Abstract
Background
Spasmolytic polypeptide-expressing metaplasia (SPEM) is considered a precursor lesion of intestinal metaplasia and intestinal-type gastric cancer (GC), but little is known about microRNA (miRNA) alterations during metaplasia and GC developments. Here, we investigate miR-30a expression in gastric lesions and identify its novel target gene which is associated with the intestinal-type GC.
Methods
We conducted in situ hybridization and qRT-PCR to determine miR-30a expression in gastric tissues. MiR-30a functions were determined through induction or inhibition of miR-30a in GC cell lines. A gene microarray was utilised to confirm miR-30a target genes in GC, and siRNA-mediated target gene suppression and immunostaining were performed. The Cancer Genome Atlas (TCGA) data were utilised to validate gene expressions.
Results
We found down-regulation of miR-30a during chief cell transdifferentiation into SPEM. MiR-30a level was also reduced in the early stage of GC, and its level was maintained in advanced GC. We identified a novel target gene of miR-30a, ITGA2, and our results showed that either ectopic expression of miR-30a or ITGA2 knockdown suppressed GC cell proliferation, migration and tumorigenesis. Levels of ITGA2 inversely correlated with levels of miR-30a in human intestinal-type GC.
Conclusion
We found down-regulation of miR-30a in preneoplastic lesions and its tumor suppressive functions by targeting ITGA2 in GC. The level of ITGA2, which functions as an oncogene, was up-regulated in human GC. The results of this study suggest that coordination of the miR-30a-ITGA2 axis may serve as an important mechanism in the development of gastric precancerous lesions and intestinal-type GC.
Keywords: miR-30a, ITGA2, metaplasia, gastric cancer, tumor suppressor
INTRODUCTION
Gastric cancer (GC) is the fifth most common cancer and third leading cause of cancer-related mortality worldwide [1]. Aetiologically, gastric carcinogenesis can be induced by Helicobacter pylori (H. pylori) and Epstein-Barr virus infections, as well as hereditary and environmental factors [2]. GC is divided into two histological types according to the Lauren classification: the intestinal type and the diffuse type. The incidence of the intestinal type of GC is higher than that of the diffuse type, and the main cause of intestinal-type GC is intestinalization of normal gastric glands with mucosal atrophy induced by H. pylori infection [3]. Intestinal-type gastric carcinogenesis is a multistep process of cell progression evolving from chronic and atrophic gastritis, metaplasia and dysplasia [4]. In particular, metaplasia consists of two types, spasmolytic polypeptide (TFF2)-expressing metaplasia (SPEM) and intestinal metaplasia (IM), which are considered precursors of intestinal-type GC [5, 6].
MicroRNAs (miRNAs), which are small non-coding RNA molecules consisting of approximately 22 nucleotides, are highly conserved and post-transcriptionally suppress gene expression by binding to the 3’ untranslated region (UTR) of target mRNAs. miRNAs can have oncogenic or tumor-suppressive roles in cancer by directly targeting genes and regulating various cancer cell properties, such as cell growth and migration [7–9]. Therefore, deregulated miRNAs are considered promising biomarkers and therapeutic targets for cancer. miRNAs have also been investigated as important biomarkers related to GC progression and prognosis [10]. Although many studies have reported that miRNAs and their target genes are associated with GC cell progression and metastasis, little is known about the association between miRNAs and development of the intestinal type of GC from precancerous lesions. Indeed, the intestinal type of GC frequently develops from precancerous lesions, and our recent study reported that miR-135b is up-regulated in the process of gastritis-mediated gastric carcinogenesis [11]. Moreover, conserved miRNA signatures in gastric tumorigenesis were recently investigated using human GC and adjacent normal tissues as well as a mouse dysplasia model [12]. It will be now important to reveal which miRNAs are conserved from precancerous lesions to GC and which miRNAs play key roles in tumorigenesis.
To understand miRNAs involved in gastric metaplasia that may evolve into GC, our previous study analysed miRNA profiles using human metaplastic tissue samples obtained from patients with intestinal-type GC and showed that down-regulation of miR-30a contributes to stomach intestinalization [13]. Furthermore, miR-30a-knockout mice infected with H. pylori displayed gastritis, dysplasia and gastric adenocarcinoma [14]. According to several previous studies focusing on global miRNA changes in GC, miR-30a is a significantly down-regulated miRNA in GC [10, 15–17], and a particular miRNA signature including miR-30a was found to be useful for predicting GC patient survival, with a low risk score for maintained expression of miR-30a [18]. In fact, miR-30a is known as a tumor suppressor that inhibits GC cell proliferation, invasion, and drug resistance by targeting oncogenes such as Vimentin or IGF1R [19–23]. However, the role of miR-30a in GC initiation and development has not yet been elucidated.
In this study, we sought to investigate whether miR-30a down-regulation is an early event in metaplasia by tracing its expression during development of SPEM, a potential precursor leading to IM as well as gastric carcinogenesis [24–26], and is maintained in GC through regulation of GC cell properties. We examined the changes in miR-30a expression from normal glands to SPEM lesions in both mouse and human stomach tissues and validated its down-regulation in GC using an integrated analysis of previous miRNA profiles and our microarray data set. In addition, we identified integrin α2 (ITGA2) as being a novel target gene of miR-30a, the expression of which shows an inverse correlation with that of miR-30a in both SPEM and GC. The biological functions of both miR-30a and ITGA2 were evaluated using GC cell lines to determine whether the miR-30a-ITGA2 axis is significantly correlated, especially in the intestinal type of GC.
MATERIALS AND METHODS
Cell lines and tissue specimens
Three human GC cell lines, SNU-216, SNU-601, and MKN28, and the human embryonic kidney cell line, HEK293, were obtained from the Korean Cell Line Bank (Seoul, Korea) and maintained in RPMI 1640 (Welgene, Daegu, Korea) with 10 % foetal bovine serum (Gibco, Invitrogen, UK) and 1 % penicillin-streptomycin (Gibco, Invitrogen, UK). HSC44Luc and 44As3Luc cell lines were provided by Kazuyoshi Yanagihara, National Cancer Center Research Institute, Japan. The cells were cultured and maintained under the same conditions as those for the GC cell lines. Thirty pairs of primary GC tissues and matched non-cancer mucosa (NM) were used to analyse expression levels of miRNA or mRNA. All tissue specimens were obtained from the Seoul National University Hospital (SNUH), Korea and Vanderbilt University Medical Center. Clinicopathological information is provided in Supplementary Table S1. The present study was approved by the Institutional Review Boards of Seoul National University Hospital (IRB No. 1308-122-517).
In vivo experiments
For SPEM mouse models, 350 mg/kg of L635 (n = 3) or 350 mg/kg of DMP-777 (n = 3) was administered to female or male C57BL/6 mice at 8 weeks of age, as previously described, [27, 28] This animal experiment followed protocols approved by the Institutional Animal Care and Use Committees (IACUC) of Vanderbilt University.
Female BALB/c nu/nu mice at 6–8 weeks of age (Orient Bio., Sungnam, Korea) were used to establish tumor xenograft models. For the tumorigenicity assay, we subcutaneously injected empty vector-transfected (n = 8) or stable miR-30a-overexpressing (n = 8) SNU-601 cells into the right flank of the mice; a total of 1 × 107 cells in 200 μl of medium was injected into each mouse. Tumor size was measured twice every week using callipers, and the tumor volume was calculated by length (L) × width (W) × height (H). On day 63, all mice were sacrificed, and the tumor mass was pathologically assessed by haematoxylin and eosin (H&E) staining. This animal experiment was approved by the IACUC of the Clinical Research Institute at Seoul National University Hospital [14–0183-C1A0(1)].
