Abstract
Despite their essential role in chromosome segregation in most eukaryotes, centromeric histones (CenH3s) evolve rapidly and are subject to gene turnover. We previously identified four instances of gene duplication and specialization of Cid, which encodes for the CenH3 in Drosophila. We hypothesized that retention of specialized Cid paralogs could be selectively advantageous to resolve the intralocus conflict that occurs on essential genes like Cid, which are subject to divergent selective pressures to perform multiple functions. We proposed that intralocus conflict could be a widespread phenomenon that drives evolutionary innovation in centromeric proteins. If this were the case, we might expect to find other instances of coretention and specialization of centromeric proteins during animal evolution. Consistent with this hypothesis, we find that most mosquito species encode two CenH3 (mosqCid) genes, mosqCid1 and mosqCid2, which have been coretained for over 150 My. In addition, Aedes species encode a third mosqCid3 gene, which arose from an independent gene duplication of mosqCid1. Like Drosophila Cid paralogs, mosqCid paralogs evolve under different selective constraints and show tissue-specific expression patterns. Analysis of mosqCid N-terminal protein motifs further supports the model that mosqCid paralogs have functionally diverged. Extending our survey to other centromeric proteins, we find that all Anopheles mosquitoes encode two CAL1 paralogs, which are the chaperones that deposit CenH3 proteins at centromeres in Diptera, but a single CENP-C paralog. The ancient coretention of paralogs of centromeric proteins adds further support to the hypothesis that intralocus conflict can drive their coretention and functional specialization.
Keywords: centromeric proteins, intralocus conflict, positive selection, gene duplication
Introduction
Centromeric proteins represent an evolutionary paradox. Their critical role in cell division and chromosome segregation makes them essential for viability throughout eukaryotic life (Stoler et al. 1995; Howman et al. 2000; Blower and Karpen 2001). However, centromeric proteins evolve rapidly in plants and animals (Malik and Henikoff 2001; Talbert et al. 2004; Schueler et al. 2010) despite their essential function. This centromere paradox (Henikoff et al. 2001) is exemplified by the centromeric histone (CenH3), which is the foundational centromeric protein in most eukaryotes. CenH3 is essential for chromosome segregation in protists, fungi, plants, and most animals (Stoler et al. 1995; Buchwitz et al. 1999; Howman et al. 2000; Blower and Karpen 2001). Nevertheless, it is subject to rapid evolution in plants and animal species that undergo asymmetric female meiosis (Talbert et al. 2004; Zedek and Bures 2016), but not in species that lack asymmetric female meiosis (Baker and Rogers 2006). Thus, asymmetry in female meiosis may provide an opportunity for centromeres to act as selfish genetic elements and bias their transmission to the next generation, in a process termed “centromere drive” (Henikoff and Malik 2002; Kursel and Malik 2018). In this model, the rapid evolution of CenH3 proteins has been hypothesized to suppress harmful cheating behavior of selfish centromeres (Henikoff et al. 2001; Henikoff and Malik 2002; Malik 2009; Kursel and Malik 2018).
CenH3’s hypothesized role as a suppressor of centromere-drive is distinct from its essential role in mitotic and meiotic cell divisions. Moreover, CenH3 may perform additional specialized germline functions in males and females. For example, CenH3 inheritance in sperm chromatin, which undergoes a histone-to-protamine transition (Gaucher et al. 2010), is essential for epigenetic inheritance of centromere identity of paternal chromosomes postfertilization (Raychaudhuri et al. 2012). Our previous research (Kursel and Malik 2017, 2019) suggested that optimality of these multiple functions of CenH3 proteins might not be simultaneously achievable by a single CenH3 gene. As a result, we proposed that proteins like CenH3 might be subject to intralocus conflict, which is hypothesized to occur when two or more divergent functions are carried out by a single gene. Under these circumstances when both functions cannot be optimally carried out by a single gene, selection would favor evolutionary retention and subsequent specialization of duplicate genes (Des Marais and Rausher 2008; Gallach and Betran 2011). Resolution of intralocus conflict has been proposed to be the underlying mechanism to account for the duplication and specialization of sperm-specific mitochondrial proteins in flies (Gallach et al. 2010) and proteins involved in pigment biosynthesis in plants (Des Marais and Rausher 2008).
CenH3’s sex- and tissue-specific functions as well as its rapid evolution despite essential function make CenH3 a prime candidate for intralocus conflict. However, our previous investigation of CenH3 duplication events in Drosophila is the only study so far that has investigated CenH3 evolution and function in this light (Kursel and Malik 2017). Prior to this, the only known instances of CenH3 duplications in animals were two recent duplications in nematode species (Monen et al. 2005, 2015), and several CenH3 duplications in Bovidae (cows and sheep), most of which have become pseudogenized (Li and Huang 2008). Our analysis of CenH3 (Cid) in Drosophila identified five independent Cid gene duplication events and revealed that the majority of Drosophila species encode two or three Cid paralogs, including some that have been coretained for over 40 My (Kursel and Malik 2017; Teixeira et al. 2018). We hypothesized that these duplicate Cid genes perform nonredundant, specialized functions based on the fact that Cid paralogs evolve under distinct evolutionary constraints, and some paralogs have germline restricted expression patterns (Kursel and Malik 2017). Moreover, our cytological analysis of Cid1 and Cid5 in Drosophila virilis revealed that Cid1 and Cid5 acquired specialized gametic localization patterns; Cid1 is the primary CenH3 in the oocyte whereas Cid5 is the primary CenH3 in mature sperm (Kursel and Malik 2019). These results suggest that Cid paralogs in Drosophila are common, long-lived and perform specialized gametic functions, possibly to resolve intralocus conflict.
If selection to resolve intralocus conflict favors retention of specialized CenH3 paralogs, we would expect to find recurrent instances of germline-specialized CenH3s outside of Drosophila, including in other Dipteran species. We took advantage of recent genome sequencing efforts in another Dipteran family, Culicidae (mosquitoes) (Giraldo-Calderon et al. 2015; Neafsey et al. 2015) to investigate whether we could discover additional instances of CenH3 duplication and specialization. In line with our hypothesis, we find that most mosquito species encode two CenH3 paralogs that diverged over 150 Ma; the oldest CenH3 duplication identified so far. We designate these as mosquito Cid (mosqCid) to distinguish them from Drosophila Cid paralogs. We find that mosqCid paralogs encode divergent N-terminal tails and evolve under different evolutionary constraints. Furthermore, some mosqCid paralogs show biased expression patterns during oogenesis and early embryogenesis. Finally, we report that Anopheles mosquitoes also encode two paralogs of the CAL1 gene, which encodes the CenH3 chaperone in Drosophila (Chen et al. 2014). Like the duplication of CENP-C seen previously in Drosophila species (Teixeira et al. 2018), our findings suggest that multiple inner kinetochore proteins in Diptera have undergone gene duplications, potentially to resolve intralocus conflict (Des Marais and Rausher 2008; Gallach and Betran 2011). Our findings further support the hypothesis that centromeric proteins may perform divergent functions in soma versus germline, which selects for retention of specialized duplicate genes.
