalene-degrading bacteria
The isolation of naphthalene-degrading bacteria was performed in the case of both aerobic and oxygen-limited enrichments. Bacterial isolations occurred during the first transfer (after 1 or 2 weeks of incubation) and at the end of the enrichment period (after 6 weeks of incubation).
Prior to isolation, from the respective microcosms, 1 ml was used for the preparation of tenfold serial dilutions in physiological salt solution (0.9 % m/v; up to 10−6 dilution). One hundred microliters of serially diluted aerobic and oxygen-limited samples was spread on the surface of (i) R2A agar plates, (ii) R2A agar plates amended with naphthalene, or (iii) Bushnell-Haas-Broth plates solidified with gellan gum and supplemented with naphthalene. The composition per liter of R2A agar was the following: proteose peptone 0.5 g, casamino acids 0.5 g, yeast extract 0.5 g, dextrose 0.5 g, soluble starch 0.5 g, dipotassium phosphate 0.3 g, MgSO4·7H2O 0.05 g, sodium pyruvate 0.3 g, agar 15 g, pH 7 ± 0.2. Bushnell-Haas-Broth mineral salt medium was contained in 1000 ml of H2O 0.002 g of CaCl2·2H2O, 1 g of MgSO4·7H2O, 1 g of NH4NO3, 1 g of KH2PO4, 1 g of K2HPO4, 0.005 g of FeCl3·6H2O, and 8 g of gellan gum, and the pH was set to 7 ± 0.2. Naphthalene had been spread on the surface of solidified media in the form of naphthalene-methanol solution (100 μl, 10 g l−1). After methanol evaporation, white crystals of naphthalene appeared and then inoculation of the bacterial suspension took place (100 μl of each member of the tenfold dilutions). After inoculation, Petri dishes were thoroughly sealed with parafilm to reduce evaporation of naphthalene. Plates were incubated at 15 °C for 2 weeks (incubation temperature common to in situ conditions of the groundwater). Developed colonies, showing different morphologies, were purified by streak plating and maintained on R2A agar slants at 4 °C, as well as at − 80 °C in a glycerol solution (15% v/v). All chemicals, if not stated otherwise, were purchased from Sigma-Aldrich, Germany.
Extraction of nucleic acids
Total community DNA from the biofilm sample (0.5 g) was extracted by using the DNeasy® PowerSoil Kit (Qiagen, Hilden, Germany). Community DNA of the enrichment cultures and genomic DNA of the isolates were extracted by using the DNeasy® UltraClean® Microbial Kit (Qiagen, Hilden, Germany). In the case of microcosms, 49 ml of enrichment cultures was centrifuged (2360×g, 15 min, Rotanta 460 R centrifuge—Hettich, Germany) and the DNA was extracted from the pellet. Genomic DNA isolation was performed from overnight cultures of bacteria. The manufacturer’s instructions were followed in all cases.
Gene amplifications
The species-level identification of cultivable biofilm bacteria took place on the basis of 16S rRNA gene–based sequence analysis. PCR amplification of the gene was performed as described earlier. Universal bacterial primers 27F (5′-AGAGTTTGATC(A/C)TGGCTCAG-3′) and 1492R (5′-TACGG(C/T)TACCTTGTTACGAC TT-3′) were used (Benedek et al. 2016).
For Illumina paired-end 16S rRNA amplicon sequencing, the variable V3 and V4 regions of the gene were amplified as described in our previous study (Benedek et al. 2018). The forward and reverse primers with Illumina adapter overhanging nucleotide sequences (written in bold) were the following: forward 5′-TCGTCGGCAGCGTCAGATGTG TATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and reverse 5′-GTCTCGTGGGCT CGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′ (Klindworth et al. 2013). PCR conditions and reagent concentrations were the same as described earlier (Benedek et al. 2018).
CODEHOP primer design for the detection of NDO
A new set of COnsensus DEgenerate Hybrid Oligonucleotide Primers has been designed to assess the PAH biodegradation ability of the initial biofilm community, and the enriched cultures, as well as of the isolated strains (CODEHOP, Rose et al. 2003, Staheli et al. 2011). During the primer design, we focused on the reductase component of the naphthalene 1,2-dioxygenase gene found in Gram-negative bacteria (NDO; EC 1.14.12.12). NDO is a member of the ring-hydroxylating dioxygenase (RHD) family of bacterial enzymes that play a pivotal role in the degradation of aromatics, such as PAHs (naphthalene, fluorene, anthracene, and benzo[a]pyrene). The enzyme is comprised of a multicomponent system, containing a reductase that is an iron-sulfur (2Fe-2S) flavoprotein (FAD; EC 1.18.1.3, ferredoxin-NAD+ reductase), an iron-sulfur oxygenase, and ferredoxin (Ensley and Gibson 1983; Jouanneau et al. 2006).
Primer design was done on the basis of conserved blocks of amino acids within aligned 2Fe-2S reductase protein sequences. Amino acid sequences were retrieved from the GenBank and aligned with ClustalW (MEGA version 7.0). For primer design, the aligned multiple protein sequences were subjected to j-CODEHOP design software, an integrated tool into Base-by-Base (Tu et al. 2018). The length of the non-degenerate 5′ clamp region was set to 18 base pairs (bp). The length of degenerate 3′ region was set to 4 amino acids (aa). Max degeneracy was set to 64, while min AA conservation to 80%.
Amplifications with the newly obtained CODEHOP primers were performed in 50 μl reaction mixture. The mixture contained 5 μl 10x DreamTaqTM buffer (ThermoFisher Scientific, Lithuania) with MgCl2 (2 mM), 0.2 mM of each dNTP, 0.1 μM of each primer, 1 U DreamTaqTM DNA Polymerase (ThermoFisher Scientific, Lithuania), 1 μl (~ 40 ng) extracted DNA, and nuclease-free water up to the final reaction volume. The following amplification conditions were applied: 95 °C for 3 min, then 32 cycles of 94 °C for 30 s, annealing temperature 66 °C for 30 s and 72 °C for 1 min, and then a final extension at 72 °C for 10 min. Amplifications were conducted on (i) isolate-derived genomic and (ii) community-derived DNA samples (initial biofilm and enrichment samples). PCR amplifications were expected to yield PCR products of ~ 750 bps. To check the specificity of the CODEHOP-primer pair for PCR amplification of the target sequence, positive and negative control genomic DNA samples were used. According to the whole genome sequences, Z. oleivorans strain BUC-1T (SDKK00000000), R. pyridinivorans AK37 (NZ_AHBW00000000), and C. basilensis OR16 (NZ_AHJE00000000) do not have NDO-related 2Fe-2S reductase genes in their genomes. Therefore, genomic DNA originating from those isolates was used as negative control. Since Malikia spinosa strain AB6 contains NDO reductase in its genome (VYSV00000000), its genomic DNA was used as a positive control during the PCR amplifications. As a further act of validation, all CODEHOP PCR products (originating either from the community or from isolate DNA) were sequenced and homology BLAST searches were made in the NCBI GenBank database.
All amplifications were carried out in a ProFlex PCR System (Applied Biosystems by Life Technologies, USA). Amplicons were analyzed under UV light after electrophoresis in 1% (w/v) agarose gel stained with EtBr. All amplicons, prior to subsequent analyses, were purified by NucleoSpin® Gel and PCR Clean-up set (Macherey-Nagel, Düren, Germany).
Cloning of NDO reductase component genes
The diversity of NDO-related 2Fe-2S reductase genes, from aerobic and oxygen-limited enrichment samples of the sixth week, was assessed by molecular cloning as described by Táncsics et al. (2012). The ~ 750-bp-long PCR products were cloned into pCR 2.1 vector (Invitrogen, Carlsbad, USA) following the manufacturer’s instructions. Standard blue-white selection method was applied for the selection of transformants. Plasmid DNA was extracted with heat shock (98 °C, 5 min); inserts were reamplified using M13 primer set (forward 5′-GTA AAA CGA CGG CCA G-3′, reverse 5′-CAG GAA ACA GCT ATG ACC-3e).
DNA sequencing
Illumina 16S rDNA amplicon sequencing was performed to compare the bacterial community composition of the initial biofilm to the selected aerobic (NAF_A_A.6) and oxygen-limited enrichments (NAF_H_A.6) of the final week. Illumina 16S rDNA (V3-V4 region) sequencing was performed as described previously (Benedek et al. 2018). Primary data analysis (base-calling) was carried out with Bbcl2fastq^ software (v2.17.1.14, Illumina). Reads were quality and length trimmed in CLC Genomics Workbench Tool 9.5.1 using an error probability of 0.05 (Q13) and a minimum length of 50 nucleotides as threshold. Trimmed sequences were processed using mothur v1.41.1 (Schloss et al. 2009) as recommended by the MiSeq SOP page (http://www.mothur.org/wiki/MiSeq_SOP) (Kozich et al. 2013). Sequences were assorted based on the alignment using SILVA 132 SSURef NR99 database (Quast et al. 2013). Chimera detection was performed with mothur’s uchime command (Edgar et al. 2011), and ‘split.abund’ command was also used to remove singleton reads according to Kunin et al. (2010). The standard 97% similarity threshold was used to determinate operational taxonomic units (OTUs) as suggested by Tindall et al. (2010) for prokaryotic species delineation. Raw sequence reads were deposited in NCBI SRA under BioProject ID PRJNA562625. Abundant OTUs were also identified by applying EzBioCloud 16S rDNA database (Yoon et al. 2017).
The nucleotide sequence determination of 16S rRNA and NDO-related 2Fe-2S reductase genes was performed by Sanger sequencing using BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, USA). Sequences were analyzed with ABI 3130 Genetic Analyzer (Life Technologies, USA), edited and assembled using MEGA version 7.0 (Kumar et al. 2016). Homology BLAST searches (Altschul et al. 1997) were made in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). On the basis of 16S rRNA genes, EzTaxon-e server carried out the determination of the closest type strains of isolates (http://eztaxon-e.ezbiocloud.net/; Kim et al. 2012). For DNA sequencing, both forward and reverse sequencing primers were used. In the case of NDO-related 2Fe-2S reductase genes for the sequencing reaction, a new set of CODEHOP primers was used without the degenerate core.
16S rRNA gene sequence data obtained in this study was deposited in the GenBank under the accession numbers MN197554-MN197593. Putatively NDO-related 2Fe-2S reductase component gene sequences derived from uncultured bacteria were deposited in the GenBank under the accession numbers MN370456-MN370503 (aerobic enrichment) and MN370504-MN370547 (oxygen-limited enrichment). NDO-related reductase component gene sequences, which were derived from the biofilm-related bacterial isolates, were deposited under the accession numbers (MN420520-MN420545).
