Abstract
Crab-eating (Cerdocyon thous) and Pampas foxes (Lycalopex gymnocercus) are wild canids distributed in South America. Domestic dogs (Canis lupus familiaris) and wild canids may share viral pathogens, including rabies virus (RABV), canine distemper virus (CDV), and canine parvovirus 2 (CPV-2). To characterize the virome of these wild canid species, the present work evaluated the spleen and mesenteric lymph node virome of 17 crab-eating and five Pampas foxes using high-throughput sequencing (HTS). Organ samples were pooled and sequenced using an Illumina MiSeq platform. Additional PCR analyses were performed to identify the frequencies and host origin for each virus detected by HTS. Sequences more closely related to the Paramyxoviridae, Parvoviridae and Anelloviridae families were detected, as well as circular Rep-encoding single-stranded (CRESS) DNA viruses. CDV was found only in crab-eating foxes, whereas CPV-2 was found in both canid species; both viruses were closely related to sequences reported in domestic dogs from southern Brazil. Moreover, the present work reported the detection of canine bocavirus (CBoV) strains that were genetically divergent from CBoV-1 and 2 lineages. Finally, we also characterized CRESS DNA viruses and anelloviruses with marked diversity. The results of this study contribute to the body of knowledge regarding wild canid viruses that can potentially be shared with domestic canids or other species.
Keywords: Canid, Fox, HTS, Wildlife, Metagenomics, Virus
1. Introduction
Crab-eating (Cerdocyon thous) and Pampas foxes (Lycalopex gymnocercus) are Canidae members with distribution ranges that overlap extensively in South America. These wild canids are widely found in the farms of southern Brazil, on the border regions of Uruguay and Argentina. The vulnerability of these animals is in part due to the destruction of their natural environment, either by deforestation for extending the agricultural borders, or spreading of urban communities on the natural environment, as well as the habitat fragmentation caused by the roads. These foxes seem to be tolerant to human disturbance and are frequently seen in rural areas and close to urban regions. These wild canids have nocturnal scavenger habits and live in close proximity with domestic animals, which may be notable, as domestic host species can play a role in the transmission of infectious agents to wild animals (Alves et al., 2018a; Antunes et al., 2018; Ferreyra et al., 2009; Hübner et al., 2010).
It is known that domestic dogs (Canis lupus familiaris) and wild canids may share viral pathogens, including the rabies virus (RABV) (Antunes et al., 2018; Rocha et al., 2017), canine distemper virus (CDV) (Conceição-Neto et al., 2017; Ferreyra et al., 2009; Hübner et al., 2010; Megid et al., 2009), canine parvovirus 2 (CPV-2) (de Almeida Curi et al., 2010; Hübner et al., 2010), canine coronavirus (Alfano et al., 2019), canine adenoviruses 1 and 2 (Dowgier et al., 2018), and canine astrovirus (Alves et al., 2018a). However, the knowledge about sanitary conditions of these animals are still scarce.
The enhanced availability and application of high-throughput sequencing (HTS) technologies has facilitated the detection of known and unknown viruses (Goodwin et al., 2016; Paim et al., 2019). Thus, the knowledge of the virus genetic diversity present in different host species can be improved. HTS sequencing has been applied in the knowledge of the virome of domestic dogs (Moreno et al., 2017; Weber et al., 2018a, Weber et al., 2018b), but reports of its application in wild canids remain scarce. Therefore, the present study aimed to evaluate and characterize the spleen and mesenteric lymph node virome of crab-eating and Pampas foxes from southern Brazil and Uruguay using HTS. The results outline the viral agents that compound the microbiota of these wild dogs, helping to elucidate the viral population present in these wild canid species.
2. Materials and methods
2.1. Study design and sample sources
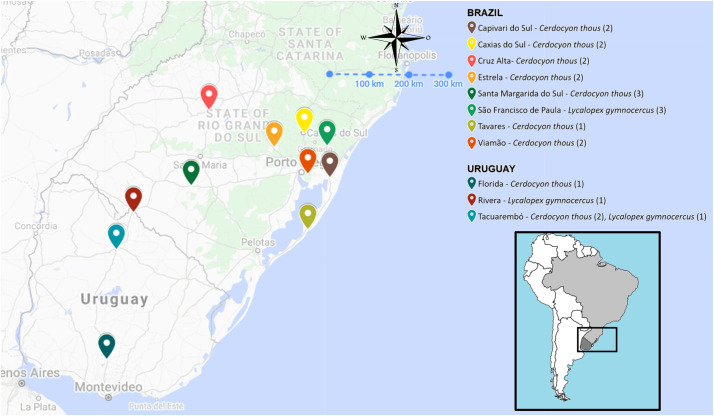
Five Pampas foxes (Lycalopex gymnocercus) and seventeen crab-eating foxes (Cerdocyon thous) run over by cars were collected by the Veterinary Pathology sector of Universidade Federal do Rio Grande do Sul and Plataforma de Salud Animal of Instituto Nacional de Investigación Agropecuaria Tacuarembó using an active search on highways between November 2017 and September 2019. Fig. 1 shows the location on the foxes sampled in the present study. The mesenteric lymph nodes and spleen of the 22 animals were collected, macerated and diluted to 20% (w/v) in phosphate-buffered saline (PBS) (pH 7.2), centrifuged at low speed (1800 ×g for 30 min), filtered through a 0.45-μm filter for removal of small debris and stored at −80 °C for subsequent analysis. These organs were selected since they concentrate antigens in order to presentation for immune system, which could increase the chance of detecting some viral agents.
Fig. 1.
Spatial distribution of the crab-eating (Cerdocyon thous) and Pampas foxes (Lycalopex gymnocercus) sampled. The quantity of each wild canid specie are shown on the map.
The authorization for the collection of the samples from Brazil was registered in SISBio/ICMBio under number 67053. Samples obtained of dead animals do not require authorization for molecular analysis in Uruguay.
2.2. Viral metagenomics and HTS
The 22 wild canid spleens and mesenteric lymph nodes (17 crab-eating foxes and five Pampas foxes) were assembled into one pool containing 500 μL of each sample. A total of 11 mL was passed through a 0.22-μm filter and subsequently centrifuged on a 25% sucrose cushion at 150,000g for 3 h at 4 °C in a Sorvall AH629 rotor. The pellet containing the viral particles was incubated for 1.5 h with DNase and RNase enzymes (Thermo Fisher Scientific, Waltham, MA, USA) (Thurber et al., 2009). Subsequently, the total RNA and DNA were isolated using TRIzol™ LS reagent (Thermo Fisher Scientific) and a standard phenol–chloroform protocol (Sambrook and Russel, 2001), respectively. The viral DNA was enriched using the Genoplex® Complete Whole Genome Amplification (WGA) kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's recommendations. Furthermore, the viral RNA was reverse-transcribed using the TransPlex® Complete Whole Transcriptome Amplification Kit (Sigma-Aldrich) following the manufacturer's recommendations. The DNA products from these enrichment protocols were pooled in equimolar amounts and purified using the PureLink™ Quick Gel Extraction and PCR Purification Combo Kit (Thermo Fisher Scientific). The quality and quantity of the DNA were assessed through spectrophotometry and fluorometry performed with NanoDrop™ (Thermo Fisher Scientific) and Qubit™ (Thermo Fisher Scientific), respectively. The DNA libraries were further prepared with 1 ng of purified DNA using the Nextera XT DNA sample preparation kit and sequenced using a MiSeq Reagent kit v2 300 (2 × 150 paired-end) at the MiSeq platform (Illumina®).
2.3. Bioinformatic analysis
The quality of the generated sequences was evaluated using FastQC. Furthermore, the sequences with bases possessing a Phred quality score < 20 were trimmed with the aid of Geneious software (version 9.0.5). Subsequently, the paired-end sequence reads were de novo assembled into contigs with SPAdes Assembler version 3.11.1 (Bankevich et al., 2012). All assemblies were confirmed by mapping reads to contigs produced by the SPAdes Assembler using Geneious software. Thereafter, the assembled contigs were examined for similarities with known sequences through BLASTX software using Blast2GO (Gotz et al., 2008). Sequences with E-values ≤10−3 were classified as likely to have originated from eukaryotic viruses, bacteria, bacteriophages, or unknown sources, a conclusion reached based on the taxonomic origin of the sequence with the best E-value. Gene and protein comparisons were performed with BLASTN and BLASTP programs (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sequences representative of viruses belonging to circular Rep-encoding single-stranded (CRESS) DNA viruses and the families Anelloviridae, Parvoviridae and Paramyxoviridae were obtained from GenBank and aligned with the sequences identified in the present study with MAFFT software (Katoh and Standley, 2013). Phylogenetic trees were constructed using MEGA6 (Tamura et al., 2013).
2.4. PCR and RT-PCR
The 22 animals were screened individually for CPV-2 (Buonavoglia et al., 2001), CDV (Fischer et al., 2013), canine circovirus (CaCV) (Li et al., 2013), and RABV (Soares et al., 2002). Positive samples in the applied CDV-RT-nested PCR were submitted to an additional assay to amplify a fragment of CDV-hemagglutinin (An et al., 2008; Riley and Wilkes, 2015) for genotyping.
