Abstract
Transient receptor potential melastatins (TRPMs) are most well known as cold and menthol sensors, but are in fact broadly critical for life, from ion homeostasis to reproduction. Yet, the evolutionary relationship between TRPM channels remains largely unresolved, particularly with respect to the placement of several highly divergent members. To characterize the evolution of TRPM and like channels, we performed a large-scale phylogenetic analysis of >1,300 TRPM-like sequences from 14 phyla (Annelida, Arthropoda, Brachiopoda, Chordata, Cnidaria, Echinodermata, Hemichordata, Mollusca, Nematoda, Nemertea, Phoronida, Priapulida, Tardigrada, and Xenacoelomorpha), including sequences from a variety of recently sequenced genomes that fill what would otherwise be substantial taxonomic gaps. These findings suggest: 1) the previously recognized TRPM family is in fact two distinct families, including canonical TRPM channels and an eighth major previously undescribed family of animal TRP channel, TRP soromelastatin; 2) two TRPM clades predate the last bilaterian–cnidarian ancestor; and 3) the vertebrate–centric trend of categorizing TRPM channels as 1–8 is inappropriate for most phyla, including other chordates.
Keywords: transient receptor potential, phylogenetics, channel evolution, TRPM, TRPS, ced-11
Introduction
Transient receptor potential (TRP) channels are a superfamily of ion channel commonly characterized by their six transmembrane (TM) segments and broad sensory capacity. Among animals, TRP channels have been canonically divided into seven families (Venkatachalam and Montell 2007; Peng et al. 2015): TRPA (ankyrin), TRPC (canonical), TRPM (melastatin), TRPML (mucolipin), TRPN (no mechanoreceptor potential C or nompC), TRPP (polycystin or polycystic kidney disease), and TRPV (vanilloid and a proposed sister family, TRPVL). These TRP channels vary substantially, but TRPM has arguably diversified the most with respect to function, participating in at least cardiac activity (Yue et al. 2015), magnesium homeostasis (Schlingmann et al. 2007; Hofmann et al. 2010), egg activation (Carlson 2019), sperm thermotaxis (De Blas et al. 2009), cell adhesion (Su et al. 2006), apoptosis (Driscoll et al. 2017), inflammation (Ramachandran et al. 2013), and most famously, cold (Bautista et al. 2007; Turner et al. 2016), and menthol (McKemy et al. 2002; Peier et al. 2002; Himmel et al. 2019) sensing.
TRPM channels are also ancient, predating the emergence of metazoans (>1,000 Ma) (Peng et al. 2015; Himmel et al. 2019). In some species, single proteins have multiple functions (e.g., Drosophila melanogaster Trpm), whereas in others, functions are compartmentalized in a set of diverse paralogs, which themselves may be multifunctional (e.g., human TRPM1-8). However, little is known about the evolutionary history of TRPMs, or to what degree channels are related across taxa. Our understanding of TRPM evolution is additionally clouded by the existence of several highly divergent putative TRPM channels with uncertain origins (Teramoto et al. 2005; Peng et al. 2015; Kozma et al. 2018; Himmel et al. 2019). Here, we made use of the rapidly growing body of genomic data in order to better characterize the evolution of the TRPM family.
Via a stringent screening process, we assembled a database of >1,300 predicted TRPM-like sequences from 14 diverse eumetazoan phyla (fig. 1). In this database, we gave particular attention to underrepresented taxa as well as included TRP genes identified in a number of recently sequenced genomes (supplementary table S1, Supplementary Material online; including, but not limited to, acoel flatworm, moon jelly, and great white shark). Herein, we elucidate the evolutionary history and familial organization of both TRPM and a previously unrecognized sister family that predates the Cnidaria–Bilateria split, TRP soromelastatin (TRPS).
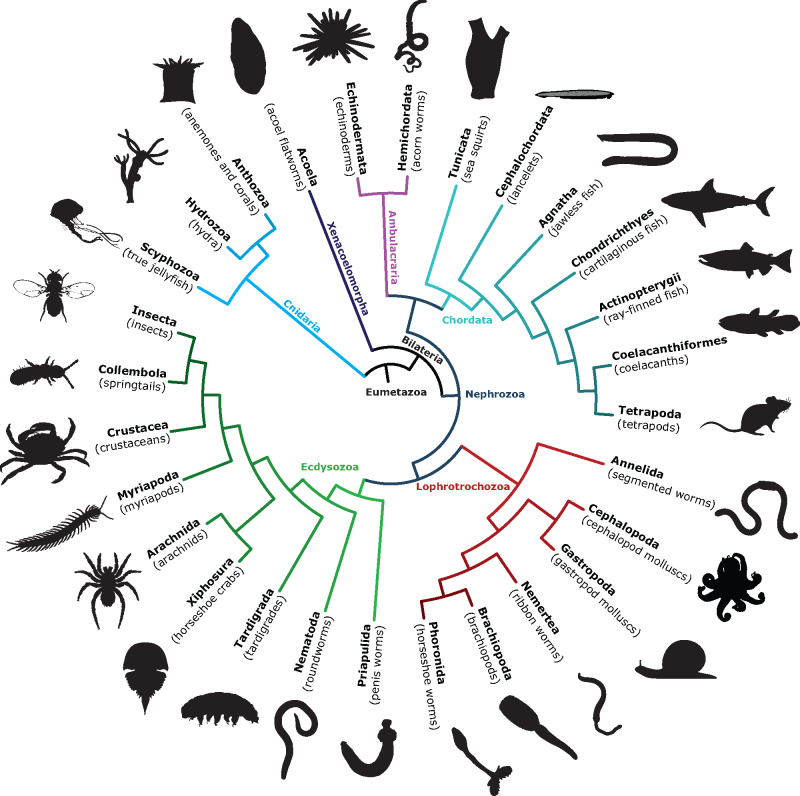
Fig. 1.
Major, widely recognized taxa included in these phylogenetic analyses and relationships assumed throughout. TRPM and TRPM-like sequences were collected for 318 species, from 14 diverse eumetazoan phyla (Annelida, Arthropoda, Brachiopoda, Chordata, Cnidaria, Echinodermata, Hemichordata, Mollusca, Nematoda, Nemertea, Phoronida, Priapulida, Tardigrada, and Xenacoelomorpha).