Results
Mosquito Genomes Harbor Ancient Cid Paralogs
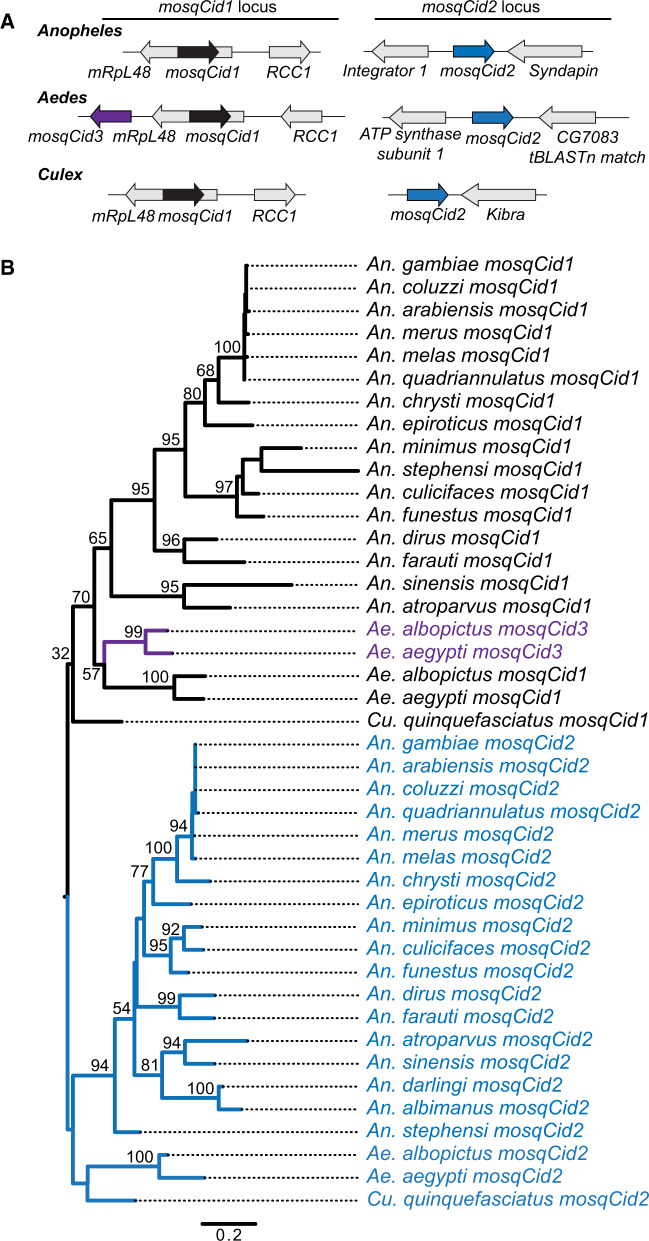
The recent publication of multiple high quality Anopheles genomes (Neafsey et al. 2015) provides a set of densely sampled, closely related Dipteran genomes that are well suited to phylogenomic analyses (Giraldo-Calderon et al. 2015). We used these genomes for phylogenomic analyses of centromeric proteins, starting with the CenH3 (or Cid) genes. To identify mosquito Cid (mosqCid) homologs, we used Drosophila melanogaster Cid as a query and performed TBlastN against 21 mosquito genomes including 18 Anophelinae mosquitoes and three Culicinae species (two Aedes and one Culex). We found that Anopheles gambiae encodes two mosqCid paralogs in distinct genomic loci (fig. 1A); both genes are encoded by a single exon. The mosqCid1 paralog is located in the intron of the mRpL48 gene, whereas mosqCid2 is found between the Integrator complex subunit 1 (Integrator 1) and Syndapin genes (fig. 1A, supplementary table S1, Supplementary Material online). The An. gambiae mosqCid paralogs are highly divergent and share only 51% amino acid identity in their histone fold domains; this divergence is similar to the Cid paralogs in Drosophila. Next, we extended our analyses to genomes of other Anopheles mosquitoes. Many of the mosqCid genes are not yet annotated in the public databases and the mosqCid open reading frame required manual curation in several cases (supplementary table S1, Supplementary Material online). Nevertheless, we found both mosqCid paralogs in the same shared syntenic location in nearly all other species. The only exceptions were in Anopheles albimanus and Anopheles darlingi where we only found the mosqCid2 gene (supplementary table S1, Supplementary Material online). In these species, we were able to find the shared syntenic locus containing the mRpL48 gene, but this location was missing mosqCid1 and contained no identifiable pseudogene. Moreover, we did not find any other mosqCid sequences in these genomes, suggesting these two species lack mosqCid1 and only encode a single mosqCid2 CenH3 gene.
Fig. 1.
Identification and evolution of mosquito Cid paralogs. (A) The genomic context of representative mosquito Cid paralogs identified by TBlastN is schematized for Anopheles, Aedes, and Culex. In total, we found three mosquito Cid genes: mosquitoCid1 (mosqCid1, black arrow) is present in the intron of mRpL48 in Anopheles, Aedes, and Culex mosquitoes. MosqCid2 (blue arrow) is found in Anopheles between the genes Integrator 1 and Syndapin. In Aedes, mosqCid2 is located between ATP synthase subunit 1 and a gene with homology to Drosophila melanogaster CG7083. In Culex, mosqCid2 is in a genomic locus next to the Kibra gene. MosqCid3 (purple arrow) is an Aedes-specific paralog that is also present in the mosqCid1 locus. Arrows colored in gray represent genes that define the syntenic locus of each paralog and are named based on the D. melanogaster gene name. (B) We performed maximum likelihood phylogenetic analyses using PhyML with a nucleotide alignment of the histone fold domain of all mosqCid paralogs. We found that mosqCid1 (black) forms a monophyletic clade from which mosqCids3 (purple) arose, indicating that mosqCid3 is derived from a mosqCid1 gene duplication event. All mosqCid2 genes form a monophyletic clade. This suggests that even though mosqCid2 genes are in a different syntenic location in Anopheles, Aedes, and Culex, they are likely orthologous. Bootstrap values >50 and values at key nodes are shown. The tree is rooted on the common ancestor of Anopheles, Aedes, and Culex mosquitoes. Scale bar represents nucleotide substitutions per site.
Next, we investigated the mosqCid genes in two Aedes species: Aedes aegypti and Aedes albopictus. To our surprise, we were able to identify three mosqCid paralogs in these species. These included mosqCid1 orthologs located in the same shared syntenic location as in Anopheles species, that is, in the intron of mRpL48 (fig. 1A, supplementary table S1, Supplementary Material online). We also identified mosqCid3 located in close proximity to mosqCid1. Finally, we identified a third Aedes mosqCid paralog located between the ATP synthase subunit 1 and CG7083 genes. Although the unique syntenic location suggested an independent mosqCid gene duplication, phylogenetic analyses (below) confirmed that this gene is likely to be an ortholog of the Anopheles mosqCid2 gene (fig. 1A); we therefore named this gene as mosqCid2. Based on the syntenic conservation of the mosqCid2 gene location in Anopheles (Integrator1-Syndapin) and in Aedes (ATP synthase 1-CG7083), we propose that a single gene transposition may account for the different locations of mosqCid2.
Finally, we examined the mosqCid genes present in Culex quinquefasciatus. We found that C. quinquefasciatus contained two mosqCid paralogs, including mosqCid1 in the mRpL48 intron (fig. 1A, supplementary table S1, Supplementary Material online) and a second mosqCid gene in a distinct syntenic location, adjacent to a gene that shares homology with D. melanogaster Kibra. Once again, we relied on phylogenetic analyses to confirm that the second gene corresponds to mosqCid2 despite its distinct genomic location from either the Anopheles or Aedes orthologs.
The presence of mosqCid genes in a shared syntenic location across species is a strong indicator that they are likely orthologous. Based on this criterion, we predicted that all mosqCid1 genes are orthologs and that mosqCid1 was likely present in the common ancestor of all mosquitoes but subsequently lost in the ancestor of An. albimanus and An. darlingi. In contrast to mosqCid1, we were unable to assign the other mosqCid genes into orthologous groups based on shared synteny alone. To clarify their evolutionary relationships to each other and to mosqCid1, we performed phylogenetic analyses based on maximum likelihood using a nucleotide alignment of the histone fold domain of all mosqCid genes (fig. 1B, supplementary data S1, Supplementary Material online). We found that mosqCid1 and mosqCid3 group together, suggesting that mosqCid3 arose from a mosqCid1 duplication event in the common ancestor of Ae. aegypti and Ae. albopictus (fig. 1B). Furthermore, we found that the mosqCid2 genes from all 21 mosquito species examined formed a monophyletic clade and are likely to be orthologous despite being found in distinct syntenic contexts in Aedes and Culex (fig. 1B). Finally, we found that the subtrees formed by the high-confidence branches for each mosqCid paralog mirrors the mosquito species tree (supplementary fig. S1, Supplementary Material online), supporting our conclusion of orthology.
Overall, our synteny and phylogenetic analyses identify two independent duplications of mosqCid genes during the 150 My history of mosquito evolution that we have investigated. We conclude that mosqCid1 and mosqCid2 were present in the common ancestor of all examined mosquito species and have been largely coretained for over 150 My, making them the oldest and most diverged CenH3 paralogs identified in any lineage. The only exception to this coretention was the loss of mosqCid1 in the common ancestor of An. albimanus and An. darlingi, suggesting that at least in this pair of species, mosqCid2 is capable of carrying out all centromeric functions. Thus, in mosquito species, coretention of ancient paralogs of CenH3s is the rule, rather than the exception.
Even though mosqCid1 is retained in the same, shared syntenic context whereas mosqCid2 is not, this does not imply that mosqCid1 is older than mosqCid2. The numbering of mosqCid1 and mosqCid2 is thus arbitrary and not an indication of ancestry. In order to assess the relative age of the two ancient mosqCid paralogs, we compared the phylogenetic relationships of mosqCid1 and mosqCid2 proteins to the putative CenH3 from the outgroup Mochlonyx cinctipes based on a multiple alignment of their conserved histone fold domains (supplementary fig. S2 and data S1, Supplementary Material online). This analysis revealed low-confidence (bootstrap support = 52) grouping of M. cinctipes CenH3 with Aedes mosqCid2 (supplementary fig. S2, Supplementary Material online). Therefore, we tentatively conclude that mosqCid2 is the ancestral mosqCid and that mosqCid1 arose from a gene duplication event in the common ancestor of Aedes and Anopheles mosquitoes.
