Abstract
Soil salinity is a global major abiotic stress threatening crop productivity. In salty conditions, plants may suffer from osmotic, ionic, and oxidative stresses, resulting in inhibition of growth and development. To deal with these stresses, plants have developed a series of tolerance mechanisms, including osmotic adjustment through accumulating compatible solutes in the cytoplasm, reactive oxygen species (ROS) scavenging through enhancing the activity of anti-oxidative enzymes, and Na+/K+ homeostasis regulation through controlling Na+ uptake and transportation. In this review, recent advances in studies of the mechanisms of salt tolerance in plants are described in relation to the ionome, transcriptome, proteome, and metabolome, and the main factor accounting for differences in salt tolerance among plant species or genotypes within a species is presented. We also discuss the application and roles of different breeding methodologies in developing salt-tolerant crop cultivars. In particular, we describe the advantages and perspectives of genome or gene editing in improving the salt tolerance of crops.
Keywords: Salinity, Osmotic stress, Ionic stress, Oxidative stress, Salt tolerance
1. Introduction
Soil salinity is one of the major abiotic stresses restricting global crop productivity. Moreover, the area undergoing salinization is still expanding. It is predicted that nearly half of all arable land in the world will be under salinization by 2050 (Butcher et al., 2016), raising a huge threat to sustainable agriculture development and food security. The development and planting of crop cultivars with high salt tolerance is the most efficient approach for fighting soil salinity. Therefore, it is important to reveal salt tolerance mechanisms and identify relevant genotypes or genes.
2. Salt toxicity in crop plants
Salt stress greatly affects crop growth and development. High concentration of soluble salt ions is a main factor resulting in salt stress. The most direct damage to crops caused by salt stress is due to high osmotic pressure resulting from a high concentration of salinity ions (i.e., Na+ and Cl−). Therefore, osmotic stress and ionic stress are severe conditions for crops exposed to salt stress. The superposition of these two stresses can lead to secondary oxidative stress (Munns and Tester, 2008).
2.1. Osmotic stress
Osmotic stress occurs when plant roots are subjected to a certain salt level. For most plants the threshold level is around 40 mmol/L NaCl (Munns and Tester, 2008). The onset of osmotic stress decreases the ability of plants to take up water. Dehydration may occur when the osmotic pressure of the external salt solution is higher than that of the root cells (Horie et al., 2012). This phase lasts for a short time, and leads to stomatal closure and inhibition of cell expansion in shoots (Isayenkov and Maathuis, 2019). The formation and development of regenerative organs of cereal crops are greatly affected by salt stress, which reduces the rate of panicle formation and changes flowering and maturity time (Munns and Rawson, 1999). Osmotic stress causes physiological water deficiency. In cereals, the major effect of salinity on shoots is a reduction in tillers per plant (Munns and Tester, 2008). For dicotyledonous species, the major effect is often a dramatic curtailing of leaf growth or a reduced number of branches (Munns and Tester, 2008).
2.2. Ionic stress
Under salt stress, excessive uptakes of Na+ and Cl− through transpiration streams cause cyto-toxicity and nutritional imbalance in plants, namely long-term ionic stress. Large amounts of salt ions, especially Na+, accumulated in shoots can reach toxic levels, inhibiting plant growth. The old leaves cannot sufficiently dilute the toxic ions by leaf expansion, thereby accelerating senescence. NaCl is a major component of saline soil. For some plants, such as citrus, Cl− is a more toxic ion than Na+ (Moya et al., 2003). However, for most crops, Na+ appears to have a lower toxic concentration threshold than Cl− (Tester and Davenport, 2003). The inhibition of K+ by Na+ in plants results in a significant reduction in K+ uptake and accumulation, and a significant increase in the Na+/K+ ratio (Chen et al., 2007). The significant reduction of K+ uptake may be connected to the phenomenon of salt stress-induced K+ loss (Chen et al., 2007; Wu et al., 2014). Too great an accumulation of Na+ in shoots causes a decrease in other essential metal cations such as Ca2+ and Mg2+, which in turn affects physiological and biochemical activities in plants (Chen et al., 2005; Munns, 2005; Wu et al., 2013a).
2.3. Oxidative stress
Salt stress is in general also accompanied by oxidative stress because of excessive generation of reactive oxygen species (ROS), including O2 −, H2O2, ·OH, and HO2· (Zhu, 2001; Munns and Tester, 2008; Isayenkov, 2012). The excess ROS cannot be eliminated fast enough by antioxidant enzymes, leading to the formation of lipid free radicals, degradation of proteins and metabolites, and inhibition of biological metabolism. Eventually, under long-term salt stress, the stabilities of cell membranes and organelles are destroyed, photosynthesis is severely inhibited, and biosynthesis and nutrient transport are largely blocked (Zhu, 2001; Zahra et al., 2014). Therefore, in the middle and late stages of salt stress, oxidative stress will cause severe damage to plants. Usually, halophytic plants contain high amounts of antioxidant compounds in their tissues, which is one adaptive strategy enabling them to survive in harsh environments (Munns and Tester, 2008).
In short, salt stress causes severe inhibition of growth and development of crop plants through osmotic, ionic, and oxidative stresses.
3. Mechanisms of salt tolerance in plants
In the process of combating salt stress, plants have evolved a set of physiological and molecular mechanisms to adapt to the different stages of salt stress (Rajendran et al., 2009; Roy et al., 2014). Recently, “omic” methodologies have been widely used in studies of abiotic stress tolerance in plants (Nawrot et al., 2016; Meena et al., 2017). Multi-omic methods have proved effective and powerful for understanding complex molecular networks in plants (Nawrot et al., 2016; Wang et al., 2016). The responses of plant crops to salt stress have been intensively investigated at physiological and molecular levels. The ionome, proteome, metabolome, and transcriptome have been studied to understand the molecular responses involved in osmotic adjustment, ROS scavenging, energy metabolism, Na+ detoxification, and ion homeostasis (Wu et al., 2013b; Shen et al., 2017, 2018; Huang et al., 2018).
3.1. Osmotic adjustment
To cope with osmotic stress, plants synthesize and accumulate compatible osmolytes to maintain turgor and osmotic potential, thereby increasing leaf stomatal conductance to conserve water and alleviate plant growth inhibition (Passioura and Munns, 2000; Fricke and Peters, 2002; Roy et al., 2014). Compatible solutions include N-containing compounds, sugars, straight-chain polyhydric polyols, and cyclic polyhydric alcohols (Hare et al., 1998). Most glycophytes synthesize compatible solutions to fight against osmotic stress, but this strategy is “expensive” with the inhibition of growth rate as a cost (Munns and Gilliham, 2015). Metabolomic and proteomic analyses have revealed that amino acids like proline, and polyols like inositol, play important roles in developing salt tolerance in barley roots (Wu et al., 2013b; Shen et al., 2018). Similarly, results from studies of the maize (Zea mays) metabolome showed that salt stress leads to the enhanced formation of sugars, contributing to the mitigation of osmotic stress (Guo et al., 2017). Inorganic ions such as Na+ also can be used as osmolytes provided that they are compartmentalized into vacuoles to reduce their cytotoxicity (Flowers and Colmer, 2008). Metabolomic and transcriptomic analyses revealed that sea barley (Hordeum marinum) accession H559 did not increase compatible solutions such as proline and maltose more than did barley (Hordeum vulgare) XZ113 or “Golden Promise,” but accumulated Na+ and K+ as the “cheapest” form of osmotic adaptation in roots (Huang et al., 2018). For halophytes or salt-tolerant taxa, the uptake, transport, and compartmentalization of inorganic ions as osmo-regulators may be an economical and efficient way to cope with osmotic stress.
