Significance
Dysregulation of MYC protein levels is associated with most human cancers. MYC is regulated by both transcription and protein stability. BRD4, a driver of oncogenesis that activates Myc transcription, is being investigated as a therapeutic target in MYC-driven cancers. We report that BRD4 directly destabilizes MYC protein by phosphorylating it at a site leading to ubiquitination and degradation, thereby maintaining homeostatic levels of MYC protein. While JQ1, an inhibitor which releases BRD4 from chromatin and reduces MYC transcription has no effect on MYC protein stability, MZ1, which degrades BRD4 has the paradoxical effect of decreasing MYC transcription but increasing MYC stability. Our findings demonstrating BRD4-mediated MYC degradation are likely to have significant translational implications.
Keywords: MYC phosphorylation, BRD4 kinase, BRD4 histone acetyltransferase, ERK1, MYC stability
Abstract
The protooncogene MYC regulates a variety of cellular processes, including proliferation and metabolism. Maintaining MYC at homeostatic levels is critical to normal cell function; overexpression drives many cancers. MYC stability is regulated through phosphorylation: phosphorylation at Thr58 signals degradation while Ser62 phosphorylation leads to its stabilization and functional activation. The bromodomain protein 4 (BRD4) is a transcriptional and epigenetic regulator with intrinsic kinase and histone acetyltransferase (HAT) activities that activates transcription of key protooncogenes, including MYC. We report that BRD4 phosphorylates MYC at Thr58, leading to MYC ubiquitination and degradation, thereby regulating MYC target genes. Importantly, BRD4 degradation, but not inhibition, results in increased levels of MYC protein. Conversely, MYC inhibits BRD4’s HAT activity, suggesting that MYC regulates its own transcription by limiting BRD4-mediated chromatin remodeling of its locus. The MYC stabilizing kinase, ERK1, regulates MYC levels directly and indirectly by inhibiting BRD4 kinase activity. These findings demonstrate that BRD4 negatively regulates MYC levels, which is counteracted by ERK1 activation.
MYC is a transcription factor that globally enhances expression of transcribing genes, including those vital for cell cycle, growth, proliferation, and survival in normal and cancer cells (1–7). MYC stimulates transcription initiation, pause release, and elongation through interactions with various factors, including the transcription elongation factor PTEFb and the PAF1 complex which facilitates transcription through nucleosome barriers (8–11). MYC indirectly regulates chromatin structure through recruitment of HATs such as p300, GCN5/TRAPP, HDACs, and chromatin remodelers such as SWI/SNF (12–14).
MYC protein levels are tightly regulated at multiple levels—chromatin decompaction (15), transcription initiation and elongation (16), and mRNA half-life (17). Posttranslational modifications that regulate MYC stability include phosphorylations at Ser62 and Thr58 (18, 19). Phosphorylation at Ser62, primarily by ERK, stabilizes MYC (20). MYC phosphorylation at Thr58 leads to destabilization by recruitment of the ubiquitin ligase Fbw7 and subsequent ubiquitin-mediated proteolysis (19, 21). While kinases other than ERK phosphorylate Ser62, GSK3β has been the only kinase known to phosphorylate MYC at Thr58 (22).
MYC pathogenicity does not primarily result from mutations but rather from aberrant levels of expression, resulting from increased transcription, chromosomal amplification, or rearrangement. Chromosomal rearrangements leading to constitutive MYC overexpression and uncontrolled cellular proliferation are the drivers of Burkitt lymphoma and multiple myeloma while changes in MYC expression underlie other cancers (23). MYC is one of the most highly expressed oncogenes in human cancer (24). In many cancer cell types, its transcription is regulated by the bromodomain protein 4 (BRD4). BRD4 is a chromatin and transcriptional regulator that plays a critical role in many cellular functions, including transcription, replication, and DNA repair (25). A variety of hematopoietic malignancies and solid tumors depend on the expression of BRD4, making BRD4 a therapeutic target. Until recently relatively little was known about its mechanisms of action. BRD4 is now known to have intrinsic histone acetyltransferase (HAT) and kinase activities located at its C-terminal and N-terminal ends, respectively (15, 26). BRD4 regulates chromatin remodeling by acetylating H3K122, causing destabilization and eviction of nucleosomes from chromatin. The resulting chromatin decompaction allows access to transcriptional machinery and activates transcription (15). BRD4 kinase directly regulates transcription by phosphorylating the RNA Pol II C-terminal domain (CTD), activating Topoisomerase I and pause release (26, 27). BRD4 regulates transcription indirectly through recruitment and phosphorylation of the transcription elongation factor, PTEFb (28, 29).
BRD4 contributes to reactivation of MYC transcription at the end of mitosis (30–33) which requires its HAT activity to mediate chromatin decompaction around the MYC gene locus (15). Thus, through its regulation of MYC transcription, BRD4 contributes to maintaining cell proliferation and growth. Preliminary reports suggested that BRD4 also coimmunoprecipitated with MYC protein (11, 34), raising the possibility that, in addition to regulating MYC transcription, BRD4 directly contributes to maintenance of homeostatic levels of MYC protein. Here we report that BRD4 binds MYC protein directly and phosphorylates Thr58, resulting in MYC destabilization. ERK1, which phosphorylates MYC at Ser62 and stabilizes it, forms a trimeric complex with BRD4 and MYC. MYC inhibits BRD4 HAT activity, whereas ERK1 inhibits BRD4 kinase activity. We propose a model in which these interactions create a regulatory network that maintains homeostatic levels of MYC.
