Abstract
Advancements in the field of next generation sequencing lead to the generation of ever-more data, with the challenge often being how to combine and reconcile results from different OMICs studies such as genome, epigenome and transcriptome. Here we provide an overview of the standard processing pipelines for ChIP-seq and RNA-seq as well as common downstream analyses. We describe popular multi-omics data integration approaches used to identify target genes and co-factors, and we discuss how machine learning techniques may predict transcriptional regulators and gene expression.
Keywords: ChIP-seq, RNA-seq, NGS, Data integration, Multi-omics, Transcriptional regulation
1. Introduction
Within an organism, all cells contain the same genome, but have vastly different roles. These tissue and cell type specific functions are largely conferred by transcriptional regulators that control gene expression and thereby define cell identity. Transcriptional regulators include trans-acting factors, such as transcription factors (TFs), cis-regulatory elements (promoters and enhancers), as well as the chromatin structure (DNA-accessibility, nucleosome structures and chromatin looping) and epigenetic marks (histone modifications and DNA methylation).
Specific elements in this gene regulatory machinery can be studied by different genome-wide analyses (Fig. 1). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) [1] has become the method of choice to explore protein-DNA interactions such as TF binding and histone modifications (HMs). Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) [2] measures genome-wide chromatin accessibility, and RNA-sequencing (RNA-seq) [3], [4] identifies the transcriptome. Additionally, Hi-C [5], Capture-C [6] and other methods analyze the 3-dimensional chromosome structure by capturing chromatin interactions.
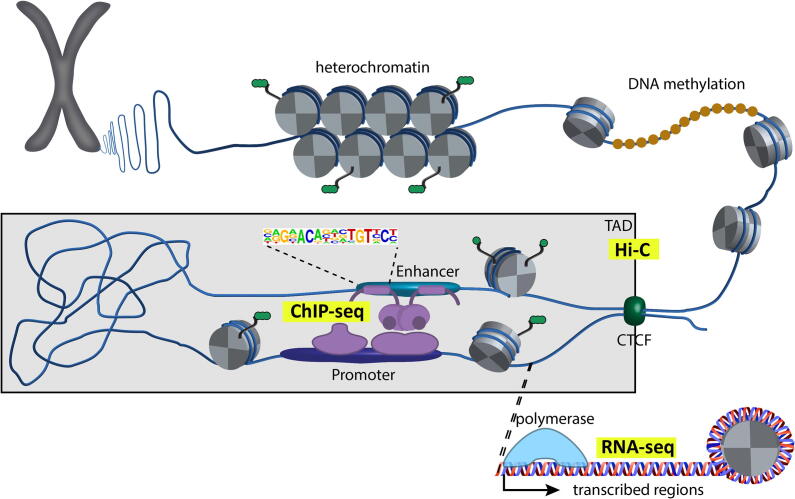
Fig. 1.
Schematic representation of the transcriptional machinery. Cis-regulatory elements (enhancers or promoters), trans-regulatory elements (transcription factors) as well as epigenetic modifications and 3D chromatin structure are known to influence gene expression. TAD: Topologically associated domain.
Epigenetic modifications [7], [8] are a fundamental network controlling transcriptional outcomes. Since 2003, the Encyclopedia of DNA Elements (ENCODE) consortium [9] has systematically built a compendium of functional elements in the human genome. ENCODE also performs data curation and offers standardized processing pipelines [1] for various assay types online (https://www.encodeproject.org/), with regular updates. ENCODE includes thousands of datasets on gene expression (RNA-seq, Cap Analysis of Gene Expression (CAGE) and RNA-pet), ChIP-seq (TF binding, HMs) and chromatin accessibility (ATAC-seq, DNase-seq) from several cell types [10]. Within the cancer research field, the Cancer Genome Atlas [11] offers a vast collection of genetic, epigenetic, transcriptional and proteomics data on 33 different cancer types, which can be accessed through the Genomic Data Commons Data Portal [12]. The Roadmap Epigenomics project [13] and the BLUEPRINT project [14] are further large-scale undertakings that systematically collect data to characterize the human epigenome. Their datasets can be accessed through the IHEC [15] data portal. With ever more data being generated (current high-throughput systems can sequence up to 6000 gigabases per run), the bottleneck has shifted from data generation towards their analysis, posing new challenges for bioinformaticians.
In this review, we provide an overview of the standard processing pipelines for ChIP-seq and RNA-seq as well as common downstream analyses. Furthermore, we discuss popular approaches for data integration and point out shortcomings along the way. Specifically, we show how ChIP-seq and RNA-seq data can be used to identify the target genes of a TF as well as coregulators for transcription, and we review methods that leverage chromatin assays to predict gene expression. Finally, we discuss how new developments in the field of machine learning contribute to the understanding of gene regulation.
2. Experimental design
2.1. General experimental considerations
ChIP-seq experiments assess the interactions of a protein of interest (such as TFs or modified histones) with DNA on a genome-wide level [1]. Depending on the samples submitted for sequencing, this can answer different questions. The classic experiment is to determine the interactions within a certain cell-type at steady state. More often however, it is of interest how these interactions change in response to a perturbation. Changing the expression level of a gene through overexpression, knock-down or knock-out experiments, can lead to changes in DNA binding of molecularly connected factors. Comparing sequencing results of these samples with baseline data can reveal new insights on the relationship between these components. Similarly, introducing a treatment condition that changes the levels of the TF itself is used to determine the target genes by comparing DNA binding in the treatment condition with the steady state. Furthermore, mutating either the gene for the TF itself, or the DNA sequence it binds to, can validate putative targets with additional wet lab experiments.
The same steady state and/or perturbed samples assayed in ChIP-seq, can also be submitted for RNA-seq, with the readout being the effect on gene expression. The advantage of combining RNA-seq and ChIP-seq in the same experiment is to link a change in occupancy with a change in transcription, which allows inference of which peaks are functional binding sites. In this review, we will discuss a number of methods that combine multiple ChIP-seq datasets and/or RNA-seq data to answer this and additional questions.
2.2. ChIP-seq specific considerations
The quality of ChIP-seq results is dependent on the specificity and the sensitivity of the chosen antibody. These factors should be taken into consideration when comparing data generated with different antibodies or the same antibody in different samples [1]. “Hyper-chippable” regions [16], GC rich regions [17] and non-random fragmentation [18] can introduce various biases or background. Therefore, “input controls” or “IgG controls” are crucial to accurately identify ‘real’ peak signals.
To ensure reproducibility of the results, it is recommended to submit biological replicates of the samples for sequencing. In most cases, two replicates can be sufficient and little information is gained by further increasing the number of replicates [19]. Ranking peaks and comparing them between replicates can then be used to assess the agreement of the results and to determine their irreproducible discovery rate (IDR). Analogous to the concept of FDR, setting the IDR to be no bigger than a predefined significance level ɑ, can control for the rate of irreproducible peaks [20].
