Abstract
Glycosylation is one of the most important post-translational modifications (PTMs) with essential physiological functions, including protein folding, cell signaling and immune response. Thus, various qualitative and quantitative glycomics analysis strategies have been developed. Recently, isobaric multiplex reagents for carbonyl containing compound (SUGAR) tag was developed for quantitative glycomics with multiplexing capacity and increased reporter ion yield. To further improve quantification efficiency and enable quantifying low abundance species, mass defect based triplex SUGAR (mdSUGAR) tag has been designed. In addition, we also introduce additional reaction sites for mdSUGAR at terminal sialic acid by periodate oxidation of polyhydroxy chain to extend the mass difference and lower the requirement for resolving power. As a result, mdSUGAR tags show complete labeling efficiency, improved fragmentation pattern and accurate quantification. Moreover, the quantitative performance of the mdSUGAR tags in complex system has been systematically evaluated and demonstrated reliable results.
Keywords: Mass defect SUGAR tags, quantitative glycomics, N-glycans, periodate oxidation, mass spectrometry, relative quantification
INTRODUCTION
Glycosylation is a prevalent post-translational modification (PTM) with essential structural and biological functions including protein folding, cell signaling and immunity response.1 Abnormal glycosylation could lead to multiple diseases, such as immunological disorders, cardiovascular diseases and various cancers.2 Due to important functions of glycans, precise quantification approach is essential to understand complex biological processes. As glycan molecules cannot be easily detected by fluorescence or mass spectrometer due to lack of chromophore or poor ionization efficiency, derivatization is crucial for glycan analysis. Over the past few decades, various qualitative and quantitative glycomics analysis strategies have been developed. In addition to fluorescence labeling, such as 2-aminobenzoic acid (2-AA) and 2-aminobenzamide (2-AB),3 using stable isotope labeling method to improve the detection by mass spectrometry (MS) has become popular as it has the potential for highly multiplexed analysis with glycan structure elucidation capability.
The quantitative analysis with isotope labeling strategy relies on either reporter ion generated upon tandem MS fragmentation or peak area in full MS spectra. Isobaric tags, for example, aminoxyTMT,4 iART,5 QUANTITY6 and SUGAR,7 demonstrate the extensive sample multiplexing capacity for quantitative glycomics. To obtain quantitative result, the precursor needs to be isolated and fragmented to generate reporter ions. However, poor reporter ion yield was observed occasionally.8 Moreover, the need for additional fragmentation further limits the quantification rate, especially for low abundance glycans using commonly employed data dependent acquisition strategy. Isotopic9–13,14 and mass defect15–16 labeling strategies such as IDAWG metabolic labeling14 and DiPyrO tag labeling16 rely on peak area for relative quantification. Data interpretation can be problematic for isotopic labeling due to increased spectral complexity. By contrast, the subtle mass difference between mass defect tags enables relative quantification by high resolving power MS without increasing the spectral complexity.
The isobaric multiplex reagents for carbonyl containing compound (SUGAR) tag has been developed for quantitative glycomics recently.7 The simple building blocks of naturally occurring amino acids make it a cost-effective chemical probe with extensive multiplexing capacity. The tertiary amine structure in the SUGAR tag enhances ionization efficiency while stepwise reductive amination with hydrazide group enables complete labeling to further increase the detection sensitivity. With higher-energy collisional dissociation (HCD), reporter ions can be generated from SUGAR labeled glycans. Thus, the relative quantification can be obtained by comparing the intensities of reporter ions. Although SUGAR tags significantly improve the performance of isobaric tags for glycan analysis, they still suffer from drawbacks associated with the isobaric tagging approach: quantification rate is limited due to the required MS2 scans for quantification and low abundance glycan species tend to be difficult to quantify with lack of MS2 scans. Here we report the development of mass defect SUGAR (mdSUGAR) tags to eliminate the need for additional fragmentation for quantitative analysis, which is especially beneficial for quantitative analysis of low abundance glycans. Moreover, additional conjugation sites at sialic acid by periodate oxidation17–19 on polyhydroxy chain further diminish instrument resolving power requirement and improve fragmentation patterns of labeled N-glycans. The excellent quantification performance with broad dynamic range offered by the mdSUGAR tag has been demonstrated by labeling glycoprotein and complex human serum samples.
