Highlights
-
•
With the increasing number of people suffering from COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is a dire need to look for effective remedies against this pandemic.
-
•
In this study, we propose a single drug – paromomycin – against two targets of SARS-CoV-2, i.e., spike protein (S1) and protease domain. Based upon docking scores and molecular dynamics simulation, paromomycin was found to have strong binding affinity for both of the targets of coronavirus.
-
•
15 anti-malarial drugs including chloroquine were also investigated against both targets using in-silico approach resulting in no effective binding.
-
•
Paromomycin, a broad-spectrum aminoglycoside antimicrobial drug that was originally used to treat acute and chronic intestinal infections, may also be of potential use in the treatment of COVID-19, as one of the major symptoms in COVID-19 includes gastrointestinal disturbance.
-
•
Clinical trials are needed to confirm the efficacy of paromomycin against SARS-CoV-2.
Keywords: COVID-19, Drug repurposing, Chloroquine, Protease, Spike, MD simulation
Abstract
Objectives
With the increasing number of people suffering from coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is a dire need to look for effective remedies against this pandemic. Drug repurposing seems to be the solution for the current situation.
Methods
In a quest to find a potential drug against this virus, 15 antimalarial drugs (including chloroquine) and 2413 US Food and Drug Administration-approved drugs were investigated for activity against both the protease and spike proteins of SARS-CoV-2 using an in silico approach. Molecular docking analysis followed by molecular dynamics simulation was performed to estimate the binding and stability of the complexes.
Results
This study identified a single drug – paromomycin – with activity against two targets of SARS-CoV-2, i.e., spike protein (S1) and protease domain. Paromomycin was found to have strong binding affinity for both targets of coronavirus. The results also showed that no antimalarial drug exhibited effective binding for either S1 or protease.
Conclusions
This study found that paromomycin may be an effective dual targeting drug against coronavirus, as it binds not only to the protease domain of the virion, but also to the spike domain, with high stability. Furthermore, none of the antimalarial drugs showed strong binding affinity for either protease or the receptor binding domain (RBD).
Introduction
First reported in December 2019 in Wuhan District of China, the coronavirus disease 2019 (COVID-19) outbreak has spread all over the world and is a menace to the world community. Birds and wild animals have been suggested as the origin of spread of the causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020, Zhou et al., 2020). According to World Health Organization (WHO) statistics from April 12, 2020, a total 1 696 588 confirmed cases of COVID-19 and 105 952 associated deaths had been reported globally (WHO, 2020). Currently no vaccine or effective drug is available for the treatment of this disease, and clinicians are treating patients with neutralizing antibodies against severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) to target the spike protein of this virus (Chen et al., 2020a, Duan et al., 2020). As suggested in recent research, the spike protein, which is present on the surface of SARS-CoV-2, interacts with host targets such as CD26 and ACE2, and is thus considered a potential and logical therapeutic target (Hoffmann et al., 2020, Zhang et al., 2020a).
Recent studies have shown the effectiveness of chloroquine against SARS-CoV-2, both in vitro and in clinical trials in China. In addition, some modifications of chloroquine, such as its sulfate and phosphate salts, have also proved beneficial (Gao et al., 2020, Hu et al., 2020, Wang et al., 2020). However, the US Food and Drug Administration (FDA) has not yet approved this drug for the treatment of COVID-19, and there is a dire need for further clinical trials conducted on geographically different populations to ascertain the effect of this drug.
Human coronaviruses are single-stranded positive-sense RNA viruses comprised of two groups of proteins: (1) structural proteins, which include S (spike) proteins, M (matrix) proteins, N (nucleocapsid) proteins, and E (envelope) proteins, and (2) non-structural proteins, which include proteases such as nsp3 and nsp5, and RdRP such as nsp12. The spike protein is present in homo-trimeric form on the outer surface of the virion particle. Two or three proteases are encoded by the viral RNA, which are potential drug targets against the coronaviruses (Chen et al., 2020b, Vankadari and Wilce, 2020).
It has been established that the spike protein plays a vital role in attachment and viral entry into the host cell. Moreover, the coronavirus spike protein enters the host cell via angiotensin converting enzyme 2 (ACE2) receptors present on lung epithelial cells and intestinal cells, causing lung disease and diarrhea. The S1 and S2 receptor-binding domain (RBD) of the spike protein attaches to the ACE2 receptor, thus the viral envelope attaches to the membrane and becomes internalized (Ge et al., 2013, Wan et al., 2020). Another potential target in treating COVID-19 is the main protease enzyme of SARS-CoV-2, considered a central enzyme in regulating viral replication and transcription (Jin et al., 2020).
This study had two main objectives: to determine the efficacy of antimalarial drugs against SARS-CoV-2 using an in silico approach and to purpose a single potential drug that acts against SARS-CoV-2 spike protein (S1) and also against the catalytic domain of its protease protein.
Methods
Structure retrieval and pre-processing
The X-ray crystal structure of unliganded protease and the RBD of S1 of the novel SARS-CoV-2 were retrieved from the Protein Data Bank (PDB) repository: ID 6y84 (resolution 1.39 Å) and ID 6vw1 (resolution 2.68 Å), respectively. Both structures were checked for errors and quality using the SAVES server (Structure Analysis and Verification Server). Discovery Studio Visualizer (v19.10.18287, 2019; Dassault Systèmes BIOVIA, San Diego, CA, USA) was used to examine the structural aspects of both proteins, specifically of 6vw1 for the receptor binding residues. After defining the ligand (A chain: angiotensin converting enzyme 2; ACE2) and receptor (E chain: RBD), the interacting residues of the A chain and E chain of 6vw1 were listed for further use in grid generation and docking analysis.
Molecular docking analysis
Selection and preparation of ligands
3D structures of 15 antimalarial drugs (Table 1 ) were retrieved from the NCBI PubChem compound database (https://pubchem.ncbi.nlm.nih.gov/), while 3D structures of 2413 FDA-approved drugs were obtained from DrugBank (https://www.drugbank.ca/).
Table 1.