Results
BRD4 and MYC Interact Directly in the Nucleus.
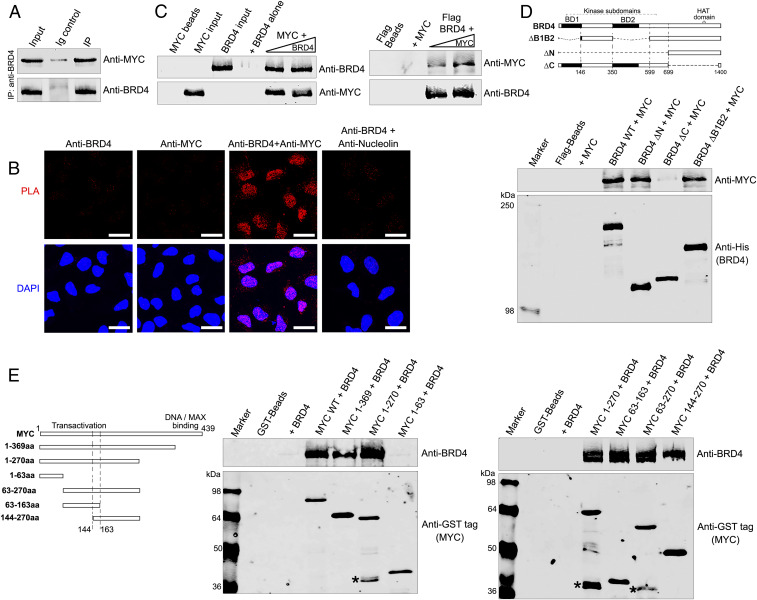
Since earlier studies suggested MYC and BRD4 coimmunoprecipitate (11, 34), we determined whether they occur in a complex. Immunoprecipitation of BRD4 from HeLa nuclear extracts coimmunoprecipitated MYC (Fig. 1A and SI Appendix, Fig. S1A), demonstrating that they coexist in a single complex. To visualize the MYC-BRD4 interaction in situ, we conducted proximity ligation assays (PLAs). The results revealed a robust nuclear colocalization between BRD4 and MYC; control PLAs did not generate signals (Fig. 1B). Thus, endogenous BRD4 and MYC coexist in a complex in situ.
Fig. 1.
BRD4 and MYC interact in the nucleus. (A) BRD4 associates with MYC. BRD4 was immunoprecipitated from HeLa nuclear extract using anti-BRD4 and immunoblotted with anti-MYC. (Anti-BRD4 specificity is shown in SI Appendix, Fig. S1A) (B) BRD4 colocalizes with MYC in the nucleus. PLAs with anti-BRD4 and anti-MYC on fixed HeLa cells. PLA with anti-BRD4 and nucleolin as a negative control. PLA, red; DAPI nuclei staining, blue. (Scale bars, 20 µM.) (C) BRD4 binds MYC directly. (Left) rBRD4 (0.5 and 0.75 µg) was pulled down with 0.2 µg rMYC immobilized on anti-MYC-agarose beads. (Right) rMYC (0.2 and 0.4 µg) was pulled down with 0.5 µg rBRD4 immobilized on Flag-beads (Right). Beads with or without prey protein are controls; input was the same for both pull-downs. (D) MYC binds BRD4 C-terminal region. (Top) Map of BRD4 and deletion mutants. (Bottom) Anti-MYC immunoblot of 0.2 µg rMYC recovered by pull-down with 0.5 µg wild-type His-BRD4-Flag or equimolar amounts of His-BRD4-Flag mutants on Flag beads. Anti-His immunoblot shows BRD4 retained on Flag beads; beads with or without rMYC are controls. (E) BRD4 binds MYC transactivation domain. (Left) Map of MYC and deletion mutants. (Middle) Anti-BRD4 immunoblots of 0.5 µg BRD4 recovered by pull-down with 0.25 µg wild-type GST-MYC or equimolar amounts of GST-MYC mutants on GST-Sepharose beads. Anti-GST immunoblots show MYC on beads; beads with or without BRD4 are controls. (Right) As in Middle, using additional MYC deletion mutants to fine map BRD4-binding region on MYC. MYC degradation products (*).
To determine if the interaction is direct, purified recombinant BRD4 and MYC proteins were subjected to reciprocal in vitro pull-down assays (Fig. 1C). BRD4 was pulled down by MYC bound to anti-MYC agarose beads. Conversely, MYC was pulled down by Flag-BRD4 immobilized on Flag beads. These results demonstrate that BRD4 and MYC directly interact. Domain organizations of both MYC and BRD4 have been extensively mapped. MYC is a helix–loop–helix protein with four regions of homology boxes (MBI to MBIV), three of which are located within the N-terminal transactivation domain and a DNA/MAX protein binding region at its C-terminal end (35). BRD4 is a bromodomain protein with two bromodomains and a kinase domain located at its N-terminal half; the C-terminal half contains the HAT domain and a carboxy terminal segment (CTD) that is the interaction site for various factors, including PTEFb and HPV E2 (25, 36). To characterize the interaction between BRD4 and MYC, we mapped the BRD4 region that interacted with MYC using a series of recombinant BRD4 mutant proteins in pull-down assays with purified MYC (Fig. 1D). BRD4 mutants lacking either both bromodomains (ΔB1B2) or the entire N-terminal half of BRD4 (ΔN) were unaffected in their ability to pull down MYC. In contrast, a BRD4 mutant (ΔC) lacking the C-terminal region of BRD4 spanning its HAT domain but retaining the last 30 amino acids (aa) through which BRD4 interacts with PTEFb failed to bind MYC (Fig. 1D). Further mapping of the BRD4 interaction domain within its C terminus using peptides generated from the segment between aa 699 and 1,400 demonstrated that MYC binds within aa 823 to 1,044, N-terminal to the HAT domain (SI Appendix, Fig. S1B). Thus, MYC binds the BRD4 C terminus independent of either its HAT domain or CTD.
