Abstract
Background and purpose:
Codon optimization has been considered as a powerful strategy to increase the expression level of protein therapeutics in mammalian cells. As an empirical approach to study the effects of the codon usage and GC content on heterologous gene expression in suspension adapted Chinese hamster ovary (CHO-s) cells, we redesigned the recombinant human interferon beta (rhIFN- β) gene based on the codon preference of the CHO cell in a way to increase the GC content in the third position of each codon.
Experimental approach:
The nucleotide sequence of the codon-optimized rhIFN-β was synthesized in parallel with the wild-type and expressed transiently in CHO-s cells using Epstein-Bar virus (EBV)-based expression system. The protein expression of the rhIFN-β by codon-optimized and wild-type genes were quantified using ELISA test.
Findings / Results:
The results indicated a 2.8-fold increase in the expression level of the biologically active form of the rhIFN-β by codon-optimized sequence.
Conclusion and implications:
These results shed light on the capability of codon optimization to create a stable CHO cell for scaling up the production of recombinant therapeutics such as rhIFN-β.
Keywords: Codon optimization, CHO-s cells, EBV-based expression system, Human interferon beta, Recombinant protein production
INTRODUCTION
Interferons (IFNs) are secretory proteins with multiple biological properties such as antiviral, antiproliferative, and immunomodulatory activities (1,2). Due to the therapeutic potential, IFNs are currently licensed for the treatment of various viral infections, malignancies, and immune diseases (3). Human IFN-β (hIFN-β) is a 166-amino acid glycoprotein with a single native N-linked glycan attached to the Asn80 residue (4,5,6). Recombinant hIFN-β (rhIFN-β) is considered as a gold standard therapeutic for treatment of patients with multiple sclerosis and is widely used as a first-line treatment to slow the progression of disabilities possibly through its anti-inflammatory properties (4,7,8).
The rhIFN-β was initially produced in Escherichia coli as a non-glycosylated form, in which Met1 was removed and Cys17 was replaced by Ser to increase its stability (9,10). Prokaryotic expression systems have some great advantages such as rapid growth, high productivity, easy manipulation, and low cost (11), however they are not suitable for production of complex glycoproteins due to their lack of post-translational modification machinery (3).
It has been demonstrated that the oligosaccharide content plays decisive roles in stability, solubility, immunogenicity, inhibition of aggregation of rhIFN-β in vitro as well as improves the in vivo biological activity (3,12,13). Therefore, different mammalian expression systems are preferably used as hosts for production of rhIFN-β to confer suitable post-translational modifications and correct biological functions (14,15). Commercially, two forms of rhIFN-β are produced; the non- glycosylated form, IFN-β lb, is produced in Escherichia coli and the glycosylated type, IFN-β la, with higher bioactivity, produced in Chinese hamster ovary (CHO) cells (10).
Despite the availability of numerous mammalian cell lines, nearly 70% of recombinant therapeutic proteins are produced in CHO cells that can easily adapt to growth in the serum-free and suspension culture conditions (16,17,18). Although different CHO expression platforms have been developed for recombinant protein expression, their relatively low productivity remains still challenging compared to the prokaryotic expression systems (17). Re-engineering to match the coding sequence of a heterologous gene to the codons frequently found in the host highly expressed genes is an appropriate strategy to achieve higher level of heterologous gene expression (19). A heterologous gene sequence that uses rare codons and is not expressed at high levels, can be converted to a gene with high expression level by replacing rare codons with high frequently used codons in a host cell (20). Codon adaptation index (CAI) is a universal measure of codon usage bias (21). The CAI index equals one means that the optimal codon for each amino acid is used (22). Codon optimization has been employed successfully to improve the expression level of a variety of proteins, from antibodies to cytokines and the promising results have been obtained (15,19,23).
In this study, coding sequence of the hIFN-β was optimized based on the codon preference of the host CHO cells to achieve efficient expression of the rhIFN-β. The expression level of the optimized hIFN-β gene was analyzed and compared to that of the wild-type in a serum free and suspension adapted CHO cell line (CHO-s) using Epstein-Bbar virus (EBV)- based expression system.
MATERIALS AND METHODS
Design and synthesis of the codon-optimized hIFN-β sequence
The hIFN-β coding sequence was retrieved from NCBI (Accession number: NM_002176) and subjected for codon adaptation using optimizer software (http://genomes.urv.es/OPTIMIZER/) based on the codon usage table of Chinese hamster (Cricetulus griseus). The optimized sequence was then validated by GenScript (www.genscript.com). The codon optimization of the gene was carried out in a way to introduce either G or C in the third position of each codon. A Kozak consensus sequence with an ATG codon for efficient initiation of translation was also added at the 5’ end of the optimized sequence. An additional stop codon was also added at the 3’ end of the gene. The designed sequence was then synthesized and cloned into the pGH plasmid. The recombinant plasmid harboring the codon- optimized sequence was named pGH-IFNβ-opt.