Temporal dynamics assessed by T-RFLP
Terminal restriction fragment length polymorphism (T-RFLP, Liu et al. 1997) was used to assess the alterations of the initial biofilm community throughout the whole enrichment period. For phylogenetic T-RFLP analysis, 16S rRNA genes from the initial biofilm and from the enrichments DNA were PCR amplified by using VIC fluorescently labelled 27F and non-labelled 1492R primers. Reagent concentrations and PCR conditions were the same as described earlier (Benedek et al. 2014). The obtained PCR products were digested with 1 U AluI (AG↓CT) restriction endonuclease (Fermentas, Lithuania) for 1.5 h at 37 °C. The generated fluorescently-labelled terminal restriction fragments (T-RFs) were purified by ethanol precipitation. Fragments were separated by capillary gel electrophoresis. T-RFs were detected using a Model 3130 Genetic Analyzer (Applied Biosystems, USA). GeneScanTM 1200 LIZTM internal size standard was used (Applied Biosystems, USA). The resulted T-RFLP electropherograms were analyzed with the GeneMapper Software version 4.0 (Applied Biosystems, USA). For consensus profiles, runs of two replicates were aligned with the T-Align program and 0.5-bp confidence interval was used (Smith et al. 2005).
To determine temporal dynamics of the initial biofilm community, due to the different enrichments, the T-align originated comparison results files were evaluated with the PAST software (Hammer et al. 2001). PCA (principal component analysis) was used to visualize the results.
Testing naphthalene degradation potential of biofilm isolates by GC-MS
Naphthalene biodegradation ability of isolates was assessed through microcosm experiments by gas chromatography coupled to mass spectrometry (GC-MS). Microcosms were set up in triplicates in hermetically closed, crimp-sealed vials containing 50 ml BBH mineral salt solution and naphthalene in a final concentration of 1 mg l−1. Naphthalene was introduced into the bottles in the form of methanol-naphthalene stock solution (1 g l−1) in a 50-μl volume. Microcosms were inoculated with 100 μl of bacterial cell suspensions made in physiological saline solution (OD600nm = 1). The concentration of naphthalene at the beginning of the experiment, as well as after 20 h of incubation (28 °C, 150 rpm), was determined from the headspace using an SPME polydimethylsiloxane fiber assembly (Supelco) for sampling and a Trace 1300 gas chromatograph coupled to ISQ Single Quadrupole mass spectrometer (Thermoscientific) for analysis. During the analysis, injector and detector temperatures were maintained at 240 °C and 270 °C, respectively. The oven temperature program was set to 40 °C for 3 min and then ramped at a rate of 70 °C min−1 to 230 °C and finally held for 5 min. Helium was used as the carrier gas at a flow rate of 1.2 ml min−1. SLB TM − 5 ms Fused Silica capillary column was used for separation (30 m × 0.25 mm × 0.25 μm, Sigma-Aldrich, Supelco). The mass spectrometer (MS) operated at full scan mode.
Testing biofilm-forming ability of isolates
Biofilm-forming ability of isolated strains was determined as described in our previous study (Benedek et al. 2018). Briefly, 250 μl bacterial cell suspensions in R2A medium were inoculated in triplicates into chimney well cell culture polystyrene microplates (greiner bio-one, Germany). After incubation at 28 °C for 24, 48, and 72 h, wells were decanted and rinsed three times with phosphate buffer (250 μl). Subsequently, biofilms that adhered to well surfaces were fixed with methanol (99%, 250 μl) and stained with crystal violet (0.5%, 250 μl). After cell resuspension with 33% glacial acetic acid, the absorbance of the solution (A) at 550 nm (absorbance of crystal violet) was determined by using a BioTek ELx800 Absorbance Microplate Reader (BioTek, USA). Evaluation of the results was done according to Stepanović et al. (2000):
non-adherent Asample ≤ AC
weakly adherent AC < Asample ≤ 2 × AC
moderately adherent 2 × AC < Asample ≤ 4 × AC
strongly adherent 4 AC < Asample
AC represents the cutoff absorbance value defined as three standard deviations above the mean absorbance of the negative control.
Results
Bacterial diversity as assessed by cultivation-dependent technique
From the enrichment cultures, a set of 40 bacterial strains were isolated (Table 1). Irrespective of the isolation method, the majority of the isolates belonged to the genus Pseudomonas (75%). The highest number of Pseudomonas spp.–related strains was obtained during the last isolation step (20 isolates). In the aerobic enrichment, Pseudomonas spp.–related strains mostly affiliated with P. silesiensis (1st week) and P. umsongensis (6th week). Mainly P. veronii–related strains were obtained from the oxygen-limited enrichments. Pseudomonas spp. were followed by Acidovorax (4 isolates, Gammaproteobacteria), Rhodococcus (3, Actinobacteria), Achromobacter (2, Gammaproteobacteria), and Massilia spp. (1 isolate, Gammaproteobacteria). Acidovorax- and Rhodococcus-related isolates were obtained only during the first isolation procedure and only from the aerobic enrichment. Achromobacter-affiliated isolates were obtained only from the oxygen-limited enrichment.
Table 1.
Taxonomic affiliation and naphthalene biodegradation potential of biofilm isolates obtained during the study
| Strain designation* | 16S rDNA accession numbers | Isolation source/type of enrichment | Nearest cultured neighbor upon 16S rRNA | Phylogenetic affiliation | bp | 16S rRNA sequence homology (%) | Naphthalene biodegradation as assessed by GC-MS (1 mg l−1) |
Presence of NDO-related 2Fe-2S genes |
|---|---|---|---|---|---|---|---|---|
| R1.1_4 | MN197554 | Aerobic 1st week | Pseudomonas silesiensis A3(T) | Gammaproteobacteria | 1422 | 99.7 | + | + |
| R1.1_5 | MN197555 | Aerobic 1st week | Acidovorax delafieldii DSM 64(T) | ¥Gammaproteobacteria | 1413 | 99.7 | + | + |
| R1.1_7 | MN197556 | Aerobic 1st week | Pseudomonas arsenicoxydans CECT 7543(T) | Gammaproteobacteria | 1440 | 99.2 | + | + |
| RN1.1_1 | MN197557 | Aerobic 1st week | Pseudomonas silesiensis A3(T) | Gammaproteobacteria | 1442 | 99.6 | − | − |
| RN1.1_3 uj | MN197558 | Aerobic 1st week | Pseudomonas silesiensis A(T) | Gammaproteobacteria | 1431 | 99.9 | + | + |
| RN1.1_4F | MN197559 | Aerobic 1st week | Rhodococcus jostii DSM 44719(T) | Actinobacteria | 1416 | 99.9 | + | − |
| RN1.1_5 | MN197560 | Aerobic 1st week | Pseudomonas silesiensis A(T) | Gammaproteobacteria | 1475 | 99.3 | + | + |
| RN1.1_7 | MN197561 | Aerobic 1st week | Massilia aurea AP13(T) | ¥Gammaproteobacteria | 1358 | 99.0 | − | − |
| BGN1.1_2 | MN197562 | Aerobic 1st week | Acidovorax facilis CCUG 2113(T) | ¥Gammaproteobacteria | 1355 | 99.6 | − | − |
| BGN1.1_3 | MN197563 | Aerobic 1st week | Acidovorax delafieldii DSM 64(T) | ¥Gammaproteobacteria | 1364 | 99.5 | + | − |
| BGN1.1_4 | MN197564 | Aerobic 1st week | Acidovorax delafieldii DSM 64(T) | ¥Gammaproteobacteria | 1432 | 99.9 | + | − |
| BGN1.1_5 | MN197565 | Aerobic 1st week | Rhodococcus jostii DSM 44719(T) | Actinobacteria | 1407 | 99.6 | + | − |
| BGN1.1_6 | MN197566 | Aerobic 1st week | Pseudomonas mandelii NBRC 103147(T) | Gammaproteobacteria | 1355 | 99.7 | + | + |
| BGN1.1_8 | MN197567 | Aerobic 1st week | Rhodococcus jostii DSM 44719(T) | Actinobacteria | 1393 | 99.8 | + | − |
| R3.6_1 | MN197568 | Aerobic 6e week | Pseudomonas umsongensis DSM 16611(T) | Gammaproteobacteria | 1430 | 99.7 | − | − |
| R3.6_2 | MN197569 | Aerobic 6th week | Pseudomonas umsongensis DSM 16611(T) | Gammaproteobacteria | 1438 | 99.5 | + | + |
| R3.6_3 | MN197570 | Aerobic 6th week | Pseudomonas laurylsulfatiphila AP3_16(T) | Gammaproteobacteria | 1376 | 99.4 | + | + |
| R3.6_4 | MN197571 | Aerobic 6th week | Pseudomonas laurentiana GSL-010(T) | Gammaproteobacteria | 1439 | 99.6 | + | + |
| RN3.6_1 | MN197572 | Aerobic 6th week | Pseudomonas umsongensis DSM 16611(T) | Gammaproteobacteria | 1446 | 99.0 | − | − |
| RN3.6_3 | MN197573 | Aerobic 6th week | Pseudomonas laurentiana GSL-010(T) | Gammaproteobacteria | 1436 | 99.2 | + | + |
| RN3.6_4 | MN197574 | Aerobic 6th week | Pseudomonas silesiensis A3(T) | Gammaproteobacteria | 1425 | 99.8 | − | − |
| BGN3.6_1 | MN197575 | Aerobic 6th week | Pseudomonas umsongensis DSM 16611(T) | Gammaproteobacteria | 1425 | 99.9 | − | − |
| BGN3.6_2 | MN197576 | Aerobic 6th week | Pseudomonas reidholzensis CCOS 865(T) | Gammaproteobacteria | 1437 | 99.5 | − | − |
| BGN3.6_3 | MN197577 | Aerobic 6th week | Pseudomonas frederiksbergensis JAJ28(T) | Gammaproteobacteria | 1434 | 99.5 | + | + |
| BGN3.6_4 | MN197578 | Aerobic 6th week | Pseudomonas umsongensis DSM 16611(T) | Gammaproteobacteria | 1438 | 99.9 | + | + |
| BGN3.6_5 | MN197579 | Aerobic 6th week | Pseudomonas laurentiana GSL-010(T) | Gammaproteobacteria | 1352 | 99.3 | + | + |
| BGN3.6_6 | MN197580 | Aerobic 6th week | Pseudomonas frederiksbergensis JAJ28(T) | Gammaproteobacteria | 1441 | 99.5 | + | + |
| HR1.1_1 | MN197581 | Oxygen-limited 2nd week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1433 | 99.8 | + | + |
| HR1.1_2 | MN197582 | Oxygen-limited 2nd week | Pseudomonas laurentiana GSL-010(T) | Gammaproteobacteria | 1432 | 99.6 | + | + |
| HR1.1_5 | MN197583 | Oxygen-limited 2nd week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1384 | 99.8 | + | + |
| HBGN1.1_3 | MN197584 | Oxygen-limited 2nd week | Pseudomonas laurentiana GSL-010(T) | Gammaproteobacteria | 1429 | 99.6 | + | + |
| HBGN1.1_4 | MN197585 | Oxygen-limited 2nd week | Achromobacter anxifer LMG 26857 | ¥Gammaproteobacteria | 1420 | 99.9 | − | − |
| HR3.3_1 | MN197586 | Oxygen-limited 6th week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1392 | 99.6 | + | + |
| HR3.3_3 | MN197587 | Oxygen-limited 6th week | Pseudomonas marginalis ATCC 10844(T) | Gammaproteobacteria | 1360 | 99.3 | + | + |
| HR3.3_4 | MN197588 | Oxygen-limited 6th week | Pseudomonas marginalis ATCC 10844(T) | Gammaproteobacteria | 1447 | 98.7 | + | + |
| HR3.3_5 | MN197589 | Oxygen-limited 6th week | Achromobacter spanius LMG 5911(T) | ¥Gammaproteobacteria | 1430 | 99.7 | − | − |
| HRN3.3_1 | MN197590 | Oxygen-limited 6th week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1441 | 99.6 | + | + |
| HRN3.3_3 | MN197591 | Oxygen-limited 6th week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1391 | 99.4 | + | + |
| HRN3.3_4 | MN197592 | Oxygen-limited 6th week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1440 | 99.8 | + | + |
| HBGN3.3_1 | MN197593 | Oxygen-limited 6th week | Pseudomonas veronii DSM11331(T) | Gammaproteobacteria | 1438 | 99.6 | + | + |
*R—direct plating on R2A agar; RN—direct plating on R2A agar, onto which 100 μl of naphthalene suspension had been previously plated (10 mg/mL); BGN—direct plating on BBH medium solidified with gellan gum, onto which 100 μl of naphthalene suspension had been previously plated (10 mg/mL); HR, HRN, HBGN—isolates originating from the oxygen-limited enrichments; “+”—designates either naphthalene degradation as assessed by GC-MS or the possession of naphthalene-dioxygenase reductase component involved in naphthalene biodegradation as detected through CODEHOP PCR
¥Formerly classified as Betaproteobacteria, which now can be found within Gammaproteobacteria in SILVA as order Betaproteobacteriales, Parks et al. 2018
It was found that ~ 90% of the isolated strains belonged to the group of Gram-negative bacteria. Only three isolates, Rhodococcus spp., belonged to the Gram-positive group (7.5%) (Table 1).