CBoV were searched applying two independent PCR protocols in the 22 individual wild canid samples, where the first one was designed against one of the contigs obtained in the HTS in order to amplify a 154-bp fragment from the nonstructural protein of CBoV-1 (Supplementary Table). The second PCR protocol using primers CBoV-QFX1-f1 and CBoV-QFX1-r2 amplifies a 311-bp fragment of the VP2 gene of CBoV-1 (Kapoor et al., 2012)
The sequences related to anelloviruses detected through HTS higher than 400 amino acids in ORF1 were investigated individually to define the host origin (Lycalopex gymnocercus or Cerdocyon thous). The primer pairs (Supplementary Table 1) were selected using Primer3 software (Untergasser et al., 2007). The PCRs for anelloviruses were conducted using [1×] PCR buffer, 1 mM MgCl2, 0.5 mM dNTP mix, 0.2 mM each specific primer pair, and 1 unit of GoTaq® DNA Polymerase (Promega, Madison, WI, USA). Reactions were performed in a Veriti 60-well Thermal Cycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: 3 min at 95 °C followed by 35 cycles of 45 s at 95 °C, 45 s at 50 °C and 45 s at 72 °C with a final extension at 72 °C for 7 min.
2.5. Sanger sequencing
All positive samples in PCR and RT-PCR were submitted for Sanger sequencing to confirm the specificity of the tests. The PCR products were purified using the PureLink™ Quick PCR Purification Kit (Invitrogen, Carlsbad, CA, USA). Both DNA strands were sequenced with an ABI PRISM 3100 Genetic Analyzer utilizing a BigDye Terminator v.3.1 cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Furthermore, overlapping fragments were aligned and assembled using Geneious software.
3. Results
3.1. HTS overview
One DNA library consisting of 22 pooled wild canid spleens and mesenteric lymph nodes (17 crab-eating foxes and five Pampas foxes) was generated and sequenced using paired-end 2 × 150 base runs on the Illumina MiSeq platform, which generated a total of 138,850 reads. The 13,250 assembled sequence contigs produced with SPAdes Assembler version 3.11.1 (Bankevich et al., 2012) were compared with the viral reference database and the GenBank nonredundant protein database through a BLASTX search conducted with an E-value cut-off of 10−5 in Blast2GO (Gotz et al., 2008). The exogenous eukaryotic virus-related sequences comprised 1.38% (1809/138,850) of the reads and 1.37% (182/13,250) of the contigs.
Furthermore, eukaryotic exogenous virus-related sequences with single-stranded DNA (ssDNA) genomes belonging to two viral families (Anelloviridae and Parvoviridae) and CRESS DNA viruses were observed, as well as with single-stranded RNA (ssRNA) genomes belonging to the Paramyxoviridae family (Table 1 ). Moreover, the majority of the viral sequences studied shared a high degree of identity with known animal viruses (CPV-2, CDV and CBoV), while others (anellovirus-like and CRESS DNA virus) exhibited a high degree of divergence to genomes already recorded in GenBank. Information regarding the sequences obtained is described in the following sections. All 22 samples analyzed using CaCV (Li et al., 2013) and RABV-specific PCR (Soares et al., 2002) were negatives.
Table 1.
Summary of sequences that matched with the animal viruses present in the pooled spleen and mesenteric lymph nodes of Cerdocyon thous and Lycalopex gymnocercus sample.
| Best blast hit (BLASTX, E-value < 1 × 10−3) | No. of hits | No. of readsa | Contigs lengths | Amino acid identity rangea |
|---|---|---|---|---|
| Canine bocavirus | 3 | 22 | 267–343 | 93.5–100% |
| Carnivore protoparvovirus 1 | 2 | 12 | 435–530 | 99.3–100% |
| Canine morbillivirus | 3 | 10 | 109–251 | 99.8–100% |
| Anelloviridae | 171 | 1678 | 886–2415 | 42.3–64.8% |
| CRESS DNA virus | 7 | 87 | 327–852 | 34.1–55.9% |
Analysis performed in BLASTX tool, from GenBank database.
3.2. Canine distemper virus (CDV)
Three contigs closely related to CDV were obtained in the pooled wild canid organs sequenced by HTS (Table 1). Applying the RT-nested PCR protocol to amplify the CDV nucleocapsid (Fischer et al., 2013), 18.18% (4/22) of the samples were positive, and all the positive samples were from Cerdocyon thous from Brazil. The amplification products from the positive samples were Sanger sequenced and shared 99.29–100% nucleotide identity between them. In the nucleotide BLAST search, the samples detected in this study presented 99.6 to 100% identity with CDV strains detected in domestic dogs in Brazil (GenBank accession numbers MH382872 and KU725677). The partial N gene sequences obtained in the present study presented the same single nucleotide polymorphisms (SNP) that characterize South America I lineage described previously (Fischer et al., 2013). These sequences were deposited in GenBank under accession numbers MT002482- MT002485.
Positive samples were submitted to an additional PCR assay to amplify a fragment of CDV-hemagglutinin (An et al., 2008; Riley and Wilkes, 2015) followed by Sanger sequencing for genotyping (Budaszewski et al., 2014). The sequences presented 99.06 to 99.87% nucleotide identity. In the nucleotide BLAST search, the samples detected in this study presented 99.3 to 99.6% identity with CDV strains detected in domestic dogs in Brazil (GenBank accession numbers KC879049 and KC879050). The partial CDV-hemagglutinin phylogenetic reconstruction (Fig. 2 ) presented 14 well-supported branches corresponding to the 14 genotypes analyzed. All the sequences generated in the present study grouped into the Europe/South America-1 genotype cluster supported by a 72% bootstrap value. The sequences grouped in the same node of samples detected in domestic dogs from southern Brazil (GenBank accession numbers KC879049 and KC879050).
Fig. 2.
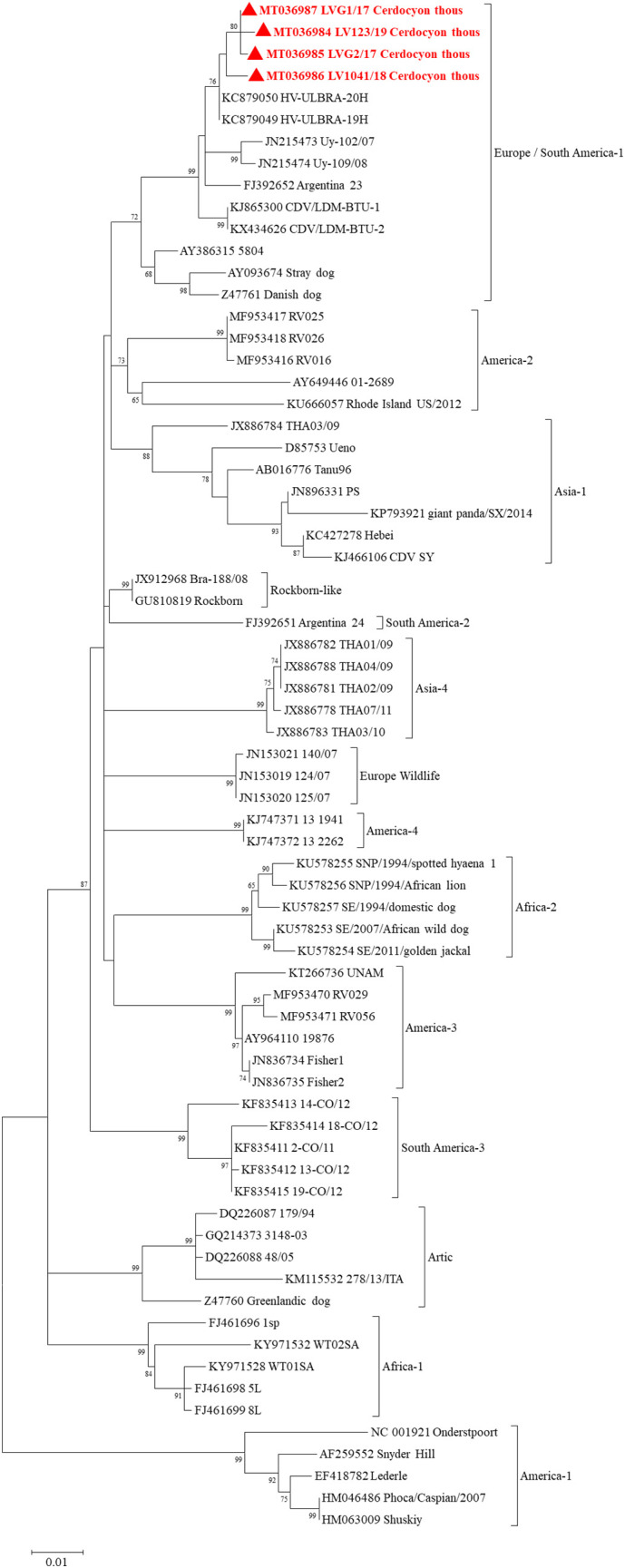
Nucleotide phylogenetic reconstruction of the partial hemagglutinin of canine morbillivirus species members highlighting the sequences detected in the present study. Sequences were analyzed through maximum-likelihood method applied with the GTR + G + I model. All analyses were conducted with 1000 bootstrap replicates, and the percentage of replicate trees in which the sequences clustered together have been depicted adjacent to the branches. Bootstrap values for each node have been demonstrated if they were > 50%. The sequence detected in the present study has been highlighted with ●. GenBank accession numbers are described in the phylogenetic tree.
