Highlights
-
•
Centralised laboratory PCR testing for COVID-19 is associated with long delays.
-
•
The FebriDx POCT can detect an antiviral host response protein (MxA) in 10 min.
-
•
We evaluated the diagnostic accuracy of FebriDx in 251 patients hospitalised with suspected COVID-19 during the first wave.
-
•
Compared to PCR sensitivity of FebriDx was 93% and specificity was 86%.
-
•
FebriDx could be used as a rapid front door triage tool in hospitals.
Keywords: Point of care testing, Host response, MxA, Accuracy, COVID-19, SARS-CoV-2
Abstract
Introduction
Management of the COVID-19 pandemic is hampered by long delays associated with centralised laboratory PCR testing. In hospitals this leads to poor patient flow and nosocomial transmission and so rapid, accurate diagnostic tests are urgently required. The FebriDx is a point-of-care test that detects an antiviral host response protein in finger prick blood within 10 min, but its accuracy for the identification of COVID-19 is unknown.
Methods
We performed a real-world diagnostic accuracy study of FebriDx in hospitalised patients during the first wave of the pandemic. Measures of diagnostic accuracy were calculated based on FebriDx results compared to the reference standard of SARS-CoV-2 PCR on combined nose and throat swabs. A multivariable predictive model including FebriDx, age, sex, and clinical characteristics was developed and underwent internal validation.
Results
FebriDx was performed on 251 patients and gave a valid result in 248. 118 of 248 (48%) were PCR positive for COVID-19. FebriDx results were available after 10 min compared with 1.7 (1.6 to 2.1) hours with point-of-care PCR testing and 23.4 (17.2 to 31.1) hours with laboratory PCR testing. Sensitivity of FebriDx for the identification of COVID-19 was 93% (110/118; 95% CI 87 to 97%) and specificity was 86% (112/130; 95%CI 79 to 92%). Positive and negative likelihood ratios were 6.73 (95%CI 4.37 to 10.37) and 0.08 (95%CI 0.04 to 0.15) respectively. In the multivariate model age, sex and other clinical features did not contribute significantly to the effect of the FebriDx result in distinguishing patients with and without COVID-19.
Conclusions
During the first wave of the pandemic, FebriDx had high accuracy for the identification of COVID-19 in hospitalised adults and could be deployed as a front door triage tool.
Trial registration
ISRCTN14966673
Introduction
The management of COVID-19 in secondary care is severely hampered by the long turnaround times of centralised laboratory PCR testing, which can take up to several days to generate results. In acute hospitals this leads to poor patient flow though acute areas, as suspected patients are held in assessment cohort areas until their results are available. In addition, lack of single occupancy rooms means that COVID-19 negative patients cohorted in these assessment areas may acquire infection from positive patients before test results are available. Rapid, accurate diagnostic tests are therefore urgently required. Molecular point-of-care testing (POCT) for COVID-19 may mitigate this situation but regulatory requirements for new POCT assays and difficulties in rapidly upscaling manufacture mean that the availability of these tests is currently severely limited. Alternative diagnostic solutions are therefore urgently required.
FebriDx (Lumos diagnostics, Sarasota, Florida, US) is a CE-marked POCT that detects two host response proteins, Myxovirus resistance protein A (MxA) and C reactive protein (CRP), in finger prick blood samples, and is designed to distinguish viral from bacterial respiratory infection.1, 2, 3, 4, 5 MxA is a marker of interferon-induced antiviral host response and in our previous work, the detection of MxA by FebriDx had high sensitivity for the detection of Influenza in hospitalized adults, during influenza season.6 MxA levels are also likely to be significantly elevated in patients with COVID-19 but the diagnostic accuracy of FebriDx in this situation is currently unknown. FebriDx is a low cost, instrument-free, disposable POCT system and if sensitive for the detection of COVID-19, could be rapidly deployed across the NHS and other healthcare systems to help mitigate the long delays in diagnostic testing and provide a rapid triage tool. The aim of this study was therefore to evaluate the real-world diagnostic accuracy of MxA detection by FebriDx for the identification of COVID-19 in hospitalised adults during the first wave of the pandemic, and to develop and internally validate a multivariable model for diagnosis of COVID-19 that includes FebriDx result and other easily obtained clinical measures.