2.3. RNA-seq specific considerations
Compared to ChIP-seq, the number of replicates is very important for the detection of differentially expressed genes. As resources are limited, a thorough experimental design also includes decisions on sample sizes and on technical parameters, such as read depth [21], [22], [23]. Power analysis can be used to decide the study's optimal sample size and its impact, for the test to be performed. In the case of RNA-seq studies, given the common statistical assumption of the most reliable differential expression methods DESeq2 and edgeR [24], power analysis is based on the theory of negative binomial count regression [25], [26]. Deciding on sample size is also influenced by biological heterogeneity, and significantly, the required minimum fold change to be detectable between the conditions at the given significance level. Various approaches, including simulation based models [24], [27] are compared and benchmarked in [28].
3. Data processing
3.1. Systematic literature search
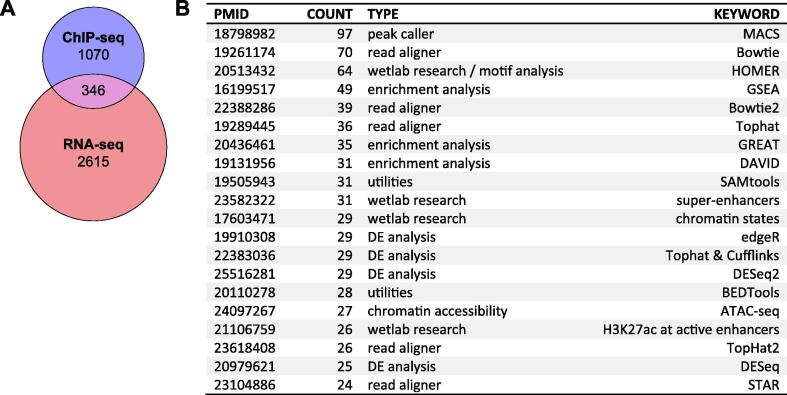
We investigated how most research groups approach data integration and whether there was a specific tool or strategy taking hold in the scientific community, by performing a systematic literature search. Gene Expression Omnibus (GEO) is an online database hosted by the National Center for Biotechnology Information (NCBI), archiving microarray and next generation sequencing (NGS) genomics data. We used the package GEOmetadb (1.44.0) within R version 3.5.2 to query all submitted entries matching the Dataset types “Expression profiling by high throughput sequencing” and “Genome binding/occupancy profiling by high throughput sequencing” performed in humans or mice. After filtering for those entries linked to Pubmed IDs, we checked what publications submitted both expression and genome binding data. Out of 4377 Pubmed IDs, 346 included datasets of both assay types (Fig. 2A). Quantifying what references those 346 studies shared revealed a number of frequently cited peak calling algorithms, read alignment tools and gene set enrichment approaches, to which we will refer in the corresponding section. Importantly, no tool designed to integrate RNA-seq and ChIP-seq data came up in our search. Hence, despite genome occupancy profiling and gene expression frequently being employed in the same project, no specialized tools for integrating their results have established themselves.
Fig. 2.
Systematic literature search on publications combining gene expression and DNA binding data. (A) Numbers of Pubmed IDs associated with RNA-seq and ChIP-Seq data submissions (retrieved on 01/22/2020). (B) Top 20 most commonly referenced citations from publications in the intersection of the Venn diagram shown in A. PMID: Pubmed ID.
3.2. ChIP-seq
Covalent modifications of histone tails are essential determinants of nucleosome positioning and gene regulation [29], [30]. Different types of HMs such as acetylation, phosphorylation, methylation or ubiquitination, can change the interaction strength of DNA with histones, which in turn influences transcription. Specific epigenetic marks are associated with gene activation, others with repression [31]. ChIP-seq offers a way to investigate HMs as well as interactions of TFs with their DNA binding sites.
To identify the genomic sequences a transcription factor is binding to, crosslinked chromatin is fragmented, and an antibody specific to the target protein is used to purify the DNA-protein complex by immunoprecipitation. After de-crosslinking, the DNA is purified and prepared for NGS. Similarly, antibodies directed against various histone residues can be employed. Recent advances have further improved the technique by significantly increasing resolution and reducing background noise, as in ChIPexo [32] or CUT&Tag [33], for example.
3.2.1. Preprocessing & read alignment
Upon successful completion of such an NGS experiment, the read quality needs to be assessed by checking the quality of the base-calls, the duplication rate, GC content and adapter content. The tool FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) evaluates these and additional criteria, and returns an overview of the sample metrics. Depending on the results, removal of adapter sequences [34] and removal of low quality bases by read trimming might be desirable before mapping them to the reference genome. Contrary to RNA-seq, aligners for ChIP-seq reads do not need to be splice-aware, since they do not contain exon boundaries. Commonly used tools include bwa [35], bowtie and its successor Bowtie2 [36] (Fig. 2B).
3.2.2. Peak calling
Most peak-calling algorithms have been developed for TF binding data and consequently were optimized for narrow peaks. Few HMs (such as H3K4me3) also fall into this category. The most commonly used peak caller (Fig. 2B) is the second version of Model-based Analysis of ChIP-seq data (MACS) [37]. MACS2 considers local biases by using a dynamic Poisson distribution when determining the fold enrichment during peak calling.
On the other hand, most histone modifications or DNA methylation patterns show broad enrichments without clear peaks, so-called domains. Methods such as histoneHMM [38] specifically identify enriched domains, and some tools that were developed for narrow peaks offer parameter adjustments to accommodate domain calling (i.e. MACS2).
MACS2 is a reliable choice for TF binding data [39], but Bayesian Change-Point (BCP) [40] and MUltiScale enrichment Calling for ChIP-seq (MUSIC) [41] slightly outperform it when calling broad peaks. For methods with higher signal-to-noise ratios such as CUT&RUN or CUT&Tag, standard peak callers may generate high false-positive rates, making specialized tools like SEACR [42] more appropriate. For an overview of statistical methods and their underlying models, see Supplementary Table 1.
Quality metrics after mapping and peak calling include the percentage of mappable reads, the library complexity, percentage of reads in peaks and strand cross-correlation ([1], [43]). As discussed in section 2.2, the robustness of the results can further be assessed using IDR [20].
3.2.3. Differential binding analysis
Experimental questions answered by ChIP-seq may include qualitative or quantitative comparisons of multiple samples, i.e. whether the same peaks are present in different conditions or whether the strength of the peak signal differs. Immunoprecipitation efficiencies can vary between samples, potentially influencing the fraction of reads in peaks. Together with the signal to noise ratio, these factors may affect differential binding analyses [44].