The BRD4-interaction segment of MYC was mapped using a series of N-terminal and C-terminal truncations. MYC truncation mutants either deleted of the DNA/MAX binding region (1 to 369 aa) or spanning the N terminus (1 to 270 aa) of the molecule pulled down BRD4, whereas a fragment extending from 1 to 63 aa did not, mapping the binding region between 63 and 270 aa (Fig. 1 E, Left and Center). The binding region was further refined using mutants containing central sections of MYC—63 to 270 aa, 63 to 163 aa, and 144 to 270 aa—which bound BRD4 (Fig. 1 E, Right). Since BRD4 bound both 63 to 163 aa and 144 to 270 aa MYC fragments, binding depended on the 19-aa overlapping region (144 to 163 aa) located within MYC’s transactivation domain. In summary, BRD4 and MYC directly interact through BRD4’s C terminus between 823 and 1,044 aa and the MYC transactivation domain between 144 and 163 aa.
BRD4 Phosphorylates MYC at Threonine 58.
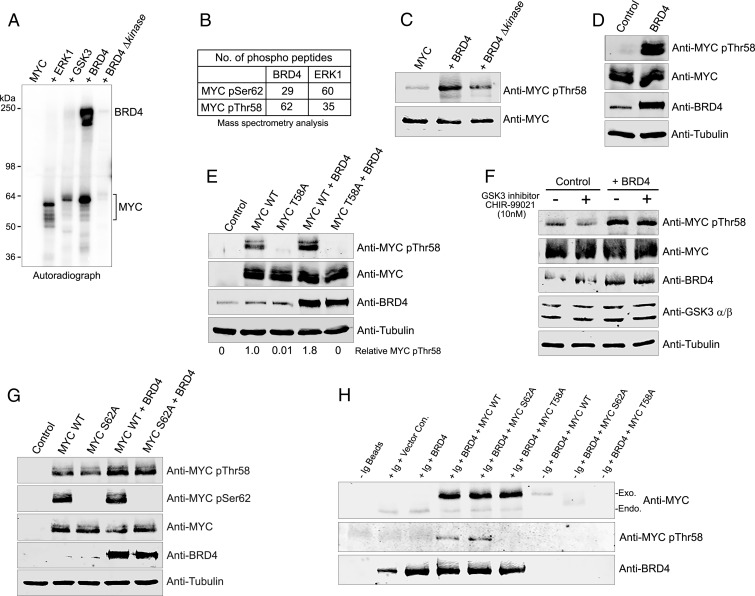
BRD4 has intrinsic kinase activity that phosphorylates the carboxyl-terminal domain of RNA polymerase II, as well as transcription factors such as PTEFb/CDK9 and TAF7 (26, 29). The interaction between MYC and BRD4 led us to consider that either BRD4 phosphorylates MYC or, conversely, MYC affects BRD4 kinase activity. Whereas MYC did not affect BRD4 kinase activity, BRD4 robustly phosphorylated MYC in vitro (Fig. 2A and SI Appendix, Fig. S2A). A kinase-deficient mutant of BRD4 (BRD4Δkinase) bound MYC efficiently but was unable to phosphorylate it (Fig. 2A and SI Appendix, Fig. S2 A and B). The extent of MYC phosphorylation by BRD4 was at least as great as that of ERK1 and GSK3β, two kinases known to phosphorylate MYC. Therefore, BRD4 is a novel MYC kinase.
Fig. 2.
BRD4 phosphorylates MYC at threonine 58. (A) BRD4 phosphorylates MYC. Autoradiograph of kinase assays with BRD4 (0.3 µg) or equimolar amounts of BRD4 ∆kinase, ERK1, or GSK3 with 0.5 µg rMYC substrate. (B) BRD4 preferentially phosphorylates MYCThr58. rMYC phosphorylated by rBRD4 (0.3 µg) or equimolar amounts of rERK1 was analyzed by mass spectrometry. Table shows number of MYC pSer62 and pThr58 containing phosphopeptides are identified. (C) BRD4 phosphorylates MYCThr58. rMYC (0.5 µg) phosphorylated by BRD4 (0.3 µg) or equimolar amounts of BRD4 ∆kinase in in vitro kinase assays was immunoblotted with anti-MYC pThr58 or anti-MYC. (D) BRD4 mediates endogenous MYCThr58 phosphorylation. MYCpThr58, MYC, BRD4, and tubulin immunoblots of whole cell extracts (WCEs) from HeLa cells transfected with 3 µg BRD4 or vector (control). (E) MYCpThr58 antibody specifically recognizes exogenous MYC phosphorylation by BRD4. MYCpThr58, MYC, BRD4, and tubulin immunoblots of WCEs from HeLa cells transfected with 3 µg vector, MYC WT, or MYC T58A mutant with or without 3 µg BRD4. Relative MYC pThr58 levels, normalized to tubulin, are shown. (F) BRD4 phosphorylation of endogenous MYC pThr58 does not depend on GSK3β. MYC pThr58, MYC, BRD4, GSK3β, and tubulin immunoblots of WCEs from HeLa cells transfected with 3 µg vector or BRD4 and treated with DMSO or GSK3β inhibitor CHIR-99021 overnight. (G) BRD4 phosphorylation of exogenous MYCThr58 is independent of MYCSer62. MYC pThr58, pSer62, MYC, BRD4, and tubulin immunoblots of WCEs from HeLa cells transfected with 3 µg vector, MYC WT, or MYC S62A mutant with or without 3 µg BRD4. (H) BRD4 interacts with MYCWT, MYCT58, and MYCS62 and phosphorylates Thr58. BRD4, MYC, and pT58 immunoblots of WCEs from HeLa cells transfected with 3 µg MYCT58, MYCS62 mutants, or vector, with or without 3 μg BRD4 and immunoprecipitated with anti-BRD4.