3.3. Canine parvovirus 2 (CPV-2)
In the 22 pooled wild dog organs submitted for HTS, the presence of two contigs closely related to CPV-2 was observed (Table 1). The application of the specific CPV PCR protocol (Buonavoglia et al., 2001) revealed 13.64% (3/22) to be positive (one Lycalopex gymnocercus and two Cerdocyon thous from Brazil).
All the CPV-2-positive PCR products were sequenced for CPV-2 type definition (CPV-2a, 2b, or 2c). Furthermore, samples were assigned to one CPV-2 type based on the presence of a deduced asparagine (2a), aspartic acid (2b), or glutamic acid (2c) at amino acid position 426 (Buonavoglia et al., 2000). The three tested samples shared 99.03 to 99.23% nucleotide identity. In the nucleotide BLAST search, the samples detected in this study presented 99.6 to 100% identity with CPV-2 strains detected in domestic dogs in southern Brazil (GenBank accession numbers MK344466, MK344470, and JF796206). One sample detected in Cerdocyon thous was classified as CPV-2b, while the two other samples detected in the present study (one sample from a Cerdocyon thous and one sample from a Lycalopex gymnocercus) were classified as CPV-2c. The CPV-2 sequences generated in the present study were deposited in the GenBank database under accession number MN997122-MN997124.
3.4. Canine bocavirus (CBoV)
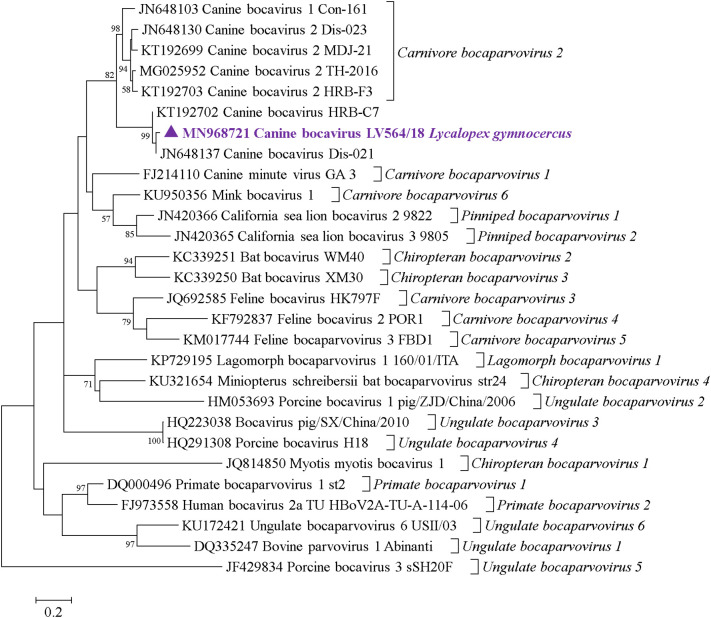
Three contigs more closely related to CBoV were obtained from the pooled wild dog organs submitted for HTS (Table 1). One of the 22 (4.55%) samples was positive in both assays and was from a Lycalopex gymnocercus sampled in Brazil. The two amplification products were submitted for Sanger sequencing. The sequence obtained using the primers designed in the present study (Supplementary Table) enabled us to identify the same sequence obtained in HTS.
In a nucleotide BLAST search, the VP2 fragment (Kapoor et al., 2012) named LV564/18 presented 96.9% identity with CBoV isolate Dis-021 reported in a domestic dog presenting respiratory disease in the United States (GenBank accession number JN648137).
To present the genetic relation between LV564/18 and other Bocaparvovirus members, a partial VP2 gene phylogenetic tree applying maximum-likelihood inference, GTR + G + I statistical method, and 1000 bootstrap was constructed in MEGA6 (Tamura et al., 2013) (Fig. 3 ). The sequence LV564/18 obtained from the Lycalopex gymnocercus sample clustered in the same node of other genetically different CBoVs (Dis-021 and HRB-C7) reported in domestic dogs from the United States and China, respectively (GenBank accession numbers JN648137 and KT192702, respectively), supported by a 99% bootstrap value. This genetically divergent CBoV cluster evolved from the same common ancestor that originated the CBoV-1 and CBoV-2 sequences, as supported by an 82% bootstrap value.
Fig. 3.
Nucleotide phylogenetic reconstruction of the partial VP2 of subfamily Parvovirinae members highlighting the bocaparvovirus detected in the present study. Sequences were analyzed through maximum likelihood method applied with the GTR + G + I model. All analyses were conducted with 1000 bootstrap replicates, and the percentage of replicate trees in which the sequences clustered together have been depicted adjacent to the branches. Bootstrap values for each node have been demonstrated if they were > 50%. The sequence detected in the present study has been highlighted with a ▲. GenBank accession numbers are described in the phylogenetic tree.
3.5. Anelloviruses
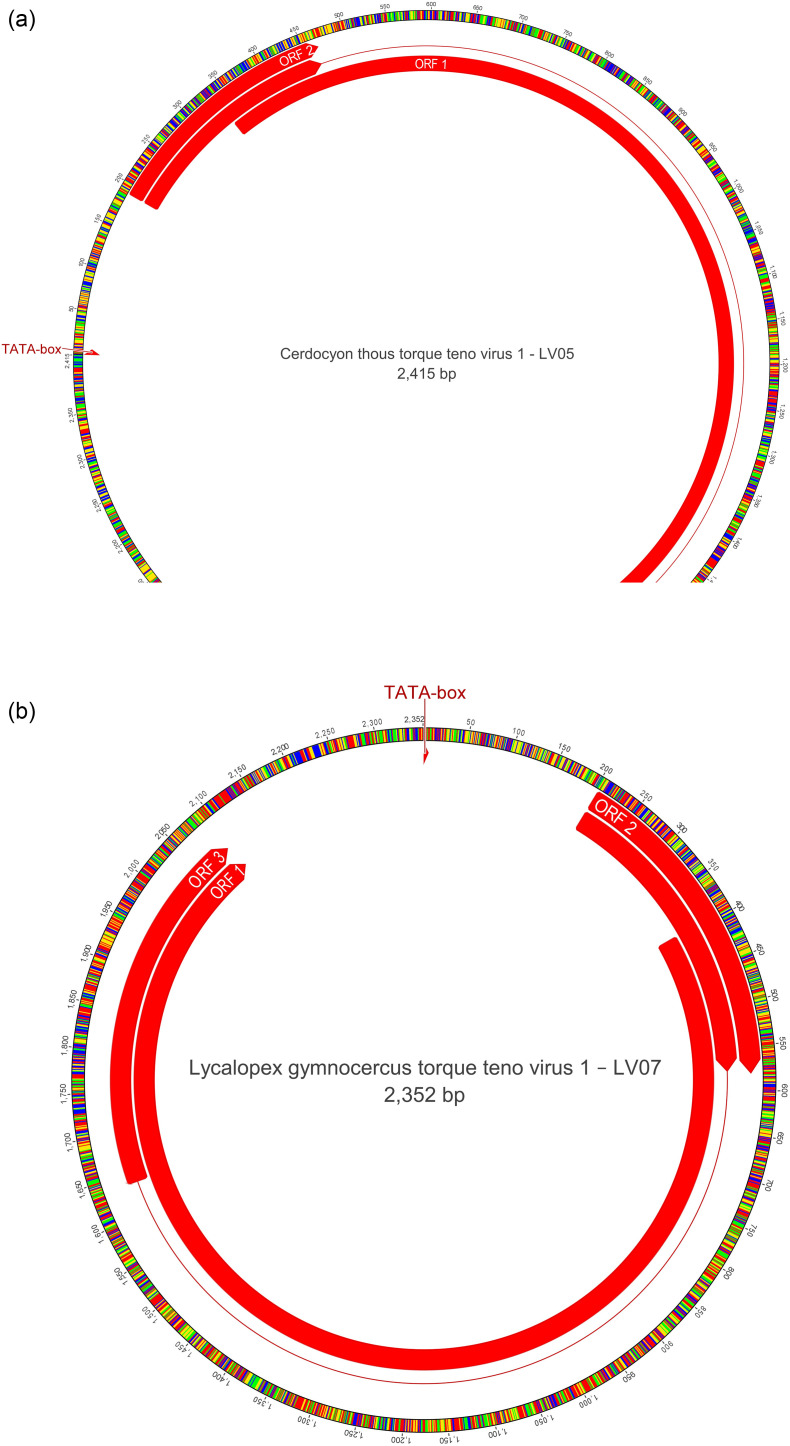
A total of 171 contigs with a closer association with Anelloviridade members were also observed (Table 1). The contigs ranged between 2415 and 886 nt in length. It was possible to obtain two complete genomes (Fig. 4A and B) and five additional sequences displaying the complete ORF1 gene (Fig. 4C).
