Abstract
Background
Genome-wide association studies have identified common genetic risk variants in many loci associated with multiple cancers. We sought to systematically evaluate the utility of these risk variants in identifying high-risk individuals for eight common cancers.
Methods
We constructed polygenic risk scores (PRS) using genome-wide association studies–identified risk variants for each cancer. Using data from 400 812 participants of European descent in a population-based cohort study, UK Biobank, we estimated hazard ratios associated with PRS using Cox proportional hazard models and evaluated the performance of the PRS in cancer risk prediction and their ability to identify individuals at more than a twofold elevated risk, a risk level comparable to a moderate-penetrance mutation in known cancer predisposition genes.
Results
During a median follow-up of 5.8 years, 14 584 incident case patients of cancers were identified (ranging from 358 epithelial ovarian cancer case patients to 4430 prostate cancer case patients). Compared with those at an average risk, individuals among the highest 5% of the PRS had a two- to threefold elevated risk for cancer of the prostate, breast, pancreas, colorectal, or ovary, and an approximately 1.5-fold elevated risk of cancer of the lung, bladder, or kidney. The areas under the curve ranged from 0.567 to 0.662. Using PRS, 40.4% of the study participants can be classified as having more than a twofold elevated risk for at least one site-specific cancer.
Conclusions
A large proportion of the general population can be identified at an elevated cancer risk by PRS, supporting the potential clinical utility of PRS for personalized cancer risk prediction.
Globally, cancer is the second leading cause of death, following cardiovascular diseases. It is estimated that approximately 18.1 million cancer case patients were diagnosed and 9.6 million individuals died of cancer in 2018 worldwide (1). Over the years, a large number of deleterious germline mutations have been identified in cancer susceptibility genes (2-5). Among them, high-penetrance mutations have been associated with notably elevated cancer risk, but they explain only a small fraction of cancer events, because of their extremely low prevalence in the general population. For example, carriers of deleterious mutations in the BRCA1 gene are estimated to have around an 80% lifetime risk of developing breast cancer (6,7), but only 0.24% of women in the general population carry BRCA1 pathogenetic mutations (8). More recently, moderate-penetrance mutations have been identified in multiple cancer predisposition genes. Most of them are associated with a two- to threefold elevated risk of cancer, and some have been included in genetic testing (5,9). For example, the CHEK2 mutation 1100delC is associated with about a twofold increased risk of breast or colorectal cancer, and deleterious ATM mutations are associated with a two- to threefold elevated risk of breast cancer (10-12). However, the frequency of these mutations is low in the general population (0.71% for CHEK2 1100delC, 1% to 2% for deleterious ATM mutations) (11,13).
Since 2005, genome-wide association studies (GWAS) have identified a large number of common genetic variants in relation to multiple site-specific cancers. Although the risk associated with each variant is small, individuals who carry multiple risk variants can be at a considerably elevated risk for the disease (14). Polygenic risk scores (PRS) can be used to summarize the combined effect of multiple variants to identify individuals at a high risk of site-specific cancer (15). Several studies have investigated the utility of PRS in risk prediction and stratification, but they typically study only one cancer at a time (16-18). Furthermore, many of the previous studies were limited by a small sample size. Herein, we evaluated the utility of PRS in predicting cancer risk for eight site-specific cancers (prostate, breast, pancreas, colorectal, kidney, bladder, lung, and ovary), which account for approximately 59% of the estimated number of all new cancer case patients that will be diagnosed in the United States in 2019 (19). Furthermore, we sought to estimate the percentage of individuals in the general population who can be predicted to have an at least twofold elevated risk of cancer, a risk level comparable to the risk associated with many moderate-penetrance mutations in known cancer predisposition genes that are currently included in clinical genetic testing.
Methods
Study Subjects, Genotype, and Imputation
The UK Biobank is a population-based cohort study, which has recruited more than 500 000 adults across England, Scotland, and Wales. The design and methods of the UK Biobank study have been previously described (20). Data and diagnoses on site-specific incident cancers were provided by the National Health Service Information Centre for participants from England and Wales (follow-up through March 31, 2016) and by the NHS Central Register Scotland for participants from Scotland (follow-up through October 31, 2015). Cancers were coded by the International Classification of Diseases, Ninth Revision (ICD-9) or the International Classification of Diseases, Tenth Revision (ICD-10). Histological subtypes were classified according to the International Classification of Diseases for Oncology (ICD-O). Included in this study are investigations of the following cancers: prostate (ICD-9 = 185 or ICD-10 = C61), breast (ICD-9 = 174 or ICD-10 = C50), pancreas (ICD-9 = 157 or ICD-10 = C25), colorectal (ICD-9 = 153, 154.1 or ICD-10 = C18, C20), kidney (ICD-9 = 189.0 or ICD-10 = C64), bladder (ICD-9 = 188 or ICD-10 = C67), lung (ICD-9 = 162.2–162.9 or ICD-10 = C34), and epithelial ovary (ICD-9 = 183.0 or ICD-10 = C56; ICD-O: 8441, 8460, 8462, 8380, 8381, 8470, 8471, 8472, 8473, 8480, 8310, 8140, 8260, 8440, 8450, 9000, 8000, and 8010) (21). Epithelial ovarian cancer was selected because it was the most common ovarian cancer, and the vast majority of risk variants identified to date for ovarian cancer is from GWAS restricting to epithelial ovarian cancer.
