Abstract
Plant breeding is well recognized as one of the most important means to meet food security challenges caused by the ever-increasing world population. During the past three decades, plant breeding has been empowered by both new knowledge on trait development and regulation (e.g., functional genomics) and new technologies (e.g., biotechnologies and phenomics). Gene editing, particularly by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) and its variants, has become a powerful technology in plant research and may become a game-changer in plant breeding. Traits are conferred by coding and non-coding genes. From this perspective, we propose different editing strategies for these two types of genes. The activity of an encoded enzyme and its quantity are regulated at transcriptional and post-transcriptional, as well as translational and post-translational, levels. Different strategies are proposed to intervene to generate gene functional variations and consequently phenotype changes. For non-coding genes, trait modification could be achieved by regulating transcription of their own or target genes via gene editing. Also included is a scheme of protoplast editing to make gene editing more applicable in plant breeding. In summary, this review provides breeders with a host of options to translate gene biology into practical breeding strategies, i.e., to use gene editing as a mechanism to commercialize gene biology in plant breeding.
Keywords: Gene editing, Expression regulation, Novel allele, Trait development, Plant breeding
1. Introduction
Plant breeding is widely expected to help meet the challenge of feeding a population of 10 billion people on earth (Hickey et al., 2019). The fulfillment of this magnificent task will rely on advances and breakthroughs in all fields related to plant breeding, including new techniques yet to be innovated. Gene editing has recently emerged as such a breakthrough technology. It was not even dreamed of ten years ago in this field. Gene editing started with zinc-finger nucleases (ZFNs) (Lloyd et al., 2005) and later, transcription activator-like effector nucleases (TALENs) (Cermak et al., 2011; Mahfouz et al., 2011). It is now dominated by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) technology (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). Although the CRISPR/Cas technology has been in being for only seven years, its development as a new technology has been unprecedented. Many CRISPR variant technologies have already been developed and quickly deployed for basic and applied research, including plant breeding (Li et al., 2013; Chavez et al., 2015; Piatek et al., 2015; Xie et al., 2015; Zetsche et al., 2015; Endo et al., 2016; Kleinstiver et al., 2016; Lin et al., 2016; Minkenberg et al., 2017; Shimatani et al., 2017; Tang L et al., 2017; Hu et al., 2018; Nishimasu et al., 2018; Ren et al., 2019).
The progress on and perspectives of genomic editing in plant breeding have already been extensively reviewed recently (Zhang H et al., 2017; Pandiarajan and Grover, 2018; Zaidi et al., 2018; Eş et al., 2019; Hua et al., 2019; Mao et al., 2019; Molla and Yang, 2019; Kausch et al., 2019; Zhang YX et al., 2019; Zimny et al., 2019). Some reviews were published almost simultaneously but in different journals, and hence the key information, insights, and perspectives conveyed in these reviews overlapped to some extent. These reviews mainly covered topics such as: (1) the ever-developing gene editing technologies adapted to plant systems; (2) the applications of gene editing in plant breeding for different breeding objectives; (3) challenges and perspectives of gene editing in plant breeding. In summary, it is now well recognized that gene editing has a broad spectrum of applications, from knocking out a gene to quickly generating a recessive but desired trait (such as fragrance, early maturity) (Shan et al., 2015; Xu et al., 2016; Soyk et al., 2017), to generating new alleles by nucleotide substitution to create a novel trait (Li et al., 2016; Hua et al., 2018; Zong et al., 2018), to quantitatively regulating the expression of a particular gene to increase stress tolerance or content of desired metabolites (Li et al., 2012; Čermák et al., 2015; Jia et al., 2017), to name a few.
In parallel with rapid technical advance in plant research, study on the mechanism of how a gene exerts its function and how a gene is regulated, i.e., gene biology, has also experienced tremendous progress in the past two decades. However, due to the lack/incomprehension of such knowledge, and in most cases lack of proper techniques to make use of, plant breeders at large have not taken advantage of this knowledge in their breeding programs. Although some of the mechanisms have already been integrated into gene editing for generation of novel traits, a comprehensive examination of how to make use of gene biology in gene editing-facilitated plant breeding has been lacking. In this perspective paper, we review the current state of plant gene biology research, from gene structure to transcription and post-transcription regulations, to translation and post-translation modifications, and propose various gene editing strategies that could be deployed to generate novel alleles of a given gene for a particular purpose in plant breeding. That is, to use gene editing to translate gene biology into practical breeding strategies, i.e., to use gene editing as a mechanism to commercialize gene biology in plant breeding.
2. A brief introduction to gene biology: type of genes, functioning mechanisms, and regulation at various levels
Our understanding of a gene and how it functions has evolved drastically since the term “gene” was first introduced into biology. Nowadays, genes are divided into two basic types: coding and non-coding. Proteins encoded by coding genes also vary greatly in their function and can be divided into different categories: (1) enzymes involved in biochemical catalytic reactions in metabolism (classical enzymes) and (2) enzymes specialized in modification of proteins, RNAs, and DNAs; (3) non-enzyme proteins such as transporters of ions and macromolecules and (4) non-enzyme proteins working together with other proteins in biochemical process or gene regulation, to name a few. Non-coding genes can also be further classified into long non-coding genes and small RNA genes. Genes of different types function in distinct ways to confer a phenotype. Consequently, different strategies could be developed to manipulate their function to achieve a specific objective in plant breeding.
For a coding gene to confer a phenotype in plants, it should first be transcribed into RNA, processed into messenger RNA (mRNA), and translated into a peptide, which should be modified before becoming active, or be transported into proper organelles for functioning. Therefore, the function of a coding gene could be regulated at transcriptional, post-transcriptional, and translational levels, and further modified post-translationally. For non-coding genes, their products are often involved in regulation of other genes through distinct or as yet unknown mechanisms, and hence their function could also be regulated or modified. In the following sections, the biological basis of gene expression and regulation is elaborated in more detail with practical examples when available, and respective ways to modify traits by gene editing are proposed for plant breeding.