Fig. 4.
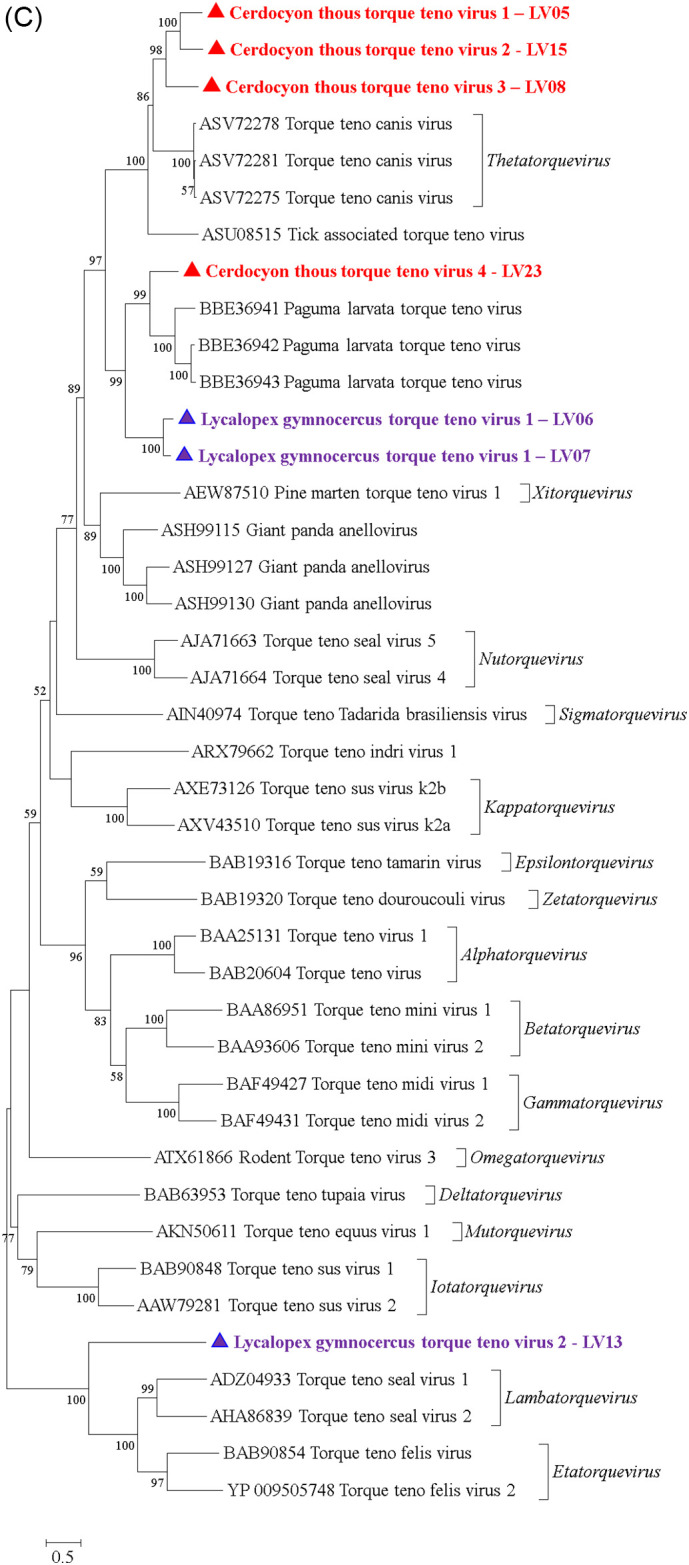
Genetic characterization of Anelloviridae family members detected in the present study. (A) Genomic organization of LV05 detected in Cerdocyon thous in the present study. (B) Genomic organization of LV07 detected in Lycalopex gymnocercus in the present study. (C) Amino acid phylogenetic tree of complete ORF1 gene highlighting the sequences obtained in the present study. The sequences were analyzed through the maximum-likelihood method with JTT + G + I model. All analyses were conducted with 1000 bootstrap replicates, and the percentage of replicate trees in which the sequences clustered together have been presented adjacent to the branches. Bootstrap values for each node have been shown if they were > 50%. The sequences detected in the present study has been highlighted with ● for sequences detected in Cerdocyon thous, and with ▲ for sequences reported in Lycalopex gymnocercus. The sequences detected in the present study were deposited in GenBank database: CtTTV-1 LV05 (MT010524), CtTTV-2 LV15 (MT010525), CtTTV-3 LV08 (MT010526), CtTTV-4 LV23 (MT010527), LgTTV-1 LV06 (MT010528), LgTTV-1 LV07 (MT010529), and LgTTV-2 LV13 (MT010530).
To identify the host of origin (pampas or crab-eating fox), we applied specific PCR protocols designed using the sequences identified in HTS (Supplementary Table). No sequence was identified in either Cerdocyon thous or Lycalopex gymnocercus. The sequences were named the putative Anelloviridae family species Cerdocyon thous torque teno virus 1 (CtTTV-1), CtTTV-2, CtTTV-3, CtTV-4, Lycalopex gymnocercus torque teno virus 1 (LgTTV-1), and LgTTV-2, where two different sequences displaying 81.21% nucleotide identity (Table 2 ) were classified as LgTTV-1 putative species. According to ICTV, nucleotide divergences in the ORF1 gene higher than 35% and 56% denote the same anellovirus species and genus, respectively (ICTV, http://www.ictvonline.org/virusTaxonomy.asp). Apparently, six putative new Anelloviridae species (CtTTV-1, CtTTV-2, CtTTV-3, CtTTV-4, LgTTV-1 and LgTTV-2) belonging to five putative new genera were observed (Fig. 4C). CtTTV-1 and CtTTV-2 can be classified in the same genus, since they present 63.15% nucleotide identity in the ORF1 gene. The relation of the sequences reported in the present study and other members of the family Anelloviridae is reported in Table 3 .
Table 2.
Summary of Anelloviridae family-related sequences reported in the present study, reporting host of origin, ORF1 size, and nucleotide identity of ORF1 gene between them.
| Sequence name | Host | ORF1 size (aa) | Nucleotide identity |
||||||
|---|---|---|---|---|---|---|---|---|---|
| LV05 | LV06 | LV07 | LV08 | LV13 | LV15 | LV23 | |||
| LV05 | Cerdocyon thous | 570 | |||||||
| LV06 | Lycalopex gymnocercus | 566 | 42.08 | ||||||
| LV07 | Lycalopex gymnocercus | 566 | 41.71 | 81.21 | |||||
| LV08 | Cerdocyon thous | 581 | 57.23 | 43.93 | 42.57 | ||||
| LV13 | Lycalopex gymnocercus | 471 | 29.36 | 31.53 | 32.10 | 29.54 | |||
| LV15 | Cerdocyon thous | 583 | 63.32 | 41.26 | 40.72 | 53.53 | 28.40 | ||
| LV23 | Cerdocyon thous | 524 | 43.14 | 51.50 | 51.97 | 42.61 | 29.88 | 42.59 | |
Table 3.
Comparison of the ORF1 sequence of anelloviruses detected in Cerdocyon thous and Lycalopex gymnocercus obtained in the present study.