Tools to determine differential binding use alternative approaches to model the data, each with their own strengths and weaknesses [45]. Some Bioconductor/R packages such as edgeR [46] and DESeq2 [47] are routinely used in RNA-seq analysis pipelines (see section 2.3.4). Others, such as csaw [48] or DiffBind (https://bioconductor.org/packages/release/bioc/html/DiffBind.html), make use of those packages in workflows specifically developed for ChIP-seq. Another popular tool called MAnorm [49] is based on the assumption that the peaks shared between samples do not differ globally, and uses them as a basis to fit a robust regression, extrapolation to all peaks and normalization. In histoneHMM [38], differential binding is formulated as an unsupervised classification problem and analyzed using a bivariate Hidden Markov Model (HMM).
In case the binding landscape changes profoundly, those assumptions do not hold true. Alternative approaches use experimental spike-ins, i.e. chromatin from a different organism, during the ChIP. The reads derived from this reference can then be used for normalization. In CUT&Tag, the small amounts of E. coli DNA remaining after transposase production, suffice as spike-in substitutes.
3.2.4. Peak annotation
For the scientist interpreting ChIP-seq results within their biological context, the positional information of putative cis-regulatory regions needs to be linked to genetic functions. An intuitive approach is to visually inspect the processed ChIP-seq data on a genome browser, such as Integrative Genomics Viewer [50] or the University of California, Santa Cruz Genome Browser [51]. The data can then be parsed in conjunction with publicly available datasets such as DNase hypersensitivity, HMs, single nucleotide polymorphisms, tissue specific gene expression etc. However, this strategy does not benefit from the myriad of tools designed to identify global patterns.
While identifying the target genes of a TF is one prime objective of ChIP-seq experiments, the fact that most peaks are not promoter proximal impedes this task. Linear proximity to the closest transcription start site is often used to identify putative target genes for a given TF peak. For example, GREAT [52] allows the user to pick from a number of association rules that assign genomic regions to their target genes. Bioconductor/R packages such as ChIPpeakAnno [53] and ChIPseeker [54] annotate large quantities of peaks simultaneously and visualize the peak distribution within certain genomic features.
One obvious shortcoming of this approach is that the three dimensional character of chromatin is discounted. For instance, distal cis-regulatory elements can physically interact with promoter regions by DNA loop formation, bringing distant regions into close spatial contacts [55]. Recent studies on the principles of phase separation have revealed a surprising complexity of 3D chromatin dynamics, which are currently challenging to study [56]. New NGS methods such as Hi-C [5] assess genome-wide chromatin interactions and should be considered when assigning peaks to their potential targets. However, Hi-C currently lacks the resolution to go beyond topology associating domains. Promoter-capture Hi-C [57] overcomes this shortcoming, but it only detects the proximity of genomic regions, which may not reflect functional interactions, as is the technical limitation of all ligation-based assays.
3.3. RNA-seq
With the advent of RNA-seq, or whole transcriptome shotgun sequencing, it became possible to screen the entire transcriptome of any organism or even single cells by NGS. Transcriptome analysis consists of the quantification of all kinds of transcripts (mRNA, microRNA, noncoding RNAs etc.), differential expression analysis, de novo transcript assembly as well as determining the transcriptional structures of genes [58], [59].
RNA-seq identifies and quantifies RNA species at a given time point (as RNA abundance is not stable over time) in biological samples. Experimentally, the RNA is extracted, randomly fragmented and reverse transcribed into cDNA with adaptors attached to one or both ends. After PCR amplification and sequencing, the raw data consists of a list of reads with associated quality scores for each sample, which are then subjected to RNA-seq data analysis.
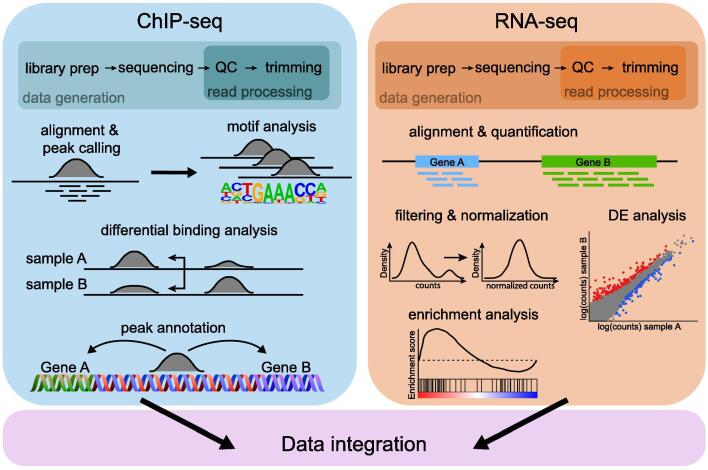
Here we focus only on the application of RNA-seq for differential gene expression analysis and we briefly summarize the most common necessary steps (Fig. 3).
Fig. 3.
Standard processing workflow of ChIP-seq and RNA-seq. In both cases, the quality of the sequenced reads is checked before performing the alignment. The ChIP-seq data analysis continues with peak calling, followed by differential binding analysis. Searching for motifs in the peak regions and peak annotation are crucial steps. For RNA-seq, the aligned reads are quantified at gene level, the raw counts are then filtered and normalized to enable further comparisons. The differential expression analysis provides a list of significant genes, from which biological meaning may be retrieved. QC: Quality control, DE: differential expression.
3.3.1. Preprocessing
The steps for preprocessing raw data are comparable to those of ChIP-seq experiments (see section 3.2.1). The downstream analysis essentially consists of mapping, quantification, filtering and normalization, detection of differentially expressed genes and finally the biological interpretation of the results.
3.3.2. Read mapping
The process of assigning reads to their best matching location in the reference is referred to as mapping. Fragments can either be mapped to a reference transcriptome or genome. In the former case, all isoforms of a gene are considered separately, whereas in the latter, reads are aligned to the underlying genes, regardless of what isoform the read stems from [60].
The most popular, splice-aware alignment tools, which rely on a reference genome are STAR [61], TopHat [62], TopHat2 [63], and Bowtie2 [36] (Fig. 2B). In the case of mapping to a transcriptome, popular efficient alignment-free tools quantify the transcripts directly, for example Kallisto [64] and Salmon [65]. Their quantification is based on k-mers, i.e. they fragment the reads into all possible k-mers and then map only the unique ones to the pre-indexed transcriptome.