EXPERIMENTAL SECTION
Materials and reagents
Leucine (L-leucine-13C, L-leucine-13C6,15N, and L-leucine-D10), glycine (glycine-d2 and glycine-13C2,15N), heavy formaldehyde (CD2O), sodium cyanoborodeuteride were purchased from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA). Acetic acid (AA), acetonitrile (ACN), dimethyl sulfoxide (DMSO), formic acid (FA), methanol (MeOH), and water were purchased from Fisher Scientific (Pittsburgh, PA). Sodium cyanoborohydride, sodium meta-periodate, triethylammonium bicarbonate buffer (TEAB, 1.0 M) and tris(2-carboxy-ethyl) phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO). PNGase F was purchased from Promega (Madison, WI). Oasis HLB 1cm3 cartridges were purchased from Waters Corporation (Milford, MA). Bovine thyroglobulin (BTG) and human serum protein (HSP) were purchased from Thermo Fisher Scientific (Rockford, IL). Microcon-30kDa centrifugal filters (30K MWCO) were purchased from Merck Millipore Ltd. (Darmstadt, Germany). PolyGLYCOPLEX A™ beads (3 μm) were purchased from PolyLC Inc. (Columbia, MD). Fused silica capillary tubing (inner diameter 75 μm, outer diameter 375 μm) was purchased from Polymicro Technologies (Phoenix, AZ). All reagents were used without additional purification.
Enzymatic release of N-glycans by Filter-Aided N-Glycan Separation (FANGS)
Enzymatic release of N-glycans from protein samples was adapted from FANGS protocol.20 Briefly, for N-glycan release without oxidation, protein samples were dissolved at a concentration of 5 μg/μL in 0.5 M TEAB buffer with 5 μL 0.5 M TCEP and heat-denatured by switching sample tubes between 95 °C and room temperature water baths every 15 seconds for 2 minutes. For N-glycan release with oxidation, protein samples were dissolved at a concentration of 5 μg/μL in 0.1 M sodium acetate buffer at pH 5.5 and heat-denatured. Then sodium meta-periodate was added to the final concentration of 1 mM. The solution was incubated at dark for 30 minutes at room temperature. For both conditions, 30 K MWCO filters were used for 3 times buffer exchange with 200 μL of 0.5 M TEAB. The prepared protein samples on the MWCO filter were then incubated with 5 μL PNGase F in 95 μL 0.5 M TEAB buffer at 37 °C overnight. The filters were washed with 100 μL 0.5 M TEAB buffer for three times. The fractions were combined and dried in vacuo. To convert glycosylamine to glycan with free reducing end, 200 μL of 1% AA was added and incubated for 4 h and dried in vacuo.
N-glycan labeling by mass-defect SUGAR tags
Released N-glycans were mixed with 1 mg mdSUGAR tag in 100 μL MeOH containing 1% FA. The solvent was removed in vacuo after 10 min incubation. The reduction was performed in 100 μL 1 M NaBH3CN in DMSO: AA (7:3 v/v) at 70 °C for 2 h.
Oasis HLB 1 cm3 cartridge was used to remove excess mdSUGAR tag and chemical reagents. The cartridge was conditioned with 1 mL 95% ACN, water, and 95% ACN respectively. The reaction mixture was loaded to the cartridge with pre-filled 1 mL of 95% ACN. Then, the cartridge was washed with 1 mL of 95% ACN for 3 times, and the mdSUGAR labeled N-glycans were eluted with 1 mL 50% ACN and 1 mL water. The eluting fractions were combined and dried in vacuo, reconstituted in 50 μL of 75% ACN, and analyzed by MALDI-MS or LC-MS/MS immediately.
Matrix-assisted laser desorption/ionization (MALDI)-MS analysis
Samples were prepared by premixing 1 μL of mdSUGAR-labeled N-glycans with 1 μL 2,5-dihydroxybenzoic acid matrix (150 mg/mL in 2% N, N-dimethylaniline, 49% MeOH and 49% water), and 1 μL of each matrix/sample mixture was spotted onto the MALDI target plate. A MALDI-LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany) was used for MALDI-MS analysis. Ionization was performed using a laser energy of 15 μJ. Spectra were acquired in the Orbitrap mass analyzer within a mass range of m/z 1,000–4,000 at a mass resolution of 30 K (at m/z 400).