3. Gene editing for transcription and post-transcription regulations
Transcription is a process in which DNA is transcribed into RNA, which involves three steps, i.e., initiation, elongation, and termination. Transcription begins with the binding of RNA polymerase to DNA at the promoter region, and RNA polymerase works together with transcriptional factor (TF) in plants. The TFs can either bind with the RNA polymerase to form complexes or directly bind with the specific sequence on the promoter region. Therefore, the transcription of a gene of interest (GOI) could be altered by editing either its sequence (e.g., promoter, terminator) or its TFs. Transcribed RNA is processed into mRNA for translation, and the stability of mRNA is dependent on sequences in the untranslated regions (UTRs) as well as others.
3.1. Editing for transcription
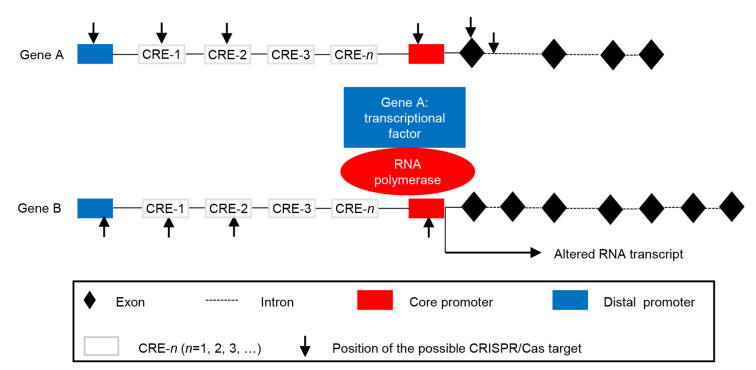
Functionally, the promoter region consists of three important components, i.e., the core/minimal, distal, and proximal promoter regions. The core/minimal promoter where the TATA box, initiator, and B recognition element (BRE) reside is the sequence that is necessary for initiating transcription. The distal promoter contains enhancers or silencers and can also influence the expression of a gene. The proximal promoter region has several TF-binding sites (TFBSs) adjacent to the core promoter sequence (Fig. 1). Cis-regulatory elements (CREs) are known to be one or more TFBSs in the proximal/distal promoter region (Pandiarajan and Grover, 2018).
Fig. 1.
Schematic presentation of a gene and its transcription, and important targets on the promoter or coding region of a gene directly confering a trait (gene B) and its transcription factor (gene A) for transcription editing
CRE: Cis regulatory element; CRISPR: clustered regularly interspaced short palindromic repeat; Cas: CRISPR-associated protein
In the promoter region, many different types of responsive elements have been reported that can induce transcription when stimulated by various biotic or abiotic stress, such as a responsive element to oxidative stress, environmental stress (light response elements (LREs), heat-shock element (HSE)), or pathogen (HSR203 responsive element (HSRE)) and responsive elements to different hormones (such as abscisic acid (ABA)-responsive element (ABRE) and gibberellin (GA)-responsive element (GARE)). The transcription proceeds for the elongation with the RNA polymerase after initiation and the template DNA strand is transcribed into RNA. At termination, RNA polymerase releases from DNA and the transcription stops. Therefore, gene transcription can be regulated by inhibiting the initial binding of the promoter with the RNA polymerase and TFs. This can be achieved by targeting the known vital elements on the promoter or unknown elements analyzed by the websites through the CRISPR/Cas technology.
Many important traits have evolved from sequence changes in the promoter region during the course of crop domestication, e.g., grain color (Espley et al., 2009; Butelli et al., 2012; Oikawa et al., 2015) and shape (Li et al., 2011; Wang et al., 2012, 2015; Xu et al., 2015), and resistance to disease (Li et al., 2017). They often resulted from altered expression levels of genes responsive to hormonal or environmental signals. In practice, it has been proven that genome editing can be used to change gene transcription by editing CREs of the GOI (gene B in Fig. 1), or by knocking out or altering transcription of its TFs (gene A in Fig. 1), thus improving stress tolerance, fruit size, and disease resistance in crops, such as rice, wheat, and tomato (Wang YP et al., 2014; Wang FJ et al., 2016; Rodriguez-Leal et al., 2017; Shi et al., 2017).
3.2. Editing of UTR
The UTR region contains the sequences ahead of the translation initiation codon ATG (5'-UTR) and after the stop codon (3'-UTR) in mature mRNA. 5'-UTR works as the entry point for the ribosome during translation of mRNA and contains elements that can interact with thermal sensors and other classes of translation or transcription attenuators, and hence plays crucial roles in the regulation of gene transcription through influencing the mRNA stability in response to environmental stimuli (Leppek et al., 2018; Al-Zeer et al., 2019). Therefore, the transcription of a GOI could theoretically be altered by targeted mutation in UTRs.
3.3. Editing for post-transcriptional regulation
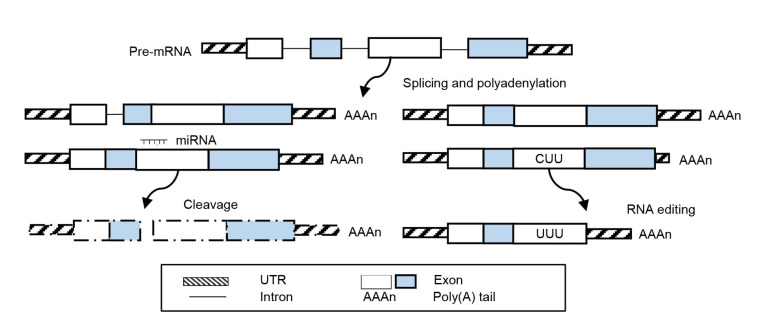
Many genes are subject to post-transcriptional regulation in plants, e.g., alternative splicing (AS), alternative polyadenylation (APA), and microRNA (miRNA) digestion (Fig. 2), which affects transcript processing and protein translation (Deng and Cao, 2017).
Fig. 2.