Antimalarial drugs against protease and RBD of SARS-CoV-2.
| List of antimalarial drugs tested |
Glide score (Grid for specified residues) |
Glide score (Global docking) |
||||
|---|---|---|---|---|---|---|
| Sr. No. | Name | PubChem CID | 6y84 | 6vw1 | 6y84 | 6vw1 |
| 1 | Amodiaquine (flavoquine) | 2165 | −4.814 | −3.211 | −5.545 | −4.200 |
| 2 | Chloroquine | 2719 | −4.111 | −2.908 | −4.831 | −3.828 |
| 3 | Primaquine | 4908 | −4.165 | −3.111 | −5.498 | −3.630 |
| 4 | Pyrimethamine | 4993 | −3.634 | −5.063 | −5.403 | −3.682 |
| 5 | Halofantrine | 37393 | −3.992 | −3.656 | −4.765 | −3.636 |
| 6 | (−)-Mefloquine | 40692 | −3.940 | −3.014 | −4.521 | −4.599 |
| 7 | Artemisinin | 68827 | −3.992 | −2.769 | −4.162 | −4.786 |
| 8 | Didesethyl chloroquine | 122672 | −4.968 | −3.018 | −6.296 | −4.050 |
| 9 | Atovaquone | 74989 | −3.386 | −2.394 | −3.727 | −3.024 |
| 10 | Clindamycin | 446598 | −5.558 | −3.466 | −5.005 | −3.866 |
| 11 | (S)-Chloroquine | 639540 | −4.111 | −2.908 | −4.831 | −3.828 |
| 12 | Quinine | 3034034 | −4.238 | −2.793 | −4.491 | −3.038 |
| 13 | Sulfonamides | 3085933 | −2.545 | 0.356 | −3.871 | −3.620 |
| 14 | Proguanil (chloroguanide) | 6178111 | −4.127 | −2.328 | −1.737 | −5.129 |
| 15 | Doxycycline | 54671203 | −5.782 | −3.749 | −6.831 | −4.869 |
RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
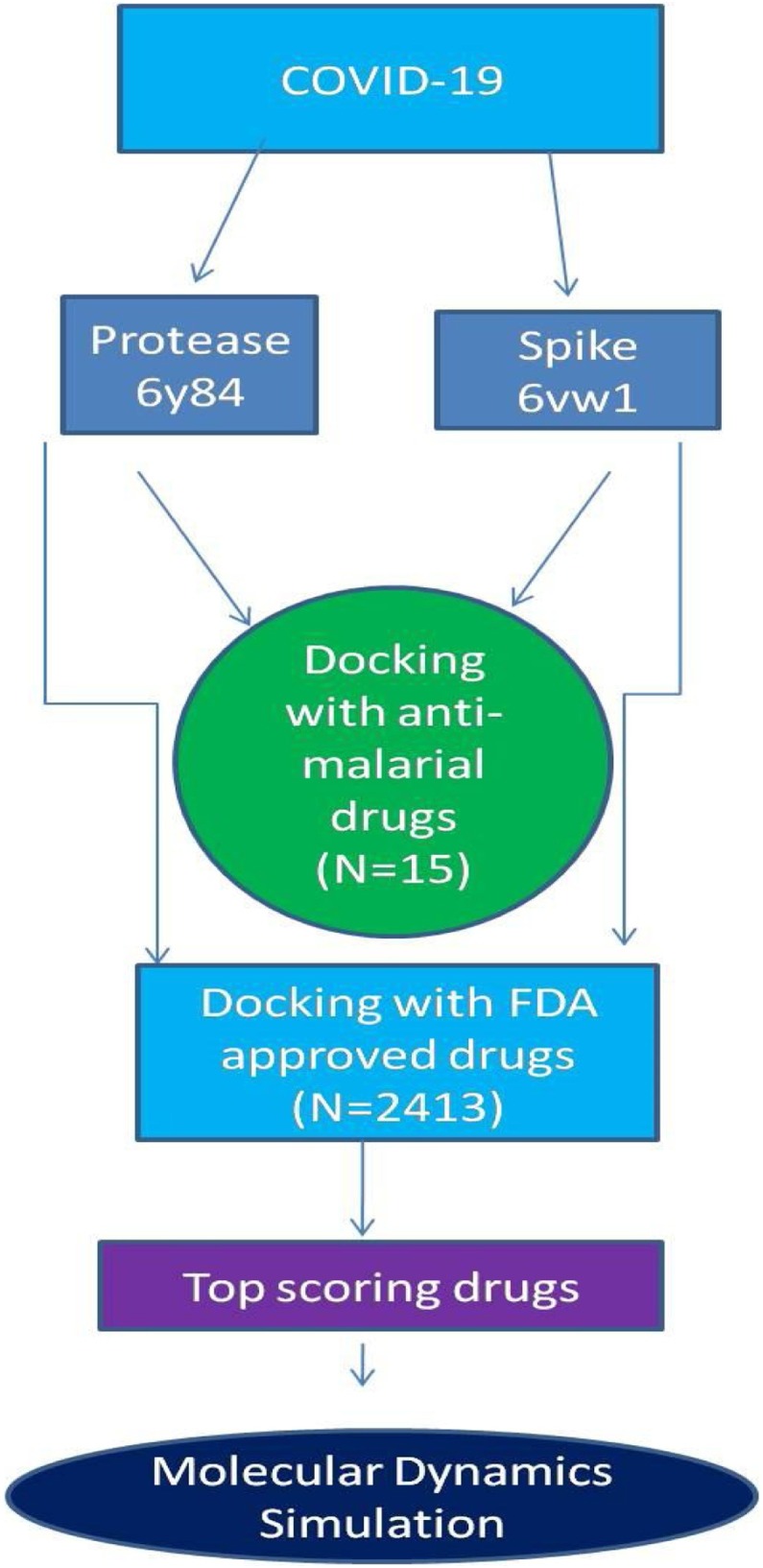
All ligand compounds were prepared using LigPrep in Schrödinger Maestro 12.2 (Release 2019-4; Schrödinger LLC, New York, NY, USA) using OPLS_2005 force field in Epik mode and applying parameters such as ‘generate possible states at target pH’ (pH 7), desalting of ligands, and tautomer generation, while retaining specific chiralities to generate at most one per ligand. The general study strategy for the analysis of these ligands is summarized in Figure 1 .
Figure 1.
Schematic representation of the study.