Imputation data of 488 377 participants were acquired from UK Biobank. Samples were genotyped using two arrays sharing a 95% marker content, the UK BiLEVE Axiom (UKBL; 807 411 markers) and the UK Biobank Axiom (UKBB; 825 927 markers), and were imputed using reference panels of the Haplotype Reference Consortium or UK10K and 1000 Genomes phase 3. We excluded individuals marked as outliers for heterozygosity, low call rates, and sex chromosome aneuploidy (n = 628). European individuals were identified from the genotype data by projecting all of the UK Biobank samples on the first two major principal components of four 1000 Genome populations (CEU, YRI, CHB, and JPT) (22). Individuals not falling in the neighborhood of the CEU cluster were excluded (n = 23 425). In the dataset from UK Biobank, a kinship coefficient was estimated for each pair of samples using KING’s robust estimator (23). We also excluded second-degree (or higher) related individuals (kinship coefficient ≥ 0.0442; n = 37 590) and participants who had been diagnosed with cancer at baseline (n = 24 944). A total of 400 812 individuals (186 376 men and 214 436 women) remained after these exclusions (not mutually exclusive).
SNP Selection
We compiled the information for the genetic variants identified by previous GWAS in association with the risk of any of the eight site-specific cancers by reviewing the GWAS catalog and previous PubMed publications. Single-nucleotide polymorphisms (SNPs) specifically associated with the risk of a specific subtype of a given cancer were not included in the analysis, except for epithelial ovarian cancer. Cancer risk variants on the X chromosome, reported exclusively from non-European populations, or in high linkage disequilibrium (r2 ≥ 0.2 in data of European ancestry in the 1000 Genomes project database), were also excluded in this study. For some previously reported risk variants that were not available in the data from UK Biobank, SNPs in high linkage disequilibrium (r2 ≥ 0.85) were selected for the study. For breast cancer, we used a PRS of 313 SNPs reported by a recent study (18), and 288 of the 313 SNPs were available in our dataset. A total of 612 SNPs were retained to build PRS after these exclusions (Table 1). A detailed list of SNPs for the PRS of each cancer is shown in Supplementary Table 1 (available online).
Table 1.
Number of cancer-associated SNPs used to construct the PRS and estimate AUC for each site-specific cancer
| Cancers | No. of SNPs | No. of Loci | PRS |
AUC (95% CI)* |
||||
|---|---|---|---|---|---|---|---|---|
| Cases, mean (SD) | Noncases, mean (SD) | P † | PRS | Family history‡ | PRS and family history§ | |||
| Prostate | 147 | 117 | 12.03 (0.68) | 11.63 (0.68) | <.001 | 0.662 (0.655 to 0.670) | 0.529 (0.522 to 0.535) | 0.669 (0.661 to 0.676) |
| Breast | 288 | 183 | 16.33 (0.60) | 16.05 (0.59) | <.001 | 0.628 (0.620 to 0.637) | 0.528 (0.521 to 0.534) | 0.633 (0.624 to 0.641) |
| Colorectal | 95 | 74 | 8.043 (0.47) | 7.859 (0.47) | <.001 | 0.609 (0.598 to 0.620) | 0.523 (0.515 to 0.532) | 0.613 (0.602 to 0.624) |
| Lung | 19 | 14 | 1.958 (0.37) | 1.886 (0.37) | <.001 | 0.591 (0.576 to 0.606) | 0.589 (0.577 to 0.602) | 0.615 (0.600 to 0.629) |
| Kidney | 15 | 14 | 2.257 (0.41) | 2.171 (0.40) | <.001 | 0.567 (0.543 to 0.591) | —‡ | —‡ |
| Bladder | 14 | 13 | 1.963 (0.35) | 1.868 (0.36) | <.001 | 0.583 (0.559 to 0.607) | —‡ | —‡ |
| Ovary | 31 | 28 | 2.478 (0.34) | 2.400 (0.32) | <.001 | 0.568 (0.537 to 0.598) | —‡ | —‡ |
| Pancreas | 22 | 18 | 3.892 (0.47) | 3.680 (0.50) | <.001 | 0.639 (0.613 to 0.664) | —‡ | —‡ |
Area under the receiver operating characteristic curve (AUC) was calculated by logistical models, adjusted for genotype array types. CI = confidence interval; PRS = polygenic risk score; SNP = single-nucleotide polymorphism.