3.2. ROS scavenging and energy metabolism
When plants are exposed to salt stress for a long time, the photosynthetic rate declines rapidly and ROS are generated, which have a strong oxidative ability and cause cytoplasmic membrane damage, irreversible metabolic dysfunction, and cell death (Miller et al., 2010). Correspondingly, plants have developed the defense system of ROS scavenging, mainly through antioxidative enzymes, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), catelase (CAT), and glutathione peroxidase (GPX) (Miller et al., 2010; Wang et al., 2016). In most cases, stress tolerance in plants is achieved at the expense of energy consumption (Munns and Gilliham, 2015). Therefore, the energy distribution of plants exposed to salt stress is important for their survival and ability to thrive. Proteomic analyses showed that dehydroascorbate reductase and peroxidases scavenging ROS in roots, and phosphoglycerate kinase, triosephosphate isomerase, sedoheptulose-1, and 7-bisphosphatase related to photosynthesis in shoots, play important roles in salt tolerance in the early stage of salt exposure in barley (Shen et al., 2017). Salt stress caused a significant reduction in metabolites involved in the glycolysis pathway and an increase in those associated with the tricarboxylic acid (TCA) cycle in the shoots of a salt-sensitive Tibetan wild barley XZ169, but little change in the salt-tolerant line XZ26 (Shen et al., 2016). Multi-omic studies also showed that XZ26 adopted strategies of root adaptation to salt stress by enhancing antioxidant ability to cope with ROS and consuming less energy to produce biomass (Shen et al., 2018). Comparative transcriptomic and alternative splicing analyses of rice and barley found that differentially expressed genes (DEGs) associated with scavenging ROS and lowering energy consumption might contribute to the higher salt tolerance of barley (Fu et al., 2019).
3.3. Na+ detoxification and ion homeostasis
Relatively salt-tolerant plant species or genotypes commonly demonstrate a lower accumulation of Na+ in shoots when under salt stress (Shen et al., 2016, 2017, 2018; Fu et al., 2018, 2019; Huang et al., 2018). On the other hand, highly tolerant species also show a high accumulation of Na+ in roots (Fu et al., 2018, 2019; Huang et al., 2018). For most plant species, the main tissue organ to suffer Na+ toxicity is the leaf, where Na+ is highly accumulated due to ion deposition through the transpiration stream (Munns, 2002). Consequently, for most crops, the main physiological response to salt stress is to restrict Na+ distribution, thereby lessening accumulation in shoots, even though the Na+ content in roots may be greatly increased. Ion homeostasis is another ubiquitous physiological strategy for achieving salt tolerance in plants. The ionomic analysis of three barley genotypes differing in salt stress tolerance revealed that the rearrangement of nutrient elements (i.e., K, Mg, and Ca) in tissue organs possibly contributed to the development of salt tolerance (Wu et al., 2013a). K and Na compete with each other for their uptake and accumulation in plants (Chen et al., 2007). The ability to retain K+ in plant tissues under salt stress is important for achieving salt tolerance (Chen et al., 2005), and the K+/Na+ ratio has been used to identify salt tolerance in different plants (Chen et al., 2007; Kronzucker et al., 2008; Shabala et al., 2010). K+ homeostasis, involving K+-related programmed cell death and K+ as a signaling moiety in metabolic pathways, is closely linked to salt tolerance (Isayenkov and Maathuis, 2019).
In short, to adapt to the different stages of stress caused by salinity, plants have evolved complex physiological and molecular mechanisms including osmotic adjustment, ROS scavenging, energy metabolism, Na+ detoxification, and ion homeostasis.
4. Genetic differences in salt tolerance among crops
Salt tolerance differs greatly among plant species (Munns and Tester, 2008). According to their tolerance and adaptive evolution, plants can be divided into glycophytes and halophytes (Flowers and Colmer, 2008). Halophytes can grow normally in an environment with substantial amounts of salt and may even benefit from salty conditions (Shabala, 2013; Shabala et al., 2014). However, most crops are glycophytes, which are sensitive to salt stress. Even if the scope is narrowed down to within the Poaceae family, salt tolerance still differs greatly among genera such as barley (H. vulgare) and rice (Oryza sativa) (Ligaba and Katsuhara, 2010; Nevo and Chen, 2010; Hamam et al., 2016). Cultivated plant species and their wild relatives may differ significantly in their salt tolerance. For example, sea barley grass (Hordeum marinum) is a wild halophyte species, while other Hordeum species are glycophytes. In a physiological study, we showed that a sea barley grass accession H559 had higher salt tolerance than barley genotypes XZ113 and “Golden Promise” (Huang et al., 2018).
Comparative studies of plant species that differ greatly in salt tolerance are an effective way to understand the mechanisms of salt tolerance in plants (Fu et al., 2018, 2019; Huang et al., 2018). There may also be distinct differences in salt tolerance among genotypes within a plant species (Wu et al., 2014). However, the domestication of crops has resulted in a narrowing of genetic variation in wild crop relatives. Therefore, closely related species of crops are valuable genetic resources for modern agriculture and breeding (Dai et al., 2018; Isayenkov, 2019). For instance, our previous study identified some Tibetan wild barley accessions, which showed higher salt tolerance than a well-recognized salt-tolerant barley cultivar CM72 (Qiu et al., 2011). The wide genetic diversity in salt tolerance among plant species and genotypes within a species could provide useful gene donors for improving the salt tolerance of crop cultivars.
Although great effort has been applied in exploiting natural variation, only modest improvements in salt stress tolerance in a few crops have been recorded for several reasons, including the complexity of salt tolerance, its interactions with other agronomic traits, and the limited understanding of the physiology and genetics of salt tolerance (Ismail and Horie, 2017).
5. Transporters involved in plant salt tolerance
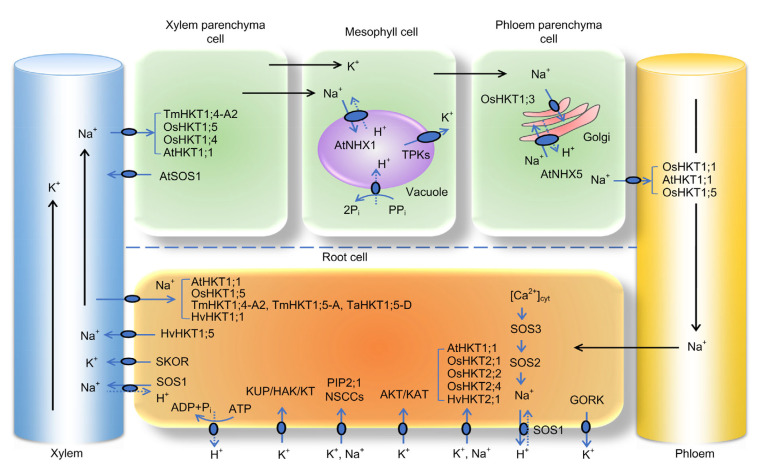
In the last two decades, many transporters related to salt tolerance have been identified in various plant species (Han et al., 2015). Among them, salt overly sensitive 1 (SOS1), high-affinity K+ transporter (HKT), and Na+/H+ exchanger (NHX) participate mainly in the processes of the uptake, long-distance transportation, and distribution of Na+ and K+. Together with the K+ uptake permease (KUP)/K+ transporter (KT) family, stelar K+ outward rectifier (SKOR) and guard cell outward rectifying K+ channel (GORK; outward-rectifying K+ channels) families, arabidopsis K+ transporter (AKT)/K+ channel in Arabidopsis thaliana (KAT; inward-rectifying K+ channel) family, two-pore K+ channels (TPKs), non-selective cation channel (NSCC), Ca2+/cation exchanger (CCX) H+-pyrophosphatase (PPase) and plasma membrane H+-adenosine triphosphatase (ATPase) pump, these transporters play an important role in maintaining Na+/K+ homeostasis under salt stress (Fig. 1).
Fig. 1.