Protein preparation and Glide docking
Protein was prepared using the Schrödinger 12.2 Protein Preparation Wizard (Release 2019-4; Schrödinger LLC), as described elsewhere (Sastry et al., 2013). Briefly, protein parameters were applied such as the addition of hydrogen atoms, assigning bond orders, creation of zero-order bonds to metal, creation of disulfide bonds, deleting waters beyond 5 Å, and generation of het states using Epik at pH 7. After pre-processing, any already attached ligands were removed and protein structures were corrected if needed by adding side chains and missing atoms, etc., followed by minimization and optimization using OPLS_2005 force field. All of the active site residues of both proteins, including the catalytic dyad and associated residues of 6y84 and RBD residues of 6vw1, were used in receptor grid generation. In the receptor grid, x,y,z coordinates were supplied according to the grid size around the mentioned residues and the grids were generated using default options. Glide molecular docking was conducted under default conditions in extra-precision mode (XP) (Release 2019-4; Schrödinger, LLC). Glide uses a series of scoring functions in XP mode to identify the optimal binding site of the ligand for acceptable poses (Friesner et al., 2006). Top ranked ligands were selected on the basis of the Glide score (in kcal/mol) in order to define the strength of the protein–ligand interaction. Protein residue interactions of docked complexes were visualized and examined in Discovery Studio Visualizer (v19.10.18287, 2019; Dassault Systèmes BIOVIA).
Molecular dynamics simulation
Molecular dynamics (MD) simulations of the top-ranked docked complexes were performed using Nano Scale Molecular Dynamics (NAMD); this works with CHARMM++ force field potential functions and parameters (Phillips et al., 2005, Brooks et al., 2009, Lee et al., 2016). The topologies and parameter files of proteins and ligand were generated in CHARMM-GUI using PDB Reader and Ligand Reader and Modeler, respectively (Jo et al., 2008, Jo et al., 2014, Kim et al., 2017). The CHARMM36 force field was applied to protein at a constant number of molecules, volume, and temperature (NVT), and the system was solvated using ‘add solvation box’ with TIP3p water. Before simulation, each system was first energy-minimized and equilibrated for 200 ps, following which the total energy was observed in multiplot. The energy-minimized systems were subsequently utilized to conduct simulations at constant temperature conditions (310 K) using Langevin dynamics parameters and under constant periodic boundary conditions for 50 ns for comparative trajectory analysis. Visual Molecular Dynamics (VMD 1.9.3) was used to analyze the trajectories and for the post simulation analysis (Humphrey et al., 1996).
Post simulation analysis of trajectories
The dynamic properties of the complexes in comparison to individual proteins were studied to investigate the stability of the interacting residues. For this purpose, binding energy, root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and per-residue hydrogen bonding interactions were calculated using VMD. The free binding energy was estimated using the Molecular Mechanics Poisson–Boltzmann Surface Area method (MMPBSA) in CaFE1.0 VMD plugin. The net energy of the system was estimated using the following equation (Liu and Hou, 2016):
| ΔGbinding = Gcomplex – (Gprotein + Gligand) |
| ΔGbind = ΔH–TΔS = {ΔEgas + ΔGpolarsol + ΔGnonpolarsol – TΔS} |
For post simulation analysis, last 1000 continuous frames were extracted without stride. For MMPBSA, data were collected for every 5 ps and the MM (NAMD), PB (APBS), and SA (VMD) values were calculated. In the APBS calculations, the interior dielectric constant was set to 2.0, with exterior dielectric constant as 80 (Wang and Kollman, 2000, Li et al., 2018).
Results
Structural aspects of 6vw1 and 6y84
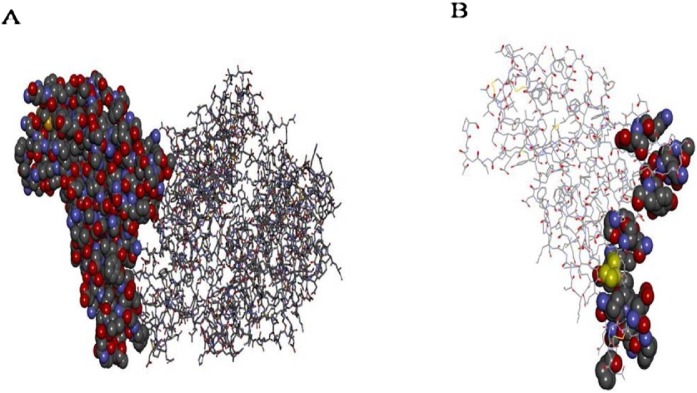
The crystal structure of 6vw1 consists of four chains – A, B, E, and F. Chains A and B belong to the human ACE2 receptor and are identical chains consisting of 571 residues each, while chains E and F belong to the virion and are identical chains comprising 217 residues each. The E chain residues interacting with ACE2 (A chain) include Arg-439, Tyr-449, Tyr-453, Leu-455, Phe-456, Ala-475, Glu-484, Phe-486, Asn-487, Tyr-489, Gln-493, Gly-496, Gln-498, Thr-500, Gly-502, and Tyr-505. In addition, ACE2 (A chain) residues interacting with the SARS-CoV-2 RBD (chain A) include Ser-19, Gln-24, Lys-31, His-34, Glu-35, Glu-37, Asp-38, Tyr-41, Gln-42, Met-82, Tyr-83, Glu-329, Lys-353, and Gly-354 (Figure 2 ).
Figure 2.
Structural aspects of 6vw1. A: RBD (E chain) of 6VW1 shown in CPK representation while ACE2 (A chain) is shown in line representation. B: Only RBD (E Chain) shown in line representation with active residues in CPK representation.
The structure of 6y84 comprises a single chain A of 306 residues, among which two residues are considered as making a catalytic dyad, i.e.; His-41 and Cys-145. Furthermore, some other residues at positions 442, 472, 479, 487, and 491 in the spike protein of SARS-CoV-2 have been reported to play an important role in inter- and intra-species transmission of the virus (Xintian et al., 2020).
