Dear Sir,
In April 2020, we published in this journal an article which highlighted the role of saliva as a reliable biological fluid to detect SARS-CoV-2.1
When we published the paper, Italy was on full lockdown at the peak of the COVID-19 epidemic.
Then, on June 3rd, 2020, the Italian government started the so-called “Phase 2″, which included the re-opening of working and social activities. In this framework, the issue of how to identify the asymptomatic individuals who, unwittingly, can spread SARS-CoV-2 infection and pose a threat to the public health has been raised worldwide.2 It is now imperative to guarantee the health and safety of the people called back to work, and to create a safety protocol in commercial and meeting spaces, which means preventing infected people from causing new epidemic outbreaks.
For this purpose, a well-established mass screening program is required to meet several needs: first, it should provide the result in a few minutes, it should be easily delivered on the territory, it should be performed in a simple way also by non-medical healthcare professionals, and eventually it should be non-invasive, repeatable and reliable.
To date, the diagnosis of SARS-CoV-2 infection is made by identifying the viral RNA in samples collected through a nasopharyngeal swab or other respiratory samples.3 This technique, however, has several limitations for its application in a mass screening, among which the most important ones are the time necessary for the diagnosis, the crowding of those centers appointed to analyze the specimens, and the non-negligible risk of viral transmission to the healthcare workers.4
The use of saliva as a diagnostic sample has several advantages, since it can be easily provided by the patient and it does not require specialized personnel for its collection.5 After these considerations, we conducted a diagnostic accuracy study to validate the use of a Rapid Salivary Test (RST) as a point-of-need antigen test suitable for a mass screening program.
Subjects who underwent the nasopharyngeal swab procedure for the diagnosis of SARS-CoV-2 infection were consecutively recruited in three independent medical areas in our hospital: the COVID-19 wards (inpatients, with the exclusion of those subjects admitted to the Intensive Care Unit), the Emergency Room (patients at high risk of disease) and the area for the healthcare workers (subjects at low risk of disease). At the same time of the nasopharyngeal swab procedure in the morning, each recruited subject provided a salivary sample of about 1 mL by the drooling technique.6
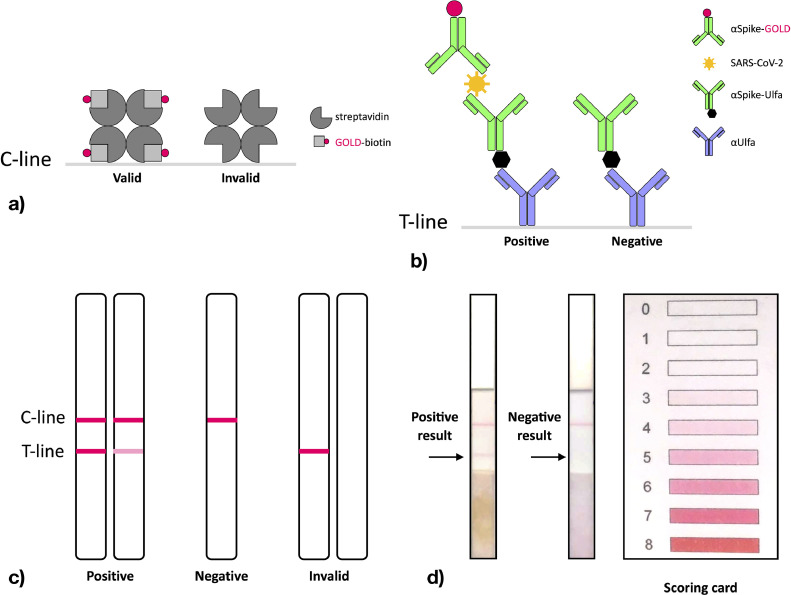
The RST consisted of an antigen test based on a customized Lateral flow assay (LFA) kit which was used to detect the presence of the virus in the saliva by identifying the viral Spike protein (Fig. 1 ).
Fig. 1.
Rapid Salivary Test based on Lateral flow technique and its interpretation. The customized sandwich LFA was designed to detect the presence of SARS-CoV-2 in salivary samples using a polyclonal antibody directed against the viral Spike protein. The same anti-Spike antibody (αSpike) was differentially conjugated in order to work as either capture antibody or detection antibody in the sandwich. Universal nitrocellulose LFA strips were used to perform the immunochromatography test. The applied sample was a mixture of diluted saliva and conjugated antibodies, added with an internal validity control (biotin). LFA results were read based on the appearance of a red “Control-line” (C-line) and a red “Test-line” (T-line) on the strip. (a) The C-line on the strip consists of immobilized streptavidin. 40 nm gold-conjugated biotin (GOLD-biotin) is added to the sample. When the flow of the sample reaches the C-line, streptavidin binds biotin with high affinity and the red C-line appears on the strip (valid test). If the flow does not reach the C-line, the test is invalid. (b) The T-line on the strip consists of immobilized anti-Ulfa-tag antibodies (αUlfa). The capture antibody is conjugated with the Ulfa-tag (αSpike-Ulfa), while the detection antibody is conjugated with 40 nm gold particles (αSpike-GOLD). When the Spike protein is present (positive test), the antibody sandwich forms and the red T-line appears on the strip. By contrast, when the Spike protein is absent (negative test), the sandwich does not form and the red T-line is not detectable. (c) The LFA strip consists of a nitrocellulose membrane, containing a “Control-line” (C-line) and a “Test-line” (T-line). The test is “positive” (presence of SARS-CoV-2) when both red lines are visible. The intensity of the T-line can be qualitatively evaluated using a scoring card. The test is “negative” (absence of SARS-CoV-2) when only the red C-line is detectable. The test is “invalid” when the red C-line is not visible, regardless of the presence of the red T-line. (d) Example of a run with a positive result (on the left) and of a run with a negative result (on the right). Both of these runs were valid since the control line appeared. The scoring card of the commercial kit (Abcam cat# ab270537) is shown on the right.