Two-sided, two-sample t tests were performed with a type I error of 0.05.
Data on family cancer history (first-degree relatives) were available for cancer of prostate, breast, lung, and colorectal only.
P < .001 for the improvement of model performance by adding family history to the PRS-based model for all cancers.
Statistical Analysis
An additive genetic model was used to calculate each cancer-specific PRS, using previously reported regression coefficients as SNP-specific weights. For each site-specific cancer, a PRS was calculated as the sum of the product of the weight and the number of risk alleles for each risk variant across all selected risk variants per individual (24). Then, we checked the difference in distribution of standardized PRS (subtracted the mean and divided by the standard deviation) between case patients and noncase patients by two-sided, two-sample tests with a type I error of 0.05. We also divided the study participants into 50 groups according to the percentile of PRS (each 2%) and calculated the cumulative risk within each group for cancers of the prostate, breast, colorectal, and lung over the follow-up period. The PRS of each site-specific cancer was then categorized into quintiles. Hazard ratios (HRs) and 95% confidence intervals (CIs) associated with PRS were estimated by Cox proportional hazard models, using age as the time scale, and adjusted for age at the baseline survey, genotype array type (UKBL or UKBB), the 10 principal components for ancestry, sex (for nonsex-specific cancers), and stratified by birth cohorts. The assumption of proportional hazard was tested by adding time-dependent interaction term. We also estimated the hazard ratios of site-specific cancers for participants within the top and bottom 1% and 5% of each PRS, using the middle quintile (40%-60%) as the reference group. Two-sided Wald tests with a type I error of 0.05 were used for trend test. Then, we estimated the proportion of study participants in the cohort with a given relative risk of each site-specific cancer (HR = 2.0, 2.5, and 3.0). The area under the receiver operating characteristic curve (AUC) of the PRS for each cancer was calculated using logistical models that were adjusted for genotype array types. AUCs for models, including family history of cancer in first-degree relatives only, or both family history and PRS, were calculated for cancers of the prostate, breast, colorectal, and lung, because data on family history of these cancers in first-degree relatives were collected from a baseline survey in the UK Biobank. We calculated the absolute risks of site-specific cancer by PRS group using incidence and mortality rates in the United Kingdom, as described previously (25,26).
Results
During a median follow-up of 5.8 years of 400 812 participants (186 376 men and 214 436 women), there were 14 584 incident case patients of cancers, including 4430 with prostate cancer, 4340 with female breast cancer, 2453 with colorectal cancer, 1508 with lung cancer, 432 with pancreatic cancer, 545 with kidney cancer, and 358 with epithelial ovarian cancer.
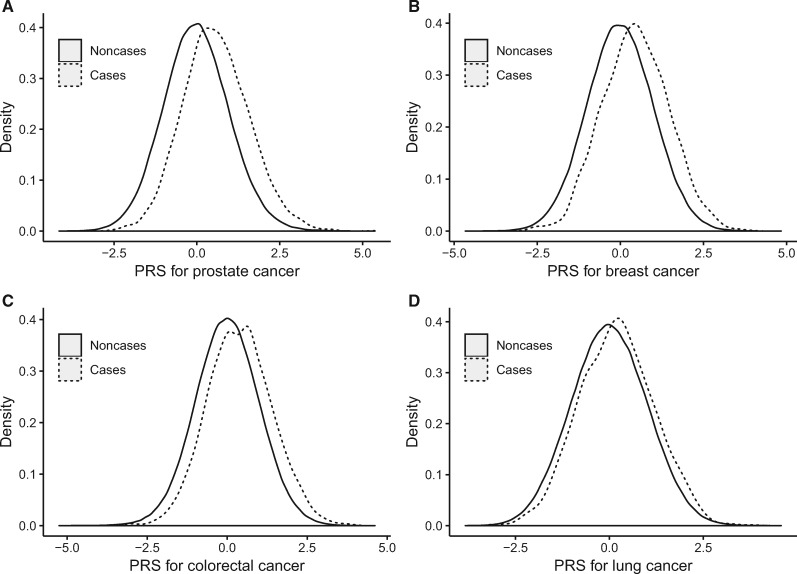
Table 1 summarizes the number of SNPs and loci for the PRS for each site-specific cancer, and Figure 1 displays the distribution of PRS for the four most common cancers: prostate, breast, colorectal, and lung. These PRS were all normally distributed, and the distribution curves for case patients were shifted to the right. Case patients had a higher mean value of each cancer-specific PRS than noncase patients, and the difference was highly statistically significant (Table 1). The PRS for prostate cancer had the largest AUC (0.662, 95% CI = 0.655 to 0.670), followed by pancreatic cancer (0.639, 95% CI = 0.613 to 0.664), and female breast cancer (0.628, 95% CI = 0.620 to 0.637). Kidney cancer has the lowest AUC (0.567, 95% CI = 0.543 to 0.591). The AUC derived from PRS was substantially higher than those derived using family history for cancers of the prostate, female breast, and colorectal, but not for lung cancer. Adding family history of cancer in first-degree relatives into the model slightly, but statistically significantly, improved the model performance in risk prediction.