Proposed working model for transmembrane transporters in regulating Na+/K+ homeostasis
The arrows in solid lines indicate the direction of Na+ and K+ flux, while dashed arrows indicate the direction of proton flux. Tm, Triticum monococcum; Os, Oryza sativa; At, Arabidopsis thaliana; HKT, high-affinity K+ transporter; SOS, salt overly sensitive; NHX, Na+/H+ exchanger; TPK, two-pore K+ channel; Pi, inorganic phosphorus; PPi, pyrophosphate; Ta, Triticum aestivum; Hv, Hordeum vulgare; SKOR, stelar K+ outward rectifier; ADP, adenosine diphosphate; ATP, adenosine triphosphate; KUP, K+ uptake permease; HAK, high-affinity potassium transporter; KT, K+ transporter; PIP, plasma membrane intrinsic protein; NSCCs, non-selective cation channels; AKT, K+ transporter; KAT, K+ channel in Arabidopsis thaliana; Cyt, cytoplasm; GORK, guard cell outward rectifying K+ channel
5.1. SOS transporters
The SOS pathway, containing a myristoylated calcium-binding protein SOS3, a serine/threonine protein kinase SOS2, and a plasma membrane Na+/H+ antiporter SOS1, has been well characterized in Arabidopsis (Shi et al., 2000, 2002; Qiu et al., 2002; Quintero et al., 2002). The main mechanism of the SOS regulatory pathway is that a salt-stress-elicited calcium signal first activates SOS3 and SOS2 to form a protein kinase complex, and then SOS1 is phosphorylated and activated by the SOS3-SOS2 complex to gain the function of achieving Na+ efflux from cells (Shi et al., 2000, 2002) (Fig. 1). It seems likely that SOS1 could also be phosphorylated in a phospholipase D (PLD) signaling pathway-dependent manner (Yu et al., 2010). A high Na+ concentration increases the enzyme activity of PLDα1, leading to fast accumulation of phosphatidic acid (PA) as a lipid second messenger. PA in turn activates mitogen-activated protein kinase 6 (MPK6), which can directly phosphorylate SOS1 (Yu et al., 2010). Apart from in Arabidopsis, the Na+/H+ antiporter SOS1 was also functionally characterized in rice (Martínez-Atienza et al., 2007; el Mahi et al., 2019), wheat (Xu et al., 2008a), and tomato (Huertas et al., 2012). The SOS1 gene expression pattern, combined with the results of ion analysis in sos1 mutant plants, suggests that SOS1 has several roles in adjusting ion homeostasis: (1) Na+ efflux from roots; (2) slowing down Na+ accumulation in the cytoplasm to allow time for Na+ storage in vacuoles; and (3) controlling long-distance Na+ transport between roots and leaves (Shi et al., 2000, 2002; Qiu et al., 2002; Quintero et al., 2002; Zhu, 2003; el Mahi et al., 2019). It has also been found that cells lacking vacuoles, such as root tip cells, exhibit high SOS1 activity. However, the disadvantage of this type of Na+ extrusion is the loss of the plasma membrane H+ gradient. Enhanced levels of SOS1 expression may increase tolerance to Na+, but plant growth remains relatively slow (Isayenkov and Maathuis, 2019). Moreover, the unusually long cytoplasmic tail of SOS1 is thought to be involved in Na+ sensing (Shi et al., 2000; Zhu, 2003). A recent study measured the acute Na+ sensitivity of sos1 plants at low NaCl concentrations to avoid the effects of osmotic stress. Roots of the sos1 mutant showed a marked down-regulation of genes, although the plants suffered from greater stress, indicating impaired stress detection or an inability to mount a comprehensive response to salinity stress without SOS1 (el Mahi et al., 2019).
5.2. NHX transporters
At the cellular level, high amounts of Na+ can be tolerated if intracellular compartmentalization maintains a cytoplasmic Na+ concentration as low as 10–30 mmol/L (Munns and Tester, 2008). Most tonoplast-localized NHXs such as AtNHX1, AtNHX2, and OsNHX5 function by sequestering Na+ into vacuoles (Fukuda et al., 2011; Bassil et al., 2012). AtNHX5, localized in golgi and trans-golgi networks, functions by trafficking Na+ into vacuoles (Bassil et al., 2011a). These processes are considered an economic strategy for adapting to salinity as they not only prevent Na+ toxicity in the cytosol, but also use Na+ as an osmolyte in the vacuole to alleviate osmotic stress (Zhu, 2003; Munns and Tester, 2008; Bassil et al., 2012). In Arabidopsis, AtNHX1 and AtNHX2 are the most abundant NHX members (Bassil et al., 2012). However, overexpression of AtNHX1 causes only a slight improvement in salt tolerance in Arabidopsis. Moreover, the tolerance level is much lower than that of transgenic plants overexpressing AtSOS1 (Yang et al., 2009). This suggests that Na+ compartmentalization has a minor effect on the engineering of salt-tolerant crops. On the other hand, NHX proteins have important roles in mediating K+ homeostasis in plants (Barragán et al., 2012; Andrés et al., 2014). Studies of transgenic tomato (Solanum lycopersicum) indicated that overexpression of vacuolar AtNHX1 or the endosomal LeNHX2 Na+/H+ and K+/H+ antiporters leads to enhanced K+ accumulation in vacuoles (Rodríguez-Rosales et al., 2008; Leidi et al., 2010). The main role of NHX-transporters may be in the maintenance of K+ homeostasis rather than the sequestration of Na+ into vacuoles. AtNHX1 and AtNHX2 antiporters control vacuolar pH and K+ homeostasis for regulating plant growth (Bassil et al., 2011b). The wheat NHX antiporter TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the capacity to retain intracellular potassium (Zhang et al., 2015).
5.3. HKT transporters
A member of the gene family of HKT was first cloned in wheat (Triticum aestivum) (Schachtman et al., 1992), and the family has since attracted significant attention because it confers robust permeability for Na+ (Schachtman and Schroeder, 1994; Rubio et al., 1995, 1999). Subsequently, the structure, localization, and expression of HKTs in crops and model plants have been successively identified, revealing substantial divergence in function. Plant HKT proteins contain four conserved P-loop domains. The serine residue (SGGG-type) in the first P-loop region primarily determines Na+ permeability for HKT sub-family 1 transporters. However, some studies showed that ion permeability is not completely consistent with the classification of the P-loop-conserved amino acid. For example, the structure of TsHKT1;2 from the Arabidopsis relative Thellungiella salsuginea belongs to subfamily 1, but the protein has K+ permeability (Rodríguez-Navarro, 2000; Mäser et al., 2002b; Ali et al., 2012; Oomen et al., 2012). More amino acid sites affecting ion transport properties in wheat have also been reported. The substitutions of amino acids A240V, L247F, Q270L, N365S, and E464Q sites in TaHKT2;1 from wheat (T. aestivum) significantly affect Na+ transport properties (Diatloff et al., 1998; Rubio et al., 1999).