Schematic overview of post-transcriptional gene regulation events that can be theoretically controlled by CRISPR/Cas9 technology in plants, including alternative splicing (AS), miRNA-mediated mRNA cleavage, RNA editing, and alternative polyadenylation (APA)
CRISPR: clustered regularly interspaced short palindromic repeat; Cas: CRISPR-associated protein; miRNA: microRNA; mRNA: messenger RNA; UTR: untranslated region
AS rearranges the pattern of intron and exon elements in mRNAs to direct the decay of a special RNA variant. This enables a gene to encode diverse protein isoforms (Filichkin et al., 2015). For example, one of the florigen genes in temperate grasses is capable of producing two splicing isoforms that play antagonistic roles in flowering control, suggestive of a pervasive phenomenon by AS to confer a gene with different cellular functions or properties (Qin et al., 2017). Most eukaryotic splicing processes of pre-mRNAs observe a GU/AG rule (Reddy et al., 2013), and thus it can be modified by a CRISPR/Cas9 technology. The recently developed Cas9-directed base editor, which can introduce desired point mutation, has been shown to be an ideally efficient way to manipulate splicing outcomes. For example, hypersensitive to ABA1 (HAB1), a phosphatase 2C gene in Arabidopsis, that can generate two isoforms playing entirely opposite roles in the GA signaling, has successfully produced a retention of intron (Xue et al., 2018). The ABA hypersensitive phenotype of transgenic mutants demonstrated that generation of HAB1 AS isoforms had been completely prevented through single-base substitutions (Xue et al., 2018). As the base editing tools with non-specific protospacer adjacent motif (PAM) properties are becoming available, manipulating pre-mRNA splicing via CRISPR-directed base editors will gain wide application in mRNA isoform functional studies in crops.
Similar to transcriptional control, the strategy of gene editing, especially by CRISPR/Cas9, has now revealed significant insights into post-transcriptional effects.
miRNAs, a class of 20-to 24-nt RNAs generated from a stem-loop structure, play regulatory roles in a wide range of biological processes in eukaryotes (Wu et al., 2010). Besides deletion of the full miRNA gene locus, making indels in miRNA precursors by CRISPR/Cas9 could also readily abolish miRNA functions, because the secondary structure of the miRNA precursor is critical for the efficiency of miRNA formation. Generally, plant miRNAs have almost perfect sequence complementarity with targets. On the one hand, it is simple to destroy the regulatory module of miRNA and targets. This is done through the introduction of indels where miRNA and target pairs, because of the matching degree at miRNA and target sites, especially in the 5' end (positions 2 to 8) and central region (positions 10 and 11) of miRNA, are crucial for the efficiency of miRNA-mediated cleavage. On the other hand, it is also possible to artificially fine-tune the target expression through adjustment of complementary miRNA and target sites by precise base editing.
In a recent study, Zhang’s group attempted to produce miR528 loss-of-function mutants through CRISPR/Cas9-based genome editing (Zhou et al., 2017). The transgenic plants with 3-bp deletion in miR528 precursor were, as expected, observed to compromise miR528 function in salt stress response. However, those with 1-bp insertion in miR528 precursor have no obvious effects. This interesting finding indicates that creating miRNA locus with large deletions is more credible than engineering miRNA precursors to obtain miRNA loss-of-function mutants via CRISPR/Cas9.
Another example is to fine-tune plant architecture by miRNA editing. Promotion of high resource use efficiency via ideal plant architecture (IPA) is an attractive way to increase crop yield potential (Jiao et al., 2010). Rice SQUAMOSA promoter-binding-like 14 (OsSPL14), an SQUAMOSA promoter-binding protein (SBP)-domain transcription factor targeted by miR156, is the locus of IPA1 and can be exploited to increase grain yield through balancing panicle size and tiller number, if it is optimally expressed (Zhang L et al., 2017). Although selection of novel promoter alleles in natural or mutagenized populations has been suggested as a useful way to control the IPA1 expression pattern, genome editing at miR156 and IPA1 target site would be an alternative way to better fine-tune IPA1 to design elite super rice varieties.
Like genomic editing, RNA editing is also a mode of nucleotide modification. However, it is usually a natural phenomenon occurring at the RNA rather than DNA nucleotides from their genome-encoded sequence, which is acting as an indirect repair mechanism that corrects DNA mutations at the RNA level (Takenaka et al., 2013). In flowering plants, mRNA editing is generally performed as a C-to-U alteration, and subsequently results in a change of protein amino acids or gene expression via AS or miRNA-mediated degradation (Takenaka et al., 2013). Since RNA editing is not always emergent, people can directly edit a DNA nucleotide sequence at an RNA editing site by CRISPR/Cas9 to artificially control the physiology caused by the occasional RNA editing event.
APA is a widespread gene regulation mechanism that generates mRNA with different 3'-UTRs, allowing them to be differentially degraded or interact with diverse sets of RNA regulators such as RNA-binding proteins and miRNAs (Hunt, 2014). Because APA generally occurs at different sites enriched with poly(A), it is conceivable to change APA events through deletion or alternation of poly(A) fragments by the CRISPR/Cas9 method to form truncated mRNA isoforms with different adenylations
In theory, gene editing could be used in all aspects of post-transcription regulation; however, there is no report yet on success of APA or RNA editing events so far by CRISPR/Cas genome editing, and even applications of gene editing on miRNA and AS are still very limited. It may well be in that the single nucleotide editing efficiency is not yet high enough. Nevertheless, with the progress made via the breakthroughs of base editing tools, genome editing promises to contribute a lot to manipulating gene expression explicitly at post-transcription levels.