LC-MS/MS analysis
A self-fabricated nano-hydrophilic interaction chromatography (HILIC) column (15 cm, 75 μm i.d., 3 μm PolyGlycoPlex A HILIC beads) was used for glycan separation. A Dionex Ultimate 3000 nanoLC system was coupled to an Orbitrap Fusion Lumos Tribrid quadrupole-ion trap-Orbitrap Mass Spectrometer with a NanoSpray Flex ion source (Thermo Scientific, Bremen, Germany) for all LC-MS/MS analyses. Mobile phase A was water with 0.1% FA, and mobile phase B was ACN with 0.1% FA. The flow rate was set at 0.3 μL/min, and the injection volume was 2 μL. The following gradient was used (time, % mobile phase B) unless otherwise specified: (0 min, 75%), (18 min, 75%), (78 min, 15%), (78.1 min, 10%), (90 min, 10%), (90.1 min, 75%), (100 min, 75%).
The following mass spectrometer parameters were used for all data acquisition. Samples were ionized in positive ion mode with a spray voltage of 3 kV. S-lens radio frequency (RF) level was set to be 30, and capillary temperature was set to be 300 °C. Full MS scans were acquired at m/z 500–2000 with resolving power of 500 K (at m/z 200). Maximum injection time of 50 ms, automatic gain control (AGC) target value of 1e5, and 1 microscan were used for full MS scans. Top 5 data-dependent MS2 analysis was performed at a resolving power of 30 K (at m/z 200) with collision-induced dissociation (CID) operating with a normalized collision energy of 30. The dynamic exclusion of acquired precursors was set to 15 sec with a ± 20 ppm tolerance.
N-glycan data analysis
The raw data was compared against an in-house database including most possible combinations of N-glycan units (Hexose (H), HexNAc (N), Fucose (F), and NeuAc (S)). mdSUGAR-labeled N-glycans were identified by accurate mass matching in full MS with a mass tolerance of 10 ppm and fragmentation match in MS/MS spectra. Peak areas for mdSUGAR-labeled glycans were used for relative quantification. Microsoft Excel and Origin were used for calculations and statistical analyses.
RESULTS AND DISCUSSION
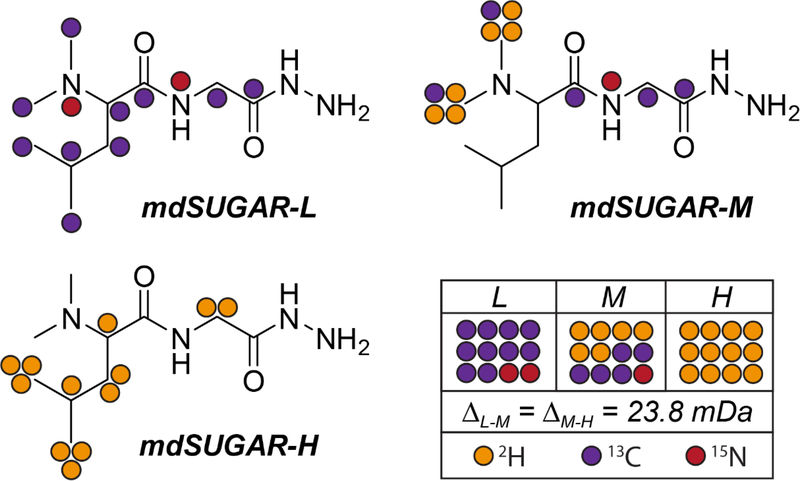
The structures of triplex mdSUGAR tags are shown in Figure 1 with three configurations (L, M and H) labeled with colored dots representing heavy isotope positions. In total, 12 heavy isotopes including 2H, 13C and 15N, have been incorporated into triplex mdSUGAR tags. The mass difference remains 23.8 mDa between adjacent channels. The synthetic route of mdSUGAR is illustrated in Figure S1. The triplex mdSUGAR tags were synthesized in three steps with commercially available heavy isotope coded starting materials respectively, and shown over 95% isotope purities in Figure S2.
Figure 1.
Structure and isotope configurations of triplex mdSUGAR tags. Orange dot: 2H, purple dot: 13C, red dot: 15N.
Due to subtle mass difference between mass defect chemical tags, ultrahigh resolving power is essential for mass defect based quantification strategy. The multiplexing capacity is often limited to duplex for quantitative glycomics with mass defect tags.15–16 To increase the multiplexing capacity for mass defect strategy, extended mass difference can effectively reduce the resolving power requirement. However, with commonly used reductive amination derivatization, only reducing end can react with mdSUGAR tag for conjugation. As larger glycans require higher resolving power for accurate quantification while they often contain one or multiple sialic acids at terminal position, periodate oxidation of polyhydroxy chain in sialic acid could be utilized to generate additional aldehyde for mdSUGAR conjugation. With multiple reaction sites to extend mass difference, the triplex mdSUGAR tags have been developed for quantitative glycomics.
