Abstract
Background and purpose:
An anticancer peptide P28, has shown to be cytolethal on various cancer cells including breast cancer. Moreover, p28 can be also used as a targeting moiety in the structure of fusion proteins. IL-24 (or its truncated form, M4) is a cytokine with anticancer activity against a wide range of tumor cells. We aimed at production of a fusion protein consisted of p28 and either IL-24 or M4 to target breast cancer. However, selection of a proper linker to join the two moieties without intervening each other’s function is a key factor in the construction of fusion proteins. In the present study, the impact of different linkers on construction of the two chimeric proteins (p28-IL-24 and p28-M4) was assessed in silico.
Experimental approach:
After selection of some linkers with different lengths and characteristics, a small library of the chimeric proteins was created and assessed. Furthermore, following selection of the most suitable linker, the three-dimensional structures and dynamic behavior of both fusion proteins were evaluated by homology modeling and molecular dynamic simulation, respectively.
Findings / Results:
Based on the results, a rigid linker having the peptide sequences of AEAAAKEAAAKA showed highest freedom of action for both moieties.
Conclusion and implications:
Between the p28-IL-24 and p28-M4 fusion proteins, the former showed better stability as well as solubility and might show stronger anticancer effects in vitro and in vivo, because its peptide moieties showed to exert their activities freely.
Keywords: p28, IL-24, fusion protein, Homology modeling, Molecular Dynamic Simulation, Breast cancer
INTRODUCTION
Cancer is a term used to describe more than one hundred malignant diseases affecting many different tissues and cell types which characterized by rapid growth of abnormal cells. It is the second most common causeof death worldwide and expected that the yearly death toll due to cancer will reach around 16 million by 2040 (1). The most common forms of cancer are breast, lung, liver, prostate, and colorectal cancers. Chemotherapy, radiotherapy, and surgery are some curative therapies available to fight cancer, among which chemotherapy is still the best strategy applied to treat advanced or metastatic cancers (1,2,3). However, most of the available chemotherapeutics are not selective for cancer cells and also affect normal cells, resulting in severe side-effects (4). Therefore, development of novel forms of anticancer drugs with high selectivity and specificity against tumor cells is obligatory (5,6). One of the approaches for pharmacotherapy of cancer is the use of peptides and fusion proteins. Properties such as good solubility, low toxicity, tumor penetration, high selectivity, application either alone or in combination with other chemotherapeutics, and the possibility of redirecting them toward different types of cancers make peptides/proteins a suitable nominee for drug design. According to the previous reports, some of these peptides can be used for a variety of functions such as anticancer activity and cell penetration as well as tumor homing capability (7,8).
A growing number of literatures have shown that a part of azurin protein, namely p28 has the ability of selective entrance into the breast cancer cells by caveolae-mediated endocytosis, therefore can be considered as an appropriate agent for preferential transitions of peptides or other cytotoxic drugs to cancer cells (9,10,11). p28 peptide has completed phase I clinical trials for the treatment of solid tumors in the United States (12).
Interleukin 24 (IL-24) is a cytokine belonging to the IL-10 cytokine family, produced by both immune and non-immune cells which plays several major antiproliferative roles (13). It can selectively induce apoptosis in cancer cells without a significant effect on normal cells (14). Cancer cell death inducing activity of IL-24 can be performed via binding to its receptor and concurrent activation of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (15,16). Previously it has been shown that intracellular IL-24 can induce endoplasmic reticulum (ER) stress in another way. IL-24 has been shown to suppress the translation of some proteins in mitochondria through ER stress leading to apoptosis (17).