The nasopharyngeal swab was analyzed by independent blinded clinicians through real-time reverse transcription (rRT)-PCR accordingly to the International guidelines.7 In addition, the salivary sample collected for the RST was also examined by rRT-PCR to provide data about the presence of the virus in the saliva and to better analyze any discrepancy between the results of the RST and the nasopharyngeal swab.
A total number of 122 patients were recruited in this study (Fig. S1 – STARD flow diagram, Appendix). The mean age was 53.5 +/- 19.8 years, and there was a M:F = 1:2 ratio (Table S1, Appendix). Three subjects were excluded from the analysis because their RST failed and was not repeated. Thus, 119 subjects were included into the analysis. The results are reported in Table 1a . The sensitivity of the RST was 0.93 (95% CI: 0.77–0.99), while its specificity was apparently low, i.e., 0.42 (95% CI: 0.32–0.53). There were not differences between the recruited subgroups or among the asymptomatic and symptomatic individuals.
Table 1a.
Assessment of sensitivity and specificity of the RST test (with 95% confidence interval) with respect to the nasopharyngeal swab, in the overall sample and stratified according to the setting of recruitment and presence of COVID-19 symptoms at the time of the swab test.
| Sensitivity assessment | Specificity assessment | |||
|---|---|---|---|---|
| n, N^ | Sensitivity (95%CI) | n, N* | Specificity (95%CI) | |
| All subjects | 26, 28 | 0.93 (0.77; 0.99) | 38, 91 | 0.42 (0.32; 0.53) |
| Setting of the nasopharyngeal swab procedure | ||||
| COVID-19 hospitalized patients | 23, 25 | 0.92 (0.74; 0.99) | 4, 13 | 0.31 (0.09; 0.61) |
| ER patients | 2, 2 | 1.0 (0.16; 1.0) | 7, 18 | 0.39 (0.17; 0.64) |
| Healthcare workers | 1, 1 | 1.0 (-) | 27, 60 | 0.45 (0.32; 0.58) |
| COVID-19 symptoms | ||||
| Any symptom | 22, 24 | 0.92 (0.73; 0.99) | 7, 17 | 0.41 (0.18; 0.67) |
| No symptoms | 4, 4 | 1.0 (0.40; 1.0) | 31, 74 | 0.42 (0.31; 0.54) |
°: 3 subjects with a technically failed RST test (1 positive and 2 negative to the nasopharyngeal swab) were excluded.
^: n=number of subjects with positive RST, N=number of subjects with positive nasopharyngeal swab.
*: n=number of subjects with negative RST, N=number of subjects with negative nasopharyngeal swab
95%Confidence Interval (CI) from exact binomial distribution. (-): not reported.
One hundred fourteen subjects had their salivary sample also analyzed by rRT-PCR (Table 1b ). A very striking feature was observed when comparing the results of the salivary rRT-PCR with those of the nasopharyngeal swab in the subjects who had been previously classified as false negatives and false positives with the RST (Fig. S2a, Appendix). The two subjects who were classified as false negatives tested also negative by salivary rRT-PCR, thus the viral RNA was not detected in the saliva.
Table 1b.
Assessment of sensitivity and specificity of the RST test (with 95% confidence interval) with respect to results recorded by salivary rRT-PCR, in the overall sample and stratified according to the setting of recruitment and presence of COVID-19 symptom at the time of the swab test.
| Sensitivity assessment | Specificity assessment | |||
|---|---|---|---|---|
| n, N^ | Sensitivity (95%CI) | n, N* | Specificity (95%CI) | |
| All subjects° | 50, 55 | 0.91 (0.80; 0.97) | 35, 58 | 0.60 (0.47; 0.73) |
| Setting of the nasopharyngeal swab procedure | ||||
| Hospitalized patients with suspect COVID-19 | 24, 24 | 1.0 (0.86; 1.00) | 6, 11 | 0.55 (0.23; 0.83) |
| ER patients | 9, 11 | 0.82 (0.48; 0.98) | 5, 8 | 0.63 (0.24; 0.91) |
| Healthcare workers | 17, 20 | 0.85 (0.62; 0.97) | 24, 39 | 0.62 (0.45; 0.77) |
| COVID-19 symptoms | ||||
| Any symptom | 25, 27 | 0.92 (0.76; 0.99) | 7, 11 | 0.64 (0.31; 0.89) |
| No symptoms | 25, 28 | 0.89 (0.72; 0.98) | 28, 47 | 0.60 (0.44; 0.74) |
°: 3 subjects with technically failed RST test (2 positive and 1 negative to the rRT-PCR), and 6 subjects with technically failed rRT-PCR value (all RST positive) were excluded.