Construction of the recombinant expression plasmids
Forward and reverse primers with 5 GCCAAGCTTTTGCCACCATGACCAAC 3’ and 5 GGACTCGAGCCCTTTTATCAG- TTCCTC3 ‘sequences, were used to amplify the codon-optimized hIFN-β sequence and incorporate HindIII and XhoI restriction sites, respectively, into the 5’ and 3’ ends of the amplified fragment. In this polymerase chain reaction (PCR), pGH-IFNP-opt plasmid, harboring the codon-optimized hIFN-β sequence, was used as template. The final PCR product equipped with recognition sites for HindIII and Xhol at the 5’ and 3’ ends was screened by Webcutter server (http://rna.lundberg.gu.se/cutter2/) to ensure that there are no extra recognition sites for those restriction enzymes. The PCR product was then digested with HindIII and XhoI restriction enzymes and inserted into the same sites in pCEP4 mammalian expression vector (Thermo Fisher Scientific, US), downstream to a cytomegalovirus (CMV) immediate early promoter/enhancer. pCEP4 expression plasmid carries EBNA1 coding sequence and the EBV origin of replication (oriP) to permit episomal amplification of plasmid in mammalian cells. The wild-type sequence was also synthesized with unique HindIII and XhoI recognition sites at the 5´/3´ ends in pGH plasmid to facilitate sub-cloning steps into pCEP4 expression vector.
The sequences of all recombinant constructs were then verified by high quality Sanger-style sequencing. All cloning steps and plasmid propagation were carried out in the DH5α strain of Escherichia coli. The recombinant plasmids were purified in amounts sufficient for transient transfection experiments using Midiprep preparation kit (Roche, Germany).
Cell line and culture condition
Suspension-adapted FreeStyle™ CHO-S cells were cultured in FreeStyle™ CHO expression medium (Thermo Fisher Scientific, USA) supplemented with 8 mM L-glutamine and placed in an incubator (37° C, 5% CO2), while shaking at 140 rpm. The cells were passaged every 2 days when the density was approximately 1-1.5 ˣ 10 6 cells/mL. In such circumstances, maximum cell densities with a dispersed single cell suspension and an overall viability of more than 90% were achieved.
Transient transfection of CHO cells
The CHO-s cells were transiently transfected with recombinant plasmids harboring either wild-type or optimized hIFN-β cDNAs using FreeStyle™ MAX reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. In brief, 24 h prior to transfection, CHO-s cells were cultured in serum free culture medium in a density of about 5-6 ˣ 105 cells/mL. Just prior to transfection, the cell density was adjusted to 106 cells/mLby adding fresh medium. For transfection complex preparation, 2.5 μg DNA and 2.5 |L FreeStyle™ MAX reagent were separately diluted in Opti-PRO™ SFM (Thermo Fisher Scientific, USA) to a final volume of 40 μL. The diluted plasmid was immediately added to the diluted reagent. The DNA-reagent complex solution was incubated for 10 min at room temperature and slowly added into plate containing cells while slowly swirling. Transfected cells were incubated at 37 °C, 5% CO2 on an orbital shaker at 140 rpm. The culture media were harvested every 24 h for three days after transfection. No medium changes or additions were made after transfection. The viability of the cells was determined by hemocytometer counting with 1:10 (v/v) dilution of cells in 0.4% trypan blue. During transfection, the pcDNA3-GFP recombinant plasmid expressing green fluorescent protein as a reporter protein was also used as a control to verify the correctness and efficiency of the transfection process.
SDS-PAGE and western blot analysis
Three days after transfection, the culture media were harvested by centrifugation of the cell suspensions at 100 g for 5 min and subjected for sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS- PAGE) and western blot analysis. The SDS- PAGE analysis was performed according to the standard method described by Laemmli in 1970 (24) using13% polyacrylamide resolving and 5% polyacrylamide stacking gels.
Electroblotting of the protein bands from polyacrylamide gel onto polyvinylidene difluoride (PVDF) membrane (Roche, Germany) was carried out using wet procedure in transfer buffer (25 mM tris, 192 mM glycine, and 20% methanol) for16 h at 86 mA. The membrane was then blocked in 5% skim milk solution (5% w/v, in tris-buffered saline). For immunoblotting, the membrane was first incubated with a 1:4000 dilution of rabbit anti- hIFN-β polyclonal antibody (Millipore Co, USA) for 1.5 h, then washed and incubated with a 1:500 dilution of horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG for one hours at room temperature (Millipore Co, USA). Immunoreactive bands were finally visualized using 4-chloronaphthol solution.
Expression quantification and biological activity determination
Human IFN-β enzyme-linked immune- osorbent assay (ELISA) kit (Invitrogen, USA) was used to quantify the expressed hIFN-β in culture media. The cell suspensions were collected every 24 h after transfection for three days and centrifuged for 5 min at 100 g. The supernatants containing the secreted rhIFN-β were subjected for ELISA test. One-step sandwich ELISA test was performed in a 96-well microplate pre-coated with polyclonal antibody specific to conformational epitopes in hIFN-β . A 1:2 serial dilution of standard hlFN- P (prepared by the kit), covering the range from 0 to 200 IU/mL, was made. The unknown samples were also 1:30 diluted. Each well of the microplate was first washed with 400 μL of washing solution and drained off thoroughly by tapping on the paper towel. The plate was then incubated for 2 h at room temperature with 50 μL of enzyme-labeled antibody and 100 μL of either diluted unknown sample or standard solution. After washing thoroughly, 100 μL of color developer was added into the each well and incubated for 30 min at room temperature while shacking. Finally, 100 μL of reaction stopper was added and the absorbance of the reaction mixture in each well was read at 450 nm. The sample was analyzed out in duplicate.
