Abstract
Neurodevelopmental abnormalities are the most common noncardiac complications in patients with congenital heart disease (CHD). Prenatal brain abnormalities may be due to reduced oxygenation, genetic factors, or less commonly, teratogens. Understanding the contribution of these factors is essential to improve outcomes. Because primary sulcal patterns are prenatally determined and under strong genetic control, we hypothesized that they are influenced by genetic variants in CHD. In this study, we reveal significant alterations in sulcal patterns among subjects with single ventricle CHD (n = 115, 14.7 ± 2.9 years [mean ± standard deviation]) compared with controls (n = 45, 15.5 ± 2.4 years) using a graph-based pattern-analysis technique. Among patients with CHD, the left hemisphere demonstrated decreased sulcal pattern similarity to controls in the left temporal and parietal lobes, as well as the bilateral frontal lobes. Temporal and parietal lobes demonstrated an abnormally asymmetric left–right pattern of sulcal basin area in CHD subjects. Sulcal pattern similarity to control was positively correlated with working memory, processing speed, and executive function. Exome analysis identified damaging de novo variants only in CHD subjects with more atypical sulcal patterns. Together, these findings suggest that sulcal pattern analysis may be useful in characterizing genetically influenced, atypical early brain development and neurodevelopmental risk in subjects with CHD.
Keywords: brain development, congenital heart disease, magnetic resonance imaging, neurodevelopment, sulcal pattern
Introduction
Congenital heart disease (CHD) is the most common birth defect and is associated with neurodevelopmental disability and structural brain abnormalities (Limperopoulos et al. 2010; Bellinger et al. 2011; Wernovsky 2012). There are many potential causes of this association. Prenatal hemodynamic alterations could result in abnormal brain structure in CHD (McQuillen et al. 2010; Khalil et al. 2016). For example, subjects with hypoplastic left heart syndrome, who have the most severe cardiovascular compromise including decreased cerebral oxygenation and perfusion in utero, have been shown to have abnormal brain structural development and high rates of neurodevelopmental delay (Goldberg et al. 2000; Sethi et al. 2013; Gaynor et al. 2014; Bellinger et al. 2015). Postnatal hypoxia, acidosis, and hypoperfusion could also contribute to abnormalities in brain structure, growth, and neurodevelopment (Mebius et al. 2017). Moreover, since much of heart and brain development occurs simultaneously and involves shared molecular pathways, genetic variants that cause CHD could affect neuronal development, patterning, and resiliency (Ising and Holsboer 2006; Fitzgerald et al. 2015; Homsy et al. 2015; Jin et al. 2017; Mebius et al. 2017) and lead to altered development of brain structures under strong genetic control.
Studies using magnetic resonance imaging (MRI) have improved our understanding of the effects of CHD on brain development, both in the prenatal and neonatal periods (Bellinger et al. 2015). In fact, fetal MRI studies have revealed delayed gyrification and atypical sulcal patterns, which preceded differences in gray/white matter volume in the second and third trimesters, emphasizing the potential role of genetic factors that act in utero (Limperopoulos et al. 2010; Clouchoux et al. 2013; Masoller et al. 2016; Ortinau et al. 2018). Recent studies have identified abnormal functional neuronal connectivity and reorganization of structural network topology in newborns with CHD in the pre-operative periods (De Asis-Cruz et al. 2018; Schmithorst et al. 2018), further supporting the likelihood that altered brain development begins in utero. Additional neonatal MRI studies have also reported findings consistent with altered in utero brain development such as alterations in white matter microstructure, reduced whole-brain volume, reduced cortical folding, and abnormal brain maturation in CHD (Mahle et al. 2002; Miller et al. 2007; Licht et al. 2009; Ortinau et al. 2012, 2013; Beca et al. 2013; Rollins et al. 2017; Karmacharya et al. 2018; Schmithorst et al. 2018).
Analysis of primary sulcal pits (the deepest local points of sulci) and folding patterns can provide important insight into structural brain development (Im and Grant 2018). Primary cortical sulci are those that develop prior to the third trimester and arise during rapid neuronal growth (Chi et al. 1977; Garel et al. 2001; Kostović and Vasung 2009; White et al. 2010). These primary cortical sulcal folds are under strong spatial-temporal genetic control and show little change with age during postnatal cortex development (Garel et al. 2001; Rakic 2004; Kostović and Vasung 2009; Hill et al. 2010; Meng et al. 2014; Rash and Rakic 2014; Sun and Hevner 2014; de Juan Romero et al. 2015; Cachia et al. 2016). The global pattern of positioning, arrangement, number, and size of sulcal pits and folds as well as their intersulcal relationships are thought to relate to optimal organization and arrangement of cortical functional areas and their white matter connections (Van Essen 1997; Klyachko and Stevens 2003; Rakic 2004; Fischl et al. 2008; Im et al. 2010; Im et al. 2011a; Sun and Hevner 2014). We have developed a novel comprehensive and quantitative analysis technique to characterize sulcal folding patterns using a graph structure with sulcal pits and catchment basins as nodes (Im et al. 2011b). It considers not only geometric features of sulcal folds themselves (position, depth, and size of sulcal pits and basins) but also their intersulcal geometric and topological relationships, emphasizing the interrelated arrangement and patterning of sulcal folds. Application of this technique has proven effective for characterizing genetically influenced cortical development in several of our previous studies (Im et al. 2011b; Im et al. 2013, 2016, 2017; Im and Grant 2018; Tarui et al. 2018). For example, similarity of sulcal patterns was significantly higher in identical twin pairs than in unrelated pairs, supporting a strong genetic influence on the global sulcal pattern (Im et al. 2011b). In addition, a recently published analysis of fetal brain MRIs revealed early-emerging atypical sulcal patterns in fetuses with CHD (Ortinau et al. 2018). The early emergence of atypical sulcal patterns in CHD suggests that genetic variants associated with both cardiac and brain development may be playing a role in altered brain development (McQuillen et al. 2010; Homsy et al. 2015; Khalil et al. 2016). However, we do not know if these atypical gyral folding patterns observed in fetal life and infancy persist into adolescence or are associated with neuropsychological outcomes.
Here, we compare the sulcal folding patterns of 115 subjects with single-ventricle CHD with 45 healthy controls using MRI studies performed at approximately 15 years of life. We use our quantitative sulcal pattern analysis technique to investigate whether atypical sulcal patterns previously identified in a cohort of fetal subjects with CHD (Ortinau et al. 2018) were also present in adolescents with CHD. Furthermore, for regions with atypical sulcal patterns, we examined the clinical significance by correlating with neuropsychological testing. To search for a genetic influence on the altered sulcal patterns, damaging de novo variants (DNVs) were analyzed for their relationship to abnormal sulcal pattern using a subset of the CHD subjects with exome sequencing available. We hypothesize that sulcal pattern analysis will capture regional brain abnormalities that are relevant to neurodevelopmental outcomes and correlate with damaging DNVs in subjects with CHD.
Materials and Methods
Subjects
The study was approved by the Boston Children’s Hospital (BCH) institutional review board, and we obtained informed consent of parent or guardian (for subjects aged < 18 years) or of subjects (aged ≥18 years), as well as assent of study participants aged < 18 years. A separate informed consent was obtained from parents of CHD subjects to obtain their DNA. We analyzed MRI data from CHD subjects previously enrolled from 2010 to 2012 at BCH (Bellinger et al. 2015). Inclusion criteria were (1) age 10 to 19 years at the time of enrollment, (2) diagnosis of single-ventricle physiology, and (3) history of Fontan procedure. Subjects were excluded if they met any of the following criteria: (1) disorders that would prevent successful completion of the planned study testing (e.g., pacemaker, metal implants preventing MRI); (2) lack of reading fluency by primary caregiver in English, which is the only language for which questionnaires have been validated; (3) foreign residence; (4) cardiac transplantation; and (5) cardiac surgery within 6 months of testing. We also recruited healthy control subjects of a similar age from local pediatric practices, from our institutional adolescent clinic, and through posted notices. We applied exclusion criteria derived from the National Institutes of Health’s MRI study of normal brain development (Evans 2006).