MYC stability and function are regulated through phosphorylation. Phosphorylation at Thr58 signals MYC degradation; Ser62 phosphorylation is associated with its stabilization and functional activation (19). To identify the site(s) on MYC of BRD4 phosphorylation, we tested if BRD4 could phosphorylate a series of MYC truncation mutants (MYC 1 to 369 aa, 1 to 270 aa, and 1 to 63 aa). MYC 1 to 369 aa and 1 to 270 aa mutants were phosphorylated as well as, or better than, full-length MYC, mapping BRD4 phosphorylation site(s) within the MYC N terminus (SI Appendix, Fig. S2C). BRD4 did not phosphorylate the 1 to 63 aa MYC fragment, consistent with its inability to bind (Fig. 1E). To identify the residues phosphorylated by BRD4, we performed mass spectrometry analyses of MYC phosphorylated in vitro by either BRD4 or by ERK1 as a control with the known target of Ser62. Strikingly, the mass spectrometry results identified Thr58 as the preferred site of phosphorylation by BRD4; Ser62 phosphorylation was detected at lower levels. As expected, ERK1 preferentially phosphorylated MYC at Ser62 with lower levels at Thr58 (Fig. 2B). BRD4 phosphorylation of MYC at Thr58 in vitro was validated by immunoblotting with a MYC pThr58-specific antibody (Fig. 2C) which detected phosphorylation at Thr58 by BRD4 but not by BRD4Δkinase. We conclude that BRD4 stably binds MYC between aa 144 and 163, phosphorylating it at Thr58.
To confirm these results in cells, pThr58 levels were compared between HeLa cells transfected with either BRD4 or a vector control. Remarkably, endogenous MYC pThr58 levels markedly increased in cells overexpressing BRD4 (Fig. 2D). Similar results were observed in HCT116 colorectal carcinoma and U2OS osteosarcoma cells, demonstrating that BRD4 phosphorylates Thr58 in multiple cell types (SI Appendix, Fig. S2D). Confirming the specificity of the MYC pThr58 antibody and the BRD4 target phosphorylation site, no phosphorylated Thr58 was detected in samples transfected with BRD4 and a Myc Thr58A mutant (Fig. 2E). The observed BRD4 phosphorylation of Thr58 is unlikely to be mediated by GSK3β since a specific inhibitor, CHIR-99021 (SI Appendix, Fig. S2E), did not inhibit in cellulo BRD4 phosphorylation of Thr58 (Fig. 2F and SI Appendix, Fig. S2E). These results demonstrate that Thr58 is a major target site of BRD4 kinase activity.
Prior phosphorylation at Ser62 is a prerequisite for subsequent GSK3β phosphorylation of MYC at Thr58 (20). Surprisingly, BRD4 phosphorylation of Thr58 did not depend on the phosphorylation status of Ser62. Cotransfection of BRD4 with either Myc or a MycSer62 point mutant (S62A) resulted in an equivalent increase in phosphorylation of Thr58 (Fig. 2G). In contrast, pSer62 levels of wild-type (WT) Myc did not increase when cotransfected with BRD4. Therefore, BRD4 neither phosphorylates MYC Ser62 in cells nor requires Ser62 to phosphorylate Thr58.
The T58A and S62A MYC mutants differentially partition within the cell (37), raising the possibility that differential localization of the MYC mutants affected BRD4’s ability to bind MYC and thus phosphorylate it. However, BRD4 efficiently coimmunoprecipitated both T58A and S62A mutants from transfected HeLa cells (Fig. 2H). Further, in these immunoprecipitates, Thr58 was phosphorylated in both the wild type and S62A mutant, but not the T58A mutant, thereby documenting that BRD4 phosphorylates Thr58 independently of Ser62 phosphorylation (Fig. 2H). We conclude that BRD4 robustly and preferentially phosphorylates MYC Thr58 in vitro and in cells independent of Ser62.
BRD4 Regulates MYC Levels and Stability.