Statistical analysis
All experiments were carried out in triplicates. Values were expressed as means ± SD. Statistical analysis were performed by oneway analysis of variance (ANOVA) followed by Scheffe’s post hoc test using SPSS version 22 software. P values less than 0.05 were considered statistically significant.
RESULTS
Codon optimization of the hIFN-β coding sequence based on CHO codon usage
In this study in order to achieve higher level of hIFN-β expression in transiently transfected CHO-s cells, the sequence encoding the hIFN- β (Accession number:NM_002176) was chosen and subjected for codon optimization. In this optimization process, 31 codons out of 166 amino acids from hIFN-β protein were modified based on the frequently used codons in Chinese hamster. The most frequently used codons in CHO cells that were chosen for codon optimization are depicted in Fig. 1.
Fig. 1.
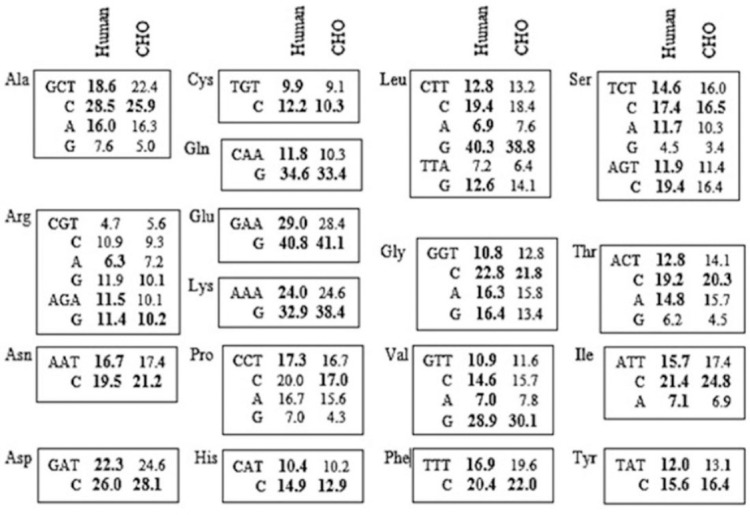
The codon usage of the highly expressed human and CHO genes. The codon usage pattern was assumed to affect the translational efficiency of the recombinant human interferon beta (IFN-β) gene, but not the nature of its amino acid sequence. The percentage frequencies of the synonymous codons are shown for each corresponding amino acid. The most prevalent codons are in bold. This was the main criteria for selection of the optimal codons. CHO-s, suspension adapted Chinese hamster ovary.
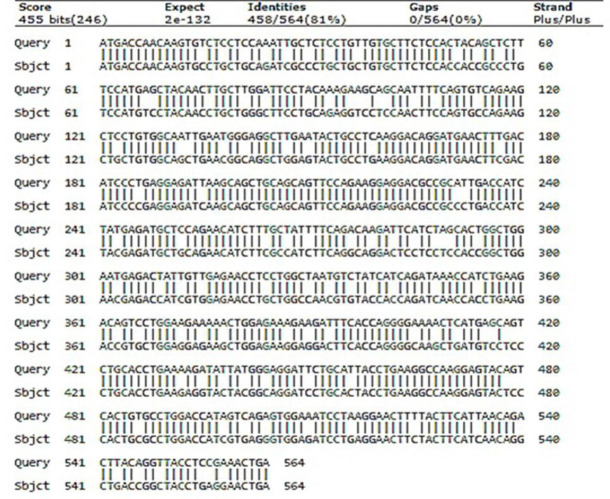
The aligned nucleotide sequences of the optimized and wild-type genes are shown in Fig. 2. The CAI scores for the wild-type and codon-optimized sequences were indicated in Table 1. The percentage of the total GC contents of the optimized sequence and its wild- type counterpart were also measured, showing a significant increase in the GC content of the optimized coding sequence (Table 1). The GC3 content (proportion of guanine and cytosine content in the third position of the codons) of the optimized hIFN-β sequence was also increased (Table 2).
Fig. 2.
Alignment of the wild-type and codon-optimized sequences encoding hIFN-β. Both coding sequences have 564 nucleotides with 458 matches, 63 transition changes (purine < - > purine / pyrimidine < - > pyrimidine), and 43 transversion changes (purines < - > pyrimidines) between wild-type (Query) and optimized (Sbjct) sequences. hIFN-β, the recombinant human interferon beta.
Table 1.
Codon adaptation index (CAI), GC percentage and Gibbs free energy calculation.
| Sequence name | CAI score | GC (%) | ΔG (kcal/mol) |
|---|---|---|---|
| Wild-type hIFN-β | 0.781 | 44.7 | -164.2 |
| Optimized-hIFN-β | 1.0 | 58.2 | -203.5 |
Table 2.