To generate additional reaction sites with mdSUGAR tag, aldehyde group could be readily introduced by mild periodate oxidation, which is known to selectively oxidize the polyhydroxy chain of sialic acid.17–19 Then, reducing end of glycan and newly generated aldehyde group were labeled with hydrazide group of mdSUGAR tag. To prevent reduction of aldehyde group prior to hydrazone formation with absence of high concentration of NaBH3CN, stepwise reductive amination was developed. The glycans were mixed with mdSUGAR tag in methanol containing 1% formic acid for 10 minutes to accelerate hydrazone formation. Then the reduction was performed in dimethyl sulfoxide and acetic acid with 1 M NaBH3CN to achieve complete irreversible conjugation. Excellent labeling efficiencies for multiple types of N-glycans including high mannose, fucosylated and sialylated N-glycans are shown in Figure S3. The workflow for mdSUGAR labeling is shown in Scheme S1.
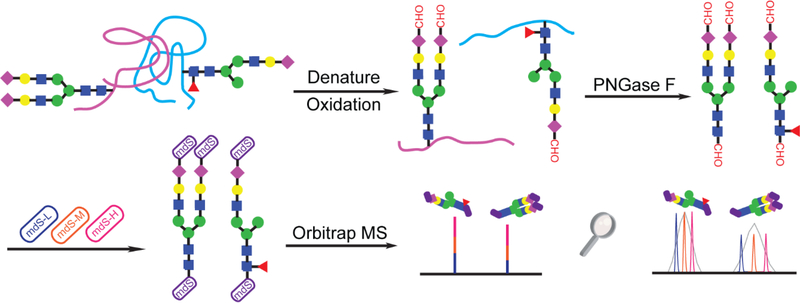
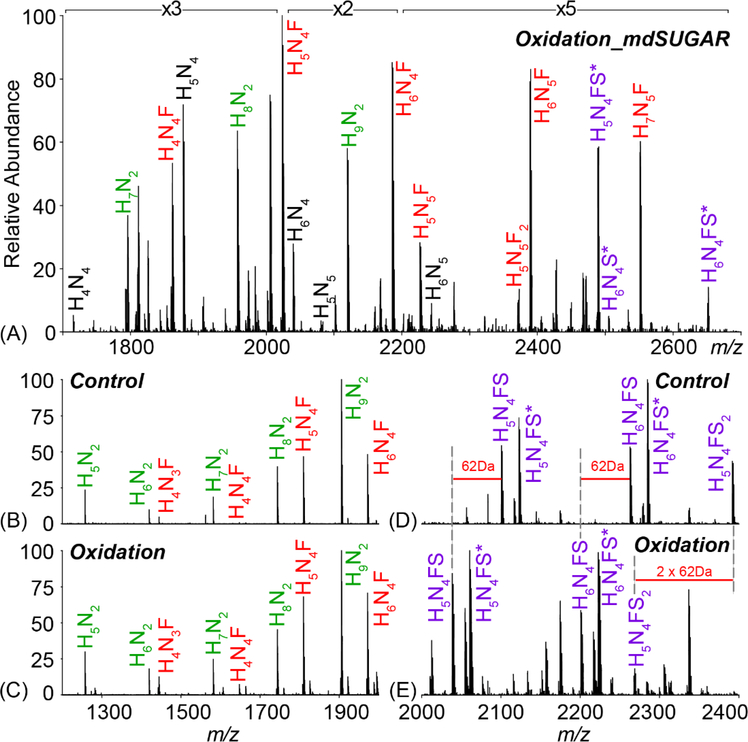
Periodate oxidation and stepwise reductive amination with mdSUGAR tags were further applied to label N-glycans released from bovine thyroglobulin (BTG) glycoprotein standard. Figure 2 depicts the workflow for quantitative glycomics with mdSUGAR tags. Mild periodate oxidation is performed prior to enzymatic release of N-glycans by PNGase F. With optimized stepwise reductive amination, N-glycans are labeled completely with mdSUGAR tags at both reducing end and terminal sialic acid. The quantitative analysis can be achieved with high resolving power MS. Figure 3A highlighted multiple types of N-glycans labeled with mdSUGAR including high mannose, fucosylated and sialylated N-glycans. The performance of periodate oxidation was also illustrated in Figure 3B–E. Almost identical spectra were observed for high-mannose and fucosylated N-glycans before and after periodate oxidation (Figure 3B–C) due to lack of polyhydroxy chain while 62 Da mass loss was observed (Figure 3D–E) for N-glycans containing single sialic acid (H5N4FS and H6N4FS) and 124 Da mass loss for H5N4FS2.