Many members of the HKT family have now been functionally characterized (Table 1). Among the dicotyledons, the model plant Arabidopsis has only one HKT gene, AtHKT1;1, which has been well documented. AtHKT1;1 has high affinity for Na+ and participates in the uptake of Na+ in roots, Na+ unloading from xylem sap, and leaf Na+ refluxing to phloem, thereby reducing shoot Na+ toxicity and protecting leaves from salt stress (Mäser et al., 2002a; Davenport et al., 2007; Møller et al., 2009) (Fig. 1). In gramineous plants, there are generally multiple copies of the HKT gene, such as 9 in rice, 5–11 in the A, B, and D genomes of wheat, and 8 in barley (Garciadeblás et al., 2003; Huang et al., 2008; Horie et al., 2009; Qiu et al., 2011; Waters et al., 2013). The functional analysis of the HKT genes in rice is relatively comprehensive, covering nine HKT genes, while only four HKT genes have been reported in wheat and only three in barley. The expression, localization, and physiological function of these reported HKT genes in rice, wheat, and barley are listed in Table 1. Among them, OsHKT1;5 in rice encodes an Na+-selective transporter that functions in K+/Na+ homeostasis under salt stress (Ren et al., 2005). Kobayashi et al. (2017) used two independent transfer DNA (T-DNA) insertion mutants of OsHKT1;5 to further reveal its physiological roles in mediating Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues under salt stress conditions (Fig. 1). A major quantitative trait locus (QTL) Nax2 was identified in Einkorn wheat (Triticum monococcum), and TmHKT1;5-A, located by map-based cloning in the region of Nax2, reduced Na+ accumulation in tomato leaves (Byrt et al., 2007; James et al., 2011; Munns et al., 2012) (Fig. 1). Furthermore, it was demonstrated that TaHKT1;5-D from bread wheat (T. aestivum) is a major gene of the Kna1 locus, which plays a role in leaf Na+ exclusion and salt tolerance (Gorham et al., 1990; Byrt et al., 2007, 2014) (Fig. 1). In barley, knock-down of HvHKT1;1 led to higher Na+ accumulation in both roots and shoots, while overexpression of HvHKT1;1 in salt-sensitive Arabidopsis mutant (hkt1-4) and loss-of-function mutants (sos1-12) resulted in significant reductions in shoot and root Na+ accumulation (Han et al., 2018). That study also showed that the constitutive expression of HvHKT1;1 did not increase Na+ influx into plant roots and that the gene may take part in the relocation of Na+ to root epidermal cells for efflux (Fig. 1). HvHKT1;5 knock-down barley lines showed higher salt tolerance accompanied by a dramatic decrease in Na+ translocation from roots to shoots and increases in the K+/Na+ ratio compared with wild-type plants under salt stress. The negative regulation of HvHKT1;5 in salt tolerance distinguishes it from other HKT1;5 members (Huang et al., 2020).
Table 1.
Gene expression, localization, and function of HKTs in rice, wheat, and barley
| Plant species | Gene | Gene expression, localization, and function | Reference |
| Oryza sativa | OsHKT1;1 | Mainly expressed in the phloem of leaves; the expression in old leaves is up-regulated by salt stress, which protects young leaves by accumulating Na+ in the old leaves | Wang et al., 2012 |
| O. sativa | OsHKT1;3 | Expressed in the root stele, phloem, and leaf vascular bundle; localized on the golgi; associated with leaf curl | Jabnoune et al., 2009 |
| O. sativa | OsHKT1;4 | Mainly expressed in the vascular tissue of shoot; Na+ was unloaded from the stem and leaf sheath during the reproductive growth stage, but this function is not significant during the vegetative growth stage | Suzuki et al., 2016 |
| O. sativa | OsHKT1;5 (SKC1) | Expressed in the plasma membrane of xylem parenchyma cells in root and leaf sheath; unloading Na+ from the xylem; reducing Na+ accumulation in leaves | Ren et al., 2005; Kobayashi et al., 2017 |
| O. sativa | OsHKT2;1 | Expressed in root epidermis and cortical cells; gene expression is reduced under salt stress; involved in root Na+ absorption | Garciadeblás et al., 2003; Kader and Lindberg, 2008 |
| O. sativa | OsHKT2;2 | Mainly expressed in root and mesophyll cells; gene expression in phloem is significantly enhanced under salt stress; K+/Na+ symporter | Kader et al., 2006; Yao et al., 2010 |
| O. sativa (cv. Nona Bokra) | No-OsHKT2;2/1 | Gene expression is induced under salt stress; increasing salt tolerance by enhancing selective absorption of K+ | Oomen et al., 2012 |
| O. sativa | OsHKT2;3 | Expressed in roots, basal stem, leaves, and sheaths, but no function; possibly pseudogenes | Garciadeblás et al., 2003 |
| O. sativa | OsHKT2;4 | Expressed in root epidermis, stems, internodes, vascular bundles in leaf sheath, and phloem; high affinity to K+, low affinity to Na+; ion affinity is inhibited by K+ | Horie et al., 2011; Sassi et al., 2012; Zhang et al., 2017 |
| Triticum monococcum | TmHKT1;4-A (Nax1) | Expressed in roots and sheaths; unloading Na+ from the vascular bundle; keeping Na+ in leaf sheath; unloading Na+ from the phloem | Lindsay et al., 2004; James et al., 2006 |
| T. monococcum | TmHKT1;5-A (Nax2) | Expressed in the plasma membrane of vascular tissues in root; unloading xylem Na+; reducing sodium accumulation in shoots | Byrt et al., 2007; Munns et al., 2012 |
| Triticum aestivum | TaHKT1;5-D (Kna1) | Expressed in the plasma membrane of stele cells in root; unloading Na+ from xylem; reducing Na+ accumulation in shoots | Byrt et al., 2014 |
| T. aestivum | TaHKT2;1 | Expressed in the plasma membrane of root cortex; Na+/K+ cotransporter under low Na+ condition; Na+ uniporter under high Na+ condition | Rubio et al., 1995; Gassmann et al., 1996 |
| Hordeum vulgare | HvHKT2;1 | Gene expression is up-regulated in the roots under low K+ condition; K+/Na+ cotransporter; when overexpressed in barley, the salt tolerance is enhanced, which may be related to the specific distribution of Na+ in leaves | Mian et al., 2011 |
| H. vulgare | HvHKT1;1 | Expressed in the plasma membrane of the root epidermis and steles; participating in the lateral transport of Na+ in the roots, and finally reducing the accumulation of Na+ in the shoots; involved in maintaining the balance of K+ and Ca2+ in the roots | Han et al., 2018 |
Oshkt2;1 mutant rice plants exhibited significantly reduced growth compared to wild-type plants under low Na+ and K+ conditions, and accumulated less Na+, but not less K+, in tissues, indicating that the OsHKT2;1 transporter mediates Na+ influx into K+-starved roots (Horie et al., 2007). HvHKT2;1 co-transports Na+ and K+ in Xenopus oocytes (Mian et al., 2011). In barley plants, overexpression of HvHKT2;1 can result in higher Na+ concentration in the xylem sap and more translocation of Na+ to leaves, indicating its role in regulating Na+ xylem transport (Mian et al., 2011). The functions of other members of HKT2 transporters in cereals are listed in Table 1. It is reported that HKT2 sub-family members have a glycine residue in that site (GGGG-type) and are permeable to Na+ or both Na+ and K+ transports (Mäser et al., 2002b; Platten et al., 2006; Rodríguez-Navarro and Rubio, 2006; Horie et al., 2009).
In conclusion, membrane transporters play an important role in the regulation of K+/Na+ homeostasis in plant tissues under salt stress, and their action is considered a key mechanism of salt tolerance in crops.
6. Genetic improvement of salt tolerance in crops
In the last few decades, many attempts have been made to develop salt-tolerant crop cultivars using various breeding methodologies, including conventional breeding, mutagenesis breeding, and genetic engineering. Great progress has been made in understanding the physiology and genetics of salt tolerance in several crops, such as rice and wheat (Ren et al., 2005; Munns et al., 2012), laying a solid foundation for further improvement of salt tolerance in crops. The genetic diversities of cultivated plant species and their wild relatives may provide new and elite genes that have yet to be effectively used (Shen et al., 2016, 2018; Huang et al., 2018). Transforming this knowledge into modern approaches through the tools of genomics and molecular breeding will enhance the development of tolerant cultivars, thereby enabling increased food production (Ismail and Horie, 2017).