Coding sequences for the wild-type (A) and codon-optimized (B) hIFN-β. The Gs and Cs in the third codon positions are bolded.
| Gene Types |
Coding sequences | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATG | ACC | AAC | AAG | TGT | CTC | CTC | CAA | ATT | GCT | CTC | CTG | TTG | TGC | TTC | |
| TCC | ACT | ACA | GCT | CTT | TCC | ATG | AGC | TAC | AAC | TTG | CTT | GGA | TTC | CTA | |
| CAA | AGA | AGC | AGC | AAT | TTT | CAG | TGT | CAG | AAG | CTC | CTG | TGG | CAA | TTG | |
| AAT | GGG | AGG | CTT | GAA | TAC | TGC | CTC | AAG | GAC | AGG | ATG | AAC | TTT | GAC | |
| ATC | CCT | GAG | GAG | ATT | AAG | CAG | CTG | CAG | CAG | TTC | CAG | AAG | GAG | GAC | |
| GCC | GCA | TTG | ACC | ATC | TAT | GAG | ATG | CTC | CAG | AAC | ATC | TTT | GCT | ATT | |
| Wild type | TTC | AGA | CAA | GAT | TCA | TCT | AGC | ACT | GGC | TGG | AAT | GAG | ACT | ATT | GTT |
| GAG | AAC | CTC | CTG | GCT | AAT | GTC | TAT | CAT | CAG | ATA | AAC | CAT | CTG | AAG | |
| ACA | GTC | CTG | GAA | GAA | AAA | CTG | GAG | AAA | GAA | GAT | TTC | ACC | AGG | GGA | |
| AAA | CTC | ATG | AGC | AGT | CTG | CAC | CTG | AAA | AGA | TAT | TAT | GGG | AGG | ATT | |
| CTG | CAT | TAC | CTG | AAG | GCC | AAG | GAG | TAC | AGT | CAC | TGT | GCC | TGG | ACC | |
| ATA | GTC | AGA | GTG | GAA | ATC | CTA | AGG | AAC | TTT | TAC | TTC | ATT | AAC | AGA | |
| CTT | ACA | GGT | TAC | CTC | CGA | AAC | TGA | TAA | |||||||
| ATG | ACC | AAC | AAG | TGC | CTG | CTG | CAG | ATC | GCC | CTG | CTG | CTG | TGC | TTC | |
| TCC | ACC | ACC | GCC | CTG | TCC | ATG | TCC | TAC | AAC | CTG | CTG | GGC | TTC | CTG | |
| CAG | AGG | TCC | TCC | AAC | TTC | CAG | TGC | CAG | AAG | CTG | CTG | TGG | CAG | CTG | |
| AAC | GGC | AGG | CTG | GAG | TAC | TGC | CTG | AAG | GAC | AGG | ATG | AAC | TTC | GAC | |
| ATC | CCC | GAG | GAG | ATC | AAG | CAG | CTG | CAG | CAG | TTC | CAG | AAG | GAG | GAC | |
| GCC | GCC | CTG | ACC | ATC | TAC | GAG | ATG | CTG | CAG | AAC | ATC | TTC | GCC | ATC | |
| Codon-optimized | TTC | AGG | CAG | GAC | TCC | TCC | TCC | ACC | GGC | TGG | AAC | GAG | ACC | ATC | GTG |
| GAG | AAC | CTG | CTG | GCC | AAC | GTG | TAC | CAC | CAG | ATC | AAC | CAC | CTG | AAG | |
| ACC | GTG | CTG | GAG | GAG | AAG | CTG | GAG | AAG | GAG | GAC | TTC | ACC | AGG | GGC | |
| AAG | CTG | ATG | TCC | TCC | CTG | CAC | CTG | AAG | AGG | TAC | TAC | GGC | AGG | ATC | |
| CTG | CAC | TAC | CTG | AAG | GCC | AAG | GAG | TAC | TCC | CAC | TGC | GCC | TGG | ACC | |
| ATC | GTG | AGG | GTG | GAG | ATC | CTG | AGG | AAC | TTC | TAC | TTC | ATC | AAC | AGG | |
| CTG | ACC | GGC | TAC | CTG | AGG | AAC | TGA | TAA | |||||||
The Genebee online program was employed to assess the free energies for secondary structures of the both wild-type and optimized sequences.
Based on the calculated free energy, the optimized sequence was more stable than the wild-type (Table 1).
The growth rates of the transfected and untransfected CHO-s cells
The CHO-s cells were transiently transfected with recombinant plasmids harboring either wild-type or codon-optimized hIFN-β sequence. The cell growth rates of transfected CHO-s cells were monitored for three days after transfection and compared with that of the non-transfected one. As depicted in Fig. 3, the cell growth was greatly affected by the transfection process. The transfected cells showed lower growth rates than their untransfected counterpart.
Fig. 3.