Figure 2.
Workflow of quantitative glycomics analysis by triplex mdSUGAR.
Figure 3.
MALDI-MS spectra of N-glycans (Na+ adduct) released from BTG: mdSUGAR labeled with periodate oxidation (A). High mannose (green label) and fucosylated (red label) N-glycans showed almost identical spectral profile prior to (B) and after (C) periodate oxidation due to lack of sialic acid. Highlighted acidic N-glycans (purple labels) showed mass loss before (D) and after (E) periodate oxidation. Mass shift of 62 Da was illustrated for each sialic acid after periodate oxidation. Star represents N-glycans as [M-H+2Na]+.
In order to maximize the separation of mdSUGAR labeled glycans with minimum ion coalescence, the resolving power and automatic gain control (AGC) were carefully optimized. Triplex mdSUGAR labeled glycan samples with 1:1:1 ratio were evaluated with resolving power at 120K, 240K and 500K (at m/z 200). At 120K, the triplex peaks were unresolved, while at 240K, most peaks were partially resolved. At 500K, baseline separation was observed for most glycans but not for larger glycans such as H5N4FS2 with only reducing end labeling (Figure S4). With periodate oxidation, additional reaction sites were introduced to label mdSUGAR for extending mass difference, all triplex peak groups were baseline resolved at 500K (Figures S5–6). Space-charge-induced ion coalescence occurs with excessively large ion populations accumulating in the orbitrap mass analyzer.21 The AGC target was evaluated to minimize ion coalescence. AGC of 1e5, 2e5, 5e5 and 1e6 were tested at a resolving power of 500K. No coalescence was observed at AGC of 1e5 while peaks started to merge together at AGC of 2e5 and unresolved at AGC of 1e6 (Figure S7). Thus, resolving power of 500K with AGC target of 1e5 was used for subsequent experiments.
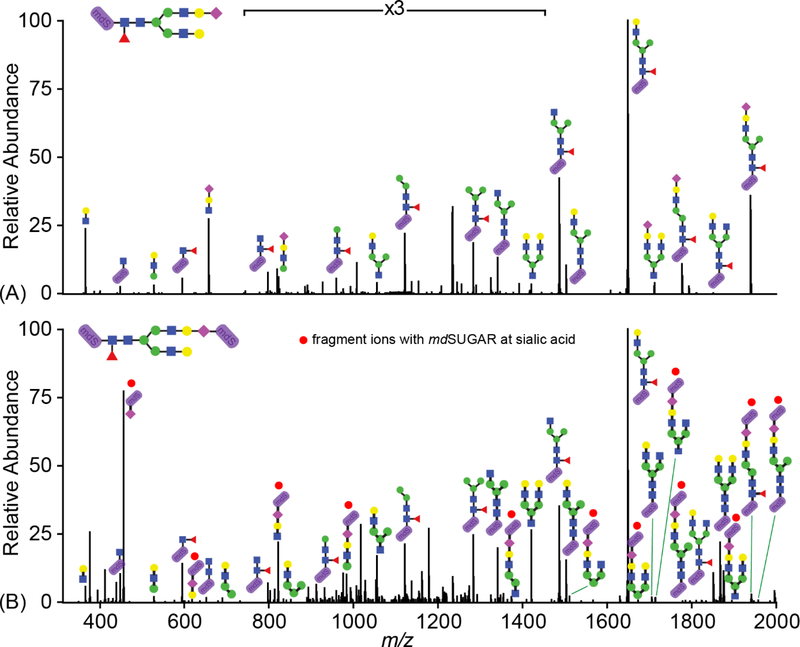
Although MS/MS fragmentation is not required for relative quantification with mass defect based strategy, it is essential for N-glycan structural elucidation. The CID-MS/MS spectrum in Figure 4A demonstrated the fragmentation pattern for H5N4FS labeled with mdSUGAR at reducing end. Due to higher proton affinity of tertiary amine in mdSUGAR structure, labeled N-glycans tend to generate y ions with mdSUGAR attached to the fragments. With additional labeling at terminal sialic acid, significant improvement of b ion (with red dots) production was observed from precursor glycan shown in Figure 4B which facilitated confident structural annotation.