Results and Discussion
An Ancient, Unrecognized Sister Family to TRPM—TRPS
Proteins within the same family typically have a high degree of sequence similarity, yet highly divergent TRPM-like proteins have been cataloged, a notable example being Caenorhabditis elegans cell death abnormal 11 (ced-11). The canonical C. elegans TRPMs gtl-1, gtl-2, and gon-2 share roughly 40% sequence identity with each other. However, ced-11—often considered a fourth C. elegans TRPM—shares ∼18% sequence identity with the three canonical paralogs. Given this substantial difference, it seemed plausible that ced-11, and like proteins, had been errantly included in the TRPM family.
The TRPC and TRPN families are typically thought to be most closely related to TRPM (Peng et al. 2015) and therefore constituted hypothetical homes for ced-11. ced-11, however, shares only 15% sequence identity with known C. elegans TRPC paralogs (trp-1 and trp-2), and 14% sequence identity with C. elegans TRPN (trp-4). Yet, sequence identity between trp-4 and its TRPC counterparts is ∼20%. In other words, ced-11 is less similar to TRPMs than TRPNs and TRPCs are to each other.
In order to clarify the relationship of ced-11-like proteins to canonical TRPM channels, we collected those sequences most similar to it from our initial TRPM-like sequence database and phylogenetically characterized them. Blasting our database with ced-11-like sequences recovered a number of sequences restricted to several protostome taxa and lancelets (Cephalochordata).
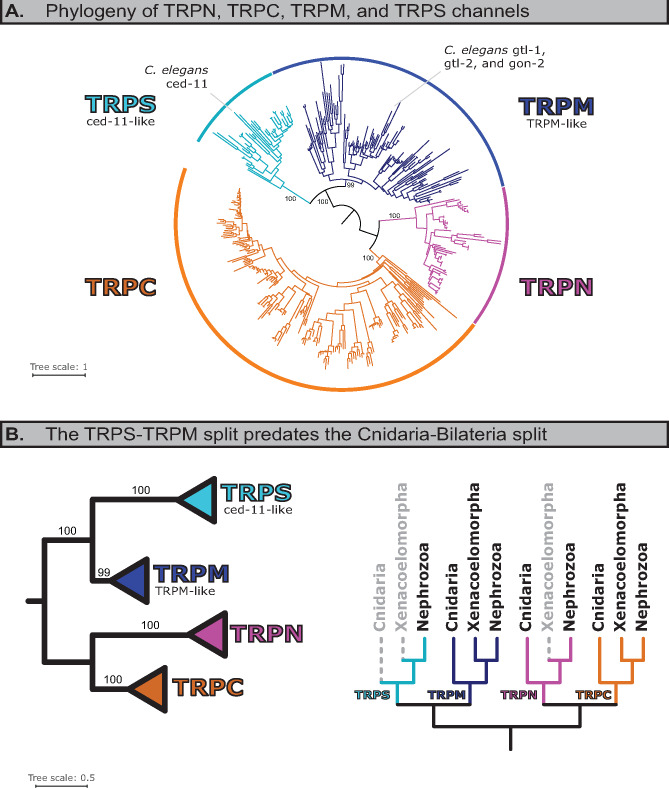
For any species with a ced-11-like protein, we assembled a database of putative TRPC and TRPN channel sequences. These sequences were then phylogenetically characterized alongside cnidarian, xenacoelomorph (basal bilaterian worms), insect (D. melanogaster), and human sequences. Cnidarians were included as the outgroup, and xenacoelomorphs as they form a sister clade to all other bilaterians (fig. 1). In the resulting tree, ced-11-like proteins formed a sister clade to the more traditional TRPM clade (fig. 2A). The traditional TRPM clade included all the cnidarian and xenacoelomorph sequences, indicating that the ced-11-like/TRPM-like split is a duplication event, and therefore that the two clades split before the Cnidaria–Bilateria split (fig. 2B and supplementary fig. S1, Supplementary Material online). Although there are no ced-11-like sequences present for Cnidaria or Xenacoelomorpha, this does not matter to the interpretation of these results. Cnidaria and Xenacoelomorpha had representatives in both clades (or had all the sequences been present in ced-11-like instead of TRPM-like), the node in question would still represent a duplication event that predates the Cnidara–Bilateria split.
Fig. 2.
Caenorhabditis elegans ced-11, and ced-11-like sequences, belong to a previously unrecognized family of TRP channels, the TRPS. TRPM-like and ced-11-like sequences form two distinct clades, with the topology suggesting divergence prior to the Cnidaria–Bilateria split. (A) Maximum likelihood tree showing the relationship between traditional TRPM and TRPS/ced-11-like sequences among those species that have TRPS/ced-11-like species. Full annotated tree available in supplementary figure S1, Supplementary Material online; alternative hypothesis testing in supplementary figures S3 and S4, Supplementary Material online. (B) Left, summary of maximum likelihood analysis and branch support (UFboot) for indicated clades. Right, presence or absence of family in indicated taxa (where Bilateria is Xenacoelomorpha+Nephrozoa). Solid lines in color indicate that the TRP family is present in the indicated taxon, whereas dashed lines in gray indicate that no sequences were found for the indicated taxon. Family specific duplication/loss events summarized in figures 4 and 5.
Two competing hypotheses could explain these findings: 1) ced-11-like proteins constitute a distinct family of TRP channel that predates the cnidarian–bilaterian split or 2) a variety of TRPM channels emerged independently in various taxa and diversified extremely rapidly, resulting in a clade which formed as a result of long-branch attraction, an artifact of many phylogenetic analyses (Bergsten 2005).