MRI Acquisition
MRI was performed on 1 of 2 systems: a 3-T General Electric system (General Electric Medical Systems) or, for subjects in whom cardiovascular devices or coils had been implanted, a 1.5-T General Electric Twin-speed magnetic resonance scanner. Subjects were scanned without sedation using a T1-weighted, 3D spoiled gradient recalled steady state sequence with the following parameters: TR/TE = 7 ms/2.8 ms, flip angle = 8°, acquisition matrix = 256 × 256, FOV = 256 mm, and slice thickness = 1 mm at 3 T; TR/TE = 40s/4s, flip angle = 20°, acquisition matrix = 256 × 192, FOV = 240 mm, and slice thickness = 1.5 mm at 1.5 T. All images were inspected by a radiologist to assure data quality and detect structural abnormalities (e.g., tumors, stroke).
Image Processing and Quantitative Sulcal Pattern Analysis
The images were processed to extract cortical surfaces using the FreeSurfer pipeline (Dale et al. 1999; Fischl et al. 1999; Fischl 2012). Once the cortical models were reconstructed, they were automatically parcellated into anatomical regions based on lobar and gyral/sulcal structure (Fischl et al. 2004; Desikan et al. 2006). We used the left and right whole hemispheres and lobar regions of the white matter surface (gray/white matter boundary) for sulcal pattern analysis in this study. The reconstruction of the white matter surface from individual MRI data and its lobe parcellation were visually inspected for each subject to ensure accuracy.
The sulcal pattern was represented as a graph structure with sulcal pits and their surrounding catchment basins as nodes. Sulcal pits are defined as the deepest local point in a sulcal catchment basin on the cortical surface. Sulcal pits and catchment basins are substructures decomposed from one primary sulcal segment (Lohmann and Von Cramon 2000; Im et al. 2010). Sulcal depth maps on the white matter surface were generated using the FreeSurfer, and sulcal pits and their surrounding sulcal catchment basins were automatically identified based on a smoothed sulcal depth map using a watershed segmentation algorithm (Im et al. 2010, 2013, 2016). If sulcal catchment basins met, sulcal pits in those basins were connected with an edge. This graph was designed to characterize the global pattern of primary sulcal folds using deep sulcal pits. Sulcal pattern graphs between different subjects were automatically compared using a spectral-based matching algorithm (Leordeanu and Hebert 2005; Im et al. 2011b). The sulcal pattern comparison was performed using geometric features of sulcal folds (3D position, depth, and area of sulcal pits and basins), their intersulcal relationships, and graph topology (the number of edges and the paths between nodes) to highlight the interrelated arrangement and patterning of sulcal folds (Im et al. 2011b). The optimal match was determined between two different sulcal graphs, and then their similarity was computed for each hemisphere and lobar region, which ranged from 0 to 1. After measuring the similarity with all features combined (sulcal position, area, depth, and graph topology), we further measured the similarity only using each individual feature by setting all weights of the other features to 0 to evaluate their relative importance on the sulcal pattern similarity (Im et al. 2011b).
Using the graph-based sulcal pattern comparison method, sulcal pattern similarities of all possible pairs in control (n = 45) and CHD (n = 115) groups (160 × 160 similarity matrix) were automatically computed for the left and right hemispheres and lobes. We first tested whether the similarity within the control group was different when compared with the similarity between control and CHD groups. For each control subject, a mean similarity with the other 44 control subjects was measured. Each CHD subject had a mean similarity with all 45 controls. Mean similarities with the controls were then compared between CHD and control groups. Since the data were acquired using either 3 T or 1.5 T, we additionally tested with control subjects if the sulcal pattern similarity values were affected by the effects of different scanners and magnets. Each control subject had mean similarities with the control set from the same scanner and with another control set from the different scanner. Mean similarities from the same scanner were then compared with the similarities from the different scanners.
Sulcal Pattern Analysis Using the Dataset of the Human Connectome Project
Because the similarity with the other controls was measured for each control within the same group, the similarity within the control group might be biased by specific characteristics of our sampling for normal controls. We additionally used a separate large dataset of normal subjects as a reference for our sulcal pattern comparison. The dataset of the Human Connectome Project (HCP), which was released in November 2014, was employed (humanconnectome.org) (Van Essen et al. 2012). We selected and included 80 normal subjects with the youngest age range (22–25 years) from the HCP dataset. Sulcal pattern similarities between our control and CHD subjects (n = 45 + 115) and the 80 HCP normal subjects were calculated (160 × 80 similarity matrix) for the left and right hemispheres and lobes, and mean similarities with the 80 HCP subjects were measured for each control and CHD subject, and compared between the two groups.
Sulcal Pattern Symmetry Analysis
For regions with unilateral significantly different sulcal pattern similarity values, we further evaluated sulcal pattern symmetry by measuring similarity between the left and right hemispheric and lobar sulcal patterns in controls and CHD subjects. Cortical surface in the left hemisphere was flipped to the right and compared with the right hemisphere cortical surface. Lower similarity between the left and right indicates asymmetric sulcal pattern between the hemispheres. Left–right sulcal pattern symmetries were measured for all control and CHD subjects and compared between the two groups.
Brain Volumes, Cortical Thickness, and Gyrification Index
To determine if sulcal pattern similarity values were confounded by brain volume or overall gyrification, we measured the volumes of the gray matter and white matter, mean cortical thickness, and gyrification index for the left and right hemispheres. Gray and white matter volumes and cortical thickness were measured using Freesurfer software. The gyrification index was computed as the ratio of the whole cortical surface to their outer convex hull surface area to measure the overall amount of cortical folding (Zilles et al. 1988).
Neurodevelopmental Assessment
The subjects’ neurodevelopmental function was assessed using tests of general intelligence, academic achievement, and executive functions as previously described (Bellinger et al. 2015). The following describes the assessments administered as well as their scoring system.
General Intelligence
The Wechsler Intelligence Scale for Children–Fourth Edition (Psychological Corporation 2003) was administered to adolescents < 17 years old, and the Wechsler Adult Intelligence Scale–Fourth Edition (Baron 2004) to adolescents 17 to 19 years of age. The end points considered were the 5 composite scores of Full-Scale IQ, verbal comprehension, perceptual reasoning, working memory, and processing speed (all with an expected mean of 100 and standard deviation (SD) of 15).
Academic Achievement
Each adolescent’s Reading and Mathematics Composite scores (both with an expected mean of 100 and SD of 15) of the Wechsler Individual Achievement Test–Second Edition (Psychological Corporation 2003) were calculated.
Executive Functions
An executive function summary score was calculated by averaging an adolescent’s scores on 5 subtests of the Delis–Kaplan Executive Function System (Delis et al. 2001): Verbal Fluency (mean score on the letter and semantic fluency trials), Design Fluency (primary combined measure), Sorting (combined conditions score), Word Context (consecutively correct score), and Tower (total achievement score). The expected mean score is 10. Three informants (parent, teacher, self) completed the Behavior Rating Inventory of Executive Function (Gioia et al. 2000; Guy et al. 2004). For each informant, the General Executive Composite score was analyzed. The expected mean score is 50 (SD, 10), with a higher score indicating less optimal function.
Exome Sequencing
Whole exome sequencing data from 34 of the 115 CHD subjects and their parents were previously reported, as well as 2 CHD subjects without parental data (Jin et al. 2017). In brief, genomic DNA was processed using the Nimblegen v.2 exome capture reagent (Roche) or Nimblegen SeqxCap EZ MedExome Target Enrichment Kit (Roche) followed by Illumina DNA sequencing. Sequence reads were mapped to the reference genome (hg19) and processed using the Genome Analysis Toolkit (GATK) Best Practices workflows. Single nucleotide variants and small indels were called with GATK HaplotypeCaller and annotated using ANNOVAR. The MetaSVM algorithm, annotated using dbNSFP v2.9, was used to predict deleteriousness of missense mutations using software defaults. De novo, rare transmitted loss of function (LOF) and rare homozygous genotypes were identified as previously described (Jin et al. 2017). In brief, GATK pass variants were further filtered for quality by requiring genotype quality ≥20 and mapping quality ≥59. Rare variants were selected for allele frequency <1E-05 in reference populations (1000 Genomes and The Genome Aggregation Database).