Figure 4.
ESI-MS/MS fragmentation comparison of mdSUGAR-labeled H5N4FS N-glycan at only reducing end (A) or reducing end and terminal sialic acid (B). Fragments with red dots are fragment ions with mdSUGAR at sialic acid generated from labeled H5N4FS with periodate oxidation.
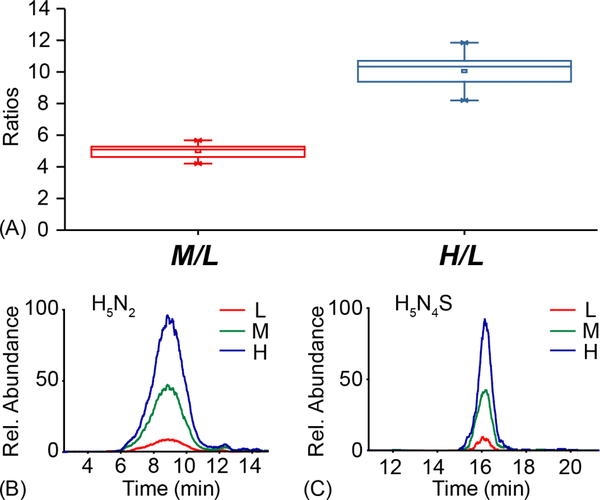
Performance of quantitative analysis with triplex mdSUGAR tags was evaluated by labeling N-glycans at known ratios. N-glycans released from BTG were aliquoted with 1:1:1, 1:5:10 and 10:5:1 ratios in triplicate and labeled with triplex mdSUGAR tags respectively. Peak areas for each N-glycan were used to calculate the experimental ratios. Figure 5A illustrated the experimental ratios with triplex mdSUGAR against theoretical ratio of 1:5:10. Representative extracted ion chromatograms are shown in Figures 5B–C for two types of N-glycans. The representative spectra for 1:1:1 are shown in Figures S3–4. For quantitative analysis with known ratios, less than 20% relative errors were observed, demonstrating the accurate quantification performance with mdSUGAR tags. In addition, no obvious LC retention time shift was noticed for triplex mdSUGAR labeled N-glycans with HILIC column separation.
Figure 5.
Relative quantification performance of triplex mdSUGAR-labeled N-glycans released from BTG. Labeled N-glycans were mixed at ratios of 1:5:10 and analyzed in triplicate. The peak areas of M/L and H/L are plotted. Box plots show the median (line), the 25th and 75th percentile (box), and the 5th and 95th percentile (whiskers) (A). Representative extracted ion chromatograms for mdSUGAR-labeled N-glycans H5N2 (B) and H5N4S (C) are shown.
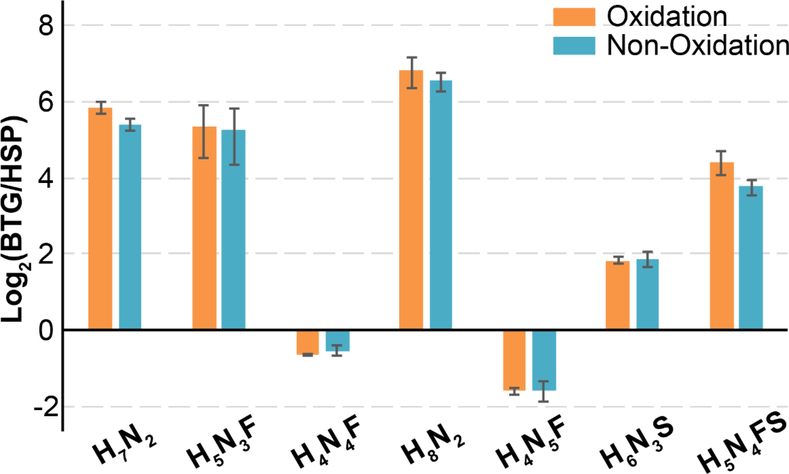
To further evaluate whether additional periodate oxidation would cause quantification bias, N-glycans released from the same amount of BTG and human serum protein (HSP) were labeled with mdSUGAR via either reducing end only labeling or reducing end and sialic acid labeling. With limited resolving power of 500K available, larger N-glycans labeled with triplex mdSUGAR at only reducing end were unable to be fully separated (Figure S4), leading to inaccurate quantification. Thus, only duplex mdSUGAR-L and H were used for the evaluation. The N-glycan ratios of BTG/HSP from different labeling strategies were plotted in Figure 6. In total, 53 N-glycans were identified and quantified with reducing end strategy while 95 N-glycans were detected with the periodate oxidation strategy. The full list of quantified N-glycans by different strategies were summarized in Table S1 and shown that multiple low abundance glycans can only be detected and quantified by mdSUGAR strategy. Most N-glycans showed significant abundance in BTG. However, several N-glycans including H4N4F and H4N5F were present at higher abundance in HSP. Although some N-glycans, for example H7N2 and H5N4FS, showed slight variation using different labeling strategies which might be due to the altered efficiency of enzymatic digestion caused by incomplete denaturation and disulfide bond reduction. Most N-glycans exhibited comparable quantification results with both labeling strategies, demonstrating the excellent quantification performance by incorporating additional periodate oxidation with mdSUGAR tags.