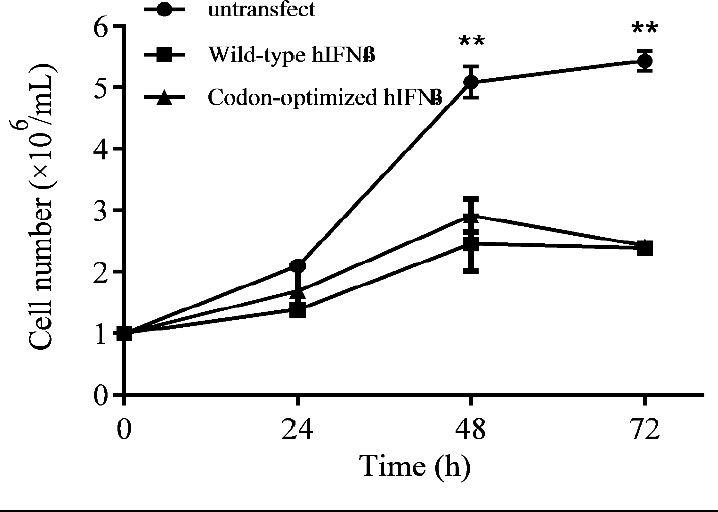
Growth curve for the CHO-s cells transfected with recombinant plasmids harboring either wild-type or codon-optimized hIFN-β sequence in comparison with the untransfected one, at transfection day (time 0) and 24, 48, and 72 h after transfection. Untransfectd cells showed higher proliferation rate in comparison with the transfected cells (** P < 0.01). CHO-s, suspension adapted Chinese hamster ovary; hIFN-β, the recombinant human interferon beta.
Quantity and quality assessment of the hIFN- β based on ELISA assay
To assess the expression level of the hIFN-β in CHO-s cells transiently transfected by either construct, the cell culture media were harvested 24-72 h after transfection and subjected for ELISA test. The results indicated higher level of hIFN-β expression by the codon-optimized sequence in comparison with the wild-type (Fig. 4).
Fig. 4.
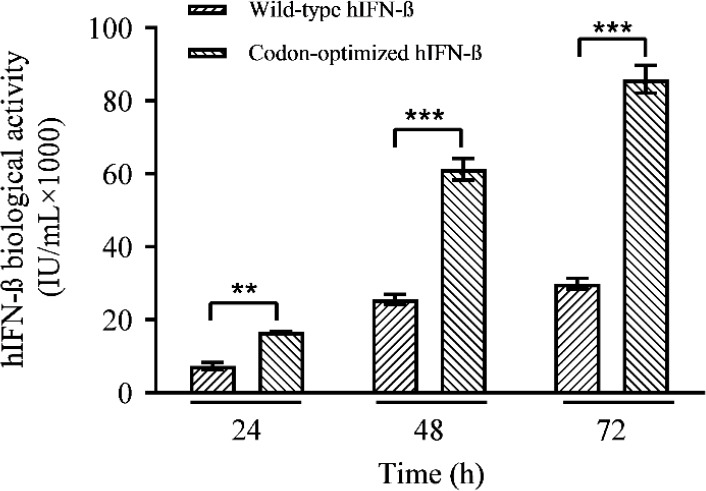
Quantification of the rhIFN-β transiently expressed by the wild-type and codon-optimized sequence in CHO-s cells at various post-transfection times, based on ELISA. **P ≤ 0.01 and ***P ≤ 0.001 indicate significant differences between 2 groups in each time. CHO-s, suspension adapted Chinese hamster ovary; hIFN-β, the recombinant human interferon beta.
The expression levels 72 h after transfection were calculated 56.282 ˣ 10 (3) and 19.976 ˣ 10 (3) IU/mL for codon-optimized and wild-type genes, respectively, showing a 2.8-fold increase in the expression level of the codon-optimized sequence (Fig. 4).
The analysis of the variance by one-way ANOVA confirmed that the differences between the expression levels of the codon- optimized and the wild-type genes were significant (P < 0.01). According to the expression profile indicated in Fig. 4 the highest protein expression levels for both constructs were observed at 72 h post-transfection. The net productions of the hIFN-β at 24, 48, and 72 h post-transfection were calculated and depicted in Fig. 5. The maximum rhIFN-β productions in the case of the wild-type and the optimized genes were achieved at 48 h after transfection. As depicted in the Fig. 5, from 48 toward 72 h post-transfection a reduction in the net production was observed in the case of both recombinant constructs. In spite of the decline in the net production of hIFN-β at 72 h post- transfection, the expression level of the optimized gene was still higher than that of the unmodified counterpart (Fig. 5).
Fig. 5.
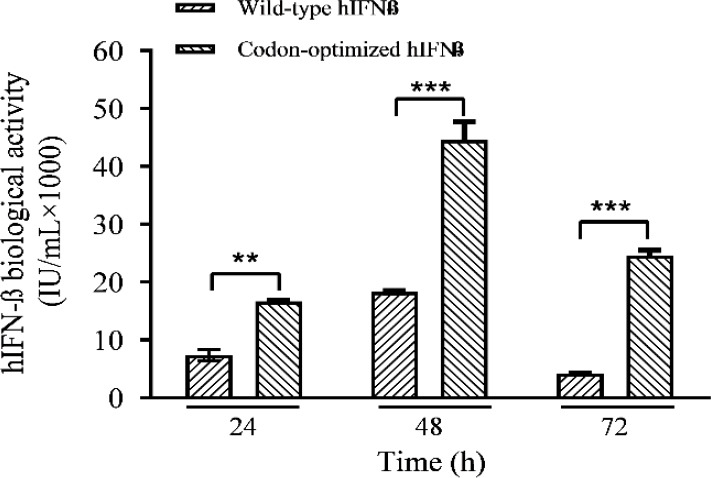
Net production of hIFN-β by the wild-type and codon-optimized constructs at 24, 48, and 72 h after transfection. **P ≤ 0.01 and ***P ≤ 0.001 indicate significant differences between 2 groups in each time. hIFN-β, the recombinant human interferon beta.