Statistical Analysis
Distribution of age at MRI, concurrent family social class (Hollingshead Four-Factor Index of Social Status), and sex ratio between CHD and control groups were assessed using independent two-sample t-tests and Fisher’s exact test. Magnet strength employed in image acquisition did not significantly correlate with sulcal pattern similarity values (P > 0.05). Sulcal pattern similarities to controls from the same scanner were compared with the similarities to other controls from the different scanner using paired t-tests. Sulcal pattern similarities and left–right symmetries between groups were compared for the combined set of all features (sulcal position, area, depth, and graph topology) using independent two-sample t-tests. Multiple testing was accounted for by calculating a false discovery rate (FDR) q-value (Benjamini and Hochberg 1995). If a statistically significant result between groups for the combined set of all features was detected, we compared the sulcal pattern similarities of individual features and locations to evaluate the impact of each feature on similarity measures using independent two-sample t-tests. Violin plots (showing the probability density of the data at different values) were used to visually compare sulcal pattern similarity by hemisphere and lobar region. Z-scores for each CHD subject sulcal pattern similarity measure were calculated by comparing the individual measure to the other CHD subject group values.
Linear regression was used to assess the associations (per SD increase in similarity) between neurodevelopmental outcome measures and sulcal pattern similarity with our control set as well as sulcal pattern similarity between left and right hemispheres, both unadjusted and adjusted for family social class, test type (Wechsler Intelligence Scale for Children [WISC] vs. Wechsler Individual Achievement Test [WIAT]), age at MRI, sex, and field strength of MRI (3 T vs. 1.5 T). An additional analysis was performed to further adjust for CHD surgical status (Norwood vs. non-Norwood).
To determine if differences in sulcal pattern similarity values between CHD and control subjects were confounded by brain volume or overall gyrification, we examined whether there were significant correlations between sulcal pattern measures and gray and white matter volumes, cortical thickness, and gyrification index using Pearson correlation coefficients.
Genes highly expressed in the brain or heart were defined by rank expression percentile > 50 among genes expressed in the E14.5 mouse embryo (Homsy et al. 2015). The overall enrichment in DNVs was calculated by comparing the observed number of de novo mutations across each functional class (synonymous, missense, deleterious missense, LOF) to the expected number under the null mutation model. One-sided t-tests were used to compare the sulcal similarity values between CHD subjects with and without rare LOF variants. Functional enrichment of genes with LOF variants was assessed using ToppGene (Chen et al. 2009).
Results
Cohort
Our study includes 115 subjects with CHD (3 T/1.5 T: 48/67) and 45 controls without CHD (3 T/1.5 T: 29/16). The age at time of imaging was not significantly different between the CHD and control groups (CHD [mean ± SD]: 14.7 ± 2.9 years, control: 15.5 ± 2.4 years; P = 0.14), and both groups showed a similar sex ratio (CHD [male/female]: 47/68, control: 19/26; P = 0.88). The CHD group had a significantly lower family social class (CHD: 48.7 ± 13.8, control: 54.1 ± 8.9; P = 0.004). Among the CHD group, 44 (38%) required the Norwood procedure.
Group Difference of Sulcal Pattern between CHD and Control Subjects
Sulcal Pattern Comparisons Between Subjects
The sulcal pattern similarities to our local control cohort were compared between CHD and control subjects using the combined features of sulcal position, depth, area, and graph topology (Supplementary Fig. 1). CHD subjects had several areas of decreased pattern similarity compared with controls (Fig. 1), including the whole left and whole right hemispheres (left: P = 0.003; right: P = 0.01). Among the lobes of the left hemisphere, all but the occipital lobe had decreased sulcal pattern similarity in CHD. Significant differences in sulcal pattern similarity were also observed in the left hemisphere for all lobes except the occipital lobe at a significance threshold of P < 0.05 (frontal: P = 0.03; temporal: P = 0.007; parietal: P = 0.03) (Fig. 1). In the right hemisphere, CHD subjects had a significant decrease in the pattern similarity compared with controls only in the frontal lobe (P = 0.02). After FDR correction of the P-values, all comparisons remained significant at a q-value < 0.05 except for the decreased sulcal pattern similarity in the left parietal and frontal lobes.
Figure 1.
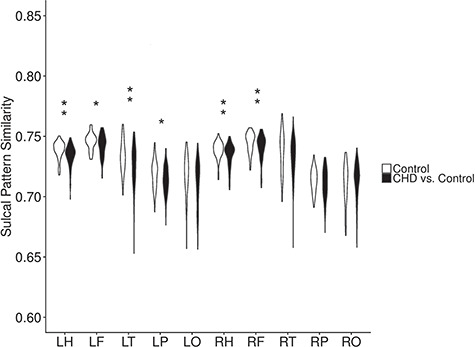
Violin plots showing the distribution of the sulcal pattern similarity among the control group (n = 45) and between CHD (n = 115) and control groups (*P < 0.05, **FDR q < 0.05, F: frontal, H: hemisphere, L: left, O: occipital, P: parietal, R: right, T: temporal).
For the regions with significantly different patterns of the combined features, the sulcal pattern similarities of individual features were also examined (Table 1). Sulcal position and sulcal area patterns were significantly less similar to controls for the left whole hemisphere and left parietal lobe (Table 1). Sulcal position, depth, and area patterns were also significantly different in the left frontal lobe. The CHD group had significantly lower pattern similarity compared with controls in the whole right hemisphere for sulcal depth and area, while having decreased pattern similarity in the right frontal lobe for sulcal area and graph topology (Table 1). However, after FDR correction, there was only significantly decreased similarity for sulcal area within the right frontal lobe, indicating the difference is due to overall sulcal pattern rather than any specific component feature.
Table 1.
Statistical comparisons of the sulcal pattern similarity among the control group (n = 45) and between CHD (n = 115) and control groups for different feature sets
| Location | Sulcal pattern feature | Control | CHD versus control | P value |
|---|---|---|---|---|
| Left hemisphere | Position | 0.7484 ± 0.0103 | 0.7437 ± 0.0118 | 0.02* |
| Depth | 0.7499 ± 0.0069 | 0.7476 ± 0.0066 | 0.05 | |
| Area | 0.9423 ± 0.0033 | 0.9404 ± 0.0042 | 0.007* | |
| Graph topology | 0.8419 ± 0.0102 | 0.8377 ± 0.0149 | 0.08 | |
| Left frontal | Position | 0.7503 ± 0.0088 | 0.7465 ± 0.0113 | 0.04* |
| Depth | 0.7554 ± 0.0084 | 0.7519 ± 0.0098 | 0.04* | |
| Area | 0.9468 ± 0.0039 | 0.9445 ± 0.0054 | 0.01* | |
| Graph topology | 0.8559 ± 0.0126 | 0.8557 ± 0.0158 | 0.94 | |
| Left temporal | Position | 0.7397 ± 0.0143 | 0.7361 ± 0.0131 | 0.14 |
| Depth | 0.7369 ± 0.0152 | 0.7312 ± 0.0192 | 0.08 | |
| Area | 0.9310 ± 0.0075 | 0.9284 ± 0.0088 | 0.08 | |
| Graph topology | 0.8607 ± 0.0181 | 0.8522 ± 0.0285 | 0.07 | |
| Left parietal | Position | 0.7624 ± 0.0101 | 0.7584 ± 0.0109 | 0.03* |
| Depth | 0.7012 ± 0.0142 | 0.6988 ± 0.0128 | 0.30 | |
| Area | 0.9225 ± 0.0080 | 0.9195 ± 0.0086 | 0.04* | |
| Graph topology | 0.8370 ± 0.0199 | 0.8334 ± 0.0217 | 0.33 | |
| Right hemisphere | Position | 0.7489 ± 0.0107 | 0.7450 ± 0.0121 | 0.06 |
| Depth | 0.7508 ± 0.0059 | 0.7485 ± 0.0062 | 0.04* | |
| Area | 0.9433 ± 0.0036 | 0.9415 ± 0.0049 | 0.02* | |
| Graph topology | 0.8396 ± 0.0096 | 0.8364 ± 0.0154 | 0.20 | |
| Right frontal | Position | 0.7490 ± 0.0119 | 0.7472 ± 0.0123 | 0.41 |
| Depth | 0.7519 ± 0.0094 | 0.7500 ± 0.0087 | 0.25 | |
| Area | 0.9482 ± 0.0045 | 0.9452 ± 0.0059 | 0.002** | |
| Graph topology | 0.8597 ± 0.0109 | 0.8527 ± 0.0214 | 0.04* |
Data are presented as mean ± SD, *P < 0.05, **FDR q < 0.05.