See supplementary materials and methods for more information.
RESULTS
Alteration of miR-30a expression in two preneoplastic metaplasias, SPEM and IM of the stomach
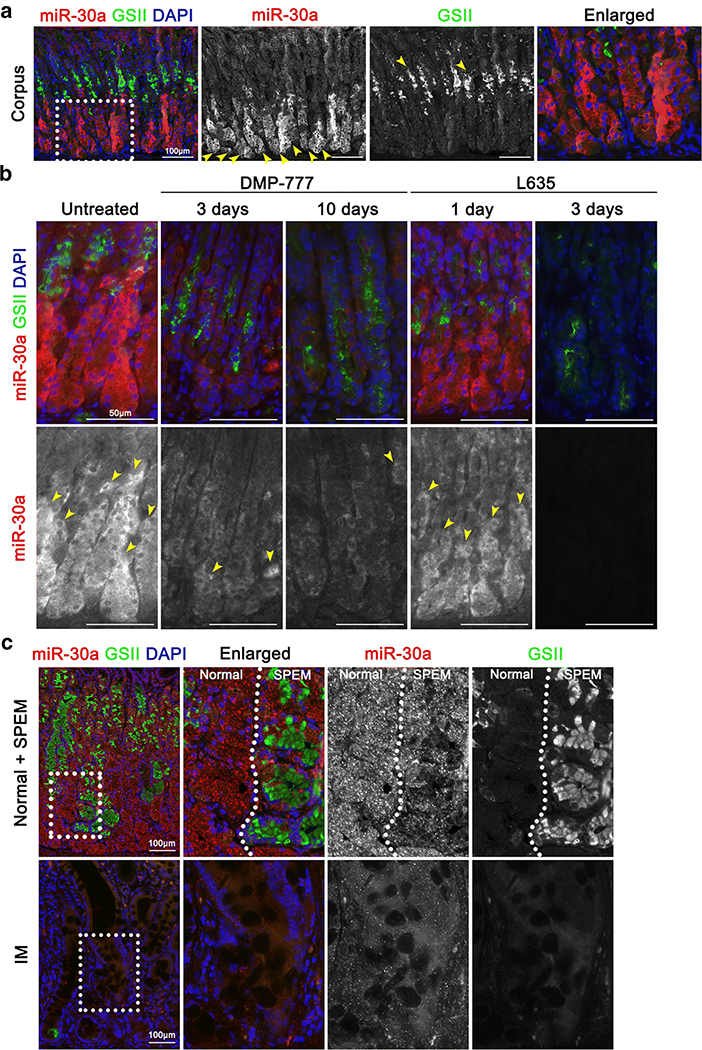
In our previous study, mRNA transcripts and miRNAs in human IM and SPEM were profiled, and two transcription factors, HNF4γ and NR2F2, and their regulatory factors, miR-30 and miR194, were identified as associated with the development of IM [13]. In the present study, we sought to expand the cellular functions and molecular mechanism of a specific miRNA, miR-30a, during gastric carcinogenesis. We first identified miR-30a localisation and expression patterns using in situ hybridization analyses of human and mouse stomach tissues with normal and/or metaplastic glands. In the normal mouse stomach, where GSII-Lectin (GSII) positive in mucous neck cells, miR-30a enrichment was observed in chief cells of corpus glands (Figure 1A), whereas miR-30a was barely or not expressed in the antrum or duodenum (Supplementary Figure S1). Next, to determine whether miR-30a expression is altered during SPEM development, we used acute oxyntic atrophy mouse models via administration of DMP-777 or L635 to induce SPEM [27, 28]. Co-staining results for miR-30a and GSII, which is a mucous neck and SPEM marker, showed reduced expression of miR-30a in the metaplastic glands after DMP-777 or L635 treatment compared to that in the untreated mouse stomach (Figure 1B). Moreover, decreases in miR-30a were observed in chief cells, which were not completely transdifferentiated into GSII-positive SPEM cells (Figure 1B).
Figure 1. miR-30a expression in normal and metaplasias of mouse and human stomachs.
(A) In situ hybridization for miR-30a (red) and immunofluorescence staining for GSII (green) and DAPI (blue) in mouse normal stomach. Scale bar = 100 μm. Yellow arrows indicate representative miR-30a- or GSII-positive glands. Dotted box indicates enlarged area. (B) In situ hybridization for miR-30a (red) and immunofluorescence staining for GSII (green) and DAPI (blue) in mouse stomachs that developed SPEM after 3 days and 10 days of DMP-777 treatment as well as after 1 day and 3 days of L635 treatment. A mouse normal stomach was used as a control. Scale bar = 50 μm. Yellow arrows indicate miR-30a-positive glands. (C) In situ hybridization for miR-30a (red) and immunofluorescence staining for GSII (green) and DAPI (blue) in human stomach tissue includes normal, SPEM and IM glands. Scale bar = 100 μm. Dotted boxes indicate enlarged area.
Next, we used clinical samples to determine miR-30a expression patterns using human normal gastric glands, SPEM and IM lesions. Similarly, in situ hybridization results showed that the miR-30a was strongly expressed in normal glands, whereas its expression was down-regulated in both GSII-positive SPEM and GSII-negative IM lesions (Figure 1C). Taken together, miR-30a is expressed in normal glands, but reduced expression was found during SPEM development and in IM. Therefore, these results indicate that down-regulation of miR-30a is associated with two preneoplastic metaplasias, SPEM and IM, as precursor lesions of intestinal-type GC.