Western blot analysis
The samples were harvested 72 h after transfection and subjected for SDS-PAGE and western blot analysis (Fig. 6). In the case of the codon-optimized sequence, the higher expression level of the hIFN-β, measured by ELISA, was further confirmed by the western blot analysis, when compared with the wild- type (Fig. 6). The bands represented the glycosylated and non-glycosylated forms with molecular weight of 23-25 and 18 kDa, respectively. Since hIFN-β is a glycoprotein with a single N-glycosylation site, it may show variability in site occupancy by N-glycan (macroheterogeneity) and result in different glycoforms with occupied and unoccupied sites.
Fig. 6.
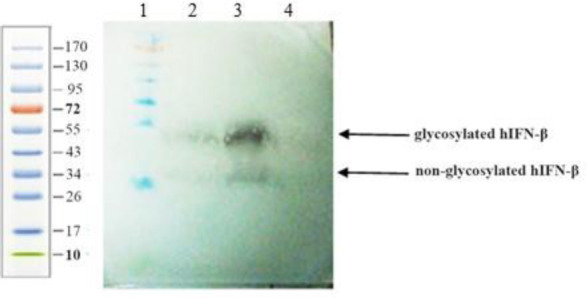
Western blot analysis of the supernatants from CHO-s cells transfected with recombinant plasmid harboring either wild-type or codon-optimized hIFN-β sequence. Lane 1, protein molecular weight marker; lane 2, hIFN-β expressed by the wild-type sequence; lane 3, hIFNβ expressed by codon-optimized sequence; and lane 4, untransfected cell culture medium as negative control. The two arrows, showing the glycosylated and non-glycosylated forms of hIFN-β, indicating the heterogeneity of the hIFN-β glycoforms. CHO-s, suspension adapted Chinese hamster ovary; hIFN-β, the recombinant human interferon beta.
DISCUSSION
Among mammalian expression systems, CHO cells are widely used for industrial-scale production of recombinant biopharmaceuticals (25). Commercially, two types of rhIFN-β have been approved for the treatment of multiple sclerosis. The non-glycosylated type, IFN-β 1b, is produced in Escherichia coli, and the glycosylated form, IFN-β 1a, with higher bioactivity is produced in CHO cells (26,27).
The heterologous protein expression in mammalian cells is significantly influenced by a variety of factors such as host cells, suitability of the promoter, position of the Shine- Dalgarno, stability and GC content of an mRNA, and codon usage bias (28). Adaptation of the codon usage of a heterologous gene to that frequently used in the host cell, improves translation. Moreover, mRNA transcription and degradation is strongly influenced by the GC content of the optimized sequence (29,30). Codon adaptation or “optimization” have been used for years to improve heterologous gene expression (22). Codon optimization strategies have resulted in the significant increase in the expression level of a variety of proteins such as erythropoietin and iCre protein (7- and 1.6-fold, respectively) (19,31).
In the current work, the effects of codon usage on the expression level of the hIFN-β in suspension adapted CHO cell line in a transient gene expression system and serum-free culture condition have been studied. The codon usage of hIFN-β was redesigned to match those frequently found in CHO cells (Figs. 1 and 2). The coding sequence was modified in a way to introduce G or C in the third position of each codon (Table 2), while the amino acid sequence of the protein remained unchanged. The codon- optimized as well as unmodified sequences were separately inserted into pCEP4 expression plasmid downstream to the CMV enhancer/ promoter to permit transgene expression in CHO-s cells. pCEP4 expression plasmid also carries the EBNA-1 gene, and oriP to support extra chromosomal plasmid replication in mammalian cells. The verified recombinant plasmids were transfected into CHO-s cells to analyze the level of the hIFN-β expression by each sequence. The results indicated that the growth rates were greatly affected by the transfection process (Fig. 3). This phenomenon can be due to the fact that the cells expressing heterologous proteins, endure more metabolic load than the untransfected ones. This metabolic load decreases the growth and proliferation rate of the recombinant cells. Moreover, the transfection reagent used for transfection in transient gene expression (TGE) systems may have an adverse effect on cell proliferation.
