Abstract
T cells isolated from the pancreatic infiltrates of nonobese diabetic mice have been shown to recognize epitopes formed by the covalent cross-linking of proinsulin and secretory granule peptides. Formation of such hybrid insulin peptides (HIPs) was confirmed through mass spectrometry, and responses to HIPs were observed among the islet-infiltrating T cells of pancreatic organ donors and in the peripheral blood of individuals with type 1 diabetes (T1D). However, questions remain about the prevalence of HIP-specific T cells in humans, the sequences they recognize, and their role in disease. We identified six novel HIPs that are recognized in the context of DRB1*04:01, discovered by using a library of theoretical HIP sequences derived from insulin fragments covalently linked to one another or to fragments of secretory granule proteins or other islet-derived proteins. We demonstrate that T cells that recognize these HIPs are detectable in the peripheral blood of subjects with T1D and exhibit an effector memory phenotype. HIP-reactive T-cell clones produced Th1-associated cytokines and proliferated in response to human islet preparations. These results support the relevance of HIPs in human disease, further establishing a novel posttranslational modification that may contribute to the loss of peripheral tolerance in T1D.
Introduction
Type 1 diabetes (T1D) is a T cell–mediated autoimmune disease in which pancreatic β-cells are selectively destroyed, leading to dependence on exogenous insulin for glycemic control. Onset of the disease is preceded and predicted by the appearance of autoantibodies, which implies the accompanying activity of autoreactive CD4+ T cells (1,2). CD4 T-cell responses to several β-cell proteins have been implicated in both human T1D and the widely used nonobese diabetic (NOD) mouse model (3). There is strong evidence (particularly in the NOD mouse model) supporting the importance of insulin-specific responses, most notably those directed against insulin B9–23 (Ins B9–23) (4). Additional work has confirmed the relevance of the Ins B9–23 epitope in human disease; in particular, Ins B9–23 is recognized by human T cells in multiple binding registers (including a low-affinity binding register) and T cells that recognize the peptide are detectable in peripheral blood using HLA class II tetramers and have been found among islet infiltrates (5–8).
In general, T-cell receptors (TCR) with inappropriately high affinity for autoreactive HLA/peptide complexes are thought to undergo deletion and/or diversion to a regulatory lineage (9,10). However, negative selection is imperfect, to the extent that even healthy individuals with HLA haplotypes that predispose to autoimmunity have been shown to have a potentially autoreactive T-cell repertoire (11). Immune escape is further facilitated by posttranslational modifications, which may be underrepresented in the thymus and have the potential to alter the affinity of HLA binding or TCR recognition (12). Several recent studies report the recognition of modified β-cell epitopes by CD4+ T cells from the peripheral blood of individuals with T1D and from pancreatic draining lymph nodes and islet infiltrates from cadaveric specimens (12,13). These posttranslationally modified epitopes include citrullinated and deamidated peptides derived from glutamic acid decarboxylase 65, insulin tyrosine phosphatase–related islet antigen 2, islet amyloid polypeptide (IAPP), and 78-kDa glucose–regulated protein (GRP78) (13–16). Important recent data document hybrid insulin peptides (HIPs) as a new class of neo-epitope (17). These peptides are formed by the covalent linkage of insulin fragments to other protein fragments (including peptides from secretory granule proteins such as chromogranin A and IAPP) through a peptide bond, an event that is thought to occur in β-cell secretory granules and which, by definition, generates non–genetically encoded peptides (18). Using MHC class II tetramers, HIP-reactive T cells have been shown to be proinflammatory, highly diabetogenic, and detectable at increasing frequencies during disease progression in NOD mice (19,20).
Here, we further probe the relevance of HIPs in human disease by investigating whether the fusion of β-cell granule–associated peptides can generate hybrid neo-epitope sequences that are bound and presented by HLA-DRB1*04:01 (DR0401) and recognized by human CD4+ T cells. This allele is part of the high-risk T1D-associated DR4/DQ8 haplotype (21). Furthermore, T cells frequently recognize insulin epitopes such as proinsulin76–90 in the context of DR0401 (22). Therefore, we aimed to evaluate DR0401-restricted HIP responses. Following a prediction-based epitope discovery process, we identified immunogenic HIPs and used HLA class II tetramers to determine whether T cells that recognize these HIPs are present within the peripheral blood of subjects with T1D. Our results support the relevance of several novel HIPs, demonstrating that HIP-reactive T cells are readily detectable in the peripheral blood of subjects with established T1D.
Research Design and Methods
Human Subjects
Peripheral blood was collected from 22 individuals with T1D and 21 healthy control subjects with DRB1*04:01 haplotypes after written consent was obtained under a study approved by the Institutional Review Board at the Benaroya Research Institute. Subject attributes are summarized in Supplementary Tables 1 and 2.
Constructing a Library of Hybrid Peptides
A library of theoretical hybrid peptide sequences was generated though the in silico combination of 86 proinsulin fragments on the N-terminal (left) side and 89 natural protein cleavage products (e.g., WE14, the N-terminus of the chromogranin A) from secretory granule– and islet-associated peptides, including insulin, islet amyloid polypeptide, chromogranin A, secretogranin I, secretogranin II, secretogranin V, neuropeptide Y, and GRP78) on the C-terminal (right) side. Supplementary Fig. 1 provides a detailed description and schematic for this process, which resulted in a database of 7,654 theoretical HIPs.
Hybrid Peptide Prediction
The probability that theoretical HIPs would be bound and presented by DR0401 was evaluated based on a previously published prediction matrix (15,23). Briefly, coefficients corresponding to each anchor residue for all possible core 9-mers within the protein were multiplied, yielding a predicted relative binding affinity score (Supplementary Table 3). Peptides within the library that shared identical core 9-mers or had a conservative single amino acid difference (e.g., Y → F) that did not change the binding score were considered redundant. Sequences were generally chosen to include at least two flanking residues on each side of the predicted minimal 9-mer motif.