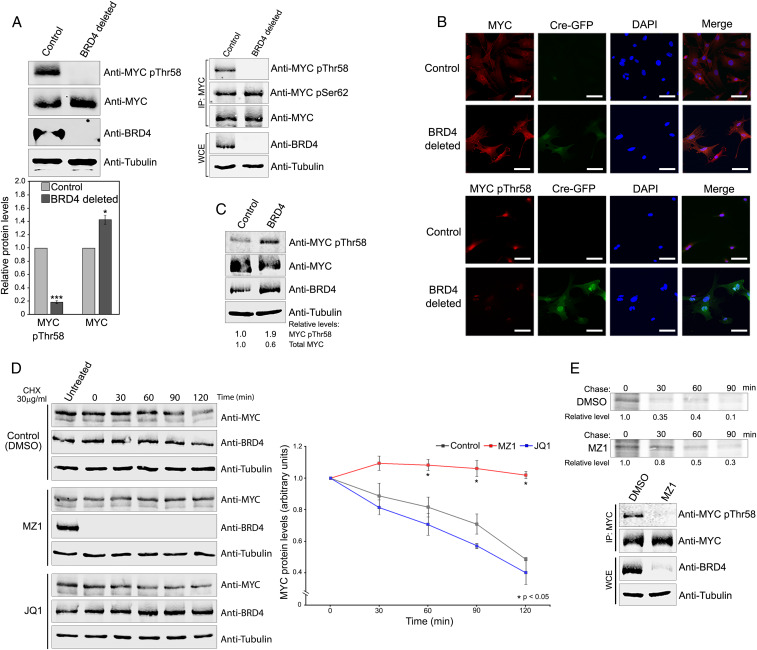
Although MYC was phosphorylated upon BRD4 overexpression, it was important to verify that BRD4 contributes to MYC phosphorylation in a physiological setting. BRD4 was deleted by CRE from a mouse embryonic fibroblast (MEF) line derived from mice homozygous for floxed BRD4 alleles. These MEFs were used to examine the effect of deletion of endogenous BRD4 on endogenous Myc phosphorylation (Fig. 3A). Remarkably, deletion of BRD4 abrogated Myc pThr58, as assessed both by immunoblotting of total MEF cell extract (Fig. 3 A, Left) and of immunoprecipitated Myc (Fig. 3 A, Upper Right). Consistent with the known role of pThr58 leading to Myc degradation, Myc protein levels significantly increased in the absence of BRD4 (Fig. 3 A, Lower). [In MEFs, BRD4 does not affect Myc mRNA levels (SI Appendix, Fig. S2F (38))]. The reduction in pThr58 levels and increase in total Myc protein upon BRD4 depletion were confirmed by in situ immunofluorescence (IF) using antibodies specific for Myc and Myc pThr58 (Fig. 3B). Conversely, overexpression of BRD4 in MEFs increased Myc Thr58 levels and reduced total Myc (Fig. 3C); GSK3β inhibition did not affect pThr58 levels in MEFs (SI Appendix, Fig. S2G). These results demonstrate that BRD4 phosphorylates endogenous Myc Thr58 which directly affects Myc protein levels.
Fig. 3.
Endogenous BRD4 kinase controls endogenous MYC Thr58 phosphorylation and ubiquitination. (A) BRD4 deletion abrogates endogenous MYC pThr58 and increases total MYC in MEFs. (Top, Left) MYCpThr58, MYC, BRD4, and tubulin immunoblots of WCEs from Brd4-floxed MEFs uninfected (control) or infected (BRD4 deleted) with Cre-GFP-expressing retrovirus. (Right) WCEs immunoprecipitated with anti-Myc antibody and immunoblotted with indicated antibodies. (Bottom) Quantification of MYCpThr58 and total MYC levels in WCEs from three independent experiments performed as above. Error bars represent ±SEM (*P < 0.05; ***P < 0.0001). (B) Endogenous MYC pThr58 and total MYC levels depend on BRD4. Brd4-floxed MEFs uninfected (control) or infected (BRD4 deleted) with a Cre-GFP-expressing retrovirus were stained with DAPI and analyzed by immunofluorescence with anti-MYC and anti-MYCpThr58 antibodies. GFP fluorescence identifies Cre-GFP-infected, BRD4-depleted cells. Representative images are shown. (Scale bars, 20 µM.) (C) BRD4 overexpression increases MYCpThr58 and decreases total endogenous MYC in MEFs. MYC pThr58, MYC, BRD4, and tubulin immunoblots of WCEs from MEFs infected with vector only or with BRD4 expressing retrovirus. Relative MYCpThr58 and total MYC levels, normalized to tubulin, are shown. (D) Degradation of endogenous BRD4, but not its inhibition by JQ1, stabilizes endogenous MYC. (Left) MYC and BRD4 immunoblots of WCEs from HCT116 cells grown in the presence of PROTAC MZ1 (0.1 µM), 500 nM JQ1, or dimethyl sulfoxide (control) overnight, treated with 30 µg/mL cycloheximide (CHX), and harvested at 0, 30, 60, 90, and 120 min after CHX treatment. Tubulin, loading control. (Right) Quantification of total MYC protein levels in WCEs from three independent experiments performed as above. Error bars represent ±SEM (*P < 0.05). (E) Degradation of BRD4 leads to endogenous MYC stabilization. (Top) Autoradiographs of MYC immunoprecipitated from HeLa cells treated with either DMSO or MZ1 (0.1 µM) for 2 h and pulse chased after labeling with [35S]methionine/cysteine for 20 min (pulse) and incubated in unlabeled complete medium for the indicated times (chase). (Bottom) MYC pThr58, MYC, BRD4, and tubulin immunoblots of immunoprecipitated MYC and WCEs from above cells.
Since BRD4 binds the kinase PTEFb, we considered the possibility that MYC phosphorylation observed in cells was mediated by PTEFb. However, we have previously reported that MEFs have barely detectable levels of PTEFb (26). Furthermore, PTEFb did not phosphorylate MYC in an in vitro kinase reaction nor did it affect BRD4 phosphorylation of MYC (SI Appendix, Fig. S2H).