Multi-mappers (i.e. reads mapping to multiple locations), represent a significant fraction of mapped reads and are bioinformatically challenging. The simplest approach is to discard ambiguously mapped reads and keep only uniquely mapped ones. Another modality is to keep all matches, which leads to an amount of mapped reads beyond the number of raw reads. It is also possible to use a scoring function to find the best possible alignment, and in case of equal scores distribute the reads randomly between loci. There is also the option to allocate ambiguous reads in relative proportion according to probabilistic inference, for example in RSEM [66] and TopHat [62]. The latter strategy might be the most applicable, as it appears to produce the least bias in inferring differential gene expression [67].
3.3.3. Gene or transcript level quantification
The counting and clustering of reads can be performed over different genomic features, such as transcripts or genes. The most common is to estimate the gene level abundances, by counting the number of reads/fragments overlapping the exons of the gene. However, even for the best annotated human or mouse data, a significant amount of the reads will map outside annotated exons [68].
Widely used quantification tools are CuffLinks [69], featureCounts [70], kallisto [64] and Salmon [65]. While featureCounts is an exon-based approach, kallisto and Salmon are transcript based approaches, which rely on an Expectation Maximization for estimating transcript abundances. In either case, the final output is a matrix of read/fragment counts, where each row corresponds to a feature of interest, while the columns represent the different samples.
3.3.4. Filtering and normalization
Importantly, the choice of normalization method has a bigger impact on the results than the mapping method or the test statistics used for finding differentially expressed genes [71], [72].
There are two types of normalization to account for biological or technical bias: within and between sample normalization. In the first case, comparisons between the features of a single sample are enabled by correcting for gene length and sequence composition, for example GC-content [73]. In the second case, for across sample feature comparisons, normalization is performed to adjust for the library size [74], [75]. To set a cutoff, zero or low count genes are omitted from the count table.
Of note, when correcting for sequencing depth, the assumption is that the total expression is similar under different conditions, so each condition is assumed to have the same amount of mRNA per cell [76]. In this case using the total count normalization, each read count will be divided by the sum of the reads of the sample [77]. The RPKM method (reads per kilobase per million mapped reads) is based on total count normalization, but accounts also for the length of the gene [78].
Other very popular methods rely on capturing information from non-changing genes. For example, the Trimmed Mean of the M-values approach implemented in the edgeR package assumes that the majority of genes are not differentially expressed and excludes those that are differential from the normalization factor [79]. It selects a reference sample for computing logarithm count ratios after trimming differential genes, and uses their mean for normalizing read counts. The DESeq normalization [47] is similar, but it computes the count ratio of a reference sample relative to the geometric mean of all other samples for each gene, then uses the median of these for scaling the reference counts.
3.3.5. Differential gene expression
After normalization, Principal Component Analysis can be used for visual data inspection to detect and remove outlier samples [80], which would distort downstream analyses. Another way to visualize the results of the read normalization and check for outliers is by heatmaps. The R package ComplexHeatmaps [81] offers highly customizable row and column annotations such as dendrograms, based on different distance functions. This way of including unsupervised clustering offers an intuitive way to interpret the overall similarity in expression across samples and genes.
Initially, RNA-seq count data was approximated with the Poisson distribution, under the assumption that reads follow a random sampling process [82], [83]. However, since the variance and mean of RNA-seq counts are not equal, the negative binomial distribution was found to be more adequate [84], [85]. The most popular approaches that were developed consequently include DESeq2 [86] and edgeR [46].
As mentioned in the previous subsection, DESeq and DESeq2 both assume a negative binomial distribution of the counts and have two parameters, the dispersion and the mean. The dispersion describes how much the variance (i.e. within-group variability) deviates from the mean (Var Kij = µij + αi µ2ij, where αi is the dispersion parameter) and it is estimated in three steps. First, with maximum likelihood, a dispersion value is estimated for every gene, then a curve model, as a function of the mean expression level, is fitted to these values. Finally, a dispersion value is assigned to every gene. In DESeq, this is computed as a function of the mean by fitting a smoothed curve to the observed values. In DESeq2, the dispersion value is assigned by using an empirical Bayes method to shrink the gene-wise dispersion estimates close to the fitted values.
When comparing the distribution of counts between different groups, DESeq2 fits a generalized linear model (GLM) for each gene, as defined by the design matrix. The coefficients represent a log2 fold change in simple case-control experiments, but more complex relations can also be modeled. After the fit, a hypothesis test for differential expression is applied on the coefficient of interest, i.e. whether they are different from 0 (the no effect case). DESeq2 offers the use of the likelihood ratio test or the Wald test, which can test individual coefficients, as well as contrasting them.
The different edgeR variants are also assuming a negative binomial distribution. In edgeR classic, the quantile-adjusted conditional maximum likelihood is used to estimate the dispersions, conditioning on the total count of the particular gene [87]. Since edgeR classic can only be used for designs with a single factor, an exact test similar to Fisher's exact test can be constructed to test for differential expression [46]. The more advanced edgeR glm [88] and edgeR robust [89] use the Cox-Reid profile-adjusted likelihood to estimate the dispersions, and fit a GLM as in DESeq, followed by a likelihood ratio test for differential expression. To reduce the influence of outliers, edgeR robust assigns weights to observations based on their Pearson residual in the GLM fit.
To identify genes that change significantly in abundance across different samples and conditions, testing methods focus on evaluating the null hypothesis that there is no difference between conditions, i.e. the log fold-changes between cases and controls are exactly zero. A threshold of 5% on these p-values would limit the number of false positives in a single test, but one still needs to account for the large numbers of tests that are typically performed in parallel. Under the assumption that the null-hypothesis is true, when performing 20.000 tests, this would lead to 1.000 false positives. To control for type I errors (i.e. incorrectly rejected null hypotheses), several methods controlling the family-wise error rate (i.e. the probability of making at least one type I error) exist. One of these, the Bonferroni correction [90], adjusts the significance threshold by dividing the significance level ɑ by the number of performed tests. In practice, this correction is too conservative, and instead of controlling the family-wise error rate, the rate of type I errors can be limited by false discovery rate (FDR) controlling procedures. The FDR is the fraction of false positives (falsely rejected null hypotheses) among all results that were declared significant (all rejected null hypotheses). Most commonly, the Benjamini–Hochberg procedure (BH step-up procedure) is implemented, which controls the FDR at a predefined level [91]. While adjusted p-values (i.e. q-values) are computed for each test, the interpretations of p-values and q-values are quite different. For p-values, a cutoff of 5% means that 5% of all tests will result in false positives, assuming that there are no differentially expressed genes (the null hypothesis is true). However, the same cutoff for q-values means that 5% of the significant tests are false positives (i.e. the rate of false discoveries is 5%). Both in edgeR and DESeq2, the p-values for each gene are adjusted for multiple testing, controlling for the false discovery rate according to the Benjamini-Hochberg procedure.