Molecular docking analysis of antimalarial drugs
Fifteen different antimalarial drugs available on the market were docked against 6vw1 and 6y84 adopting two different strategies. In the first strategy, all antimalarial drugs were docked against specific residues (mentioned above) in both proteins, while in the second strategy, the whole protein grid was used to dock each drug for performing blind docking. Glide scores were calculated and were analyzed to check the binding efficiency of these drugs (Table 1, Supplemenatry Material Tables S1–S4)
Molecular docking analysis of FDA-approved drugs
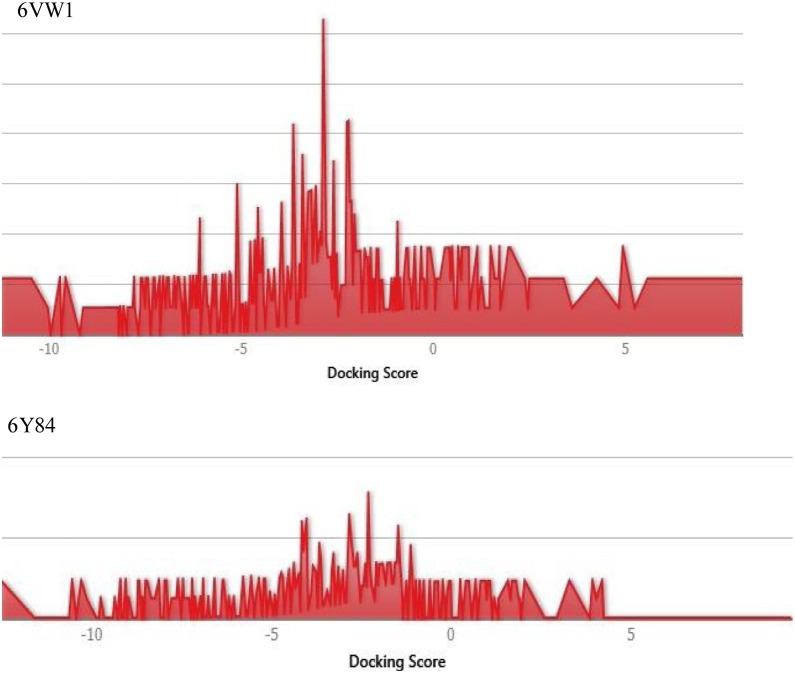
A total of 2413 FDA-approved drugs were used to study potential effective binding against the catalytic site of protease (6y84) and RBD of spike protein (6vw1) (Supplementary Material Tables S5–S10). A distribution plot of docking scores was made for the entire dataset of drugs (Figure 3 ). Top ranked drugs were selected on the basis of their Glide score (Table 2 ). The top three drugs against the protease domain were acarbose, colistin, and paromomycin, with Glide scores of −13.139, −12.63, and −11.5.79, respectively. On the other hand, the drugs framycetin, acarbose, and paromomycin showed the highest binding affinity for the RBD with Glide scores of −11.233, −10.49, and −10.01, respectively.
Figure 3.
Distribution plot of docking scores for all the FDA approved drugs' dataset used.
Table 2.
FDA-approved drug showing high binding affinities against protease and RBD of SARS-CoV-2.
| 6y84 |
6vw1 |
||||||
|---|---|---|---|---|---|---|---|
| Drug name | Formula | DrugBank ID | Glide score | Drug name | Formula | DrugBank ID | Glide score |
| Acarbose | C25H43NO18 | DB00284 | −13.139 | Framycetin | C23H46N6O13 | DB00452 | −11.233 |
| Colistin | C52H98N16O13 | DB00803 | −12.63 | Acarbose | C25H43NO18 | DB00284 | −10.649 |
| Paromomycin | C23H45N5O14 | DB01421 | −11.579 | Paromomycin | C23H45N5O14 | DB01421 | −10.01 |
| Iotrolan | C37H48I6N6O18 | DB09487 | −11.001 | Plazomicin | C25H48N6O10 | DB12615 | −9.935 |
| Indium In-111 pentetreotide | C62H80InN12O19S2 | DB11835 | −10.795 | Omadacycline | C29H40N4O7 | DB12455 | −9.178 |
| Framycetin | C23H46N6O13 | DB00452 | −10.713 | Mangafodipir | C22H30MnN4O14P2 | DB06796 | −9.089 |
| Lutetium Lu-177 dotatate | C65H87LuN14O19S2 | DB13985 | −10.607 | Flavin adenine dinucleotide | C27H33N9O15P2 | DB03147 | −8.986 |
| Rutin | C27H30O16 | DB01698 | −9.853 | Ribostamycin | C17H34N4O10 | DB03615 | −8.781 |
FDA, US Food and Drug Administration; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Tautomeric forms of paromomycin and their docking analysis
There are 11 tautomeric forms of paromomycin (I–XI) due to the alternate protonation state of its amide groups (Supplementary Material Figure S1). To elucidate further, both of the proteins (6y84 and 6vw1) were docked against these 11 tautomeric forms (Supplementary Material Tables S11 and S12). It was observed that both proteins gave the highest Glide score for form I. It was clear from the results that paromomycin I exhibits the highest binding affinity against SARS-CoV-2. Thus, these docked complexes of both proteins with paromomycin I were further utilized for MD simulation.