Sulcal Pattern Comparison with HCP Controls
Reproducibility of the sulcal pattern analysis was studied using a second cohort of control subjects from the HCP (n = 80). As with our local control cohort, significantly decreased sulcal pattern similarity to the HCP group was observed for CHD subjects using the combined features (Supplementary Fig. 2). CHD subjects had decreased similarity of the sulcal pattern compared with the controls for the left and right whole hemispheres (left: P = 0.03; right: P = 0.05). Decreased sulcal pattern similarity was also observed to be significant for left temporal (P = 0.01) and parietal (P = 0.03) lobar regions but not right lobar regions. Decreased sulcal pattern similarity in CHD subjects remained significant for the left temporal lobe after FDR correction. Decreased sulcal pattern similarity was not observed in the frontal lobes (Supplementary Fig. 2).
When analyzing individual features, the CHD group had decreased similarity in left and right hemispheric sulcal position, depth, and area patterns compared with the control group (Supplementary Table 1). Sulcal area pattern was the only significantly different measure in the parietal lobe, which did not remain significant after FDR correction. No significant differences were found in the temporal lobe in sulcal pattern similarity with individual features.
Sulcal Pattern Left–Right Symmetry
Similarity between the sulcal pattern of the left and right hemispheres was compared between individual CHD or control subjects using the combined features. Hemispheric sulcal pattern symmetry was significantly lower for individuals in the CHD group compared with individuals in the control group (P = 0.05; Fig. 2). This did not remain significant after FDR correction. In lobar analysis, lower sulcal pattern symmetry was found in the temporal (P = 0.02) and parietal (P = 0.01) lobes in CHD subjects compared with controls, both of which remained significant after FDR correction.
Figure 2.
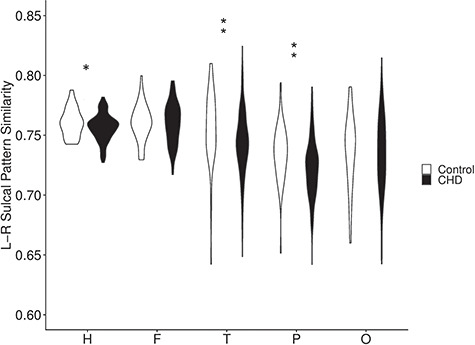
Violin plots showing the distribution of the left–right sulcal pattern symmetry within the control group (n = 45) and within the CHD group (n = 115) (*P < 0.05, **FDR q < 0.05, F: frontal, H: hemisphere, L: left, L-R: left versus right, O: occipital, P: parietal, R: right, T: temporal).
Further analysis assessed similarity between the left and right hemisphere in individual sulcal features (Table 2). Sulcal area pattern was the only sulcal feature with significantly less sulcal symmetry in CHD subjects, both for the overall hemisphere as well as for the temporal and parietal lobes. Only sulcal area pattern for the entire hemisphere remained significant after FDR correction.
Table 2.
Statistical comparisons of the left–right sulcal pattern symmetry within the control group (n = 45) and within the CHD group (n = 115) for different feature sets
| Location | Sulcal pattern feature | Control | CHD | P value |
|---|---|---|---|---|
| Hemisphere | Position | 0.7624 ± 0.0159 | 0.7590 ± 0.0151 | 0.22 |
| Depth | 0.7681 ± 0.0164 | 0.7642 ± 0.0156 | 0.17 | |
| Area | 0.9475 ± 0.0069 | 0.9439 ± 0.0063 | 0.002** | |
| Graph topology | 0.8661 ± 0.0135 | 0.8632 ± 0.0170 | 0.30 | |
| Temporal | Position | 0.7618 ± 0.0348 | 0.7514 ± 0.0310 | 0.07 |
| Depth | 0.7590 ± 0.0385 | 0.7470 ± 0.0383 | 0.08 | |
| Area | 0.9344 ± 0.0161 | 0.9269 ± 0.0186 | 0.02* | |
| Graph topology | 0.8708 ± 0.0439 | 0.8622 ± 0.0447 | 0.27 | |
| Parietal | Position | 0.7501 ± 0.0337 | 0.7418 ± 0.0336 | 0.16 |
| Depth | 0.7201 ± 0.0356 | 0.7084 ± 0.0374 | 0.07 | |
| Area | 0.9227 ± 0.0180 | 0.9132 ± 0.0223 | 0.01* | |
| Graph topology | 0.8733 ± 0.0314 | 0.8648 ± 0.0332 | 0.14 |
Data are presented as mean ± SD, *P < 0.05, **FDR q < 0.05.
Neurodevelopmental Outcomes and Sulcal Analysis
Neuropsychological Testing and Sulcal Pattern Similarity
Neurodevelopmental outcomes were correlated with sulcal patterns similarity values for the regions with significantly lower values in CHD subjects (Fig. 1). CHD subjects generally had lower neuropsychological test scores (Fig. 3a–e). Statistically significant associations were observed between sulcal pattern similarity and neuropsychological testing for all queried regions of the left hemisphere and the right frontal lobe in unadjusted analyses (Table 3; Supplementary Table 2). Associations between the left frontal sulcal pattern similarity values with working memory and processing speed remained significant after adjusting for family social class, test type, age at MRI, sex, and field strength (Table 3, Supplementary Table 3), as well as after further adjusting for whether or not the subject had a Norwood procedure (Table 3, Supplementary Table 4). Left temporal sulcal pattern similarity persisted in association with executive functioning (P = 0.02) and right frontal sulcal pattern similarity correlated with processing speed (P = 0.02) after full adjustment. All correlations remained significant after FDR correction in the fully adjusted model. Within the control cohort, no neurodevelopmental outcomes were significant correlated with sulcal pattern similarity.
Figure 3.
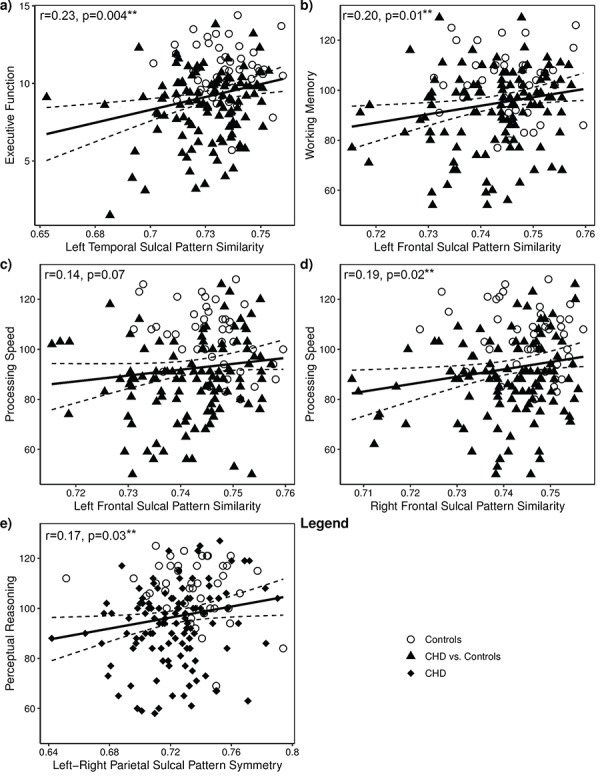
Unadjusted association between neurodevelopmental outcomes and sulcal pattern similarity for the combined sample (n = 160) of control subjects (n = 45; open circles) and CHD subjects (n = 115; black triangles). Unadjusted Pearson correlation coefficients and corresponding P values are shown. (a) Left temporal sulcal pattern similarity and executive functioning; (b) left frontal sulcal pattern similarity and working memory; (c) left frontal sulcal pattern similarity and processing speed; (d) right frontal sulcal pattern similarity and processing speed; and (e) parietal sulcal pattern symmetry and perceptual reasoning (**FDR q < 0.05.).