As noted above, pThr58 is associated with MYC destabilization. The finding that loss of BRD4 leads to decreased pThr58 and increased total MYC levels suggested that loss of BRD4 would result in increased MYC half-life. To test this prediction, HCT116 cells were treated with MZ1, a proteolysis targeting chimera (PROTAC) molecule that degrades BRD4, and then with cycloheximide to prevent de novo protein synthesis. The stability of endogenous MYC in MZ1-treated and untreated cells was compared at various times after cycloheximide treatment (Fig. 3D). As predicted, MZ1 treatment resulted in complete loss of BRD4 and a significant stabilization of MYC protein. Thus, endogenous BRD4 leads to destabilization of endogenous MYC. In contrast to MZ1, which degrades BRD4, small molecule inhibitors (e.g., JQ1) bind BRD4 bromodomains, both preventing BRD4 binding to chromatin and stabilizing it (39). The above results led to the paradoxical prediction that, whereas treatment of cells with MZ1 markedly increased MYC half-life, treatment with JQ1 should not affect MYC half-life. (JQ1 releases BRD4 from cellular chromatin but does not affect either its binding to or phosphorylation of MYC in vitro) (SI Appendix, Fig. S3 A–C). Consistent with the prediction, MYC stability was unaffected by JQ1 treatment (Fig. 3D). MZ1, but not JQ1, similarly extended Myc half-life in MEF cells (SI Appendix, Fig. S3D). Thus, the presence of BRD4 correlates with pThr58 levels and MYC stability.
Similar results were observed using a different approach. HeLa cells treated with MZ1 were pulsed with [35S]methionine and the effect of BRD4 loss on endogenous MYC stability was monitored (Fig. 3E). In the absence of BRD4, MYC levels were stabilized relative to the BRD4-proficient control. In parallel pulse-chase experiments with MEF cells, deletion of BRD4 similarly stabilized endogenous Myc half-life (SI Appendix, Fig. S3E).
BRD4 Phosphorylation of MYCThr58 Leads to MYC Ubiquitination and Degradation.
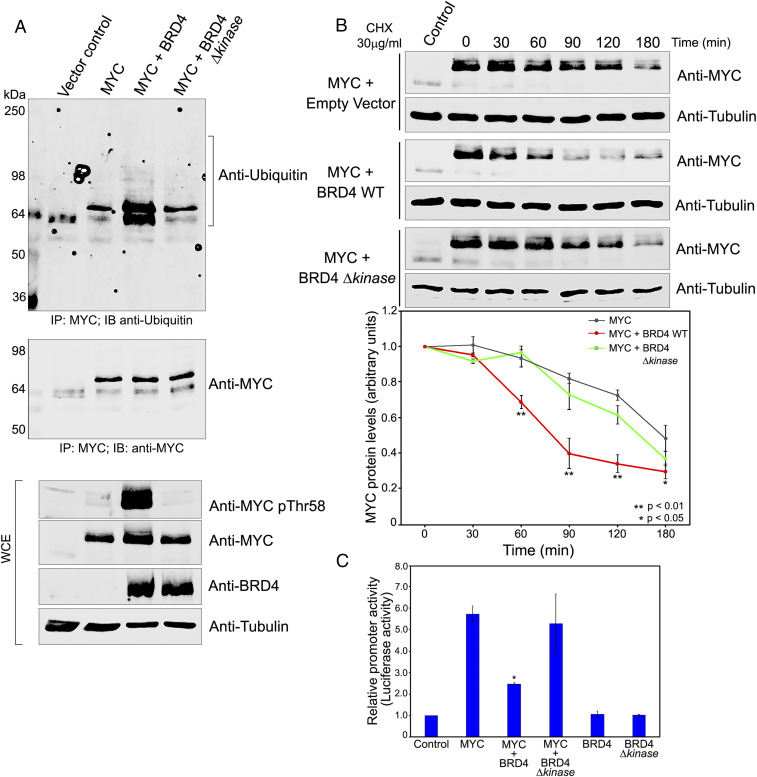
The above results demonstrate that endogenous BRD4 contributes to regulating phosphorylation of endogenous MYCThr58 and MYC protein stability. However, the role of BRD4 kinase activity was not directly demonstrated. To test this, HeLa cells were cotransfected with Myc and either WT BRD4, BRD4 Δkinase mutant, or a vector control. Increased Myc pThr58 was seen in cells overexpressing BRD4, but not the kinase-deficient mutant, BRD4 Δkinase, documenting that Thr58 phosphorylation by BRD4 depends on its kinase activity (Fig. 4 A, Lower).
Fig. 4.
MYC stability and function are regulated by BRD4 kinase. (A) BRD4 phosphorylates exogenous MYC Thr58 resulting in MYC ubiquitination. WCEs from HeLa cells transfected with 3 µg vector or MYC, with or without BRD4 and BRD4 Δkinase were immunoblotted with anti-MYC pThr58, MYC, BRD4, and tubulin antibodies (Bottom). Total MYC was immunoprecipitated from WCEs using MYC-agarose beads and immunoblotted with anti-MYC (Middle) or anti-ubiquitin (Top). (B) Exogenous MYC stability is regulated by BRD4 kinase activity. (Left) MYC immunoblots of WCEs from HeLa cells transfected with 3 µg vector or MYC with or without BRD4 and BRD4 Δkinase, treated with 30 µg/mL CHX 18 h posttransfection and harvested at 0, 30, 60, 90, 120, and 180 min after CHX treatment. Tubulin, loading control. (Right) Quantification of MYC protein levels in WCEs from three independent experiments performed as above. Error bars represent ±SEM (*P < 0.05; **P < 0.01). (C) MYC transactivation of CDK4 promoter is modulated by BRD4 kinase activity. Relative luciferase activity in HeLa cells transfected with 1 µg CDK4 E-box luciferase reporter plasmid alone (control) or cotransfected with 1 µg MYC and BRD4 or BRD4 Δkinase in combination or individually. Error bars represent ±SEM (*P < 0.05, relative to MYC alone).