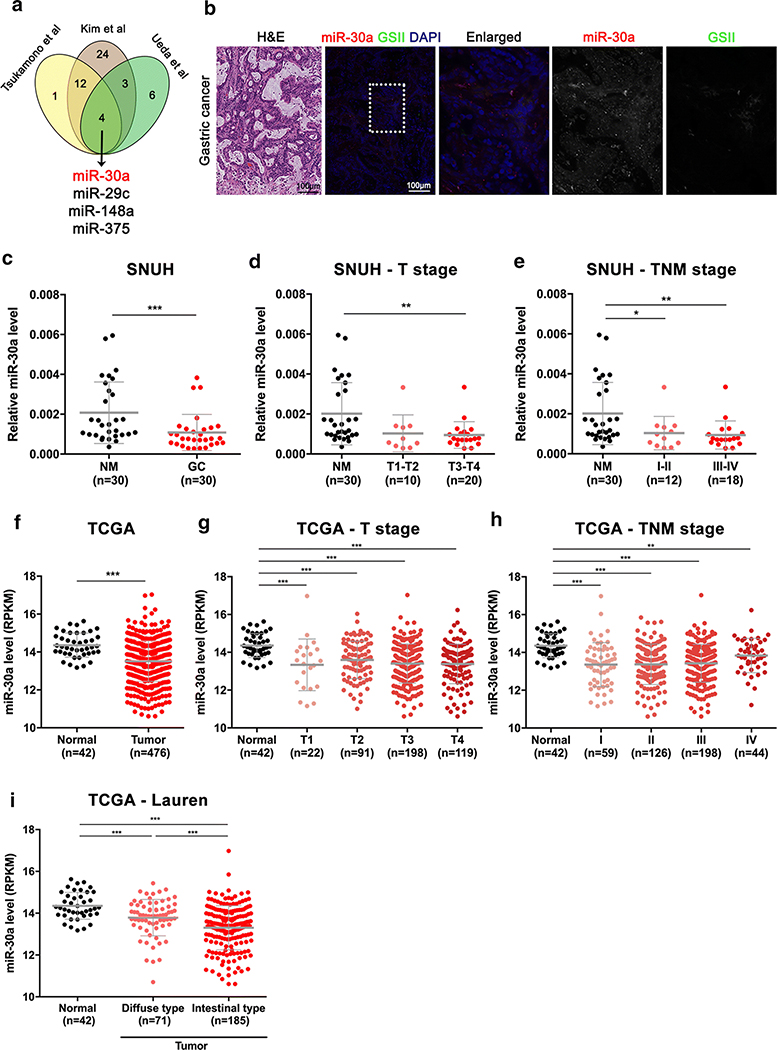
miR-30a is down-regulated in human GC tissues and related to the intestinal type of GC
To determine whether miR-30a is related to GC development, we performed integrated analysis of three independent microRNA microarray profiles for human normal and GC samples from Tsukamoto et al. [16], Ueda et al. [10], and Kim et al. [15]. As shown in Figure 2A, miR-30a was commonly down-regulated in GC tissues compared to non-tumor tissues in the three independent miRNA profiles. To confirm this observation, we analysed miR-30a expression in human GC tissues by in situ hybridization and performed qRT-PCR for miR-30a using 30 pairs of GC and adjacent non-cancer tissues from the SNUH GC cohort. We found that miR-30a was barely expressed in human GC tissues that were GSII-negative (Figure 2B) and was significantly down-regulated in 25 of 30 GC tissues (83.3%) compared to their matched non-cancer tissues (Figure 2C). Moreover, miR-30a levels were significantly reduced in early-stage (Stages I/II) as well as in advanced (Stages III/IV) GC, but there was no significant difference between early and advanced samples (Figure 2D and 2E). These results are supported by stomach adenocarcinoma patient data deposited in TCGA (Figure 2G and 2H), with significantly down-regulated levels of miR-30a in GC samples at all stages (Figure 2F). Similar results were found in an NCBI GEO database, GSE26595, which revealed a low expression level for miR-30a in stomach adenocarcinoma samples compared with non-cancer samples (Supplementary Figure S2). Interestingly, miR-30a levels were significantly lower in GC samples classified as the intestinal type, compared with the diffuse type (Figure 2I). Overall, these integrated analyses of miRNA profiles show consistently reduced-miR-30a expression in early stages of human GC, which are maintained during GC progression, and its low level is associated with the intestinal type of GC.
Figure 2. Expression level of miR-30a in GC.
(A) Identification of downregulated miRNAs in GC in three independent miRNA profiles which have been previously reported [10, 15, 16]. (B) In situ hybridization for miR-30a (red) and immunofluorescence staining for GSII (green) and DAPI (blue) in human GC tissue. Scale bar = 100 μm. Dotted box indicates enlarged area. (C-E) Expression level of miR-30a in 30 pairs of GC and adjacent non-cancer mucosa (NM) (C) and in NM and GC according to T stage (D) and TNM stage (E); samples were obtained from the Seoul National University Hospital (SNUH). (F-H) Expression status of miR-30a in 42 normal stomach tissues and 476 gastric adenocarcinoma samples (F) according to T stage (G) and TNM stage (H) from RNA sequencing data (Illumina GA, HiSeq) of TCGA. (I) miR-30a expression patterns according to Lauren classification in TCGA. *p < 0.05, **p < 0.01, ***p < 0.001, t-test
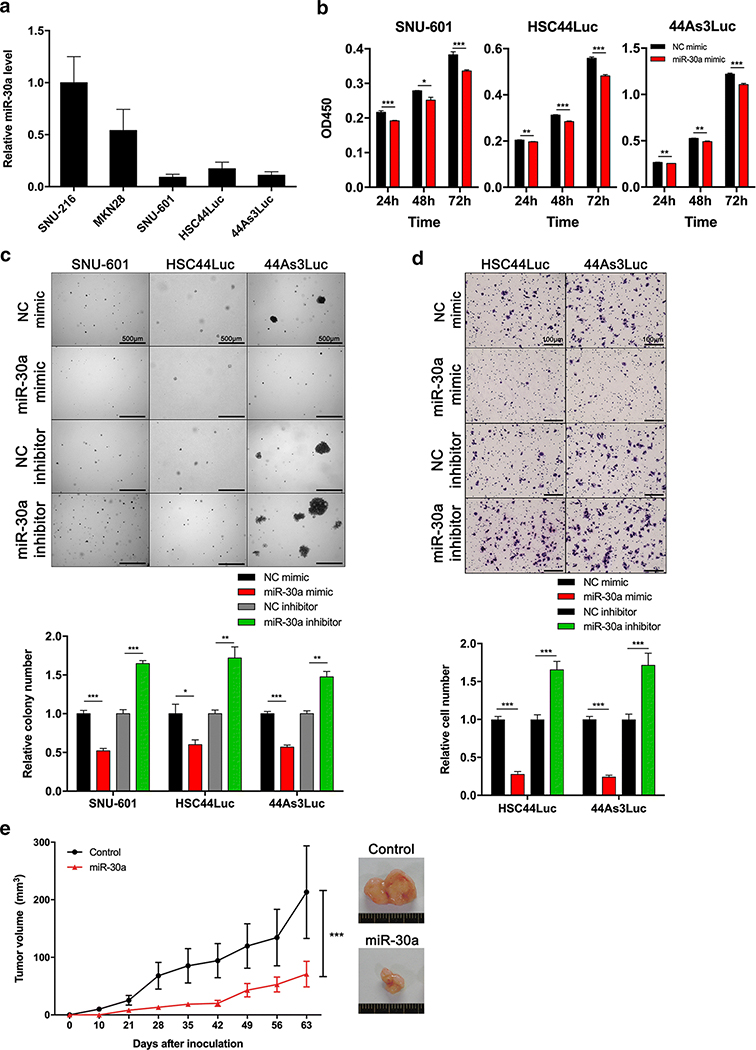
miR-30a functions as a tumor suppressor by inhibiting GC cell proliferation, migration and tumorigenic abilities
Because miR-30a expression is down-regulated during gastric carcinogenesis, including metaplasia lesions and GC, we sought to determine the cellular functions of miR-30a in a series of GC cell lines. We first investigated miR-30a expression in GC cell lines (Figure 3A). miR-30a mimic or single-stranded miR-30a-targeting inhibitor (miR-30a inhibitor) was transfected into SNU-601, HSC44Luc and 44As3Luc cells, which exhibited lower expression of miR-30a than SNU-216 and MKN28 cells, for overexpression of miR-30a or inhibition of endogenous miR-30a expression (Supplementary Figure S3). According to a proliferation assay, the miR-30a mimic significantly decreased the growth of SNU-601, HSC44Luc and 44As3Luc cells (Figure 3B). To confirm whether miR-30a regulates the tumorigenic potential of GC cells, we performed an anchorage-independent cell culture with a soft agar colony forming assay, and showed that colony number was reduced by the mimic in miR-30a-overexpressing cells, but that it was increased in miR-30a inhibitor-treated cells (Figure 3C). Next, we performed a Transwell migration assay to confirm oncogenic properties by suppressing miR-30a. Ectopic expression of miR-30a in GC cells decreased the number of migrating cells, but miR-30a inhibitor treatment increased the number of migrating cells (Figure 3D).