| Genera | Species | Accession number | Host | ORF1 size (aa) | Difference (%) within the ORF1 amino acid sequence |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LV05 | LV06 | LV07 | LV08 | LV13 | LV15 | LV23 | |||||
| Alphatorquevirus | Torque teno virus 1 | BAA25131 | Human (Homo sapiens) | 770 | 82.56 | 82.09 | 82.35 | 82.22 | 88.33 | 16.26 | 84.32 |
| Torque teno virus 2 | BAB20604 | Chimpanzee (Pan troglodytes) | 727 | 81.73 | 83.71 | 82.65 | 82.63 | 88.20 | 17.24 | 84.52 | |
| Betatorquevirus | Torque teno mini virus 1 | BAA86951 | Human (Homo sapiens) | 662 | 85.45 | 80.91 | 80.91 | 84.79 | 90.18 | 84.59 | 86.50 |
| Torque teno mini virus 2 | BAA93606 | Human (Homo sapiens) | 628 | 83.64 | 81.36 | 81.07 | 83.59 | 87.94 | 82.71 | 86.16 | |
| Deltatorquevirus | Torque teno tupaia virus | BAB63953 | Tupaia (Tupaia belangeri chinensis) | 502 | 80.65 | 81.83 | 81.83 | 81.27 | 83.54 | 82.97 | 82.99 |
| Epsilontorquevirus | Torque teno tamarin virus | BAB19316 | Cotton-top tamarin (Saguinus oedipus) | 712 | 80.84 | 81.31 | 80.63 | 80.44 | 87.65 | 79.95 | 84.38 |
| Etatorquevirus | Torque teno felis virus 1 | BAB90854 | Cat (Felis catus) | 436 | 86.95 | 86.58 | 86.74 | 87.70 | 79.61 | 86.92 | 89.26 |
| Torque teno felis virus 2 | YP_009505748 | Cat (Felis catus) | 404 | 88.71 | 88.60 | 88.11 | 88.56 | 79.68 | 88.00 | 89.63 | |
| Gammatorquevirus | Torque teno midi virus 1 | BAF49427 | Human (Homo sapiens) | 673 | 82.92 | 82.27 | 81.98 | 82.62 | 88.03 | 82.11 | 85.33 |
| Torque teno midi virus 2 | BAF49431 | Human (Homo sapiens) | 677 | 82.26 | 83.02 | 82.73 | 83.36 | 89.28 | 83.88 | 85.31 | |
| Iotatorquevirus | Torque teno sus virus 1 | BAB90848 | Pig (Sus scrofa domesticus) | 635 | 80.84 | 81.91 | 82.07 | 81.02 | 83.09 | 81.32 | 82.78 |
| Torque teno sus virus 2 | AAW79281 | Pig (Sus scrofa domesticus) | 437 | 80.45 | 82.12 | 81.52 | 80.09 | 84.73 | 80.21 | 83.09 | |
| Kappatorquevirus | Torque teno sus virus k2a | AXV43510 | Pig (Sus scrofa domesticus) | 620 | 78.98 | 81.02 | 80.12 | 79.82 | 86.76 | 79.38 | 80.82 |
| Torque teno sus virus k2b | AXE73126 | Wild boar (Sus scrofa) | 633 | 79.43 | 81.53 | 79.73 | 81.83 | 85.42 | 80.21 | 80.69 | |
| Lambdatorquevirus | Torque teno seal virus 1 | ADZ04933 | Seal (Phoca vitulina richardsi) | 469 | 86.49 | 85.88 | 85.88 | 87.48 | 79.37 | 87.26 | 87.28 |
| Torque teno seal virus 2 | AHA86839 | Seal (Phoca vitulina richardsi) | 464 | 86.63 | 86.84 | 87.50 | 86.95 | 79.85 | 87.14 | 89.07 | |
| Mutorquevirus | Torque teno equus virus 1 | AKN50611 | Horse (Equus caballus) | 435 | 81.11 | 80.83 | 79.60 | 80.59 | 86.74 | 80.96 | 79.12 |
| Nutorquevirus | Torque teno seal virus 4 | AJA71664 | Seal (Phoca vitulina richardsi) | 642 | 76.30 | 78.00 | 77.39 | 78.12 | 86.10 | 78.59 | 78.74 |
| Torque teno seal virus 5 | AJA71663 | Seal (Phoca vitulina richardsi) | 439 | 77.43 | 77.34 | 78.15 | 78.01 | 86.34 | 78.15 | 77.59 | |
| Omegatorquevirus | Rodent Torque teno virus 3 | ATX61866 | Hairy-tailed bolo mouse (Bolomys lasiurus) | 585 | 79.87 | 78.77 | 77.80 | 79.04 | 84.86 | 81.69 | 79.64 |
| Sigmatorquevirus | Torque teno Tadarida brasiliensis virus | AIN40974 | Bat (Tadarida brasiliensis) | 547 | 77.54 | 78.54 | 77.47 | 77.29 | 84.28 | 78.49 | 77.62 |
| Thetatorquevirus | Torque teno canis virus | ASV72275 | Dog (Canis lupus familiaris) | 576 | 56.29 | 71.24 | 70.05 | 56.13 | 83.78 | 56.52 | 71.85 |
| Xitorquevirus | Pine marten torque teno virus 1 | AEW87510 | Pine marten (Martes martes) | 568 | 74.71 | 74.84 | 73.52 | 73.06 | 85.97 | 76.11 | 76.27 |
| Zetatorquevirus | Torque teno douroucouli virus | BAB19320 | Douroucouli (Aotus trivirgatus) | 720 | 82.50 | 82.93 | 81.84 | 81.56 | 86.67 | 82.00 | 86.24 |
| Unassigned | Giant panda anellovirus | ASH99115 | Giant panda (Ailuropoda melanoleuca) | 559 | 71.61 | 69.30 | 68.12 | 69.19 | 84.79 | 71.57 | 71.61 |
| Paguma larvata torque teno virus | BBE36941 | Masked palm civet (Paguma larvata) | 582 | 70.59 | 64.91 | 66.23 | 69.34 | 86.83 | 71.06 | 52.51 | |
| Tick-associated torque teno virus | ASU08515 | Tick (Dermacentor variabilis) | 578 | 59.35 | 73.66 | 72.32 | 58.42 | 83.53 | 58.38 | 74.03 | |
| Torque teno indri virus | ARX79662 | Lemur (Indri indri) | 520 | 79.26 | 75.97 | 77.31 | 80.23 | 85.44 | 79.24 | 79.23 | |
Two whole genome Anelloviridae sequences were obtained by HTS: CtTTV-1 LV05 and LgTTV-1 LV07. CtTTV-1 LV05 (Fig. 4A) displayed a typical Anelloviridae organization comprised of a circular single-stranded DNA genome containing 2415 nucleotides (nt) and a 51.9% C + G content. The sequence presents an untranslated intergenic region comprising 556 nucleotides, putative ORF1 (nt 336 to 2048) and ORF2 genes (nt 190 to 468), and an ORF3 gene divided in two intervals (nt 190 to 465 and 1678 to 2034).
LgTTV-1 LV07 (Fig. 4B) also displayed a typical Anelloviridae organization comprised of a circular single-stranded DNA genome containing 2352 nucleotides (nt) and a 44.4% C + G content. The sequence presents an untranslated intergenic region comprising 460 nucleotides, putative ORF1 (nt 395 to 2095) and ORF2 genes (nt 204 to 581), and an ORF3 gene divided in two intervals (nt 204 to 578 and 1638 to 2090).
To analyze the sequences detected in the present study and representative sequences within the Anelloviridae family, a complete amino acid ORF1 phylogenetic reconstruction was performed (Fig. 4C). CtTTV-1 LV05, CtTTV-2 LV15 and CtTTV-3 LV08 were closely related and apparently emerged from the same common ancestor that originated Torque teno canis virus (genus Thetatorquevirus) that was reported in domestic dogs (Lan et al., 2011). CtTTV-4 LV23 emerged from the same common ancestor that originated unclassified anelloviruses detected in masked palm civet (Paguma larvata) (Nishizawa et al., 2018). LgTTV-1 LV06 and LV07 apparently evolved from the same common ancestor of CtTTV-4 LV23 and the unclassified anelloviruses reported in the masked palm civet. Finally, LgTTV-2 LV13 apparently presents the same common ancestor of Etatorquevirus and Lambdatorquevirus genera members reported in domestic cats (Okamoto et al., 2002) and seals (Ng et al., 2011), respectively.
3.6. Circular rep-encoding single-stranded (CRESS) DNA viruses
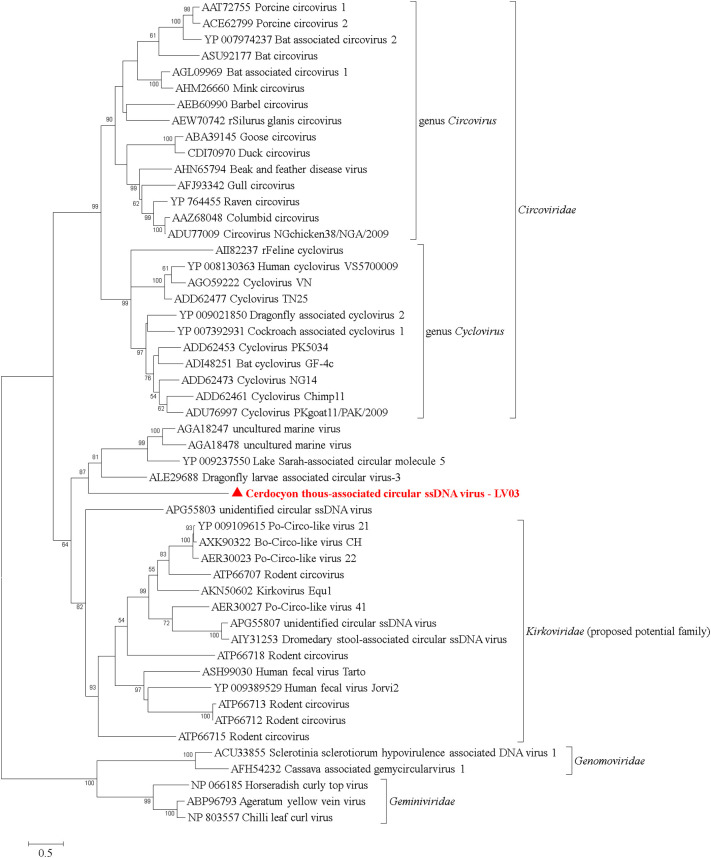
Seven contigs closely related to unclassified CRESS DNA viruses were detected by HTS in the present work (Table 1). One of the seven sequences presents the complete sequence of the putative replicase-associated (Rep) protein, which is widely used to classify CRESS DNA viruses (Zhao et al., 2019). This sequence was screened using PCR (Supplementary Table) in the 22 wild canid samples and was detected in one Cerdocyon thous from Brazil and putatively named Cerdocyon thous-associated circular ssDNA virus - LV03.