6.1. Conventional breeding
Conventional breeding of new salt-tolerant crop cultivars has been successful in recent decades (Singh et al., 2009). Since the 1970s, the International Rice Research Institute (IRRI) has bred more than 30 salt-tolerant rice cultivars through sexual hybridization. With the development of molecular markers, traditional breeding with marker-assisted selection can shorten the breeding cycle. Success in conventional breeding relies first on the proper identification of tolerant genotypes and genes. Land races and wild relatives are considered ideal breeding materials because they exhibit great genetic diversity, including large variation in their responses to salt stress (Reynolds et al., 2005, 2007). Successful breeding for salt stress tolerance also relies on the identification of QTLs responsible for tolerance and their association with linked molecular markers, which are then used for effective marker-assisted selection. Numerous studies have been conducted to map genes or QTLs. For example, Munns et al. (2012) produced two independent BC4 lines containing Nax2 (TmHKT1;5-A) from a cross between the durum wheat cultivar Marrocos and T. monococcum accession C68-101, and a backcross with durum wheat cultivar Tamaroi (James et al., 2006; Byrt et al., 2007). Field trials on saline soils demonstrated that the presence of TmHKT1;5-A significantly reduced leaf Na+ concentration and increased durum wheat grain yield by 25% compared to near-isogenic lines without the Nax2 locus (Munns et al., 2012). Saltol is a major QTL for salt tolerance in rice (Thomson et al., 2010). A marker-assisted backcross strategy was undertaken to transfer positive alleles of Saltol from FL478 into BT7, and the rice salt tolerance was successfully improved without penalizing agronomic performance (Linh et al., 2012). Although the progress of conventional breeding has been greatly accelerated through marker-assisted selection, it is still time-consuming and relies largely on high-throughput genotyping platforms.
6.2. Mutagenesis breeding
In the last century, there were many studies on the use of plant tissue and cell cultures to obtain salt-tolerant mutants. In vitro plant cells or tissues including callus, cell suspensions, and protoplasts have been used in mutagenesis breeding. The selection of salt-tolerant cultivars by in vitro culture includes spontaneous mutation and artificially induced variation. In spontaneous mutation, NaCl, seawater, polyethylene glycol (PEG), hydroxyproline (HYP), and L-azetidine-ethyl-carboxylic acid have been used to screen for the selection of salt-tolerant cultivars (Zhao et al., 1995; Du et al., 1999; Wang and Jia, 1999). Nabors et al. (1980) used nine consecutive screens to select salt-tolerant tobacco cell lines, and the regenerated salt-tolerant plants were able to stably withstand seawater irrigation for two consecutive generations. This was the first report of an artificially bred cultivar with high salt tolerance that could be inherited by future generations through sexual reproduction. Ye et al. (1987) combined anther culture techniques with hybridization to screen for salt-tolerant barley cultivars in salt-stressed media. Genetic variation was artificially induced using physical methods (i.e., X-rays, γ-rays, β-rays, or neutrons) or chemical mutagenic agents (i.e., alkylation agents or azides), which cause a higher mutation frequency than spontaneous mutation, and can create more valuable mutants in a shorter time (Li et al., 1990; Guo et al., 1997). A successful example is the production of the barley cultivar Golden Promise, which is a γ-ray-induced semi-dwarf mutant from the cultivar Maythorpe (Forster, 2001). However, the randomness of mutation and difficulties in regenerating whole plants greatly restrict the application and development of mutagenesis breeding.
6.3. Genetic engineering
Transgenic approaches have been widely used in gene functional characterization for several decades. Genomic studies in combination with transcriptomic analysis can be used to discover new genes and reveal regulatory systems and their genomic locations (Roorkiwal et al., 2014). Alternatively, genetic transformation provides opportunities for extensive application of genetic engineering across different species, regardless of their reproductive isolation. Thus, many transgenic plants with high salt tolerance have been developed across several species (Khan et al., 2015). The overexpression of type-I H+-PPase genes enhanced salt and drought tolerance in tobacco (Nicotiana tabacum), cotton (Gossypium hirsutum), and maize (Z. mays) (Gao et al., 2006; Li et al., 2008; Lv et al., 2008, 2009). Under salt stress, HvHKT1;5 knock-down transgenic barley lines showed improved salt tolerance, dramatic decreases in Na+ translocation from roots to shoots, and increases in the K+/Na+ ratio compared with wild-type plants (Huang et al., 2020). A late embryogenesis abundant (LEA) protein gene, HvA1, from barley was introduced into rice, and the transgenic rice plants showed a significant increase in their tolerance to water and salt stresses (Xu et al., 2008b). Chen et al. (2015) also demonstrated a successful application of an inducible promoter in regulating the spatial and temporal expression of HvA1 for improving rice root architecture and multiple stress tolerance without yield reduction. However, because of public concerns about transgenic crops, further research is required to reveal whether this approach is effective for developing commercial salt-tolerant cultivars.
Genome editing is defined as the precise modification of the nucleotide sequence of the genome of an organism or cell by inserting, deleting, or replacing DNA at a specific site, usually through the use of engineered nucleases (Chen and Gao, 2014; Gao, 2015). Genome editing using zinc-finger nucleases (ZFNs) (Kim et al., 1996) and transcription activator-like effector nucleases (TALENs) (Christian et al., 2010) has been used for almost two decades, but it has recently come under the spotlight through the development of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems, which provide simplicity and ease for targeted gene editing (Jinek et al., 2012; Li et al., 2013). Technologies involving targeted genome editing using sequence-specific nucleases have great potential for crop improvement (Khatodia et al., 2016). Such technologies are particularly useful when favorable alleles associated with the specific have been identified and characterized, as is the case for salt tolerance (Ismail and Horie, 2017). So far, targeted genome editing for disease resistance has been applied many times in rice (Li et al., 2012), wheat (Wang et al., 2014), and tomato (Nekrasov et al., 2017), but only a few studies have focused on the improvement of salinity tolerance in crops through genome editing. Zhang et al. (2018) enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Zhou et al. (2017) used CRISPR-Cas9-based genome editing to obtain mutants of single microRNA (miRNA) genes (OsmiR408 and OsmiR528) and miRNA gene families (miR815a/b/c and miR820a/b/c) in rice, and revealed a positive regulator of rice salt stress tolerance, OsMIR528. With the rapid development of functional genomics, and the identification and characterization of other important genes, genome editing will provide more powerful and efficient opportunities for improving the salt tolerance of crops. Certainly, more studies should be carried out on the applications of genome editing to the precision breeding of crops.
In conclusion, various breeding methodologies, including conventional breeding, mutagenesis breeding, and genetic engineering, have been commonly used in developing salt-tolerant crop cultivars and, in future, gene editing is likely to become a dominant method.