Methods
Study design and participants
This study of FebriDx was nested within the CoV-19POC study, a trial assessing the clinical impact of molecular POCT for COVID-19. Adults presenting with suspected COVID-19 were enroled; full details of the inclusion and exclusion criteria can be found in the protocol, linked below. The study was approved by the South Central - Hampshire A Research Ethics Committee: REC reference 20/SC/0138, on the 16th March 2020. The protocol is available at: https://eprints.soton.ac.uk/439309/1/CoV_19POC_Protocol_v1.1_eprints.pdf
Procedures
Combined nose and throat swabs were obtained from patients by research staff and tested immediately on the QIAstat-Dx platform using the Respiratory SARS-CoV-2 Panel,7 at the point of care. A full list of the pathogens detected by the panel is shown in Table E1 in the online data supplement.8 , 9 In addition, laboratory PCR testing for SARS-CoV-2 on combined nose and throat swabs was performed on all patients, in the on-site Public Health England (PHE) microbiology laboratory, using the PHE RdRp and E gene reference assays.10 , 11 Demographic and clinical data was collected at enrolment and outcome data collected retrospectively from case note and electronic systems, using an electronic case report form.
For this sub-study the first 266 patients enroled were consecutively approached for testing using the FebriDx host response POCT on finger prick blood samples, taken at the same time as the nose as throat swabs for PCR. Detailed instructions for use of FebriDX are provided via this link: https://www.febridx.com/how-to-use#testing but in brief; after puncture of the skin with the integral lancet, 5 microliters of blood are drawn into the blood collection tube via capillary action by placing it against a blood drop. The blood is then transferred to the lateral flow section of the device and reagents released by pressing a button. The test is read after 10 min.
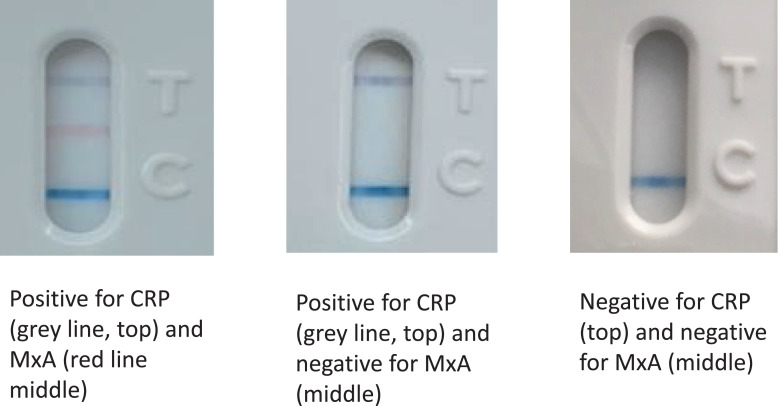
Interpretation of results
The FebriDx system generates results in the form of the presence or absence of three lines, assessed by visual inspection; the CRP line (top, grey), the MxA line (middle, red) and the control line (bottom, blue). Fig. 2 shows examples of FebriDx positive and negative tests. The colour change on the CRP and MxA lines when detected is dependant on the amount of the target protein in the sample and so there is potential variability in interpretation with weaker lines (quoted threshold for detection of CRP line 20 mg/L and MxA line of 40 ng/ml). For this reason each FebriDx result was read independently by two investigators and if there was disagreement on the results displayed this was further adjudicated by a third investigator. The results of the FebriDx were not shared with the clinical teams and the readers of the FebriDx test lines were blinded to the PCR results (FebriDx results were read and recorded before PCR results were available).
Fig. 2.
Examples of positive and negative FebriDx read-outs.
For the reference standard of PCR, the QIAstat-Dx PCR system gives a readout of positive or negative for the detection of targets including SARS-CoV-2.7 The PHE laboratory RdRp and E gene assay PCR results are also provided as a binary result, RNA detected or RNA not detected, and the detection of RNA by either assay is considered a positive result. For this study PCR detection by either QIAstat-Dx or laboratory PCR or both was considered as positive for COVID-19.
Sample size
The sample size for this sub-study was driven by consideration for estimating sensitivity of FebriDx MxA for the detection of COVID-19 (as defined by the reference standard of detection of SARS-CoV-2 by PCR on respiratory samples) and of the methods proposed by Riley et al12 for multivariable predictive models. In order to estimate a sensitivity of 85% to within +/- 8%, based on the score method for a 95% confidence interval,13 88 positive cases are required. Assuming a prevalence of Covid-19 of around 40% in those tested, 220 people are required, and to achieve an 80% chance of obtaining enough cases, this number was increased to 236. This was further increased to 266 to allow for patients who decline or are unable to undergo finger prick testing (estimated from the rates in our previous work). Full justification for the samples size in the multivariable model is provided in the supplementary appendix.
Statistical analysis
Baseline characteristics are summarised for all those recruited to the study, where data are available, and presented for the whole sample and by COVID-19 status. Diagnostic accuracy was assessed in two ways. Simple measures of sensitivity, specificity, predictive values and likelihood ratios are given for FebriDx MxA detection for the identification of COVID-19 (and as FebriDx MxA is a broad marker of antiviral host response and not limited to COVID-19 these measures are also calculated for the detection of all viruses). To further assess the diagnostic value of the test in practice, a multivariable logistic regression model was developed,14 , 15 full details are provided supplementary appendix.