Bacterial community dynamics assessed by cultivation-independent approach
Temporal dynamics during the enrichment as revealed by T-RFLP
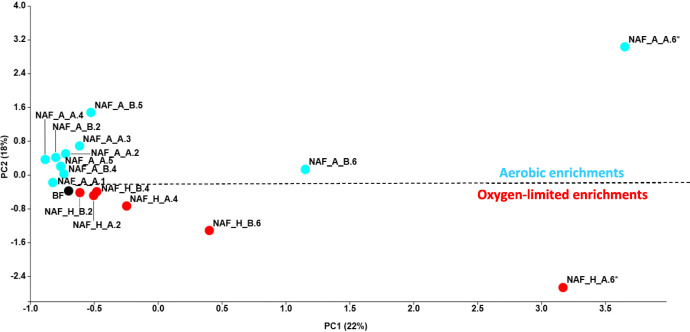
The PCA based on 16S rRNA gene T-RFLP patterns showed a clear separation of aerobic and oxygen-limited naphthalene-degrading biofilm bacterial communities throughout the enrichment period. The bacterial communities were separated along axis 1 according to time (Fig. 1), while axis 2 separated the bacterial communities according to the oxygenic conditions. The highest differences were recorded between samples of the last week (NAF_A_B.6 vs. NAF_H_B.6 or NAF_A_A.6 vs. NAF_H_A.6). In order to characterize the effect of oxygenic conditions on the bacterial community, the composition of bacterial communities—assessed by Illumina 16S rRNA gene amplicon sequencing—obtained under the aerobic enrichment at the end of the enrichment period (NAF_A_A.6) was compared to that of oxygen-limited conditions obtained at the same time (NAF_H_A.6). Additionally, the bacterial community composition of both NAF_A_A.6 and NAF_H_A.6 was compared with that of the initial biofilm community (BF).
Fig. 1.
PCA clustering of the aerobic and oxygen-limited naphthalene-amended biofilm enrichments on the basis of 16S rRNA gene–based T-RFLP electropherograms. (BF, initial biofilm (black spot); NAF_A, naphthalene-amended aerobic enrichments (blue spot); NAF_H, naphthalene-amended oxygen-limited enrichments (red spot); A/B replicates; numbers 1–6, number of weeks of enrichment; *, enrichment cultures selected for Illumina 16S rRNA amplicon sequencing together with the initial sample BF. Please note that in the case of aerobic enrichments of the first and third week, we were able to isolate the community DNA only from one replicate (NAF_A_A.1 and NAF_A_A.3, respectively)
Bacterial diversity according to the Illumina 16S rRNA gene amplicon sequencing
The Illumina 16S rRNA gene amplicon sequencing provided 63 828 reads for the initial biofilm sample (BF), 16 683 for the aerobic enrichment (NAF_A_A.6), and 59 197 for the oxygen-limited enrichment (NAF_H_A.6) samples. The rarefaction curves of the three samples indicated that the data contained enough sequence depth to ascertain the full bacterial diversity. High sequencing coverage was reached in all the three samples (Supplementary Fig. S1).
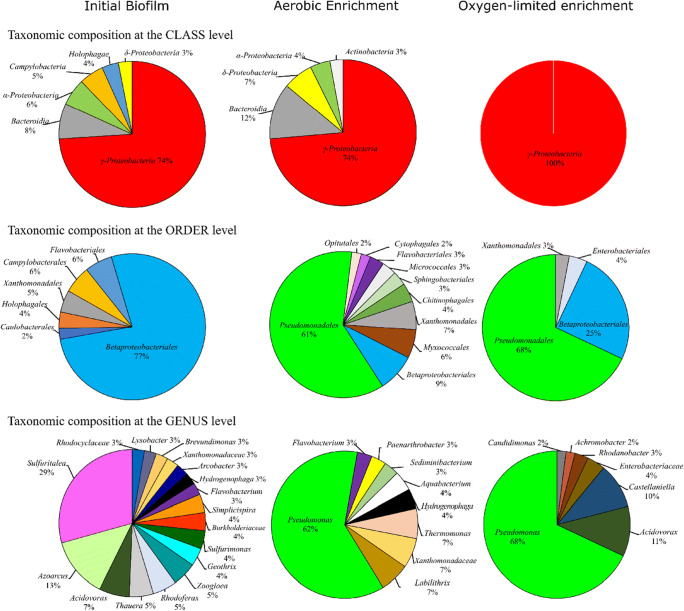
At the class level, all investigated samples were dominated by Gammaproteobacteria (including the former Betaproteobacteria class, which is now included within the Gammaproteobacteria in SILVA as Betaproteobacteriales, Parks et al. 2018) followed by Bacteroidia. At the order level, Betaproteobacteriales were the most dominant in the initial biofilm community, while in the aerobic and oxygen-limited enrichments Pseudomonadales were the most abundant. Throughout the enrichment period, the relative abundance of Betaproteobacteriales decreased remarkably to 9% (aerobic) and 25% (oxygen-limited). At the genus level, the initial biofilm was inhabited by the following well-known, strictly aerobic or facultative anaerobic chemolithotrophic or chemoorganotrophic bacteria: Sulfuritalea, Azoarcus, Acidovorax, Thauera, Rhodoferax, Zoogloea, Geothrix, etc. The genus Pseudomonas showed the highest relative abundance in the enrichment cultures of the 6th week; however, Pseudomonas-related sequences were found to be very low in the initial biofilm (0.3%). In the aerobic enrichment, the next most abundant sequences belonged to unclassified bacteria affiliating with Xanthomonadaceae and Labilithrix. In the oxygen-limited enrichment, Acidovorax and Castellaniella were the next most dominant genera. Interestingly, while at species level the aerobic enrichment of the last week was dominated by Pseudomonas laurentiana (57% relative abundance), Pseudomonas veronii/extremaustralis lineage affiliating bacteria dominated the oxygen-limited setup (64%) (Supplementary Table S1, Fig. 2).
Fig. 2.
Bacterial diversity of the initial biofilm sample (BF), as well as of aerobic (sample NAF_A_A.6) and oxygen-limited enrichments at the last week (NAF_H_A.6) assessed by Illumina 16S rRNA gene amplicon sequencing. Community members with a relative abundance ≥ 2% are shown
Bacterial community shifts in response to different enrichment conditions
A clear shift in the biofilm bacterial community composition was observed during the course of the enrichment. For instance, while at the class level Holophagae and Campylobacteria were abundant in the starting community, their relative abundance decreased to non-detectable during enrichment. Deltaproteobacteria, Alphaproteobacteria, and Bacteroidia were in notable amounts only in the initial biofilm and in the aerobic enrichment. Gammaproteobacteria were the most dominant in the oxygen-limited setup. At the order level, the most striking shift was observed in the case of Pseudomonadales: very low abundance in the initial sample and higher than 60% relative abundance after the enrichments. In contrast, while Betaproteobacteriales were dominant in the initial biofilm, their relative abundance remarkably decreased during the enrichment, reaching the lowest value in the aerobic setting. At the genus level, bacteria belonging to the genera Arcobacter, Simplicispira, Sulfurimonas, Geothrix, Zoogloea, Rhodoferax, Thauera, Azoarcus, and Sulfuritalea showed the highest relative abundance only in the starting biofilm. During the selective enrichments, their relative abundance decreased close to or under the detection limit in both enrichments. Hydrogenophaga and Flavobacterium were present only in the initial biofilm and in the aerobic enrichment. Representatives of the genera Thermomonas, Labilithrix, Paenarthrobacter, Sediminibacterium, and Aquabacterium were in considerable proportions only in the aerobic enrichment. Besides Pseudomonas spp., in the oxygen-limited enrichment, the most dominant bacteria belonged to the genera Acidovorax, Castellaniella, Rhodanobacter, and Achromobacter.
Overall, the culture-independent data was consistent with the results of the cultivation-dependent approach showing that Gram-negative bacteria, mainly belonging to the genus Pseudomonas, dominated the enrichments.