Table 3.
Association between neurodevelopmental outcomes and sulcal pattern measures of similarity or symmetry in the total sample (n = 160) using 3 linear regression models (per SD increase in measure)
| Association | Measure SD * | Unadjusted model 1 | Adjusted model 2 | Fully adjusted model 3 |
|---|---|---|---|---|
| Executive function versus L temporal sulcal pattern similarity | 0.0163 | 0.54 ± 0.19 (0.004**) | 0.47 ± 0.18 (0.01**) | 0.48 ± 0.20 (0.02**) |
| Working memory versus L frontal sulcal pattern similarity | 0.0086 | 3.18 ± 1.28 (0.01**) | 2.93 ± 1.27 (0.02**) | 3.26 ± 1.35 (0.02**) |
| Processing speed versus L frontal sulcal pattern similarity | 0.0086 | 2.50 ± 1.39 (0.07) | 3.09 ± 1.37 (0.03**) | 3.38 ± 1.40 (0.02**) |
| Processing speed versus R frontal sulcal pattern similarity | 0.0099 | 3.25 ± 1.38 (0.02**) | 4.16 ± 1.37 (0.003**) | 3.49 ± 1.43 (0.02**) |
| Perceptual reasoning versus L-R parietal sulcal pattern symmetry | 0.0254 | 2.80 ± 1.27 (0.03**) | 3.21 ± 1.19 (0.008**) | 3.09 ± 1.36 (0.03**) |
Model 1 is unadjusted; model 2 includes adjustment for family social class, test type (WISC vs. WIAT), age at MRI, sex, and field strength (3 T vs. 1.5 T); and model 3 was further adjusted for CHD surgical status (CHD Norwood vs. CHD non-Norwood vs. control). Data are presented as β ± standard error (P value). L: left, L-R: left versus right, R: right.
*SD refers to standard deviation of sulcal pattern similarity for the total sample, ** FDR q < 0.05.
Neuropsychological Testing and Sulcal Pattern Symmetry
Associations between left–right sulcal pattern symmetry and neuropsychological test scores were examined for the temporal and parietal regions, which had significantly decreased symmetry in CHD subjects (Table 2). A significant association was observed for lower parietal sulcal pattern symmetry and lower perceptual reasoning scores, as well as for lower temporal sulcal pattern symmetry and lower Full-Scale IQ, verbal comprehension, processing speed, and executive functioning in the unadjusted model (Supplementary Table 2). The association between lower parietal sulcal pattern symmetry and lower perceptual reasoning persisted after partial or full adjustment for covariates (P = 0.008 for partially adjusted and P = 0.03 for fully adjusted model; Fig. 3e, Table 3, Supplementary Tables 3–4). Adjustment resulted in a stronger association of sulcal pattern symmetry with lower perceptual reasoning (β ± SD: 2.80 ± 1.27 vs. 3.09 ± 1.36; Table 3). All correlations remained significant after FDR correction in the fully adjusted model but not for the unadjusted or partially adjusted models. No neurodevelopmental outcomes were significantly correlated with sulcal pattern symmetry for the control cohort.
Relationship between Sulcal Pattern Measures and Other MRI Measures
All brain regions with significantly decreased sulcal pattern similarity among CHD subjects were studied for correlation with other MRI measures. Gyrification index was negatively correlated with parietal lobe sulcal pattern symmetry bilaterally (correlation coefficient r = −0.29, P = 0.002 on left; r = −0.33, P = 0.001 on right), which remained significant after FDR correction of the P-value (Supplementary Table 5). However, gyrification index did not decrease the correlation between sulcal pattern measures and neurodevelopmental outcomes. The other nominally significant associations were between left hemispheric white volume and whole left hemispheric sulcal pattern similarity, and between left hemispheric cortical thickness and parietal lobe sulcal pattern symmetry but the FDR q-value was > 0.05.
Genetic Variants in Subjects with Atypical Sulcal Patterns
Previously published assessment of copy number variants (CNVs) among 71 patients in the cohort was examined for enrichment among patients having sulcal pattern similarity below the average among CHD subjects the left frontal, right frontal, and left parietal regions, as well as those having below average left–right parietal asymmetry (Bellinger et al. 2015). Presence of a CNV was not associated with below-average sulcal pattern similarity (P = 0.20–0.43 for all regions).
Thirty-four probands had trio (proband + parents) exome sequencing results available, with 30 rare DNVs identified by comparison with parental sequencing (Supplementary Data 1). Twenty-four of the DNVs were synonymous or missense variants not predicted to affect protein function. There were LOF DNVs in AKAP12 and SRRM2, as well as missense DNVs predicted to be damaging in KLF2, DSCAML1, COLGALT1, and DGKA. Three of those genes, AKAP12, SRRM2, and COLGATL1, are highly expressed in the developing heart and brain (HHB). Z-scores for sulcal pattern similarity measures for individual CHD subjects compared with the overall CHD cohort were reviewed for the 6 subjects with damaging DNVs. The CHD subject with the SRRM2 LOF DNV had markedly abnormal sulcal patterning, with z-score <−1 for sulcal pattern similarity in the right and left hemispheres overall, left frontal lobe, left temporal lobe, and right and left occipital lobes. Subjects with COLGALT1 and DGKA damaging missense DNVs had z-scores <−1 for two regions (right and left temporal lobes; and left temporal and occipital lobes, respectively). Compared with subjects without damaging DNVs, the 5 subjects with damaging DNVs had a trend toward decreased sulcal pattern similarity in the left temporal lobe (mean 0.725 vs. 0.710, P = 0.05) but not other regions. All 6 of the damaging DNVs occurred in the 32 subjects with at least 1 left cortical lobar region with sulcal pattern similarity below the mean value for the group using the local controls or HCP cohort for comparison, and none were present in the 2 subjects that had no left cortical areas of below-average sulcal pattern similarity. In subjects with lower-than-average sulcal pattern symmetry in at least 1 region, there was a 2.56-fold enrichment in DNVs compared with expected number of damaging DNVs in genes highly expressed in the developing heart based on cohort size and sequencing areas (P = 0.18). This suggests an enrichment of DNVs in CHD subjects with lower symmetry in their sulcal patterns. This enrichment was not present when grouped by the presence of right regional similarity below average in any area nor when grouped by left–right asymmetry in pattern similarity.
There were 130 rare DNV or inherited LOF variants identified in the 36 CHD subjects with trio or nontrio exome sequencing data available, of which 57 were in HHB genes (Supplementary Data 2). CHD subjects with rare LOF variants in HHB genes had trend toward lower sulcal pattern similarity in the left hemisphere overall (mean 0.736 vs. 0.731, P = 0.05) and the right frontal lobe (mean 0.747 vs. 0.740, P = 0.05) but not other regions. Within the 129 unique genes with rare transmitted LOF variants in the group with lower than average similarity compared with HCP controls, there was no enrichment for biological function, biological process, or cellular compartment when compared with the overall list of genes with LOF variants in the larger cohort.