^: n=number of subjects with positive RST, N=number of subjects with positive nasopharyngeal swab.
*: n=number of subjects with negative RST, N=number of subjects with negative nasopharyngeal swab
95%Confidence Interval (CI) from exact binomial distribution. (-): not reported.
Startingly, 57% of the false positive cases had their saliva positive also when analyzed with rRT-PCR, which means that the virus was actually present and that the nasopharyngeal swab was less sensitive in these cases. These discrepancies between the salivary rRT-PCR and the nasopharyngeal swab were also confirmed by sequencing a sample of the positive specimens (Fig. S3, Table S2, Appendix). There were no differences in the viral load values among RST True positive (median value: 472 copies/μl, IQR: 145–975) and False positive subjects (median value: 371 copies/μl, IQR: 149–727; Kruskal–Wallis test p = 0.6) nor between asymptomatic or symptomatic individuals (median values: 480 vs 195 copies/μl, respectively; Kruskal–Wallis test p = 0.6) (Fig. S2b, Table S3, Appendix).
In our study we recorded a high sensitivity (i.e., 93%) and a mediocre specificity (i.e., 42%) of the RST.
These results were explained by two reasons. Firstly, the specificity suffered the fact that the majority of the presumed false positive individuals with the RST were rather positive also by salivary rRT-PCR, giving reason to the index test. Therefore, their nasopharyngeal swab (i.e., reference standard) provided a false negative result. Secondly, a certain degree of difficulty in reading the strip was reported by the observers, especially for low-intensity signals. In these cases, the observers tended to overestimate the positivity of the test, and this accounts for most of the remaining false positive cases. This fault will be corrected in the following stage of our project, when it translates into an industrial prototype which will be tested on the general population in the next weeks.
In conclusion, the RST based on LFA to detect the presence of SARS-CoV-2 may represent an innovative step in the diagnosis of the infection and in the armamentarium against the pandemic. Accordingly, it should be part of those policies of containment of the infection that the political decision-makers have to implement on the basis of Public Health Safety.
Study protocol
A detailed report of the clinical design of the study and of the laboratory procedures can be found in the Supplementary Appendix.
Trial registration
Local Ethical Committee (Comitato Etico dell'Insubria): protocol n° 68/2020.
ClinicalTrials.gov: NCT04357327.
This study adhered to the STARD-15 Guidelines.
Declaration of Competing Interest
The authors declare the absence of any conflict of interests. Alberio Tiziana, Azzi Lorenzo, Baj Andreina, Fasano Mauro and Lualdi Marta are the co-inventors of the Rapid Salivary Test described in this paper and of the Italian patent filing number 102,020,000,006,400 registered on 2020, March 26th.
Acknowledgments
Funding
This study was funded by the Department of Medicine and Surgery, University of Insubria.
Acknowledgements
The authors gratefully acknowledge all the colleagues fighting COVID-19 at each level and the Nurse staff directed by Aurelio Filippini who actively helped in the study recruitment process.
The authors are thankful to Dr Agostino Rossi, Dr Antonio Tamborini, Dr Marco Collura, Dr Angela Superchi and Mrs Laura Biella for their precious support in the study.
Prof Marina Tettamanti supervised the revision process in the English language.
A special thank deserves NatrixLab S.r.l., the company that will convert our experimental work in an industrial prototype the next weeks.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.06.042.
Contributor Information
Lorenzo Azzi, Email: l.azzi@uninsubria.it.
Vittorio Maurino, Email: v.maurino@uninsubria.it.
Appendix. Supplementary materials
References
- 1.Azzi L., Carcano G., Gianfagna F., Grossi P.A., Gasperina D.Dalla, Genoni A.P. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandhi M., Yokoe D.S., Havlir D.V. Asymptomatic transmission, the Achille's Heel of current strategies to control Covid-19. N Engl J Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol. 2020 doi: 10.1002/jmv.25721. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng K., Poon B.H., Kiat Puar T.H., Shan Quah J.L., Loh W.J., Wong Y.J. COVID-19 and the risk to health care workers: a case report. Ann Intern Med. 2020;172:766–767. doi: 10.7326/L20-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golatowski C., Salazar M.G., Dhople V.M., Hammer E., Kocher T., Jehmlich N. Comparative evaluation of saliva collection methods for proteome analysis. Clin Chim Acta. 2013;419:42–46. doi: 10.1016/j.cca.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 7.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.