PCR detection and diversity of genes involved in naphthalene biodegradation
CODEHOP primer pair design for the detection of NDO
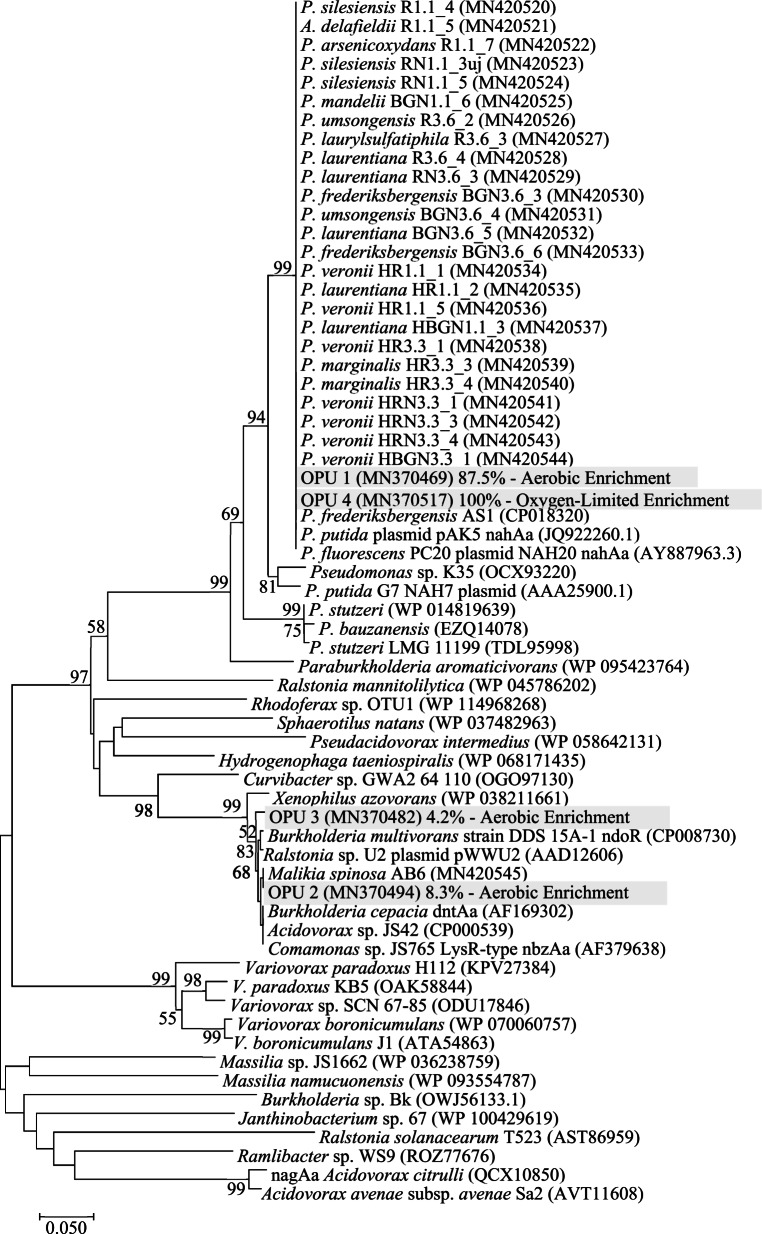
By both cultivation-dependent and cultivation-independent methods, it has been found that the naphthalene (potentially PAH)-degrading community members of the investigated biofilm predominantly belonged to the group of Gram-negative bacteria. Therefore, the CODEHOP primers have been designed on amino acid sequences of naphthalene 1,2-dioxygenase-related 2Fe-2S reductase proteins found in Gram-negative bacteria (Fig. 3).
Fig. 3.
Alignment of naphthalene 1,2-dioxygenase-related 2Fe-2S reductase component protein sequences involved in the biodegradation of PAH compounds. Sequences have been retrieved from the GenBank (NCBI). GenBank accession numbers are given in parentheses. Amino acids marked with an asterisk have consensus through the investigated sequences. Variations in the target sequence are shown with gray shading and in bold letters. Sequence sections shown in green and blue represent the 3′ degenerate core of the forward and reverse primers, respectively. Red and orange colors represent the 5′ consensus clamps
By aligning the protein sequences, a highly conserved N-terminal motif of 4 amino acids was identified (“CGTC”). A second C-terminal conserved motif, 246 amino acids downstream of the “CGTC,” was also identified (“WRAY”). To design the consensus clamps for each strand, “sense” and “anti-sense,” the most common 6 amino acids were determined upstream and downstream of the target motifs, respectively. The most frequently used codon for each of these amino acids was determined. The obtained forward (CGTC-F 32x) and reverse (WRAY-R 64x) CODEHOP primers were 5′-ATTCTTGTATGTCTGGCCGTtgyggnacntg-3′ and 5′-ATAGCAGGTGCGCCACACAGrtangcnckcca-3′, respectively. Lower case characters denote the 3′ degenerate core of the primers.
The diversity of NDO-related 2Fe-2S reductase component genes
First, by using the newly designed primers, a PCR reaction was conducted on the community DNA of the initial biofilm and the enrichment cultures. PCR amplicons of the desired size were detected only in the case of enrichments. In the case of the initial biofilm sample, no amplicons were obtained during the PCR, even though the template DNA concentration was increased up to 150 ng. However, by using high DNA concentration, the PCR amplification of 16S rRNA genes was successful.
From the aerobic and oxygen-limited enrichments, 48 and 44 clones were sequenced, respectively. The obtained clone sequences were grouped into four operational protein units (OPUs) using at least 98% protein sequence similarity. From the aerobic enrichment, OPU 1, OPU 2, and OPU 3 accounted for 87.5%, 8.3%, and 4.2% of total clone sequences, respectively. OPU 4 accounted for 100% of the total clone sequences originating from the oxygen-limited enrichment. OPU 1 and OPU 4 showed the closest amino acid sequence similarity to Pseudomonas species encoded NDO-related 2Fe-2S reductase component genes (e.g., P. fluorescens PC20, P. putida, and P. frederiksbergensis AS1; amino acid sequence similarities ranged from 98.7 to 100%). OPU 2 showed the closest similarity to the putatively NDO-related 2Fe-2S reductase component gene of Malikia spinosa strain AB6 (amino acid sequence homologies ranged from 99.2 to 100%). OPU 3 showed the closest amino acid sequence homologies either to oxidoreductase component of 2,4-DNT dioxygenase of Burkholderia cepacia (99.2% amino acid sequence homology) or to ferredoxin reductase component of Ralstonia sp. U2 (99.2%).
Amino acid sequence homologies between OPU 1 and OPU 2 ranged from 65.8 to 67.4%. In the case of OPU 1 and OPU 3, homologies ranged from 66.9 to 67.5%. Irrespective of the origin, the vast majority of the NDO-related 2Fe-2S reductase component clone sequences showed the closest homology with nahAa proteins of Pseudomonas spp. (aerobic enrichment 87.5% of total clone sequences, oxygen-limited enrichment 100%).
The majority of the isolated strains (24 Pseudomonas sp. isolates and 1 Acidovorax sp. isolate, 60%) harbored NDO-related 2Fe-2S reductase component genes involved in naphthalene biodegradation (Table 1, Fig. 4, Supplementary Fig. S2). Amino acid sequences of the amplified gene regions showed 100% amino acid sequence homology with Pseudomonas species encoded NDO-related 2Fe-2S reductase proteins (Fig. 4). By using the designed CODEHOP primers, NDO-related genes were not detected in the case of Rhodococcus, Massilia, and Achromobacter strains nor in the majority of the Acidovorax-related isolates. In the case of M. spinosa AB6, the PCR positive control, a 750-bp amplicon was detected, (Supplementary Fig. S2, Fig. 4). Non-specific PCR products were not detected in the case of the community nor in the case of genomic DNA samples.
Fig. 4.
Phylogenetic analysis of NDO-related 2Fe-2S reductase protein sequences using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated OPUs clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 66 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 222 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016)
Naphthalene biodegradation potential of isolated strains
During the microcosm experiments, 75% of the isolates (30 isolates) were able to degrade naphthalene (1 mg l−1) (Table 1). Isolates showing positive biodegradation capacities were able to quickly degrade the tested PAH compound within 20 h (the incubation time set for the microcosm experiments). Among the 30 Pseudomonas-related isolates, 24 were able to degrade naphthalene. All P. veronii, P. marginalis, and P. laurentiana strains showed naphthalene biodegradation ability. This was not always true for P. silesiensis– and P. umsongensis–affiliated isolates. All Rhodococcus spp.– and A. delafieldii–related isolates degraded naphthalene. M. aurea–, A. facilis–, P. reidholzensis–, and Achromobacter-related isolates were not able to degrade naphthalene.
It was observed that all isolates—except R. jostii RN1.1_4F, BGN1.1_5, and BGN1.1_8, and A. delafieldii BGN1.1_3 and BGN1.1_4—that degraded naphthalene also possessed naphthalene dioxygenase. It was also found that while some strains belonging to a certain bacterial species exhibited naphthalene biodegradation ability, others, belonging to the same species, did not (P. silesiensis R1.1_4, RN1.1_3_uj and RN1.1_5 versus P. silesiensis RN1.1_1 and RN3.6_4; P. umsongensis R3.6_2 and BGN3.6_4 versus P. umsongensis R3.6_1, RN3.6_1 and BGN3.6_1). Surprisingly, not only isolates possessing the NDO gene were able to degrade naphthalene, but also some of the isolates that did not carry this gene (at least as assessed by the newly developed primers) were able to degrade naphthalene as well.
In the case of bacterial strains, which genomic DNA was used as negative control during the PCR amplification of the targeted functional gene, no naphthalene biodegradation was observed (Zoogloea oleivorans BUC-1T, Rhodococcus pyridinivorans AK37, and Cupriavidus basilensis OR16). However, after 60 h of incubation, the positive control isolate M. spinosa AB6 was able to degrade naphthalene, although at a much slower rate.
Biofilm-producing potential of isolates
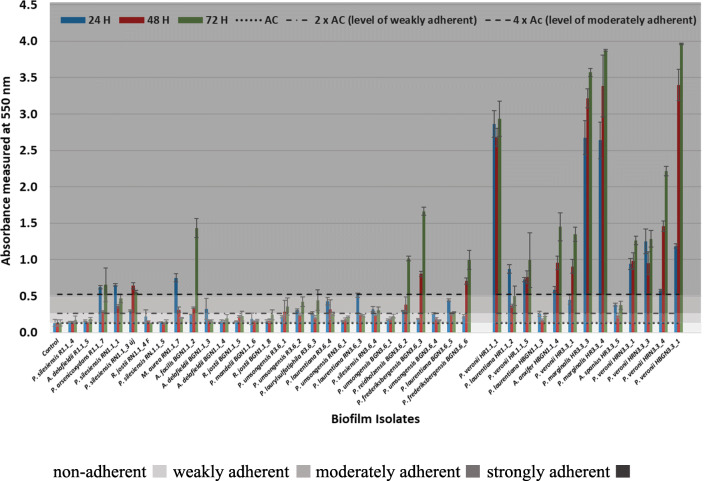
The majority of isolates was moderately or strongly adherent to the polystyrene wall of the microplate (moderately adherent 8 isolates, 20%; strongly adherent 19 isolates, 47.5%). Isolates HR1.1_1, HR3.3_3, HR3.3_4, and HBGN3.3_1 (10% of total isolates) can be viewed as extremely adherent, because after 72 h of incubation their adherence 5–7 times exceeded the level of moderately adherent. These extreme biofilm-producing organisms showed the closest similarity to species P. veronii and P. marginalis. Almost 10% of isolates proved to be non-adherent and 22.5% weakly adherent. A. delafieldii– and Rhodococcus-related isolates did not form biofilms on the tested surface (Fig. 5, Supplementary Table S2).
Fig. 5.
Biofilm-forming potential of isolates based on crystal-violet absorbance assay by using 96-well, cell culture chimney well polystyrene microplates
Discussions
PAHs are extremely toxic compounds (ATSDR 1995). Microorganisms play a crucial role in determining their fate in the environment (Duran and Cravo-Laureau 2016). From a bioremediation perspective, it is important to know which bacteria play a role in the biodegradation of these compounds under different environmental conditions.
In this study, we aimed at enrichment, identification, and isolation of naphthalene (presumably PAH)-degrading bacteria from a bacterial biofilm developed in a hydrocarbon-contaminated subsurface, freshwater ecosystem. Full aerobic and oxygen-limited selective enrichment cultures were initiated in mineral salt solution amended with high concentrations of naphthalene as sole source of energy and carbon. Oxygen-limited enrichment cultures were set up in order to mimic conditions close to in situ, namely low-oxygen availability in hydrocarbon-contaminated groundwater.