Calling a gene as being differentially expressed based on an FDR cutoff alone has the disadvantage of including results whose effect size, while being statistically significant due to the consistency of the result, is biologically insignificant. Hence an additional filter may be applied on the log2 fold change, at the risk of distorting the FDR statistics in the selected subset. Accordingly, the SEQC consortium [92] found that pipeline-dependent filters for p-value, fold-change and expression-level are necessary to reproduce results.
3.3.6. Biological interpretation of the results
Once a gene set of interest has been defined, enrichment analyses can ascribe biological meaning. Gene Set Enrichment Analysis [93] is the most widely used tool (Fig. 2B) and checks for significant over- or underrepresentation of annotated gene sets, such as Gene Ontology terms [94], within provided lists. Also, DAVID [95], [96] is an online platform which functionally annotates and classifies genes.
Other approaches determine overrepresentation of selected genes in metabolic pathways or map them to putative protein interaction networks. These analyses obviously depend on prior knowledge about those biological pathways. Gene lists can be mapped onto specific pathways diagrams, and statistically significant associations can be retrieved and visualized, for example using the Kyoto Encyclopedia of Genes and Genomes [97], Reactome [98] and WikiPathways [99]. Protein-protein interaction networks contribute to the system-level data interpretation. Known cellular interaction networks represent another source of information, since proteins that participate in the same biological process may be more likely to interact. Therefore, integrative interactomics aim to provide a similar view as pathway analyses, by exploiting large interactomes identified in model organisms [100], [101]. For example, differentially expressed genes can be mapped to protein–protein interaction data, and then the functional clusters in the networks could be determined. Important protein network databases include IntAct [102], STRING [103] and BioGRID [104].
4. Data integration
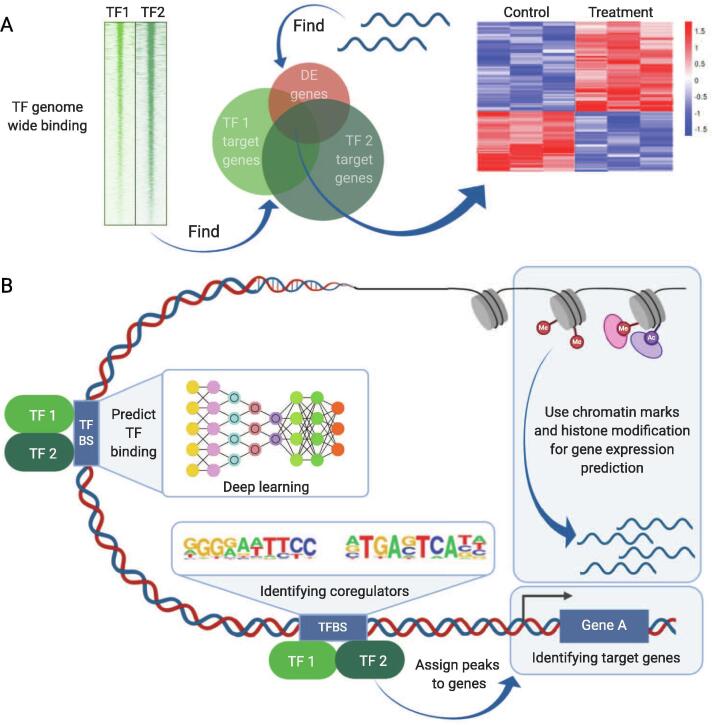
Jointly characterizing multiple omics might enable an in-depth understanding of the interplay between various cogs of the transcriptional machinery. Depending on the specific question, various flavors of data integration could be applied (Fig. 4).
Fig. 4.
Data integration approaches. (A) ChIP-seq and RNA-seq data can be integrated in a discretized fashion by determining the overlap of significantly affected genes in the 2 assays. (B) Newer approaches combine ChIP-seq data from multiple TFs and HMs together with expression data and accessibility data such as DNase-seq and ATAC-seq. They achieve data integration through various different mathematical concepts such GLMs, HMMs and deep neural networks to identify co-regulators, predict gene expression or model TF binding. DE: differential expression, TF: transcription factor. This figure was created with BioRender (biorender.com).
4.1. Identifying coregulators
TF ChIP-seq can serve to identify co-factors through motif analysis, which takes a number of sequences as input and finds motifs (usually 8–16 bp in length) that are present more frequently than would be expected [105]. In addition to the consensus motif expected for the TF targeted by the specific antibody, other binding sites for co-factors cross-talking with the protein of interest may be enriched. Furthermore, motif analyses can pinpoint the exact site within the ChIP peak that is occupied by the TF. Also, ChIP peak lists can first be narrowed down by integrating expression data before searching for distinct motifs associated with a defined transcriptional outcome.
Exploring all possible solutions to find the highest ranking motifs is still challenging. The most commonly used tool HOMER [106] determines enrichment using cumulative hypergeometric distributions. MEME-ChIP [107] applies expectation maximization and Discrover [108] uses discriminative learning based on Hidden Markov Models. Most tools perform de novo motif discovery as well as testing for the enrichment of known-motifs, which are represented as position weight matrices (PWMs). Motif databases like JASPAR [109], Cis-BP [110] and HOCOMOCO [111] store PWMs and can be used by motif analysis tools to link the discovered sequences to known consensus motifs.
4.2. Identifying epigenetic cofactors
In addition to the profiling and functional characterization of individual histone marks, comprehensive models aim to combine several dozens of epigenetic HMs [112]. For example, a multivariate HMM on the combinatorial patterns of 38 different modifications, RNA polymerase II, H2A.Z and CTCF ChIP-seq data, was used to define “chromatin states” and to systematically annotate the genome at 200 bp resolution [113].
This approach of chromatin segmentation has since been implemented and expanded by the NIH Roadmap Epigenomics Consortium [13]. The Roadmap project integrated chromatin states with DNA methylation, DNA accessibility and RNA expression to create reference epigenomes for over 100 human cell types and tissues.
4.3. Identifying target genes
Classical strategies to investigate the direct and indirect targets of a TF, are gain and loss of function experiments or specific treatments in conjunction with controls. ChIP-seq and RNA-seq data of matched samples may first be processed separately according to their respective analysis standards, and then be combined in a discretized fashion. In order to obtain comparable results, ChIP-seq peaks are usually assigned to nearest genes (see section 3.2.4). Then, one can determine whether the genes that are differentially expressed show concordant patterns of differential TF binding or epigenetic modifications. A prevalent approach to assess the similarity in changes across assays is to arrange those genes showing differential ChIP signals, and those being differentially expressed, as contingency tables and to test for overrepresentation with Fisher’s exact test. A common way to depict these numbers in publications is as a Venn diagram. The intersection, which represents genes that have differential ChIP signals and expression changes, can then further be displayed in a heatmap to visually inspect their expression pattern (Fig. 4A). The biggest shortcoming of this approach is that the results for both assays need to be binarized by setting an arbitrary threshold to split the data into significant and non-significant results.