Hypothesis 2 appears highly unlikely. Most importantly, whereas C. elegans ced-11 itself has a relatively long branch, when qualitatively compared with other clades, the branches within the ced-11-like clade were not unusually long (fig. 2 and supplementary fig. S1, Supplementary Material online). Additionally, principal component analysis (PCA) of a pairwise sequence identity matrix revealed that ced-11-like sequences cluster together independent of TRPM-like sequences, suggesting they cluster in the phylogram due to sequence similarity (fig. 3A; TM only, supplementary fig. S2, Supplementary Material online). We tested the long-branch hypothesis by estimating trees which excluded Cnidaria and Xenacoelomorpha, which had particularly long branches and could serve to exacerbate long-branch attraction, were it present. The resultant phylogram still evidenced the split between ced-11-like and TRPM-like channels, with high branch confidence (supplementary fig. S3, Supplementary Material online). Moreover, we generated a phylogram by the Graph Splitting method, which is reported to be extremely robust when faced with the possibility of long-branch attraction in superfamily level data sets (Matsui and Iwasaki 2019). This method likewise reproduced the ced-11-like-TRPM split with high edge perturbation branch support (supplementary fig. S4, Supplementary Material online).
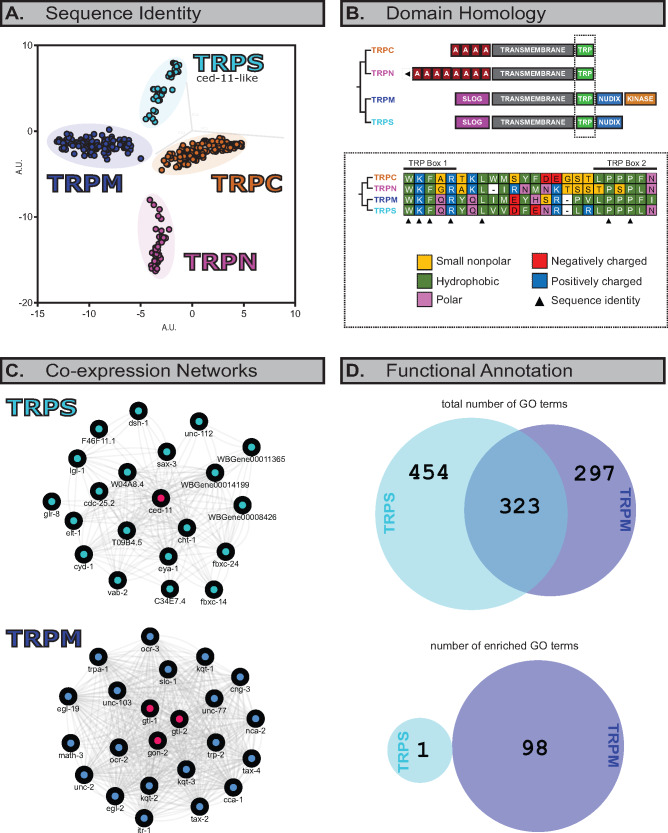
Fig. 3.
TRPS and TRPM are, putatively, structurally and functionally distinct, and their domain arrangements suggest a SLOG- and Nudix-linked, ankyrin-free TRPS–TRPM ancestor. (A) Principal component analyses of pairwise sequence identity for alignment of TRPC, TRPN, TRPM, and ced-11-like (TRPS) sequences show four distinct clusters. 3D PCA plot extracted from Jalview and plotted in two transformed, arbitrary dimensions. PCA against just TM segments in supplementary figure S2, Supplementary Material online. (B) Like TRPM channels, these TRPS channels have both SLOG and Nudix domains, but lack the kinase domain associated with a small subset of TRPM channels. Moreover, these TRPS channels have a divergent consensus sequence in the highly conserved TRP domain. (C) In Caenorhabditis elegans, TRPS (top; ced-11) and TRPM (bottom; gon-2, gtl-1, and gtl-2) have nonoverlapping coexpressed gene networks. (D) Top, different GO terms are associated with C. elegans TRPS and TRPM coexpression networks. Bottom, only a single GO term, “cell surface receptor signaling pathway” (GO: 0007166), showed statistically significant enrichment in the TRPS gene network. This term does not appear for the TRPM network. GO terms are detailed in supplementary data S2 and S3, Supplementary Material online.
These results strongly indicate that these two lineages diverged in or prior to the last cnidarian–bilaterian ancestor, and that ced-11-like proteins constitute an eighth family of metazoan TRP channel. We have thus named the ced-11-like family of TRP channels TRPS (soro-, sister) (figs. 2 and 3).
The Structure of TRPS Channels Suggests an SLOG- and Nudix-Linked Ancestor
Although the function of TRPS channels remains largely unknown, domain prediction reveals that both TRPM and TRPS channels share an N-terminal SMF/DprA-LOG (SLOG) domain and a C-terminal ADP-ribose phosphohydrolase (Nudix) domain (fig. 3B and supplementary data S1, Supplementary Material online). The SLOG domain is thought to bind a variety of nucleotides but has not been extensively studied in this capacity (Burroughs et al. 2015). Likewise, the Nudix domain has not been well characterized, but it is known to function in ADP-dependent activation of TRPM2 (Fliegert et al. 2017). TRPS channels may therefore be sensitive to ADP and possibly to a variety of other nucleotides.
These results indicate that the ancestral TRPM–TRPS channel was likely both SLOG- and Nudix-linked and are consistent with previous findings, suggesting that the TRPM ancestor was Nudix-linked (Schnitzler et al. 2008). Ankyrin repeats, which are present in all TRP families except TRPM (Schüler et al. 2015), were not detected in TRPS by either InterProScan or an HMMER query using additional Hidden Markov Models specific to Ankyrin repeats. Ankyrin repeats were therefore most likely lost prior to the TRPM–TRPS split.
The TRPM alpha kinase domain, which is only known to be present in vertebrate TRPM6 and TRPM7, is not present in these TRPS sequences. This is further evidence in support of the hypothesis that the alpha kinase domain was gained relatively recently in vertebrates (Schnitzler et al. 2008), after the TRPM–TRPS split.