The protein expression levels were then monitored using ELISA for three days after transfection and the maximum protein expression was achieved on the third day after transfection (Fig. 4). The quantification of the hIFN-β expression showed a 2.8-fold increase in the expression level of the codon-optimized sequence at 72 h post-transfection. These results were further confirmed by the western blot analysis (Fig. 6). The increased expression level of the rhIFN-β by codon-optimized sequence can be attributed to several factors such as more efficient translation, due to the use of more abundant isoacceptor tRNAs, and increased mRNA level due to the higher GC content (19). Previously it has been shown that the GC-rich versions of Hsp70, green fluorescent protein, IL2, and HIV gag genes were expressed up to 100-fold more efficiently than the GC-poor counterparts (29,32). In our study, the codon optimization increased the total GC content of the gene of interest from 44.7% to 58.2% (Table 1). The GC3 content was also increased from 60 to 100% (Table 2). The free energies (ΔGs) released upon folding of the complete transcripts were calculated using Genebee, suggesting a higher mRNA stability for the codon-optimized sequence (Table 1). The CAI of the optimized sequence was also raised from 0.78 to 1. The CAI is a simple and effective approach to measure the codon adaptation level of a gene (21).
The CAI value equals one means that the optimal codon for each amino acid was applied. Previously, it has been reported by several studies that the efficiency of recombinant protein production can be improved by raising the CAI score and GC content of an open reading frame (22). These results, therefore, indicate that the codon engineering is an effective technique to increase the expression level of the rhIFN-β in CHO-s cells. This finding makes it possible to use codon optimization approach to create a stable CHO cell lines expressing hIFN-β for further studies in bioreactors.
CONCLUSION
The sequence encoding the hIFN-β was redesigned based on the frequently used codons in hamster. The optimized gene was then transiently expressed in CHO-s cells in parallel with the unmodified sequence. Quantification of the hIFN-β expression using ELISA showed that an increase in the CAI value and GC content of the hIFN-β gene can be resulted in higher protein expression level. These results shed light on the capability of codon optimization to create a stable CHO cell for scaling up the production of recombinant therapeutics such as hIFN-β.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest for this study.
AUTHORS’ CONTRIBUTION
All authors contributed equally in this work.
ACKNOWLEDGMENTS
This research was financially supported by National Institute of Genetic Engineering and Biotechnology (Grant No. 528). The authors would like to thank Dr. Mojtaba Samoudi for his valuable technical advices.
REFERENCES
- 1.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons? Nat Rev Immunol, 2012;12(2):125–135. doi: 10.1038/nri3133. DOI: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbert K, Schlaak JF, Yang D, Dittmer U. IFN-a subtypes: distinct biological activities in anti-viral therapy. Br J Pharmacol. 2013;168(5):1048–1058. doi: 10.1111/bph.12010. DOI: 10.1111/bph.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villela Dr A, Renard G, Palma MS, Chies JM, Dalmora SL, Basso LA, et al. Human interferon B1 ser17: coding DNA synthesis, expression, purification and characterization of bioactive recombinant protein. J Microb Biochem Technol. 2010;2(5):111–117. DOI: 10.4172/1948-5948.1000034. [Google Scholar]
- 4.Rodriguez J, Spearman M, Tharmalingam T, Sunley K, Lodewyks C, Huzel N, et al. High productivity of human recombinant beta-interferon from a low- temperature perfusion culture. J Biotechnol. 2010;150(4):509–518. doi: 10.1016/j.jbiotec.2010.09.959. DOI: 10.1016/j.jbiotec.2010.09.959. [DOI] [PubMed] [Google Scholar]
- 5.Runkel L, Meier W, Pepinsky RB, Karpusas M, Whitty A, Kimball K, et al. Structural and functional differences between glycosylated and non- glycosylated forms of human interferon-beta (IFN- beta) Pharm Res. 1998;15(4):641–649. doi: 10.1023/a:1011974512425. DOI: 10.1023/a:1011974512425. [DOI] [PubMed] [Google Scholar]
- 6.Meager A, Gaines Das R. Biological standardization of human interferon beta: establishment of a replacement world health organization international biological standard for human glycosylated interferon beta. J Immunol Methods. 2005;306(1-2):1–15. doi: 10.1016/j.jim.2005.08.007. DOI: 10.1016/j.jim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Song K, Yoon IS, Kim NA, Kim DH, Lee J, Lee HJ, et al. Glycoengineering of interferon-P 1a improves its biophysical and pharmacokinetic properties. PLoS One. 2014;9(5):e96967,1–14. doi: 10.1371/journal.pone.0096967. DOI: 10.1371/journal.pone.0096967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong VW, Chabot S, Stuve O, Williams G. Interferon beta in the treatment of multiple sclerosis: mechanisms of action. Neurology. 1998;51(3):682–689. doi: 10.1212/wnl.51.3.682. DOI: 10.1212/wnl.51.3.682. [DOI] [PubMed] [Google Scholar]
- 9.Derynck R, Remaut E, Saman E, Stanssens P, De Clercq E, Content J, et al. Expression of human fibroblast interferon gene in Escherichia coli. Nature. 1980;287(5779):193–197. doi: 10.1038/287193a0. DOI: 10.1038/287193a0. [DOI] [PubMed] [Google Scholar]