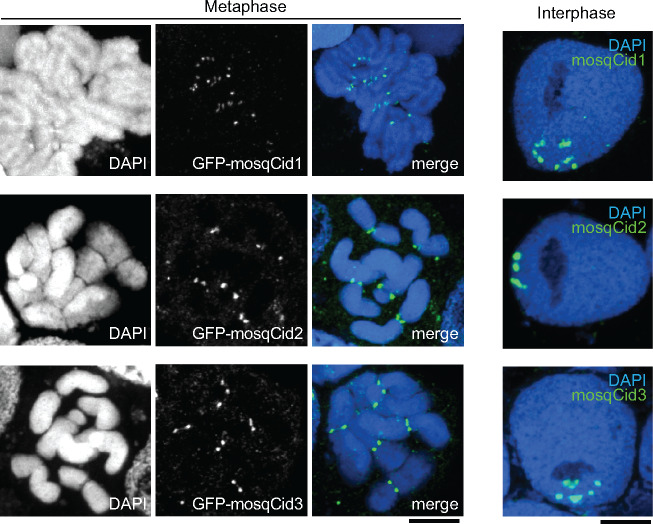
Given the long period of their coretention, we considered the possibility that at least some of the mosqCid paralogs have acquired a new, noncentromeric function. To test this possibility, we assayed the cytological localization of each of the three Ae. albopictus mosqCid paralogs in mosquito cells. We expressed GFP-tagged versions of each of Ae. albopictus mosqCid1, mosqCid2, and mosqCid3 in Ae. albopictus cell lines using transient transfections and examined the cytological location of the expressed proteins (fig. 2). We found that all three proteins localize to the primary constriction in metaphase chromosomes and to presumed centromeric foci in interphase cells, confirming that all three mosqCid paralogs localize to centromeres and therefore likely function as CenH3s.
Fig. 2.
Localization of mosqCid paralogs in an Aedes albopictus cell line. Images of GFP-tagged mosqCid paralogs from Ae. albopictus transiently expressed in Ae. albopictus cell culture. All mosqCid paralogs localize to the primary constrictions on metaphase chromosomes (left three panels) and to discrete foci in interphase cells (right). Scale bar = 2 μm.
Anopheles Mosquitoes Encode Two Paralogs of the Cid Chaperone CAL1 but a Single CENP-C Ortholog
A previous study showed that Cid duplication coincided with the duplication of CENP-C in some Drosophila species (Teixeira et al. 2018). This finding motivated us to examine if any other inner kinetochore proteins showed parallel signatures of gene duplication in mosquitoes. Unlike vertebrates, which have a complex network of inner kinetochore proteins (Hori et al. 2008), Drosophila inner kinetochores are relatively less complex, comprised primarily of Cid, CENP-C, and the Cid chaperone CAL1 (Mellone et al. 2011). Furthermore, Cid physically interacts with and is thought to co-evolve with the CenH3 chaperone, CAL1 (Chen et al. 2014; Rosin and Mellone 2016). We, therefore, investigated the possibility that the highly divergent mosqCid paralogs may require different CAL1 chaperones to aid their deposition.
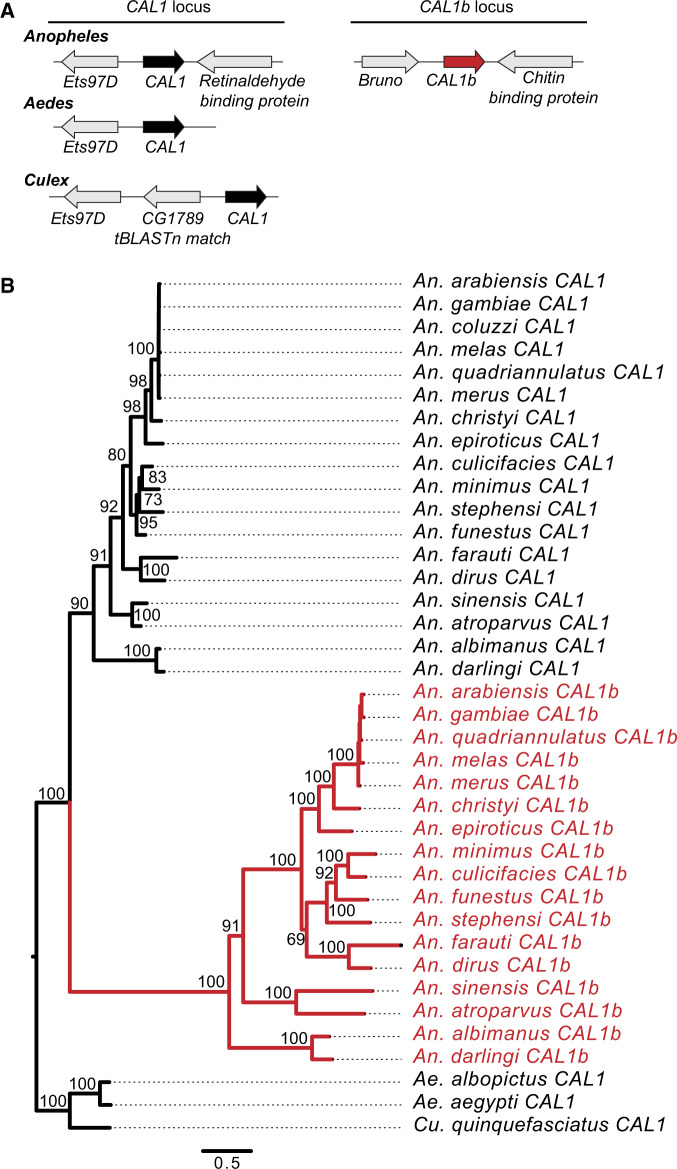
Cid homologs are relatively easy to identify due to the conservation of their histone fold domains. However, CAL1 homology is less well conserved, and we could obtain only marginal matches to a few mosquito genomes using D. melanogaster CAL1 as a BLAST query. We, therefore, adopted an iterative search strategy (see Materials and Methods) to successfully identify CAL1 in An. gambiae and Ae. aegypti, similar to a previous study (Phansalkar et al. 2012). These genes are both in the same syntenic location and share Ets97D as their 5-prime neighbor gene (fig. 3A, supplementary table S1, Supplementary Material online). When we extended our search to all Anopheles, Aedes, and Culex genomes, we found CAL1 in the same syntenic location in all species (fig. 3A, supplementary table S1, Supplementary Material online). Surprisingly, we also found a second strong BLAST hit but only in Anopheles genomes. This CAL1-related gene (CAL1b) resides in a distinct shared syntenic location between genes homologous to D. melanogaster Bruno and Chitin binding protein (supplementary table S1, Supplementary Material online). We found the presence of CAL1b in all Anopheles species. We found no additional CAL1 genes in Aedes or Culex even with other iterations of BLAST searches in which we used multiple Anopheles CAL1 or CAL1b homologs as starting queries. This suggests that CAL1b arose via a gene duplication of CAL1 in the common ancestor of Anopheles species.
Fig. 3.
Identification and evolution of mosquito CAL1 paralogs. (A) The genomic context of representative mosquito CAL1 paralogs identified by TBlastN is schematized for Anopheles, Aedes, and Culex. We found two CAL1 genes in Anopheles. The CAL1 (black arrow) syntenic locus is defined by the genes Ets97D and Retinaldehyde binding protein. The CAL1b (red arrow) syntenic locus is defined by the genes Bruno and Chitin binding protein. We only found one CAL1 gene in Aedes and Culex. Arrows colored in gray represent genes that define the shared syntenic locus of each paralog. Genes that define each syntenic locus are named based on the Drosophila melanogaster gene name. (B) We performed maximum likelihood phylogenetic analyses using PhyML with an amino acid alignment of all CAL1 and CAL1b proteins. We found that Anopheles CAL1 and CAL1b each form well-supported monophyletic clades. Aedes and Culex CAL1 form a well-supported outgroup to Anopheles CAL1 and CAL1b. This suggests that CAL1 was the ancestral chaperone and that CAL1b was born in the common ancestor of Anopheles mosquitoes. Bootstrap values >50 are shown. The tree is rooted on the common ancestor of Anopheles, Aedes, and Culex mosquitoes. Scale bar represents number of substitutions per site.