A complete Rep amino acid phylogenetic analysis was performed using Cerdocyon thous-associated circular ssDNA virus - LV03 and other representative CRESS DNA viruses (Fig. 5 ). The phylogenetic reconstruction presented six-well separated clusters corresponding to classified families Circoviridae, Genomoviridae, Geminiviridae, the proposed potential family Kirkoviridae, and two independent clades containing sequences not classified in any CRESS DNA viral family supported by bootstrap values ranging between 82 and 100%. The Cerdocyon thous-associated circular ssDNA virus LV03 grouped into one of the unclassified CRESS DNA viral families supported by an 87% bootstrap value and was more closely related to sequences reported in dragonflies, lakes and marine water (GenPept accession numbers ALE29688, YP_009237550, AGA18247, and AGA18478).
Fig. 5.
Amino acid phylogenetic reconstruction of complete putative replicase-associated (Rep) protein of CRESS DNA virus members highlighting the sequence detected in the present study. Sequences were analyzed through maximum-likelihood method applied with the JTT + G + I model. All analyses were conducted with 1000 bootstrap replicates, and the percentage of replicate trees in which the sequences clustered together have been depicted adjacent to the branches. Bootstrap values for each node have been demonstrated if they were > 50%. The sequence detected in the present study has been highlighted with ●. The sequence generated in the present study was deposited in GenBank database under accession number MT013549.
4. Discussion
The virome present in spleen and mesenteric lymph node samples obtained from 17 crab-eating foxes (Cerdocyon thous) and five Pampas foxes (Lycalopex gymnocercus) has been described using HTS and metagenomic analysis (Fig. 1). Previous works described the virome in samples obtained in domestic dogs (Canis lupus familiaris) (Li et al., 2011; Moreno et al., 2017; Weber et al., 2018a, Weber et al., 2018b), but only a limited number of works analyzed wild canids as wolves (Canis lupus signatus) (Conceição-Neto et al., 2017) and red foxes (Vulpes vulpes) (Lojkić et al., 2016). Our study, which employed nonspecific amplification, revealed the presence of commonly reported dog viruses and previously unknown viral agents (Table 1). The outstanding presence of Parvoviridae and Anelloviridae members is consistent with the findings reported in domestic dogs (Weber et al., 2018a, Weber et al., 2018b). We also observed the presence of highly prevalent viral agents in domestic dogs from Brazil, such as CPV-2 and CDV (Alves et al., 2018b), that are genetically closely related to those reported in dogs of the same Brazilian region where the wild canids were sampled.
RABV was not detected in the analyzed samples using either HTS (Table 1) or RABV-specific PCR (Soares et al., 2002). In Brazil, RABV was controlled in domestic dogs by intense public vaccination campaigns (Freire de Carvalho et al., 2018). However, the circulation of RABV in wildlife has become a major concern for public health (Antunes et al., 2018; Campos et al., 2019; Rocha et al., 2017), where biting of humans by wild animals has been reported. Moreover, crab-eating foxes were identified as reservoirs of RABV variants (Campos et al., 2019; Rocha et al., 2017). It is important to highlight that the central nervous system is the optimal sample to detect RABV, instead of the samples used in the present study, and this factor may have contributed to the non-detection of RABV in the samples analyzed.
The CDV species, renamed canine morbillivirus, was detected in four of the 17 crab-eating foxes sampled from Brazil in the present study by applying an RT-nested PCR protocol against the CDV nucleocapsid (Fischer et al., 2013). Two of these samples had a partial sequence within the CDV-hemagglutinin (An et al., 2008; Riley and Wilkes, 2015) sequenced and were genotyped as South America I/Europe (Fig. 2), which is the most prevalent CDV genotype in southern Brazil and Uruguay (Budaszewski et al., 2014; Fischer et al., 2016; Sarute et al., 2014) and is also reported in Argentina (Panzera et al., 2012). Other CDV genotypes as South America 2, 3 and 4 are frequently reported in other South American countries as Argentina, Ecuador and Colombia (Espinal et al., 2014; Panzera et al., 2014; Panzera et al., 2012; Sarute et al., 2014). The CDV sequences detected in crab-eating foxes of the present study were assigned as closely related to CDV infecting domestic dogs from southern Brazil in both nucleocapsid and hemagglutinin sequencing. CDV infection causes high mortality in domestic and wild dogs (Beineke et al., 2015), and it is a common cause of wildlife population declines (Cleaveland et al., 2000; Roelke-Parker et al., 1996; Viana et al., 2015). Additionally, CDV was determined to be the cause of death of crab-eating foxes in Brazil (Megid et al., 2009) and Argentina (Ferreyra et al., 2009). Our data reinforce that crab-eating foxes may be wild reservoirs of CDV and suggest a possible spillover between crab-eating foxes and domestic dogs.
CPV-2 was reclassified as carnivore protoparvovirus 1 (genus Protoparvovirus, subfamily Parvovirinae, family Parvoviridae) by the International Committee of Taxonomy of Viruses (ICTV) (Cotmore et al., 2014). In the present study, CPV-2 genome segments were found in one Pampas fox and two crab-eating foxes from Brazil. These sequences were subtyped as CPV-2b and -2c. CPV-2b and CPV-2c strains are more frequently found in domestic dogs from Brazil, where CPV-2c substituted CPV-2b as the most prevalent CPV-2 subtype (Pinto et al., 2012). Additionally, the CPV-2 fox sequences were closely related to those reported in domestic dogs of the same Brazilian region. Wild canids are also likely to act as reservoirs of CPV-2 infection for domestic canine populations (Truyen et al., 1998; Van Arkel et al., 2019), and CPV-2-positive serology was previously reported in Pampas and crab-eating foxes from southern Brazil (de Almeida Curi et al., 2010; Hübner et al., 2010). CPV-2 is the most frequent canine pathogen worldwide (Alves et al., 2018b; Decaro et al., 2011; Decaro and Buonavoglia, 2012) and an important cause of severe diarrhea in puppies (Decaro and Buonavoglia, 2012). Moreover, CPV-2 is highly stable in the environment and can persist in domestic dog populations due to its indirect faecal-oral transmission and circulation in susceptible dogs (Van Arkel et al., 2019). Our data reinforce that Pampas and crab-eating foxes may be wild reservoirs of CPV-2. However, more studies are required to understand the impact of CPV-2 on these two wildlife species.
CBoV was detected in one Pampas fox from Brazil in the present study (Fig. 3). This sequence was closely related to bocaparvoviruses reported in domestic dogs presenting respiratory disease (Kapoor et al., 2012). This CBoV is genetically different from CBoV-1 and CBoV-2, which were reclassified as carnivore bocaparvovirus 2 (CBPV-2) in the genus Bocaparvovirus of the subfamily Parvovirinae and family Parvoviridae (Cotmore et al., 2014). Apparently, the CBoV sequence from the present study is from a new species within the genus Bocaparvovirus. This putative new genus was only reported in domestic dogs from the United States (Kapoor et al., 2012) and China (Guo et al., 2016). To the best of our knowledge, this report is the first to describe these genetically divergent CBoVs in South America and in canids other than domestic dogs.
The Anelloviridae members represent nonenveloped ssDNA viruses comprised of more than 65 species grouped in 16 genera (ICTV, http://www.ictvonline.org/virusTaxonomy.asp), where some anelloviruses remain unassigned (Nishizawa et al., 2018). The anellovirus-related sequences were the most abundant viral sequences detected by HTS in the present study (Table 1). The predominance of anelloviruses in the mesenteric lymph nodes and spleen of both wild canid species is similar to what was reported in the serum of domestic dogs (Weber et al., 2018a, Weber et al., 2018b). Moreover, we described two complete genomes (Fig. 4A and B) and five additional sequences that displayed the complete ORF1 gene, comprising five putative new genera within the Anelloviridae family (Fig. 4C). The present study expands the host range of this viral family and suggests that novel anelloviruses of marked diversity can be found in wild canids. It is important to emphasize that anellovirus pathogenicity in canids has not been examined in depth (Lan et al., 2011; Sun et al., 2017; Weber et al., 2018a, Weber et al., 2018b).
We also detected sequences related to Rep and capsid (Cap) proteins of different circular ssDNA viruses (Table 1). One of these sequences presented the complete Rep protein that is used for the classification of the so-called CRESS-DNA viruses (Fig. 5). This sequence was genetically distinct from the other reported genomes and apparently was not classified in any assigned viral family. CRESS DNA viruses were previously described as novel circovirus-like viruses, since these viruses present Rep and Cap genes in agreement with Circoviridae members (Rosario et al., 2012b). Recently, with the recognition of its diversity and low degree of genome similarity with members of the family Circoviridae, the term CRESS-DNA viruses was proposed (Rosario et al., 2012a). These viruses have been detected in samples from a number of widely different sources, including samples from humans and other mammals, fishes, and insects, as well as from the environment (López-Bueno et al., 2016; Rosario et al., 2012a; Steel et al., 2016; Weber et al., 2018a, Weber et al., 2018b). Apparently, CRESS viruses were not reported in previous works that analyzed the virome of domestic (Moreno et al., 2017; Weber et al., 2018a, Weber et al., 2018b) and wild canids (Conceição-Neto et al., 2017; Lojkić et al., 2016), which may suggest that these viruses were not abundant in those animal species.