M4 is a truncated segment of the IL-24 consisting of its amino acid residues 102 to 206. It exhibits similar biological properties and activities to full-length IL-24. M4 shows a pro- apoptotic activity by selective reduction in viability of cancer cells through ER stress. In fact, it seems that M4 is responsible for the pro- apoptotic activity of IL-24 (18,19). Therefore, in order to provide a novel targeted anticancer remedy, we aimed at production of fusion proteins p28-IL24 and p28-M4 to take advantage of both selective delivery to and selective activity on cancer cells. However, choosing a suitable linker to join the two parts of a fusion protein is an indispensable element for successful construction of a recombinant fusion protein.
The linker must act in a manner that none of the moieties intervene the other’s function (20). Majority of the proteins in nature generally consist of several distinctive functional domains, which have been attached to each other by linker peptides. Based on properties like length, amino acid composition, hydrophobicity, and spatial conformation, linkers can be classified in 3 series of rigid, flexible, and in vivo cleavable linkers (20,21). The first step in evaluation of a new drug candidate is in silico analysis by bioinformatics approach and computational techniques such as molecular dynamics (MD) simulation. Interesting features like molecular interactions, physical phenomena’s, minima geometries of proteins, binding free energy of drugs and detailed motion of molecules or atoms during a fixed period of time can be studied via MD simulation. This technique also has been able to check the location and wrapping, distance, interaction of residues and possibly intervening of the targeting and toxic moieties in fusion protein assessment and design (22,23,24).
In the current study we designed two fusion proteins including p28-IL-24 and p28-M4 in which p28 acts as a targeting or cell penetrating moiety and IL-24/M4 functions as a toxic moiety. However, due to difference in charge between the targeting and toxic moieties the probability of salt bridge formation between the two parts is high.
This, consequently, may greatly affect their function, and therefore an appropriate linker must be applied to keep the mentioned moieties separated. Hence, following selection of some linkers with different lengths and characteristics, we created a small library of the fusion proteins connected by the selected linkers and evaluated them. All linkers assessed here were selected from common linkers used in other studies. Then, after selection of the most appropriate linker, the three-dimensional (3D) structures and dynamic behavior of both fusion proteins were evaluated by homology modeling and molecular dynamic simulation, respectively.
MATERIALS AND METHODS
Peptide sequences
A small library of linker peptides with different lengths and characteristics were selected according to literatures and then used to connect the p28 and IL-24 or M4 peptide sequences in silico. Table 1 represents the amino acid sequences and the features corresponding to each linker.
Table 1.
Sequence, length and type of the linkers used in the study.
| Linker sequences | Linker types |
|---|---|
| (PA)1-5P | Rigid linker |
| (EAAAK)1-3 | Rigid linker |
| PAPAP | Rigid linker |
| AEAAAKEAAAKA | Rigid linker |
| A(EAAAK)nA(n=2–5) | Rigid linker |
| (GGGGS)3 | Flexible linker |
| EGKSSGSGSESKST | Flexible linker |
| (GGGGS)n (n = 1,2,4) | Flexible linker |
| (Gly)6 | Flexible linker |
| GSAGSAAGSGEF | Flexible linker |
P, Proline; A, alanine; E, glutamic acid; K, lysine; G, glycine; S, serine; T, threonine; F, phenylalanine; n, the n, the number of repeats.
Homology modeling
In the first step, 3D structure of the two fusion proteins were constructed with all linkers represented in Table 1 using I-TASSER (http://zhang.bioinformatics.ku.edu/I-TASSER) online server (19). Then, to assess the possibility of salt bridge formation between functional groups of the two moieties, VMD 1.9.3 software (25) was applied. Moreover, MODELLER software 9.13 (26) was used to obtain an accurate 3D structures of the fusions. By crystal structure of the selected templates, multiple template modeling was performed. For each one of our fusion proteins, 1000 models have been made via homology modeling. The model with minimum molpdf energy was chosen as the best model (27). Quality of the models was validated through MolProbity (http://molprobity.biochem.duke.edu/), and ProSA-web (an online server which demonstrates local model quality by plotting energies as a function of amino acid sequence position) (28), and the quality of the structures was analyzed by Ramachandran plots (a plot which indicates the number of all residues located in favored regions as well as in allowed regions). These plots are used to determine the validity and quality of protein models depending on the number of residues on allowed or disallowed regions (29).