4. Gene editing for translation, post-translation modification, and protein localization
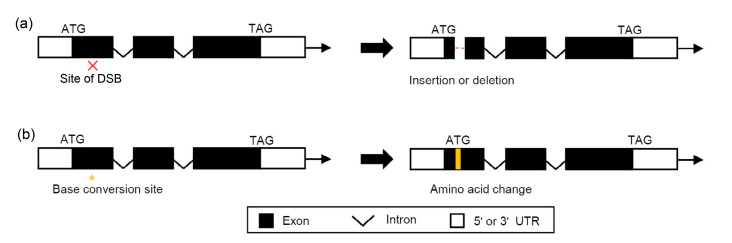
Classical gene editing often results in indel mutations in the targeted genome region, and when it occurs in the exonic region, it would completely or partially knock out a protein-coding gene. Indeed, many important traits, selected by farmers during the domestication process of various crops, are controlled by recessive alleles, such as the “green revolution” gene semi-dwarf 1 (sd1), the fragrance gene betaine aldehyde dehydrogenase 2 (badh2), and the long grain gene grain size 3 (gs3) in rice. Hence, gene editing has already been successfully applied to quickly breed a trait that is in demand, such as fragrance, waxy rice, low phytic acid, and low Cd accumulation (Shan et al., 2015; Sun et al., 2017; Tang X et al., 2017; Zhang JS et al., 2018; Zhou et al., 2018; Jiang et al., 2019; Liu et al., 2019). By using base editing techniques, the amino acid sequence could also be modified to produce partial knockout or gain-of-function mutations through various mechanisms (Fig. 3).
Fig. 3.
Generation of gain-or loss-of-function mutations using genome editing tools
(a) Frameshift mutations cause loss-of-function alleles; (b) Amino acid substitutions cause partial loss-of-function or gain-of-function alleles. DSB: double-strand break; UTR: untranslated region
4.1. Upstream ORF editing for regulating of primary ORF translation
It is now considered as a general mechanism that upstream ORFs (uORFs) control the amount of protein that is synthesized from their downstream primary ORFs (pORFs). According to von Arnim et al. (2014), from about 30% to more than 40% transcripts harbor uORFs in plants, including important crop plants such as maize and rice. By demonstrating that genome editing of endogenous uORFs could enable the modulation of translation of mRNAs from four pORFs that are involved in either development or antioxidant biosynthesis, Zhang HW et al. (2018) proposed that gene editing could be a generalized instrument for translation regulation in plants.
4.2. Editing for protein activation and degradation
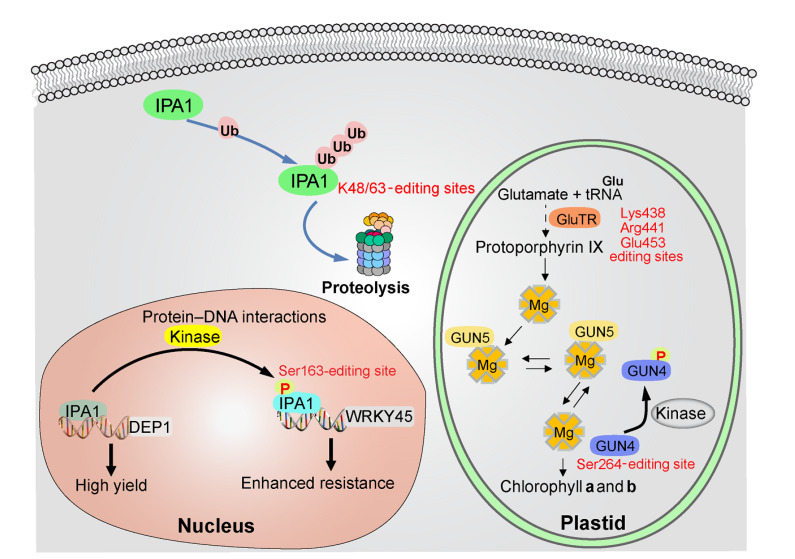
After being transcribed and translated, the complexity of genetic information encoded in DNA is largely increased by multiple post-translational modifications (PTMs) (Fig. 4). PTMs are chemical alterations to protein structure, typically catalyzed by substrate-specific enzymes, which themselves are under strict control by PTMs. Due to a lot of types of PTMs, there is a rich diversity of gene products (Deribe et al., 2010). PTMs might induce conformational changes or form a docking site to mediate molecular recognition and stabilize protein–ligand and protein–protein interactions. By reversible multisite PTMs, they can rapidly and dynamically regulate protein turnover, localization, and activation, and thus dynamically coordinate developmental processes (Huber and Hardin, 2004; Ytterberg and Jensen, 2010; Duan and Walther, 2015). PTMs play the key role in plant development stages. For example, GENOMES UNCOUPLED 4 (GUN4) is a positive regulator of light-dependent chlorophyll biosynthesis. GUN4 activates Mg chelatase (MgCh) that catalyzes the insertion of an Mg2+ ion into protoporphyrin IX. Arabidopsis thaliana GUN4 is phosphorylated at Ser264 (S264), the penultimate amino acid residue at the C terminus. Phosphorylation of GUN4 alters stimulation of MgCh activity in angiosperms (Richter et al., 2016). The IPA1 gene encodes OsSPL14, a SBP-domain transcription factor, and promotes both yield and disease resistance in rice (Zhang H et al., 2017; Wang J et al., 2018). Wang B et al. (2018) reported that ubiquitination and phosphorylation modifications could regulate IPA1 function at the post-transcriptional level. For example, the RING-finger E3 ligase stabilizes IPA1 in shoot apexes through K63-linked polyubiquitination, but it promotes the degradation of IPA1 in panicles through K48-linked polyubiquitination. Moreover, phosphorylated IPA1 at amino acid Ser163 activates the expression of pathogen defense gene WRKY45, leading to enhanced disease resistance, which indicate that fine-modifying IPA1 protein leads to an “ideal” rice plant with fewer tillers, better lodging resistance, and enhanced grain yield. Thus, new directions of research are beginning to examine how we can exploit genome editing to benefit crop breeding by targeting the key sites in PTMs. Although some technical hurdles still remain to be overcome, we probably can apply CRISPR/Cas technology to simultaneously editing of multiple phosphorylation or ubiquitination sites of IPA1 or other key regulators in a precise and efficient manner, dynamic modulating of key protein activity or abundance, redirecting plant development in a multifunctional way, and providing new insights to a new level of plant synthetic biology.