A notable difference between TRPM and TRPS channels lies in the TRP domain, a highly conserved, hydrophobic region located C-terminally to the TM domain of TRPC, TRPN, and TRPM channels (Venkatachalam and Montell 2007). Consensus sequences for the initial TRPM and TRPS data sets (fig. 2), while identical in TRP box 1, are divergent in TRP box 2 and the intermediate TRP segment (fig. 3B, bottom). The TRP domain has been associated with PIP2 binding and menthol- and cold-evoked gating (Rohács et al. 2005); differences in the TRP domain may therefore reflect differences in ligand selectively or sensory modality between TRPS and TRPM. The TRP domain has also been associated with the formation of the calcium-binding site in vertebrate TRPM2 and TRPM4 (Chen et al. 2019); as such, differences in the TRPS TRP domain may point to differences in calcium-binding capacity compared with TRPM channels. These functional consequences, however, remain speculative.
In order to gain some preliminary insight into putative functional divergence between TRPS and TRPM, we leveraged the abundance of C. elegans transcriptomic data in order to perform coexpression and gene ontology (GO) analyses (fig. 3C and 3D and supplementary data S2–S3, Supplementary Material online). The results of these analyses further indicate that TRPS and TRPM may be functionally divergent. When considered individually, the TRPS and TRPM families have completely nonoverlapping coexpressed gene networks (fig. 3C). Moreover, these gene networks do not share the majority of GO terms associated with them (fig. 3D, top), and there is no overlap when considering terms enriched to a statistically significant extent (fig. 3D, bottom). As with ced-11, however, many of the coexpressed genes are uncharacterized or understudied, which is perhaps responsible for only a single term (“cell surface receptor signaling pathway”) being enriched in the TRPS network.
The TRPS Family Is Largely Restricted to Protostomes
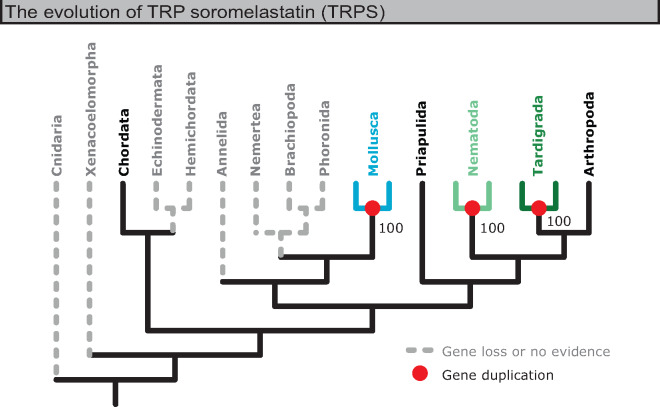
Having established that these TRPS sequences constitute a distinct set of channels, we assembled a more complete TRPS sequence database and phylogenetically characterized the channel family. These data suggest that, among Eumetazoa, TRPS genes are only present in some protostomes and lancelets (fig. 4 and supplementary fig. S5, Supplementary Material online). The lack of widespread conservation among deuterostomes (most notably vertebrates) and insects likely explains why the family had gone unnoticed until now.
Fig. 4.
Summary of the evolution of TRPS channels. The TRPS family is largely restricted to protostome lineages. Figure derived from reconciled maximum likelihood tree of TRPS sequences (supplementary fig. S5, Supplementary Material online). Red dots and colored branches indicate phylum-specific duplication events. Gray and dashed branches indicate that no TRPS sequences were found for the indicated phylum and were inferred to be loss events.
TRPS was likely lost early in deuterostome evolution—among the ambulacrarians (echinoderms and hemichordates)—and in early olfactores (tunicates and vertebrates) following the olfactore–lancelet split (supplementary fig. S6, left, Supplementary Material online). A recent study evidences that Ambulacraria and Xenacoelomorpha might form sister clades (Philippe et al. 2019)—if this is the case, it may be more likely that TRPS was lost early in so-called “xenambulacrarian” evolution (supplementary fig. S6, right, Supplementary Material online).
TRPS duplication appears to have been limited during early animal evolution. Although the number of TRPS paralogs varies by species (supplementary fig. S5, Supplementary Material online), duplication events occurred only after major taxa emerged, independently in mollusks, nematodes, tardigrades, and chelicerates (including arachnids and horseshoe crabs). In some instances, these taxon-specific TRPS expansions may be traced to whole-genome duplications (WGDs), such as in horseshoe crabs (Kenny et al. 2016) and other arachnids (Schwager et al. 2017). Mollusks have two TRPS paralogs, but present lack of evidence for lophotrochozoan TRPS outside of mollusks makes it difficult to predict at what point in spiralian evolution the duplication event occurred. The simplest explanation is that it occurred specifically in mollusks, perhaps also as a result of WGD (Yoshida et al. 2011), and that a single TRPS copy was lost among other lophotrochozoan taxa.
Among Euarthropoda, TRPS appears in chelicerates (including arachnids and horseshoe crabs) and myriapods (including centipedes), but there is no evidence for TRPS in crustaceans, springtails (a type of noninsect hexapod), or insects, suggesting that the single arthropod TRPS was lost in Pancrustacea (all crustaceans and hexapods), conserved in Myriapoda, and expanded independently in Chelicerata (supplementary fig. S7, Supplementary Material online).
Two TRPM Clades Predate the Cnidaria–Bilateria Split
We next phylogenetically characterized and reconciled TRPM sequences among major taxa. Each set of sequences was initially assessed alongside cnidarian, xenacoelomorph, Drosophila, and human sequences, and rooted with TRPS sequences.
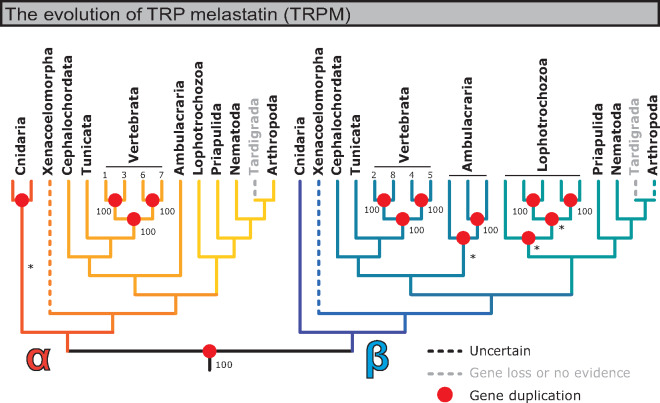
The general consensus of these analyses indicates that the TRPM family is made up of two distinct clades, here and previously deemed αTRPM and βTRPM (Himmel et al. 2019), which emerged prior to the Cnidaria–Bilateria split (fig. 5 and supplementary figs. S8–S17, Supplementary Material online). What might have constituted a previously described “basal clade” can be almost wholly explained by the discovery of TRPS (Peng et al. 2015; Himmel et al. 2019). In some of our initial phylograms, a basal or separate clade did appear, yet it always included Xenacoelomorpha and was inconsistent in its topology across analyses (supplementary figs. S8–S12, Supplementary Material online), suggesting that Xenacoelomorpha acted as a phylogenetically unstable rogue taxon (Thomson and Shaffer 2010). In order to assess this possibility, we performed a second set of analyses which excluded Xenacoelomorpha. This resulted in trees with largely consistent topology despite differing taxon sampling, indicating that xenacoelomorph sequences are in fact problematic (supplementary figs. S13–S17, Supplementary Material online).