During the early stages of the study, it became evident that the investigated biofilm harbors naphthalene (presumably PAH)-degrading community members. After 1 or 2 weeks of incubation, the naphthalene crystals disappeared completely from the hermetically closed bottles. The progressive consumption of naphthalene crystals in enrichment cultures was not unique. Disappearance of naphthalene crystals in a microaerophilic, naphthalene-amended enrichment culture was also observed by Martirani-Von Abercron et al. (2017). Surprisingly, naphthalene 1,2-dioxygenase reductase (NDO) genes for Gram-negative bacteria were not detected by PCR using the newly designed CODEHOP primers in the starting biofilm community, but only after the first or second week of enrichment incubations. Such observation suggested that the NDO gene copy number initially was low, under the PCR detection limit. However, the number of degrading organisms and the naphthalene biodegradation ability increased throughout the enrichment period. These observations suggested that, although, until the biofilm sampling, the majority of the hydrocarbon pollutants in the groundwater had been removed, the hydrocarbon, possible PAH degradation capacity of the biofilm community, remained. The swift propagation of the hidden PAH degradation capacity of the biofilm community was triggered by the presence of naphthalene.
The CODEHOP approach is a powerful tool for the detection and identification of novel and/or distantly related genes. It has been used to identify and characterize new gene orthologs and paralogs in different plant, animal, bacterial, and virus species (Rose et al. 2003; Rose 2005; Staheli et al. 2011).
The vast majority of the NDO-related 2Fe-2S reductase component gene clone sequences, originating from either the aerobic or oxygen-limited enrichments, affiliated with Pseudomonas spp. encoded genes (Fig. 4). This observation indicated a possible horizontal gene transfer (HGT) event allowing the propagation of the naphthalene degradation ability within the bacterial community. The supposed HGT was consistent with the fact that bacterial isolates not related to Pseudomonas, such as Acidovorax species, harbored the same NDO gene. The random occurrence of the NDO gene and also of naphthalene biodegradation ability in the case of different strains, affiliating with the same bacterial species (e.g., A. delafieldii, P. silesiensis, and P. umsongensis strains), further indicated a horizontal gene transfer event within the community. Due to HGT, some strains acquired the NDO gene and the naphthalene degradation ability while others, belonging to the same species, did not. These observations were in accordance with previous reports showing that PAH-degrading genes are often found on mobile genetic elements (e.g., plasmid NAH7) allowing their dispersion in the bacterial community through HGT (Obayori and Salam 2010; Ghosal et al. 2016). For example, it has been demonstrated that Pseudomonas spp. possess conjugative plasmids carrying catabolic operons located in a transposon structure allowing their spread within microbial communities (Herrick et al. 1997; Leahy and Colwell 1990). Moreover, such HGT has been found to be favored in biofilm organizations (Molin and Tolker-Nielsen 2003); the close proximity of bacterial cells facilitates cell-to-cell interactions and thus DNA exchange (Kostakioti et al. 2013).
Interestingly, the initial biofilm investigated in this study was phylogenetically similar to the biofilm from the same site described in a previous sampling campaign (March 2016, Benedek et al. 2018) suggesting that the biofilm was stable over the time. Both biofilm samples, originating from the two different sampling campaigns, were dominated by Beta- (now Gamma-, Parks et al. 2018), Alpha-, and Deltaproteobacteria (Fig. 2, Supplementary Table S1). At the genus level, Sulfuritalea, Azoarcus, Acidovorax, Thauera, Zoogloea, and Flavobacterium were amongst the most abundant genera in both cases. However, the selective enrichment, using either simple aromatic (BTEX, March 2016) or polycyclic aromatic hydrocarbons (naphthalene, January 2018) as the sole carbon source, resulted in different bacterial communities. For both studies, a temporal shift during the enrichment was observed. Both BTEX and naphthalene behaved as strong selectors, which exerted a selective pressure on the bacterial community. Although in the presence of BTEX Malikia (aerobic) and Acidovorax (oxygen-limited enrichment) species became dominant (first biofilm sampling, Benedek et al. 2018), in naphthalene-amended enrichments, Pseudomonas species dominated both enrichment types (second sampling, this study).
The remarkable presence of Pseudomonas species in the naphthalene enrichments is not in accordance with findings of Martirani-Von Abercron et al. (2017); they found the dominance of Variovorax spp. (54%) and Starkeya spp. (43%) in microaerophilic naphthalene enrichments. Surprisingly, the presence of Pseudomonas spp. in their enrichment cultures was marginal. Moreover, Pseudomonas isolates obtained during their study were barely able of naphthalene biodegradation. In contrast, Ma et al. (2006) observed the dominance of Pseudomonas spp. during the enrichment and isolation of naphthalene- and phenanthrene-degrading bacteria from Antarctic soils. In a petroleum-contaminated soil, where the main PAH pollutant was naphthalene, Alquati et al. (2005) also revealed the presence of Pseudomonas spp. Moreover, in one of our previous studies, in the case of a highly PAH-contaminated soil, the dominance of Pseudomonas spp. was observed again (~ 40% relative abundance, Benedek et al. 2013).
The high incidence of Pseudomonas species in naphthalene enrichments, as well as their PAH biodegradation ability, has been widely reported (Amini et al. 2017; Nwinyi et al. 2016; Thomas et al. 2016; Patowary et al. 2015; Wald et al. 2015; Ma et al. 2012; Obayori et al. 2008; Ma et al. 2006; Leahy and Colwell 1990; Foght and Westlake 1988). However, this is the first study reporting the naphthalene biodegradation ability of P. silesiensis, P. arsenicoxydans, P. umsongensis, P. laurylsulfatiphila, P. mandelii, and P. laurentiana; so far, no other PAH compound degradation ability of isolates affiliated with these species has been reported either. In addition, to the best of our knowledge, no information can be found in the literature regarding petroleum hydrocarbon degradation ability of P. silesiensis–, P. laurylsulfatiphila–, and P. laurentiana–related pure isolates. On the other hand, petroleum hydrocarbon degradation ability of P. arsenicoxydans and P. umsongensis pure isolates is already known (Pham et al. 2014).
According to the whole genome sequences, type strains of P. silesiensis A3T, P. umsongensis DSM16611T and P. laurylsulfatiphila AP3_16T, as well as P. arsenicoxydans strains ACM1 and E3, do not contain in their genome naphthalene 1,2-dioxygenase-related genes (Kaminski et al. 2018; Furmanczyk et al. 2018; Kwon et al. 2003; Altshuler et al. 2019). This observation further indicated that isolates obtained in this study, affiliating with the abovementioned bacterial species, acquired the naphthalene biodegradation capability through HGT.
The obtained results indicated that the aerobic enrichment was dominated by P. laurentiana, while the oxygen-limited enrichment was dominated by bacteria affiliating with the P. extremaustralis/P. veronii lineage. The type strain GSL-010T of P. laurentiana was first isolated by Wright et al. (2019) as a Mn(III)-oxidizing bacteria from the St. Lawrence Estuary. As stated above, no information can be found regarding petroleum hydrocarbon degradation ability of P. laurentiana pure cultures. In contrast, a series of studies discuss the hydrocarbon degradation ability (including both aliphatic- and aromatic) of P. extremaustralis/P. veronii lineage affiliating bacteria (Tribelli et al. 2012, 2018; Imperato et al. 2019; Morales et al. 2016; Wald et al. 2015). P. extremaustralis 14-3T was isolated, for the first time, from an Antarctic environment and was able to tolerate and degrade hydrocarbons. It can be used in extreme (cold) environments for hydrocarbon bioremediation. According to its whole genome sequence (VFET00000000.1), it harbors alkane monooxygenase (alkB), catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O), genes involved in either “orto” or “meta” cleavage of aromatic compounds. However, no genes involved in naphthalene biodegradation have been identified in its genome. Genomic analysis of P. veronii VI4T1 (NZ_MULN00000000.1) indicated the presence of the full naphthalene dioxygenase operon and also genes involved in the degradation of BTEX compounds and alkanes (alkB gene). In addition, another isolate Pseudomonas veronii strain 20a2, obtained from a PAH-contaminated lake sediment, was able to degrade a series of PAHs as indicated in Table 2 (Wald et al. 2015). The fact that P. extremaustralis/P. veronii affiliating bacteria were the most dominant in the oxygen-limited enrichment is not a coincidence. According to Tribelli et al. (2018), P. extremaustralis has a high affinity towards microaerobic hydrocarbon degradation. It was found that P. extremaustralis was able to grow under microaerobic conditions using diesel as sole carbon and energy source. Interestingly, under full aerobic conditions, no hydrocarbon degradation occurred (Tribelli et al. 2018). Moreover, Révész et al. (2019) also found the dominance of P. extremaustralis/P. veronii lineage–related bacteria in diesel fuel/crude oil mixture–amended enrichment cultures. They concluded that P. extremaustralis/P. veronii lineage affiliating bacteria are adapted to microaerobic conditions and may have an important role in alkane degradation in subsurface ecosystems. The ability to use petroleum hydrocarbons under low-oxygen conditions presents an adaptive/evolutionary advantage for this bacteria.
Table 2.
PAH degradation ability of bacterial species which showed naphthalene biodegradation capability in this study
| Species | Isolate | Source of isolation/type of selective enrichment | PAH degradationb | Reference |
|---|---|---|---|---|
| Acidovorax delafieldii | P4-1 | –a | FLU, FLN, PYR, PHE | Samanta et al. 1999 |
| TNA921 | Creosote-contaminated soil/PHE-amended MSM c | PHE | Shuttleworth and Cerniglia 1996 | |
| NA3 | PAH-contaminated soil/PHE-amended MSM | NAP, PHE, CHR, B[a]ANT, B[a]PYR | Singleton et al. 2009 | |
| Rhodococcus jostii | 016 | Chronically hydrocarbon-contaminated soil/- | NAP, and PHE and PYR co-metabolically with glucose | Bourguignon et al. 2014 |
| Pseudomonas veronii | SFI 3 | Lagoon sediment sample/FLN-amended MSM | FLN, PHE; PYR | Ben Said et al. 2007 |
| 20a2 | PAH-contaminated lake sediment sample/NAP-amended MSM |
NAP, ACE, FLU, PHE, ANT, FLN, PYR |
Wald et al. 2015 | |
| Pseudomonas frederiksbergensis | JAJ28 | Tar-polluted soil/PHE-coated minimal agar medium | PHE | Andersen et al. 2000 |
| AS1 | Arsenic-contaminated site/NAP-amended MSM | NAP | Yoon-Suk et al. 2006 | |
| Pseudomonas marginalis | PD-14B | PAH-contaminated soil/PHE-coated agar | ANT, ACY, NAP, CHR | Burd and Ward 1996 |
aNot known
bNAP naphthalene, FLU fluorene, FLN fluoranthene, PYR pyrene, PHE phenanthrene, ANT anthracene, CHR chrysene, B[a]ANT benz[a]anthracene, B[a]PYR benzo[a]pyrene, ACE acenaphthene, ACY acenaphthylene
cMSM mineral salt medium
Based on the abovementioned, it is evident that Pseudomonas spp. are good PAH-degrading organisms. The presence of this bacterial species may indicate an elevated natural attenuation capacity of a PAH-contaminated site. Most probably P. laurentiana is capable of aerobic PAH degradation while P. extremaustralis/P. veronii lineage affiliating bacteria are efficient in oxygen-limited PAH degradation.