Figure 1.
Distribution of standardized PRS between case patients and noncase patients. Distribution of standardized PRS was displayed for cancer of the (A) prostate, (B) breast, (C) colorectal, (D) and lung. Case patients (solid line) have a higher PRS value compared with noncase patients (dashed line) for all four cancers. PRS was standardized by subtracting the mean and dividing by the standard deviation. PRS = polygenic risk score.
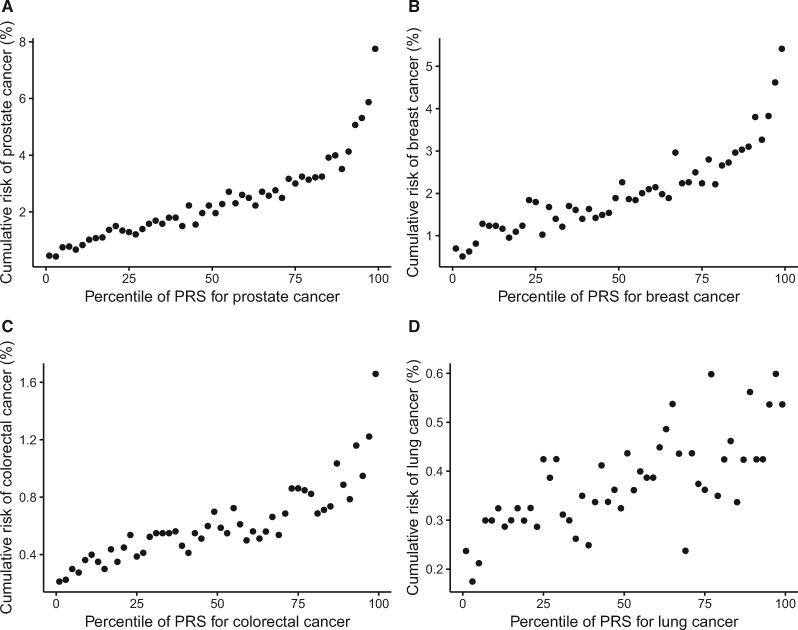
The risk of developing cancer of the prostate, breast, colorectal, or lung during the 5.8-year follow-up period increased substantially with an increased PRS (Figure 2). For example, the cumulative risk of female breast cancer increased from 0.6% in the lowest 2% of PRS to 5.4% in the highest 2% of PRS. Similar results were observed for four other less common cancers (data not shown). Using Cox regression models, hazard ratios of each site-specific cancer were estimated in association with its PRS, with the quintile of the lowest PRS as reference (Table 2). The risk of each site-specific cancer was statistically significantly associated with its PRS, following a dose-response pattern (P < .001). Compared with individuals in the lowest PRS quintile, those in the highest quintile had a greater than threefold risk for cancer of the prostate (HR = 5.63, 95% CI = 5.00 to 6.35), breast (HR = 3.77, 95% CI = 3.39 to 4.21), pancreas (HR = 3.37, 95% CI = 2.39 to 4.76), and colorectal (HR = 3.08, 95% CI = 2.68 to 3.55), and a 1.7- to 2.2-fold elevated risk was observed for other cancers. Hazard ratios estimated with the middle quintile as the reference with or without additional adjustment of cancer family history are presented in Supplementary Table 2 (available online).
Figure 2.
Cumulative risk of cancer over follow-up period by percentile of PRS. Study participants were divided into 50 groups according to the percentile of PRS (each 2%). Cumulative risk over a 5.8-year follow-up period of the UK Biobank cohort was displayed for cancers of the (A) prostate, (B) breast, (C) colorectal, (D) and lung. PRS = polygenic risk score.
Table 2.