Peptide Binding Measurements
Peptides (13- to 20-mer) representing the top 50 nonredundant theoretical HIPs (Supplementary Table 4) were synthesized (Sigma-Aldrich), and their binding to DR0401 was assessed through a competition assay as previously described (24). Briefly, increasing concentrations of each theoretical HIP were incubated in competition with a biotinylated reference influenza hemagglutinin peptide (HA306–318) at 0.02 μmol/L in wells coated with DR0401 protein. After washing, residual biotin-HA306–318 was detected using europium-conjugated streptavidin (PerkinElmer) and quantified using a Victor2 D time-resolved fluorometer (PerkinElmer). Curves were simulated using Prism software (version 5.03; GraphPad Software Inc.), and IC50 values calculated as the concentration needed to displace 50% of the reference peptide.
HLA Class II Protein and Tetramer Reagents
DR0401 protein was purified from insect cell cultures as previously described (7,25). Monomers were loaded with 0.2 mg/mL peptide at 37°C for 72 h in the presence of 0.2 mg/mL n-dodecyl-β-maltoside and 1 mmol/L Pefabloc (Sigma-Aldrich). Peptide-loaded monomers were conjugated into tetramers using R-phycoerythrin (PE) streptavidin (Invitrogen), PE-Cy5 streptavidin (BD Biosciences), or PE-CF594 streptavidin at a molar ratio of 8:1.
In Vitro Tetramer Assays and T-Cell Clone Isolation
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll underlay, resuspended in T-cell media (RPMI, 10% pooled human serum, 1% penicillin-streptomycin, 1% l-glutamine) at 4 × 106 cells/mL, and stimulated with peptides (20 µg/mL total) in 48-well plates for 14 days, with addition of medium and IL-2 starting on day 7. Cells were stained with individual tetramers for 75 min at 37°C, followed by CD4 PerCP (BD Biosciences), CD3 APC (eBioscience), and CD25 FITC (BioLegend) for 15 min at 4°C run on a FACSCalibur (BD Biosciences), and analyzed using FlowJo (Treestar Inc.). Clones were isolated by sorting single tetramer–positive CD4+ T cells using a FACSAria (BD Biosciences) and expanded in 96-well plates in the presence of 1 × 105 irradiated PBMC and 2 µg/mL phytohemagglutinin (Remel), with addition of media and IL-2 starting on day 10.
T-Cell Clone Maintenance and Characterization
Clones specific for HIP peptides were maintained in supplemented RPMI and restimulated using phytohemagglutinin (2 µg/mL; Remel) every 2 weeks. For assessment of specificity and preferential recognition of hybrid peptides, 104 T cells/well were plated with 1 × 105 irradiated DR0401+ PBMCs and stimulated in triplicate with 10 µg/mL peptide. After incubation for 48 h at 37°C and pulsing with medium containing 3[H]-thymidine (1 µCi/well), incorporation was measured 18 h later with a scintillation counter. To assess responsiveness toward human islets, we added 100 µL PBS and 100 µL trifluoroethanol to 3,000 islet equivalents in a microfuge tube. After sonicating for 5 min, the islets were briefly vortexed and heated for 10 min at 95°C with brief vortex steps every 2 min. Islets were then sonicated for another 5 min and subsequently centrifuged at 17,000g for 5 min. Supernatants were fractionated in 25 mmol/L ammonium acetate buffer by size exclusion chromatography. DR0401+ PBMCs were pulsed with fractionated primary human islets, left unpulsed, or pulsed with peptide as a positive control. The pulsed (or unpulsed) PBMCs were incubated for 4 h and then irradiated, plated in triplicate at 1 × 105 cells per well plus 104 T cells/well, and incubated and pulsed with medium containing 3[H]-thymidine, and incorporation was measured as described above. For assessment of the functional phenotype of T cells, clones were activated with 50 ng/mL PMA and 1 µg/mL ionomycin for 30 min, followed by incubation with Brefeldin A (eBioscience) for 3 h at 37°C. Cells were fixed in fixation/permeabilization buffer (eBioscience); washed in permeabilization buffer; stained with IL-4 AF488 (eBioscience), IFN-γ AF700 (eBioscience), and IL-10 BV421, IL-17A APC/Cy7, and TNF-α PerCP-Cy5.5 (all BioLegend) for 15 min at 4°C; run on an LSR II (BD Biosciences); and analyzed using FlowJo.
Ex Vivo Tetramer Analysis
Analysis of T-cell frequency was accomplished using our previously published approach (26). Briefly, 40–60 × 106 PBMCs were resuspended in a total of 400–600 µL media, divided into two or three independent tubes of 20 × 106 cells (200 µL) each, incubated with 50 nmol/L dasatinib for 10 min at 37°C, and stained with 20 µg/mL PE-labeled, PE-CF594–labeled, or PE-Cy5–labeled HIP tetramers at room temperature for 120 min (three tetramers per tube for a total of six hybrid peptide tetramers plus an nonimmunogenic control tetramer if adequate cells were available for a third staining tube). Cells were washed, incubated with PE-magnetic beads (Miltenyi Biotec) for 20 min at 4°C, and magnetically enriched; 1% of the cells were retained as a nonenriched sample. Enriched (bound) and nonenriched (precolumn) samples were stained with CD4 V500, CD14 PerCP-Cy5.5, and CD19 PerCP-Cy5.5 (eBioscience) and CD45RA AF700 (BD), CXCR3 FITC, CCR6 BV421, and CCR4 BV605 (BioLegend) for 15 min at 4°C. After washing, cells were labeled with ViaProbe (BD Biosciences) and analyzed on a FACSCanto (BD Biosciences), gating on CD4+CD14−CD19−ViaProbe− cells and plotting tetramer versus CD45RA. Frequencies were calculated as previously described (26).
Statistics
A Mann-Whitney U test was used for two group comparisons between subjects with T1D and control subjects. A Kruskal-Wallis test and Dunn multiple comparisons were used for multiple group comparisons of T-cell frequencies. The Kolmogorov-Smirnov test was used to compare the distribution of T-cell frequencies for subjects with T1D and control subjects, and an F test was used to compare variances.
Data and Resource Availability
Data and materials are available upon written request to E.A.J.