As the antiviral host response to SARS-CoV-2 would be expected to wane over time, but detection of viral RNA by PCR may persist for several weeks, a secondary analysis was pre-specified to assess diagnostic accuracy in those who have had duration of less than seven days. Basic measures of diagnostic accuracy (e.g., sensitivity, specificity) are presented separately for those with duration less than or more than seven days. A multivariable model, pre-specified to be the same as that for the full model, was also run in those with duration less than seven days.
This study was prospectively registered with the ISRCTN14966673 on the 18th March 2020.
Results
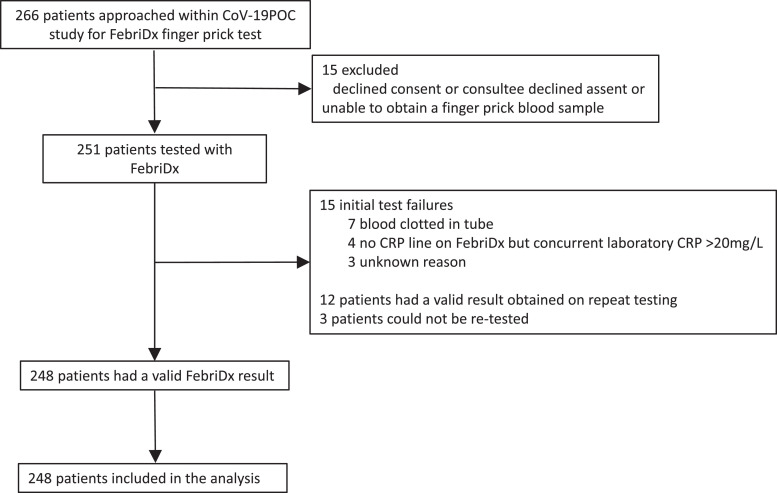
Between the 20th March and 12th April 2020 266 patients consecutive patients recruited to the CoV-19POC study were approached and FebriDx was performed in 251. 12 of 251 (5%) tests failed to give a valid result on the first test and a subsequent valid result was obtained on re-testing in 9 of 12, so that overall 248 patients had a valid FebriDx result. Flow of study participants is shown in Fig. 1 . 118 of 248 (48%) had COVID-19 (positive by PCR for SARS-CoV-2 on combined nose and throat swabs) and 92 of 116 (79%) COVID-19 positive patients had pneumonia as defined by Chest X-ray changes. PCR and FebriDx results were available for all tested patients. FebriDx results were available after 10 min compared with 1.7 (1.6 to 2.1) hours with point-of-care PCR testing and 23.4 (17.2 to 31.1) hours with laboratory PCR testing. For symptoms, 8 to 13% of data was missing and for vital signs and demographics missing data was <2.5%. Results are presented using complete-case analysis. Results including anosmia are included as an additional secondary analysis. Data collection for this was initiated after recruitment started, once it came to light in other literature that this was a relevant symptom of COVID-19; a total of 64 of 248 (26%) patients had missing data. Baseline demographics and clinical characteristic of all patients and those testing positive and negative for COVID-19, are shown in Table 1 . Patients positive for COVID-19 were less likely to be ethnically White British, had been unwell for longer (median duration of illness prior to presentation of 8 days), had a higher temperature and respiratory rate and lower oxygen saturations, compared to COVID-19 negative patients. In addition, COVID-19 positive patients had higher CRP levels, lower white cell counts and were more likely to have radiological evidence of pneumonia, shown in Table 1.
Fig. 1.
Flow of participants through the study.
Table 1.
Baseline demographics and clinical characteristics in all patients, COVID-19 positive patients and COVID-19 negative patients.