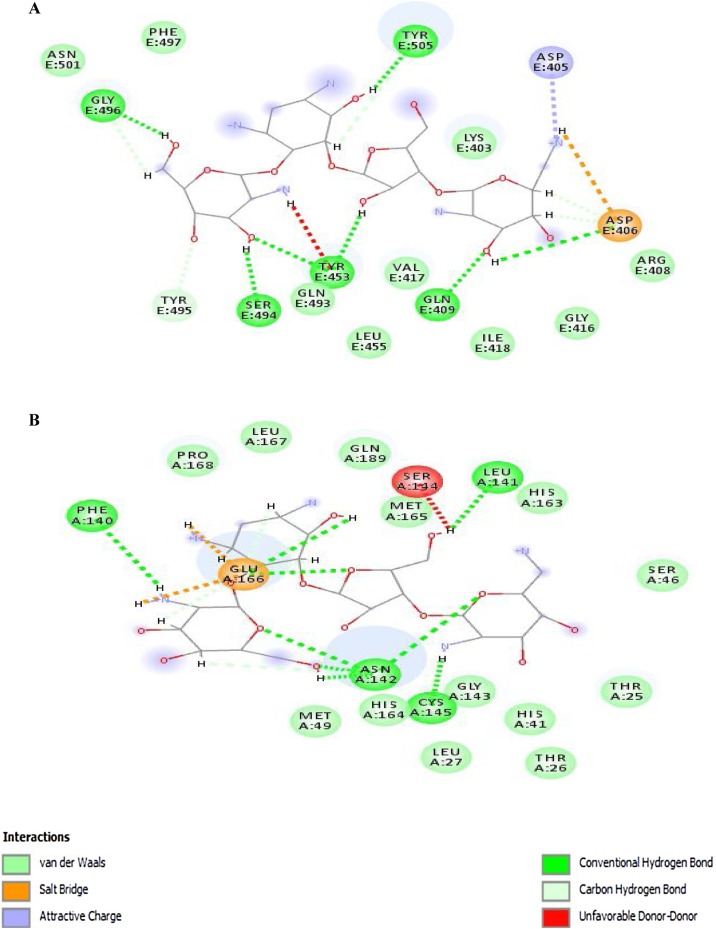
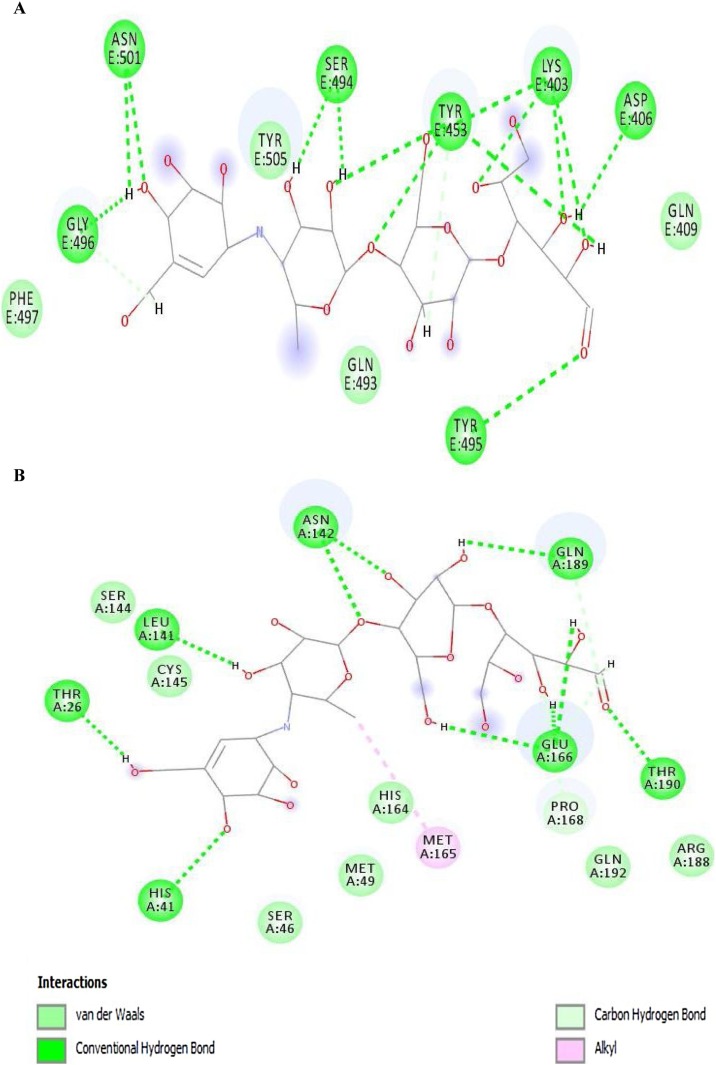
The 2D diagrams from ligand–protein interactions of docked complexes were evaluated to examine the residues involved in binding with paromomycin I. In the 2D diagram of the 6vw1–ligand complex, a total of seven hydrogen bonds, two salt bridges, five carbon–hydrogen bonds, and a few van der Waals interactions were observed (Figure 4A). Residues involved in the different interaction types include Lys-403, Asp-405, Asp-406, Arg-408, Gln-409, Ile-418, Tyr-453, Gln-493, Ser-494, Tyr-495, Gly-496, Phe-497, and Tyr-505. Figure 4. It was further observed that protease (6y84) showed a total of nine hydrogen bonds, two salt bridges, and three carbon–hydrogen bonds with a few van der Waals interactions with paromomycin I (Figure 4B). It was clearly depicted from the interaction diagram that Cys-145 makes a strong hydrogen bond with the protonated amide group of paromomycin I, while His-41 exhibits van der Waals interactions in close proximity. Besides these two catalytic residues, Glu-166 makes two hydrogen bonds, two salt bridges, and one carbon–hydrogen bond. Other residues in the interaction diagram are Leu-27, Phe-140, Leu-141, Asn-142, Ser-144, His-163, Met-165.
Figure 4.
2D diagram of (A) 6vw1-liagnd (complex-1) and (B) 6y84-ligand (complex-2).
Docking analysis of acarbose with target proteins
Acarbose is another common drug that showed strong binding affinity for both targets, i.e. 6y84 =−13.139 kcal/mol; 6vw1 = −10.649 kcal/mol (Table 2). Acarbose made a total of 13 hydrogen bonds, one carbon–hydrogen bond, and also some van der Waals interactions with 6vw1 in docked complex 3 (Figure 5 A). The interacting residues are Lys-403, Asp-406, Gln-409, Tyr-453, Gln-493, Tyr-495, Gly-496, Phe-497, Asn-501, and Tyr-505. In the case of 6y84, acarbose forms 10 hydrogen bonds, one carbon–hydrogen bond, one alkyl bond, and other non-covalent interactions in complex 4 (Figure 5B). Residues involved in these interactions are as follow: Thr-26, His-41, Ser-46, Met-49, Leu-141, Asn-142, Ser-144, Cys-145, His-164, Met-165, Glu-166, Pro-168, Arg-188, Gln-189, Thr-190, and Gln-192.
Figure 5.
2D diagram of (A) 6vw1-ligand (Complex-3) and (B) 6y84-ligand (Complex-4).