Molecular dynamic simulation
The MD simulation of p28-IL-24 and p28- M4 fusion proteins were performed as described previously (21). Briefly, for each fusion protein the best models obtained from homology modeling were subjected to MD simulation thought GROMACS 4.5.6 package (30), under Gromos force field (G43A1) which carried out at the constant temperature, constant pressure ensemble (NPT), and periodic boundary condition (31). The system neutralizing process was done by adding 4 Na+ ions and 1 Na+, 1 Cl- in p28-IL-24 and p28-M4, respectively. The solvation process of fusion proteins was carried out by a layer of at least12 Å in all directions. MD simulation was performed in 4 steps based on our previous study (21).
In the production phase or final step, 50 ns MD simulations was performed for each fusion protein and final structures were obtained. The production step was accomplished at 300 K with 2 fs time step.
RESULTS
Homology modeling
The I-TASSER server was used for prediction of 3D structures of the fusion proteins with all types of linkers described in Table 1. Among the 5 models predicted by this server, the model with minimum value of confidence scores was selected as the best predicted model. VMD software (version 1.9.2) was used for visual assessment of salt bridges within 3D structure of these models. The study of linkers with different lengths and types revealed that amongst all types of linkers mentioned in Table 1, AEAAAKEAAAKA can efficiently prevent the formation of salt bridges between functional domains of our two fusion proteins. In the next step, to improve the quality of models as well as producing more accurate 3D structures, the MODELLER software was used.
Since there is no template available for homology modeling of p28, IL-24, and M4 peptides in the protein databases, we used BLAST algorithm against protein data bank (PDB) to identify homologous sequences of the p28 peptide, AEAAAKEAAAKA linker, and IL-24 or M4 peptides, separately. For this purpose, according to maximum sequence similarity, PDB entries 2FT6, 1KYO, and 6DF3 were used as templates for p28 peptide, AEAAAKEAAAKA linker, and IL-24 or M4, respectively.
The chimera templates created via alignment of the selected templates were used for homology modeling by MODELLER.
The Ramachandran plot of the first fusion protein composed of p28-AEAAAKEAAAKA- IL-24 showed 97.5% (194/199) of all residues were in favored regions and 100.0% (199/199) of all residues were in allowed regions in the obtained model from MODELLER and there were no outliers (Fig. 1A). For the second fusion protein composed of p28- AEAAAKEAAAKA-M4, 95.9% (141/147) of residues were in favored regions, 98.0% (144/147) of residues were in allowed regions and there were 3 outliers (17 SER, 34 LYS, 43 CYS) (Fig. 1B). These results indicated appropriate quality of the models.
Fig. 1.
The Ramachandran plot of the fusion proteins from model obtained by MODELLER. (A) The fusion protein p28-AEAAAKEAAAKA-IL-24 which 97.5% (194/199) of all residues were in favored regions and 100.0% (199/199) of all residues were in allowed regions and (B) the fusion protein p28-AEAAAKEAAAKA-M4 which 95.9% (141/147) of residues were in favored regions, 98.0% (144/147) of residues were in allowed regions.
The energy analysis of the fusion proteins was applied by ProSA-web server that demonstrates local model quality by plotting energies as a function of amino acid sequence position in which positive values correspond to erroneous parts of the protein (28). Based on our results, in both models the energies of residues were negative with the exception of the N-terminus of the fusion proteins. Figure 2 shows the results of energy analysis of fusion proteins from the ProSA-web server.
Fig. 2.
Energy plot of two fusion protein structures using the ProSA-web server (demonstrates local model quality by plotting energies as a function of amino acid sequence position). (A) Energy result of the fusion protein p28-AEAAAKEAAAKA-IL-24 and (B) energy result of the fusion protein p28-AEAAAKEAAAKA-M4.