Figure 3. Functional roles of miR-30a in GC cells.
(A) miR-30a expression level in five GC cell lines. (B) Cell proliferation assay (WST assay) using SNU-601, HSC44Luc and 44As3Luc cells treated with either negative control (NC) mimic or miR-30a mimic. The absorbance of 450 nm was measured at 72 h after transient transfection. (C) Soft agar colony formation assay using SNU-601, HSC44Luc and 44As3Luc cell lines treated with either NC mimic or miR-30a mimic, and colony numbers were counted in each group. Scale bar = 500 μm. (D) Transwell migration assay using HSC44Luc and 44As3Luc cells treated with either NC mimic or miR-30a mimic, and migrated cell numbers were counted in each group. Scale bar = 100 μm. *p < 0.05, ** p <0.01, *** p < 0.001, unpaired t-test. (E) In vivo tumorigenesis assay using BALB/c nude mice. SNU-601 cells stably overexpressing either empty vector (control) or miR-30a were subcutaneously injected into mice (each group; n = 8). *** p < 0.001, ANOVA.
Based on the regulation of oncogenic properties by miR-30a in vitro, we next examined the role of miR-30a in tumorigenicity in vivo. We subcutaneously injected SNU-601 control cells or cells stably expressing miR-30a into BALB/c nude mice and examined tumor volume for 9 weeks. Interestingly, tumor volume was significantly reduced in the miR-30a-overexpressing group compared with the control group (Figure 3E), suggesting that miR-30a acts as a tumor suppressor by inhibiting GC cell proliferation, migration and tumorigenicity.
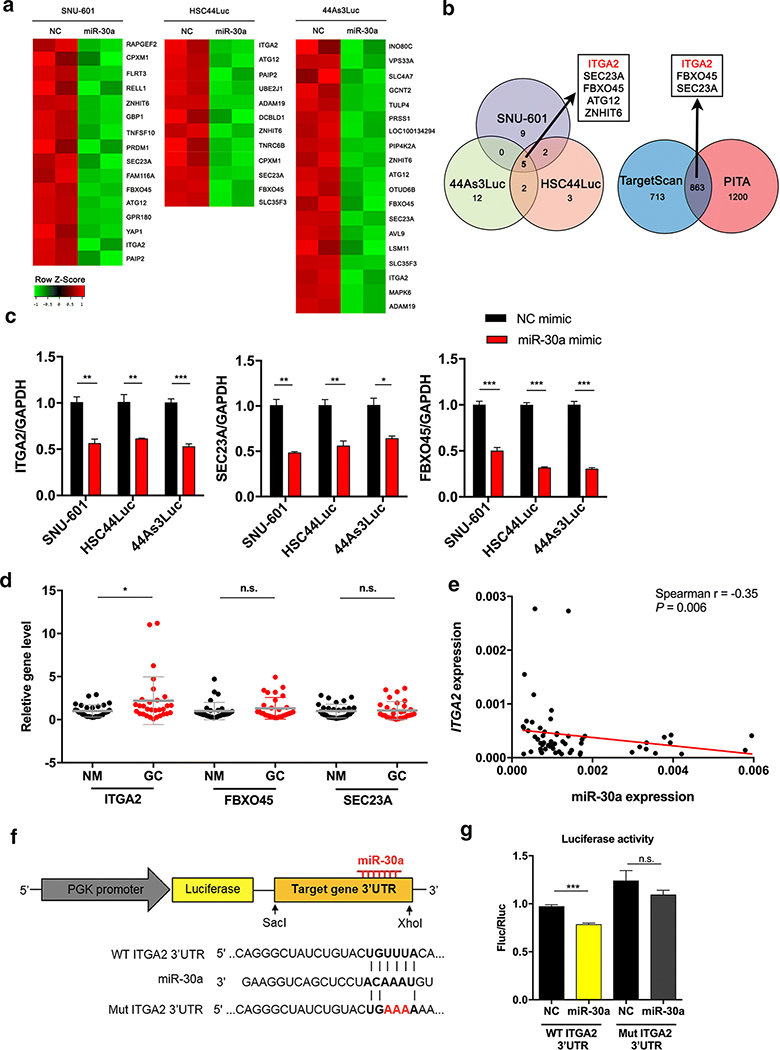
ITGA2 is a novel target gene of miR-30a in GC
To better understand the tumor-suppressive functions of miR-30a, a microarray analysis was conducted to identify potential target genes of miR-30a in GC. MiR-30a mimic or negative control mimic was transfected into three GC cell lines, SNU-601, HSC44Luc and 44As3Luc, after which total RNAs were extracted and microarray profiling performed. As shown in Figure 4A, expression of several genes was significantly decreased in the three cell lines after miR-30a mimic treatment. Among them, five genes (ITGA2, SEC23A, FBXO45, ATG12 and ZNHIT6) were commonly down-regulated in the three GC cell lines following miR-30a overexpression (Figure 4B, left), suggesting that they are putative target genes. Next, two independent target prediction programs (TargetScan7.1 and PITA) revealed 863 genes as common predicted targets of miR-30a, and an integrated analysis of both microarray and prediction programs identified three genes, which were integrin alpha 2 (ITGA2), F-Box Protein 45 (FBXO45) and Sec23 homolog A (SEC23A) (Figure 4B, right). To validate the down-regulation of these candidate target genes in miR-30a mimic-treated GC cells, we performed qRT-PCR for each gene and found that the expression levels of all three were significantly reduced in miR-30a mimic-treated cells compared to negative control cells (Figure 4C).
Figure 4. Microarray-based identification of ITGA2 as a miR-30a target gene.
(A) Heat map for downregulated genes in miR-30a mimic-treated cells (miR-30a) compared to NC mimic-treated cells (NC) in SNU-601, HSC44Luc and 44As3Luc cells. The samples were prepared in duplicate. (B) Venn diagram showing the overlapping downregulated genes between three miR-30a mimic-treated cells compared to NC mimic-treated cells (left). Using two miRNA target gene prediction programmes, we identified whether these genes have miR-30a binding sites, and the overlapping genes that have miR-30a binding sites are indicated in a Venn diagram (right). (C) Validation of expression levels of ITGA2, SEC23A and FBXO45 in either NC mimic- or miR-30a mimic-treated cell lines. (D) Analysis of ITGA2, FBXO45 and SEC23A expression in 30 pairs of GC tissues (GC) and adjacent non-cancer mucosa (NM) using qRT-PCR. (E) Correlation between miR-30a and ITGA2 expression in SNUH tissues. The red line represents the linear regression line. (F) Reporter constructs containing the wild-type (WT) or the mutant (Mut) of ITGA2 3’ UTR. (G) A luciferase reporter assay showed direct binding of miR-30a to the WT ITGA2 3’ UTR but not the Mut sequences. Fluc, firefly luciferase; Rluc, Renilla luciferase (right). *p < 0.05, ** p <0.01, *** p < 0.001, n.s., not significant. t-test.