Fig. 4.
Potential post-translationally modified sites involved in phosphorylation and ubiquitination processes using genome editing
IPA1: ideal plant architecture 1; Glu: glutamate; Lys: lysine; Arg: arginine; Ser: serine; tRNA: transfer RNA; GluTR: Glu-tRNA reductase; GUN: GENOMES UNCOUPLED; K48/63: lysine 48/63; DEP1: dense and erect panicle 1; Ub: ubiquitination
4.3. Editing for modification of protein–protein interaction
The term “protein–protein interaction” encompasses a variety of events, such as transient and stable complexes. The interaction between the proteins can be relevant to a variety of biological processes including metabolic and signaling pathways, stress responses, plant defense, and organismal systems (Morsy et al., 2008; Jorrín-Novo et al., 2009). For example, the nicotinamide adenine dinucleotide phosphate (NADPH)-dependent reduction of glutamyl-transfer RNA (tRNA) catalyzed by glutamyl-tRNA reductase (GluTR) is the rate-limiting enzyme of 5-aminolevulinic acid (ALA) formation and chlorophyll biosynthesis (Beale, 1999). Three proteins are known to regulate GluTR activity in chloroplasts, fluorescent (FLU) protein (Meskauskiene et al., 2001; Goslings et al., 2004; Kauss et al., 2012), the GluTR-binding protein (GBP, OsGBP) (Czarnecki et al., 2011), and the caseinolytic protease (Clp) (Apitz et al., 2016). The tetratricopeptide repeat (TPR)-containing protein FLU is a negative regulator of chlorophyll biosynthesis in plants. It directly interacts through its TPR domain with GluTR (Zhang et al., 2015). Three sites (Arg450, Arg485, and Arg500) in the C terminal of GluTR and three sites (Glu284, Asp291, and Asp307) in the TPR3 domain of FLU play important roles in protein–protein interaction in Arabidopsis (Zhang et al., 2015). We can use a genome editing tool for single-base substitution to edit these sites to vary the interaction ability between GluTR and FLU in crops, which may change the ALA biosynthesis, chlorophyll biosynthesis rate, or even plant stress resistance. This paves the way for achieving functional modifications rather than total knock-out.
5. Protoplast editing: making gene editing more applicable in plant breeding
In animals, genome editing is often DNA-free because guide RNA (gRNA) and Cas9 were delivered into cells in the form of pre-assembled ribonucleoproteins (RNPs) via electroporation (Kim et al., 2014; Liang et al., 2015). However, in plants, the presence of a cell wall makes it impossible to use transfection or electroporation for nucleic acid and/or protein delivery. Transient protoplast transfection has also been used for evaluation of gene editing reagents in various crops such as rice and tomato (Shan et al., 2014; Čermák et al., 2015; Woo et al., 2015; Lin et al., 2018), but because of the lack of a reasonably efficient protoplast regeneration system for almost all crop plants, gene editing via the protoplast seems not feasible yet for plant breeding. However, RNP-based protoplast editing has numerous advantages over the current transgenic plant-based method, and hence efforts should continue to be made to make it work in crops.
5.1. Advantages of RNP-based protoplast editing
Transient delivery systems for transgene-free and DNA-free genome editing show a promising advantage in crop genetic modification. Based on lots of published data the mutagenesis efficiency in the protoplast looks higher than that in the plant-or tissue-based genome editing, which relies on traditional genetic engineering techniques (originally including gene guns, electroporation, and agrobacterium (Li et al., 2013; Lin et al., 2018)). To avoid some disadvantages of the expression of Cas9 and gRNA from genome-inserted plasmid DNA, an efficient plasmid-free genome editing system has been developed using pre-assembled RNPs (containing Cas9/single-guide RNA (sgRNA)) in animal and plants (Liang et al., 2015; Woo et al., 2015). The new strategy achieved almost comparable efficiency to the plasmid-based transgenic systems with low off-target frequency. After transfection, the RNP can cleave the target immediately. The transcriptional or translational machinery is not required. Importantly, the RNP complex is degraded quickly, to achieve another valuable advantage with fewer off-target mutations. The rate of regenerated mutants was up to 46% in lettuce, according to the published data (Woo et al., 2015). Sheen lab made progress with a DNA-free genome editing system with higher efficiency, approximately 99% (Sheen, pers. comm.).
Multiplex gene editing by CRISPR/Cas9 provides a powerful tool for targeting members of multigene families. Although previous studies have shown that multiplex gene editing in plants is possible with CRISPR/Cas9 (Xie et al., 2015) or CRISPR/centromere and promoter factor 1 (Cpf1) (Wang et al., 2017), the Cas9 system requires large constructs to express multiple sgRNA cassettes, which is more laborious to construct and may cause instability and reduce transformation efficiency. Engineered CRISPR/Cpf1 with a simple short direct repeat (DR)-guide array still has the disadvantage of low cleave efficiency and off-target mutation. For pre-assembled RNPs one can synthesize multiplex sgRNAs, with, theoretically, no limitation for the RNP complex. This will demonstrate the feasibility of high-efficiency multiplex gene editing in the protoplast.
Currently, most advances in CRISPR/Cas application in crops focus on single or multigene knockouts, and chromosomal deletions, which exclude some “bad” genes or Cis-elements. In wheat, mutations of three mildew-resistance locus O (MLO) homeologs by CRISPR/Cas9 result in improved resistance to Blumeria graminis f. sp. tritici infection (Wang et al., 2014). Also editing of an α-Kafirin gene family increases digestibility and protein quality in sorghum (Li et al., 2018). These strategies are based on CRISPR/Cas-targeted modification of susceptibility genes in crop species. Compared with the great progress of genome editing in animals, the big challenge in plants is in-frame gene knock-ins by the CRISPR/Cas system, which can produce a “gain of function” and facilitate breeding by introducing new alleles faster or generating allelic variants that do not exist naturally. Also, knock-in can be used to alter multiple elite traits by stacking genes in a single variety, which have great value for crop trait improvement (Chen et al., 2019). Apparently, gene knock-in with a protoplast system shows more advantages in higher efficiency and accuracy than the traditional method. The Sheen lab at Harvard University successfully in-frame integrated the exogenous tags in Arabidopsis and the tobacco genomes (Sheen, pers. comm.), because using preassembled RNP complexes consisting of Cas9 protein and sgRNAs, the complexes were introduced directly into protoplasts using a polyethylene glycol-based method and have shown precise and efficient gene knock-out (Woo et al., 2015) or knock-in (Sheen, pers. comm.). The previous advantages and pitfalls give us a perspective on CRISPR/Cas that might be safely used for the design of genetic modified crops as with hybrid crops.