Hazard ratios (95% CI) of cancers by quintile of PRS, UK Biobank*
| Cancer site | Q1 (low) | Q2 | Q3 | Q4 | Q5 | P trend † |
|---|---|---|---|---|---|---|
| Prostate | ||||||
| No. of cases | 316 | 566 | 795 | 1037 | 1716 | |
| HR (95% CI) | 1.00 (Referent) | 1.78 (1.55 to 2.05) | 2.54 (2.23 to 2.89) | 3.34 (2.94 to 3.79) | 5.63 (5.00 to 6.35) | <.001 |
| Breast | ||||||
| No. of cases | 413 | 639 | 774 | 996 | 1518 | |
| HR (95% CI) | 1.00 (Referent) | 1.56 (1.38 to 1.76) | 1.89 (1.68 to 2.13) | 2.45 (2.18 to 2.74) | 3.77 (3.39 to 4.21) | <.001 |
| Colorectum | ||||||
| No. of cases | 257 | 399 | 460 | 554 | 788 | |
| HR (95% CI) | 1.00 (Referent) | 1.56 (1.33 to 1.82) | 1.80 (1.54 to 2.10) | 2.16 (1.87 to 2.51) | 3.08 (2.68 to 3.55) | <.001 |
| Lung | ||||||
| No. of cases | 221 | 266 | 300 | 342 | 379 | |
| HR (95% CI) | 1.00 (Referent) | 1.20 (1.01 to 1.44) | 1.36 (1.15 to 1.62) | 1.54 (1.30 to 1.82) | 1.71 (1.45 to 2.02) | <.001 |
| Kidney | ||||||
| No. of cases | 76 | 100 | 99 | 122 | 148 | |
| HR (95% CI) | 1.00 (Referent) | 1.32 (0.98 to 1.77) | 1.30 (0.97 to 1.76) | 1.61 (1.21 to 2.14) | 1.96 (1.48 to 2.58) | <.001 |
| Bladder | ||||||
| No. of cases | 62 | 89 | 99 | 126 | 137 | |
| HR (95% CI) | 1.00 (Referent) | 1.43 (1.03 to 1.97) | 1.6 (1.16 to 2.19) | 2.04 (1.5 to 2.76) | 2.21 (1.64 to 2.99) | <.001 |
| Ovary | ||||||
| No. of cases | 60 | 55 | 62 | 73 | 108 | |
| HR (95% CI) | 1.00 (Referent) | 0.92 (0.64 to 1.32) | 1.03 (0.72 to 1.47) | 1.22 (0.86 to 1.71) | 1.81 (1.32 to 2.48) | <.001 |
| Pancreas | ||||||
| No. of cases | 42 | 60 | 71 | 119 | 140 | |
| HR (95% CI) | 1.00 (Referent) | 1.43 (0.96 to 2.12) | 1.70 (1.16 to 2.48) | 2.84 (2.00 to 4.03) | 3.37 (2.39 to 4.76) | <.001 |
Cutoff points for quintiles were based on the distribution of all study participants. Hazard ratios (HRs) were estimated using Cox regression and adjusted for age, birth cohort, genotyping array, top 10 principal components for ancestry, and sex (for nonsex-specific cancer only). CI = confidence interval; PRS = polygenic risk score.
Two-sided Wald tests were performed with a type I error of 0.05.
Compared with individuals in the middle quintile, individuals in the top 1% of the PRS had a threefold or higher risk of cancers of the prostate, breast, and colorectal, whereas individuals in the bottom 1% of the PRS had a 70% or greater reduced risk of these cancers (Table 3). Substantially elevated or reduced risks of other cancers in these extreme risk groups were also observed.
Table 3.
Hazard ratios (95% CI) of cancers in the top or bottom PRS groups compared with those with an average risk in the population, UK Biobank*
| Cancer site | PRS groupsHR (95% CI) |
|||
|---|---|---|---|---|
| Top 5% | Top 1% | Bottom 5% | Bottom 1% | |
| Prostate | 3.20 (2.88 to 3.56) | 4.39 (3.70 to 5.20) | 0.22 (0.16 to 0.29) | 0.15 (0.07 to 0.34) |
| Breast | 2.74 (2.45 to 3.07) | 3.52 (2.93 to 4.24) | 0.31 (0.24 to 0.41) | 0.54 (0.35 to 0.83) |
| Colorectum | 2.36 (2.03 to 2.75) | 3.02 (2.35 to 3.89) | 0.41 (0.30 to 0.55) | 0.30 (0.14 to 0.63) |
| Lung | 1.54 (1.24 to 1.91) | 1.31 (0.83 to 2.06) | 0.55 (0.40 to 0.77) | 0.46 (0.22 to 0.98) |
| Kidney | 1.53 (1.05 to 2.23) | 1.61 (0.78 to 3.32) | 0.87 (0.54 to 1.39) | 1.64 (0.80 to 3.36) |
| Bladder | 1.54 (1.06 to 2.23) | 1.57 (0.76 to 3.23) | 0.57 (0.32 to 0.99) | 0.62 (0.20 to 1.96) |
| Ovary | 2.42 (1.61 to 3.64) | 2.59 (1.24 to 5.41) | 0.69 (0.37 to 1.32) | 0.62 (0.15 to 2.55) |
| Pancreas | 2.31 (1.57 to 3.39) | 1.98 (0.91 to 4.33) | 0.45 (0.21 to 0.92) | —‡ |
Hazard ratios (HRs) were estimated using Cox regression and adjusted for age, birth cohort, genotyping array, top 10 principal components for ancestry, and sex (for nonsex-specific cancer only), with middle quintile (40%-60%) as the reference group. CI = confidence interval; PRS = polygenic risk score.