Fig. 5.
Summary of the evolution of TRPM channels. The TRPM family is widely conserved and is present in all phyla surveyed except Tardigrada. Figure derived from consensus topologies in reconciled maximum likelihood trees generated against TRPM database sequences, with branch support for duplication branches extracted from phylograms without Xenacoelomorpha (supplementary figs. S13–S17, Supplementary Material online). Asterisk (*) indicates that a duplication branch most frequently formed due to rearrangement (initial UFboot branch support <95). Here, phyla were expanded/collapsed to more easily show TRPM diversification: Chordata was expanded into Cephalochordata (lancelets), Tunicata, and Vertebrata; Echinodermata and Hemichordata were collapsed into Ambulacraria; and Annelida, Nemertea, Brachiopoda, Phoronida, and Mollusca were collapsed into Lophotrochozoa. Red dots indicate duplication events. Dashed lines in color indicate sequences considered incertae sedis. Dashed lines in gray indicate that no sequences were found for the indicated taxon and were inferred to be loss events.
Due to the overwhelming consistency of trees with different taxon sampling, and the inconsistency seen in trees including Xenacoelomorpha, xenacoelomorph TRPM sequences were treated as rogue taxa. In addition, an extremely small subset of arthropod TRPMs (12 sequences restricted to chelicerates and crustaceans; supplementary figs. S12 and S17, Supplementary Material online) may be part of a previously described Crustacea-specific TRPM subfamily (Kozma et al. 2018). These trees suggest that these sequences are βTRPMs and related to a subset of cnidarian sequences, yet this Cnidaria-inclusive clade is not strongly evidenced in phylograms with different taxon samplings. Like xenacoelomorph sequences, the evolutionary histories of these cnidarian and arthropod sequences are left incertae sedis.
In summary, these results strongly support two duplication events predating the Cnidaria–Bilateria split: the TRPS–TRPM split and the α–β TRPM split.
TRPM1-8 Expansion Occurred Early in Vertebrate Evolution and Constitutes a Poor Standard for TRPM Family Organization
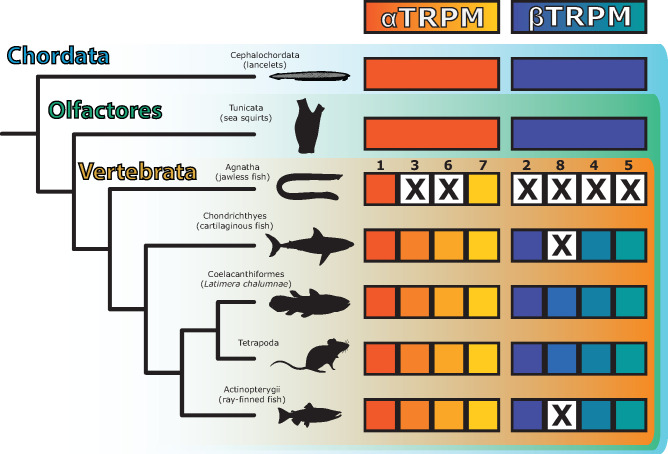
The vertebrate TRPM1-8 expansion has been the focus of the majority of TRPM literatures and has been the principal basis for characterizing TRPM channels (Samanta et al. 2018; Zhang et al. 2018; Chen et al. 2019). However, these trees evidence that the TRPM1-8 expansion occurred after the vertebrate–tunicate split (figs. 5 and 6 and supplementary fig. S13, Supplementary Material online). As agnathans (jawless fish; lampreys and hagfish) have no representative in the βTRPM clade, it is possible that βTRPM expanded after agnathans and other vertebrates split. However, two WGDs likely occurred before the common ancestor of vertebrates (Sacerdot et al. 2018). WGD may therefore be the basis of these TRPM expansions. As such, the most parsimonious hypothesis is that both α- and βTRPM diversified before agnathans split from the ancestor of all other vertebrates.
Although immunohistochemical evidence has previously suggested that TRPM8 is present in teleost fish (Majhi et al. 2015), we found no evidence of it in available sequences for ray-finned fish, nor did we find any evidence in cartilaginous fish or agnathans (fig. 6 and supplementary figs. S14 and S18, Supplementary Material online). The most parsimonious hypothesis prima facia would be that TRPM8 did not emerge until lobe-finned fish emerged. If this were the case, the TRPM8 clade would be a branch off of the vertebrate TRPM2 clade. This is not the case. Instead, these reconciled trees (fig. 6 and supplementary figs. S14 and S18, Supplementary Material online) show that TRPM8 forms a sister clade to TRPM2, the former not containing any nonlobe-finned sequences, and the latter containing lobe-finned and non-lobe-finned sequences. This indicates that TRPM8 was independently lost in ray-finned fish, cartilaginous fish, and agnathans but was conserved in the lobe-finned vertebrate lineage (including tetrapods).
Fig. 6.
TRPM1-8 diversified soon after the vertebrate–tunicate split, and there is no evidence for TRPM8 in jawless, cartilaginous, or ray-finned fish. Figure derived from reconciled tree of chordate TRPM sequences (supplementary fig. S18, Supplementary Material online; see supplementary fig. S14, Supplementary Material online, for more visually accessible phylogram, which lacks ray-finned fish). An “X” indicates inferred gene loss, or lack of evidence of that gene in the indicated taxon.