A possible approach to avoid arbitrary cutoffs when integrating the results of different experiments was proposed by Roider and colleagues [114]. It was originally developed for a combination of ChIP-chip and affinity data, but could be applied to combine p-values of ChIP-seq and expression data as well. This method transforms the results into ranked lists and systematically adjusts the threshold to find the optimal cutoff, yielding the most significant enrichment as measured by hypergeometric testing.
A way to avoid setting a hard p-value threshold on one of the datasets is by performing gene set testing. The results of one platform are hereby ranked according to a test-statistic of choice, and the positions of the elements in a gene set on that ranked list, such as the significant hits of another platform, are determined [115]. E.g. RNA-seq results can be ranked based on a test statistic representing the degree of differential expression between two samples (such as the t-value), and the genes with significant ChIP-seq peaks can be indexed on the ranked list. This can then be used to test whether the genes with ChIP-seq peaks tend to be more differentially expressed than genes without ChIP-seq peaks.
In order to prevent setting thresholds altogether, the log2 fold changes of expression and peak intensities can be tested for correlation. Those genes that show alterations in ChIP-seq and RNA-seq are likely direct or indirect targets of the TF.
More formally, BETA [116] assigns a regulatory potential to each gene based on the number and proximity of TF binding sites to its transcription start site, and, determines if the TF is mainly an activator or a repressor in conjunction with the gene expression data. Direct targets are selected using a rank product between the RNA and ChIP data. Interestingly, BETA and Discrover [108] also return differential motifs, which again might identify co-regulators.
4.4. Predicting gene expression
It is still an open question whether TF binding strength (ChIP-Seq) can be used to predict gene expression levels (RNA-Seq). A study using the quantitative ChIP-seq signal of TFs around the transcription start site could explain 67% of the variation in CAGE data, i.e. nascent transcription, but performed poorly for total RNA [117].
Conversely, chromatin marks and histone modifications (for example H3K27ac) are more established predictors, with a small number of HMs at promoter regions being sufficient to correlate well with gene expression (Fig. 4B). It appears that the relationship between the chromatin landscape and RNA expression can be generalized across different cell types [118].
Finally, IMAGE pinpoints transcriptional regulators by utilizing PWMs to model the activity of a certain motif. This information is then used to infer causality by modelling the contribution of the motif to expression levels [119].
4.5. Predicting TF binding
Modelling cell-type specific gene expression on TF binding data remains difficult, as the available ChIP-seq datasets for any given cell type are still limited. In an attempt to predict gene expression with fewer assays, tools hinging on chromatin accessibility in combination with PWMs were developed.
4.5.1. Classical approaches
DNase-seq and ATAC-seq find cell type specific open regulatory regions in the genome, which are prone to DNase I and Tn5 activity, respectively [120], [121]. TF occupancy protects short sequences from these cleavage enzymes, causing dips in the accessibility signal. Matching the protected sequence of these footprints with known PWMs can identify the bound TF [122]. The presence of TF motifs within proximal and distal DNase I hypersensitive sites can be quantified and used to generate scores or footprints for regression models classifying tissue specific expression patterns [123], [124].
The CENTIPEDE [125] algorithm uses DNase-seq and HM data as prior information to predict TF binding with hierarchical mixture models. HINT [126] also uses accessibility data and HMs to calculate active TF binding sites based on HMMs. This algorithm was later extended by HINT-ATAC [127] to identify footprints in ATAC-seq data, while correcting for transposase specific artifacts.
Continued interest in predicting in vivo TF binding for various tissue types sparked the ENCODE-DREAM challenge which now serves as a benchmarking study (https://www.synapse.org/#!Synapse:syn6131484/wiki/402031).
4.5.2. Deep learning approaches
The availability of large amounts of training data and breakthroughs in high-performance computing such as the use of graphical processing units (GPUs) have triggered a comeback of neural networks in the analysis of genomic data [128].
Two methods based on convolutional neural networks (CNNs), are DeepSEA [129] and DeepBind [130]. DeepSEA predicts chromatin features such as HMs, DNase I hypersensitive sites and TF binding sites, and calculates how sequence alterations can affect chromatin. DeepBind applies deep CNNs to predict both binding affinity from sequence and the binding in vivo (as measured by ChIP-seq).
The performance of deep learning is evidenced by FactorNet placing in the top 3 of the DREAM challenge, predicting TF binding from DNA sequences. Their convolutional-recurrent neural networks can predict cell type specific TF binding [126], leveraging binding in a reference cell type and chromatin accessibility from the cell type of interest.
Moreover, ExPecto [131] uses a deep CNN to predict HMs, TF binding and other transcriptional regulators from DNA sequence alone by training on ENCODE and Roadmap Epigenomics data. These features are then transformed and fed into a cell type-specific linear model to predict gene expression (whereas DeepSEA only predicted the effect of non-coding variants on chromatin).
CNNs are also used in BPNet [132], which takes a DNA sequence as input and directly predicts ChIPexo signals at single base resolution to elucidate how TF binding is influenced by the motif syntax. This way, no information is lost in the intermediary peak calling process which usually precedes motif discovery in a standard analysis pipeline, and the regulatory elements of multiple TFs can be assessed simultaneously.
5. Conclusions & outlook
Taken together, the integration of multi-omics data can contribute to decrypting transcriptional regulatory codes. With new techniques and data forms constantly emerging, novel data integration methods are evolving. Besides data analysis tools, databases provide meaningful biological interpretation.
Overall, the exact molecular mechanisms of TF binding, histone modifications and transcriptional regulation are far from understood. The field has moved from individual genes and factors towards a higher dimensional view, integrating epigenetic marks, distal regulatory elements and the 3D structure. Furthermore, live cell imaging coupled with single-cell RNA-seq is on the rise.
Advancements in the development of experimental methods in combination with novel analysis tools hold great potential. As such, pooled CRISPR screening combined with single-cell RNA-seq is a powerful method to investigate distinct perturbations in thousands of individual cells. For example, Perturb-seq [133] couples gene inactivation using CRISPR with single-cell RNA-seq to study phenotypic alterations in parallel in many cells. scMAGeCK [134] is now able to find genes and enhancers which play a role in cell proliferation, simply by associating common proliferation markers.
Despite the progress in bioinformatics, the identification of functional enhancer-promoter interactions remains challenging, and in silico predictions still require high-throughput experimental validation. While STARR–seq [135] (self-transcribing-active-regulatory- region-sequencing) creates genome-wide quantitative enhancer activity maps, the recently published enCRISPRa and enCRISPRi [136] epigenetic editing systems allow for functional interrogation of enhancers in situ and in vivo.