Results
Identification of Theoretical HIPs Presented by DR0401
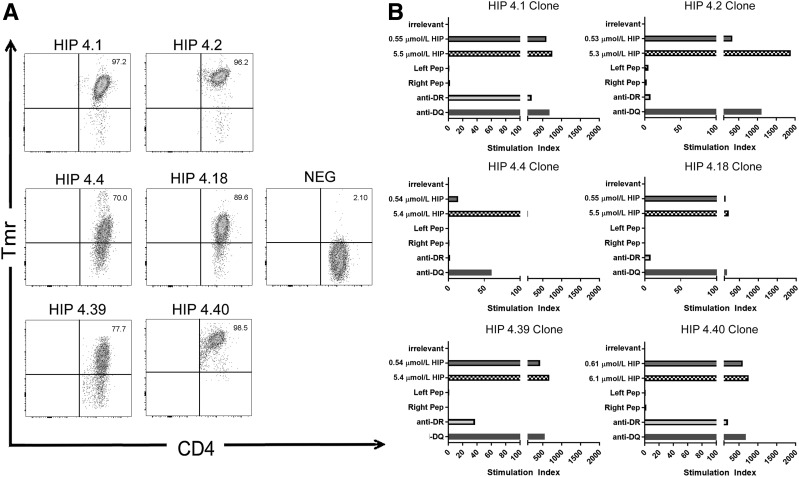
To identify HIP sequences that can be bound and presented by DR0401, we used a two-step strategy: first, nonredundant sequences within a library of 7,654 possible HIPs (Supplementary Fig. 1) were scored and predicted using a previously published approach (23,27); the top 50 peptides were synthesized and their binding to recombinant DR0401 protein was assessed using a competition assay (15,23). This narrowed our list of theoretical HIPs to 30 peptides that bound to DR0401 with appreciable affinity (Supplementary Table 4). We then produced DR0401 tetramers for each of the 30 peptides and used these to investigate the ability of each peptide to elicit CD4+ T-cell responses in vitro. PBMCs from eight unique subjects with T1D were stimulated with pools of HIP peptides for 2 weeks and subsequently stained with the corresponding individual tetramers (Supplementary Fig. 2). Six of the peptides (summarized in Table 1) elicited detectable populations of tetramer-positive T cells in multiple subjects with T1D (with response rates that varied between 25% and 62.5%), whereas the remaining staining results were consistently negative (representative negative results are shown for HIP-4.7 and HIP-4.47). Tetramer-positive T-cell clones were isolated from at least two different subjects for each of these epitopes to facilitate further studies (tetramer staining of representative clones shown in Fig. 1A).
Table 1.
Sequences and binding affinities for antigenic HIP peptides
| Peptide | Amino acid sequencea,b | Left peptide source | Right peptide source | IC50 (µmol/L)c | Response rated |
|---|---|---|---|---|---|
| HIP-4.1 | VCGERGFFEELVARSE | Ins B | Secretogranin I | 2.8 | 37.5 (3 of 8) |
| HIP-4.2 | HLVEALYLEELVARSE | Ins B | Secretogranin I | 0.44 | 25 (2 of 8) |
| HIP-4,4 | ICSLYQLEFVNQHLCG | Ins A | Ins B | 10 | 25 (2 of 8) |
| HIP-4.18 | SLQKRGIVEELVARSE | C-peptide/Ins A | Secretogranin I | 1.8 | 62.5 (5 of 8) |
| HIP-4.39 | CSLYQLENSVPHFSDE | Ins A | Secretogranin V | 0.24 | 25 (2 of 8) |
| HIP-4.40 | QPLALEGSALSSQHQA | C-peptide | GRP78 | 2.4 | 62.5 (5 of 8) |
The residues that originate from the left peptide source are bolded in each sequence.
The predicted minimal epitope is underlined.
IC50 represents the peptide concentration that displaces half of the reference peptide.
In vitro responses were examined in a total of eight subjects with established T1D. The response rate (percentage of subjects with a positive response) is noted, and the fraction of responders is listed in parentheses.
Figure 1.
Recognition of HIP peptides by T-cell clones from subjects with T1D. A: T-cell clones that recognize HIPs were isolated from peripheral blood after peptide-specific expansion by sorting of tetramer-positive T cells. In each case, the clones were stained by the corresponding HIP-loaded tetramer (but not a tetramer loaded with an irrelevant peptide). Each staining result shown is representative of clones isolated from at least two different subjects with established T1D. B: Proliferation of HIP-specific T-cell clones in response to an irrelevant peptide (∼5 μmol/L, corresponding to 10 µg/mL), their cognate peptide (∼0.5 and ∼5 μmol/L, corresponding to 1 and 10 µg/mL), control nonhybrid left and right peptides (∼5 μmol/L, corresponding to 10 µg/mL), or the cognate peptide (∼5 μmol/L, corresponding to 10 µg/mL) in the presence of an anti–HLA-DR blocking antibody or an irrelevant HLA-DQ blocking antibody. Data are represented as stimulation index values, calculated by normalizing the proliferation of each clone based on [3H]-thymidine incorporation of unstimulated wells. Each clone did not proliferate (stimulation index <3) in response to the nonhybrid peptides but exhibited robust proliferation (stimulation index >10) in response to 1 µg/mL cognate hybrid peptide, which was further increased at 10 µg/mL. The majority of the response was blocked through addition of the anti-DR antibody (ranging from 73% blocking for HIP-4.40 to 99% blocking for HIP-4.2) but not the irrelevant anti-DQ antibody. NEG, negative; Pep, peptide; Tmr, tetramer.
We next examined whether these T-cell responses were directed exclusively against the hybrid peptide sequences or whether T cells could respond to extended versions of the corresponding nonhybrid peptides. Each “left” peptide included a C-terminal extension and each “right” peptide included an N-terminal extension to replace the hybrid portion of the peptide (Supplementary Table 5). We assessed the HLA binding of the extended nonhybrid left and right peptides, and in all cases, the nonhybrid left and right peptides were not able to bind to DR0401. Furthermore, isolated T-cell clones exhibited proliferation above background in response to the full hybrid peptide but failed to proliferate in response to equivalent concentrations of the corresponding nonhybrid peptides (Fig. 1B). Proliferation was completely or partially blocked by an anti–HLA-DR antibody (the degree of blocking ranged from 73% to 99%) but not by a control anti–HLA-DQ antibody, confirming the restriction of the response (Fig. 1B). Finally, additional binding assays with arginine-substituted peptides verified that the minimal HLA-DR0401–restricted epitope within each of the six peptides spans the hybrid peptide junction (Supplementary Table 6). Therefore, we concluded that the epitopes recognized by these T-cell clones are hybrid peptides and focused our subsequent experiments on these six HIPs.