7. Conclusions
Many studies have shown that Na+ accumulation in tissues, especially in shoots, is a major factor affecting salt tolerance, which in turn depends on Na+ uptake, long-distance transportation, and distribution in plants mediated by ion transporters like SOS1, HKT, and NHX. There are dramatic differences in salt tolerance among plant species and genotypes within a species. The wide genetic diversity paves the way for further improvement of salt tolerance in crops. Development of salt-tolerant crop cultivars relies on advanced breeding methodologies, including molecular marker-assisted breeding, mutagenesis breeding, and genetic transformation, while recently developed genome or gene editing provides a more powerful and efficient tool for crop breeding. Commonly regarded as a non-genetically modified (GM) technology, gene or genome editing will become a focus of study and use in crop breeding.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31620103912), the China Agriculture Research System (No. CARS-05), the Fundamental Research Funds for the Central Universities (No. 2019FZA6011), and the Jiangsu Collaborative Innovation Centre for Modern Crop Production (No. JCIC-MCP), China
Contributors: Lu HUANG performed literature search, figure design, writing and editing of the manuscript. De-zhi WU and Guo-ping ZHANG performed the study design and revision of the manuscript. All authors have read and approved the final manuscript. Therefore, all authors have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Lu HUANG, De-zhi WU, and Guo-ping ZHANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Ali Z, Park HC, Ali A, et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 2012;158(3):1463–1474. doi: 10.1104/pp.111.193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrés Z, Perez-Hormaeche J, Leidi EO, et al. Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci USA. 2014;111(17):E1806–E1814. doi: 10.1073/pnas.1320421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barragán V, Leidi EO, Andrés Z, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis . Plant Cell. 2012;24(3):1127–1142. doi: 10.1105/tpc.111.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassil E, Ohto MA, Esumi T, et al. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell. 2011;23(1):224–239. doi: 10.1105/tpc.110.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassil E, Tajima H, Liang YC, et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23(9):3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassil E, Coku A, Blumwald E. Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot. 2012;63(16):5727–5740. doi: 10.1093/jxb/ers250. [DOI] [PubMed] [Google Scholar]
- 7.Butcher K, Wick AF, Desutter T, et al. Soil salinity: a threat to global food security. Agron J. 2016;108(6):2189–2200. doi: 10.2134/agronj2016.06.0368. [DOI] [Google Scholar]
- 8.Byrt CS, Platten JD, Spielmeyer W, et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1 . Plant Physiol. 2007;143(4):1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrt CS, Xu B, Krishnan M, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 2014;80(3):516–526. doi: 10.1111/tpj.12651. [DOI] [PubMed] [Google Scholar]
- 10.Chen KL, Gao CX. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 2014;33(4):575–583. doi: 10.1007/s00299-013-1539-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen YS, Lo SF, Sun PK, et al. A late embryogenesis abundant protein HVA1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotechnol J. 2015;13(1):105–116. doi: 10.1111/pbi.12241. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Newman I, Zhou M, et al. Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant Cell Environ. 2005;28(10):1230–1246. doi: 10.1111/j.1365-3040.2005.01364.x. [DOI] [Google Scholar]
- 13.Chen ZH, Pottosin II, Cuin TA, et al. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 2007;145(4):1714–1725. doi: 10.1104/pp.107.110262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai F, Wang XL, Zhang XQ, et al. Assembly and analysis of a qingke reference genome demonstrate its close genetic relation to modern cultivated barley. Plant Biotechnol J. 2018;16(3):760–770. doi: 10.1111/pbi.12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davenport RJ, Muñoz-Mayor A, Jha D, et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis . Plant Cell Environ. 2007;30(4):497–507. doi: 10.1111/j.1365-3040.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 17.Diatloff E, Kumar R, Schachtman DP. Site directed mutagenesis reduces the Na+ affinity of HKT1, an Na+ energized high affinity K+ transporter. FEBS Lett. 1998;432(1-2):31–36. doi: 10.1016/S0014-5793(98)00833-3. [DOI] [PubMed] [Google Scholar]
- 18.Du LQ, Li YX, Li HJ, et al. Screening of salt tolerant watercress variants on natural seawater contained medium. Acta Bot Sin. 1999;41(6):633–639. doi: 10.3321/j.issn:1672-9072.1999.06.014. [DOI] [Google Scholar]
- 19.el Mahi H, Pérez-Hormaeche J, de Luca A, et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019;180(2):1046–1065. doi: 10.1104/pp.19.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flowers TJ, Colmer TD. Salinity tolerance in halophytes. New Phytol. 2008;179(4):945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 21.Forster BP. Mutation genetics of salt tolerance in barley: an assessment of golden promise and other semi-dwarf mutants. Euphytica. 2001;120(3):317–328. doi: 10.1023/A:1017592618298. [DOI] [Google Scholar]
- 22.Fricke W, Peters WS. The biophysics of leaf growth in salt-stressed barley. A study at the cell level. Plant Physiol. 2002;129(1):374–388. doi: 10.1104/pp.001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu LB, Shen QF, Kuang LH, et al. Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol Biochem. 2018;130:248–257. doi: 10.1016/j.plaphy.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Fu LB, Shen QF, Kuang LH, et al. Transcriptomic and alternative splicing analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Environ Exp Bot, 166:103810. 2019 doi: 10.1016/j.envexpbot.2019.103810. [DOI] [Google Scholar]
- 25.Fukuda A, Nakamura A, Hara N, et al. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta. 2011;233(1):175–188. doi: 10.1007/s00425-010-1289-4. [DOI] [PubMed] [Google Scholar]
- 26.Gao CX. Genome editing in crops: from bench to field. Natl Sci Rev. 2015;2(1):13–15. doi: 10.1093/nsr/nwu054. [DOI] [Google Scholar]
- 27.Gao F, Gao Q, Duan XG, et al. Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot. 2006;57(12):3259–3270. doi: 10.1093/jxb/erl090. [DOI] [PubMed] [Google Scholar]
- 28.Garciadeblás B, Senn ME, Bañuelos MA, et al. Sodium transport and HKT transporters: the rice model. Plant J. 2003;34(6):788–801. doi: 10.1046/j.1365-313X.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- 29.Gassmann W, Rubio F, Schroeder JI. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 1996;10(5):869–882. doi: 10.1046/j.1365-313X.1996.10050869.x. [DOI] [PubMed] [Google Scholar]
- 30.Gorham J, Jones RGW, Bristol A. Partial characterization of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta. 1990;180(4):590–597. doi: 10.1007/BF02411458. [DOI] [PubMed] [Google Scholar]
- 31.Guo FQ, Li Q, Gu RQ. Mutation, selection and comparison of several saline-tolerant wheat strains. Acta Agric Nucl Sin. 1997;11(1):1–8. (in Chinese) [Google Scholar]
- 32.Guo R, Shi LX, Yan CR, et al. Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol, 17:41. 2017 doi: 10.1186/s12870-017-0994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamam AM, Britto DT, Flam-Shepherd R, et al. Measurement of differential Na+ efflux from apical and bulk root zones of intact barley and Arabidopsis plants. Front Plant Sci, 7:272. 2016 doi: 10.3389/fpls.2016.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, Yin SY, Huang L. Towards plant salinity tolerance-implications from ion transporters and biochemical regulation. Plant Growth Regul. 2015;76(1):13–23. doi: 10.1007/s10725-014-9997-6. [DOI] [Google Scholar]
- 35.Han Y, Yin SY, Huang L, et al. A sodium transporter HvHKT1;1 confers salt tolerance in barley via regulating tissue and cell ion homeostasis. Plant Cell Physiol. 2018;59(10):1976–1989. doi: 10.1093/pcp/pcy116. [DOI] [PubMed] [Google Scholar]
- 36.Hare PD, Cress WA, van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998;21(6):535–553. doi: 10.1046/j.1365-3040.1998.00309.x. [DOI] [Google Scholar]
- 37.Horie T, Costa A, Kim TH, et al. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007;26(12):3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horie T, Hauser F, Schroeder JI. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009;14(12):660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horie T, Brodsky DE, Costa A, et al. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 2011;156(3):1493–1507. doi: 10.1104/pp.110.168047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horie T, Karahara I, Katsuhara M. Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice, 5:11. 2012 doi: 10.1186/1939-8433-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang L, Kuang LH, Li X, et al. Metabolomic and transcriptomic analyses reveal the reasons why Hordeum marinum has higher salt tolerance than Hordeum vulgare . Environ Exp Bot. 2018;156:48–61. doi: 10.1016/j.envexpbot.2018.08.019. [DOI] [Google Scholar]