Molecular dynamic simulation
Fifty ns MD simulation was carried out to evaluate the dynamic behavior of the fusion proteins. Table 2 shows the results. Parameters including root mean square deviation (RMSD) of backbone relative to initial positions, temperature, kinetic energies, average of the potential, radius of gyration (Rg), Cα atoms root mean square fluctuation (RMSF), minimum distance between of two peptides (p28-IL-24 and p28-M4), H-bond pro-sol, solvent accessible surface area (SASA), and ratio of the total energy drift to average of total energy during 50 ns for both fusion proteins were evaluated and the results demonstrated in Table 2.
Table 2.
Results of the last 50 ns molecular dynamic simulation for the fusion proteins. Data represent mean ± SD.
| Parameters | p28-Linker-IL-24 fusion protein | p28-Linker-M4 fusion protein |
|---|---|---|
| Protein backbone RMSD | 0.98 ± 0.01 | 1.36 ± 0.08 |
| Protein backbone RMSF | 0.43 ± 0.29 | 0.418 ± 0.21 |
| Rg | 1.67 ±0.006 | 2.001 ± 0.082 |
| Temperature (Kº) | 300.07 ± 1.51 | 299.9 ± 1.66 |
| Potential (KJ/mol) | -488416 ± 732.03 | -398516 ± 659.01 |
| Total energy (KJ/mol) | -397398.264 ± 900.94 | -324549 ± 813.10 |
| Kinetic (KJ/mol) | 91017.55 ± 460.83 | 73969.55 ± 409.59 |
| H-bond pro-sol | 369.11 ± 11.56 | 299.30 ± 11.09 |
| Solvent accessible surface | 99.69 ± 1.76 | 91.62 ± 2.0291 |
| Minimum distance between of two peptides | 0.17 ± 0.01 | 0.20 ± 0.020 |
RMSD, Root mean square deviation; RMSF, root mean square fluctuation; Rg, radius of gyration.
The RMSD value of backbone atoms during MD simulation for fusion proteins are shown in Fig. 3. The p28-AEAAAKEAAAKA-IL-24 and p28-AEAAAKEAAAKA-M4 fusion proteins reached a stable state after 30 ns and 20 ns of the MD simulation. Also, Rg after 20 ns for the p28- AEAAAKEAAAKA-IL-24 fusion protein and after 30 ns for the p28-AEAAAKEAAAKA-M4 fusion protein reached the plateau phase (Fig. 3). Our results suggest that for stabilizing the fusion proteins, 50 ns MD simulation was sufficient.
Fig. 3.
(A) RMSD and (B) radius of gyration of the fusion proteins, p28-AEAAAKEAAAKA-M4 and p28-AEAAAKEAAAKA-IL-24, during 50 ns molecular dynamic simulation. RMSD, Root mean square deviation; IL, interleukin.
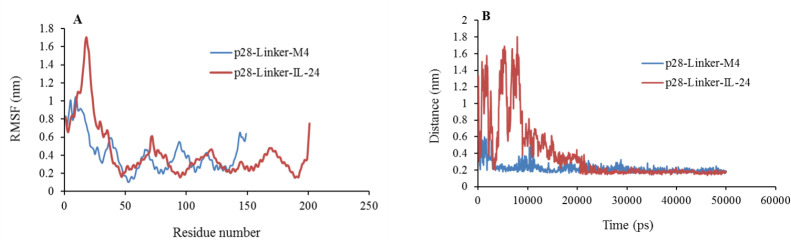
The flexibility of the fusion proteins was evaluated by Cα RMSF (Fig. 4). It was observed that the p28 -AEAAAKEAAAKA-IL- 24 fusion protein has a bit more fluctuations than the p28-AEAAAKEAAAKA-M4 fusion protein. The average Ca RMSF for all residues in the p28-AEAAAKEAAAKA-IL-24 and p28-AEAAAKEAAAKA-M4 fusion proteins was shown in Fig. 4. We also calculated the minimum distance between p28 from IL-24 and M4 peptides in the structure of our fusion proteins. The average of this distance between p28 from IL-24 peptides in the p28- AEAAAKEAAAKA-IL-24 and between p28 from M4 peptides in the p28- AEAAAKEAAAKA-M4 fusion proteins were shown in Fig. 4.