Moreover, the 1–8 nomenclature may underdescribe TRPMs among one of the most abundant vertebrate clades—the teleost fish. Although basal ray-finned fish (e.g., Erpetoichthys calabaricus, the freshwater snakefish, or reedfish) have a TRPM topology that closely matches other vertebrates, the emergence of teleosts came with TRPM expansion. For example, there are as many as three teleost TRPM4 paralogs (supplementary fig. S18, Supplementary Material online).
Conclusions
The evolutionary history of TRPM channels has been clouded by divergent sequences, making it uncertain if a basal clade of TRPMs had survived in species like C. elegans, or if these species had independently evolved rapidly changing TRPM paralogs (Teramoto et al. 2005; Peng et al. 2015; Kozma et al. 2018; Himmel et al. 2019). By taking advantage of the abundance of publicly available genomic data, we have instead demonstrated that the difficulty in phylogenetically characterizing TRPM channels is the result of an ancient, hidden family of channels that appeared before the Cnidaria–Bilateria split—the TRPS. By recognizing and characterizing this family, we now better understand not only the evolution and diversification of TRPM but also the evolution of the broader TRP superfamily.
Although some have been careful in describing TRP channels in taxon-specific ways (Saito and Shingai 2006; Hofmann et al. 2010; Peng et al. 2015), the current findings are the strongest challenge to the pervasive, vertebrate–centric dogma that the TRPM family is constituted by eight distinct paralogs organized into four subfamilies (Samanta et al. 2018; Zhang et al. 2018; Chen et al. 2019). These results instead support that the eumetazoan TRPM family consists of two distinct radiations (αTRPM and βTRPM) which themselves predate the Cnidaria–Bilateria split. Importantly, these findings support that TRPM diversification occurred independently among cnidarians, ambulacrarians, lophotrochozoans, and other taxa, and that the TRPM1-8 expansion is specific to vertebrates. Based on these findings, we conclude that the TRPM1-8 nomenclature is at best evolutionarily uninformative (e.g., insect channels being simply TRPM1- or 3-like), and at worst grossly inaccurate (e.g., cnidarian TRPMs belonging to the TRPM2/8 subfamily) for describing members of this diverse family of critically important ion channels.
Materials and Methods
Data Collection and Curation
Starting with previously characterized TRPM sequences from human (NCBI CCDS), mouse (NCBI CCDS), D. melanogaster (fruit fly, FlyBase), and Caenorhabditis elegans (nematode worm, WormBase), a TRPM-like protein sequence database was assembled by performing Blastp against NCBI collections of nonredundant protein sequences, with D. melanogaster Trpm (isoform RE, FlyBase ID: FBtr0339077) serving as the bait sequence. In order to maximize useful phylogenetic information, only Blast hits >300 amino acids in length with an E value less than 1E−30 were retained. As we were interested in the origins of TRPM channels, and in less-studied taxa, only three tetrapod sequence sets were included, from human, mouse, and chicken.
In order to expand the taxa sampled, tBlastn and Blastp were used to search publicly available, genomically informed gene models for 11 cnidarians, 2 xenacoelomorphs, 1 hemichordate (acorn worm), 1 nemertean (ribbon worm), 1 phoronid (horseshoe worm), 2 agnathans (hagfish and lamprey), and 4 chondrichthyes (cartilaginous fish) (supplementary table S1, Supplementary Material online).
We used several methods in order to validate and improve the quality of the initial database. First, CD-HIT (threshold 90% similarity) was used to identify and remove duplicate sequences and predicted isoforms, retaining the longest isoform in order to maximize available phylogenetic information (Li and Godzik 2006; Huang et al. 2010; Fu et al. 2012). Phobius was then used to predict TM topology (Käll et al. 2004, 2007); sequences which did not have at least six predicted TM segments, which is typical of TRP channels, were removed. Sequences with more than the six predicted TM segments were analyzed via InterProScan (Mitchell et al. 2019), and those with more than one ion-transport domain were removed. More than 90% of the remaining sequences contained a highly conserved glycine residue in the predicted TM domain (corresponding to D. melanogaster G-1049); the vast majority of those missing this residue had large gaps in an initial alignment and were subsequently removed.
Searches for TRPS (ced-11-like), TRPN, and TRPC sequences followed the same protocol. For TRPS, sequences from C. elegans, Strigamia maritima (centipede), and Octopus vulgaris (common octopus) were used as bait. For TRPN and TRPC data sets, D. melanogaster nompC (isoform PA, FlyBase ID: FBpp0084879) and Trp (isoform PA, FlyBase ID: FBpp0084879) served as bait sequences, respectively.
Principal Component Analysis
PCA was used to help resolve protein families. TRPC, TRPN, and TRPM/TRPS database sequences were aligned by MAFFT. A pairwise sequence identity matrix was then computed, and PCA performed against it in Jalview (Waterhouse et al. 2009). PCA was performed on a full-length alignment and on an alignment trimmed to only include the TM segments. Data were exported from Jalview and visualized and edited in GraphPad Prism and Adobe Illustrator CS6.
Domain Identification
TRPS protein domains were primarily identified by InterProScan (Mitchell et al. 2019). As previous research has indicated that ankyrin repeats might go undetected in a variety of TRP sequences (Schüler et al. 2015), we performed a secondary analysis using HMMER (Wheeler and Eddy 2013) and custom Hidden Markov Models, specific to ankyrin repeats (models generated by Gonzalo Parra et al. 2015).
Coexpression and GO Analyses
C. elegans gene coexpression networks were generated by GeneMANIA (Warde-Farley et al. 2010) and visualized in Cytoscape (Shannon et al. 2003). Associated and enriched GO terms were then retrieved using g: Profiler, with a significance threshold of 0.05, using the g: SCS method to compute multiple testing corrections (Reimand et al. 2007). The GO Venn diagrams were generated using BioVenn (Hulsen et al. 2008) and edited in Adobe Illustrator CS6.