Next, we performed phylogenetic analyses on all mosquito CAL1 and CAL1b genes. We made an amino acid-based alignment of all CAL1 homologs and used PhyML to make a maximum likelihood phylogeny with 100× resampling (fig. 3B, supplementary data S2, Supplementary Material online). We found that Anopheles CAL1 and CAL1b each form monophyletic sister clades within Anopheles, with Aedes and Culex CAL1 proteins as an outgroup. This supports our hypothesis that CAL1 is the ancestral chaperone and that CAL1b arose from a CAL1 gene duplication event in the common ancestor of Anopheles mosquitoes ∼100 Ma (fig. 3B) and has been strictly coretained since. To the best of our knowledge, this is the first time a CenH3 chaperone duplication has been reported in any eukaryote.
Having found paralogs for both mosqCid and CAL1 in mosquito genomes, we next examined if they also encoded paralogs of the third conserved inner kinetochore protein CENP-C. At the sequence level, CENP-C is even less conserved than CAL1. Only the C-terminal cupin domain is a reliable bioinformatic marker for assigning CENP-C homology (Talbert et al. 2004; Cohen et al. 2008; Orr and Sunkel 2011; Kral 2015). Therefore, we used the D. melanogaster CENP-C cupin domain to identify all mosquito homologs of CENP-C (supplementary data S3, Supplementary Material online, see Materials and Methods). In each case, we were able to find CENP-C orthologs with high confidence based on the conserved cupin domain. However, we discovered no putative paralogs even while using the mosquito CENP-C proteins as a query for iterative BLAST searches.
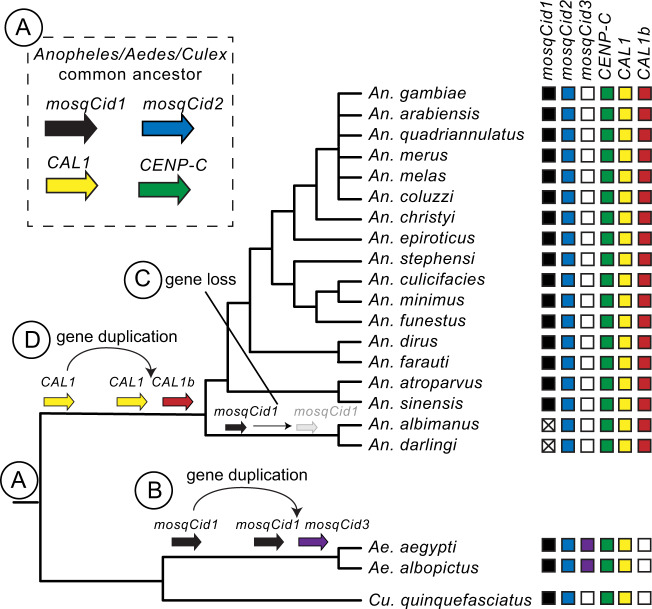
Thus, our analyses reveal that the CenH3 mosqCid and the CenH3 chaperone CAL1 underwent ancient gene duplications in mosquitoes, whereas CENP-C did not. We compared the duplication histories of mosqCid and CAL1 to examine the possibility that two duplications may be causally related (fig. 4). If this were the case, we would expect that the mosqCid and CAL1 duplications and retention patterns would coincide, that is, they would be born and lost along the same branches. In contrast to this expectation, we find that the mosqCid duplication in all mosquito species preceded the CAL1b duplication (which is found only in Anopheles species). Moreover, even after the loss of mosqCid1 in An. albimanus and An. darlingi, CAL1 and CAL1b are still coretained, arguing against a one-for-one specialization of the two chaperones with the two mosqCid paralogs in Anopheles species (fig. 4).
Fig. 4.
Summary of mosquito Cid, CAL1, and CENP-C evolution. A mosquito species tree is presented with boxes to the right of each species indicating the presence (color-filled box) or absence (white box) of each mosquito Cid, CAL1, and CENP-C gene. A white box with an “X” indicates that a given paralog is absent but was likely lost based on its presence in other species. Genes present in the same vertical column are hypothesized to be orthologous. (A) The common ancestor of Anopheles, Aedes, and Culex mosquitoes likely had two mosqCid genes, mosqCid1 (black arrow), and mosqCid2 (blue arrow), making these paralogs over 150 million years old. The Anopheles/Aedes/Culex common ancestor also had single copy of CAL1 (yellow arrow) and CENP-C (green arrow). (B) mosqCid1 duplicated in the common ancestor of Aedes mosquitoes to give rise to mosqCid3 (purple arrow) 20–60 Ma. (C) mosqCid1 was lost in the common ancestor of Anopheles albimanus and Anopheles darling (gray arrow and white box with an “X”). (D) CAL1 (yellow arrow) duplicated in the common ancestor of Anopheles mosquitoes to give rise to CAL1b (red arrow) ∼60 Ma.
Tissue-Specific Expression Pattern of mosqCid and CAL1 Paralogs
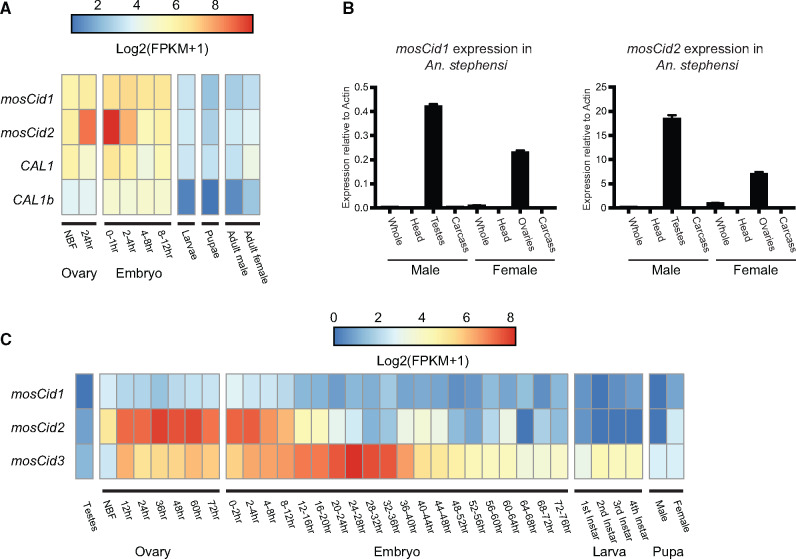
The ancient coretention of mosqCid and CAL1 paralogs in mosquito genomes raised the possibility that these paralogs have acquired specialized functions, potentially via tissue-specific expression patterns, similar to Drosophila Cid paralogs (Kursel and Malik 2017). We began by analyzing previously published genome-wide RNA-seq analyses to discern any evidence for tissue-specific expression. A previous study investigated gene expression in Anopheles stephensi at several life stages including early embryos, larvae, pupae, adult males, adults females, and ovaries both prior to blood feeding and 24 h after blood feeding (Biedler et al. 2014). We examined the expression of mosqCid1, mosqCid2, CAL1, and CAL1b in each of these tissues. We found that mosqCid1, mosqCid2, CAL1, and CAL1b are all expressed at relatively high levels in ovaries and the early embryo but at fairly low levels in larvae, pupae, and adults (fig. 5A, supplementary table S2, Supplementary Material online). Interestingly, however, we find that mosqCid2 expression increases 6-fold compared with the non-blood fed (NBF) ovary 24 h after blood feeding, which induces oogenesis in mosquitoes. Furthermore, mosqCid2 expression continues to increase in the 0–1 h embryo, ultimately reaching expression levels >10-fold higher than in the NBF ovary. This suggests that mosqCid2 plays an important function in female meiosis or female gamete development. In contrast to mosqCid2, blood feeding did not alter expression of mosqCid1, CAL1, and CAL1b. Our finding of significantly elevated expression of a CenH3 paralog but not its chaperone is unexpected. This discrepancy could either suggest that the bulk of mosqCid2 transcripts are either not immediately translated or that mosqCid2 proteins are not immediately incorporated into centromeric chromatin during oogenesis.
Fig. 5.
Expression of mosqCid and CAL1 paralogs. (A) Heatmap displaying Log2(FPKM+1) values for Anopheles stephensi mosqCid1, mosqCid2, CAL1, and CAL1b at various developmental stages. RNAseq data were re-analyzed from Biedler et al. (2014). mosqCid2 expression increases in the ovary after blood feeding. (B) RT-qPCR for mosqCid1 and mosqCid2 from dissected tissues from A. stephensi males and females revealed that both mosqCid1 and mosqCid2 are expressed in testes and ovaries but mosqCid1 is expressed at higher levels than mosqCid2. All RT-qPCR was normalized using Actin as a control. Error bars represent standard deviation calculated from three technical replicates. (C) Heatmap displaying Log2(FPKM+1) values for Aedes aegypti mosqCid1, mosqCid2, and mosqCid3 at various developmental stages. Original RNAseq data are from Akbari et al. (2013).