Fig. 4.
(A) RMSF values and (B) minimum distance of the fusion proteins, p28-AEAAAKEAAAKA-M4 and p28-AEAAAKEAAAKA-IL-24, during 50 ns molecular dynamic simulation. RMSF, root mean square fluctuation; IL, interleukin.
The solubility of the fusion proteins were evaluated by H-bond pro-sol and SASA parameters (Fig. 5). The average of H-bond pro-sol for the p28 -AEA AAKEAAAKA-IL-24 and p28-AEAAAKEAAAKA-M4 fusion proteins was shown in Fig. 5. We also assessed solubility by the SASA parameter. As demonstrated in Fig. 5 the average SASA in the p28-AEAAAKEAAAKA-IL-24 and between p28 from M4 peptides in the p28- AEAAAKEAAAKA-M4 fusion proteins were shown in Fig. 5. According to our results, p28- AEAAAKEAAAKA-IL-24 has better solubility in comparison with p28- AEAAAKEAAAKA-M4.
Fig. 5.
(A) H-bond pro-sol values and (B) SASA of fusion proteins, p28-AEAAAKEAAAKA-M4 and p28-AEAAAKEAAAKA-IL-24, during 50 ns molecular dynamic simulation. SASA, Solvent accessible surface area; IL, interleukin.
DISCUSSION
Because of the advanced nature of tumor cells, they escape all regulation of growth inhibition. The generation of multi-functional fusion proteins has become a very interesting research field in developing anticancer reagents. In the current study, we in silico designed two fusion proteins consisting of p28 as a cell-penetrating bacterial protein, and either IL-24 or its truncated form, M4, as a pro- apoptotic cytokine for targeted induction of cancer cell death.
Here, our data clearly predicted the best model with the minimum value of confidence scores. To the construction of effective fusion protein, two parts of each fusion protein have to be joint together via an appropriate linker. Linkers are natural protein structures in multidomain proteins to maintain protein domains in a suitable distance (32). For instance, proline- and hydroxylamine-rich cellulase usually keep protein domains in an extended conformation and also due to O- glycosylation they are protected from enzymatic hydrolysis and proteolysis (33). Interestingly, in our study, the AEAAAKEAAAKA linker efficiently hindered the formation of salt bridges between the functional domains of the two fusion proteins. Similarly, this linker has widely been used to construct other fusion proteins. For instance, Zhao et al. indicated that this linker was capable to separate human serum albumin and interferon-a2b effectively in a fusion protein (34).