Discussion
Here, we describe the first sulcal pattern analysis of adolescent subjects with single-ventricle CHD and demonstrate important correlations between sulcal pattern and neurodevelopmental outcomes and variants in genes expressed during development. In CHD subjects, the bilateral hemispheric sulcal patterns had reduced similarity compared with control subjects. The left hemisphere demonstrated more atypical sulcal patterns than the right, with the temporal and parietal lobes most consistently affected. When evaluating individual sulcal features, sulcal area was only major feature with significant differences in the sulcal patterns in our CHD cohort, indicating that the differences in sulcal pattern seem to be driven by the composite measure of sulcal pattern rather than any individual feature. Our novel findings extend previous reports of leftward-biased early abnormalities in sulcal folding in fetuses and infants with CHD (Ortinau et al. 2013; Ortinau et al. 2018) to show that similar differences exist in adolescents with CHD with relevant neurodevelopmental and genetic associations.
Atypical Sulcal Patterns and Neurodevelopment in Patients with CHD
Altered sulcal area patterns in CHD may result from variants in developmentally expressed genes that cause early abnormalities in organization and arrangement of cortical functional areas and their connectivity (McQuillen et al. 2010; Homsy et al. 2015; Khalil et al. 2016). Our recent study found atypical global patterns in the earliest emerging sulci in CHD fetuses using the same sulcal pattern comparison approach (Ortinau et al. 2018). Our adolescent study suggests that the atypical sulcal pattern we observed here may originate in fetal life and persist long after birth.
Correlations have been previously demonstrated between brain MRI measures and neurodevelopment in patients with CHD. These correlations were not affected by other MRI measures such as volume, cortical thickness, or gyrification index. Mathematical and visual–spatial skills, as well as executive function, have been shown to correlate with reduced regional white matter microstructure in 16-year-old subjects with d-transposition of the great arteries, as measured by fractional anisotropy (Rollins et al. 2014). Specifically, decreased white matter microstructure anisotropy in the right frontal region was correlated with poorer visual–spatial skills, and decreased left parietal white matter microstructure was correlated with poorer mathematical skills and increased inattention/hyperactivity. Notably, these regions were also found to have the most significant and consistent differences in sulcal pattern among the subjects with single-ventricle physiology included in the current study. The consistency of abnormal brain structure in patients with transposition of the great arteries and hypoplastic left heart syndrome argues that features common to both cohorts, such as hypoxia and genetic factors, are likely to contribute. As genetic background is known to influence sulcal patterning, and most CHD genes are known to cause several phenotypes, genetic variants are likely to impact sulcal patterning among patients with CHD (Im et al. 2011b; Jin et al. 2017; Le Guen et al. 2018). However, larger studies are needed to systematically determine if the genetic risks are different between the two types of CHD.
It is interesting that among the CHD cohort, bilateral frontal sulcal pattern similarity was correlated with processing speed, while left frontal sulcal pattern alone correlated with working memory. Processing speed has previously been associated with white matter volume, but not with a specific brain region (Magistro et al. 2015). Working memory has been previously correlated with frontal lobe activity (Christodoulou et al. 2001). Thus, sulcal pattern similarity may serve as a sensitive and specific measure of neuroanatomical effects on function.
High Asymmetry of Sulcal Patterns in CHD
Hemispheric asymmetries of cortical structure are common. Left–right asymmetries in cortical folding pattern during normal development have been identified in several regions such as superior and inferior temporal, inferior frontal, and intraparietal regions (Blanton et al. 2001; Ochiai et al. 2004). In particular, the temporal lobe asymmetries arise during typical fetal brain development (Kasprian et al. 2011; Habas et al. 2012). However, among our CHD cohort, we observed increased sulcal pattern asymmetry due to increased sulcal pattern disruption in the left hemisphere, particularly the temporal and parietal lobes. Our finding of sulcal pattern abnormalities primarily in the left hemisphere may suggest amplification of the typical asymmetric cortical development. Similar findings were reported among fetuses and infants with CHD, where abnormalities of sulcal folding depth and global pattern were also found in more left-sided regions than right-sided regions (Ortinau et al. 2013; Ortinau et al. 2018). Expanding sulcal analysis to other forms of CHD with differing effects on fetal cerebral hemodynamics will clarify the contribution of perfusion abnormalities to changes in left hemispheric sulcal patterns. Further, as the molecular basis for the development of structural left–right asymmetry of the brain is poorly understood, studying subjects with CHD where such asymmetries are perturbed could provide insight into the molecular and physiologic mechanisms regulating asymmetric brain development in humans.
Association between Damaging Genetic Variants and Sulcal Patterns in CHD
Many genetic factors are known to influence left–right patterning during embryonic development, though specific understanding of the genetic determinants of sulcal patterning in humans remains incomplete (Levin 2005; Roussigné et al. 2012; Le Guen et al. 2018). Previous analysis of this cohort by comparative genomic hybridization identified likely pathogenic CNVs in just 7% of subjects (Bellinger et al. 2015). Only 6 CHD subjects had damaging DNVs, but two affected genes that are expressed in the fetal brain and/or heart (AKAP12, DSCAML1). Dysregulation of genes co-expressed in the developing brain and heart could directly or indirectly affect cerebral patterning, blood flow, and metabolism leading to alterations in sulcal patterns (Rajagopalan et al. 2011). However, larger studies are needed to rigorously assess the correlation between rare, damaging genetic variants and sulcal pattern abnormalities in CHD.
Limitations of the current study include a single developmental time point and genetic analysis available for only a subset of subjects. With a larger sample size and increased genetic characterization, we may be able to detect correlations between genetic variants and degree or location of sulcal pattern asymmetry. Testing over several points of development would help disentangle prenatal from postnatal effects on sulcal features. Strengths include a relatively homogeneous cohort of subjects with CHD who were cared for at a single center and all required a Fontan procedure early in life. However, they are heterogeneous for operative course, degree of extracardiac disease, and anesthetic exposure.
In conclusion, we have identified significant differences in sulcal patterning among school-aged subjects with CHD who had undergone a Fontan palliative procedure compared with controls. The left hemisphere was more significantly affected, consistent with observations in patients with d-transposition of the great arteries, raising the possible role of genetic influences on sulcal pattern formation as these two cohorts share both fetal cerebral hypoxia and similar genetic risks. These differences in sulcal pattern similarity are correlated with meaningful neurodevelopmental outcomes including memory and executive function. Finally, damaging DNVs were identified only among CHD subjects with greater than average sulcal pattern abnormalities in at least one region. These findings, together with our observation of similar sulcal pattern differences in fetuses with CHD, suggest that sulcal pattern abnormalities develop in fetal life and persist until adolescence with relevant associations to neurodevelopmental outcomes. The association of abnormal sulcal patterns with damaging DNVs suggest that such variants influence in utero brain development. Future longitudinal studies or a larger cohort beginning in utero is needed to confirm these findings. If confirmed, such measures of sulcal patterning may serve as early markers of those at risk for neurodevelopmental complications and of success of interventions.
Supplementary Material
Funding
Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (R21HD083956 to K.I.); National Institute of Biomedical Imaging and Bioengineering (R01-HD065762, U01-HD087211, and R01EB017337 to P.E.G.); National Heart, Lung and Blood Institute R01HL096825; Pediatric Cardiac Genomics Consortium (U01-HL098188, U01-HL098147, U01-HL098153, U01-HL098163, U01-HL098123, and U01-HL098162); Howard Hughes Medical Institute (to C.E.S.).
Notes
The authors are enormously grateful to the patients and families who participated in this research. Conflict of Interest: None declared.