Phylogenetic Tree Estimation
For the maximum likelihood approach, sequences were first aligned using MAFFT with default settings (Rozewicki et al. 2019). Gap-rich sites and poorly aligned sequences were trimmed with TrimAl (Capella-Gutiérrez et al. 2009). IQ-Tree (Nguyen et al. 2015) was then used to generate trees by the maximum likelihood approach, using the best models automatically selected by ModelFinder (Kalyaanamoorthy et al. 2017). Branch support was calculated by ultrafast bootstrapping (UFBoot, 2,000 bootstraps) (Hoang et al. 2018).
In order to test the alternative hypothesis that some trees formed due to long-branch attraction, gs2 was used to generate trees by the Graph Splitting method (Matsui and Iwasaki 2019). Branch support values were computed by the packaged edge perturbation method (EP, 2,000 iterations). All trees were visualized and edited in iTOL and Adobe Illustrator CS6.
Tree Reconciliation
In order to identify duplication events, TRPS and TRPM phylograms were reconciled using NOTUNG 2.9.1 (Durand et al. 2006; Vernot et al. 2008; Stolzer et al. 2012). Edge weight threshold was set to 1.0, and the costs of duplications and losses were set to 1.5 and 1.0, respectively. In order to formulate the most parsimonious interpretation of the resulting trees, weak branches were rearranged (UFboot 95 cutoff) against a cladogram based in an NCBI taxonomic tree, wherein we placed Xenacoelomorpha (represented by acoel flatworms) as the sister group to all other bilaterians (Cannon et al. 2016), and Priapulida (penis worms) as an outgroup to all other ecdysozoans (Yamasaki et al. 2015). All other polytomies were randomly resolved using the ape package in R (Paradis and Schliep 2019).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Charles Derby and Mihika Kozma (Georgia State University) for critically assessing the original manuscript, Dr Surajit Bhattacharya (Children’s Research Institute) for bioinformatics advice, four anonymous peer reviewers for feedback during the peer-review process, and the PhyloPic repository, the source for many of the animal silhouettes used throughout (distributed in public domain). We also thank all the investigators who made sequence information public, which made this work possible. This work is supported by the National Institute of Neurological Disorders and Stroke at the NIH (R01NS115209 to D.N.C.), the National Institute of General Medical Sciences at the NIH (R25GM109442-01A1), a GSU Brains and Behavior Fellowship (to N.J.H.), a GSU Brains and Behavior Seed (to D.N.C.), and a Kenneth W. and Georgeanne F. Honeycutt Fellowship (to N.J.H.).
Author contributions
Conceptualization and methodology, N.J.H.; sequence collection, N.J.H.; database curation, N.J.H. and T.R.G.; domain homology analysis, N.J.H. and T.R.G.; phylogenetic and other formal analyses, N.J.H.; prepared the original draft, N.J.H.; reviewed and edited the final draft, N.J.H., T.R.G., and D.N.C.; visualization, N.J.H.; supervision, D.N.C.; funding acquisition, D.N..C.
The TRPN, TRPC, TRPM, and TRPS sequence databases have been deposited on Dryad in the FASTA format (doi:10.5061/dryad.kwh70rz03).
References
- Bautista DM, Siemens J, Glazer JM, Tsuruda PR, Basbaum AI, Stucky CL, Jordt S-E, Julius D.. 2007. The menthol receptor TRPM8 is the principal detector of environmental cold. Nature. 448(7150):204–208. [DOI] [PubMed] [Google Scholar]
- Bergsten J. 2005. A review of long-branch attraction. Cladistics. 21(2):163–193. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schäffer DE, Iyer LM, Aravind L.. 2015. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43(22):10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JT, Vellutini BC, Smith J, Ronquist F, Jondelius U, Hejnol A.. 2016. Xenacoelomorpha is the sister group to Nephrozoa. Nature. 530(7588):89–93. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics (Oxford, England). 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson AE. 2019. Mechanical stimulation activates Drosophila eggs via Trpm channels. Proc Natl Acad Sci U S A. 116(38):18757–18758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang X, Yang T, Bi R, Huang Z, Ding H, Li J, Zhang J.. 2019. Emerging structural biology of TRPM subfamily channels. Cell Calcium. 79:75–79. [DOI] [PubMed] [Google Scholar]
- De Blas GA, Darszon A, Ocampo AY, Serrano CJ, Castellano LE, Hernández-González EO, Chirinos M, Larrea F, Beltrán C, Treviño CL.. 2009. TRPM8, a versatile channel in human sperm. PLoS One. 4(6):e6095–e6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll K, Stanfield GM, Droste R, Horvitz HR.. 2017. Presumptive TRP channel CED-11 promotes cell volume decrease and facilitates degradation of apoptotic cells in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 114(33):8806–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Halldórsson BV, Vernot B.. 2006. A hybrid micro–macroevolutionary approach to gene tree reconstruction. J Comput Biol. 13(2):320–335. [DOI] [PubMed] [Google Scholar]
- Fliegert R, Watt JM, Schöbel A, Rozewitz MD, Moreau C, Kirchberger T, Thomas MP, Sick W, Araujo AC, Harneit A, et al. 2017. Ligand-induced activation of human TRPM2 requires the terminal ribose of ADPR and involves Arg1433 and Tyr1349. Biochem J. 474(13):2159–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics (Oxford, England). 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel NJ, Letcher JM, Sakurai A, Gray TR, Benson MN, Cox DN.. 2019. Drosophila menthol sensitivity and the Precambrian origins of transient receptor potential-dependent chemosensation. Philos Trans R Soc B: Biol Sci. 374(1785):20190369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann T, Chubanov V, Chen X, Dietz AS, Gudermann T, Montell C.. 2010. Drosophila TRPM channel is essential for the control of extracellular magnesium levels. PLoS One. 5(5):e10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W.. 2010. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics (Oxford, England). 26(5):680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W.. 2008. BioVenn: a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 9(1):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer E.. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 338(5):1027–1036. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer E.. 2007. Advantages of combined transmembrane topology and signal peptide prediction: the Phobius web server. Nucleic Acids Res. 35:W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, Chan TF, Kwan HS, Holland PW, Chu KH, et al. 2016. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Hereditary. 116(2):190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma MT, Schmidt M, Ngo-Vu H, Sparks SD, Senatore A, Derby CD.. 2018. Chemoreceptor proteins in the Caribbean spiny lobster, Panulirus argus: expression of ionotropic receptors, gustatory receptors, and TRP channels in two chemosensory organs and brain. PLoS One. 13(9):e0203935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Majhi RK, Saha S, Kumar A, Ghosh A, Swain N, Goswami L, Mohapatra P, Maity A, Kumar Sahoo V, Kumar A, et al. 2015. Expression of temperature-sensitive ion channel TRPM8 in sperm cells correlates with vertebrate evolution. PeerJ. 3:e1310–e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Iwasaki W.. 2019. Graph splitting: a graph-based approach for superfamily-scale phylogenetic tree reconstruction. Syst Biol. syz049. [DOI] [PubMed] [Google Scholar]