5.2. Paradigm-changing technique for protoplast regeneration
There is a problem in crop regeneration after gene editing in the protoplast. Though some papers claimed that they regenerated some crops from the protoplast, as is known the regeneration efficiency is far from providing a feasible application. How to regenerate a plant from a single protoplast? Actually, in animals, pluripotent stem cells (also known as induced pluripotent stem (iPS) cells) are derived directly from adult tissues, and induced by some kinases, transcription factors, or other components (Takahashi and Yamanaka, 2006). Unlike animals, plant stem cells are innately undifferentiated cells located in the meristems of plants during the whole life cycle (Pierre-Jerome et al., 2018). However, the tissue is limited and surrounded by differentiated cells. Importantly, these innate stem cells are changed at the cell characteristics, cell polarity, epigenetic modification, and physiological levels after removing the cell wall. Compared with animal cells, plant cells also can be induced to homogeneous embryonic stem cells in Arabidopsis and tobacco. With these cells, we can manipulate the genome rewriting easily and improve the regeneration at higher efficiency in Arabidopsis and tobacco (data not shown; rice is a potential model, which is shown in Fig. 5). Whether it works well in main crops such as rice, maize, wheat, soybean, and barley remains to be seen. It remains challenging but would be rewarding if achieved. Another question is what the protoplast-regenerated plants look like? The plants germinate from a seed and experience programed genetic and epigenetic modifications to produce environmentally adaptable plants. If we produce a somatic regenerated plant, the agronomic traits and adaptability will be doubtful, even though the reported protoplasts in regenerated tobacco look good (Lin et al., 2018). Another group found that regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability (Fossi et al., 2019), so more detailed information is needed before application. CRISPR/Cas is a sophisticated toolkit, but still under development and evolution. More and more researchers are working on the improvement and application of the toolkit, and such efforts should eventually lead to the implement of precision agriculture in the future.
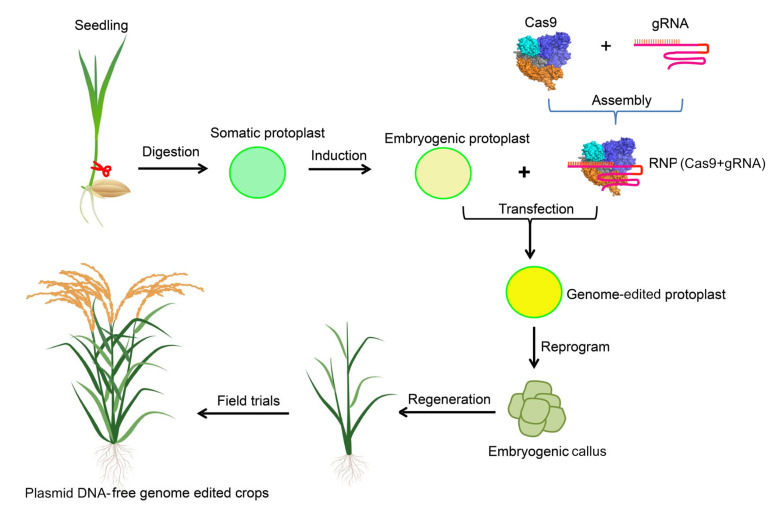
Fig. 5.
Workflow of potential protoplast-based gene editing and regeneration for plant breeding (rice as a model)
Cas: clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein; gRNA: guide RNA; RNP: ribonucleoprotein
Acknowledgments
We thank Dr. Jen SHEEN (Department of Molecular Biology in Massachusetts General Hospital, Harvard University, USA) for sharing her working progress on the protoplast editing in lettuce, Arabidopsis, and tobacco.
Footnotes
Project supported by the Zhejiang Provincial S&T Project on Breeding Agricultural (Food) Crops (No. 2016C02050-2) and the National Natural Science Foundation of China (No. 31701394)
Contributors: Qing-yao SHU contributed to the overall structure of the review. Yuan-yuan TAN wrote the part of editing for transcription and edited the manuscript. Yan-hua LIU was responsible for the part of editing of untranslated regions (UTRs). Liang WU and Xia WU wrote the part of editing for post-transcriptional regulation. Shi-yong SONG and Meng JIANG wrote the part of editing for translation, post-translation modification, and protein localization. Hao DU summarized the protoplast editing in this review. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the review and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Yuan-yuan TAN, Hao DU, Xia WU, Yan-hua LIU, Meng JIANG, Shi-yong SONG, Liang WU, and Qing-yao SHU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of authors.