- 10.Rudick RA, Goelz SE. Beta-interferon for multiple sclerosis. Exp Cell Res. 2011;317(9):1301–1311. doi: 10.1016/j.yexcr.2011.03.002. DOI: 10.1016/j.yexcr.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Sadeghian-Rizi T, Ebrahimi A, Moazzen F, Yousefian H, Jahanian-Najafabadi A. Improvement of solubility and yield of recombinant protein expression in E coli using a two-step system. Res Pharm Sci. 2019;14(5):400–407. doi: 10.4103/1735-5362.268200. DOI: 104103/1735-5362268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matasci M, Hacker DL, Baldi L, Wurm FM. Recombinant therapeutic protein production in cultivated mammalian cells: current status and future prospects. Drug Discov Today Technol. 2008;5(2- 3):e37–42. doi: 10.1016/j.ddtec.2008.12.003. DOI: 10.1016/j.ddtec.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Devasahayam M. Factors affecting the expression of recombinant glycoproteins. Indian J Med Res. 2007;126(1):22–27. [PubMed] [Google Scholar]
- 14.Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv. 2009;27(3):297–306. doi: 10.1016/j.biotechadv.2009.01.008. DOI: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Chung BK, Yusufi FN, Mariati, Yang Y, Lee DY. Enhanced expression of codon optimized interferon gamma in CHO cells. J Biotechnol. 2013;167(3):326–333. doi: 10.1016/j.jbiotec.2013.07.011. DOI: 10.1016/j.jbiotec.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Kim JY, Kim YG, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93(3):917–930. doi: 10.1007/s00253-011-3758-5. DOI: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- 17.Jayapal KP, Wlaschin KF, Hu WS, Yap MGS. Recombinant protein therapeutics from CHO cells-20 years and counting. Chem Eng Prog. 2007;103(10):40–47. [Google Scholar]
- 18.Bandaranayake AD, Almo SC. Recent advances in mammalian protein production. FEBS Lett. 2014;588(2):253–260. doi: 10.1016/j.febslet.2013.11.035. DOI: 10.1016/j.febslet.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim CH, Oh Y, Lee TH. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene. 1997;199(1-2):293–301. doi: 10.1016/s0378-1119(97)00384-3. DOI: 10.1016/s0378-1119(97)00384-3. [DOI] [PubMed] [Google Scholar]
- 20.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22(11):1393–1398. doi: 10.1038/nbt1026. DOI: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 21.Sharp PM, Li WH. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. DOI: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotsopoulou E, Bosteels H, Chim YT, Pegman P, Stephen G, Thornhill SI, et al. Optimised mammalian expression through the coupling of codon adaptation with gene amplification: maximum yields with minimum effort. J Biotechnol. 2010;146(4):186–193. doi: 10.1016/j.jbiotec.2010.02.004. DOI: 10.1016/j.jbiotec.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Carton JM, Sauerwald T, Hawley-Nelson P, Morse B, Peffer N, Beck H, et al. Codon engineering for improved antibody expression in mammalian cells. Protein Expr Purif. 2007;55(2):279–286. doi: 10.1016/j.pep.2007.05.017. DOI: 10.1016/j.pep.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. DOI: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Poulain A, Mullick A, Massie B, Durocher Y. Reducing recombinant protein expression during CHO pool selection enhances frequency of high- producing cells. J Biotechnol. 2019;296:32–41. doi: 10.1016/j.jbiotec.2019.03.009. DOI: 10.1016/j.jbiotec.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Samoudi M, Minuchehr Z, Harcum SW, Tabandeh F, Omid Yeganeh N, Khodabandeh M. Rational design of glycoengineered interferon-P analogs with improved aggregation state: experimental validation. Protein Eng Des Sel. 2017;30(1):23–30. doi: 10.1093/protein/gzw058. DOI: 10.1093/protein/gzw058. [DOI] [PubMed] [Google Scholar]
- 27.Han YK, Koo TY, Lee GM. Enhanced interferon-beta production by CHO cells through elevated osmolality and reduced culture temperature. Biotechnol Prog. 2009;25(5):1440–1447. doi: 10.1002/btpr.234. DOI: 10.1002/btpr.234. [DOI] [PubMed] [Google Scholar]
- 28.Ghavim M, Abnous K, Arasteh F, Taghavi S, Nabavinia MS, Alibolandi M, et al. High level expression of recombinant human growth hormone in Escherichia coli: crucial role of translation initiation region. Res Pharm Sci. 2017;12(2):168–175. doi: 10.4103/1735-5362.202462. DOI: 10.4103/1735-5362.202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180, 0933–0942. doi: 10.1371/journal.pbio.0040180. DOI: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman ZR, Young JM, Ingolia NT, Barton GM. Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proc Natl Acad Sci U S A. 2016;113(10):E1362–E1371. doi: 10.1073/pnas.1518976113. DOI: 10.1073/pnas.1518976113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32(1):19–26. doi: 10.1002/gene.10023. DOI: 10.1002/gene. 10023. [DOI] [PubMed] [Google Scholar]
- 32.Graf M, Bojak A, Deml L, Bieler K, Wolf H, Wagner R. Concerted action of multiple cis-acting sequences is required for Rev dependence of late human immunodeficiency virus type 1 gene expression. J Virol. 2000;74(22):10822–10826. doi: 10.1128/jvi.74.22.10822-10826.2000. DOI: 10.1128/jvi.74.22.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]