T Cells That Recognize HIPs Are Present in T1D and Can Cross-react With Homologous Sequences
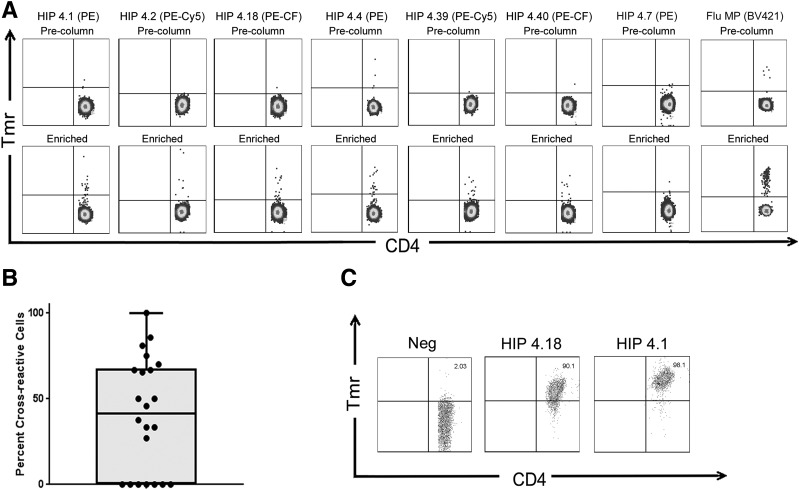
For evaluation of the relevance of HIP-reactive T cells in human subjects, we applied a direct tetramer enrichment approach (26) to measure their frequency in the peripheral blood of subjects with T1D. To assess the frequency of T cells that recognize all six theoretical HIPs of interest, we simultaneously labeled individual HIP specificities with PE-, PE-CF594–, or PE-Cy5–labeled tetramers, staining two separate aliquots of cells to assess all six HIPs (representative results for a subject with T1D shown in Fig. 2A). For some individuals, a third aliquot of cells was available and was stained using a negative control tetramer loaded with the nonimmunogenic HIP-4.7 peptide. In each case, the number of T cells stained by the HIP-4.7 tetramer was near or below the limit of detection for our assay (determined to be 0.5 cells per million in a previous study using a serial dilution method [28]). As we previously reported in our tetramer-based study of influenza-specific T-cell responses before and after vaccination (29), when homologous epitopes are included in the same staining tube and labeled with different fluorophores, their cross-reactivity can be estimated by evaluating the number of T cells that are double labeled by both tetramers. Applying this strategy, we observed that 15 of the 22 subjects with T1D had a substantial proportion of T cells that were costained by HIP-4.1 and HIP-4.18 tetramers (Fig. 2B). The remaining 7 subjects had no double-labeled cells, suggesting that cross-staining was not likely to be merely caused by an experimental artifact. On average, 40% of T cells that were labeled by the HIP-4.18 tetramer were also stained by the HIP-4.1 tetramer. Given that the sequences of these two peptides share considerable homology (Table 1), such cross-reactivity was not altogether unexpected. We further confirmed cross-recognition of these homologous epitopes by staining T-cell clones that had originally been isolated using the HIP-4.1 tetramer with HIP-4.18 tetramer, observing reduced but clearly positive staining (Fig. 2C).
Figure 2.
CD4+ T cells specific for novel HIPs are directly detectable, and T cells specific for homologous HIPs can cross-react. A magnetic enrichment procedure was used to enumerate HIP-reactive T cells directly ex vivo. A: Two independent tetramer stains per subjects were performed with PE-, PE-Cy5–, or PE-CF594–labeled tetramers to enumerate CD4+ T cells for each of the six individual HIP specificities and a BV421-labeled influenza control tetramer (Flu MP). If sufficient cells were available, an additional tetramer stain with a control (HIP-4.7 loaded) tetramer was also performed. Each upper panel shows a precolumn fraction, used to determine the total number of CD4+ T cells in the unmanipulated sample and to set a threshold for positive tetramer staining. Each lower panel shows the corresponding enriched fraction, used to determine the total number of epitope-specific CD4+ T cells in the sample. Cells were gated based on size, viability and lack of CD14/CD19 expression, and CD4 expression and then displayed on a CD4 versus tetramer (PE, PE-Cy5, or PE-CF594 [PE-CF] labeled) as shown. B: Tetramer staining revealed significant cross-reactivity between HIP-4.1– and HIP-4.18–reactive T cells, in that an average of 42% of the tetramer-positive cells were stained by both tetramers. C: Cross-reactivity between these epitopes was further confirmed by demonstration of cross-staining of HIP-4.1–reactive T-cell clones with HIP-4.18 tetramer. Neg, negative; Tmr, tetramer.
T Cells Specific for HIPs Exhibit Diverse Profiles in Subjects With T1D
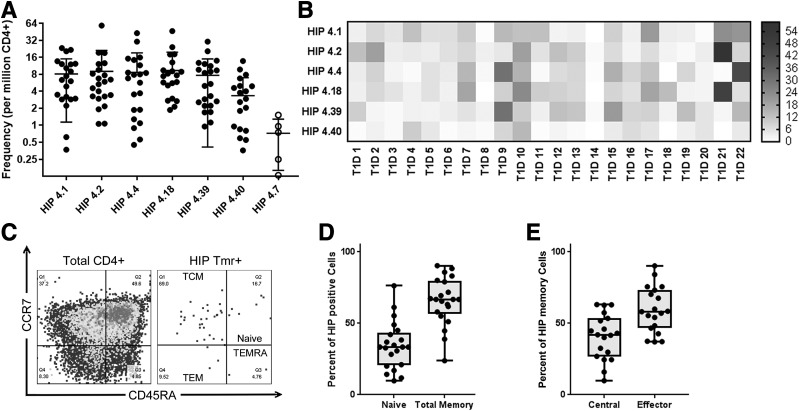
Based on the observed cross-reactivity, we included dual-labeled cross-reactive T cells in our estimates of the frequencies of HIP-4.1– and HIP-4.18–reactive T cells. In total our analysis showed that each of the six novel HIP specificities was present at a frequency that was above the limit of detection for our assay in most subjects (Fig. 3A). To draw further inferences about the significance of HIP-reactive T cells in subjects with T1D, we reorganized these T-cell frequency data into a heat map to visualize patterns of HIP-specific T cells in different individuals (Fig. 3B). From this analysis, it was evident that some subjects had high frequencies for multiple HIPs (e.g., participant 9 and participant 21), whereas others had very few HIP-specific T cells (e.g., participant 8 and participant 14). Given this observed diversity, we asked whether the frequency of HIP-reactive T cells might be correlated with characteristics such as disease duration, age at diagnosis, or number of autoantibodies. We observed a significant negative correlation (P = 0.0048) between age and the combined frequency of HIP-reactive T cells (Supplementary Fig. 3A). No other associations were significant, except that the two subjects who were insulin autoantibody negative (IAA−) had significantly lower frequencies of HIP-reactive T cells than the remaining subjects (P = 0.0173), who were all IAA+ (Supplementary Fig. 3B). However, it should be noted that since IAA were tested after diagnosis, antibodies in some subjects could be directed against exogenous insulin rather than being true autoantibody responses against self-insulin.