Stability of complexes by MD analysis
Molecular Mechanics Poisson–Boltzmann Surface Area method (MMPBSA)
MD simulation of complex 1 (6vw1–ligand) and complex 2 (6y84–ligand) along with individual proteins without ligands (6vw1: protein-1; 6y84: protein-2) was conducted to record the trajectories for 50 ns. The MMPBSA model was generated using CaFE1.0 VMD plugin. The net binding energy for complex 1 and complex 2 were found to be −18.3624 kcal/mol and −234.0711 kcal/mol, respectively, showing both complexes to be stable (Table 3 ). The stability of complex 1 is favored by electrostatic (−25.7727 kcal/mol), van der Waals (−18.0870 kcal/mol), and non-polar energies (−22.0747 kcal/mol). In the case of complex 2, the net binding energy is highly contributed by electrostatic energy (−451.4246 kcal/mol) and less by van der Waals (−37.7393 kcal/mol).
Table 3.
MMPBSA model for complex 1 and complex 2.
| MMPBSA model | Complex 1 | Complex 2 |
|---|---|---|
| Elec | −25.7727 ± 14.0375 | −451.4246 ± 16.3534 |
| Vdw | −18.0870 ± 2.9519 | −37.7393 ± 3.4501 |
| PB | 29.4849 ± 6.9718 | 261.2760 ± 8.1962 |
| SA | −3.9877 ± 0.1210 | −6.1831 ± 0.1261 |
| Gas | −43.8596 ± 13.5039 | −489.1639 ± 16.2019 |
| Sol | 25.4972 ± 6.9007 | 255.0928 ± 8.1390 |
| Pol | 3.7122 ± 8.5699 | −190.1487 ± 9.4145 |
| Npol | −22.0747 ± 2.9860 | −43.9224 ± 3.4572 |
| ΔGbind | −18.3624 ± 7.5918 | −234.0711 ± 8.9385 |
MMPBSA, Molecular Mechanics Poisson–Boltzmann Surface Are
RMSD and RMSF calculations
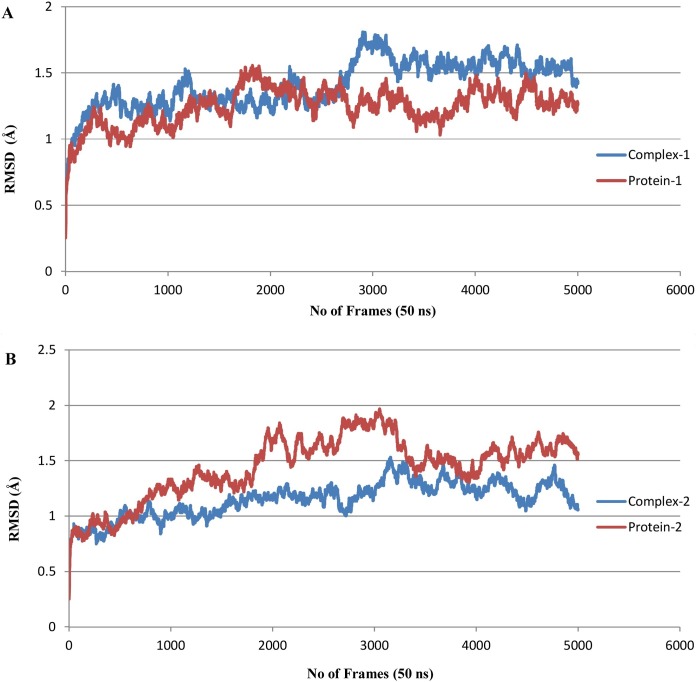
In order to determine time-dependent conformational changes, RMSD analysis was performed in the RMSD trajectory tool of VMD. RMSD graphs were generated with respect to 5000 frames (50 ns) using the backbone of their initial structures as reference. The average RMSD of complex 1 was found to be 1.413 ± 0.180 Å, while the average RMSD of protein-1 was calculated to be 1.245 ± 0.140 Å. It was observed that the maximum RMSD value never exceeded 1.8 Å during simulation, which means that complex 1 remained stable for the entire simulation period. For complex 1, the RMSD gradually increased up until 28 ns, with a drift at 30 ns suggesting some structural changes, but the complex remained stable and converged after 35 ns (Figure 6 A). For complex 2, the average RMSD was calculated as 1.150 ± 0.157 Å, while the average RMSD of protein-2 was calculated to be 1.443 ± 0.280 Å. Complex 2 was also found to be stable, as maximum RMSD never exceeded 2 Å for the entire simulation period. In the case of complex 2, the RMSD value increased up until 32 ns and converged afterwards, with minor fluctuations (Figure 6B).
Figure 6.
RMSD plot. (A) Complex-1 in comparison to Protein-1 and (B) Complex-2 in comparison to Protein-2.