References
- 1.Al-Zeer MA, Dutkiewicz M, von Hacht A, et al. Alternatively spliced variants of the 5'-UTR of the ARPC2 mRNA regulate translation by an internal ribosome entry site (IRES) harboring a guanine-quadruplex motif. RNA Biol. 2019;16(11):1622–1632. doi: 10.1080/15476286.2019.1652524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apitz J, Nishimura K, Schmied J, et al. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 2016;170(4):2040–2051. doi: 10.1104/pp.15.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beale SI. Enzymes of chlorophyll biosynthesis. Photosynth Res. 1999;60(1):43–73. doi: 10.1023/a:1006297731456. [DOI] [Google Scholar]
- 4.Butelli E, Licciardello C, Zhang Y, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24(3):1242–1255. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Čermák T, Baltes NJ, Čegan R, et al. High-frequency, precise modification of the tomato genome. Genome Biol, 16:232. 2015 doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen KL, Wang YP, Zhang R, et al. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 9.Czarnecki O, Hedtke B, Melzer M, et al. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell. 2011;23(12):4476–4491. doi: 10.1105/tpc.111.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Cao XF. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr Opin Plant Biol. 2017;35:45–53. doi: 10.1016/j.pbi.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 12.Duan GY, Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11(2):e1004049. doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo A, Masafumi M, Kaya H, et al. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida . Sci Rep, 6:38169. 2016 doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eş I, Gavahian M, Marti-Quijal FJ, et al. The application of the CRISPR-Cas9 genome editing machinery in food and agricultural science: current status, future perspectives, and associated challenges. Biotechnol Adv. 2019;37(3):410–421. doi: 10.1016/j.biotechadv.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Espley RV, Brendolise C, Chagné D, et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21(1):168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filichkin S, Priest HD, Megraw M, et al. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr Opin Plant Biol. 2015;24:125–135. doi: 10.1016/j.pbi.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Fossi M, Amundson K, Kuppu S, et al. Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 2019;180:78–86. doi: 10.1104/pp.18.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goslings D, Meskauskiene R, Kim C, et al. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40(6):957–967. doi: 10.1111/j.1365-313x.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 19.Hickey LT, Hafeez AN, Robinson H, et al. Breeding crops to feed 10 billion. Nat Biotechnol. 2019;37(2):744–754. doi: 10.1038/s41587-019-0152-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua K, Tao XP, Zhu JK. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol J. 2018;17(2):499–504. doi: 10.1111/pbi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua K, Zhang JS, Botella JR, et al. Perspectives on the application of genome-editing technologies in crop breeding. Mol Plant. 2019;12(8):1047–1059. doi: 10.1016/j.molp.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Huber SC, Hardin SC. Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol. 2004;7(3):318–322. doi: 10.1016/j.pbi.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Hunt AG. The Arabidopsis polyadenylation factor subunit CPSF30 as conceptual link between mRNA polyadenylation and cellular signaling. Curr Opin Plant Biol. 2014;21:128–132. doi: 10.1016/j.pbi.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Jia HG, Zhang YZ, Orbović V, et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J. 2017;15(7):817–823. doi: 10.1111/pbi.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang M, Liu YH, Li RQ, et al. A suppressor mutation partially reverts the xantha trait via lowered methylation in the promoter of genomes uncoupled 4 in Rice. Front Plant Sci, 10:1003. 2019 doi: 10.3389/fpls.2019.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao YQ, Wang YH, Xue DW, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 28.Jorrín-Novo JV, Maldonado AM, Echevarria-Zomeno S, et al. Plant proteomics update (2007–2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics. 2009;72(3):285–314. doi: 10.1016/j.jprot.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Kausch AP, Nelson-Vasilchik K, Hague J, et al. Edit at will: genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 2019;281:186–205. doi: 10.1016/j.plantsci.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Kauss D, Bischof S, Steiner S, et al. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Lett. 2012;586(3):211–216. doi: 10.1016/j.febslet.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinstiver BP, Tsai SQ, Prew MS, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34(8):869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leppek K, Das R, Barna M. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell. 2018;19(3):158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li AX, Jia SG, Yobi A, et al. Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018;177(4):1425–1438. doi: 10.1104/pp.18.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JF, Norville JE, Aach J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Liu B, Spalding MH, et al. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Liu B, Chen CY, et al. TALEN-mediated homologous recombination produces site-directed DNA base change and herbicide-resistant rice. J Genet Genomic. 2016;43(5):297–305. doi: 10.1016/j.jgg.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Li WT, Zhu ZW, Chern M, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170(1):114–126. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Li YB, Fan CC, Xing YZ, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 40.Liang XQ, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 41.Lin CS, Hsu CT, Yang LH, et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J. 2018;16(7):1295–1310. doi: 10.1111/pbi.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin S, Zhao YY, Zhu YF, et al. An effective and inducible system of TAL effector-mediated transcriptional repression in Arabidopsis . Mol Plant. 2016;9(11):1546–1549. doi: 10.1016/j.molp.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Liu SM, Jiang J, Liu Y, et al. Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low Cd rice. Rice Sci. 2019;26(2):88–97. doi: 10.1016/j.rsci.2019.01.002. [DOI] [Google Scholar]
- 44.Lloyd A, Plaisier CL, Carroll D, et al. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis . Proc Natl Acad Sci USA. 2005;102(6):2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahfouz MM, Li LX, Shamimuzzaman M, et al. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011;108(6):2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao YF, Botella JR, Liu YG, et al. Gene editing in plants: progress and challenges. Natl Sci Rev. 2019;6(3):421–437. doi: 10.1093/nsr/nwz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meskauskiene R, Nater M, Goslings D, et al. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2001;98(22):12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minkenberg B, Xie KB, Yang YN. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 2017;89(3):636–648. doi: 10.1111/tpj.13399. [DOI] [PubMed] [Google Scholar]