Similarly, overexpression of WT BRD4 should reduce MYC stability through its phosphorylation of Thr58, while the BRD4 Δkinase should have no effect on MYC. To test this prediction, HeLa cells were cotransfected with MYC and either WT BRD4, BRD4 Δkinase mutant, or an empty vector. After 16 h, cycloheximide was added and MYC stability was monitored over a 3-h period (Fig. 4B). As predicted, WT BRD4 led to destabilization of MYC, whereas the BRD4 Δkinase mutant had no significant effect on MYC stability (Fig. 4B). Thus, MYC phosphorylation and stability both depend on BRD4 kinase activity.
Phosphorylation of MYC at Thr58 triggers its ubiquitination and subsequent degradation (21, 39). The robust phosphorylation of MYC Thr58 by BRD4 should result in increased MYC ubiquitination. Accordingly, MYC was immunoprecipitated from extracts of HeLa cells transfected with BRD4 or BRD4 Δkinase mutant and probed for ubiquitin by immunoblotting (Fig. 4 A, Upper). MYC ubiquitination substantially increased in cells cotransfected with WT BRD4 relative to cells transfected with MYC alone or with control vector (Fig. 4 A, Upper). In contrast, MYC ubiquitination was unaffected by overexpression of the BRD4 Δkinase mutant.
When HeLa cells were cotransfected with ubiquitin, BRD4 and either MYC, MYC S62A, or MYC T58A mutants, MYC immunoprecipitates from these cells showed increased ubiquitination in cells with WT MYC or MYC S62A but not in cells transfected with MYC T58A (SI Appendix, Fig. S3F). Conversely, anti-HA-ubiquitin immunoprecipitated WT MYC, but not MYC T58A from HeLa cells cotransfected with BRD4 and WT MYC or MYC T58A (SI Appendix, Fig. S3G). Although MAX, the MYC partner protein, was coimmunoprecipitated with MYC, BRD4 does not interact with it directly (SI Appendix, Fig. S3H). Thus, BRD4 kinase activity regulates MYC ubiquitination and degradation through its phosphorylation of MYC Thr58.
Since BRD4 destabilizes MYC, BRD4 overexpression should reduce transcriptional activation of MYC target promoters in cells. Among the best characterized target promoters are those, such as CDK4, that contain the MYC/E box DNA element (40). The effect of BRD4 on MYC transactivation activity was assessed using a luciferase reporter driven by the CDK4 core promoter with four E boxes. HeLa cells were cotransfected with the CDK4 promoter reporter and Myc alone, or in combination with either BRD4 or BRD4Δkinase (Fig. 4C). Myc transactivation of the promoter was significantly reduced by cotransfection with BRD4 but not BRD4Δkinase (Fig. 4C). Therefore BRD4 regulates Myc stability through its phosphorylation of Thr58, resulting in reduced Myc protein levels and reduced transactivation of target promoters. The effect of BRD4 on endogenous Myc targets remains to be directly demonstrated.
MYC Inhibits BRD4 HAT Activity.
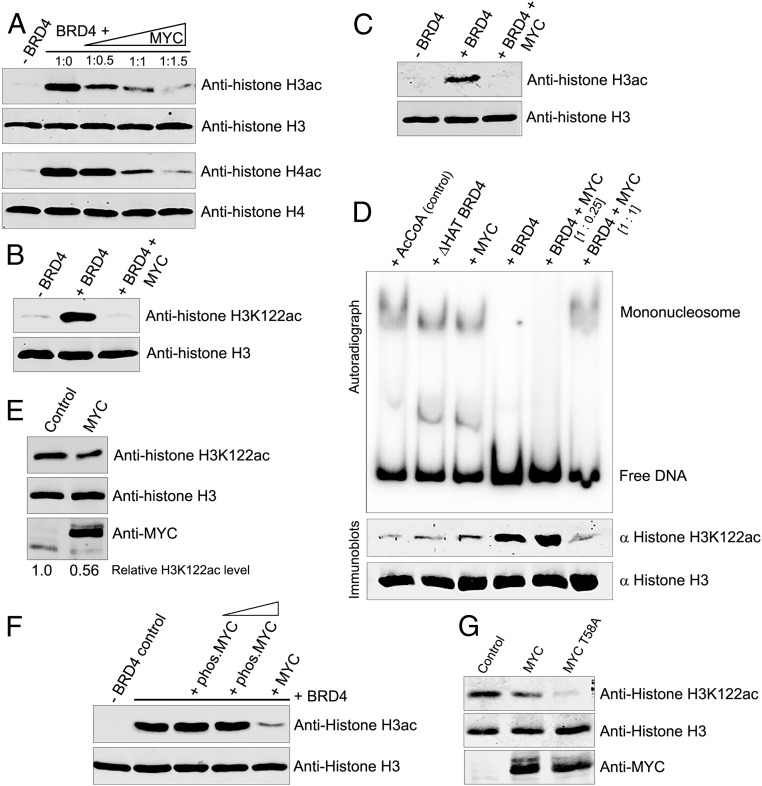
BRD4 also has HAT activity that acetylates histones. BRD4-mediated acetylation remodels chromatin and activates transcription at the MYC locus and other gene loci (15, 41). Remarkably, we find that MYC inhibits BRD4’s HAT activity, as assessed in HAT assays with H3 and H4 (Fig. 5A). BRD4 acetylates the histone H3 core lysine K122, in the globular domain, resulting in eviction of histone octamers from nucleosomes (15). Accordingly, MYC suppressed acetylation of H3K122 by BRD4 in vitro (Fig. 5B). MYC is not a competitive substrate for BRD4 HAT activity since BRD4 does not acetylate MYC but does autoacetylate in the presence of MYC (SI Appendix, Fig. S4A). Thus, MYC is not a general inhibitor of BRD4 acetyltransferase activity. Additionally, MYC does not compete with BRD4 for binding histone H3 or reduce the efficiency with which BRD4 binds H3 (SI Appendix, Fig. S4 B and C). Consistently, MYC does not bind the BRD4 HAT domain (aa 1,157 to 1,197) or the AcCoA binding site (aa 1,097 to 1,102). Rather it binds within aa 823 to 1,044 (SI Appendix, Fig. S1B) and can bind BRD4 lacking the HAT domain (BRD4 [ΔHAT]) (SI Appendix, Fig. S4D). We speculate MYC inhibition of BRD4 HAT may be due to steric hindrance or a conformational change in BRD4 resulting from MYC binding.