5. Conclusion
The present study categorized the virome of crab-eating and Pampas foxes. We reported viruses commonly detected in domestic dogs, including the pathogenic and prevalent ones CDV and CPV-2, that may impact wildlife. Moreover, the reported CDV and CPV-2 sequences were closely related to genomes reported in domestic dogs from southern Brazil. The present study also expands the host range of CBoV, CRESS DNA viruses and anelloviruses of marked diversity. The results of this study contribute to the body of knowledge regarding the wild canid virome and viral agents that are potentially transmitted through domestic and wild canids.
Declaration of Competing Interest
None declared.
Acknowledgements
The authors are grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Finance Code 001, Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Pró-Reitoria de Pesquisa (PROPESQ-UFRGS) for supporting this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104421.
Appendix A. Supplementary data
Supplementary material
References
- Alfano F., Dowgier G., Valentino M.P., Galiero G., Tinelli A., Decaro N., Fusco G. Identification of pantropic canine coronavirus in a wolf (Canis lupus italicus) in Italy. J. Wildl. Dis. 2019;55:504–508. doi: 10.7589/2018-07-182. [DOI] [PubMed] [Google Scholar]
- Alves C.D.B.T., Budaszewski R.F., Cibulski S.P., Weber M.N., Quoos F.M., Bianchi M.V., Zafalon-Silva B., Konradt G., Slaviero M., Sonne L., Driemeier D., Alievi M.M., Canal C.W. Mamastrovirus 5 detected in a crab-eating fox (Cerdocyon thous): expanding wildlife host range of astroviruses. Comp. Immunol. Microbiol. Infect. Dis. 2018;58:36–43. doi: 10.1016/j.cimid.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves C.D.B.T., Granados O.F.O., Budaszewski R. da F., Streck A.F., Weber M.N., Cibulski S.P., Pinto L.D., Ikuta N., Canal C.W. Identification of enteric viruses circulating in a dog population with low vaccine coverage. Braz. J. Microbiol. 2018 doi: 10.1016/j.bjm.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D.J., Yoon S.H., Park J.Y., No I.S., Park B.K. Phylogenetic characterization of canine distemper virus isolates from naturally infected dogs and a marten in Korea. Vet. Microbiol. 2008;132:389–395. doi: 10.1016/j.vetmic.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Antunes K.D., Matos J.C.C., Mol L.P., Oliveira M.A., Arcebispo T.L.M., Santos V.G., Oliveira T.M., Fontes C.C., Reis C.H.L., Diniz S.A., Pereira P.L.L., Silva M.X. Descriptive analysis of rabies in wild animals in the state of Sergipe, Brazil. Arq. Bras. Med. Vet. e Zootec. 2018;70:169–173. doi: 10.1590/1678-4162-9574. [DOI] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beineke A., Baumgärtner W., Wohlsein P. Cross-species transmission of canine distemper virus — an update. One Heal. 2015;1:49–59. doi: 10.1016/j.onehlt.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budaszewski R. da F., Pinto L.D., Weber M.N., Caldart E.T., Alves C.D.B.T., Martella V., Ikuta N., Lunge V.R., Canal C.W. Genotyping of canine distemper virus strains circulating in Brazil from 2008 to 2012. Virus Res. 2014;180:76–83. doi: 10.1016/j.virusres.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Buonavoglia D., Cavalli A., Pratelli A., Martella V., Greco G., Tempesta M., Buonavoglia C. Antigenic analysis of canine parvovirus strains isolated in Italy. New Microbiol. 2000;23:93–96. [PubMed] [Google Scholar]
- Buonavoglia C., Martella V., Pratella A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- Campos A.A.S., dos Santos R.N., Benavides J.A., Batista H.B.C.R., Finoketti F., Wagner P.G.C., Zafalon-Silva B., Alievi M., da Silva F.B., Witt A., Tartarotti A., de da Silva A.C.R., Ferreira K.C.S., Frazzon A.P.G., Roehe P.M., Franco A.C. Rabies surveillance in wild mammals in South of Brazil. Transbound. Emerg. Dis. 2019:1–8. doi: 10.1111/tbed.13415. [DOI] [PubMed] [Google Scholar]
- Cleaveland S., Appel M.G.J., Chalmers W.S.K., Chullingworth C., Kaare M., Dye C. Serological and demographic evidence of domestic dogs as a source of canine distemper virus infection for Seringeti wildlife. Vet. Microbiol. 2000;72:217–227. doi: 10.1016/s0378-1135(99)00207-2. [DOI] [PubMed] [Google Scholar]
- Conceição-Neto N., Godinho R., Álvares F., Yinda C.K., Deboutte W., Zeller M., Laenen L., Heylen E., Roque S., Petrucci-Fonseca F., Santos N., Van Ranst M., Mesquita J.R., Matthijnssens J. Viral gut metagenomics of sympatric wild and domestic canids, and monitoring of viruses: insights from an endangered wolf population. Ecol. Evol. 2017;7:4135–4146. doi: 10.1002/ece3.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-Mckenna M., Chiorini J.A., Dmitry V.M., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J. The family Parvoviridae. Arch. Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Curi N.H., Araújo A.S., Campos F.S., Lobato Z.I.P., Gennari S.M., Marvulo M.F.V., Silva J.C.R., Talamoni S.A. Wild canids, domestic dogs and their pathogens in Southeast Brazil: disease threats for canid conservation. Biodivers. Conserv. 2010;19:3513–3524. doi: 10.1007/s10531-010-9911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. Canine parvovirus - a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet. Microbiol. 2012;155:1–12. doi: 10.1016/j.vetmic.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Billi M., Mari V., Elia G., Cavalli A., Martella V., Buonavoglia C. Western European epidemiological survey for parvovirus and coronavirus infections in dogs. Vet. J. 2011;187:195–199. doi: 10.1016/j.tvjl.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowgier G., Lahoreau J., Lanave G., Losurdo M., Varello K., Lucente M.S., Ventriglia G., Bozzetta E., Martella V., Buonavoglia C., Decaro N. Sequential circulation of canine adenoviruses 1 and 2 in captive wild carnivores, France. Vet. Microbiol. 2018;221:67–73. doi: 10.1016/j.vetmic.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinal M.A., Díaz F.J., Ruiz-Saenz J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet. Microbiol. 2014;172:168–176. doi: 10.1016/j.vetmic.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Ferreyra H., Calderón M.G., Marticorena D., Marull C., Leonardo B.C. Canine distemper infection in crab-eating fox (Cerdocyon thous) from Argentina. J. Wildl. Dis. 2009;45:1158–1162. doi: 10.7589/0090-3558-45.4.1158. [DOI] [PubMed] [Google Scholar]
- Fischer C.D.B., Ikuta N., Canal C.W., Makiejczuk A., Allgayer M. da C., Cardoso C.H., Lehmann F.K., Fonseca A.S.K., Lunge V.R. Detection and differentiation of field and vaccine strains of canine distemper virus using reverse transcription followed by nested real time PCR (RT-nqPCR) and RFLP analysis. J. Virol. Methods. 2013;194:39–45. doi: 10.1016/j.jviromet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C.D.B., Gräf T., Ikuta N., Lehmann F.K.M., Passos D.T., Makiejczuk A., Silveira M.A.T., Fonseca A.S.K., Canal C.W., Lunge V.R. Phylogenetic analysis of canine distemper virus in South America clade 1 reveals unique molecular signatures of the local epidemic. Infect. Genet. Evol. 2016;41:135–141. doi: 10.1016/j.meegid.2016.03.029. [DOI] [PubMed] [Google Scholar]
- Freire de Carvalho M., Vigilato M.A.N., Pompei J.A., Rocha F., Vokaty A., Molina-Flores B., Cosivi O., Vilas V.J.D.R. Rabies in the Americas: 1998-2014. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S., Garcia-Gomez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talon M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Wang Z., Yao S., Li C., Geng Y., Wang E., Zhao X., Su M., Wei S., Wang X., Feng L., Chang Y. fu, Sun D. Epidemiological investigation reveals genetic diversity and high co-infection rate of canine bocavirus strains circulating in Heilongjiang province, Northeast China. Res. Vet. Sci. 2016;106:7–13. doi: 10.1016/j.rvsc.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner S.O., Pappen F.G., Ruas J.L., Vargas G.D., Fischer G., Vidor T. Exposure of pampas fox (Pseudalopex gymnocercus) and crab-eating fox (Cerdocyon thous) from the southern region of Brazil to canine distemper virus (CDV), canine parvovirus (CPV) and canine coronavirus (CCoV) Braz. Arch. Biol. Technol. 2010;53:593–597. doi: 10.1590/S1516-89132010000300012. [DOI] [Google Scholar]