References
- Baron IS. 2004. Delis–Kaplan Executive Function System. Child Neuropsychol. San Antonio, TX: Psychological Assessment Resources Inc; 10(2):147–152. [Google Scholar]
- Beca J, Gunn JK, Coleman L, Hope A, Reed PW, Hunt RW, Finucane K, Brizard C, Dance B, Shekerdemian LS. 2013. New white matter brain injury after infant heart surgery is associated with diagnostic group and the use of circulatory arrest. Circulation. 127:971–979. [DOI] [PubMed] [Google Scholar]
- Bellinger DC, Watson CG, Rivkin MJ, Robertson RL, Roberts AE, Stopp C, Dunbar-Masterson C, Bernson D, DeMaso DR, Wypij D. 2015. Neuropsychological status and structural brain imaging in adolescents with single ventricle who underwent the Fontan procedure. J Am Heart Assoc. 4:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger DC, Wypij D, Rivkin MJ, Demaso DR, Robertson RL, Dunbar-Masterson C, Rappaport LA, Wernovsky G, Jonas RA, Newburger JW. 2011. Adolescents with d-transposition of the great arteries corrected with the arterial switch procedure: neuropsychological assessment and structural brain imaging. Circulation. 124:1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. . [Google Scholar]
- Blanton RE, Levitt JG, Thompson PM, Narr KL, Capetillo-Cunliffe L, Nobel A, Singerman JD, McCracken JT, Toga AW. 2001. Mapping cortical asymmetry and complexity patterns in normal children. Psychiatry Res Neuroimaging. 107:29–43. [DOI] [PubMed] [Google Scholar]
- Cachia A, Borst G, Tissier C, Fisher C, Plaze M, Gay O, Rivière D, Gogtay N, Giedd J, Mangin JF. 2016. Longitudinal stability of the folding pattern of the anterior cingulate cortex during development. Dev Cogn Neurosci. 19:122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37:W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JG, Dooling EC, Gilles FH. 1977. Gyral development of the human brain. Ann Neurol. 1:86–93. [DOI] [PubMed] [Google Scholar]
- Christodoulou C, DeLuca J, Ricker JH, Madigan NK, Bly BM, Lange G, Kalnin AJ, Liu WC, Steffener J, Diamond BJ. 2001. Functional magnetic resonance imaging of working memory impairment after traumatic brain injury. J Neurol Neurosurg Psychiatry. 71:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouchoux C, du Plessis AJ, Bouyssi-Kobar M, Tworetzky W, McElhinney DB, Brown DW, Gholipour A, Kudelski D, Warfield SK, McCarter RJ. 2013. Delayed cortical development in fetuses with complex congenital heart disease. Cereb Cortex. 23:2932–2943. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 9:179–194. [DOI] [PubMed] [Google Scholar]
- De Asis-Cruz J, Donofrio MT, Vezina G, Limperopoulos C. 2018. Aberrant brain functional connectivity in newborns with congenital heart disease before cardiac surgery. Neuroimage Clin. 17:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan Romero C, Bruder C, Tomasello U, Sanz-Anquela JM, Borrell V. 2015. Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 34:1859–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis DC, Kaplan B, Kramer JH. 2001. Delis-Kaplan Executive Function System. San Antonio, TX: The Psychological Corporation. [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gryal based regions of interest. Neuroimage. 31:968–980. [DOI] [PubMed] [Google Scholar]
- Evans AC. 2006. The NIH MRI study of normal brain development. Neuroimage. 30:184–202. [DOI] [PubMed] [Google Scholar]
- Fischl B. 2012. FreeSurfer. Neuroimage. 62(2):774–781 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BTT, Mohlberg H, Amunts K, Zilles K. 2008. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 18:1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. Neuroimage. 9:195–207. [DOI] [PubMed] [Google Scholar]
- Fischl B, Van Der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D. 2004. Automatically parcellating the human cerebral cortex. Cereb Cortex. 14:11–22. [DOI] [PubMed] [Google Scholar]
- Fitzgerald TW, Gerety SS, Jones WD, Van Kogelenberg M, King DA, McRae J, Morley KI, Parthiban V, Al-Turki S, Ambridge K. 2015. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 519:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel C, Chantrel E, Brisse H, Elmaleh M, Luton D, Oury JF, Sebag G, Hassan M. 2001. Fetal cerebral cortex: normal gestational landmarks identified using prenatal MR imaging. Am J Neuroradiol. 22:184–189. [PMC free article] [PubMed] [Google Scholar]
- Gaynor JW, Ittenbach RF, Gerdes M, Bernbaum J, Clancy RR, McDonald-Mcginn DM, Zackai EH, Wernovsky G, Nicolson SC, Spray TL. 2014. Neurodevelopmental outcomes in preschool survivors of the Fontan procedure. J Thorac Cardiovasc Surg. 147:1276–1283.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia GA, Isquith PK, Guy SC, Kenworthy L. 2000. Behavior Rating Inventory of Executive Function. Odessa, FL: Psychological Assessment Resources Inc. [Google Scholar]
- Goldberg CS, Schwartz EM, Brunberg JA, Mosca RS, Bove EL, Schork MA, Stetz SP, Cheatham JP, Kulik TJ, Goldberg CS. 2000. Neurodevelopmental outcome of patients after the Fontan operation: a comparison between children with hypoplastic left heart syndrome and other functional single ventricle lesions. J Pediatr. 137:646–652. [DOI] [PubMed] [Google Scholar]
- Guy SC, Isquith PK, Gioia GA. 2004. Behavior Rating Inventory of Executive Function. Self-Report Version. Odessa, FL: Psychological Assessment Resources Inc. [Google Scholar]
- Habas PA, Scott JA, Roosta A, Rajagopalan V, Kim K, Rousseau F, Barkovich AJ, Glenn OA, Studholme C. 2012. Early folding patterns and asymmetries of the normal human brain detected from in utero MRI. Cereb Cortex. 22:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. 2010. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci U S A. 107:13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J. 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 350:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Choi YY, Yang JJ, Lee KH, Kim SI, Grant PE, Lee JM. 2011a. The relationship between the presence of sulcal pits and intelligence in human brains. Neuroimage. 55:1490–1496. [DOI] [PubMed] [Google Scholar]
- Im K, Pienaar R, Lee JM, Seong JK, Choi YY, Lee KH, Grant PE. 2011b. Quantitative comparison and analysis of sulcal patterns using sulcal graph matching: a twin study. Neuroimage. 57:1077–1086. [DOI] [PubMed] [Google Scholar]
- Im K, Grant PE. 2018. Sulcal pits and patterns in developing human brains. Neuroimage. 185:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Guimaraes A, Kim Y, Cottrill E, Gagoski B, Rollins C, Ortinau C, Yang E, Grant PE. 2017. Quantitative folding pattern analysis of early primary sulci in human fetuses with brain abnormalities. Am J Neuroradiol. 38:1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Jo HJ, Mangin JF, Evans AC, Kim SI, Lee JM. 2010. Spatial distribution of deep sulcal landmarks and hemispherical asymmetry on the cortical surface. Cereb Cortex. 20:602–611. [DOI] [PubMed] [Google Scholar]
- Im K, Pienaar R, Paldino MJ, Gaab N, Galaburda AM, Grant PE. 2013. Quantification and discrimination of abnormal sulcal patterns in polymicrogyria. Cereb Cortex. 23:3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K, Raschle NM, Smith SA, Ellen Grant P, Gaab N. 2016. Atypical sulcal pattern in children with developmental dyslexia and at-risk kindergarteners. Cereb Cortex. 26:1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising M, Holsboer F. 2006. Genetics of stress response and stress-related disorders. Dialogues Clin Neurosci. 8:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, Depalma SR, Zeng X, Qi H, Chang W, Sierant MC. 2017. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 49:1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmacharya S, Gagoski B, Ning L, Vyas R, Cheng HH, Soul J, Newberger JW, Shenton ME, Rathi Y, Grant PE. 2018. Advanced diffusion imaging for assessing normal white matter development in neonates and characterizing aberrant development in congenital heart disease. Neuroimage Clin. 19:360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprian G, Langs G, Brugger PC, Bittner M, Weber M, Arantes M, Prayer D. 2011. The prenatal origin of hemispheric asymmetry: an in utero neuroimaging study. Cereb Cortex. 21:1076–1083. [DOI] [PubMed] [Google Scholar]