Figure 6.
Selected N-glycan relative quantification released from equal amount of BTG and HSP. Ratios represent peak areas for mdSUGAR-labeled N-glycans. Error bars represent the standard deviation of three replicates.
CONCLUSIONS
In summary, triplex mdSUGAR tags with periodate oxidation were developed in this study for quantitative glycomics. Periodate oxidation at polyhydroxy chain provided additional conjugation sites to extend mass difference and minimize instrument requirement. Stepwise reductive amination was utilized to enable complete labeling with hydrazide group of mdSUGAR tags. The mdSUGAR labeled glycans demonstrated improved fragmentation pattern to generate abundant b ions for structural elucidation. Triplex mdSUGAR tags also showed accurate quantification with broad dynamic range. Moreover, mdSUGAR tags with periodate oxidation strategy was applied to compare the abundance of N-glycans released from BTG and HSP. The successful development of mdSUGAR approach offers a platform for N-glycan analysis to generate multiple conjugation sites, especially for mass defect labeling with limited resolving power instrument. In conclusion, we anticipate that the triplex mdSUGAR with periodate oxidation strategy can be widely applied in multiple biological and clinical studies.
Supplementary Material
ACKNOWLEDGMENTS
Support for this research was provided in part by the NIH grants U01CA231081, R01 DK071801, R21 AG055377, RF1 AG052324, and a Robert Draper Technology Innovation Fund grant with funding provided by the Wisconsin Alumni Research Foundation (WARF). The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and the University of Wisconsin-Madison, Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. LL acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website
REFERENCES
- 1.Varki A; Gagneux P, Biological Functions of Glycans In Essentials of Glycobiology, rd; Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Darvill AG; Kinoshita T; Packer NH; Prestegard JH; Schnaar RL; Seeberger PH, Eds. Cold Spring Harbor; (NY), 2015; pp 77–88. [PubMed] [Google Scholar]
- 2.Varki A; Kannagi R; Toole B; Stanley P, Glycosylation Changes in Cancer In Essentials of Glycobiology, rd; Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Darvill AG; Kinoshita T; Packer NH; Prestegard JH; Schnaar RL; Seeberger PH, Eds. Cold Spring Harbor; (NY: ), 2015; pp 597–609. [Google Scholar]
- 3.Ruhaak LR; Zauner G; Huhn C; Bruggink C; Deelder AM; Wuhrer M, Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem 2010, 397 (8), 3457–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahne H; Neubert P; Kuhn K; Etienne C; Bomgarden R; Rogers JC; Kuster B, Carbonyl-reactive tandem mass tags for the proteome-wide quantification of N-linked glycans. Anal Chem 2012, 84 (8), 3716–24. [DOI] [PubMed] [Google Scholar]
- 5.Yang S; Yuan W; Yang W; Zhou J; Harlan R; Edwards J; Li S; Zhang H, Glycan analysis by isobaric aldehyde reactive tags and mass spectrometry. Anal Chem 2013, 85 (17), 8188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S; Wang M; Chen L; Yin B; Song G; Turko IV; Phinney KW; Betenbaugh MJ; Zhang H; Li S, QUANTITY: An Isobaric Tag for Quantitative Glycomics. Sci Rep 2015, 5, 17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y; Chen B; Yu Q; Zhong X; Frost DC; Ikonomidou C; Li L, Isobaric Multiplex Labeling Reagents for Carbonyl-Containing Compound (SUGAR) Tags: A Probe for Quantitative Glycomic Analysis. Anal Chem 2019, 91 (4), 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B; Zhong X; Feng Y; Snovida S; Xu M; Rogers J; Li L, Targeted MultiNotch MS(3) Approach