- Kapoor A., Mehta N., Dubovi E.J., Simmonds P., Govindasamy L., Medina J.L., Street C., Shields S., Ian Lipkin W. Characterization of novel canine bocaviruses and their association with respiratory disease. J. Gen. Virol. 2012;93:341–346. doi: 10.1099/vir.0.036624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan D., Hua X., Cui L., Luo X., Liu Z., San T., Zhu C.X., Zhao W., Yang Z. Sequence analysis of a torque Teno canis virus isolated in China. Virus Res. 2011;160:98–101. doi: 10.1016/j.virusres.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Li L., Pesavento P.A., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., McGraw S., Zhu K., Leutenegger C.M., Marks S.L., Kubiski S., Gaffney P., Dela Cruz F.N., Wang C., Delwart E., Pesavento P.A. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg. Infect. Dis. 2013;19:534–541. doi: 10.3201/eid1904.121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lojkić I., Bidin M., Prpić J., Šimić I., Krešić N., Bedeković T. Faecal virome of red foxes from peri-urban areas. Comp. Immunol. Microbiol. Infect. Dis. 2016;45:10–15. doi: 10.1016/j.cimid.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A., Mavian C., Labella A.M., Castro D., Borrego J.J., Alcami A., Alejo A. Concurrence of Iridovirus, Polyomavirus, and a unique member of a new Group of Fish Papillomaviruses in Lymphocystis disease-affected Gilthead Sea bream. J. Virol. 2016;90:8768–8779. doi: 10.1128/JVI.01369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megid J., De Souza V.A.F., Teixeira C.R., Cortez A., Amorin R.L., Heinemman M.B., Cagnini D.Q., Richtzenhain L.J. Canine distemper virus in a crab-eating fox (Cerdocyon thous) in Brazil: case report and phylogenetic analyses. J. Wildl. Dis. 2009;45:527–530. doi: 10.7589/0090-3558-45.2.527. [DOI] [PubMed] [Google Scholar]
- Moreno P.S., Wagner J., Mansfield C.S., Stevens M., Gilkerson J.R., Kirkwood C.D. Characterisation of the canine faecal virome in healthy dogs and dogs with acute diarrhoea using shotgun metagenomics. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Wheeler E., Greig D., Waltzek T.B., Gulland F., Breitbart M. Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J. Gen. Virol. 2011;92:1318–1323. doi: 10.1099/vir.0.029678-0. [DOI] [PubMed] [Google Scholar]
- Nishizawa T., Sugimoto Y., Takeda T., Kodera Y., Hatano Y., Takahashi M., Okamoto H. Identification and whole genome characterization of novel anelloviruses in masked palm civets (Paguma larvata): segregation into four distinct clades. Virus Res. 2018;256:183–191. doi: 10.1016/j.virusres.2018.08.015. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Takahashi M., Nishizawa T., Tawara A., Fukai K., Muramatsu U., Naito Y., Yoshikawa A. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 2002;83:1291–1297. doi: 10.1099/0022-1317-83-6-1291. [DOI] [PubMed] [Google Scholar]
- Paim W.P., Weber M.N., Cibulski S.P., da Silva M.S., Puhl D.E., Budaszewski R.F., Varela A.P.M., Mayer F.Q., Canal C.W. Characterization of the viral genomes present in commercial batches of horse serum obtained by high-throughput sequencing. Biologicals. 2019;61:1–7. doi: 10.1016/j.biologicals.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Panzera Y., Calderón M.G., Sarute N., Guasco S., Cardeillac A., Bonilla B., Hernández M., Francia L., Bedó G., La Torre J., Pérez R. Evidence of two co-circulating genetic lineages of canine distemper virus in South America. Virus Res. 2012;163:401–404. doi: 10.1016/j.virusres.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Panzera Y., Sarute N., Carrau L., Aldaz J., Pérez R. Review genetic diversity of canine distemper virus in South America. Br. J. Virol. 2014;1:48–53. [Google Scholar]
- Pinto L.D., Streck A.F., Gonçalves K.R., Souza C.K., Corbellini Â.O., Corbellini L.G., Canal C.W. Typing of canine parvovirus strains circulating in Brazil between 2008 and 2010. Virus Res. 2012;165:29–33. doi: 10.1016/j.virusres.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M.C., Wilkes R.P. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the United States associated with disease in wildlife and domestic canine populations. Virol. J. 2015;12 doi: 10.1186/s12985-015-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S.M., de Oliveira S.V., Heinemann M.B., Gonçalves V.S.P. Epidemiological profile of wild rabies in Brazil (2002−2012) Transbound. Emerg. Dis. 2017;64:624–633. doi: 10.1111/tbed.12428. [DOI] [PubMed] [Google Scholar]
- Roelke-Parker M.E., Munson L., Packer C., Kock R., Cleaveland S., Carpenter M., O’Brien S.J., Pospischil A., Hofman-Lehmann R., Lutz H., Mwamengele G.L.M., Mgasa M.N., Machange G.A., Summers B.A., Appel M.J.G. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Dayaram A., Marinov M., Ware J., Kraberger S., Stainton D., Breitbart M., Varsani A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta) J. Gen. Virol. 2012;93:2668–2681. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch. Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. 3rd ed. 2001. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, USA) [Google Scholar]
- Sarute N., Pérez R., Aldaz J., Alfieri A.A., Alfieri A.F., Name D., Llanes J., Hernández M., Francia L., Panzera Y. Molecular typing of canine distemper virus strains reveals the presence of a new genetic variant in South America. Virus Genes. 2014;48:474–478. doi: 10.1007/s11262-014-1054-z. [DOI] [PubMed] [Google Scholar]
- Soares R.M., Bernardi F., Sakamoto S.M., Heinemann M.B., Cortez A., Alves L.M., Meyer A.D., Ito F.H., Richtzenhain L.J. A heminested polymerase chain reaction for the detection of Brazilian rabies isolates from vampire bats and herbivores. Mem. Inst. Oswaldo Cruz. 2002;97:109–111. doi: 10.1590/S0074-02762002000100019. [DOI] [PubMed] [Google Scholar]
- Steel O., Kraberger S., Sikorski A., Young L.M., Catchpole R.J., Stevens A.J., Ladley J.J., Coray D.S., Stainton D., Dayaram A., Julian L., van Bysterveldt K., Varsani A. Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand. Infect. Genet. Evol. 2016;43:151–164. doi: 10.1016/j.meegid.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Sun W., Xie C., Liang C., Zheng M., Zhao G., Zhang P., Han J., Jing J., Wen S., Xiao P., Cui Z., Zhang J., Ren J., Liu H., Lu H., Jin N. Molecular detection and genomic characterization of torque Teno canis virus in domestic dogs in Guangxi Province, China. J. Biotechnol. 2017;252:50–54. doi: 10.1016/j.jbiotec.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R.V., Haynes M., Breitbart M., Wegley L., Rohwer F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Truyen U., Müller T., Heidrich R., Tackmann K., Carmichael L.E. Survey on viral pathogens in wild red foxes (Vulpes vulpes) in Germany with emphasis on parvoviruses and analysis of a DNA sequence from a red fox parvovirus. Epidemiol. Infect. 1998;121:433–440. doi: 10.1017/S0950268898001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J. a M. Primer3 plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arkel A., Kelman M., West P., Ward M.P. The relationship between reported domestic canine parvovirus cases and wild canid distribution. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana M., Cleaveland S., Matthiopoulos J., Halliday J., Packer C., Craft M.E., Hampson K., Czupryna A., Dobson A.P., Dubovi E.J., Ernest E., Fyumagwa R., Hoare R., Grant J., Hopcraft C., Horton D.L., Kaare M.T., Kanellos T., Lankester F., Mentzel C., Mlengeya T., Mzimbiri I., Takahashi E., Willett B., Haydon D.T., Lembo T. Dynamics of a morbillivirus at the domestic–wildlife interface: canine distemper virus in domestic dogs and lions. PNAS. 2015;112:1464–1469. doi: 10.1073/pnas.1411623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M.N., Cibulski S.P., Olegário J.C., da Silva M.S., Puhl D.E., Mósena A.C.S., Alves C.D.B.T., Paim W.P., Baumbach L.F., Mayer F.Q., Fernandes A.R.F., Azevedo S.S., Canal C.W. Characterization of dog serum virome from Northeastern Brazil. Virology. 2018;525:192–199. doi: 10.1016/j.virol.2018.09.023. [DOI] [PubMed] [Google Scholar]
- Weber M.N., Cibulski S.P., Silveira S., Siqueira F.M., Mósena A.C.S., da Silva M.S., Olegário J.C., Varela A.P.M., Teixeira T.F., Bianchi M.V., Driemeier D., Pavarini S.P., Mayer F.Q., Roehe P.M., Canal C.W. Evaluation of the serum virome in calves persistently infected with Pestivirus a, presenting or not presenting mucosal disease. Virus Genes. 2018 doi: 10.1007/s11262-018-1599-3. [DOI] [PubMed] [Google Scholar]
- Zhao L., Rosario K., Breytbart M., Duffy S. In: Advances in Virus Research. Mettenleiter T., Kielian M., Roossinck M.J., editors. Elsevier; London: 2019. Eukaryotic circular rep-encoding single-stranded DNA (CRESS DNA) viruses: Ubiquitous viruses with small genomes and a diverse host range; pp. 71–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material