Figure 3.
CD4+ T cells specific for novel HIPs exhibit diverse frequency profiles and have an effector memory phenotype. A: The observed frequency hierarchy for HIP-reactive T cells in subjects with T1D (accounting for cross-reactive double-stained T cells). B: A heat map analysis of the same T-cell frequencies shown in A, indicating patterns of reactivity that differed between subjects. In the heat map, each column represents one subject and each column reflects one HIP specificity, with each square color coded to indicate the observed T-cell frequency in cells per million. C: Quadrant boundaries were defined for CD45RA and CCR7 surface expression based on the attributes of total CD4+ T cells. These were applied to classify HIP-reactive T cells as naive (CD45RA+CCR7+), TCM (CD45RA−CCR7−), and TEM (CD45RA−CCR7−). Terminal effectors (CD45RA−CCR7−) were rarely seen among HIP-reactive cells. D: Across all specificities, the phenotype of HIP-specific CD4− T cells was predominantly memory as opposed to naive. E: Among CD45RA− T cells, HIP-specific CD4− T cells were more like TEM (CCR7−) as opposed to TCM. Tmr, tetramer.
CD4+ T Cells That Recognize HIPs Exhibit an Effector Phenotype
We next evaluated the surface marker expression of HIP-reactive T-cells to draw inferences about their phenotype. Our staining panel was designed to address whether HIP-reactive cells were naive or antigen experienced and to assess their effector versus central memory status based on coexpression of CD45RA and CCR7. We defined naive cells as CD45RA+CCR7+, central memory (TCM) as CD45RA−CCR7+, effector memory (TEM) as CD45RA−CCR7−, and terminal effector-like cells (TEMRA) as CD45RA+CCR7− (Fig. 3C). Indeed, in subjects with T1D, the majority of HIP-reactive T cells had a memory phenotype (Fig. 3D) and the majority of these had an effector-like status, evidenced by lack of CCR7 expression (Fig. 3E). To further assess whether HIP-responsive T cells exhibit effector function, we performed intracellular cytokine staining of HIP-reactive T-cell clones following activation with PMA/ionomycin and observed substantial levels of IFN-γ (Supplementary Fig. 4). TNF-α was also observed in all but one of the clones, and significant levels of IL-4 were present within some clones.
CD4+ T Cells That Recognize HIPs Respond to Human Islets
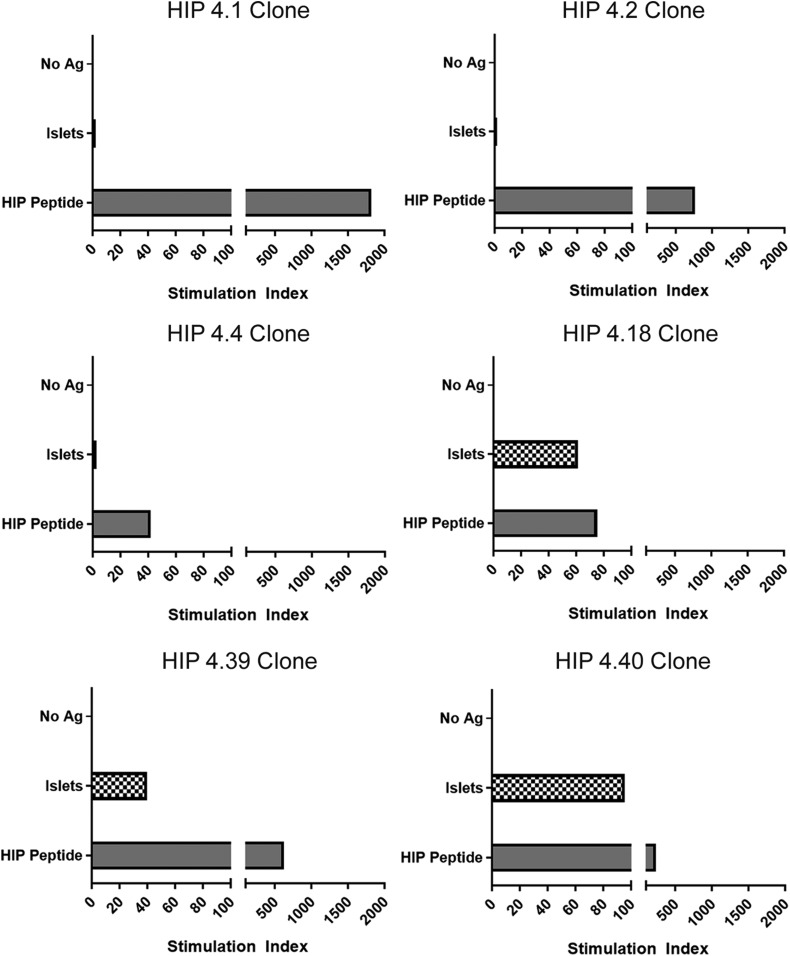
To further support the relevance of these putative HIP epitopes, we next evaluated whether epitopes corresponding to these peptides are present within human islets. HIP-reactive T-cell clones corresponding to each novel DR0401-restricted HIP peptide were activated in vitro using DR0401+ T cell–depleted PBMCs pulsed with fractionated primary human islets (using unpulsed cells as a negative control and peptide pulsed cells as a positive control), and their proliferation was assessed through thymidine incorporation. For HIP-4.1–, HIP-4.2–, and HIP-4.4–reactive T-cell clones, the levels of proliferation in response to human islets were much lower than the levels observed for peptide and not above the unpulsed control condition (Fig. 4). In contrast, the HIP-4.18, HIP-4.39, and HIP-4.40 T-cell clones exhibited proliferation in response to islets that was well above the unpulsed control condition, strongly suggesting that epitopes that correspond to these hybrid peptides are present within human islets (or that equivalent epitopes can form through additional processing within antigen-presenting cells).