To determine the relationship of these downstream target genes with miR-30a, we evaluated their expression in 30 pairs of SNUH cohort and TCGA data of the same cohorts. Expression of only ITGA2 was increased in GC compared to non-cancer samples in both datasets (Figure 4D and Supplementary Figure S4A). In addition, ITGA2 expression was inversely correlated with miR-30a expression in both datasets (Figure 4E and Supplementary Figure S4B). To confirm whether miR-30a directly interacts with the ITGA2 3’-UTR, we generated reporter gene constructs containing wild-type (WT) or mutant (Mut) ITGA2 3’-UTR sequences fused with the firefly luciferase coding sequence (Figure 4F). When the miR-30a mimic was co-transfected in HEK293 cells with the ITGA2 WT 3’-UTR or Mut 3’-UTR reporter vector, luciferase activity was significantly lower in the WT ITGA2 3’-UTR group than in the negative control group. Conversely, miR-30a did not interact with the mutant-type ITGA2 3’ UTR (Figure 4G). We additionally observed upregulation of ITGA2 transcript in miR-30a inhibitor-treated GC cells (Supplementary Figure S4C). According to these results, ITGA2 is a novel direct target of miR-30a in GC.
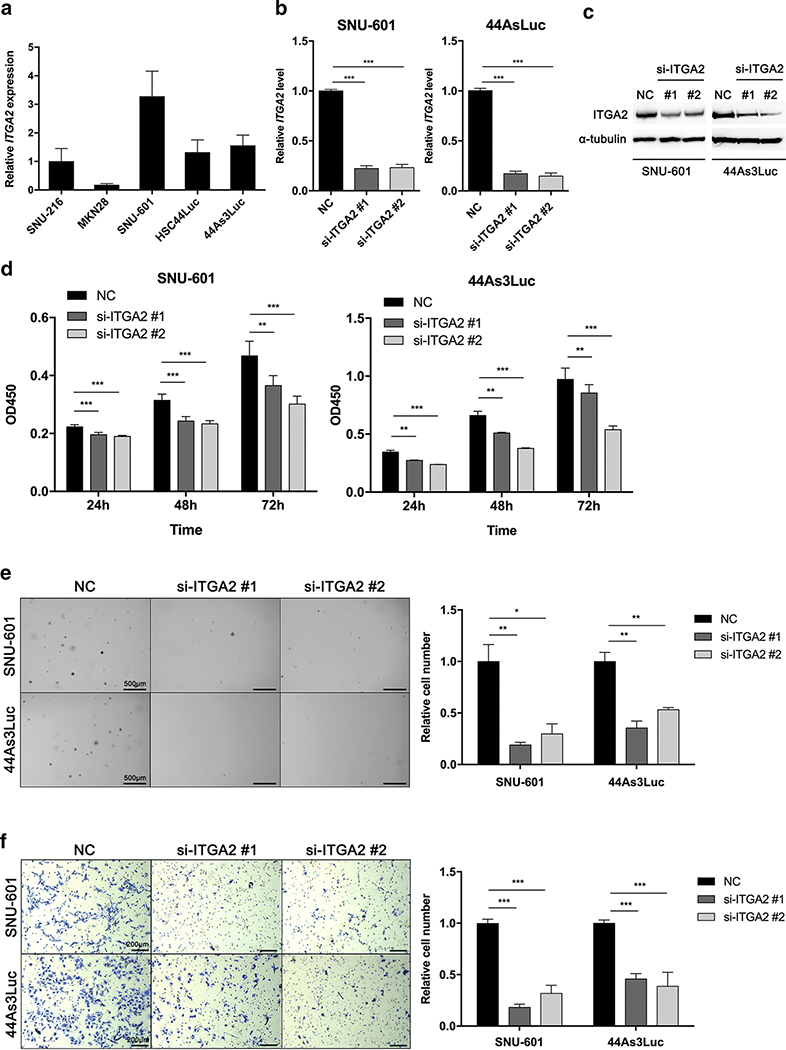
Suppression of ITGA2 inhibits GC cell proliferation, colony formation and migration abilities
We next treated GC cell lines with siRNAs against ITGA2 to verify the observed effects on cell proliferation, colony formation and motility by ITGA2 knockdown. We first assessed ITGA2 expression in five GC cell lines (Figure 5A). We then transfected two types of siRNAs with different binding sequences targeting the ITGA2 transcript into SNU-601 and 44As3Luc cells, which highly express ITGA2. Both mRNA and protein levels were decreased by the ITGA2 siRNAs compared with the control (Figure 5B and C), and cell proliferation was significantly reduced in these two cell lines at 24 h, 48 h and 72 h following siRNA treatments (Figure 5D). To confirm whether the decreased proliferation by si-ITGA2 can be rescued by ITGA2 overexpression, we established an ITGA2 gene-inserted plasmid DNA (IRES-EGFP-ITGA2). We transfected the IRES-EGFP-ITGA2 at 48 h following siRNA treatment at 24 h in 44As3Luc cells. We validated that ITGA2 gene expression levels were significantly changed after either ITGA2 knockdown by si-ITGA2 or overexpression by the IRES-EGFP-ITGA2 (Supplementary Figure S5A). We next performed a proliferation assay using the transfected cells. While cell proliferation was decreased after the inhibition of ITGA2 gene expression, it was significantly increased after the ITGA2 gene overexpression (Supplementary Figure S5B). Additionally, based on soft-agar colony forming assays, anchorage-independent cell growth was significantly suppressed in siRNA-treated cells compared to control cells (Figure 5E). Furthermore, we measured the cells migrating through a Transwell chamber after ITGA2 siRNA treatment and found that ITGA2 knockdown led to a significant reduction in migrating cells compared to control cells (Figure 5F). These data reveal that ITGA2 knockdown inhibits the proliferative and migratory characteristics of GC cells and it is consistent with miR-30a re-expression results.
Figure 5. siRNA-mediated ITGA2 knockdown in GC cell lines.
(A) ITGA2 transcript levels in five GC cell lines. (B-C) ITGA2 mRNA (B) and protein (C) knockdown by two siRNAs in SNU-601 and 44As3Luc cells (si-ITGA2 #1 and #2) compared to negative control siRNA (NC). (D) Cell proliferation assay (WST-assay) using SNU-601 and 44As3Luc cells treated with ITGA2 siRNAs. The absorbance at 450 nm was measured at 24 h, 48 h and 72 h after siRNA treatment. (E) Soft agar colony formation assay using SNU-601 and 44As3Luc cells after siRNA treatments. The anchorage-independent cell growth was measured by counting relative colony numbers. (F) Transwell migration assay using SNU-601 and 44As3Luc cells after siRNA treatments. Migrated cells were counted and compared to those in the NC group after siRNA treatment. *p < 0.05, ** p < 0.01, *** p < 0.001, unpaired t-test.