Our previous analyses showed that many Drosophila Cid paralogs show testis-biased expression (Kursel and Malik 2017). However, the published expression analysis (Biedler et al. 2014) study did not investigate gene expression in testes. To address this, we dissected adult tissues (including testes and ovaries) from An. stephensi mosquitoes and investigated the expression of mosqCid1 and mosqCid2 by RT-qPCR (fig. 5B). We found that relative expression of both mosqCid genes was highest in testes and ovaries. However, expression of mosqCid2 was nearly 40 times higher than mosqCid1 in testes and ∼20 times higher than mosqCid1 in ovaries, relative to the Actin controls. Our RT-qPCR analysis is consistent with our previous expression studies that show highly enriched mosqCid2 expression in ovaries upon blood feeding. However, neither mosqCid paralog appears to have a testes-specific expression. Our findings suggest that mosqCid2 may be the predominant germline-specific CenH3 gene expressed in both males and females. However, our RT-qPCR analyses cannot address the possibility that mosqCid1 and mosqCid2 are specialized for specific germline cell types, as is the case for Cid1 and Cid5 in D. virilis (Kursel and Malik 2019). For example, if mosqCid1 were specialized for or retained on sperm, RNA-seq or RT-qPCR analyses of whole testes would lack the resolution to identify this.
We extended our survey of expression to include the three mosqCid paralogs in Ae. aegypti, mosqCid1, mosqCid2, and the Aedes-specific mosqCid3. Using previously published RNA-seq data (Akbari et al. 2013), we found that Ae. aegypti mosqCid2 expression in the ovary dramatically increased after blood feeding, then gradually decreased over the first 24 h of embryonic development (fig. 5C) paralleling the observed expression pattern of mosqCid2 in An. stephensi (fig. 5A). This conserved pattern of increased expression after blood feeding in divergent Aedes and Anopheles species further supports our conclusion of orthology between these two mosqCid2 genes, which appear to be the primary CenH3 expressed in mosquito ovaries. Furthermore, we found that the expression of the Aedes-specific paralog, mosqCid3, increased during the first 28 h of embryogenesis, concurrent with the decrease in mosqCid2 expression. We found that mosqCid3 expression peaked after 24–28 h and then decreased from 28 to 76 h after the onset of embryogenesis (fig. 5C). This suggests that mosqCid3 might have a specialized function in early embryogenesis in Aedes species. Expression of Ae. aegypti mosqCid1 is consistently low at all stages. However, without cytological analyses in vivo, we cannot rule out the possibility that mosqCid1 could be the predominant CenH3 protein in certain tissues in the soma or germline. Notably, all Aedes mosqCid paralogs are expressed at low levels in testes. Thus, in contrast to Drosophila, where Cid paralogs have acquired testis-biased expression patterns, mosqCid paralogs have acquired a predominantly ovary-biased expression pattern in mosquitoes.
Distinct Selective Pressures Act on mosqCid Paralogs
Our phylogenomic analyses reveal that mosqCid and CAL1 paralogs have been largely coretained for >100 My of Anopheles evolution. This suggests that in most Anopheles species, both pairs of paralogs perform nonredundant functions. We investigated whether these nonredundant functions could have led to different selective constraints, as was found for Cid paralogs in Drosophila (Kursel and Malik 2017).
We focused our attention on a subset of Anopheles species for two reasons. First, these species are the most densely sampled mosquito genus. Second, these species are moderately diverged from one another, which allowed us to evaluate selective constraints without the confounding effect of saturated synonymous site substitutions. We used maximum likelihood methods in the PAML suite of programs to analyze the selective constraints, comparing rates of nonsynonymous (dN) to synonymous (dS) substitutions for each codon in full-length alignments of mosqCid1, mosqCid2, CAL1, and CAL1b. We found strong evidence for recurrent positive selection having acted on mosqCid1 (table 1, mosqCid1, M7 vs. M8 P-value = 0.008, M8a vs. M8 P-value = 0.004, supplementary data S4, Supplementary Material online) but not mosqCid2. Indeed, we found that nearly one-quarter (22%) of the codons in mosqCid1 have evolved with an average dN/dS of 2.5. Our finding of positive selection acting only on mosqCid1 suggests that mosqCid1 is involved in a genetic conflict and may evolve rapidly to suppress the deleterious effects of centromere-drive in mosquito genomes, as we previously hypothesized for Cid in Drosophila species (Malik and Henikoff 2001). This finding is also consistent with the intralocus conflict hypothesis, which posits that gene duplication followed by specialization allows one mosqCid paralog to evolve rapidly without compromising essential centromeric function mediated by the other paralog (Des Marais and Rausher 2008; Gallach and Betran 2011). Our observations are highly reminiscent of our previous findings in Drosophila, where one of multiple Cid paralogs usually showed signatures of positive selection (Kursel and Malik 2017).
Table 1.
PAML Tests for Positive Selection on mosqCid and CAL1 Paralogs.
| Number of Sequences | Alignment Length | M1 versus M2P-Value | M7 versus M8P-Value | M8a versus M8P-Value | Omega (% Sites) | Tree Length | |
|---|---|---|---|---|---|---|---|
| mosqCid1 | 7 | 705 | 0.01 | 0.008 | 0.004 | 2.5 (22%) | 2.04 |
| mosqCid2 | 7 | 777 | 1.00 | 1 | 0.90 | n.a. | 1.64 |
| CAL1 | 8 | 1671 | 1.00 | 0.71 | 0.92 | n.a. | 1.86 |
| CAL1b | 7 | 1740 | 1.00 | 0.003 | 0.99 | n.a. | 4.33 |
Note.—Using the PAML suite (Yang 2007), we tested whether NsSites models that permitted a subset of codons to evolve under positive selection (M8) were a more likely fit to the data than those models (M7, M8a) that disallowed it. Tree length refers to the number of nucleotide substitutions per codon, giving an indication of the divergence of the data set. The results we present are from codeml runs using the F3x4 codon frequency model and initial omega (dN/dS) of 0.4. This test was performed with multiple initial omega values and codon frequency models and the results were consistent with those shown. Summary table of M1 versus M2, M7 versus M8 and M8a versus M8 PAML results for mosqCid1, mosqCid2, CAL1, and CAL1b. P-values <0.05 are indicated in bold text.
Our analyses thus far suggest that mosqCid2 is the ancestral CenH3 paralog. It is expressed at higher levels than mosqCid1 and it evolves under a higher degree of selective constraint in species with both mosqCid paralogs. In contrast, the putatively younger mosqCid1 is expressed at low levels but shows unmistakable signatures of recurrent positive selection, implicating it in a genetic conflict. Despite these differences in expression and selective constraint, both mosqCid paralogs have been almost completely coretained for 150 My. The only exception is the loss of mosqCid1 in two sister species, An. albimanus and An. darlingi. This loss may have created a change in the selective pressure on the remaining mosqCid2 gene, which presumably performs all centromeric function in these species. To test this possibility, we used the RELAX method (Wertheim et al. 2015) to evaluate whether the loss of mosqCid1 precipitated a change in the selective constraints acting on mosqCid2 in these two species relative to other mosqCid2 orthologs in Anopheles species. Although the RELAX method is not suitable explicitly for identifying positive selection, it is highly suitable for finding shifts in selective constraints. We found significant evidence (P = 0.042) for intensification of selection (K = 1.58) due to an increase in the strength of positive selection acting on a subset of sites in An. albimanus and An. darlingi mosqCid2 following the loss of mosqCid1 (supplementary fig. S3, table S3 and data S1, Supplementary Material online). Our analyses suggest that, in the absence of mosqCid1, mosqCid2 may take over the genetic conflict-associated function of mosqCid1 (e.g., as a suppressor of centromere-drive) leading to mosqCid2 now being subject to more rapid evolution, consistent with the predictions of the intralocus conflict hypothesis.
We performed similar tests for positive selection on CAL1 and CAL1b. We found no evidence of positive selection acting on CAL1 (table 1, CAL1 M7 vs. M8 P-value = 0.71, M8a vs. M8 P-value = 0.92). Our findings are consistent with previous analyses that found no evidence for positive selection acting on CAL1 in Drosophila (Phansalkar et al. 2012). For CAL1b, although the M7 versus M8 comparison did indicate positive selection (table 1, CAL1b M7 vs. M8 P-value = 0.003), the M8a versus M8 comparison was not significant (P-value = 0.99). Therefore, we attribute the majority of the positive selection signal identified by M7 versus M8 to codons evolving close to neutrality with dN/dS = 1. Indeed, the longer branch lengths on the CAL1b phylogeny relative to CAL1 suggest that CAL1b evolves under more relaxed constraint (fig. 3). RELAX analyses also found significant evidence (P = 0.000) for relaxed selection (K = 0.77) on CAL1b compared with CAL1 (supplementary fig. S3, table S3 and data S2, Supplementary Material online). Taken together, these data suggest that despite its strict retention in Anopheles species, CAL1b evolves under more relaxed constraints than ancestral CAL1.