To understand complex biological systems, specialized tools merging different omics data sets like genomics, transcriptomics, metabolomics, proteomics etc. and ultimately integrating not only transcriptional data, will yield unprecedented insights into the feature space of systems biology.
CRediT authorship contribution statement
Barbara Höllbacher: Conceptualization, Writing - original draft, Visualization, Formal analysis. Kinga Balázs: Conceptualization, Writing - original draft, Visualization. Matthias Heinig: Conceptualization, Writing - review & editing, Funding acquisition, Supervision. N. Henriette Uhlenhaut: Conceptualization, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We sincerely thank S. Regn, I. Guderian and F. Quagliarini, for their contributions to this manuscript. We apologize to all authors whose work could not be cited due to space constraints. This article was supported by the ERC StG SILENCE 638573 to NHU, the Helmholtz Association ICEMED and the joint research school "Munich School for Data Science" (MUDS).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.05.018.
Contributor Information
Matthias Heinig, Email: matthias.heinig@helmholtz-muenchen.de.
N. Henriette Uhlenhaut, Email: henriette.uhlenhaut@helmholtz-muenchen.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buenrostro J., Wu B., Chang H., Greenleaf W. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol Ed Frederick M Ausubel Al. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu Y., Corey D.R. RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther. 2012;22:271–274. doi: 10.1089/nat.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 7.Philip M., Fairchild L., Sun L., Horste E.L., Camara S., Shakiba M. Chromatin states define tumor-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling C., Rönn T. Epigenetics in human obesity and Type 2 diabetes. Cell Metab. 2019;29:1028–1044. doi: 10.1016/j.cmet.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Jensen M.A., Zenklusen J.C. A practical guide to the cancer genome atlas (TCGA) Methods Mol Biol Clifton NJ. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 12.Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams D., Altucci L., Antonarakis S.E., Ballesteros J., Beck S., Bird A. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 15.Bujold D., de Morais D.A. de L., Gauthier C., Côté C., Caron M., Kwan T. The international human epigenome consortium data portal. Cell Syst. 2016;3:496–499. doi: 10.1016/j.cels.2016.10.019. e2. [DOI] [PubMed] [Google Scholar]
- 16.Teytelman L., Thurtle D.M., Rine J., van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohm J.C., Lottaz C., Borodina T., Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auerbach R.K., Euskirchen G., Rozowsky J., Lamarre-Vincent N., Moqtaderi Z., Lefrançois P. Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci U S A. 2009;106:14926–14931. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozowsky J., Euskirchen G., Auerbach R.K., Zhang Z.D., Gibson T., Bjornson R. PeakSeq: systematic scoring of ChIP-Seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Brown J.B., Huang H., Bickel P.J. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 21.Baccarella A., Williams C.R., Parrish J.Z., Kim C.C. Empirical assessment of the impact of sample number and read depth on RNA-Seq analysis workflow performance. BMC Bioinf. 2018;19:423. doi: 10.1186/s12859-018-2445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Zhou J., White K.P. RNA-seq differential expression studies: more sequence or more replication? Bioinforma Oxf Engl. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hart S.N., Therneau T.M., Zhang Y., Poland G.A., Kocher J.-P. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20:970–978. doi: 10.1089/cmb.2012.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ching T., Huang S., Garmire L.X. Power analysis and sample size estimation for RNA-Seq differential expression. RNA. 2014;20:1684–1696. doi: 10.1261/rna.046011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S., Li C.-I., Guo Y., Sheng Q., Shyr Y. RnaSeqSampleSize: real data based sample size estimation for RNA sequencing. BMC Bioinf. 2018;19:191. doi: 10.1186/s12859-018-2191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C.-I., Shyr Y. Sample size calculation based on generalized linear models for differential expression analysis in RNA-seq data. Stat Appl Genet Mol Biol. 2016;15:491–505. doi: 10.1515/sagmb-2016-0008. [DOI] [PubMed] [Google Scholar]
- 27.Wu H., Wang C., Wu Z. PROPER: comprehensive power evaluation for differential expression using RNA-seq. Bioinforma Oxf Engl. 2015;31:233–241. doi: 10.1093/bioinformatics/btu640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poplawski A., Binder H. Feasibility of sample size calculation for RNA-seq studies. Brief Bioinform. 2018;19:713–720. doi: 10.1093/bib/bbw144. [DOI] [PubMed] [Google Scholar]
- 29.Allfrey V.G., Faulkner R., Mirsky A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat Rev Mol Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- 31.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 32.Rhee H.S., Pugh B.F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aronesty E. ea-utils : “Command-line tools for processing biological sequencing data”. HttpsgithubcomExpressionAnalysisea-Utils. 2011 [Google Scholar]
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinig M., Colomé-Tatché M., Taudt A., Rintisch C., Schafer S., Pravenec M. histoneHMM: Differential analysis of histone modifications with broad genomic footprints. BMC Bioinf. 2015;16:60. doi: 10.1186/s12859-015-0491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas R., Thomas S., Holloway A.K., Pollard K.S. Features that define the best ChIP-seq peak calling algorithms. Brief Bioinform. 2017;18:441–450. doi: 10.1093/bib/bbw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing H., Mo Y., Liao W., Zhang M.Q. Genome-wide localization of protein-DNA binding and histone modification by a bayesian change-point method with ChIP-seq data. PLOS Comput Biol. 2012;8:e1002613. doi: 10.1371/journal.pcbi.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmanci A., Rozowsky J., Gerstein M. MUSIC: identification of enriched regions in ChIP-Seq experiments using a mappability-corrected multiscale signal processing framework. Genome Biol. 2014;15:474. doi: 10.1186/s13059-014-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meers M.P., Tenenbaum D., Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;12:42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakato R., Shirahige K. Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation. Brief Bioinform. 2017;18:279–290. doi: 10.1093/bib/bbw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao Y., Vinciotti V., Wit E., ’t Hoen P.A. Accounting for immunoprecipitation efficiencies in the statistical analysis of ChIP-seq data. BMC Bioinf. 2013;14:169. doi: 10.1186/1471-2105-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu S., Shao Z. An introduction to computational tools for differential binding analysis with ChIP-seq data. Quant Biol. 2017;5:226–235. doi: 10.1007/s40484-017-0111-8. [DOI] [Google Scholar]
- 46.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lun A.T.L., Smyth G.K. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1191. e45 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao Z., Zhang Y., Yuan G.-C., Orkin S.H., Waxman D.J. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 55.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoenfelder S., Javierre B.-M., Furlan-Magaril M., Wingett S.W., Fraser P. Promoter capture Hi-C: high-resolution, genome-wide profiling of promoter interactions. J Vis Exp JoVE. 2018 doi: 10.3791/57320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morozova O., Hirst M., Marra M.A. Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 59.Wolf J.B.W. Principles of transcriptome analysis and gene expression quantification: an RNA-seq tutorial. Mol Ecol Resour. 2013;13:559–572. doi: 10.1111/1755-0998.12109. [DOI] [PubMed] [Google Scholar]