- 49.Molla KA, Yang YN. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37(10):1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Morsy M, Gouthu S, Orchard S, et al. Charting plant interactomes: possibilities and challenges. Trends Plant Sci. 2008;13(4):183–191. doi: 10.1016/j.tplants.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Nekrasov V, Staskawicz B, Weigel D, et al. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(8):691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 52.Nishimasu H, Shi X, Ishiguro S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361(6408):1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oikawa T, Maeda H, Oguchi T, et al. The birth of a black rice gene and its local spread by introgression. Plant Cell. 2015;27(9):2401–2414. doi: 10.1105/tpc.15.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandiarajan R, Grover A. In vivo promoter engineering in plants: are we ready? Plant Sci. 2018;277:132–138. doi: 10.1016/j.plantsci.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 55.Piatek A, Ali Z, Baazim H, et al. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13(4):578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 56.Pierre-Jerome E, Drapek C, Benfey PN. Regulation of division and differentiation of plant stem cells. Annu Rev Cell Dev Biol. 2018;34(1):289–310. doi: 10.1146/annurev-cellbio-100617-062459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin ZR, Wu JJ, Geng SF, et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nat Commun, 8:14320. 2017 doi: 10.1038/ncomms14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy ASN, Marquez Y, Kalyna M, et al. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25(10):3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren B, Liu L, Li SF, et al. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant. 2019;12(7):1015–1026. doi: 10.1016/j.molp.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Richter AS, Hochheuser C, Fufezan C, et al. Phosphorylation of GENOMES UNCOUPLED 4 alters stimulation of Mg chelatase activity in angiosperms. Plant Physiol. 2016;172(3):1578–1595. doi: 10.1104/pp.16.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Leal D, Lemmon ZH, Man J, et al. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–480E8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 62.Shan QW, Wang YP, Li J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 63.Shan QW, Wang YP, Li J, et al. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9(10):2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- 64.Shan QW, Zhang Y, Chen KL, et al. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J. 2015;13(6):791–800. doi: 10.1111/pbi.12312. [DOI] [PubMed] [Google Scholar]
- 65.Shi JR, Gao HR, Wang HY, et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15(2):207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimatani Z, Kashojiya S, Takayama M, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 67.Soyk S, Müller NA, Park SJ, et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49(1):162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- 68.Sun YW, Jiao GA, Liu ZP, et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci, 8:298. 2017 doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 70.Takenaka M, Zehrmann A, Verbitskiy D, et al. RNA editing in plants and its evolution. Annu Rev Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- 71.Tang L, Mao BG, Li YK, et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep, 7:14438. 2017 doi: 10.1038/s41598-017-14832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang X, Lowder LG, Zhang T, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3(3):17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- 73.von Arnim AG, Jia QD, Vaughn JN. Regulation of plant translation by upstream open reading frames. Plant Sci. 2014;214:1–12. doi: 10.1016/j.plantsci.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 74.Wang B, Smith SM, Li JY. Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018;69(1):437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- 75.Wang FJ, Wang CL, Liu PQ, et al. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922 . PLoS ONE. 2016;11(4):e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Zhou L, Shi H, et al. A single transcription factor promotes both yield and immunity in rice. Science. 2018;361(6406):1026–1028. doi: 10.1126/science.aat7675. [DOI] [PubMed] [Google Scholar]
- 77.Wang MG, Mao YF, Lu YM, et al. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant. 2017;10(7):1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Wang SK, Wu K, Yuan QB, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 79.Wang SK, Li S, Liu Q, et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet. 2015;47(8):949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- 80.Wang YP, Cheng X, Shan QW, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 81.Woo JW, Kim J, Kwon SI, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 82.Wu L, Zhou HY, Zhang QQ, et al. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38(3):465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 83.Xie KB, Minkenberg B, Yang YN. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112(11):3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu CJ, Liu Y, Li YB, et al. Differential expression of GS5 regulates grain size in rice. J Exp Bot. 2015;66(9):2611–2623. doi: 10.1093/jxb/erv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu RF, Yang YC, Qing RY, et al. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genomics. 2016;43(8):529–532. doi: 10.1016/j.jgg.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 86.Xue CX, Zhang HW, Lin QP, et al. Manipulating mRNA splicing by base editing in plants. Sci China Life Sci. 2018;61(11):1293–1300. doi: 10.1007/s11427-018-9392-7. [DOI] [PubMed] [Google Scholar]
- 87.Yang RX, Li PC, Mei HL, et al. Fine-tuning of miR528 accumulation modulates flowering time in rice. Mol Plant. 2019;12(8):1103–1113. doi: 10.1016/j.molp.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 88.Ytterberg AJ, Jensen ON. Modification-specific proteomics in plant biology. J Proteomics. 2010;73(11):2249–2266. doi: 10.1016/j.jprot.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 89.Zaidi SSA, Mukhtar MS, Mansoor S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018;36(9):898–906. doi: 10.1016/j.tibtech.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 90.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang H, Zhang JS, Lang ZB, et al. Genome editing-principles and applications for functional genomics research and crop improvement. Plant Sci. 2017;36(4):291–309. doi: 10.1080/07352689.2017.1402989. [DOI] [Google Scholar]
- 92.Zhang HW, Si XM, Ji X, et al. Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol. 2018;36(9):894–898. doi: 10.1038/nbt.4202. [DOI] [PubMed] [Google Scholar]
- 93.Zhang JS, Zhang H, Botella JR, et al. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol. 2018;60(5):369–375. doi: 10.1111/jipb.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Yu H, Ma B, et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat Commun, 8:14789. 2017 doi: 10.1038/ncomms14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang M, Zhang FL, Fang Y, et al. The non-canonical tetratricopeptide repeat (TPR) domain of fluorescent (FLU) mediates complex formation with glutamyl-tRNA reductase. J Biol Chem. 2015;290(28):17559–17565. doi: 10.1074/jbc.M115.662981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang YX, Malzahn AA, Sretenovic S, et al. The emerging and uncultivated potential of CRSIPR technology in plant science. Nat Plants. 2019;5(8):778–794. doi: 10.1038/s41477-019-0461-5. [DOI] [PubMed] [Google Scholar]
- 97.Zhou JP, Deng KJ, Cheng Y, et al. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front Plant Sci, 8:1598. 2017 doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou X, Deng L, Wang Q, et al. Breeding of waxy rice by genome editing. Mol Plant Breed. 2018;16(17):5608–5615. (in Chinese) [Google Scholar]
- 99.Zimny T, Sowa S, Tyczewska A, et al. Certain new plant breeding techniques and their marketability in the context of EU GMO legislation–recent developments. New Biotechnol. 2019;51:49–56. doi: 10.1016/j.nbt.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Zong Y, Song QN, Li C, et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol. 2018;36(10):950–953. doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]