Protein Motif Analyses Provide Insights into mosqCid Specialization and CAL1 Origins
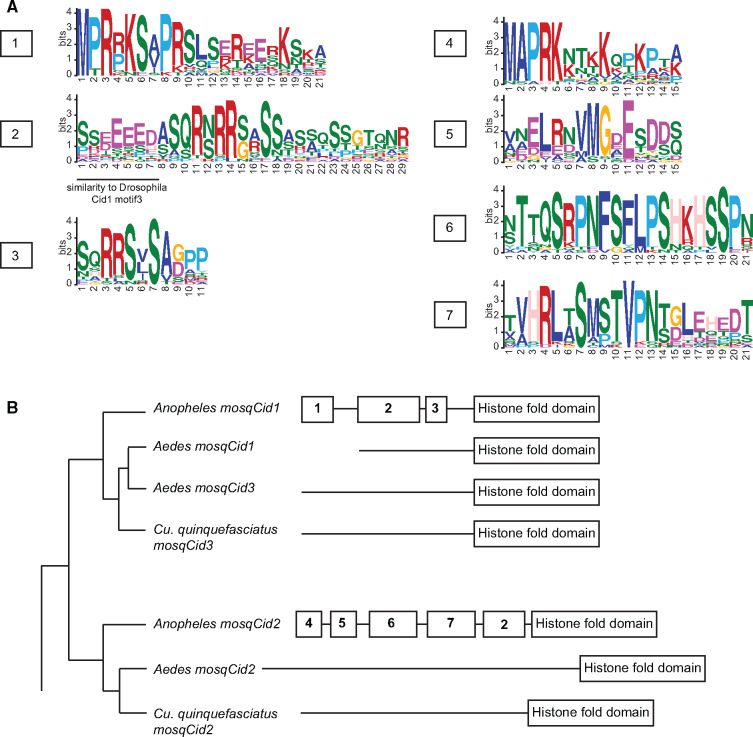
We previously showed that Drosophila Cid paralogs acquired and lost N-terminal motifs (Kursel and Malik 2017) that might mediate protein–protein interactions with other kinetochore proteins. Therefore, we asked if Anopheles mosqCid1 and mosqCid2 protein sequences contained the motifs we previously identified in the N-terminal tail of D. melanogaster Cid paralogs. The only modest hit we found was to motif 3 found in Drosophila Cid proteins (Kursel and Malik 2017), but this “hit” could be attributed to a stretch of acidic amino acids. We conclude that the Drosophila Cid motifs are generally not conserved in mosquito Cid paralogs.
We next investigated whether mosqCids have their own unique set of N-terminal tail motifs. The de novo discovery of protein motifs requires a minimum number of orthologs. Thus, we are only able to discover motifs within the mosqCid paralogs in Anopheles genomes. We subsequently ascertained whether these motifs were conserved in the Aedes and Culex species’ mosqCid paralogs. We used the motif generator algorithm MEME to identify conserved protein motifs in the N-terminal tails of all Anopheles mosqCid1s and separately in all Anopheles mosqCid2s. We identified three protein motifs (called motifs 1–3) conserved in all Anopheles mosqCid1 orthologs (fig. 6A, B). Similarly, we identified four short motifs present in the N-terminal tails of all Anopheles mosqCid2 proteins (motifs 4–7, fig. 6A, B). We used the motif search program MAST to search for mosqCid1 motifs in mosqCid2 sequences and vice versa. This analysis revealed almost no significant matches of motifs 1–3 in mosqCid2 and no significant matches of motifs 4–7 in mosqCid1. This confirms that mosqCid1 and mosqCid2 in Anopheles have almost distinct N-terminal tails with essentially nonoverlapping motifs. The only common motif was mosqCid1 motif2, which is likely a result of the acidic patch similar to D. melanogaster motif 3 (fig. 6B).
Fig. 6.
Analysis of N-terminal motifs in mosquito Cid proteins. (A) Logos generated by MEME for mosqCid consensus motifs 1–7. Drosophila Cid1 motif 3 shared similarity with mosqCid1 motif 2 in that they both contain a stretch of negatively charged amino acids (underlined). (B) A mosquito species tree with a schematic of N-terminal tail motifs identified by MEME and MAST displayed to right of each species or species group. Each number represents a unique motif that does not statistically match any other motif.
Next, we looked for all mosqCid motifs in Aedes and Culex mosqCid paralogs. We found no significant matches to Aedes or Culex N-terminal tails using the Anopheles motifs as a query. Aedes and Culex almost certainly have their own set of N-tail motifs but identification of Aedes- or Culex-specific motifs would require additional sequencing efforts. It is not surprising that the Anopheles motifs do not match the Aedes or Culex mosqCid sequences because Anopheles and Aedes shared a common ancestor over 150 Ma, far more ancient than the ∼60 million-year-old divergence we previously analyzed in Drosophila species (Kursel and Malik 2017).
In summary, our motif analysis revealed that mosqCid1 and mosqCid2 are subject to distinct selective pressures and have highly divergent N-terminal tails. Although the function of the N-terminal tail remains mostly unknown, we hypothesize that differences in N-terminal tail motifs may be indicative of different protein–protein interactions, possibly due to functional specialization.
Discussion
Although CenH3 duplications were thought to be rare and short-lived in animal species, we previously identified multiple, ancient duplications of Cid paralogs during Drosophila evolution (Kursel and Malik 2017). Indeed, the majority of Drosophila species likely encode more than one Cid paralog. In addition to harboring two Cid paralogs (Cid1 and Cid5), the Drosophila subgenus also contains two CENP-C paralogs, CENP-C1 and CENP-C2, which have been coretained for at least 40 My without loss (Teixeira et al. 2018). Our cytological analysis of Cid1 and Cid5 in D. virilis further revealed that Cid1 and Cid5 likely perform specialized functions in the oocyte and mature sperm, respectively. These findings suggest that centromeric protein duplicates might be both more common as well as more long-lived than previously believed. This led us to hypothesize that retention of CenH3 gene duplicates and subsequent functional specialization may be advantageous in order to resolve intralocus conflict. In the present study, we further confirm this hypothesis by finding that most mosquito species encode two mosqCid paralogs (mosqCid1 and mosqCid2) that diverged at least 150 Ma. In addition, all Anopheles species encode two paralogs of CAL1, which is the CenH3 chaperone in Diptera. Thus, coretention of CenH3, and other centromeric protein paralogs appears to be a frequent occurrence in insect genomes.
The ancient coretention of multiple mosqCid paralogs suggests that they must have diverged in some aspect of their centromeric function. Indeed, we find multiple pieces of evidence suggestive of mosqCid specialization similar to our previous findings for Cid paralogs in Drosophila (Kursel and Malik 2017). First, mosqCid1 evolves under recurrent positive selection whereas mosqCid2 does not. Second, we find that both mosqCid1 and mosqCid2 have different motifs conserved in their N-terminal tails, presumably for different protein–protein interactions with other kinetochore factors. Third, the expression patterns of the two paralogs are distinct; mosqCid1 has low ubiquitous expression whereas mosqCid2 is abundantly expressed in germline tissues, especially ovaries.
What might be the function of mosqCid paralogs? The best evidence for mosqCid1 function comes from our analysis of positive selection. We found that mosqCid1 evolves rapidly under recurrent positive selection in Anopheles species but mosqCid2 does not (table 1). This suggests that mosqCid1 may be involved in a genetic conflict, perhaps as a suppressor of centromere-drive. The best evidence for mosqCid2 function comes from our analysis of tissue-specific expression (fig. 5). MosqCid2 expression increases 10-fold in the ovary and early embryo following blood feeding (fig. 5A). Given that blood-feeding provides the cue to initiate oogenesis in female mosquitoes, we hypothesize that mosqCid2 is important for female germ cell development or for early embryonic cell divisions. Interestingly, our finding that mosqCid1 was lost in An. albimanus and An. darlingi suggests that mosqCid2 is capable of performing all CenH3 functions, at least in these two Anopheles species. Our analysis of selection on mosqCid2 in An. albimanus and An. darlingi revealed that mosqCid2 evolves more rapidly in these species than species encoding both mosqCid1 and mosqCid2. This suggests that mosqCid2 has acquired a drive suppressor function in these two species. These findings are consistent with the predictions of the intralocus conflict model; if gene duplication can resolve the functional dilemma of divergent functions, loss of one paralog can reimpose this dilemma on single copy genes. Finally, like mosqCid2, mosqCid3 also appears to play an important role in oogenesis and embryogenesis based on its expression pattern (fig. 5C). Ultimately, dissection of the specific function of the various paralogs will require tools to closely examine their cytological localization and for genetic knockout of individual paralogs, ideally in multiple mosquito species. The recent development of robust Cas9-mediated techniques for genetic knockouts in mosquito species (Kistler et al. 2015; Chaverra-Rodriguez et al. 2018) should facilitate these studies in the future.