No cases observed for the bottom 1% of PRS for pancreatic cancer.
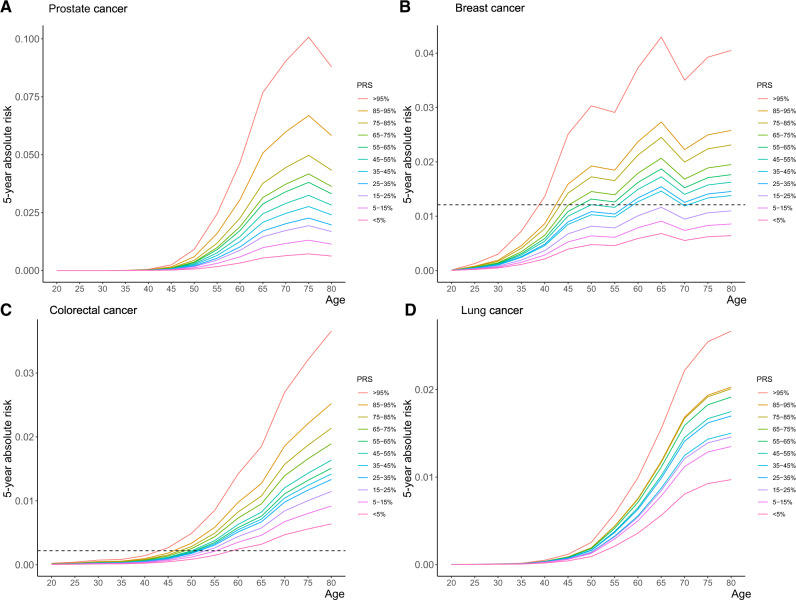
Figure 3 shows the estimated 5-year absolute risk of cancer of the breast, prostate, colorectal, and lung by PRS groups. In the United Kingdom, people become eligible for colorectal testing starting at the age of 60 years (50 years in Scotland), and people become eligible for breast cancer screening starting at the age of 50 years (47 years in parts of England) (27,28). The 5-year absolute risk is 1.2% for breast cancer and 0.2% for colorectal cancer for an individual aged 50 years in the United Kingdom with a median PRS (45-55 percentile group). However, people in the highest PRS group (>95%) reached the risk levels by ages 38 and 43 years for breast cancer and colorectal cancer, respectively, much earlier than the average-risk group.
Figure 3.
Five-year absolute risks of site-specific cancers by PRS groups. Five-year absolute risk of developing cancer of (A) prostate, (B) breast, (C) colorectal, (D) and lung. The horizontal lines show the estimated 5-year risk for individuals with median PRS (45%-55%) at the age of 50 years for (B) breast cancer or (C) colorectal cancer. PRS = polygenic risk score.
We estimated the proportion of the study participants at a given elevated risk (HR ≥ 2.0, 2.5, or 3.0; Table 4). Compared with the middle quintile, 40.4% of participants were at a greater than twofold risk of at least one type of cancer, including prostate cancer (28% of men), female breast cancer (20% of women), pancreatic cancer (9%), colorectal cancer (10%), kidney cancer (0.6%), lung cancer (0.1%), and epithelial ovarian cancer (8% of women). A total of 5.7% of the study participants were at a greater than threefold risk (HR ≥ 3.0) of at least one type of these cancers.
Table 4.