- 42.Huang L, Kuang LH, Wu LY, et al. The HKT transporter HvHKT1;5 negatively regulates salt tolerance. Plant Physiol. 2020;182(1):584–596. doi: 10.1104/pp.19.00882.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang SB, Spielmeyer W, Lagudah ES, et al. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J Exp Bot. 2008;59(4):927–937. doi: 10.1093/jxb/ern033. [DOI] [PubMed] [Google Scholar]
- 44.Huertas R, Olías R, Eljakaoui Z, et al. Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 2012;35(8):1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x. [DOI] [PubMed] [Google Scholar]
- 45.Isayenkov SV. Physiological and molecular aspects of salt stress in plants. Cytol Genet. 2012;46(5):302–318. doi: 10.3103/S0095452712050040. [DOI] [Google Scholar]
- 46.Isayenkov SV. Genetic sources for the development of salt tolerance in crops. Plant Growth Regul. 2019;89(1):1–17. doi: 10.1007/s10725-019-00519-w. [DOI] [Google Scholar]
- 47.Isayenkov SV, Maathuis FJM. Plant salinity stress: many unanswered questions remain. Front Plant Sci, 10:80. 2019 doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol. 2017;68:405–434. doi: 10.1146/annurev-arplant-042916-040936. [DOI] [PubMed] [Google Scholar]
- 49.Jabnoune M, Espeout S, Mieulet D, et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009;150(4):1955–1971. doi: 10.1104/pp.109.138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James RA, Davenport RJ, Munns R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2 . Plant Physiol. 2006;142(4):1537–1547. doi: 10.1104/pp.106.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.James RA, Blake C, Byrt CS, et al. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J Exp Bot. 2011;62(8):2939–2947. doi: 10.1093/jxb/err003. [DOI] [PubMed] [Google Scholar]
- 52.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kader MA, Lindberg S. Cellular traits for sodium tolerance in rice (Oryza sativa L.) Plant Biotechnol. 2008;25(3):247–255. doi: 10.5511/plantbiotechnology.25.247. [DOI] [Google Scholar]
- 54.Kader MA, Seidel T, Golldack D, et al. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot. 2006;57(15):4257–4268. doi: 10.1093/jxb/erl199. [DOI] [PubMed] [Google Scholar]
- 55.Khan MS, Ahmad D, Khan MA. Trends in genetic engineering of plants with (Na+/H+) antiporters for salt stress tolerance. Biotechnol Biotechnol Equip. 2015;29(5):815–825. doi: 10.1080/13102818.2015.1060868. [DOI] [Google Scholar]
- 56.Khatodia S, Bhatotia K, Passricha N, et al. The CRISPR/Cas genome-editing tool: application in improvement of crops. Front Plant Sci, 7:506. 2016 doi: 10.3389/fpls.2016.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi NI, Yamaji N, Yamamoto H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017;91(4):657–670. doi: 10.1111/tpj.13595. [DOI] [PubMed] [Google Scholar]
- 59.Kronzucker HJ, Szczerba MW, Schulze LM, et al. Non-reciprocal interactions between K+ and Na+ ions in barley (Hordeum vulgare L.) J Exp Bot. 2008;59(10):2793–2801. doi: 10.1093/jxb/ern139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leidi EO, Barragán V, Rubio L, et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010;61(3):495–506. doi: 10.1111/j.1365-313X.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- 61.Li B, Wei AY, Song CX, et al. Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol J. 2008;6(2):146–159. doi: 10.1111/j.1467-7652.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 62.Li CL, Hou QM, Zeng LH, et al. Preliminary study on induction of salt-tolerant cell lines in wheat by radiation mutagenesis combined with tissue culture. J Nucl Agric Sci. 1990;1990(01):8–12. (in Chinese) [Google Scholar]
- 63.Li JF, Norville JE, Aach J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li T, Liu B, Spalding MH, et al. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 65.Ligaba A, Katsuhara M. Insights into the salt tolerance mechanism in barley (Hordeum vulgare) from comparisons of cultivars that differ in salt sensitivity. J Plant Res. 2010;123(1):105–118. doi: 10.1007/s10265-009-0272-2. [DOI] [PubMed] [Google Scholar]
- 66.Lindsay MP, Lagudah ES, Hare RA, et al. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol. 2004;31(11):1105–1114. doi: 10.1071/FP04111. [DOI] [PubMed] [Google Scholar]
- 67.Linh LH, Linh TH, Xuan TD, et al. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int J Plant Genomics. 2012;2012(66):949038. doi: 10.1155/2012/949038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lv SL, Zhang KW, Gao Q, et al. Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol. 2008;49(8):1150–1164. doi: 10.1093/pcp/pcn090. [DOI] [PubMed] [Google Scholar]
- 69.Lv SL, Lian LJ, Tao PL, et al. Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta. 2009;229(4):899–910. doi: 10.1007/s00425-008-0880-4. [DOI] [PubMed] [Google Scholar]
- 70.Martínez-Atienza J, Jiang XY, Garciadeblas B, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143(2):1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mäser P, Eckelman B, Vaidyanathan R, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1 . FEBS Lett. 2002;531(2):157–161. doi: 10.1016/S0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- 72.Mäser P, Hosoo Y, Goshima S, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA. 2002;99(9):6428–6433. doi: 10.1073/pnas.082123799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meena KK, Sorty AM, Bitla UM, et al. Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci, 8:172. 2017 doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mian A, Oomen RJFJ, Isayenkov S, et al. Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 2011;68(3):468–479. doi: 10.1111/j.1365-313X.2011.04701.x. [DOI] [PubMed] [Google Scholar]
- 75.Miller G, Suzuki N, Ciftci-Yilmaz S, et al. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33(4):453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 76.Møller IS, Gilliham M, Jha D, et al. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis . Plant Cell. 2009;21(7):2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moya JL, Gómez-Cadenas A, Primo-Millo E, et al. Chloride absorption in salt-sensitive Carrizo citrange and salt-tolerant Cleopatra mandarin citrus rootstocks is linked to water use. J Exp Bot. 2003;54(383):825–833. doi: 10.1093/jxb/erg064. [DOI] [PubMed] [Google Scholar]
- 78.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25(2):239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 79.Munns R. Genes and salt tolerance: bringing them together. New Phytol. 2005;167(3):645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 80.Munns R, Rawson HM. Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley. Aust J Plant Physiol. 1999;26(5):459–464. doi: 10.1071/PP99049. [DOI] [Google Scholar]
- 81.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 82.Munns R, Gilliham M. Salinity tolerance of crops–what is the cost? New Phytol. 2015;208(3):668–673. doi: 10.1111/nph.13519. [DOI] [PubMed] [Google Scholar]
- 83.Munns R, James RA, Xu B, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol. 2012;30(4):360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- 84.Nabors MW, Gibbs SE, Bernstein CS. NaCl-tolerant tobacco plants from cultured cells. Z Pflanzenphysiol. 1980;97(1):13–17. doi: 10.1016/S0044-328X(80)80061-4. [DOI] [Google Scholar]
- 85.Nawrot R, Barylski J, Lippmann R, et al. Combination of transcriptomic and proteomic approaches helps to unravel the protein composition of Chelidonium majus L. milky sap. Planta. 2016;244(5):1055–1064. doi: 10.1007/s00425-016-2566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nekrasov V, Wang CM, Win J, et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep, 7:482. 2017 doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nevo E, Chen GX. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010;33(4):670–685. doi: 10.1111/j.1365-3040.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 88.Oomen RJFJ, Benito B, Sentenac H, et al. HKT2;2/1, a K+-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 2012;71(5):750–762. doi: 10.1111/j.1365-313X.2012.05031.x. [DOI] [PubMed] [Google Scholar]
- 89.Passioura JB, Munns R. Rapid environmental changes that affect leaf water status induce transient surges or pauses in leaf expansion rate. Aust J Plant Physiol. 2000;27(10):941–948. doi: 10.1071/PP99207. [DOI] [Google Scholar]
- 90.Platten JD, Cotsaftis O, Berthomieu P, et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006;11(8):372–374. doi: 10.1016/j.tplants.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 91.Qiu L, Wu DZ, Ali S, et al. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor Appl Genet. 2011;122(4):695–703. doi: 10.1007/s00122-010-1479-2. [DOI] [PubMed] [Google Scholar]