The overwhelming dominance of Pseudomonas species in the selective enrichments may be explained with the theory of r/K selection. r-strategists (e.g., all Gammaproteobacteria including Pseudomonas spp.) undergo rapid changes when an environment is perturbed and can respond with large population increases. K-strategists dominate in stable non-perturbed environments (De Lei et al. 1995; Brzeszcz et al. 2016). Being r-strategists, the high adaptability to the changing environment and fast reproduction rate made Pseudomonas spp. the most dominant in the naphthalene-amended selective enrichments; pseudomonads could grow without hindrance. Related to the aforementioned, the r/K selection may have been responsible also for the dominance of Gram-negative bacteria over Gram-positives, as it has been proven by both cultivation-dependent and cultivation-independent phylogenetic studies, although Gram-positive bacteria are known to have great potential for the biotransformation and biodegradation of organic compounds (Larkin et al. 2005) when detected are never dominant (Kaplan and Kitts 2004). This may be explained by the fact that Gram-positive bacteria broadly represent K-strategists or oligotrophic organisms with slower growth rates than Gram-negatives (de Vries and Shade 2013; de Vries and Griffiths 2018).
Besides Pseudomonas spp., isolates affiliating with the genera Acidovorax and Castellaniella also deserved attention. In this study, A. defluvii and C. caenii were found to be in considerable amounts in naphthalene-amended oxygen-limited enrichment (Supplementary Table S1). Therefore, the oxygen-limited PAH biodegradation ability of these two species can also be assumed. The occurrence of Acidovorax in petroleum hydrocarbon–contaminated environments, with elevated concentration of PAHs, is already known just like the PAH bioremediation potential of the genus (Singleton et al. 2018). On the other hand, the implication of Castellaniella spp. in PAH biodegradation is not known; only one article discusses weak anthracene degradation ability of the genus (Ntougias et al. 2015).
Based on literature data, it can be assumed that naphthalene was a good choice for the selection and isolation of bacteria capable of degrading other PAHs too. Studies of Wald et al. (2015) and Nwinyi et al. (2013) revealed that the simplest PAH (naphthalene) induced the selection, and allowed the isolation of bacteria capable of degrading besides naphthalene acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, and chrysene. Moreover, based on Table 2, it can be highly assumed that naphthalene-degrading species obtained during this study, particularly P. veronii, A. delafieldii, and P. marginalis, are most probably able to degrade PAHs other than naphthalene. For instance, Pseudomonas veronii strain 20a2, enriched on naphthalene from a PAH-contaminated lake sediment sample, was capable of degrading PAHs with three or four rings (high-molecular-weight PAHs, Table 2). In addition, A. delafieldii Na3, enriched from a PAH-contaminated soil by using phenanthrene as sole source of carbon and energy, was capable of degrading naphthalene, phenanthrene, chrysene, benz[a]anthracene, and benzo[a]pyrene. Based on the aforementioned, it can be highly assumed that naphthalene-degrading isolates can be representative of bacterial species capable of degrading other PAHs too (Table 2). Nevertheless, the availability of solely naphthalene-degrading organisms is already of a high significance since depending on the quality in the gasoline on average naphthalene contributes 97 ± 1% of the total concentration of the PAHs.
In this study, at least four bacterial strains were isolated with extremely prolific biofilm production ability, isolates HR3.3_3 and HR3.3_4 identified as P. marginalis, and isolates HR1.1_1 and HBGN3.3_1 affiliated with P. veronii. These organisms were also able to rapidly degrade naphthalene (within only 20 h) and possessed the NDO (2Fe-2S reductase) gene. P. marginalis isolates are usually identified as postharvest plant pathogens causing soft rot in fruits and vegetables (Brown 1918; Kudela et al. 2010; El Hassan et al. 2014). Biofilm formation, and petroleum hydrocarbon, as well as PAH biodegradation ability of P. marginalis and P. extremaustralis/P. veronii lineage–related isolates have been reported before (Kim et al. 2018; Daniel et al. 2016; Ghazy et al. 2012; Tribelli and López 2011; Lagacé et al. 2006; Burd and Ward 1996).
The ability of biofilm production is essential for microorganisms. The EPS allows bacteria to attach to particles (soil/sand or other biological or non-biological surfaces) and at the same time protects them against harsh environmental conditions. From a bioremediation perspective, these two traits are extremely important. While the first prevents washout from the contaminated zone of the introduced bacteria with the desired metabolic ability, the second protects the inoculated organisms from both the adverse effects of the endogenous bacterial population and the new environment. In the case of biofilm-based biobarriers, biofilm-producing ability of the used microorganisms is of particular importance (Carreghini et al. 2013). Moreover, according to recent studies, the biofilm production ability may accelerate petroleum hydrocarbon degradation potential of microbes as well. Omarova et al. (2019) proved that microbial biofilms aid in the stabilization of dispersed oil droplets through the formation of biofilm at the oil-water interface. Stabilization of dispersed oil droplets may indirectly accelerate the process of oil biodegradation. According to Dasgupta et al. (2013), biofilm-supported batch cultures of Pseudomonas isolates were able to degrade crude oil more effectively than the planktonic cells. As found by Shimada et al. (2012), biofilm-associated P. stutzeri T102 cells were more efficient in naphthalene degradation than planktonic cultures.
Eventually, it is worth mentioning that the results obtained in this study may be valid under the applied enrichment conditions. It may happen that under circumstances closer to in situ environmental conditions different results could have been obtained. Similarly to the theory of “great plate count anomaly” (Staley and Konopka 1985) under in vitro conditions, one can hardly mimic the entire in situ conditions. Therefore, during the selective enrichment, the propagation of only those bacteria was promoted which proliferation was favored by the applied enrichment conditions. In this study, the presence of naphthalene and different oxygen levels were only two out of many other selectors originally found in contaminated groundwaters.
Based on the results, it can be concluded that, besides the isolation of BTEX-degraders, the Bugyi-biofilm sample also allowed the isolation of efficient naphthalene-degrading microorganisms. In both aerobic and oxygen-limited naphthalene-amended enrichments, Pseudomonas spp. became the most dominant as it has been proven by both cultivation-dependent and cultivation-independent approaches. Pseudomonas spp. may be considered bio-indicator species applicable in the detection of the natural attenuation capacity of PAH- or naphthalene-contaminated sites. We supported our previous findings indicating that oxygen availability is one of the main driving factors in the hydrocarbon-contaminated groundwater environments. Two different microbial communities developed during the enrichments. P. laurentiana can be an important member of a PAH-degrading community under aerated circumstances while P. veronii/P. extremaustralis-, Acidovorax-, and Castellaniella-related bacteria are most probably capable of PAH biodegradation under oxygen limitation.
A molecular biological tool was developed for the detection of the NDO (naphthalene 1,2-dioxygenase-related 2Fe-2S reductase) genes of Gram-negative bacteria in either genomic or community DNA. The newly designed primer pairs and the developed CODEHOP-PCR technique may be used for monitoring the natural attenuation capacity of PAH-contaminated sites. A bacterial strain collection with prolific biofilm-producing and effective naphthalene-degrading organisms was established. Subsequently, the obtained strain collection can potentially be applied in the development of biofilm-based bioremediation systems for the elimination of PAHs (e.g., biofilm-based biobarriers or SBPs).
Electronic supplementary material
(PDF 334 kb)
Acknowledgments
The authors are grateful for the employees of the Agruniver Holding Ltd. for providing help during the biofilm samplings. The authors acknowledge the grammar and spellchecking of Stephen John Howard from SpainBCN Language School.
Author Contribution Statement
TB, AT, and BK conceived and designed the research. TB wrote the paper. FSZ conducted the experiments. MF analyzed the next-generation sequencing data. ISZ provided help in biofilm sampling and site description. RD contributed the new CODEHOP method.
Funding information
Open access funding provided by Szent István University (SZIE). This work was supported by the Higher Education Institutional Excellence Program awarded by the Ministry of Human Capacities within the framework of water-related researches of Szent István University (NKFIH-1159-6/2019). Tibor Benedek was supported by the National Research, Development and Innovation Office—PD 128831. Flóra Szentgyörgyi was supported by the ÚNKP-19-3-I, New National Excellence Program of the Ministry of Innovation and Technology.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Key Points
• Aerobic and oxygen-limited naphthalene-degrading biofilm bacteria have been identified.
• P. laurentiana aerobic and P. veronii/P. extremaustralis oxygen-limited PAH degrader.
• Tool developed for monitoring natural attenuation capacity of PAH-contaminated sites.