To date various bioinformatics methods and software have been developed to predict protein structure. MODELLER, as a strong and free software, is a comparative protein structure modeling tool based on homology and stereo chemical restraints such as bond length and bond angle (27). Therefore, in order to improve the quality of prediction, the MODELLER software was used. Due to the lack of identical protein with high similarity, the maximum similarity was determined between our model and 2FT6, 1KYO, and 6DF3 for p28 peptide, AEAAAKEAAAKA linker, and IL-24/M4, respectively. The possible predicted models were then evaluated by the Ramachandran plot. This plot is particularly used to identify the structure of proteins. It emphasizes on the torsional (Φ, Ψ) angles along the backbone of the polypeptide and protein chain (35). Our results indicated that the p28- AEAAAKEAAAKA-IL-24 and p28-AEAAA- KEAAAKA-M4 have 97.5 and 100.0% of allowed residues, respectively. Similarly, the Ramachandran plot has been largely accepted to predict fusion protein and other synthetic proteins (36,37,38). Docking algorithms are often far from considering the dynamic effects of natural molecules because docking studies work usually on the rigid structure of proteins retrieved from the PDB. But, dynamic based models such as MD simulation assume molecules flexible and adjust their conformation in different microenvironments (39). Our MD analysis totally confirmed the validity of both fusion proteins. However, p28- AEAAAKEAAAKA-IL-24 fusion protein was predicted to be more flexible than the p28- AEAAAKEAAAKA-M4 fusion protein. All valid protein structures have to be checked by the energy plot. Our data also confirmed that the energies of all residues were favorably negative with the exception of the N-terminal part of the fusion proteins. The last part of the prediction is solubility. Due to the aqueous manner of biological fluids, these chemical properties of fusion proteins highlight their function. The results of the present study also showed that p28- AEAAAKEAAAKA-IL-24 is more soluble than the p28- AEAAAKEAAAKA-M4.
CONCLUSION
In conclusion, the present study highlighted the in silico prediction of the two fusion proteins. Taken together our findings showed that p28-AEAAAKEAAAKA-IL-24 has better stability and solubility than the p28-M4 protein. Therefore, this fusion protein is anticipated to show better anticancer efficacy. However in vitro and in vivo studies are needed to assess their biological activity and cytotoxic effects. Studies are undergoing for further assessments of these two fusion proteins.
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
R. Ghavimi and A. Jahanian-Najafabadi designed the study. R. Ghavimi, E. Mohammadi, and F. Shafiee contributed to the experiments and the results were verified by R. Ghavimi. The results were analyzed by R. Ghavimi, V. Akbari, and A. Jahanian- Najafabadi. R. Ghavimi prepared first draft of the manuscript. All the authors read the manuscript and it was finalized by A. Jahanian- Najafabadi. R. Ghavimi and A. Jahanian- Najafabadi finalized the manuscript corresponding to the comments raised by the reviewers.
ACKNOWLEDGMENTS
The content of this paper is extracted from Ph.D thesis, submitted by R. Ghavimi which was financially (Grant No. 395907) supported by the Vice Chancellor of Research of Isfahan University of Medical Sciences, Isfahan, I.R. Iran.
REFERENCES
- 1.Shafiee F, Aucoin MG, Jahanian-Najafabadi A. Targeted diphtheria toxin-based therapy: a review article. Front Microbiol. 2019;10:2340–2362. doi: 10.3389/fmicb.2019.02340. DOI: 10.3389/fmicb.2019.02340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31(1):100–110. doi: 10.1093/carcin/bgp263. DOI: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. DOI: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 4.Saini RK, Chouhan R, Bagri LP, Bajpai AK. Strategies of targeting tumors and cancers. J Can Res Updates. 2012;1(1):129–152. DOI: 10.6000/1929-2279.2012.01.01.19. [Google Scholar]