- 92.Qiu QS, Guo Y, Dietrich MA, et al. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99(12):8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quintero FJ, Ohta M, Shi HZ, et al. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA. 2002;99(13):9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rajendran K, Tester M, Roy SJ. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2009;32(3):237–249. doi: 10.1111/j.1365-3040.2008.01916.x. [DOI] [PubMed] [Google Scholar]
- 95.Ren ZH, Gao JP, Li LG, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37(10):1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 96.Reynolds MP, Mujeeb-Kazi A, Sawkins M. Prospects for utilising plant-adaptive mechanisms to improve wheat and other crops in drought- and salinity-prone environments. Ann Appl Biol. 2005;146(2):239–259. doi: 10.1111/j.1744-7348.2005.040058.x. [DOI] [Google Scholar]
- 97.Reynolds M, Dreccer F, Trethowan R. Drought-adaptive traits derived from wheat wild relatives and landraces. J Exp Bot. 2007;58(2):177–186. doi: 10.1093/jxb/erl250. [DOI] [PubMed] [Google Scholar]
- 98.Rodríguez-Navarro A. Potassium transport in fungi and plants. Biochim Biophys Acta. 2000;1469(1):1–30. doi: 10.1016/S0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 99.Rodríguez-Navarro A, Rubio F. High-affinity potassium and sodium transport systems in plants. J Exp Bot. 2006;57(5):1149–1160. doi: 10.1093/jxb/erj068. [DOI] [PubMed] [Google Scholar]
- 100.Rodríguez-Rosales MP, Jiang XY, Gálvez FJ, et al. Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 2008;179(2):366–377. doi: 10.1111/j.1469-8137.2008.02461.x. [DOI] [PubMed] [Google Scholar]
- 101.Roorkiwal M, Nayak SN, Thudi M, et al. Allele diversity for abiotic stress responsive candidate genes in chickpea reference set using gene based SNP markers. Front Plant Sci, 5:248. 2014 doi: 10.3389/fpls.2014.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 103.Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270(5242):1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 104.Rubio F, Schwarz M, Gassmann W, et al. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J Biol Chem. 1999;274(11):6839–6847. doi: 10.1074/jbc.274.11.6839. [DOI] [PubMed] [Google Scholar]
- 105.Sassi A, Mieulet D, Khan I, et al. The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiol. 2012;160(1):498–510. doi: 10.1104/pp.112.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schachtman DP, Schroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370(6491):655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- 107.Schachtman DP, Schroeder JI, Lucas WJ, et al. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science. 1992;258(5088):1654–1658. doi: 10.1126/science.8966547. [DOI] [PubMed] [Google Scholar]
- 108.Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot. 2013;112(7):1209–1221. doi: 10.1093/aob/mct205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shabala S, Shabala S, Cuin TA, et al. Xylem ionic relations and salinity tolerance in barley. Plant J. 2010;61(5):839–853. doi: 10.1111/j.1365-313X.2009.04110.x. [DOI] [PubMed] [Google Scholar]
- 110.Shabala S, Bose J, Hedrich R. Salt bladders: do they matter? Trends Plant Sci. 2014;19(11):687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 111.Shen QF, Fu LB, Dai F, et al. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genomics, 17:889. 2016 doi: 10.1186/s12864-016-3242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen QF, Fu LB, Qiu L, et al. Time-course of ionic responses and proteomic analysis of a Tibetan wild barley at early stage under salt stress. Plant Growth Regul. 2017;81(1):11–21. doi: 10.1007/s10725-016-0180-0. [DOI] [Google Scholar]
- 113.Shen QF, Yu JH, Fu LB, et al. Ionomic, metabolomic and proteomic analyses reveal molecular mechanisms of root adaption to salt stress in Tibetan wild barley. Plant Physiol Biochem. 2018;123:319–330. doi: 10.1016/j.plaphy.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 114.Shi HZ, Ishitani M, Kim C, et al. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi HZ, Quintero FJ, Pardo JM, et al. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14(2):465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singh RK, Redoña E, Refuerzo L. Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice. In: Pareek A Sopory SK, Bohnert BH et al., editors. Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation. Springer, Dordrecht; 2009. pp. 387–415. [Google Scholar]
- 117.Suzuki K, Yamaji N, Costa A, et al. OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol, 16:22. 2016 doi: 10.1186/s12870-016-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91(5):503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thomson MJ, de Ocampo M, Egdane J, et al. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3(2):148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- 120.Wang FB, Liu JC, Zhou LJ, et al. Senescence-specific change in ROS scavenging enzyme activities and regulation of various SOD isozymes to ROS levels in psf mutant rice leaves. Plant Physiol Biochem. 2016;109:248–261. doi: 10.1016/j.plaphy.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 121.Wang H, Zhang MS, Guo R, et al. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.) BMC Plant Biol, 12:194. 2012 doi: 10.1186/1471-2229-12-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y, Jia JF. Selection and characteristics analysis of NaCl-tolerant cell line from Astragalus adsurgens callus cultures. Chin J Appl Environ Biol. 1999;5(6):547–550. (in Chinese) [Google Scholar]
- 123.Wang YP, Cheng X, Shan QW, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 124.Waters S, Gilliham M, Hrmova M. Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: structural insights to probe differences in ion selectivity. Int J Mol Sci. 2013;14(4):7660–7680. doi: 10.3390/ijms14047660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu DZ, Shen QF, Cai SG, et al. Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 2013;54(12):1976–1988. doi: 10.1093/pcp/pct134. [DOI] [PubMed] [Google Scholar]
- 126.Wu DZ, Cai SG, Chen MX, et al. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS ONE. 2013;8(1):e55431. doi: 10.1371/journal.pone.0055431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu DZ, Shen QF, Qiu L, et al. Identification of proteins associated with ion homeostasis and salt tolerance in barley. Proteomics. 2014;14(11):1381–1392. doi: 10.1002/pmic.201300221. [DOI] [PubMed] [Google Scholar]
- 128.Xu D, Duan X, Xue QZ, et al. Transformation of rice with agronomically useful genes toward production of insect-resistant and water stress-tolerant plants. Proceedings of the 3rd International Rice Genetics Symposium, Manila, Philippines; 2008. pp. 796–803. [Google Scholar]
- 129.Xu HX, Jiang XY, Zhan KH, et al. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch Biochem Biophys. 2008;473(1):8–15. doi: 10.1016/j.abb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 130.Yang Q, Chen ZZ, Zhou XF, et al. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis . Mol Plant. 2009;2(1):22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yao X, Horie T, Xue SW, et al. Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol. 2010;152(1):341–355. doi: 10.1104/pp.109.145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ye JM, Kao KN, Harvey BL, et al. Screening salt-tolerant barley genotypes via F1 anther culture in salt stress media. Theor Apple Genet. 1987;74(4):426–429. doi: 10.1007/BF00289816. [DOI] [PubMed] [Google Scholar]
- 133.Yu LJ, Nie JN, Cao CY, et al. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana . New Phytol. 2010;188(3):762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- 134.Zahra J, Nazim H, Cai SG, et al. The influence of salinity on cell ultrastructures and photosynthetic apparatus of barley genotypes differing in salt stress tolerance. Acta Physiol Plant. 2014;36(5):1261–1269. doi: 10.1007/s11738-014-1506-z. [DOI] [Google Scholar]
- 135.Zhang C, Li HJ, Wang JY, et al. The rice high-affinity K+ transporter OsHKT2;4 mediates Mg2+ homeostasis under high-Mg2+ conditions in transgenic Arabidopsis . Front Plant Sci, 8:1823. 2017 doi: 10.3389/fpls.2017.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y, Massel K, Godwin ID, et al. Applications and potential of genome editing in crop improvement. Genome Biol, 19:210. 2018 doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang YM, Zhang HM, Liu ZH, et al. The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the retention capacity of intracellular potassium. Plant Mol Biol. 2015;87(3):317–327. doi: 10.1007/s11103-014-0278-6. [DOI] [PubMed] [Google Scholar]
- 138.Zhao RT, Gao SG, Qiao YK, et al. Studies on the application of anther culture in salt-tolerance breeding in wheat (Triticum aestivum L.) Acta Agron Sin. 1995;21(2):230–234. (in Chinese) [Google Scholar]
- 139.Zhou JP, Deng KJ, Cheng Y, et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front Plant Sci, 8:1598. 2017 doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6(2):66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 141.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6(5):441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]