Figure 4.
HIP-reactive CD4+ T-cell clones proliferate in response to human islets. Proliferation of HIP-specific T-cell clones in response to nonpulsed antigen-presenting cells, antigen-presenting cells pulsed with human islet preparations, or their cognate peptide (∼5 μmol/L, corresponding to 10 µg/mL). Data are represented as stimulation index values, calculated by normalizing the proliferation of each clone based on [3H]-thymidine incorporation of the control nonpulsed antigen-presenting cell–stimulated wells. HIP-4.1–, HIP -4.2–, and HIP-4.4–specific clones did not proliferate (stimulation index <3) in response to antigen-presenting cells pulsed with human islet fractions. HIP-4.18–, HIP-4.39–, and HIP-4.40–specific clones did proliferate (stimulation index = 61, 40, and 95, respectively) in response to antigen-presenting cells pulsed with human islet fractions. Ag, antigen.
T Cells Specific for HIPs Are Present at Elevated Frequencies in Subjects With T1D
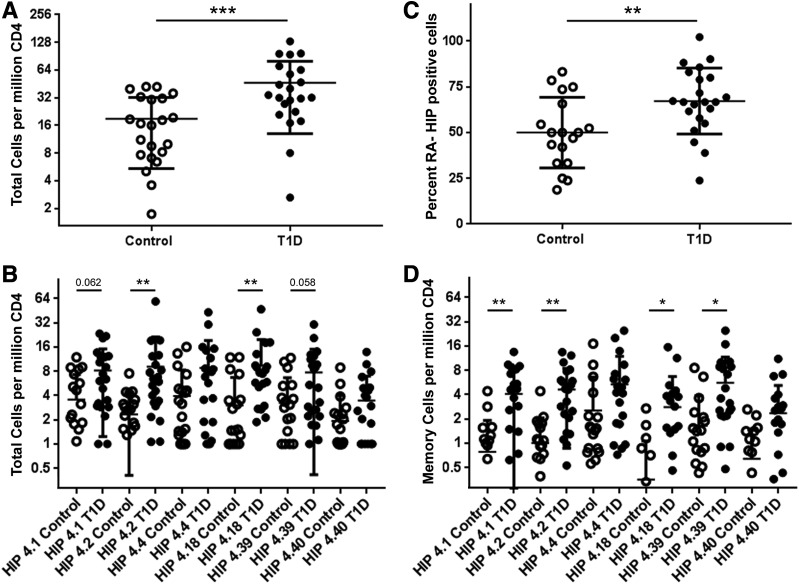
To assess the relevance of HIP-reactive T cells, we also measured their frequency and assessed their phenotype in the peripheral blood of HLA-matched control subjects. Notably, some HIP-reactive CD4+ T cells were present in control subjects. However, we observed that the combined frequency of HIP-reactive CD4+ T cells in subjects with T1D was significantly higher than in control subjects (P = 0.0012) (Fig. 5A). Although there is some overlap between the HIP-reactive T-cell frequencies observed for control subjects and subjects with T1D, the frequency distributions were significantly different (Kolmogorov-Smirnov test, P = 0.0082), as was the variance observed in each group (F test, P < 0.0001). Individual T-cell frequencies for HIP-4.2– and HIP-4.18–reactive T cells were significantly higher in subjects with T1D than in control subjects (P = 0.0074 and P = 0.0051, respectively) and trended toward being higher for HIP-4.1– and HIP-4.39–reactive T cells (Fig. 5B). Using CD45RA as a marker to distinguish antigen-experienced (CD45RA−) versus naive T cells (CD45RA+), we found that subjects with T1D had a higher proportion of HIP-reactive T cells that were antigen experienced than control subjects (P = 0.0124) (Fig. 5C). Notably, although memory HIP-reactive T cells mainly had an effector status in subjects with T1D, the opposite trend was observed in control subjects (Supplementary Fig. 5). TEMRA cells were rarely observed in T1D patients and were essentially absent in control subjects. Correspondingly, memory T-cell frequencies for HIP-4.1–, HIP-4.2–, HIP-4.18–, and HIP-4.39–reactive T cells were significantly higher in subjects with T1D than in control subjects (P = 0.0065, P = 0.0026, P = 0.011, and P = 0.029, respectively) (Fig. 5D). Thus, subjects with established T1D had significantly greater numbers of HIP-reactive CD4+ T cells than control subjects, and their HIP-reactive T cells showed increased signs of antigen exposure.
Figure 5.
CD4+ T cells specific for DR4-restricted HIPs are more frequent in subjects with T1D. A: Comparison of total HIP-reactive CD4+ T-cell frequencies in the peripheral blood of 21 healthy control subjects and 22 subjects with T1D. The total frequencies of HIP-reactive T cells (HIPs 4.1, 4.2, 4.4, 4.18, 4.39, and 4.40 combined) were significantly higher (P = 0.0004) in subjects with T1D than in HLA-matched control subjects. B: The individual frequencies were significantly higher in subjects with T1D (filled circles) than in healthy control subjects (open circles) for HIP-4.2 and HIP-4.18 (P = 0.0075 and P = 0.005, respectively). Individual frequencies for HIP-4.1 and HIP-4.39 also trended toward having higher frequencies (P = 0.062 and P = 0.058, respectively) but did not reach statistical significance. C: HIP-specific CD4+ T cells in subjects with T1D had a significantly higher (P = 0.0065) proportion of memory (CD45RA−) T cells than healthy control subjects. D: Correspondingly, the memory frequencies were significantly higher in subjects with T1D (filled circles) than in healthy control subjects (open circles) for HIP-4.1, HIP-4.2, HIP-4.18, and HIP-4.39 (P = 0.0065, P = 0.0026, P = 0.011, and P = 0.029, respectively). *P < 0.05, **P < 0.01, ***P < 0.001. RA-, CD45RA−.