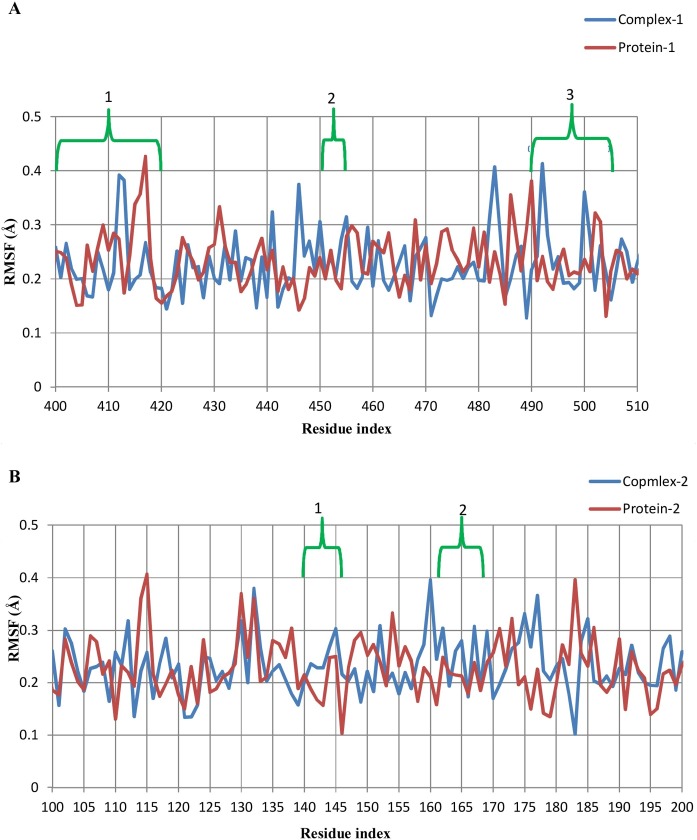
Per residue RMSFs were calculated for the last 1000 frames in the timeline plugin of VMD using a comparative approach (Figure 7 ). In the case of complex 1, the binding pocket region (residues 400–505) was divided into three interacting regions: (1) residues 403–418, (2) residues 453–455, and (3) residues 493–505. In the first region of complex 1, Pro-412 (0.39/0.27 Å, complex 1/protein-1) and Gly-413 (0.38/0.17 Å, complex 1/protein-1) showed drift in RMSF values that could be due to two hydrogen bond formations by Asp-406 and Gln-409 with the ligand in this region. In region 2, Arg-454 and Leu-455 showed higher RMSFs than protein-1, possibly due to the contribution of two hydrogen bonds by Tyr-453 and van der Waals interactions by Leu-455. Prior to region 3, Pro-492 exhibited very much higher fluctuation in complex 1 (0.41 Å) in addition to Ser-494 (0.21 Å) and Thr-500 (0.36 Å) inside the region in comparison to the respective positions in protein-1. All other interacting residues, except those mentioned above, had comparable or even lower fluctuations as compared to unliganded protein. The lower RMSF values of these residues could be related to the rigidity of these residues for any motion due to the presence of the ligand that contributes stability and loss of flexibility (Chen et al., 2018). In the case of complex 2, four interacting regions were found: (1) residues 140–146, (2) residues 163–168, (3) residues 25–27, and (4) residues 41–49. In region 1, Cys-145 showed higher fluctuation in complex 2 (0.3 Å) than in protein-2 (0.25 Å). Glu-166 did not show any prominent variation in complex 2, but its neighboring residues Met-165 and Leu-167 had higher residual fluctuations. Regions 3 and 4 contributed through van der Waals interactions with the ligand and showed no major residual fluctuations. Overall, both of the complexes exhibited lower RMSF values, referring to less motion in the binding region and conferring their stability.
Figure 7.
Comparative RMSF plot of (A) complex-1 and protein-1; and (B) complex-2 and protein-2.
Hydrogen bond interactions
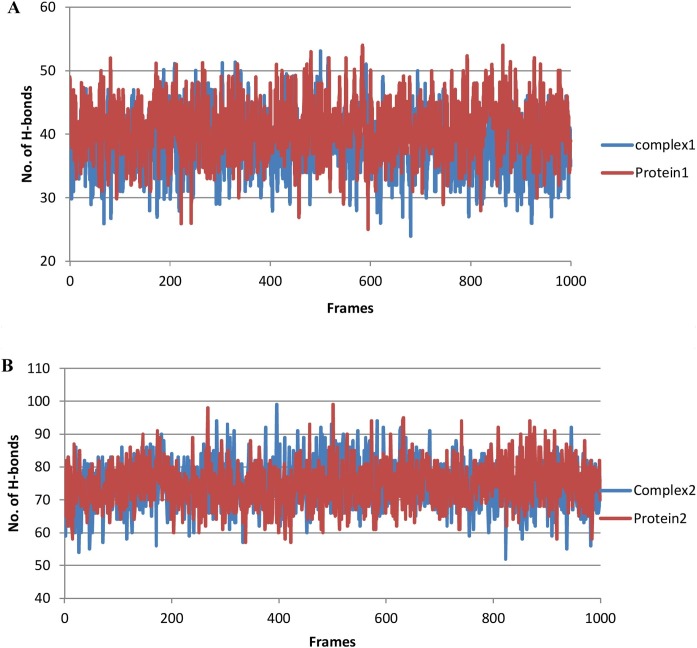
The hydrogen bond interactions of the complexes were calculated to validate affinity of the ligand to inhibit the proteins predicted by the docking simulation studies. For this purpose, two strategies were adopted. In the first strategy, the total number of hydrogens in the complexes and proteins were examined for comparative purposes (Figure 8 ). Based on the result, complex 2 had a higher number of hydrogen atoms than protein-1, while complex 1 and protein-1 exhibited almost comparable hydrogen bonds. In the second strategy, the number of hydrogen bonds between the receptor and ligand in complexes (acceptor/donor) were calculated and matched for identity with the hydrogen bond residues predicted in the docking analysis (Table 4 ). It was confirmed that the residues involved in hydrogen bonding during post simulation analysis of trajectories were the same residues contributing to hydrogen bonding during the docking analysis.
Figure 8.
Total hydrogen bonds during the time course of MD simulation.
Table 4.
Total hydrogen bonds between receptor and ligand determined from MD trajectories.
| Complex 1 |
Complex 2 |
||
|---|---|---|---|
| Donor | Acceptor | Donor | Acceptor |
| UNK900-Side | Asp406-Side | UNK1-Side | Glu166-Main |
| Tyr505-Side | UNK900-Side | UNK1-Side | GluU166-Side |
| UNK900-Side | Gln09-Side | Glu166-Main | UNK1-Side |
| Tyr453-Side | UNK900-Side | Asn142-Side | UNK1-Side |
| Gln409-Side | UNK900-Side | UNK1-Side | Asn142-Side |
| UNK1-Side | Phe140-Main | ||
| UNK1-Side | Gln189-Side | ||
| UNK1-Side | Cys145-Side | ||
| UNK1-Side | Ser46-Side | ||
| UNK1-Side | Leu141-Main | ||
| Gln189-Side | UNK1-Side | ||
| UNK1-Side | Ser144-Side | ||
| UNK1-Side | Thr25-Side | ||
| UNK1-Side | His41-Side | ||
MD, molecular dynamics.
Discussion
In this study, it was observed that none of the antimalarial drugs showed strong binding affinity with the active sites of either protease or the RBD. Considering the novelty of the virus, blind docking was performed with antimalarial drugs for further elucidation. Again, no significant binding was observed, as evident from the Glide scores (Table 1; Supplementary Material Tables S1–S4). This study confirmed that no antimalarial drug shows strong binding affinity for SARS-CoV-2 using molecular docking analysis.