ITGA2 is upregulated in two preneoplastic metaplasias and is associated with intestinal-type GC
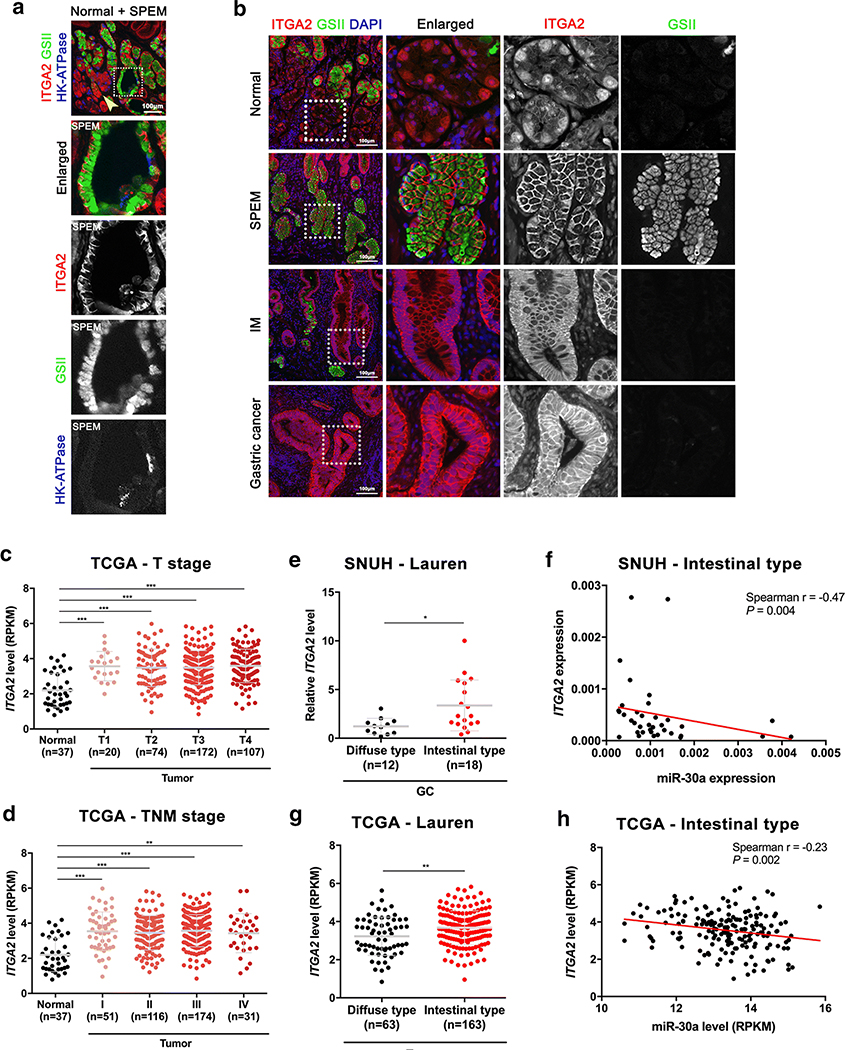
Since miR-30a downregulation was observed during SPEM development both in mouse and human, we next examined whether ITGA2 protein expression is also changed during SPEM development. We immunostained for ITGA2, GSII, a mucus neck cell and SPEM cell marker, and HK-ATPase, a parietal cell marker, in human normal donor stomach sections containing both normal and SPEM glands side-by-side. We observed strong membranous expression of ITGA2 in SPEM glands, which express GSII at the base of the glands (Figure 6A). However, the ITGA2 was localized in cytoplasm or barely expressed in normal glands, which contain GSII-positive mucus neck cells in the neck region and many HK-ATPase-positive parietal cells (Figure 6A, yellow arrow).
Figure 6. ITGA2 expression in human GC.
(A) Immunofluorescence staining for ITGA2 (red), GSII (green) and HK-ATPase (blue) in human normal donor stomach tissue includes normal and SPEM glands side-by-side. Scale bar = 100 μm. Yellow arrow indicates normal glands. Dotted boxes indicate an enlarged SPEM gland. (B) Immunofluorescence staining for ITGA2 (red), GSII (green) and DAPI (blue) in human GC patient tissues, including normal, SPEM, IM and GC regions. Scale bar = 100 μm. Dotted boxes indicate enlarged area. (C-D) ITGA2 expression levels in normal versus tumor samples according to T stage (C) and TNM stage (D) in TCGA data. (E-F) ITGA2 expression levels according to Lauren classification (E) and correlation with miR-30a expression levels in the intestinal type of GC (F) in SNUH cohort. (G-H) ITGA2 expression levels according to Lauren classification (G) and correlation with miR-30a expression levels in the intestinal type of GC (H) in TCGA cohort. The red line represents the linear regression line. *p < 0.05, ** p < 0.01, *** p < 0.001, unpaired t-test.
To determine whether expression of ITGA2 correlates with the down-regulation of miR-30a in gastric carcinogenesis, we examined the expression patterns of ITGA2 by immunofluorescence staining in normal glands, SPEM, IM and cancer lesions of the same GC patient. In the normal stomach, we detected low expression of ITGA2 at the base of normal glands and relatively strong expression of ITGA2 in GSII-positive SPEM glands, GSII-negative IM and GC lesions (Figure 6B). Furthermore, analysis of ITGA2 expression according to cancer stage in a large set of TCGA data indicated that its expression is already up-regulated in T1 and stage I GC tissues compared to non-cancer tissues and that elevated expression of ITGA2 is conserved in both early and advanced cancers (Figure 6C and 6D). Next, to determine whether ITGA2 gene expression is associated with the intestinal type of GC, we used the SNUH cohort to compare ITGA2 expression levels in each diffuse and intestinal type of GC tissue. Interestingly, ITGA2 was found to be highly expressed in the intestinal type compared to the diffuse type of GC (Figure 6E). Moreover, ITGA2 expression correlated inversely with miR-30a expression in the intestinal type of GC (Figure 6F). Similarly, TCGA data analysis showed up-regulation of ITGA2 in the intestinal type of GC compared to the diffuse type (Figure 6G), and there was a significant negative correlation between miR-30a and ITGA2 in the intestinal type of GC (Figure 6H). Collectively, these data provide compelling evidence that suppression of miR-30a mediates the development of the intestinal type of GC by up-regulating the ITGA2 expression with oncogenic functions.
DISCUSSION
In the current study, we showed that miR-30a is down-regulated in metaplasias and GC. Our study (i) to demonstrate the suppression of miR-30a in SPEM-induced animal models, human SPEM and GC, (ii) to use a series of cell culture and animal models to identify the tumor-suppressive roles of miR-30a in the stomach, (iii) to identify ITGA2 as a target gene of miR-30a, (iv) to examine inverse correlations between miR-30a and ITGA2 levels in the intestinal type of GC, and (v) to provide evidence for a potential early GC marker and therapeutic for the intestinal type of GC.
Indeed, this is the first study to employ an in situ hybridization method to provide localisation of miR-30a expression in SPEM and gastric neoplasia. Enrichment of miR-30a was found in normal corpus glands, but not in intestinal-type cells such as the pyloric gland and duodenum (Figure 1A and Supplementary Figure S1). Furthermore, we observed that intestinalization of the stomach in SPEM-induced mouse models and human SPEM samples was associated with the loss of miR-30a in corpus glands (Figure 1B and 1C). Therefore, transdifferentiation of chief cells or intestinalization of gastric glands leads to suppressed miR-30a expression, suggesting that loss of miR-30a is an early event in inducing precancerous lesions of the stomach.
MiR-30a regulatory mechanisms during GC development have been reported. For example, Liu et al. found a binding site for the upstream region of miR-30a and investigated a consensus binding sequence of Runt-related transcription factor 3 (RUNX3) ‘TGTGGT’ (RBS) between the nucleotides −3854 to −3861 using a ChIP assay [22]. The research group also observed that overexpression of RUNX3 increases miR-30a levels in GC cells; therefore, RUNX3, as a transcription factor, is a positive regulator of miR-30a. However, it has been reported that RUNX3 expression is frequently down-regulated in precancerous gastric lesions and GC by DNA hypermethylation, H. pylori and phosphorylation by Src kinase [29]. This evidence suggests that inactivation of RUNX3 may affect miR-30a expression in the stomach during the precancerous state. Interestingly, Ito et al. observed that Runx3 is expressed most highly in the pepsinogen-positive chief cells at the bottom of the corpus gland in mice, whereas it was rarely expressed in TFF2-expressing neck cells or pyloric glands [30]. Nonetheless, Runx3−/− mice display induced hyperplasia in the gastric mucosa. Our findings also show similar expression patterns; miR-30a was strongly expressed in chief cells but not in pyloric glands and precancerous lesions of the stomach. Furthermore, we found a positive correlation between RUNX3 and miR-30a levels in the intestinal type of STAD TCGA data set (Supplementary Figure S6A). Based on these previous reports and our results, we suggest that inactivation of RUNX3 may be associated with the stomach intestinalization through the regulation of the downstream gene, miR-30a and its targets.