• A bacterial strain collection for biobarrier development was established.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alquati C, Papacchini M, Riccardi C, Spicaglia S, Bestetti G. Diversity of naphthalene-degrading bacteria from a petroleum contaminated soil. Ann Microbiol. 2005;55:237–242. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler I, Hamel J, Turney S, Magnuson E, Lévesque R, Gree CW, Whyte LG (2019) Species interactions and distinct microbial communities in high arctic permafrost affected cryosols are associated with the CH4 and CO2 gas fluxes. Environ Microbiol. 10.1111/1462-2920.14715 [DOI] [PubMed]
- Amini I, Tahmouespour A, Abdollahi A. Biodegradation of polycyclic aromatic hydrocarbons by Pseudomonas species. Pollution. 2017;3:9–19. [Google Scholar]
- Andersen SM, Johnsen K, Sørensen J, Nielsen P, Jacobsen CS. Pseudomonas frederiksbergensis sp. nov., isolated from soil at a coal gasification site. Int J Syst Evol Microbiol. 2000;50:1957–1964. doi: 10.1099/00207713-50-6-1957. [DOI] [PubMed] [Google Scholar]
- ATSDR (1995) Agency for Toxic Substance and Disease Registry. Department of Health and Human Services, Public Health Service. Public Health statement-Polycyclic aromatic hydrocarbons (PAHs): https://www.atsdr.cdc.gov/ToxProfiles/tp69-c1-b.pdf
- Ben Said O, Goni-Urriza MS, El Bour M, Dellali M, Aissa P, Duran R. Characterization of aerobic polycyclic aromatic hydrocarbon-degrading bacteria from Bizerte lagoon sediments, Tunisia. J Appl Microbiol. 2007;104:987–997. doi: 10.1111/j.1365-2672.2007.03621.x. [DOI] [PubMed] [Google Scholar]
- Ben Said O, Louati H, Soltani A, Preud’homme H, Cravo-Laureau C, Got P, Pringault O, Aissa P, Duran R. Changes of benthic bacteria and meiofauna assemblages during bio-treatments of anthracene-contaminated sediments from Bizerta lagoon (Tunisia) Environ Sci Pollut Res. 2015;22:15319–15331. doi: 10.1007/s11356-015-4105-7. [DOI] [PubMed] [Google Scholar]
- Benedek T, Vajna B, Táncsics A, Márialigeti K, Sz L, Máthé I. Remarkable impact of PAHs and TPHs on the richness and diversity of bacterial species in surface soils exposed to long-term hydrocarbon pollution. W J Microbiol Biotechnol. 2013;29:1989–2002. doi: 10.1007/s11274-013-1362-9. [DOI] [PubMed] [Google Scholar]
- Benedek T, Táncsics A, Szilágyi N, Tóth I, Farkas M, Szoboszlay S, Krifaton C, Hartman M, Kriszt B. Analysis of biofilm bacterial communities responsible for carbon removal through a reactor cascade treating wastewater. W J Microbiol Biotechnol. 2014;30:977–987. doi: 10.1007/s11274-013-1516-9. [DOI] [PubMed] [Google Scholar]
- Benedek T, Táncsics A, Szabó I, Farkas M, Szoboszlay S, Fábián K, Maróti G, Kriszt B. Polyphasic analysis of an Azoarcus-Leptothrix-dominated bacterial biofilm developed on stainless stell surface in a gasoline-contaminated hypoxic groundwater. Environ Sci Pollut Res. 2016;23:9019–9035. doi: 10.1007/s11356-016-6128-0. [DOI] [PubMed] [Google Scholar]
- Benedek T, Szentgyörgyi F, Szabó I, Kriszt B, Révész F, Radó J, Maróti G, Táncsics A. Aerobic and oxygen-limited enrichment of BTEX-degrading biofilm bacteria: dominance of Malikia versus Acidovorax species. Environ Sci Pollut Res. 2018;25:32178–32195. doi: 10.1007/s11356-018-3096-6. [DOI] [PubMed] [Google Scholar]
- Bordenave S, Fourçans A, Blanchard S, Goñi MS, Caumette P, Duran R. Structure and functional analyses of bacterial communities changes in microbial mats following petroleum exposure. Ophelia. 2004;58:195–203. [Google Scholar]
- Bordenave S, Jézéque R, Fourçans A, Budzinski H, Merlin FX, Fourel T, Goñi-Urriza M, Guyoneaud R, Grimaud R, Caumette P, Duran R. Degradation of the “Erika” oil. Aquat Living Resour. 2004;17:261–267. [Google Scholar]
- Bourguignon N, Isaac P, Alvarez H, Amoroso MJ, Ferrero MA. Enhanced polyaromatic hydrocarbon degradation adapted cultures of actinomycetes strains. J Basic Microbiol. 2014;54:1–7. doi: 10.1002/jobm.201400262. [DOI] [PubMed] [Google Scholar]
- Brown NA. Some bacterial diseases of lettuce. J Agr Res. 1918;13:367–388. [Google Scholar]
- Brzeszcz J, Steliga T, Kapusta P, Turkiewicz A, Pawel K. r-strategist versus K-strategist for the application in bioremediation of hydrocarbon-contaminated soils. Int Biodeter Biodegr. 2016;106:41–52. [Google Scholar]
- Burd G, Ward OP. Involvement of a surface-active high molecular weight factor in degradation of polycyclic aromatic hydrocarbons by Pseudomonas marginalis. Can J Microbiol. 1996;42:791–797. doi: 10.1139/m96-099. [DOI] [PubMed] [Google Scholar]
- Candeli A, Morozzi G, Paolacci A. Analysis using thin layer and gas-liquid chromatography of polycyclic aromatic hydrocarabons in the exhaust products from a European car running on fuels containing a range of concentration of these hydrocarbons. L Atmos Environ. 1975;9:843–849. [Google Scholar]
- Carreghini A, Saponaro D, Sezenna E. Biobarriers for groundwater treatment: a review. Water Sci Technol. 2013;67:453–468. doi: 10.2166/wst.2012.599. [DOI] [PubMed] [Google Scholar]
- Cerniglia CE, Heitkamp MA. Microbial degradation of polycyclic aromatic hydrocarbons (PAHs) in the aquatic environment. In: Varanasi U, editor. Metabolism of Polycyclic Aromatic Hyrocarbons in the Aquatic Environment. Boca Raton: CRC Press; 1989. pp. 41–68. [Google Scholar]
- Cravo-Laureau C, Duran R (2014) Marine coastal sediments microbial hydrocarbon degradation processes: contribution of experimental ecology in the omics’era. Front Microbiol 5. 10.3389/fmicb.2014.00039 [DOI] [PMC free article] [PubMed]
- Cravo-Laureau C, Hernandez-Raquet G, Vitte I, Jézéquel R, Bellet V, Godon JJ, Caumette P, Balaguer P, Duran R. Role of environmental fluctuations and microbial diversity in degradation of hydrocarbons in contaminated sludge. Res Microbiol. 2011;162:888–895. doi: 10.1016/j.resmic.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Daniel MA, Barrionuevo MR, Doyle SR, Vullo DL. Kinetics of Pseudomonas veronii 2E biofilm development under different nutritional conditions for a proper bioreactor design. Biochem Eng J. 2016;105:150–158. [Google Scholar]
- Dasgupta D, Ghosh R, Sengupta TK. Biofilm-mediated enhanced crude oil degradation by newly isolated pseudomonas species. ISRN Biotechnol. 2013;2013:1–13. doi: 10.5402/2013/250749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Wagelmans M. Polycyclic aromatic hydrocarbons in soil – practical options for remediation. Clean - Soil, Air, Water. 2016;44:648–653. [Google Scholar]
- de Lei F, Whipps JM, Fenlon JS, Lynch JM. Impact of field release of genetically modified Pseudomonas fluorescens on indigenous microbial populations of wheat. Appl Environ Microbiol. 1995;61:3443–3453. doi: 10.1128/aem.61.9.3443-3453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, Griffiths . Impacts of climate change of soil microbial communities and their functioning. In: Horwath WR, Kuzyakov Y, editors. Climate change impacts on soil processes and ecosystem properties. Amsterdam: Elsevier; 2018. [Google Scholar]
- de Vries FT, Shade A. Controls on soil microbial community stability under climate change. Front Microbiol. 2013;4:265. doi: 10.3389/fmicb.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran R, Cravo-Laureau C. Role of environmental factors and microorganisms in determining the fate of polycyclic aromatic hydrocarbons in the marine environment. FEMS Microbiol Rev. 2016;40:814–830. doi: 10.1093/femsre/fuw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran R, Bonin P, Jezequel R, Dubosc K, Gassie C, Terrisse F, Abella J, Cagnon C, Militon C, Michotey V, Gilbert F, Cuny P, Cravo-Laureau C. Effect of physical sediments reworking on hydrocarbon degradation and bacterial community structure in marine coastal sediments. Environ Sci Pollut R. 2015;22:15248–15259. doi: 10.1007/s11356-015-4373-2. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;15:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassan A, Sadik S, Kahkahi RE, Benbouazza A, Mazouz H. First report on Pseudomonas marginalis bacterium causing soft rot of onion in Morocco. Atlas J Biol. 2014;3:218–223. doi: 10.5147/ajb.2014.0136. [DOI] [Google Scholar]
- Ensley BD, Gibson DT. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J Bacteriol. 1983;155:505–511. doi: 10.1128/jb.155.2.505-511.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy A, McGenity TJ, Timmis KN, Ball AS. Heterogeneous aerobic benzene-degrading communities in oxygen- depleted groundwaters. FEMS Microbiol Ecol. 2006;58:260–270. doi: 10.1111/j.1574-6941.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Foght JM, Westlake DWS. Degradation of polycyclic aromatic hydrocarbons and aromatic heterocycles by a Pseudomonas species. Can J Microbiol. 1988;34:1135–1141. doi: 10.1139/m88-200. [DOI] [PubMed] [Google Scholar]
- Furmanczyk EM, Lipinski L, Dziembowski A, Sobczak A. Genomic and functional characterization of environmental strains of SDS-degrading spp., providing a source of new sulfatases. Front Microbiol. 2018;9:1795. doi: 10.3389/fmicb.2018.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy EA, Elmokadem MT, Gadallah M, Mahmoud MN, Abo Elsoud MM (2012) Combating biocorrosions induced by biofilm forming bacterial isolate Pseudomonas marginalis. Middle East J Sci Res 12:1017–1025
- Ghosal D, Shreya G, Dutta TK, Ahn Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front Microbiol. 2016;7:1369. doi: 10.3389/fmicb.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe H, Omori T. Genetic of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci Biotechnol Biochem. 2003;67:225–243. doi: 10.1271/bbb.67.225. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9. [Google Scholar]
- Harley RA, Coulter-Burke SC, Yeung TS. Relating liquid fuel and headspace vapor composition for California reformulated gasoline samples containing ethanol. Environ Sci Technol. 2000;34:4088–4094. [Google Scholar]
- Harvey RG. Polycyclic aromatic hydrocarbons: chemistry and carcinogenicity. Cambridge: Cambridge University Press; 1991. [Google Scholar]
- Herrick JB, Stuart-Keil KG, Ghiorse WC, Madsen EL. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperato V, Portillo-Estrada M, McAmmond BM, Douwen Y, Van Hamme JD, Gawronski SW, Vangronsveld J, Thijs S (2019) Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of Bóbrka (Poland). Genes. 10.3390/genes10060443 [DOI] [PMC free article] [PubMed]
- Jia C, Batterman S. A critical review of naphthalene sources and exposures relevant to indoor and outdoor air [published correction appears in Int J Environ Res Public Health. 2011 Aug;8(8):3191] Int J Environ Res Public Health. 2011;7:2903–2939. doi: 10.3390/ijerph7072903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau Y, Meyer C, Jakoncic J, Stojanoff V, Gaillard J. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry. 2006;45:12380–12391. doi: 10.1021/bi0611311. [DOI] [PubMed] [Google Scholar]
- Kaminski MA, Furmanczyk EM, Sobczak A, Dziembowski A, Lipinski L. Pseudomonas silesiensis sp. nov. strain A3T isolated from a biological pesticide sewage treatment plant and analysis of the complete genome sequence. Syst Appl Microbiol. 2018;41:13–22. doi: 10.1016/j.syapm.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Kaplan CW, Kitts CL. Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol. 2004;70:1777–1786. doi: 10.1128/AEM.70.3.1777-1786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na PSC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim H, Beuchat LR, Ryu JH. Inhibition of Escherichia coli O157:H7 on stainless steel using Pseudomonas veronii biofilms. Lett Appl Microbiol. 2018;66:394–399. doi: 10.1111/lam.12866. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res:41 e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed]
- Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic Era. Cold Spring Harb Perspect Med. 2013;3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudela V, Krejzar V, Pánková I. Pseudomonas corrugata and Pseudomonas marginalis associated with the collapse of tomato plant in Rockwool slab hydroponic culture. Plant Protect Sci. 2010;46:1–11. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Kuppusamy S, Thavamani P, Venkateswarlu K, Lee YB, Naidu R, Megharaj M. Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: technological constraints, emerging trends and future directions. Chemosphere. 2016;168:944–968. doi: 10.1016/j.chemosphere.2016.10.115. [DOI] [PubMed] [Google Scholar]
- Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, Lim CK, Go SJ. Pseudomonas koreensis sp. nov, Pseudomonas umsongensis sp. nov, and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int J Syst Evol Microbiol. 2003;53:21–27. doi: 10.1099/ijs.0.02326-0. [DOI] [PubMed] [Google Scholar]
- Lagacé L, Jaques M, Mafu AA, Roy D. Biofilm formation and biocides sensitivity of Pseudomonas marginalis isolated from a maple sap collection system. J Food Prot. 2006;69:2411–2416. doi: 10.4315/0362-028x-69.10.2411. [DOI] [PubMed] [Google Scholar]
- Larkin MJ, Kulakov LA, Allen CC. Biodegradation and Rhodococcus– masters of catabolic versatility. Curr Opin Biotechnol. 2005;16:282–290. doi: 10.1016/j.copbio.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Leahy JG, Colwell RR. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–315. doi: 10.1128/mr.54.3.305-315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphism of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Wang L, Shao Z. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol. 2006;8:455–465. doi: 10.1111/j.1462-2920.2005.00911.x. [DOI] [PubMed] [Google Scholar]
- Ma J, Xu L, Jia L. Degradation of polycyclic aromatic hydrocarbons by Pseudomonas sp. JM2 isolated from active sewage sludge of chemical plant. J Environ Sci. 2012;24:2141–2148. doi: 10.1016/s1001-0742(11)61064-4. [DOI] [PubMed] [Google Scholar]
- Marr LC, Kirchstetter TW, Harley RA, Miguel AH, Hering SV, Hammond SK. Characterization of polycyclic aromatic hydrocarbons in motor vehicle fuels and exhaust emissions. Environ Sci Technol. 1999;33:3091–3099. [Google Scholar]
- Martirani-Von Abercron SM, Marin P, Solsona-Ferraz M, Castaneda-Catana MA, Marques S. Naphthalene biodegradation under oxygen-limiting conditions: community dynamics and the relevance of biofilm-forming capacity. Microbiol Biotechnol. 2017;10:1781–1796. doi: 10.1111/1751-7915.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe O, Kurzbaum E. Small-bioreactor platform technology as a municipal wastewater additive treatment. Water Sci Technol. 2014;69:504–510. doi: 10.2166/wst.2013.709. [DOI] [PubMed] [Google Scholar]
- Militon C, Jézéquel R, Gilbert F, Corsellis Y, Sylvi L, Cravo-Laureau C, Duran R, Cuny P. Dynamics of bacterial assemblages and removal of polycyclic aromatic hydrocarbons in oil-contaminated coastal marine sediments subjected to contrasted oxygen regimes. Environ Sci Pollut R. 2015;22:15260–15272. doi: 10.1007/s11356-015-4510-y. [DOI] [PubMed] [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilization of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Morales M, Sentchilo V, Bertelli C, Komljenovic A, Kryuchkova-Mostacci N, Bourdilloud A, Linke B, Goesmann A, Harshman K, Segers F, Delapierre F, Fiourucci D, Seppey M, van der Meer JR (2016) The genome of the toluene-degrading Pseudomonasveronii strain 1YdBTEX2 and its differential gene expression in contaminated sand. Plos One. 10.1371/journal.pone.0165850 [DOI] [PMC free article] [PubMed]
- National Center for Biotechnology Information. PubChem Database, accessed on Apr. 15, 2020
- Ntougias S, Melidis P, Navrozidou E, Tzegkas F. Diversity and efficiency of anthracene-degrading bacteria isolated from a denitrifying activated sludge system treating municipal wastewater. Int Biodeter Biodegr. 2015;97:151–158. [Google Scholar]
- Nwinyi O, Picardal FW, An TT, Amund OO. Aerobic degradation of naphthalene, fluoranthene, pyrene and chrysene using indigenous strains of bacteria isolated from a former industrial site. CJPAS. 2013;7:2303–2314. [Google Scholar]
- Nwinyi OC, Ajayi OO, Amund OO. Degradation of polynuclear aromatic hydrocarbons by two strains of Pseudomonas. Braz J Microbiol. 2016;47:551–562. doi: 10.1016/j.bjm.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayori OS, Salam LB. Degradation of polycyclic aromatic hydrocarbons: Role of plasmids. Sci Res Essays. 2010;5:4093–4106. [Google Scholar]
- Obayori OS, Ilori MO, Adebusoye S, Oyetibo GO, Amund O. Pyrene-degradation potentials of Pseudomonas species isolated from polluted tropical soils. W J Microbiol Biotechnol. 2008;24:2639–2646. [Google Scholar]
- Omarova M, Swientoniewski T, Tsengam IKM, Blake DA, John V, McCormick A, Bothun GD, Raghavan SR, Bose A. Biofilm formation by hydrocarbon-degrading marine bacteria and its effects on oil dispersion. ACS Sustainable Chem Eng. 2019;7:14490–14499. [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;39:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- Patowary K, Kalita MC, Deka S. Degradation of polycyclic aromatic hydrocarbons (PAHs) employing biosurfactant producing Pseudomonas aeruginosa KS3. Ind J Biotechnol. 2015;14:208–215. [Google Scholar]
- Pham VHT, Kim J, Jeong SW. Enhanced isolation and culture of highly efficient psychrophilic oil-degrading bacteria from oil-contaminated soils in South Korea. J Environ Microbiol. 2014;35:1145–1149. [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Révész F, Figueroa-Gonzalez PA, Probst AJ, Kriszt B, Banerjee S, Szoboszlay S, Maróti G, Táncsics A (2019) Microaerobic conditions caused the overwhelming dominance of Acinetobacter spp. and the marginalization of Rhodococcus spp. in diesel fuel/crude oil mixture-amended enrichment cultures. Arch Microbiol. 10.1007/s00203-019-01749-2 [DOI] [PMC free article] [PubMed]
- Rose TM. CODEHOP-mediated PCR – a powerful technique for the identification and characterization of viral genomes. Virol J. 2005;2:xxx–xxx. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Samanta S, Chakraborti A, Jain R. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl Microbiol Biotechnol. 1999;53:98–107. doi: 10.1007/s002530051621. [DOI] [PubMed] [Google Scholar]
- Schauer JJ, Kleeman MJ, Cass GR, Simoneit BRT. Measurement of emissions from air pollution sources. 5. C-1- C-32 organic compounds from gasoline-powered motor vehicles. Environ Sci Technol. 2002;36:1169–1180. doi: 10.1021/es0108077. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source platform-independent community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Itoh Y, Washio K, Morikawa M. Efficacy of forming biofilms by naphthalene degrading Pseudomonas stutzeri T102 toward bioremediation technology and its molecular mechanisms. Chemosphere. 2012;87:226–233. doi: 10.1016/j.chemosphere.2011.12.078. [DOI] [PubMed] [Google Scholar]
- Shuttleworth KL, Cerniglia CE. Bacterial degradation of low concentrations of phenanthrene and inhibition by naphthalene. Microb Ecol. 1996;31:305–317. [PubMed] [Google Scholar]
- Singleton DR, Ramirez LG, Aitken MD. Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in phenanthrene-degrading Acidovorax strain. Appl Environ Microbiol. 2009;75:2613–2620. doi: 10.1128/AEM.01955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Lee J, Dickey AN, Stroud A, Scholl EH, Wright FA, Aitken MD. Polyphasic characterization of four soil-derived phenanthrene-degrading Acidovorax strains and proposal of Acidovorax carolinensis sp. nov. Syst Appl Microbiol. 2018;41:460–472. doi: 10.1016/j.syapm.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Danilowicz BS, Clear AK, Costello FJ, Wilson B, Meijer WG. T-Align, a web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol Ecol. 2005;54:375–380. doi: 10.1016/j.femsec.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Staheli JP, Boyce R, Kovarik D, Rose TM (2011) CODEHOP PCR and CODEHOP PCR primer design. In: Park D (eds) PCR Protocols. Methods in molecular biology (Methods and Protocols), vol 687, Humana Press [DOI] [PubMed]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Stepanović S, Vuković D, Dakić I, Savić B, Čvabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40:175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- Táncsics A, Szoboszlay S, Szabó I, Farkas M, Kovács B, Kukolya J, Mayer Z, Kriszt B. Quantification of subfamily I.2.C catechol 2,3-dioxygenase mRNA transcripts in groundwater samples of an oxygen-limited BTEX-contaminated site. Environ Sci Technol. 2012;46:232–240. doi: 10.1021/es201842h. [DOI] [PubMed] [Google Scholar]
- Terrisse F, Cravo-Laureau C, Noël C, Cagnon C, Dumbrell AJ, McGenity TJ, Duran R (2017) Variation of oxygenation conditions on a hydrocarbonoclastic microbial community reveals Alcanivorax and Cycloclasticus ecotypes. Front Microbiol, 8, art. no. 1549 [DOI] [PMC free article] [PubMed]
- Thomas F, Lorgeoux C, Faure P, Billet D, Cébron A. Isolation and substrate screening of polycyclic aromatic hydrocarbon degrading bacteria from soil with long history of contamination. Int Biodeter Biodegr. 2016;107:1–9. [Google Scholar]
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- Tribelli PM, López NI. Poly(3-hydroxybutyrate) influences biofilm formation and motility in the novel Antarctic species Pseudomonas extremaustralis under cold conditions. Extremophiles. 2011;15:541–547. doi: 10.1007/s00792-011-0384-1. [DOI] [PubMed] [Google Scholar]
- Tribelli PM, di Martino C, López NI, Raiger Iustman LJ. Biofilm lifestyle enhances diesel bioremediation and biosurfactant production in the Antarctic polyhydroxyalkanoate producer Pseudomonas extremaustralis. Biodegr. 2012;23:645–651. doi: 10.1007/s10532-012-9540-2. [DOI] [PubMed] [Google Scholar]
- Tribelli PM, Rossi L, Ricardi MM, Gomez-Lozano M, Molin S, Raiger Lustman LJ, López NI. Microaerophilic alkane degradation in Pseudomonas extremaustralis: a transcriptomic and physiological approach. J Ing Microbiol Biotechnol. 2018;45:15–23. doi: 10.1007/s10295-017-1987-z. [DOI] [PubMed] [Google Scholar]
- Tu SL, Staheli JP, McClay C, McLeod K, Rose TM, Upton C (2018) Base-By-Base version 3: New comparative tools for large virus genomes. Viruses 10. 10.3390/v10110637 [DOI] [PMC free article] [PubMed]
- Vitte I, Duran R, Jézéquel R, Caumette P, Cravo-Laureau C. Effect of oxic/anoxic switches on bacterial communities and PAH biodegradation in an oil-contaminated sludge. Environ Sci Pollut R. 2011;18:1022–1032. doi: 10.1007/s11356-010-0435-7. [DOI] [PubMed] [Google Scholar]
- Wald J, Hroudova M, Jansa J, Vrchotova B, Macek T, Uhlik O. Pseudomonads rule degradation of polyaromatic hydrocarbons in aerated sediment. Front Microbiol. 2015;6:1268. doi: 10.3389/fmicb.2015.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Hanna JG, Pica DA, Maclean Tebo B. Pseudomonas laurentiana sp. nov., an Mn(III)-oxidizing bacterium isolated from the St. Lawrence Estuary. Pharmacogn Comm. 2019;8:153–157. [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon-Suk K, Kim YJ, Jeon CO, Park W. Characterization of naphthalene-degrading Pseudomonas species isolated from pollutant-contaminated sites. Oxidative stress during their growth on naphthalene. J Microbiol Biotechnol. 2006;16:1819–1825. [Google Scholar]
- Zoccolillo L, Babi D, Felli M. Evaluation of polycyclic aromatic hydrocarbons in gasoline by HPLC and GC-MS. Chromatographia. 2000;52:373–376. [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. Edited in Evolving Genes and Proteins by V. Bryson and H.J. Vogel, pp. 97-166. Academic Press, New York. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 334 kb)