Our finding that mosqCid2, the germline-expressed CenH3 in mosquitoes, does not evolve under positive selection, whereas mosqCid1 does, may provide unique insight into the genetic conflicts that spur the recurrent rapid evolution of centromeric proteins. The centromere-drive model was previously proposed to explain the rapid evolution of centromeric DNA and proteins in a two-step evolutionary process (Henikoff et al. 2001; Henikoff and Malik 2002; Malik 2009; Kursel and Malik 2018). In the first step, centromeres compete with each other by recruiting centromeric proteins during asymmetric female meiosis, in which only one of four meiotic products is chosen to be the oocyte nucleus. “Winning” centromeres recruit more centromeric proteins and are more likely to be transmitted to the oocyte. Several tenets of this first step of the centromere-drive model have now been elegantly demonstrated in experiments that take advantage of meiotic transmission biases in mouse oocytes (Chmatal et al. 2017; Kursel and Malik 2018). These studies demonstrate that centromeric satellite DNA expansions recruit more centromeric proteins (Iwata-Otsubo et al. 2017) allowing them to out-compete homologs in female meiosis (Chmatal et al. 2014) by biased recruitment of microtubule-destabilizing kinases (Akera et al. 2019) to exploit an intrinsic asymmetry of the spindle cytoskeletal apparatus in oocytes (Chmatal et al. 2015; Akera et al. 2017). In the second step of the centromere-drive model, centromere-drive incurs (unknown) fitness costs to the rest of the genome. Consequently, genes encoding centromeric proteins could rapidly evolve to suppress these fitness costs. We originally hypothesized that male meiosis may bear the brunt of the costs of unsuppressed centromere-drive. However, there is currently a paucity of evidence supporting this hypothesis (Henikoff et al. 2001; Henikoff and Malik 2002; Malik 2009; Kursel and Malik 2018). If it were true that unsuppressed centromere-drive negatively impacted male meiosis, we might expect that testis-specific CenH3 paralogs or paralogs of other centromeric proteins would be most likely to undergo positive selection. In Drosophila, we did not find sufficient discrimination in selective signatures between Cid paralogs to support this hypothesis. For example, all three Cid genes in the montium group (including the ubiquitous Cid4 and germline-specific Cid1 and Cid3 paralogs) evolve under positive selection (Kursel and Malik 2017). In contrast, we found no evidence for positive selection in either the ubiquitously expressed Cid1 or testis-specific Cid5 paralogs in the Drosophila subgenus (Kursel and Malik 2017). However, a subsequent study with greater species sampling suggested that the testis-specific Cid5 may evolve under a subtle signature of recurrent positive selection whereas the ubiquitously expressed Cid1 does not (Teixeira et al. 2018). Our findings of positive selection in the ubiquitous mosqCid1 but not the germline-expressed mosqCid2 strongly suggests that somatic rather than germline centromeric function might bear the brunt of the deleterious consequences of centromere-drive and that rapid evolution of mosqCid1 may suppress these deleterious effects.
Although our initial identification of CAL1b raised the intriguing possibility that each CAL1 paralog may have specialized interactions with each mosqCid paralog, the pattern of retention and duplication in both genes does not strongly support this hypothesis (fig. 4). MosqCid duplication in the common ancestor of Anopheles and Aedes preceded CAL1 duplication by >50 My, and loss of mosqCid1 did not lead to loss of either CAL1 paralog in An. albimanus and An. darlingi. However, the lack of a completely corresponding pattern of gene gain and loss does not rule out the possibility of specialized interaction between CAL1 and mosqCid paralogs. Given that we expect the CenH3, and not its chaperone, to be subject to intralocus conflict, it seems reasonable that duplication and specialization of mosqCid would precede specialization of its interacting partners, CAL1 and CENP-C. Again, better cytological and genetic tools will allow us to precisely define these interactions in future studies.
In summary, our data suggest that mosquitoes employ multiple, specialized CenH3s to carry out centromere function. We hypothesize that mosqCid paralogs have acquired specialized germline functions, either as a suppressor of the deleterious effects of centromere-drive (mosqCid1) or in oogenesis (mosqCid2). This is in line with our findings in Drosophila where CenH3 paralogs have acquired specialized functions in the male and female germline. Taken together, our findings in mosquitoes and in Drosophila support the hypothesis that the germline and soma functions of CenH3 are at odds with one another, creating intralocus conflict. In future studies, mosquito species, in addition to Drosophila species, present an excellent opportunity to study the functional specialization of centromeric proteins, in turn providing insight into their multiple functions.
Materials and Methods
Identification of CenH3, CENP-C, and CAL1 Orthologs and Paralogs
Mosquito Cid genes were identified in previously sequenced genomes. We used D. melanogaster Cid1 histone fold domain to query mosquito genomes using TBlastN implemented in Vectorbase (Giraldo-Calderon et al. 2015). Many mosqCid BLAST hits were not annotated genes or were misannotated and required manual curation of the mosqCid gene open reading frame (supplementary table S1, Supplementary Material online). To identify CenH3 in the outgroup Mochlonyx cinctipes, we used Ae. aegypti mosqCid1, mosqCid2, and mosqCid3 as well as An. gambiae mosqCid1 and mosqCid2 as queries in a BLASTp search of M. cinctipes whole-genome shotgun assembly ASM101484v1 implemented on NCBI BLAST suite. To identify mosquito CAL1 homologs, we employed an iterative TBlastN search strategy in which we first used D. melanogaster CAL1 to identify homologs in an intermediate branching Dipteran species, Glossina morsitans. Subsequently, we used G. morsitans CAL1 to identify CAL1 homologs in all mosquito species using BLASTp and TBlastN searches (supplementary table S1, Supplementary Material online and supplementary data S2, Supplementary Material online). To identify CENP-C homologs, we relied on the C-terminal cupin domain as a reliable bioinformatic marker. We used the D. melanogaster CENP-C cupin domain to do BLASTp searches of the predicted mosquito proteomes to first identify putative CENP-C orthologs in the well annotated An. gambiae and Ae. aegpyti genomes. We then used these predicted mosquito proteins as queries in iterative BLASTp and TBlastN searches to identify all mosquito homologs of CENP-C. We also recorded the syntenic locus (3′ and 5′ flanking genes) of each gene hit as annotated in Vectorbase genome browser track and by homology (using BLASTp) to genes in D. melanogaster (supplementary table S1, Supplementary Material online). Each mosqCid and CAL1 gene was named according to its shared syntenic location and phylogenetic relationship to other paralogs if present.
Phylogenetic Analyses
MosqCid nucleotide sequences were aligned using the ClustalW (Larkin et al. 2007) translation align function in the Geneious software package (version 6) (Kearse et al. 2012). Alignments were further refined manually by removing of poorly aligned regions. Maximum likelihood phylogenetic trees of Cid nucleotide sequences were generated using the HKY85 substitution model in PhyML (Guindon and Gascuel 2003), implemented in Geneious, using 100 bootstrap replicates for statistical support. Amino acid alignments of CAL1 and CAL1like were generated using ClustalW function in the Geneious software package. Neighbor-Joining phylogenetic trees (Saitou and Nei 1987) of CAL1 and CAL1b protein sequences were generated using the Jukes–Cantor model for genetic distance and implemented in the Geneious tree builder in the Geneious software package. For visualization of phylogenies, we used the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/).
Positive Selection and RELAX Analyses
We used the PAML suite of programs (Yang 2007) to test for positive selection on mosqCid1, mosqCid2, CAL1, and CAL1b in Anopheles. Alignments for each gene paralog were generated and refined as described above. We chose a subset of Anopheles species (An. coluzzi, An. gambiae, An. arabiensis, An. melas, An. merus, An. quadriannulatus, and An. chrysti) for these analyses in order to maintain high-confidence alignments across the full length of each gene. Alignments and gene trees were used as input into the CODEML NSsites model of PAML (supplementary data S4, Supplementary Material online). To determine whether each mosqCid or CAL1 paralog evolves under positive selection, we compared a model that does not allow dN/dS to exceed 1 (M8a) to a model that allows dN/dS > 1 (M8). Positively selected sites were classified as those sites with a M8 Bayes Empirical Bayes posterior probability > 95%. We used the RELAX method (Wertheim et al. 2015) to test for relaxed or intensified selection on mosqCid2 in An. albimanus and An. darlingi (test branches) compared with mosqCid2 in all other Anopheles species (reference branhes). We also used the RELAX method to test for relaxed or intensified selection in Anopheles CAL1b (test branches) using Anopheles CAL1 sequences as a reference. For both analyses, we aligned mosqCid2 or CAL1 and CAL1b nucleotide sequences using the translation align function in the Geneious software package. Test branches and reference branches were indicated as in supplementary figure S3, Supplementary Material online and RELAX analyses was run with default parameters on datamonkey.org/relax (Weaver et al. 2018).