| All patients n = 248 |
COVID-19 positive patients n = 118 |
COVID-19 negative patients n = 130 |
p value* | |
|---|---|---|---|---|
| Age, years | 70 (52 to 81) | 67 (49 to 79) | 72 (58 to 81) | 0.13 |
| Sex | ||||
| Male | 135/248 (54) | 69/118 (58) | 66/130 (51) | 0.25 |
| Female | 113/248 (46) | 49/118 (42) | 64/130 (49) | – |
| White British | 219/245 (89) | 94/116 (81) | 125/129 (97) | <0.0001 |
| Current smoker | 21/211 (10) | 6/102 (6) | 15/109 (14) | 0.067 |
| Influenza vaccine† | 127/189 (67) | 63/92 (69) | 63/97 (65) | 0.53 |
| Pregnant | 4/46 (9) | 2/22 (9) | 2/25 (8) | 0.99 |
| Duration of symptoms, days‡ | 3 (1 to 7) | 8 (5 to 12) | 4 (2 to 11) | 0.0006 |
| Comorbidity | ||||
| Hypertension | 97/246 (39) | 48/117 (41) | 49/129 (38) | 0.69 |
| Cardiovascular disease | 79/247(32) | 35/118 (30) | 44/129 (34) | 0.49 |
| Respiratory disease | 98/248 (40) | 37/118 (31) | 61/130 (47) | 0.014 |
| Renal disease | 20/245 (8) | 8/118 (7) | 12/127 (9) | 0.49 |
| Liver disease | 8/247 (3) | 4/118 (3) | 4/129 (3) | 1.0 |
| Diabetes mellitus | 53/247 (22) | 25/118 (21) | 28/129 (22) | 0.88 |
| Cancer | 16/247 (7) | 8/118 (7) | 8/129 (6) | 1.0 |
| Immunosuppression | 10/244 (4) | 5/117 (4) | 5/127 (4) | 1.0 |
| Dementia | 26/148 (11) | 9/118 (8) | 17/130 (13) | 0.21 |
| Observations at admission | ||||
| Temperature, °C | 36.9 (36.5 to 37.8) | 37.2 (36.6 to 38.2) | 36.7 (36.4 to 37.4) | 0.0042 |
| Temperature ≥38 °C | 55/242 (23) | 36/114 (32) | 19/128 (15) | 0.0021 |
| Pulse rate, bpm | 92 (81 to 108) | 92 (92 to 109) | 92 (81 to 108) | 0.91 |
| Respiratory rate, bpm | 24 (20 to 28) | 25 (20 to 28) | 23 (19 to 28) | 0.023 |
| Oxygen saturations,% | 96 (94 to 97) | 95 (92 to 97) | 96 (94 to 98) | 0.030 |
| Supplementary O2 | 90/246 (37) | 45/116 (39) | 44/128 (34) | 0.51 |
| Blood pressure, mmHg | ||||
| Systolic | 130 (120 to 146) | 130 (120 to 145) | 132 (118 to 150) | 0.52 |
| Diastolic | 75 (66 to 83) | 75 (68 to 83) | 74 (65 to 84) | 0.60 |
| NEWS2 score | 5 (3 to 6) | 5 (3 to 7) | 4 (2 to 6) | 0.017 |
| Laboratory and radiology at admission | ||||
| CRP, mg/L | 60 (14 to 128) | 83 (32 to 136) | 33 (9 to 114) | 0.0003 |
| WCC, 109/L | 8.5 (6.1 to 12.7) | 6.6 (5.0 to 8.7) | 11.1 (7.9 to 15.4) | <0.0001 |
| CXR performed | 241/245 (98) | 116/116 (100) | 125/129 (97) | 0.12 |
| Final Diagnosis | ||||
| Pneumonia | 143 (58) | 92/116 (79) | 51/129 (40) | <0.0001 |
| Other | 102 (42) | 24/116 (21) | 78/129 (60) | – |
All data are given as n/n (%) and median (inter-quartile range). COVID-19 positive and negative status is defined by PCR positivity.
or negativity for SARS-CoV-2. *P value relate to comparison between COVID-19 positive and negative patients. NEWS=National.
Early Warning Score 2, CRP=C reactive protein, WCC=White cell count. †For the most recent influenza season. ‡Duration of symptoms.
prior to presentation, evaluated in 215 patients.
Diagnostic accuracy
Compared to the reference standard of detection of SARS-CoV-2 RNA by PCR on respiratory samples, the sensitivity of FebriDx MxA detection for identification of COVID-19 was 93% (110/118; 95%CI 87 to 97%). Specificity in this cohort was 86% (112/130; 95%CI 79 to 92%). Positive predictive value (PPV) was 86% (110/128; 95% CI 79 to 91%) and negative predictive value (NPV) was 93% (112/120; 95% CI 87 to 97%). Overall accuracy was 90% (222/248; 95%CI 86 to 93%), shown in Table 2 . As detection of MxA is not specific for COVID-19 the diagnostic accuracy of FebriDx MxA detection for the identification of all respiratory viruses is shown in Table 3 . Of the 11 patients with positive FebriDx MxA but who were PCR negative for SARS-CoV-2 and other viruses, 7 of 11 (63%) had classic radiological features of COVID-19.
Table 2.