- 5.Dong X, Mumper RJ. Nanomedicinal strategies to treat multidrug-resistant tumors: current progress. Nanomedicine (Lond) 2010;5(4):597–615. doi: 10.2217/nnm.10.35. DOI: 10.2217/nnm.10.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty S, Rahman T. The difficulties in cancer treatment. Ecancermedicalscience. 2012;6:ed16,1–6. doi: 10.3332/ecancer.2012.ed16. DOI: 10.3332/ecancer.2012.ed16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. DOI: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 8.Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J Amino Acids. 2012;2012:967347–967359. doi: 10.1155/2012/967347. DOI: 10.1155/2012/967347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Punj V, Bhattacharyya S, Saint-Dic D, Vasu C, Cunningham EA, Graves J, et al. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene. 2004;23(13):2367–2378. doi: 10.1038/sj.onc.1207376. DOI: 10.1038/sj.onc.1207376. [DOI] [PubMed] [Google Scholar]
- 10.Yamada T, Fialho AM, Punj V, Bratescu L, Gupta TK, Chakrabarty AM. Internalization of bacterial redox protein azurin in mammalian cells: entry domain and specificity. Cell Microbiol. 2005;7(10):1418–1431. doi: 10.1111/j.1462-5822.2005.00567.x. DOI: 10.1111/j. 1462-5822.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- 11.Soleimani M, Sadeghi HM, Jahanian-Najafabadi A. A Bi-functional targeted P28-NRC chimeric protein with enhanced cytotoxic effects on breast cancer cell lines. Iran J Pharm Res. 2019;18(2):735–744. doi: 10.22037/ijpr.2019.2392. DOI: 10.22037/ijpr.2019.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada T, Das Gupta TK, Beattie CW. p28- Mediated activation of p53 in G2-M phase of the cell cycle enhances the efficacy of DNA damaging and antimitotic chemotherapy. Cancer Res. 2016;76(8):2354–2365. doi: 10.1158/0008-5472.CAN-15-2355. DOI: 10.1158/0008-5472.CAN-15-2355. [DOI] [PubMed] [Google Scholar]
- 13.Persaud L, De Jesus D, Brannigan O, Richiez-Paredes M, Huaman J, Alvarado G, et al. Mechanism of action and applications of interleukin 24 in immunotherapy. Int J Mol Sci. 2016;17(6):869–881. doi: 10.3390/ijms17060869. DOI: 10.3390/ijms17060869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pourhadi M, Jamalzade F, Jahanian-Najafabadi A, Shafiee F. Expression, purification, and cytotoxic evaluation of IL24-BR2 fusion protein. Res Pharm Sci. 2019;14(4):320–328. doi: 10.4103/1735-5362.263556. DOI: 10.4103/1735-5362.263556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282(28):20059–20063. doi: 10.1074/jbc.R700016200. DOI: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 16.Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Lebedeva IV, Dent P, et al. MDA-7/IL-24: novel cancer growth suppressing and apoptosis inducing cytokine. Cytokine Growth Factor Rev. 2003;14(1):35–51. doi: 10.1016/s1359-6101(02)00074-6. DOI: 10.1016/s1359-6101(02)00074-6. [DOI] [PubMed] [Google Scholar]
- 17.Dent P, Yacoub A, Hamed HA, Park MA, Dash R, Bhutia SK, et al. The development of MDA-7/IL-24 as a cancer therapeutic. Pharmacol Ther. 2010;128(2):375–384. doi: 10.1016/j.pharmthera.2010.08.001. DOI: 10.1016/j.pharmthera.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher P, Gupta P. MDA-7 protein variants having antiproliferative activity Google Patents WO/2006/060680. 2006 [Google Scholar]
- 19.Dash R, Bhutia SK, Azab B, Su ZZ, Quinn BA, Kegelmen TP, et al. Mda-7/IL-24: a unique member of the IL-10 gene family promoting cancer-targeted toxicity. Cytokine Growth Factor Rev. 2010;21(5):381–391. doi: 10.1016/j.cytogfr.2010.08.004. DOI: 10.1016/j.cytogfr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357–1369. doi: 10.1016/j.addr.2012.09.039. DOI: 10.1016/j.addr .2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soleimani M, Mahnam K, Mirmohammad-Sadeghi H, Sadeghi-Aliabadi H, Jahanian-Najafabadi A. Theoretical design of a new chimeric protein for the treatment of breast cancer. Res Pharm Sci. 2016;11(3):187–199. [PMC free article] [PubMed] [Google Scholar]