Heatmaps Generated from Previously Published RNAseq Experiments
To visualize the expression of Ae. aegypti CenH3 paralogs across multiple developmental time points we generated a heatmap in R using FPKM values from Akbari et al. (2013). All Ae. aegypti CenH3 paralogs were already annotated, so we looked up the corresponding FPKM values in Akbari et al. supplementary data, Supplementary Material online. To examine the expression of CenH3 and Cal1 paralogs in An. stephensi, we used RNAseq data from Biedler et al. (2014). We manually added an annotation for mosqCid1 to the An. stephensi GFF3 file and then calculated FPKM values for all four genes (mosqCid1, mosqCid2, Cal1, and Cal1b) using cufflinks (Trapnell et al. 2012). Heatmaps for Anopheles expression data were generated in R.
RT-qPCR Expression Analyses
Adult An. stephensi mosquitoes were obtained from the Center for Infectious Disease in Seattle, WA. RNA was extracted from whole bodies, and dissected tissues (heads, germline and the remaining carcasses). All samples were DNase treated (Ambion) and then used for cDNA synthesis (SuperScript III, Invitrogen). During cDNA synthesis, a “No RT” control was generated for each RNA extraction in which the reverse transcriptase was excluded from the reaction. RT-qPCR was performed according to the standard curve method using the Platinum SYBR Green reagent (Invitrogen) and primers designed to each mosqCid paralog and to Actin. Reactions were run on an ABI QuantStudio 5 qPCR machine using the following conditions: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of (95 °C for 15 s, 60 °C for 30 s). We ensured that all primer pairs had similar amplification efficiencies using a dilution series of genomic DNA. Three technical replicates were performed for each cDNA sample. Transcript levels of each gene were normalized to Actin.
Cloning GFP-mosqCid Fusion Proteins
MosqCid genes from Ae. albopictus (mosqCid1, mosqCid2, and mosqCid3) were amplified from genomic DNA extracted from the C6/36 cell line and cloned into pENTR/D-TOPO (ThermoFisher). We used LR clonase II (ThermoFisher) to directionally recombine each mosqCid gene into a destination vector from the Drosophila Gateway Vector Collection, generating N-terminal Venus (pHVW) fusion under the control of the D. melanogaster heat-shock promoter.
Transfection and Imaging of Ae. albopictus Tissue Culture Cells
The Ae. albopictus cell line C6/36 (a gift from Alan Goodman) was used for all transfection experiments. One microgram of plasmid DNA was transfected using Xtremegene HP transfection reagent (Roche) according to the manufacturer’s instructions. Cells were heat-shocked at 37 °C for 1 h 24 h after transfection to induce expression of the mosqCid fusion protein. Cells were transferred to a glass coverslip 24 h after heat shock and were treated with 0.5% sodium citrate for 10 min, then centrifuged on a Cytospin III (Shandon) at 1,900 rpm for 1 min to remove cytoplasm. Cells were fixed in 4% PFA for 5 min and blocked with PBSTx (0.3% Triton) plus 3% BSA for 30 min at room temperature. Coverslips with cells were incubated with primary antibodies at 4 °C overnight at the following concentration. We used the chicken anti-GFP (Abcam AB13970) at 1:1,000 dilution. Coverslips with cells were incubated with secondary antibodies for 1 h at room temperature at the following concentration. We used a goat antichicken (Invitrogen Alexa Fluor 488, A-11039) secondary at 1:5,000 dilution. DNA was stained with DAPI. Images were acquired from the Leica TCS SP5 II confocal microscope using a 63× objective with LASAF software.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to past and present Malik lab members for valuable discussions and advice. We would like to thank Tera Levin and Courtney Schroeder for their comments on the manuscript. We also thank Sean Shadle for assistance with RNAseq analysis and Alan Goodman for the Ae. albopictus C6/36 tissue culture cells. This study was supported by funding from the National Institutes of Health training grants T32 HG000035 and T32 GM007270 (to L.E.K.), Summer Undergraduate Research Program funded by Cancer Center Support Grant (CCSG) CURE Supplement: NCI 3P30CA015704 (to F.C.W.) and R01 GM074108 (to H.S.M.). The funders played no role in study design, data collection, and interpretation, or the decision to publish this study. H.S.M. is an Investigator of the Howard Hughes Medical Institute.
References
- Akbari OS, Antoshechkin I, Amrhein H, Williams B, Diloreto R, Sandler J, Hay BA.. 2013. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 3:1493–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA.. 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358(6363):668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T, Trimm E, Lampson MA.. 2019. Molecular strategies of meiotic cheating by selfish centromeres. Cell 178(5):1132–1144.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, Rogers K.. 2006. Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174(3):1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler JK, Qi Y, Pledger D, Macias VM, James AA, Tu Z.. 2014. Maternal germline-specific genes in the Asian malaria mosquito Anopheles stephensi: characterization and application for disease control. G3 (Bethesda) 5:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH.. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 3(8):730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S.. 1999. A histone-H3-like protein in C. elegans. Nature 401(6753):547–548. [DOI] [PubMed] [Google Scholar]
- Chaverra-Rodriguez D, Macias VM, Hughes GL, Pujhari S, Suzuki Y, Peterson DR, Kim D, McKeand S, Rasgon JL.. 2018. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun. 9(1):3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG.. 2014. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 204(3):313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA.. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 24:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmatal L, Schultz RM, Black BE, Lampson MA.. 2017. Cell Biology of cheating-transmission of centromeres and other selfish elements through asymmetric meiosis. Prog Mol Subcell Biol. 56:377–396. [DOI] [PubMed] [Google Scholar]
- Chmatal L, Yang K, Schultz RM, Lampson MA.. 2015. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr Biol. 25:1835–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell. 19(10):4480–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD.. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454(7205):762–765. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betran E.. 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 26(5):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E.. 2010. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S.. 2010. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 277(3):599–604. [DOI] [PubMed] [Google Scholar]
- Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, VectorBase C, Madey G, et al. 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43:D707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Malik HS.. 2002. Centromeres: selfish drivers. Nature 417(6886):227–227. [DOI] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135(6):1039–1052. [DOI] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA.. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 97(3):1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE.. 2017. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27(15):2365–2373.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler KE, Vosshall LB, Matthews BJ.. 2015. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral L. 2015. Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000Research 4:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2017. Recurrent gene duplication leads to diverse repertoires of centromeric histones in Drosophila species. Mol Biol Evol. 34(6):1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2018. The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol. 52:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel LE, Malik HS.. 2019. Gametic specialization of centromeric histone paralogs in Drosophila virilis. biorXiv. 530295. doi: 10.1101/530295. [DOI] [PMC free article] [PubMed]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang JF.. 2008. Identification and molecular evolution of cow CENP-A gene family. Mamm Genome. 19(2):139–143. [DOI] [PubMed] [Google Scholar]
- Malik HS. 2009. The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol. 48:33–52. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S.. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157(3):1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH.. 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 7(5):e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Hattersley N, Muroyama A, Stevens D, Oegema K, Desai A.. 2015. Separase cleaves the N-tail of the CENP-A related protein CPAR-1 at the meiosis i metaphase-anaphase transition in C. elegans. PLoS One 10(4):e0125382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C-elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 7(12):1248–1255. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arca B, Arensburger P, Artemov G, et al. 2015. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347(6217):1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B, Sunkel CE.. 2011. Drosophila CENP-C is essential for centromere identity. Chromosoma 120(1):83–96. [DOI] [PubMed] [Google Scholar]
- Phansalkar R, Lapierre P, Mellone BG.. 2012. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 20(5):493–504. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF.. 2012. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 10(12):e1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin L, Mellone BG.. 2016. Co-evolving CENP-A and CAL1 domains mediate centromeric CENP-A deposition across Drosophila species. Dev Cell. 37(2):136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Program NCS, Green ED.. 2010. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 27(7):1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M.. 1995. A Mutation in Cse4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell-cycle arrest at mitosis. Genes Dev. 9(5):573–586. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S.. 2004. Adaptive evolution of centromere proteins in plants and animals. J Biol. 3(4):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira JR, Dias GB, Svartman M, Ruiz A, Kuhn GCS.. 2018. Concurrent duplication of drosophila cid and Cenp-C genes resulted in accelerated evolution and male germline-biased expression of the new copies. J Mol Evol. 86(6):353–364. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 35(3):773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bures P.. 2016. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Sci Rep. 6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.