- Khalil A, Bennet S, Thilaganathan B, Paladini D, Griffiths P, Carvalho JS. 2016. Prevalence of prenatal brain abnormalities in fetuses with congenital heart disease: a systematic review. Ultrasound Obstet Gynecol. 48:296–307. [DOI] [PubMed] [Google Scholar]
- Klyachko VA, Stevens CF. 2003. Connectivity optimization and the positioning of cortical areas. Proc Natl Acad Sci USA. 100:7937–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostović I, Vasung L. 2009. Insights from in vitro fetal magnetic resonance imaging of cerebral development. Semin Perinatol. 33:220–233. [DOI] [PubMed] [Google Scholar]
- Le Guen Y, Auzias G, Leroy F, Noulhiane M, Dehaene-Lambertz G, Duchesnay E, Mangin J-F, Coulon O, Frouin V. 2018. Genetic influence on the Sulcal pits: on the origin of the first cortical folds. Cereb Cortex. 28:1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leordeanu M, Hebert M. 2005. A spectral technique for correspondence problems using pairwise constraints. Proceedings of the IEEE International Conference on Computer Vision. 1482–1489. [Google Scholar]
- Levin M. 2005. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 122:3–25. [DOI] [PubMed] [Google Scholar]
- Licht DJ, Shera DM, Clancy RR, Wernovsky G, Montenegro LM, Nicolson SC, Zimmerman RA, Spray TL, Gaynor JW, Vossough A. 2009. Brain maturation is delayed in infants with complex congenital heart defects. J Thorac Cardiovasc Surg. 137:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limperopoulos C, Tworetzky W, McElhinney DB, Newburger JW, Brown DW, Robertson RL, Guizard N, McGrath E, Geva J, Annese D. 2010. Brain volume and metabolism in fetuses with congenital heart disease: evaluation with quantitative magnetic resonance imaging and spectroscopy. Circulation. 121:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann G, Von Cramon DY. 2000. Automatic labelling of the human cortical surface using sulcal basins. Med Image Anal. 4:179–188. [DOI] [PubMed] [Google Scholar]
- Magistro D, Takeuchi H, Nejad KK, Taki Y, Sekiguchi A, Nouchi R, Kotozaki Y, Nakagawa S, Miyauchi CM, Iizuka K. 2015. The relationship between processing speed and regional white matter volume in healthy young people. PLoS One. 10: e0136386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahle WT, Tavani F, Zimmerman RA, Nicolson SC, Galli KK, Gaynor JW, Clancy RR, Montenegro LM, Spray TL, Chiavacci RM. 2002. An MRI study of neurological injury before and after congenital heart surgery. Circulation. 106:I109–I114. [PubMed] [Google Scholar]
- Masoller N, Sanz-Cortés M, Crispi F, Gómez O, Bennasar M, Egaña-Ugrinovic G, Bargalló N, Martínez JM, Gratacós E. 2016. Severity of fetal brain abnormalities in congenital heart disease in relation to the main expected pattern of in utero brain blood supply. Fetal Diagn Ther. 39:269–278. [DOI] [PubMed] [Google Scholar]
- McQuillen PS, Goff DA, Licht DJ. 2010. Effects of congenital heart disease on brain development. Prog Pediatr Cardiol. 29:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebius MJ, Kooi EMWW, Bilardo CM, Bos AF. 2017. Brain injury and neurodevelopmental outcome in congenital heart disease: a systematic review. Pediatrics. 140:e20164055. [DOI] [PubMed] [Google Scholar]
- Meng Y, Li G, Lin W, Gilmore JH, Shen D. 2014. Spatial distribution and longitudinal development of deep cortical sulcal landmarks in infants. Neuroimage. 100:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SP, McQuillen PS, Hamrick S, Xu D, Glidden DV, Charlton N, Karl T, Azakie A, Ferriero DM, Barkovich AJ. 2007. Abnormal brain development in Newborns with congenital heart disease. N Engl J Med. 357:1928–1938. [DOI] [PubMed] [Google Scholar]
- Ochiai T, Grimault S, Scavarda D, Roch G, Hori T, Rivière D, Mangin JF, Régis J. 2004. Sulcal pattern and morphology of the superior temporal sulcus. Neuroimage. 22:706–719. [DOI] [PubMed] [Google Scholar]
- Ortinau C, Rollins C, Gholipour A, Yun H, Marshall M, Gagoski B, Afacan O, Friedman K, Tworetzky W, Warfield S. 2018. Early emerging sulcal patterns are atypical in fetuses with congenital heart disease. Cereb Cortex. doi: 10.1093/cercor/bhy235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinau C, Alexopoulos D, Dierker D, Van Essen D, Beca J, Inder T. 2013. Cortical folding is altered before surgery in infants with congenital heart disease. J Pediatr. 163:1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinau C, Beca J, Lambeth J, Ferdman B, Alexopoulos D, Shimony JS, Wallendorf M, Neil J, Inder T. 2012. Regional alterations in cerebral growth exist preoperatively in infants with congenital heart disease. J Thorac Cardiovasc Surg. 143:1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychological Corporation 2003. WISC-IV. Psychological Corp.
- Rajagopalan V, Scott J, Habas PA, Kim K, Corbett-Detig J, Rousseau F, Barkovich AJ, Glenn OA, Studholme C. 2011. Local tissue growth patterns underlying normal fetal human brain gyrification quantified in utero. J Neurosci. 31:2878–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 2004. Genetic control of cortical convolutions. Science. 303:1983–1984. [DOI] [PubMed] [Google Scholar]
- Rash BG, Rakic P. 2014. Genetic resolutions of brain convolutions. Science. 343:744–745. [DOI] [PubMed] [Google Scholar]
- Rollins CK, Asaro LA, Akhondi-Asl A, Kussman BD, Rivkin MJ, Bellinger DC, Warfield SK, Wypij D, Newburger JW, Soul JS. 2017. White matter volume predicts language development in congenital heart disease. J Pediatr. 181:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins CK, Watson CG, Asaro LA, Wypij D, Vajapeyam S, Bellinger DC, Demaso DR, Robertson RL, Newburger JW, Rivkin MJ. 2014. White matter microstructure and cognition in adolescents with congenital heart disease. J Pediatr. 165:936–944.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigné M, Blader P, Wilson SW. 2012. Breaking symmetry: the zebrafish as a model for understanding left-right asymmetry in the developing brain. Dev Neurobiol. 72:269–281. [DOI] [PubMed] [Google Scholar]
- Schmithorst VJ, Votava-Smith JK, Tran N, Kim R, Lee V, Ceschin R, Lai H, Johnson JA, De Toledo JS, Blüml S. 2018. Structural network topology correlates of microstructural brain dysmaturation in term infants with congenital heart disease. Hum Brain Mapp. 39:4593–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V, Tabbutt S, Dimitropoulos A, Harris KC, Chau V, Poskitt K, Campbell A, Azakie A, Xu D, Barkovich AJ. 2013. Single-ventricle anatomy predicts delayed microstructural brain development. Pediatr Res. 73:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Hevner RF. 2014. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 15:217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarui T, Madan N, Farhat N, Kitano R, Ceren Tanritanir A, Graham G, Gagoski B, Craig A, Rollins CK, Ortinau C. 2018. Disorganized patterns of sulcal position in fetal brains with agenesis of corpus callosum. Cereb Cortex. 28:3192–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. 1997. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 385:313–318. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW. 2012. The Human Connectome Project: a data acquisition perspective. Neuroimage. 62:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernovsky G. 2012. Neurodevelopmental Outcomes in Children with Congenital Heart Disease. Avery’s Dis Newborn (Ninth Ed. 126). [DOI] [PMC free article] [PubMed]
- White T, Su S, Schmidt M, Kao CY, Sapiro G. 2010. The development of gyrification in childhood and adolescence. Brain Cogn. 72:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Armstrong E, Schleicher A, Kretschmann HJ. 1988. The human pattern of gyrification in the cerebral cortex. Anat Embryol (Berl). 179:173–179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