Discussion
Published studies support a role for various classes of modified and nonconventional epitopes in T1D (12,14,15,30,31). In the current study we provide evidence showing that HIP-reactive T cells are present in human subjects. This builds on recently published work showing for the first time that HIP-reactive T cells can be observed in the peripheral blood of newly diagnosed T1D patients (32). In that study, HIP-reactive T cells were detectable in peripheral blood by IFN-γ enzyme-linked immune absorbent spot assay, and HIP-specific T-cell responses were shown to persist over time. Notably, multiple HIP-reactive T-cell clones isolated from these subjects were shown to be HLA-DR restricted (32). In the current study, HIP-reactive cells were detected in the peripheral blood of subjects with T1D using HLA-DRB1*04:01 tetramers and exhibited an effector-like surface phenotype (evidenced by lack of CCR7) and some evidence of effector function (e.g., secretion of IFN-γ and TNF-α). Among subjects with T1D, HIP-reactive T-cell frequencies were diverse; average frequencies were near 8 cells per million CD4+ T cells in peripheral blood for HIP-4.1, HIP-4.2, HIP-4.4, HIP-4.18, and HIP-4.39 (only HIP-4.40 stood out as having a lower mean frequency), but frequencies in individual subjects varied from <1 cell to >50 cells per million CD4+ T cells. While such frequencies are modest in comparison with those observed for vaccine antigens, they meet or exceed those observed for commonly studied CD4+ T-cell islet epitopes (5,22). Surprisingly, although the cross-sectional cohort of subjects we studied was fairly well distributed with respect to characteristics such as age at diagnosis and disease duration, the only significant association we observed was between age and the frequency of HIP-reactive T cells. Anecdotally, the two subjects in our cohort who were IAA− had low frequencies of HIP-reactive T cells, suggesting a possible link between T-cell responses to HIPs and insulin antibodies.
HIP-reactive T cells were also present in HLA-matched control subjects, indicating incomplete central and/or peripheral tolerance. Furthermore, HIP-reactive T cells in healthy subjects were not completely limited to a naive phenotype. These observations are in accord with a recent tetramer-based study of autoreactive CD8+ T cells and prior studies that used in vitro assays and can be inferred to suggest that autoreactive T cells are present in the T-cell repertoire of healthy subjects with disease-susceptible HLA and can undergo limited expansion in the absence of autoimmune disease (11,33,34). However, in spite of detectable frequencies in control subjects, HIP-reactive T cells were significantly more frequent in subjects with T1D. Admittedly, the immunologic significance of this two- to threefold difference in frequency is not entirely clear. Once possibility is that these modestly elevated frequencies are reflective of recent T-cell activation and turnover. For example, in a previous study, T-cell frequencies for conserved influenza epitopes were two to threefold higher following influenza exposure (35). HIP-reactive T cells in subjects with T1D exhibited a memory phenotype that was biased toward having an effector status, implying disease-associated expansion of HIP-reactive T cells to form an autoreactive memory pool. The presence of elevated frequencies of HIP-reactive memory T cells in subjects with T1D supports their relevance, but further elaborating the significance of HIP-reactive cells in disease is challenging. However, the anecdotal observation that insulin antibody–negative subjects appear to have reduced numbers of HIP-reactive T cells raises the intriguing possibility that HIP responses and insulin antibodies may be linked in some way. Longitudinal studies or larger and more diverse sample sets could shed further light on the role that HIP-reactive T cells play in disease development.
Our study does have limitations. Notably, although HIP-reactive T-cell clones proliferated in response to human islets, we have not yet been able to verify the formation of these putative HIP peptides in β-cells by mass spectrometry. Therefore, the peptides identified through our study could be mimotopes, which activate T cells that recognize other homologous sequences that are present in human islets. It could be argued that a more stringent negative control (e.g., a similarly prepared extract from the same donor using tissue devoid of islets) should have been included to exclude the possibility that HIP-reactive T-cell clones proliferated in response to protein fragments that were generated during the isolation and/or fractionation process. Likewise, it is possible that peptides or protein fragments from the islet fractions may have undergone additional processing within antigen-presenting cells during the pulsing and incubation steps of our assay. In that scenario, it would be necessary to elute peptides from pulsed antigen-presenting cells to identify the exact peptide sequences recognized by each of these T-cell clones. Until precise hybrid peptide sequences are known, it will be difficult to draw clear conclusions about cross-reactivity. It is likely that additional HIPs beyond the six putative HIPs that we studied are also recognized in human subjects with T1D. Given that HIP-reactive T cells appear to be present at much higher frequencies in some subjects, an important unanswered question is whether abundant HIP responses are reflective of a specific disease state or endotype. Likewise, because the precise mechanisms through which HIPs are formed within the secretory granules of β-cells remain unknown, it is unclear whether stressed β-cells are more prone to generating HIPs. Future studies to address these questions would be of value.
In all, our findings support a potential role for HIP-reactive T cells in general and, more specifically, a new class of DRB1*04:01-restricted HIPs in T1D and suggest that these epitopes activate pathogenic CD4+ T cells that are not effectively eliminated by negative selection or controlled by peripheral tolerance. These results and prior observations in murine models support the notion that the pathways by which HIP epitopes are generated may be highly conserved among species. Indeed, recent mass spectrometric analyses confirmed the presence of other HIPs in both mouse and human pancreatic islets (36). Our data, then, support the hypothesis that the formation of HIPs in human β-cells generates a potentially diverse set of epitopes, some of which are recognized in the context of DR0401, that contribute to autoreactive responses. Given that other HIP-reactive T cells have been found among islet-infiltrating T cells, it will be interesting to ascertain whether T cells that recognize these novel DR0401-restricted HIPs are present within disease proximal tissues.
Article Information
Acknowledgments. The authors thank Aru Arumaganathan (Benaroya Research Institute) for flow cytometry support; Jenna Snavely, McKenzie Lettau, and Jani Klein (Benaroya Research Institute) for assisting with subject recruitment; and members of the laboratory of W.W.K. (Benaroya Research Institute) for helpful discussions.