Measures of diagnostic accuracy of FebriDx MxA for identification of COVID-19, compared to the reference standard of PCR positivity, n = 248.
| n/n | Value (95%CI) | |
|---|---|---|
| Prevalence of Covid-19 | 118/248 | 48 (41 to 54) |
| Sensitivity | 110/118 | 93 (87 to 97) |
| Specificity | 112/130 | 86 (79 to 92) |
| Positive predictive value | 110/128 | 86 (79 to 91) |
| Negative predictive value | 112/120 | 93 (87 to 97) |
| Positive likelihood ratio | 6.73 (4.37 to 10.37) | |
| Negative likelihood ratio | 0.08 (0.04 to 0.15) | |
| Overall accuracy | 222/248 | 90 (86 to 93) |
CI=Confidence interval.
Table 3.
Measures of diagnostic accuracy of FebriDx MxA for identification of any respiratory virus, compared to the reference standard of PCR positivity, n = 248.
| n/n | Value (95%CI) | |
|---|---|---|
| Prevalence of any virus* | 135/248 | 54 (48 to 61) |
| SARS-CoV-2 (COVID-19) | 118/248 | 48 (41 to 54) |
| Rhino/enterovirus | 10/248 | 4 (2 to 7) |
| Human Metapneumovirus | 4/248 | 2 (1 to 4) |
| Human Coronavirus OC43 | 2/248 | 2 (1 to 4) |
| Human Coronavirus HKU1 | 1/248 | 1 (0 to 2) |
| Human Coronavirus NL63 | 1/248 | 1 (0 to 2) |
| Sensitivity† | 117/135 | 87 (80 to 92) |
| Specificity | 102/113 | 90 (83 to 95) |
| Positive predictive value | 117/128 | 91 (86 to 94) |
| Negative predictive value | 112/120 | 85 (78 to 90) |
| Positive likelihood ratio | 8.9 (5.1 to 15.7) | |
| Negative likelihood ratio | 0.15 (0.1 to 0.23) | |
| Overall accuracy | 219/248 | 88 (83 to 92) |
CI=Confidence interval. *One patient had co-detection of SARS-CoV-2 and HCoV HKU1. † Viral detections associated with false negative MxA were: SARS-CoV-2 × 8, Rhino/enterovirus x 7 and Human Coronavirus x 3.
Using the calculated performance of FebriDx MxA for the detection of COVID-19 to estimate PPV and NPV at different disease prevalence, a prevalence of COVID-19 of 20% would give a PPV of 63% (95%CI 52 to 72%) and NPV of 99% (95%CI 96 to 99%). At a prevalence of 10% the PPV would be 43% (95%CI 33 to 53%) and the NPV would be 99% (95%CI 98 to 100%).
Measures of diagnostic accuracy according to duration of symptoms are shown in Table 4 . Although direct comparison of accuracy was not possible due to samples size, the sensitivity, specificity and positive and negative predictive values of FebriDx appeared comparable for patients presenting before and after 7 days of symptom duration.
Table 4.
Measures of diagnostic accuracy of FebriDx MxA for identification of COVID-19, according to duration of illness.
| n/n | ≤7 days of symptoms n = 129 Value (95% CI) |
n/n | >7 days of symptoms n = 86 Value (95% CI) |
|
|---|---|---|---|---|
| Median (IQR) duration, days | 4 (1 to 6) | 14 (8 to 14) | ||
| Prevalence of COVID-19 | 52/129 | 40 (32 to 49) | 53/86 | 62 (51 to 71) |
| Sensitivity | 49/52 | 94 (84 to 98) | 48/53 | 91 (80 to 96) |
| Specificity | 65/77 | 84 (75 to 91) | 29/33 | 88 (73 to 95) |
| Positive predictive value | 49/61 | 80 (69 to 88) | 48/52 | 92 (82 to 97) |
| Negative predictive value | 65/68 | 96 (88 to 99) | 29/34 | 85 (71 to 93) |
| Positive likelihood ratio | 6.05 (3.58 to 10.0) | 7.47 (2.96 to 19.0) | ||
| Negative likelihood ratio | 0.07 (0.02 to 0.21) | 0.11 (0.5 to 0.25) |
IQR=Interquartile range, CI=Confidence interval.
Predictive model
A total of 201 participants contributed data to the multivariable model, with symptoms being the main contributor to missing data. With the possible exception of cough (12% missing data in those without COVID-19 versus 6% with), missingness of data appeared unrelated to outcome. All continuous variables (age, respiratory rate and temperature) were mean-centred and assessed for a non-linear relationship with outcome using restricted cubic splines, following visual assessment and using Bayesian Information Criterion values (where smaller values indicate a more parsimonious model), all were entered in the model on the assumption that a linear relationship with the log odds of having COVID-19 was appropriate. All variables were entered into the multivariable model with no variable selection. Model results are given in Table 5 , alongside adjusted estimates.
Table 5.