MiR-30 family members, such as miR-30a, miR-30b, miR-30c, miR-30d and miR-30e, have been reported to function as negative regulators of tumor growth and metastasis in various types of cancer, such as breast cancer, colorectal cancer, ovarian cancer, prostate cancer and GC [31–37]. However, among these family members, only miR-30a was predicted in our study to be an overlapping putative tumor suppressor for GC in three different miRNA microarray profi les (Figure 2A); additionally, the expression patterns of miR-30 family members in GC were validated by utilizing RNA sequencing data from the STAD TCGA data set. Consistently, miR-30a was significantly down-regulated in tumors compared to normal tissues (Supplementary Figure S7 and Supplementary Table S3).
In our previous miRNA sequencing analysis, miR-30a was one of the downregulated miRNAs in immortalized mouse SPEM cells (ImSPEM) compared to immortalized mouse chief cells (ImChief) [38][39], and gene regulation by miR-30a during metaplasia development and progression was also reported in human [13]. MiR-30a acts as a tumor suppressor by regulating cancer cell proliferation, invasion, metastasis and drug resistance through multiple downstream target genes in many types of cancer [40] [41]. Accordingly, regulatory networks linking key molecules such as p21 and Snai1 are involved in miR-30a-mediated cancer pathogenesis and progression [42] [43]. Although tumor suppressive roles of miR-30a have been investigated in many cancer cells, it still remains obscure whether its downregulation is involved in gastric carcinogenesis. We identified novel putative target genes for miR-30a, including ITGA2, FBXO45 and SEC23A, which might be associated with gastric carcinogenesis. Among these target genes, ITGA2 was selected because it was the only gene highly expressed in GC samples of two independent dataset. Furthermore, ITGA2 expression levels correlated inversely with miR-30a in both TCGA and our cohorts (Figure 4E and Supplementary Figure S4B). Importantly, based on TCGA data, RUNX3, a positive regulator of miR-30a, also correlated negatively with ITGA2 expression in intestinal-type GC samples (Supplementary Figure S6B). Thus, we focused on ITGA2 as a promising target of miR-30a in intestinal-type GC. In particular, many cancer cells of epithelial origin express high levels of integrin α2 [44], and increased levels of integrin α2 can promote development of cancers, including GC [45]. Although we clearly observed decreased GC cell proliferation and migration after ITGA2 knockdown (Figure 5), we did not observe proliferation changes after transient overexpression of the ITGA2 gene (Supplementary Figure S5B). The proliferation of GC cells, which already highly express ITGA2, might not be affected by the ITGA2 overexpression. ITGA2 is a member of the integrin family, which consists of transmembrane receptors for cell adhesion that form heterodimers with integrin beta subunits. Many studies support the functional roles of α2β1 integrin in the regulation of various processes of cancer development and progression [45]. It is well known that α2β1 integrin contributes to cancer cell invasion, metastasis and angiogenesis, and there is emerging evidence that it also promotes cancer initiation and progression. According to these studies, α2β1 integrin contributes to the tumorigenesis of human papilloma virus-induced squamous carcinoma by promoting dysplasia [46] and is often associated with prostate and colorectal cancer stem cells [47] [48]. In addition, type IV collagen binds to α2β1 integrin and then induces colorectal cancer cell proliferation by regulating the cell cycle [49].
According to the TCGA dataset, ITGA2 mRNA expression was strongly associated with the intestinal type of GC, and we thus investigated the levels of ITGA2 protein expression by tissue microarray (TMA) using 450 tissues of gastric adenocarcinomas and analysed associations between ITGA2 level and Lauren classification. In our immunohistochemistry analyses, we identified both membranous and cytoplasmic expression of ITGA2. Expression in each sample was individually scored using a range of negative (score 0) to strongly positive (score 3), as shown in Supplementary Figure S8A. We categorised each sample according to the Lauren classification and statistically compared the scores. Interestingly, stronger staining intensities of cytoplasmic ITGA2 were observed in the intestinal type than in the diffuse type of GC, whereas membranous ITGA2 expression was not significantly different between the two types of GC (Supplementary Figure S8B). These results indicate that the cytoplasmic ITGA2 expression is associated with intestinal type of GC. Previously, integrin endocytosis has been suggested to constitute an important event for regulating cancer cell properties such as migration and invasion [50, 51]. The endocytosis process is tightly controlled in normal cells to maintain integrin activity, but it is abnormally regulated in cancer cells [52]. This evidence suggests that detection of ITGA2 in the cytosol is associated with oncogenic properties and that its accumulation may be associated with intestinal-type GC development.
In conclusion, we found that miR-30a is down-regulated in epithelial cells in SPEM, IM and GC, indicating a key role for this miRNA in maintaining homeostasis of gastric glands. Furthermore, miR-30a plays tumor-suppressive functions in the stomach by targeting ITGA2, contributing to the inhibition of preneoplastic lesions and intestinal-type GC development. Therefore, miR-30a may represent an effective therapeutic target for preventing early gastric carcinogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kazuyoshi Yanagihara for providing HSC44Luc and 44As3Luc cell lines.
FUNDING
This work was supported by grants from the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI14C3426 and HI13C2148), and National Research Foundation of Korea, which is funded by the Ministry of Science and ICT (NRF-2017R1C1B2012268) and the KRIBB Research Initiative Program. This study was supported by grants from DOD W81XWH-17-1-0257 (to E.C), from a Department of Veterans Affairs Merit Review Award IBX000930 and NIH RO1 DK071590 and RO1 DK101332 (to J.R.G). T.S. was the recipient of JSPS Postdoctoral Fellowships for Research Abroad. This work was supported by core resources of the Vanderbilt Digestive Disease Center (NIH P30 DK058404) and imaging in the Vanderbilt Digital Histology Shared supported by a VA Shared Instrumentation grant (1IS1BX003097).
Footnotes
CONFLICTS OF INTEREST: The authors declare that they have no conflict of interest.