Proportions of subjects in the UK Biobank participants estimated to have a hazard ratio of no less than 2.00, 2.50, or 3.00 of site-specific cancer*
| Cancers† | HR ≥ 2.00 |
HR ≥ 2.50 |
HR ≥ 3.00 |
|---|---|---|---|
| No. of subjects (%) | No. of subjects ( %) | No. of subjects ( %) | |
| Prostate | 52 185 (28.0) | 26 091 (14.0) | 13 045 (7.0) |
| Breast | 42 887(20.0) | 15 010 (7.0) | 4288 (2.0) |
| Colorectal | 40 072 (10.0) | 12 024 (3.0) | 3206 (0.8) |
| Lung | 400 (0.1) | 200 (0.05) | 0 (0) |
| Kidney | 2404 (0.6) | 1603 (0.4) | 1202 (0.3) |
| Bladder | 0 (0) | 0 (0) | 0 (0) |
| Ovary | 17 152 (8.0) | 8575 (4.0) | 0 (0) |
| Pancreas | 36 070 (9.0) | 16 031 (4.0) | 1202 (0.3) |
| Total ‡ | 161 723 (40.4) | 74 261 (18.5) | 22 638 (5.7) |
Hazard ratios (HRs) were estimated using Cox regression and adjusted for age, birth cohort, genotyping array, top 10 principal components for ancestry, and sex (for nonsex-specific cancer only), with middle quintile (40%-60%) as reference group.
Percentage of cancer of the prostate, breast, or ovary was calculated among men and women separately.
Number of subjects who had an elevated risk of at least one site-specific cancer. Individuals who were at high risk of multiple cancers were only counted once.
Discussion
This is the first large prospective cohort study to systematically evaluate the performance of PRS that were constructed using GWAS-identified cancer risk variants for multiple major cancers. Although the discriminatory ability of PRS in classifying cancer case patients and noncase patients for any of the cancers evaluated in the study was moderate, as measured using AUCs ranging from 0.568 (for epithelial ovarian cancer) to 0.662 (for prostate cancer), these PRS can identify a large number of individuals in the general population who are at a twofold or higher risk of developing cancer, which is a risk level that is often regarded as clinically actionable for some cancers (eg, breast and colorectal cancers). For example, 28% of men and 20% of women were identified to have a greater than twofold risk of prostate cancer or female breast cancer, respectively. Overall, approximately 40.4% and 5.7% of study participants in the UK Biobank cohort had a greater than two- or threefold elevated risk of at least one site-specific cancer, compared with those who had an average risk of cancer in the total population.
For risk prediction, instead of obtaining information on the family history of cancer and other cancer risk factors, which could change over time, cancer-specific PRS can be assessed at any time in life. In the United Kingdom, current recommendations are to start breast cancer and colorectal cancer (in Scotland) screening at age 50 years (27,28). In the United States, the US Preventive Services Task Force also recommends starting breast and colorectal cancer screening at age 50 years (29,30). These guidelines are based on the average risk for the general population. Our results show that people in the highest PRS group reached the same risk level much earlier than the recommended age to start screening for the general population, which suggests that cancer-specific PRS can identify high-risk individuals who should receive screening tests earlier than the general population. The utility of PRS in identifying high-risk women for cost-effective breast cancer screening programs is currently under investigation in several ongoing studies (31,32). In addition, PRS can also be used to identify individuals at a low risk of cancer to postpone the age of receiving regular screenings to reduce the costs and complications related to cancer screenings. Certainly, further studies are needed to address issues related to the efficacy and ethics of selected screenings before they can be implemented in the general population.
In this study, the PRS was constructed based on the most updated GWAS-identified risk variants for each cancer. The AUC derived using PRS was substantially higher than that using family history for each site-specific cancer except for lung cancer. Fewer SNPs were included in the PRS for lung cancer compared with the PRS for prostate, breast, or colorectal cancers. In addition, the family history of lung cancer may reflect some shared smoking status in a family, which is a strong risk factor for lung cancer. Several previous studies have evaluated the association of various cancer-specific PRS. For example, Mavaddat et al. (18) built a PRS for breast cancer using 313 SNPs, with a P value less than 10-5 for hard-threshold stepwise forward regression and reported an AUC of 0.630 for overall breast cancer risk. For prostate cancer, Schumacher et al. (16) reported that men in the top 1% of PRS had a relative risk of 5.71 (95% CI: 5.04, 6.48) when compared with men in the 35-75 PRS percentiles. For epithelial ovarian cancer, Yang et al. (17) showed that women in the top 5% PRS category had a 1.78-fold and 2.14-fold elevated risk of overall epithelial and serous subtype ovarian cancer, respectively. These results are, in general, comparable to what we found in this study.
There are some limitations in our study. We built and evaluated PRS among individuals of European descent only. The clinical utility of PRS needs to be evaluated among populations of other races and ethnicities. A recent study has shown that PRS for breast cancer derived from non-Hispanic European-ancestry women performed well in Latina women (33). We also anticipate that the performance of PRS in cancer risk prediction and stratification will be further improved when combined with other lifestyle risk factors. In addition, the follow-up time (median = 5.8 years) of this cohort was relatively short, and a longer follow-up of this cohort could provide additional data to further evaluate cancer-associated PRS in the future. Nevertheless, our study suggests that we could start to consider using PRS to identify high-risk individuals for cost-efficient screenings.