- McKemy DD, Neuhausser WM, Julius D.. 2002. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 416(6876):52–58. [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, et al. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47(D1):D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Schliep K.. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35(3):526–528. [DOI] [PubMed] [Google Scholar]
- Parra RG, Espada R, Verstraete N, Ferreiro DU.. 2015. Structural and energetic characterization of the ankyrin repeat protein family. PLoS Comput Biol. 11(12):e1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peier AM, Moqrich A, Hergarden AC, Reeve AJ, Andersson DA, Story GM, Earley TJ, Dragoni I, McIntyre P, Bevan S, et al. 2002. A TRP channel that senses cold stimuli and menthol. Cell. 108(5):705–715. [DOI] [PubMed] [Google Scholar]
- Peng G, Shi X, Kadowaki T.. 2015. Evolution of TRP channels inferred by their classification in diverse animal species. Mol Phylogenet Evol. 84:145–157. [DOI] [PubMed] [Google Scholar]
- Philippe H, Poustka AJ, Chiodin M, Hoff KJ, Dessimoz C, Tomiczek B, Schiffer PH, Müller S, Domman D, Horn M, et al. 2019. Mitigating anticipated effects of systematic errors supports sister-group relationship between Xenacoelomorpha and Ambulacraria. Curr Biol. 29(11):1818–1826.e1816. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Hyun E, Zhao L, Lapointe TK, Chapman K, Hirota CL, Ghosh S, McKemy DD, Vergnolle N, Beck PL, et al. 2013. TRPM8 activation attenuates inflammatory responses in mouse models of colitis. Proc Natl Acad Sci U S A. 110(18):7476–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J.. 2007. g: profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 5:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohács T, Lopes CM, Michailidis I, Logothetis DE.. 2005. PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat Neurosci. 8(5):626–634. [DOI] [PubMed] [Google Scholar]
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K.. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47:W5–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Shingai R.. 2006. Evolution of thermoTRP ion channel homologs in vertebrates. Physiol Genomics. 27(3):219–230. [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Louis A, Bon C, Berthelot C, Roest Crollius H.. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A, Hughes TET, Moiseenkova-Bell VY.. 2018. Transient receptor potential (TRP) channels. Subcell Biochem. 87:141–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlingmann KP, Waldegger S, Konrad M, Chubanov V, Gudermann T.. 2007. TRPM6 and TRPM7: Gatekeepers of human magnesium metabolism. Biochim Biophys Acta. 1772(8):813–821. [DOI] [PubMed] [Google Scholar]
- Schnitzler MY, Wäring J, Gudermann T, Chubanov V.. 2008. Evolutionary determinants of divergent calcium selectivity of TRPM channels. FASEB J. 22:1540–1551. [DOI] [PubMed] [Google Scholar]
- Schüler A, Schmitz G, Reft A, Özbek S, Thurm U, Bornberg-Bauer E.. 2015. The rise and fall of TRP-N, an ancient family of mechanogated ion channels, in metazoa. Genome Biol Evol. 7(6):1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, Buffry AD, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, Durand D.. 2012. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics (Oxford, England). 28(18):i409–i415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L-T, Agapito MA, Li M, Simonson WTN, Huttenlocher A, Habas R, Yue L, Runnels LW.. 2006. TRPM7 regulates cell adhesion by controlling the calcium-dependent protease calpain. J Biol Chem. 281(16):11260–11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto T, Lambie EJ, Iwasaki K.. 2005. Differential regulation of TRPM channels governs electrolyte homeostasis in the C. elegans intestine. Cell Metabol. 1(5):343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson RC, Shaffer HB.. 2010. Sparse supermatrices for phylogenetic inference: taxonomy, alignment, rogue taxa, and the phylogeny of living turtles. Syst Biol. 59(1):42–58. [DOI] [PubMed] [Google Scholar]
- Turner HN, Armengol K, Patel AA, Himmel NJ, Sullivan L, Iyer SC, Bhattacharya S, Iyer EPR, Landry C, Galko MJ, et al. 2016. The TRP channels Pkd2, NompC, and Trpm Act in cold-sensing neurons to mediate unique aversive behaviors to noxious cold in Drosophila. Curr Biol. 26(23):3116–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C.. 2007. TRP channels. Annu Rev Biochem. 76(1):387–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Stolzer M, Goldman A, Durand D.. 2008. Reconciliation with non-binary species trees. J Comput Biol. 15(8):981–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(suppl_2):W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England). 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR.. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 29(19):2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Fujimoto S, Miyazaki K.. 2015. Phylogenetic position of Loricifera inferred from nearly complete 18S and 28S rRNA gene sequences. Zool Lett. 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida MA, Ishikura Y, Moritaki T, Shoguchi E, Shimizu KK, Sese J, Ogura A.. 2011. Genome structure analysis of molluscs revealed whole genome duplication and lineage specific repeat variation. Gene. 483(1–2):63–71. [DOI] [PubMed] [Google Scholar]
- Yue Z, Xie J, Yu AS, Stock J, Du J, Yue L.. 2015. Role of TRP channels in the cardiovascular system. Am J Physiol Heart Circ Physiol. 308(3):H157–H182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tóth B, Szollosi A, Chen J, Csanády L.. 2018. Structure of a TRPM2 channel in complex with Ca2+ explains unique gating regulation. eLife. 7:e36409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.