Results for multivariable model.
| Variable | Regression coefficient (odds ratio) |
95% confidence interval |
p-value | Adjusted coefficient (odds ratio) |
|---|---|---|---|---|
| FebriDx MxA result | 92.3 | 31.6 to 269.6 | <0.0001 | 49.8 |
| Cough | 2.1 | 0.67 to 6.9 | 0.21 | 1.9 |
| Fever | 0.9 | 0.3 to 2.9 | 0.85 | 0.9 |
| Shortness of breath | 0.7 | 0.2 to 2.1 | 0.48 | 0.7 |
| Supplementary O2 | 0.7 | 0.3 to 1.9 | 0.49 | 0.7 |
| Sex | 1.0 | 0.4 to 2.7 | 0.92 | 1.0 |
| Age | 1.0 | 1.0 to 1.0 | 0.74 | 1.0 |
| Respiratory rate | 1.1 | 1.0 to 1.2 | 0.03 | 1.1 |
| Temperature | 1.2 | 0.7 to 1.9 | 0.52 | 1.1 |
| Intercept | 0.1 | 0.02 to 0.27 | <0.0001 | 0.1 |
Nagelkerke's R2 = 0.71; Brier score 0.09. Unless otherwise indicated, all reference categories for categorical variables are no/absence.
The overall results suggested that the addition of the patient and clinical characteristics do not add significantly to the diagnostic accuracy compared to using FebriDx MxA result alone. The apparent area under the receiver-operating characteristic (ROC) curve (AUC; equivalent to the C-statistic) for the multivariable model was 0.93, suggesting very good discrimination between those with and without PCR-confirmed COVID-19. The calibration plot shows good calibration (supplementary appendix), with points lying near the 45-degree line representing perfect fit.
The model underwent internal validation, using bootstrap resampling with 500 samples following the same model development process, to estimate optimism and provide a basis for adjusting model coefficients and the C-statistic. The calibration slope suggests the model tended to estimate probabilities that are too extreme and that if the model were to be used externally, the original multivariable regression coefficients should be adjusted by the value of the calibration slope (the shrinkage factor) to adjust for optimism. The C-statistic was also adjusted for optimism, but remained very high (0.90) suggesting the model would still perform well outside of the current sample.
Secondary analyses
The multivariable model was rerun with anosmia; a total of n = 164 were included in this analysis. Conclusions for the other covariates did not change, and there was little change in discrimination results (area under the curve, sensitivity and specificity at the Youden index, Brier score) nor other measures of fit such as Nagelkerke's R2; anosmia itself had an estimated odds ratio of 4.0 (95% CI: 0.7 to 22; p = 0.11) suggesting a potentially useful diagnostic indicator, though this conclusion is hampered by the uncertainty in the estimate. Lastly, the multivariable model (excluding anosmia) was also run in the subgroup of participants who presented within 7 days of symptom onset. Although the odds ratio for FebriDx increased (190), the uncertainty of this effect was large (95% CI 26 to 1404). There was little change in model performance with regard discrimination and model fit.
Test failures
As detailed above, some FebriDx tests failed to give a valid result on the first attempt. Tests could fail by blood clotting in the sample collection tube so that no blood would pass onto the lateral flow strip.
Safety
There were no adverse events seen with FebriDx testing. A single patient suffered brief epistaxis following nasal swabbing for PCR testing.
Discussion
In this large, real-world study we have shown that the detection of the antiviral host response protein MxA using the FebriDx point-of-care test had high sensitivity for the identification of COVID-19 during the first wave of the pandemic. Although there were differences in baseline characteristics between COVID-19 positive and negative patients, the multivariate predictive model results showed that clinical features did not contribute significantly to the effect of the FebriDx MxA result in distinguishing these groups. MxA detection is a marker of antiviral host response and is not specific for SARS-CoV-2 and so the high specificity of FebriDx MxA for COVID-19 seen in this study reflects the low levels of other respiratory viruses circulating during the study which will change with the seasonal circulation of other viruses such as influenza. It is now recognised that the diagnostic accuracy of PCR on upper respiratory tract samples is suboptimal and that patients presenting with COVID-19 often initially test falsely negative when compared to serial PCR testing or CT scanning.16, 17, 18 This means that our reference standard was likely to be suboptimal and it is notable that several of the patients in our study with positive MxA results and negative PCR for viruses (ie those considered to be false positive MxA) had classical radiological changes of COVID-19, strongly suggesting that they had the disease despite negative PCR results. The calculated specificity of FebriDx MxA for the detection of COVID-19 in the first wave was therefore likely to have been an underestimate.