Funding
This research was supported in part by funds provided by National Institutes of Health grants R01CA202981 and R01CA235553, as well as Anne Potter Wilson chair endowment at Vanderbilt University.
Notes
Role of the funder: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that no conflicts of interest exist.
Acknowledgements: This research has been conducted using the UK Biobank Resource under application number 40685. The authors would also like to thank Marshal Younger at Department of Epidemiology, Vanderbilt University Medical Center, for assistance with editing and manuscript preparation. He did not receive additional compensation besides his usual salary.
Author contributions: GJ contributed to the data curation, formal analysis, original draft preparation, review and editing; YLu and YLi contributed to the data curation, review and editing; WW contributed to the methodology of analysis; WZ contributed to the conceptualization, review and editing; JL, RT, BL, JCD, and X-OS contributed to the review and editing.
Supplementary Material
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Yurgelun MB, Kulke MH, Fuchs CS, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35(10):1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nielsen FC, van Overeem Hansen T, Sørensen CS.. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16(9):599–612. [DOI] [PubMed] [Google Scholar]
- 4. Carlo MI, Mukherjee S, Mandelker D, et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol. 2018;4(9):1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Easton DF, Pharoah PDP, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mersch J, Jackson M, Park M, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121(2):269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review (1975–2014), November 2016 SEER data submission. National Cancer Institute, https://seer.cancer.gov/csr/1975_2014/. Published April 2017. Accessed October 2019.
- 8. Whittemore AS, Gong G, John EM, et al. Prevalence of BRCA1 mutation carriers among U.S. non-Hispanic whites. Cancer Epidemiol Biomarkers Prev. 2004;13(12):2078–2083. [PubMed] [Google Scholar]
- 9. Tung N, Domchek SM, Stadler Z, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13(9):581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–59. [DOI] [PubMed] [Google Scholar]
- 11. Ma X, Zhang B, Zheng W.. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut. 2014;63(2):326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–875. [DOI] [PubMed] [Google Scholar]
- 13. Jerzak KJ, Mancuso T, Eisen A.. Ataxia-telangiectasia gene (ATM) mutation heterozygosity in breast cancer: a narrative review. Curr Oncol. 2018;25(2):e176–e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park J-H.. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45(4):400–405; 405e1–405e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schumacher FR, Al Olama AA, Berndt SI, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X, Leslie G, Gentry-Maharaj A, et al. Evaluation of polygenic risk scores for ovarian cancer risk prediction in a prospective cohort study. J Med Genet. 2018;55(8):546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2019. CA A Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 20. Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma X, Beeghly-Fadiel A, Shu X-O, et al. Anthropometric measures and epithelial ovarian cancer risk among Chinese women: results from the Shanghai Women’s Health Study. Br J Cancer. 2013;109(3):751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M.. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang N, Lu Y, Khankari NK, et al. Evaluation of genetic variants in association with colorectal cancer risk and survival in Asians. Int J Cancer. 2017;141(6):1130–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng W, Wen W, Gao Y-T, et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J Natl Cancer Inst. 2010;102(13):972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen W, Shu X, Guo X, et al. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast Cancer Res. 2016;18(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Research UK. Breast screening. https://www.cancerresearchuk.org/about-cancer/breast-cancer/screening/breast-screening. Accessed October 2019.
- 28.Cancer Research UK. Bowel cancer screening. https://www.cancerresearchuk.org/about-cancer/bowel-cancer/getting-diagnosed/screening. Accessed October 2019.
- 29.Final Recommendation Statement: Breast Cancer: Screening-US Preventive Services Task Force. https://www.uspreventiveservicestaskforce.org/Page/Document/RecommendationStatementFinal/breast-cancer-screening1. Accessed November 4, 2019.
- 30. Bibbins-Domingo K, Grossman DC, Curry SJ, et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA. 2016;315(23):2564–2575. [DOI] [PubMed] [Google Scholar]
- 31. Esserman LJ; WISDOM Study and Athena Investigators. The WISDOM Study: breaking the deadlock in the breast cancer screening debate. NPJ Breast Cancer. 2017;3(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MyPeBS. Randomized comparison of risk-stratified versus standard breast cancer screening in European women aged 40–70 (MyPeBS); 2018. https://cordis.europa.eu/project/id/755394. Accessed October 2019.
- 33. Shieh Y, Fejerman L, Lott PC, et al. A polygenic risk score for breast cancer in US Latinas and Latin American Women. J Natl Cancer Inst. 2020;112(6):djz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.