for Relative Quantification of N-Glycans Using Multiplexed Carbonyl-Reactive Isobaric Tags. Anal Chem 2018, 90 (2), 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker SH; Budhathoki-Uprety J; Novak BM; Muddiman DC, Stable-isotope labeled hydrophobic hydrazide reagents for the relative quantification of N-linked glycans by electrospray ionization mass spectrometry. Anal Chem 2011, 83 (17), 6738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker SH; Taylor AD; Muddiman DC, Individuality Normalization when Labeling with Isotopic Glycan Hydrazide Tags (INLIGHT): a novel glycan-relative quantification strategy. J Am Soc Mass Spectrom 2013, 24 (9), 1376–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecht ES; McCord JP; Muddiman DC, Definitive Screening Design Optimization of Mass Spectrometry Parameters for Sensitive Comparison of Filter and Solid Phase Extraction Purified, INLIGHT Plasma N-Glycans. Anal Chem 2015, 87 (14), 7305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z; Zhong X; Tie C; Chen B; Zhang X; Li L, Development of a hydrophilic interaction liquid chromatography coupled with matrix-assisted laser desorption/ionization-mass spectrometric imaging platform for N-glycan relative quantitation using stable-isotope labeled hydrazide reagents. Anal Bioanal Chem 2017, 409 (18), 4437–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei L; Cai Y; Yang L; Zhang Y; Lu H, Duplex Stable Isotope Labeling (DuSIL) for Simultaneous Quantitation and Distinction of Sialylated and Neutral N-Glycans by MALDI-MS. Anal Chem 2018, 90 (17), 10442–10449. [DOI] [PubMed] [Google Scholar]
- 14.Orlando R; Lim JM; Atwood JA 3rd; Angel PM; Fang M; Aoki K; Alvarez-Manilla G; Moremen KW; York WS; Tiemeyer M; Pierce M; Dalton S; Wells L, IDAWG: Metabolic incorporation of stable isotope labels for quantitative glycomics of cultured cells. J Proteome Res 2009, 8 (8), 3816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atwood JA 3rd; Cheng L; Alvarez-Manilla G; Warren NL; York WS; Orlando R, Quantitation by isobaric labeling: applications to glycomics. J Proteome Res 2008, 7 (1), 367–74. [DOI] [PubMed] [Google Scholar]
- 16.Chen B; Feng Y; Frost DC; Zhong X; Buchberger AR; Johnson J; Xu M; Kim M; Puccetti D; Diamond C; Ikonomidou C; Li L, Quantitative Glycomic Analysis by Mass-Defect-Based Dimethyl Pyrimidinyl Ornithine (DiPyrO) Tags and High-Resolution Mass Spectrometry. Anal Chem 2018, 90 (13), 7817–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bank PA; Kellam B; Kendall DA; Shakesheff KM, Surface engineering of living myoblasts via selective periodate oxidation. Biotechnol Bioeng 2003, 81 (7), 800–8. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y; Ramya TN; Dirksen A; Dawson PE; Paulson JC, High-efficiency labeling of sialylated glycoproteins on living cells. Nat Methods 2009, 6 (3), 207–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrientos RC; Zhang Q, Isobaric Labeling of Intact Gangliosides toward Multiplexed LC-MS/MS-Based Quantitative Analysis. Anal Chem 2018, 90 (4), 2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdul Rahman S; Bergstrom E; Watson CJ; Wilson KM; Ashford DA; Thomas JR; Ungar D; Thomas-Oates JE, Filter-aided N-glycan separation (FANGS): a convenient sample preparation method for mass spectrometric N-glycan profiling. J Proteome Res 2014, 13 (3), 1167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorshkov MV; Fornelli L; Tsybin YO, Observation of ion coalescence in Orbitrap Fourier transform mass spectrometry. Rapid Commun Mass Spectrom 2012, 26 (15), 1711–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.