The current reliance on centralised laboratory PCR for the diagnosis of COVID-19 with its prolonged turnaround time, leads to long delays in identification of positive and negative patients, with subsequent nosocomial transmissions, poor patient flow and reduced operational capacity in hospitals. As FebriDx is a low cost (£12 per test), rapid and highly scalable test these findings suggest that FebriDx could be rapidly deployed in hospitals and urgent care centres to be used as a front door triage tool. Because of the high sensitivity and negative predictive value, MxA negative patients being admitted to hospitals could be rapidly directed to non COVID-19 areas whilst MxA positive patients could be moved to assessment areas whilst awaiting confirmation or exclusion of COVID-19 positive status by PCR or CT scanning.
The manufacturers of FebriDx state that it is intended for use in patients aged over two years of age who have had respiratory symptoms for less than or equal to 7 days and/or fever for 48 h duration or less. In this study we have demonstrated that MxA detection by FebriDx remains accurate for the detection of COVID-19 beyond this period, presumably due to a very strong and persistent antiviral host response to SARS-CoV-2.
There are several limitations to the generalisability of our study. As the study was performed in almost exclusively immunocompetent adults the results cannot be applied to immune-compromised patients or to children and separate evaluations should be urgently undertaken in these important patient groups. In addition the findings of our study cannot be extrapolated to community dwelling patients including those who are infected but asymptomatic or pauci-symptomatic, as it uncertain whether their antiviral host response would be comparable to hospitalised patients. The diagnostic accuracy of FebriDx reported in this study is in the context of a very high disease prevalence and as we have shown the performance characteristics will change as the disease prevalence drops, with implications for how the test could be used. In particular the positive predictive value will decrease now that the first wave is subsiding, although the negative predictive value will increase further meaning that a negative FebriDx will remain a useful rule-out test. As MxA detection is a marker of antiviral host response and is not specific for COVID-19, the calculated test specificity will be lower when other viruses such as influenza are circulating. As early identification of influenza alongside COVID-19 will remains vitally important in hospitals, a rapid triage tool that detects both influenza and COVID-19 is likely to be of utility in the coming winter months. Finally, as this was a single centre study the diagnostic accuracy of FebriDx MxA for COVID-19 should be external validated in other centres.
In summary, FebriDx MxA had high accuracy for the identification of COVID-19 during the first wave of the pandemic and could be rapidly deployed in secondary care settings as a real-time triage tool to address the current problems of delayed diagnosis with laboratory PCR.
Declaration of Competing Interest
TWC has received speaker fees, honoraria, travel reimbursement, and equipment and consumables free of charge for the purposes of research outside of this submitted study, from BioFire diagnostics LLC and BioMerieux. TWC has received consultancy fees from Synairgen research Ltd, Randox laboratories Ltd and Cidara therapeutics. He a member of an advisory board for Roche and a member of two independent data monitoring committees for trials sponsored by Roche. He has acted as the UK chief investigator for an IMP study sponsored by Janssen. All other authors have completed the Unified Competing Interest form (available on request from the corresponding author) and declare: no support from any organisation for the submitted work no financial relationships with any organisations that might have an interest in the submitted work in the previous three years, no other relationships or activities that could appear to have influenced the submitted work.
Acknowledgments
Author contributions
TWC reviewed the medical literature, conceived of and designed the study, oversaw the conduct of the study, participated in the analysis and interpretation or results, drafted and wrote the manuscript. NJB assisted with the design of the study, screened and recruited patients and collected data. SP, CM, NN, VN, HW and LP screened and recruited patients and collected data. SE analysed the data and created and internally validated the multivariate model. All authors reviewed and contributed to the manuscript during its development.
Funding and support
This study was funded by University Hospital Southampton Foundation NHS trust. The laboratory work, nursing costs and consumables were supported by the NIHR Southampton Biomedical Research Centre. Statistical analysis was supported by Cancer Research UK core funding and NIHR CTU support funding at the Southampton Clinical Trials Unit. The FebriDx kits were purchased independently from a UK distributer and the manufacturer (Lumos Diagnostics, Sarasota, Florida, USA) had no role in the study conception, design, data analysis or manuscript preparation. The parent study is supported by Qiagen in the form of discounted equipment and consumables. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Acknowledgments
We would like to acknowledge and gives thanks to all the patient who kindly participated in this study and to all the clinical staff at University Hospital Southampton who cared for them. We would also like to give thanks to Richard Poole for assistance in data management.
We would like to acknowledge the NIHR Southampton Clinical Research Facility (CRF) laboratory, project and nursing teams; the UHSFT research nursing team; and the NIHR Southampton Biomedical Research Centre staff and project teams for their support in the set up of this study.
This report is independent research supported by the National Institute for Health Research (NIHR Post Doctorial Fellowship, Dr Tristan Clark, PDF 2016–09–061). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.06.051.