ETHICAL APPROVAL: All institutional and national guidelines for the care and use of laboratory animals were followed.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Bray F, et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2018. 68(6): p. 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, et al. , Gastric cancer. Lancet, 2016. 388(10060): p. 2654–2664. [DOI] [PubMed] [Google Scholar]
- 3.Rugge M, Fassan M, and Graham DY, Epidemiology of Gastric Cancer, in Gastric Cancer. 2015. p. 23–34.25209115 [Google Scholar]
- 4.Yuasa Y, Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer, 2003. 3(8): p. 592–600. [DOI] [PubMed] [Google Scholar]
- 5.Correa P, Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res, 1992. 52(24): p. 6735–40. [PubMed] [Google Scholar]
- 6.Schmidt PH, et al. , Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest, 1999. 79(6): p. 639–46. [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R, Marcucci G, and Croce CM, Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov, 2010. 9(10): p. 775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes J, Peruzzi PP, and Lawler S, MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med, 2014. 20(8): p. 460–9. [DOI] [PubMed] [Google Scholar]
- 9.Lee YS and Dutta A, MicroRNAs in cancer. Annu Rev Pathol, 2009. 4: p. 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda T, et al. , Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol, 2010. 11(2): p. 136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han TS, et al. , Interleukin 1 Upregulates MicroRNA 135b to Promote Inflammation-associated Gastric Carcinogenesis in Mice. Gastroenterology, 2018. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, et al. , Integrated Analysis of Mouse and Human Gastric Neoplasms Identifies Conserved microRNA Networks in Gastric Carcinogenesis. Gastroenterology, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa JF, et al. , miR-30-HNF4gamma and miR-194-NR2F2 regulatory networks contribute to the upregulation of metaplasia markers in the stomach. Gut, 2016. 65(6): p. 914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, et al. , miR-30a acts as a tumor suppressor by double-targeting COX-2 and BCL9 in H. pylori gastric cancer models. Sci Rep, 2017. 7(1): p. 7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YK, et al. , Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res, 2009. 37(5): p. 1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto Y, et al. , MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14–3-3zeta. Cancer Res, 2010. 70(6): p. 2339–49. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S, et al. , A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A, 2006. 103(7): p. 2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. , Survival prediction of gastric cancer by a seven-microRNA signature. Gut, 2010. 59(5): p. 579–85. [DOI] [PubMed] [Google Scholar]
- 19.Du X, et al. , miR-30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp Ther Med, 2018. 15(1): p. 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, et al. , MiR-30a Decreases Multidrug Resistance (MDR) of Gastric Cancer Cells. Med Sci Monit, 2016. 0: p. 0. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, et al. , MicroRNA-30a-5p inhibits the proliferation and invasion of gastric cancer cells by targeting insulin-like growth factor 1 receptor. Exp Ther Med, 2017. 14(1): p. 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu Z, et al. , RUNX3 regulates vimentin expression via miR-30a during epithelial-mesenchymal transition in gastric cancer cells. J Cell Mol Med, 2014. 18(4): p. 610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LL, et al. , MiR-30a increases cisplatin sensitivity of gastric cancer cells through suppressing epithelial-to-mesenchymal transition (EMT). Eur Rev Med Pharmacol Sci, 2016. 20(9): p. 1733–9. [PubMed] [Google Scholar]
- 24.Nam KT, et al. , Amphiregulin-deficient mice develop spasmolytic polypeptide expressing metaplasia and intestinal metaplasia. Gastroenterology, 2009. 136(4): p. 1288–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi E, et al. , Expression of Activated Ras in Gastric Chief Cells of Mice Leads to the Full Spectrum of Metaplastic Lineage Transitions. Gastroenterology, 2016. 150(4): p. 918–30 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldenring JR, et al. , Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology, 2010. 138(7): p. 2207–10, 2210 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura S, et al. , Alterations in gastric mucosal lineages induced by acute oxyntic atrophy in wild-type and gastrin-deficient mice. Am J Physiol Gastrointest Liver Physiol, 2005. 288(2): p. G362–75. [DOI] [PubMed] [Google Scholar]
- 28.Petersen CP, et al. , Macrophages promote progression of spasmolytic polypeptide-expressing metaplasia after acute loss of parietal cells. Gastroenterology, 2014. 146(7): p. 1727–38 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li WQ, et al. , RUNX3 methylation and expression associated with advanced precancerous gastric lesions in a Chinese population. Carcinogenesis, 2011. 32(3): p. 406–10. [DOI] [PubMed] [Google Scholar]
- 30.Ito K, et al. , Loss of Runx3 is a key event in inducing precancerous state of the stomach. Gastroenterology, 2011. 140(5): p. 1536–46 e8. [DOI] [PubMed] [Google Scholar]
- 31.Cao JM, et al. , MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed Pharmacother, 2017. 93: p. 554–560. [DOI] [PubMed] [Google Scholar]
- 32.Jia W, et al. , MicroRNA-30c-2* expressed in ovarian cancer cells suppresses growth factor-induced cellular proliferation and downregulates the oncogene BCL9. Mol Cancer Res, 2011. 9(12): p. 1732–45. [DOI] [PubMed] [Google Scholar]
- 33.Liao WT, et al. , MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol, 2014. 232(4): p. 415–27. [DOI] [PubMed] [Google Scholar]
- 34.Lin ZY, et al. , MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Cancer, 2017. 16(1): p. 48. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Liu K, et al. , SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol Cancer, 2017. 16(1): p. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian SB, et al. , MiR-30b suppresses tumor migration and invasion by targeting EIF5A2 in gastric cancer. World J Gastroenterol, 2015. 21(31): p. 9337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, et al. , MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene, 2014. 33(24): p. 3119–28. [DOI] [PubMed] [Google Scholar]
- 38.Weis VG, et al. , Establishment of novel in vitro mouse chief cell and SPEM cultures identifies MAL2 as a marker of metaplasia in the stomach. Am J Physiol Gastrointest Liver Physiol, 2014. 307(8): p. G777–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu T, et al. , Decrease in MiR-148a Expression During Initiation of Chief Cell Transdifferentiation. Cell Mol Gastroenterol Hepatol, 2020. 9(1): p. 61–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang LH, Zhang HD, and Tang JH, MiR-30a: A Novel Biomarker and Potential Therapeutic Target for Cancer. J Oncol, 2018. 2018: p. 5167829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Chen Y, and Chen L, The Versatile Role of microRNA-30a in Human Cancer. Cell Physiol Biochem, 2017. 41(4): p. 1616–1632. [DOI] [PubMed] [Google Scholar]
- 42.Kumarswamy R, et al. , MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer, 2012. 130(9): p. 2044–53. [DOI] [PubMed] [Google Scholar]
- 43.Liu M, et al. , Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res, 2015. 75(21): p. 4593–604. [DOI] [PubMed] [Google Scholar]
- 44.Madamanchi A, Santoro SA, and Zutter MM, alpha2beta1 Integrin. Adv Exp Med Biol, 2014. 819: p. 41–60. [DOI] [PubMed] [Google Scholar]
- 45.Naci D, Vuori K, and Aoudjit F, Alpha2beta1 integrin in cancer development and chemoresistance. Semin Cancer Biol, 2015. 35: p. 145–53. [DOI] [PubMed] [Google Scholar]
- 46.Tran T, et al. , Loss of the alpha2beta1 integrin alters human papilloma virus-induced squamous carcinoma progression in vivo and in vitro. PLoS One, 2011. 6(10): p. e26858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrawala L, et al. , Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res, 2007. 67(14): p. 6796–805. [DOI] [PubMed] [Google Scholar]
- 48.Kirkland SC, Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br J Cancer, 2009. 101(2): p. 320–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honore S, et al. , Alpha2beta1-integrin signaling by itself controls G1/S transition in a human adenocarcinoma cell line (Caco-2): implication of NADPH oxidase-dependent production of ROS. Exp Cell Res, 2003. 285(1): p. 59–71. [DOI] [PubMed] [Google Scholar]
- 50.Paul NR, Jacquemet G, and Caswell PT, Endocytic Trafficking of Integrins in Cell Migration. Curr Biol, 2015. 25(22): p. R1092–105. [DOI] [PubMed] [Google Scholar]
- 51.Shin S, Wolgamott L, and Yoon SO, Integrin trafficking and tumor progression. Int J Cell Biol, 2012. 2012: p. 516789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamidi H and Ivaska J, Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer, 2018. 18(9): p. 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.