Molecular docking analysis of FDA-approved drugs showed that paromomycin could bind both protease and RBD effectively, so it was selected for subsequent studies. Previous studies have used a single target approach for targeting coronavirus, either using protease or spike protein (RBD) as the target (Ortega et al., 2020, Xia et al., 2020). In the present study, both the catalytic and RBD domains were targeted side by side using a single drug, i.e. paromomycin. This dual targeted approach could have many advantages over a single target approach. The potential benefit of this strategy is that it may keep the virus from binding to the receptor (ACE2) and at the same time halt replication of the virus by binding to the catalytic domain of protease.
In the catalytic domain, paromomycin not only interacts with His-41 and Cys-145, but also with Glu-166. Besides the two key catalytic residues, Glu-166 is also considered to play a central role in maintaining the shape of the protease (S1) catalytic pocket, rendering it in active form (Zhang et al., 2020b). Further analysis of complex 2 using MD validated its stability. It is also evident from the binding energies of complex 2 that it is highly stable in the presence of the ligand. Further validation of high binding energy was supported by the hydrogen bond analysis of MD trajectories. Complex 2 showed 14 hydrogen bonds between the ligand and protease, which is well supported by the values of free binding energy.
In the case of spike protein, complex 1 showed strong binding affinity with the said residues. Some residues such as Asn-439, Asn-501, Gln-493, Gly-485, Phe-486, Arg-408, Gln-409, Thr-445, Val-417, Leu-461, Asp-467, Ser-469, Leu-491, Asn-492, Asp-493, Tyyr-494, Thr-497, Thr-150, and Tyr-504 are also considered important during interaction with receptors (ACE2 and CD26) (Lu et al., 2020, Vankadari and Wilce, 2020). So these residues must also be taken into account when suggesting a drug targeting the RBD. In this study, the paromomycin binding site with RBD showed interactions involving many of these residues. Overall, the binding energy and the number of hydrogen bonds in complex 1 were lower than those in complex 2, but it still showed good stability in the presence of ligand and can be considered well inhibited by it.
In addition to paromomycin, acarbose, an anti-diabetic drug, was also found to show strong binding affinity for both target proteins. As this drug is used for the diabetic population, we propose that it could be tested at the clinical level in diabetic patients suffering from COVID-19.
Paromomycin, a broad-spectrum aminoglycoside antimicrobial that was originally used to treat acute and chronic intestinal infections, may also be of potential use to treat COVID-19. We propose that paromomycin should be tested to reposition its therapeutic target for the treatment of COVID-19.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical approval and consent to participate
Not applicable.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgements
The authors acknowledge Schrödinger (https://www.schrodinger.com/) for providing a free license for Maestro suites 2019-4 on a humanitarian basis, particularly for this work. We also thank DrugBank (https://www.drugbank.ca) for permitting us to access the list of FDA-approved drugs.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.06.063.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Brooks B.R., III C.L.B., Jr A.D.M., Nilsson L., Petrella R.J., Roux B. CHARMM: The Biomolecular Simulation Program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis [Internet] 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen X., Luo G., Zhang X., He W., Li G. Discovery of potential inhibitors of squalene synthase from traditional Chinese medicine based on virtual screening and in vitro evaluation of lipid-lowering effect. Molecules. 2018;23(5):1040. doi: 10.3390/molecules23051040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol [Internet] 2020;92(4):418–423. doi: 10.1002/jmv.25681. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020:1–7. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Ge X., Li J., Yang X., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature [Internet] 2013;503:535–538. doi: 10.1038/nature12711. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-weber H., Schroeder S., Ger N.K., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T.Y., Frieman M., Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol [Internet] 2020:1–3. doi: 10.1038/s41565-020-0674-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey William, Dalke Andrew, VMD K.S. visual molecular dynamics. J Mol Graph [Internet] 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. http://www.ks.uiuc.edu/Research/vmd/ Available from: [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature [Internet] 2020 doi: 10.1038/s41586-020-2223-y. Available from: [DOI] [PubMed] [Google Scholar]
- Jo S., Cheng X., Islam S.M., Huang L. CHARMM-GUI PDB manipulator for advanced modeling and simulations of proteins containing nonstandard residues. Adv Protein Chem Struct Biol [Internet] 2014;96:235–265. doi: 10.1016/bs.apcsb.2014.06.002. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. Software news and updates CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem [Internet] 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. http://www.charmm-gui.org/ Available from: [DOI] [PubMed] [Google Scholar]
- Kim S., Lee J., Jo S., Iii L.B. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J Comput Chem [Internet] 2017;38(21):1879–1886. doi: 10.1002/jcc.24829. http://www.charmm-gui.org/?doc=input/pdbreader Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12(1):405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cong Y., Feng G., Zhong S., Zhang J.Z.H., Sun H. The impact of interior dielectric constant and entropic change on HIV-1 complex binding free energy prediction. Struct Dyn. 2018;5(6) doi: 10.1063/1.5058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hou T. CaFE: a tool for binding affinity prediction using end-point free energy methods. Bioinformatics. 2016;32(14):2216–2218. doi: 10.1093/bioinformatics/btw215. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet [Internet] 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: the main protease as molecular target. EXCLI J. 2020;19(Savarino 2005):400–409. doi: 10.17179/excli2020-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W.E.I., Gumbart J., Tajkhorshid E., Villa E. Scalable molecular dynamics with NAMD. J Comput Chem [Internet] 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. http://www.ks.uiuc.edu/Research/namd/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging Wuhan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect ISSN. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Kollman P.A. Free energy calculations on dimer stability of the HIV protease using molecular dynamics and a continuum solvent model. J Mol Biol. 2000;303(4):567–582. doi: 10.1006/jmbi.2000.4057. [DOI] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease 2019 (COVID-19) Situation Report – 83. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xintian X., Ping C., Jingfang W., Jiannan F., Hui Z., Xuan L. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin – converting enzyme 2 (ACE2) as a SARS – CoV 2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med [Internet] 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α -ketoamide inhibitors. Science (80-) 2020;3405:1–9. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature [Internet] 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, LLC; New York, NY: 2020. Schrödinger Release 2019-4: Protein Preparation Wizard; Epik, Schrödinger, Prime. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.