Funding. This work was supported by the National Institutes of Health grants R01-DK-081166 (to K.H.) and R21AI133059 (to R.L.B.), JDRF grant 2-SRA-2018-551-S-B (to E.A.J.), and the American Diabetes Association Pathway to Stop Diabetes 1-15-ACE-14 (to T.D.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.A.-L. performed peptide binding and tetramer assays, analyzed data, and cowrote the manuscript. P.G. assisted with peptide binding, performed T-cell clone characterization, and prepared figures for the manuscript. T.D. assembled the theoretical hybrid peptide database, assisted with study design, and edited the manuscript. M.D. purified human islet preps for T-cell assays and assisted with experimental design. I.-T.C. produced HLA-DR protein, provided technical support on ex vivo tetramer staining, and edited the manuscript. C.S. and C.J.G. were responsible for subject selection and characterization, contributed important ideas, and edited the manuscript. K.H. and R.L.B. assisted with study design, contributed important ideas, and edited the manuscript. W.W.K. developed experimental methods, contributed important ideas, and edited the manuscript. E.A.J. designed the research, summarized data, provided technical training to D.A.-L. and P.G. cowrote the manuscript. E.A.J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/db20-4567/suppl.12103698.
References
- 1.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol 2007;148:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med 2012;2:a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGinty JW, Marré ML, Bajzik V, Piganelli JD, James EA. T cell epitopes and post-translationally modified epitopes in type 1 diabetes. Curr Diab Rep 2015;15:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama M, Abiru N, Moriyama H, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005;435:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spanier JA, Sahli NL, Wilson JC, et al. Increased effector memory insulin-specific CD4+ T cells correlate with insulin autoantibodies in patients with recent-onset type 1 diabetes. Diabetes 2017;66:3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durinovic-Belló I, Wu RP, Gersuk VH, Sanda S, Shilling HG, Nepom GT. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes Immun 2010;11:188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Chow IT, Sosinowski T, et al. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A 2014;111:14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michels AW, Landry LG, McDaniel KA, et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes 2017;66:722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams CB, Engle DL, Kersh GJ, Michael White J, Allen PM. A kinetic threshold between negative and positive selection based on the longevity of the T cell receptor-ligand complex. J Exp Med 1999;189:1531–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol 2007;178:7032–7041 [DOI] [PubMed] [Google Scholar]
- 11.Danke NA, Koelle DM, Yee C, Beheray S, Kwok WW. Autoreactive T cells in healthy individuals. J Immunol 2004;172:5967–5972 [DOI] [PubMed] [Google Scholar]
- 12.James EA, Pietropaolo M, Mamula MJ. Immune recognition of β-cells: neoepitopes as key players in the loss of tolerance. Diabetes 2018;67:1035–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babon JA, DeNicola ME, Blodgett DM, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med 2016;22:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Lummel M, Duinkerken G, van Veelen PA, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes 2014;63:237–247 [DOI] [PubMed] [Google Scholar]
- 15.McGinty JW, Chow IT, Greenbaum C, Odegard J, Kwok WW, James EA. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes 2014;63:3033–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marre ML, McGinty JW, Chow IT, et al. Modifying enzymes are elicited by ER stress, generating epitopes that are selectively recognized by CD4+ T cells in patients with type 1 diabetes. Diabetes 2018;67:1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016;351:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiles TA, Delong T, Baker RL, et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J Autoimmun 2017;78:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker RL, Jamison BL, Wiles TA, et al. CD4 T cells reactive to hybrid insulin peptides are indicators of disease activity in the NOD mouse. Diabetes 2018;67:1836–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito Y, Ashenberg O, Pyrdol J, et al. Rapid CLIP dissociation from MHC II promotes an unusual antigen presentation pathway in autoimmunity. J Exp Med 2018;215:2617–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erlich H, Valdes AM, Noble J, et al.; Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blahnik G, Uchtenhagen H, Chow IT, et al. Analysis of pancreatic beta cell specific CD4+ T cells reveals a predominance of proinsulin specific cells. Cell Immunol 2019;335:68–75 [DOI] [PubMed] [Google Scholar]
- 23.James EA, Rieck M, Pieper J, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014;66:1712–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ettinger RA, Kwok WW. A peptide binding motif for HLA-DQA1*0102/DQB1*0602, the class II MHC molecule associated with dominant protection in insulin-dependent diabetes mellitus. J Immunol 1998;160:2365–2373 [PubMed] [Google Scholar]
- 25.Chow IT, Yang J, Gates TJ, et al. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121-140 and GAD65 250-266 in T1D development. PLoS One 2014;9:e112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok WW, Roti M, Delong JH, et al. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol 2010;125:1407–1409.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rims C, Uchtenhagen H, Kaplan MJ, et al. Citrullinated Aggrecan epitopes as targets of autoreactive CD4+ T cells in patients with rheumatoid arthritis. Arthritis Rheumatol 2019;71:518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok WW, Tan V, Gillette L, et al. Frequency of epitope-specific naive CD4+ T cells correlates with immunodominance in the human memory repertoire. J Immunol 2012;188:2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchtenhagen H, Rims C, Blahnik G, et al. Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat Commun 2016;7:12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannering SI, Harrison LC, Williamson NA, et al. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med 2005;202:1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kracht MJ, van Lummel M, Nikolic T, et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 2017;23:501–507 [DOI] [PubMed] [Google Scholar]
- 32.Baker RL, Rihanek M, Hohenstein AC, et al. Hybrid insulin peptides are autoantigens in type 1 diabetes. Diabetes 2019;68:1830–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culina S, Lalanne AI, Afonso G, et al.; ImMaDiab Study Group . Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol 2018;3:eaao4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, James EA, Sanda S, Greenbaum C, Kwok WW. CD4+ T cells recognize diverse epitopes within GAD65: implications for repertoire development and diabetes monitoring. Immunology 2013;138:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, James E, Gates TJ, et al. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol 2013;25:447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiles TA, Powell R, Michel R, et al. Identification of hybrid insulin peptides (HIPs) in mouse and human islets by mass spectrometry. J Proteome Res 2019;18:814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]