Appendix. Supplementary materials
Pathogens tested for by QIAstat-Dx Respiratory SARS-CoV-2 Panel
Sample size justification and methods for multivariable predictive model
Calibration plot for multivariate model
References
- 1.Nakabayashi M., Adachi Y., Itazawa T., Okabe Y., Kanegane H., Kawamura M., Tomita A., Miyawaki T. MxA-based recognition of viral illness in febrile children by a whole blood assay. Pediatr Res. 2006;60:770–774. doi: 10.1203/01.pdr.0000246098.65888.5b. [DOI] [PubMed] [Google Scholar]
- 2.Engelmann I., Dubos F., Lobert P.E., Houssin C., Degas V., Sardet A., Decoster A., Dewilde A., Martinot A., Hober D. Diagnosis of viral infections using myxovirus resistance A (MxA) Pediatrics. 2015;135:e985–e993. doi: 10.1542/peds.2014-1946. [DOI] [PubMed] [Google Scholar]
- 3.Self W.H., Rosen J., Sharp S.C., Filbin M.R., Hou P.C., Parekh A.D., Kutz M.C., Shapiro N.I. Diagnostic accuracy of FebriDx: a rapid test to detect immune responses to viral and bacterial upper respiratory infections. J Clin Med. 2017;6:E94. doi: 10.3390/jcm6100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro N.I., Self W.H., Rosen J., Sharo S.C., Filbin M.R., Hou P.C., Parekh A.D., Kurz M., Sambursky R. A prospective, multi-centre US clinical trial to determine accuracy of FebriDx point-of-care testing for acute upper respiratory infections with and without a confirmed fever. Ann Med. 2018;18:1–10. doi: 10.1080/07853890.2018.1474002. [DOI] [PubMed] [Google Scholar]
- 5.Shirley M. FebriDx: a Rapid Diagnostic Test for Differentiating Bacterial and Viral Aetiologies in Acute Respiratory Infections. Mol Diagn Ther. 2019;23:803–809. doi: 10.1007/s40291-019-00433-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard K., Chan C., Mills S., Poole S., Brendish N.J., Clark T.W. Evaluation of the Febridx Host Response Point-of-Care Test to Differentiate Viral From Bacterial Aetiology in Adults Hospitalized with Acute Respiratory Illness During Influenza Season. Open Forum Infect Dis. 2019;6:S300–S301. [Google Scholar]
- 7.https://www.fda.gov/media/136571/download. Accessed 09th April 2020
- 8.Parčina M., Schneider U.V., Visseaux B., Jozić R., Hannet I., Lisby J.G. Multicenter evaluation of the QIAstat Respiratory Panel-A new rapid highly multiplexed PCR based assay for diagnosis of acute respiratory tract infections. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0230183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leber A.L., Lisby J.G., Hansen G., Relich R.F., Schneider U.V., Granato P., Young S., Pareja J., Hannett I. Multicenter Evaluation of the QIAstat-Dx Respiratory panel for the detection of viruses and bacteria in nasopharyngeal swab specimens. J Clin Microbiol. 2020;58:e00155. doi: 10.1128/JCM.00155-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D., Bleicker T., Brünink S., Schneider J., Luisa Schmidt M., Mulders D., Haagmans B., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.https://www.england.nhs.uk/coronavirus/wpcontent/uploads/sites/52/2020/03/guidance-and-sop-covid-19-virus-testing-in-nhs-laboratories-v1.pdf. Accessed 09th April 2020
- 12.Riley R.D., Snell K.I., Ensor J., Burke D.L., Frank E.H., Moons K.G.M., Collins G.S. Minimum sample size for developing a multivariable prediction model: PART II - binary and time-to-event outcomes. Stat Med. 2019;38:1276–1296. doi: 10.1002/sim.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julious S.A. Two-sided confidence intervals for the single proportion: comparison of seven methods by Robert G. Newcombe. Statist Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. Stat Med 2005;24:3383-4. [DOI] [PubMed] [Google Scholar]
- 14.Wynants L., Van Calster B., Bonten M.M.J., Collins G.S., Debray T.P.A., De Vos M., Haller M.C., Heinze G., Moons K.G.M., Riley R.D., Schuit E., Smits L.J.M., Snell K.I.E., Steyerberg E.W., Wallisch C., van Smeden M. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrell F.E., Jr, Lee K.L., Califf R.M., Pryor D.B., Rosati R.A. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 16.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J.L., Luo L., Luo Z.D., Lyu J.X., Ng M.Y., Shen X.P., Wen Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir Med. 2020;168 doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucirka L.M., Lauer S., Laeyendecker O., Boon D., Lessler J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Ann Intern Med. 2020:M20–1495. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pathogens tested for by QIAstat-Dx Respiratory SARS-CoV-2 Panel
Sample size justification and methods for multivariable predictive model
Calibration plot for multivariate model