- 22.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. DOI: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahnam K, Saffar B, Mobini-Dehkordi M, Fassihi A, Mohammadi A. Design of a novel metal binding peptide by molecular dynamics simulation to sequester Cu and Zn ions. Res Pharm Sci. 2014;9(1):69–82. [PMC free article] [PubMed] [Google Scholar]
- 24.Wadood A, Ahmed N, Shah L, Ahmad A, Hassan H, Shams S. In-silico drug design: an approach which revolutionarised the drug discovery process. OA drug design & delivery. 2013;1(1):3–7. DOI: 10.13172/2054-4057-1-1-1119. [Google Scholar]
- 25.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. DOI: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 26.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;(5) doi: 10.1002/0471250953.bi0506s15. Unit-5.6.(1-47). DOI: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein Structure Modeling with MODELLER. In: Kobe B, Guss M, Huber T, editors. Structural Proteomics. Methods in Molecular Biology™. Vol. 426. Humana Press; 2008. pp. 145–159. [DOI] [PubMed] [Google Scholar]
- 28.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three- dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. doi: 10.1093/nar/gkm290. DOI: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooft RWW, Sander C, Vriend G. Objectively judging the quality of a protein structure from a Ramachandran plot. Bioinformatics. 1997;13(4):425–430. doi: 10.1093/bioinformatics/13.4.425. DOI: 10.1093/bioinformatics/ 13.4.425. [DOI] [PubMed] [Google Scholar]
- 30.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 45: a high- throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. DOI: 101093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. DOI: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 32.Yu K, Liu C, Kim BG, Lee DY. Synthetic fusion protein design and applications. Biotechnol Adv. 2015;33(1):155–164. doi: 10.1016/j.biotechadv.2014.11.005. DOI: 10.1016/j .biotechadv.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Rizk M, Antranikian G, Elleuche S. End-to-end gene fusions and their impact on the production of multifunctional biomass degrading enzymes. Biochem Biophys Res Commun. 2012;428(1):1–5. doi: 10.1016/j.bbrc.2012.09.142. DOI: 10.1016/j.bbrc.2012.09.142. [DOI] [PubMed] [Google Scholar]
- 34.Zhao HL, Yao XQ, Xue C, Wang Y, Xiong XH, Liu ZM. Increasing the homogeneity, stability and activity of human serum albumin and interferon-a2b fusion protein by linker engineering. Protein Expr Purif. 2008;61(1):73–77. doi: 10.1016/j.pep.2008.04.013. DOI: 10.1016/j.pep.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 35.DasGupta D, Kaushik R, Jayaram B. From Ramachandran maps to tertiary structures of proteins. J Phys Chem B. 2015;119(34):11136–11145. doi: 10.1021/acs.jpcb.5b02999. DOI: 10.1021/acs.jpcb.5b02999. [DOI] [PubMed] [Google Scholar]
- 36.Shamriz S, Ofoghi H, Moazami N. Effect of linker length and residues on the structure and stability of a fusion protein with malaria vaccine application. Comput Biol Med. 2016;76:24–29. doi: 10.1016/j.compbiomed.2016.06.015. DOI: 10.1016/j.compbiomed.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Fahimi H, Sadeghizadeh M, Mohammadipour M. In silico analysis of an envelope domain III-based multivalent fusion protein as a potential dengue vaccine candidate. Clin Exp Vaccine Res. 2016;5(1):41–49. doi: 10.7774/cevr.2016.5.1.41. DOI: 10.7774/cevr.2016.5.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad A, Javed MR, Rao AQ, Khan MAU, Ahad A, Salah UD, et al. In-silico determination of insecticidal potential of Vip3Aa-Cry1Ac fusion protein against Lepidopteran targets using molecular docking. Front Plant Sci. 2015;6:1081–1088. doi: 10.3389/fpls.2015.01081. DOI: 10.3389/fpls.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hospital A, Goni JR, Orozco M, Gelpi JL. Molecular dynamics simulations: advances and applications. Adv Appl Bioinform Chem. 2015;8:37–47. doi: 10.2147/AABC.S70333. DOI: 10.2147/AABC.S70333. [DOI] [PMC free article] [PubMed] [Google Scholar]