- 60.Babarinde I.A., Li Y., Hutchins A.P. Computational methods for mapping, assembly and quantification for coding and non-coding transcripts. Comput Struct Biotechnol J. 2019;17:628–637. doi: 10.1016/j.csbj.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 65.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vijay N., Poelstra J.W., Künstner A., Wolf J.B.W. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol. 2013;22:620–634. doi: 10.1111/mec.12014. [DOI] [PubMed] [Google Scholar]
- 68.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 71.Bullard J.H., Purdom E., Hansen K.D., Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinf. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., Brock G.N., Rouchka E.C., Cooper N.G.F., Wu D., O’Toole T.E. A comparison of per sample global scaling and per gene normalization methods for differential expression analysis of RNA-seq data. PLoS ONE. 2017;12:e0176185. doi: 10.1371/journal.pone.0176185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamini Y., Speed T.P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 75.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans C., Hardin J., Stoebel D.M. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions. Brief Bioinform. 2017;19:776–792. doi: 10.1093/bib/bbx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dillies M.-A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 78.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 79.Robinson Mark D., Oshlack Alicia. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merino G.A., Fresno C., Netto F., Netto E.D., Pratto L., Fernández E.A. The impact of quality control in RNA-seq experiments. J Phys Conf Ser. 2016;705 doi: 10.1088/1742-6596/705/1/012003. [DOI] [Google Scholar]
- 81.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma Oxf Engl. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 82.Li J., Witten D.M., Johnstone I.M., Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostat Oxf Engl. 2012;13:523–538. doi: 10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 84.Hulse A.M., Cai J.J. Genetic variants contribute to gene expression variability in humans. Genetics. 2013;193:95–108. doi: 10.1534/genetics.112.146779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu M., Zhu Y., Taylor J.M.G., Liu J.S., Qin Z.S. Using poisson mixed-effects model to quantify transcript-level gene expression in RNA-Seq. Bioinforma Oxf Engl. 2012;28:63–68. doi: 10.1093/bioinformatics/btr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson M.D., Smyth G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostat Oxf Engl. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 88.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou X., Lindsay H., Robinson M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bland J.M., Altman D.G. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 92.A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control consortium. Nat Biotechnol 2014;32:903–14. https://doi.org/10.1038/nbt.2957. [DOI] [PMC free article] [PubMed]
- 93.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith B., Williams J., Steffen S.-K. The ontology of the gene ontology. AMIA Annu Symp Proc. 2003;2003:609–613. [PMC free article] [PubMed] [Google Scholar]
- 95.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 96.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinf. 2017;18:142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kelder T., Pico A.R., Hanspers K., van Iersel M.P., Evelo C., Conklin B.R. Mining biological pathways using WikiPathways web services. PLoS ONE. 2009;4:e6447. doi: 10.1371/journal.pone.0006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berger B., Peng J., Singh M. Computational solutions for omics data. Nat Rev Genet. 2013;14:333–346. doi: 10.1038/nrg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hawe J.S., Theis F.J., Heinig M. Inferring interaction networks from multi-omics data. Front Genet. 2019;10. doi: 10.3389/fgene.2019.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zambelli F., Pesole G., Pavesi G. Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Brief Bioinform. 2013;14:225–237. doi: 10.1093/bib/bbs016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinforma Oxf Engl. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maaskola J., Rajewsky N. Binding site discovery from nucleic acid sequences by discriminative learning of hidden Markov models. Nucleic Acids Res. 2014;42:12995–13011. doi: 10.1093/nar/gku1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weirauch M.T., Yang A., Albu M., Cote A., Montenegro-Montero A., Drewe P. Determination and inference of Eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Sharipov R.N., Fedorova A.D., Rumynskiy E.I. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 113.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roider H.G., Manke T., O’Keeffe S., Vingron M., Haas S.A. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang S., Sun H., Ma J., Zang C., Wang C., Wang J. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng C., Alexander R., Min R., Leng J., Yip K.Y., Rozowsky J. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22:1658–1667. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karlić R., Chung H.-R., Lasserre J., Vlahoviček K., Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Madsen J.G.S., Rauch A., Hauwaert E.L.V., Schmidt S.F., Winnefeld M., Mandrup S. Integrated analysis of motif activity and gene expression changes of transcription factors. Genome Res. 2018;28:243–255. doi: 10.1101/gr.227231.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2010;2010:pdb.prot5384. https://doi.org/10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed]
- 121.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boyle A.P., Song L., Lee B.-K., London D., Keefe D., Birney E. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Natarajan A., Yardımcı G.G., Sheffield N.C., Crawford G.E., Ohler U. Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt F., Gasparoni N., Gasparoni G., Gianmoena K., Cadenas C., Polansky J.K. Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res. 2017;45:54–66. doi: 10.1093/nar/gkw1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gusmao E.G., Dieterich C., Zenke M., Costa I.G. Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modifications. Bioinformatics. 2014;30:3143–3151. doi: 10.1093/bioinformatics/btu519. [DOI] [PubMed] [Google Scholar]
- 127.Li Z., Schulz M.H., Look T., Begemann M., Zenke M., Costa I.G. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45. doi: 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eraslan G., Avsec Ž., Gagneur J., Theis F.J. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet. 2019;20:389–403. doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
- 129.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 131.Zhou J., Theesfeld C.L., Yao K., Chen K.M., Wong A.K., Troyanskaya O.G. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet. 2018;50:1171–1179. doi: 10.1038/s41588-018-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Avsec Ž., Weilert M., Shrikumar A., Alexandari A., Krueger S., Dalal K. Deep learning at base-resolution reveals motif syntax of the cis-regulatory code. BioRxiv. 2019;737981 doi: 10.1101/737981. [DOI] [Google Scholar]
- 133.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866. doi: 10.1016/j.cell.2016.11.038. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang L., Zhu Y., Yu H., Cheng X., Chen S., Chu Y. scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol. 2020;21 doi: 10.1186/s13059-020-1928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arnold C.D., Gerlach D., Stelzer C., Boryń Ł.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 136.Li K., Liu Y., Cao H., Zhang Y., Gu Z., Liu X. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat Commun. 2020;11:1–16. doi: 10.1038/s41467-020-14362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.