Fig. 5.
MYC inhibits BRD4 histone acetylation and nucleosome disassembly. (A) MYC inhibits BRD4 acetylation of histones H3 and H4. Histone H3ac, H3, H4ac, and H4 immunoblots of 1 µg H3 and H4 subjected to HAT assays with 0.5 µg BRD4 in the presence or absence of MYC, at BRD4:MYC molar ratios of 1:0.5, 1:1, and 1:1.5. (B) MYC inhibits BRD4 acetylation of H3K122. Histone H3K122ac and H3 immunoblots of 1 µg H3 subjected to HAT assays with 0.5 µg BRD4 alone or with 0.2 µg MYC. (C) MYC inhibits BRD4 acetylation of histone H3 in assembled mononucleosomes. H3ac and H3 immunoblots of mononucleosomes (10 pmol), assembled in vitro with 5S rDNA and subjected to a HAT assay with 0.5 µg BRD4 alone or with 0.2 µg MYC (1:1 molar ratio). (D) BRD4-mediated nucleosome disassembly is inhibited by MYC. Mononucleosomes (10 pmol) assembled with radiolabeled 5S rDNA were subjected to HAT assays with or without 0.5 µg BRD4 or BRD4 ΔHAT mutant in the presence or absence of MYC added at 1:0.25 and 1:1 BRD4:MYC molar ratios. Reactions with only AcCoA or MYC were controls. Free radiolabeled DNA was resolved from mononucleosomes by electrophoresis (autoradiogram, Upper). Identical HAT assays were done in parallel with nonradiolabeled mononucleosomes and the extent of H3K122 acetylation was determined by immunoblotting; histone H3 immunoblot served as control (Lower). (E) Exogenous MYC inhibits acetylation of histone H3K122. Histone H3, H3K122ac, and MYC immunoblots of WCEs from HeLa cells transfected with 3 µg MYC or vector (control). Relative H3K122ac levels, normalized to total histone H3, are shown. (F) BRD4 HAT activity is not inhibited by MYC prephosphorylated by BRD4. Histone H3ac and H3 immunoblots of 1 µg histones H3 subjected to HAT assays with 0.5 µg BRD4 in the presence or absence of purified, BRD4 prephosphorylated MYC (0.2 and 0.4 µg), or unphosphorylated MYC (0.2 µg). (G) Exogenous MYC T58A mutant inhibits acetylation of histone H3K122. H3K122ac, histone H3, and MYC immunoblots of WCEs from HeLa cells transfected with 3 µg MYC, MYC T58A mutant, or vector (control).
MYC similarly inhibited BRD4 acetylation of in vitro reconstituted nucleosomes. Whereas BRD4 acetylated nucleosomal histone H3, addition of MYC completely blocked acetylation (Fig. 5C). While acetylation of H3K122 by BRD4, but not BRD4 (ΔHAT), released free DNA from nucleosomes, both H3K122 acetylation and release of free DNA were increasingly inhibited by the addition of increasing amounts of MYC (Fig. 5D). Thus, MYC binding to BRD4 inhibits its HAT activity and ability to dissociate nucleosomes.
To confirm these findings, H3K122ac levels were compared between cells transiently transfected with Myc or a vector control. In both HeLa and U2OS osteosarcoma cells, H3K122ac levels were reduced >40% in cells overexpressing Myc (Fig. 5E and SI Appendix, Fig. S4E). These results indicate that MYC suppresses BRD4-mediated histone H3K122 acetylation in cells. The only HAT other than BRD4 known to acetylate H3K122, p300, has been reported to bind MYC (42). However, MYC did not markedly inhibit HAT activity of p300, reducing it ≤20% (SI Appendix, Fig. S4F). Therefore, reduction in H3K122ac following MYC overexpression (Fig. 5E) is attributable to inhibition of BRD4 HAT.
These studies demonstrated that MYC inhibits BRD4 HAT activity, whereas BRD4 phosphorylates MYC resulting in its degradation, leading to the question of whether phosphorylation of MYC affected its ability to inhibit BRD4 HAT activity. While unphosphorylated MYC drastically reduced BRD4 acetylation of histone H3, MYC prephosphorylated by BRD4 had no effect even at high concentrations (Fig. 5F). Importantly, whereas overexpression of WT MYC resulted in a modest inhibition of H3K122Ac, overexpression of the MYC T58A mutant, which is not phosphorylated by BRD4, markedly inhibited H3K122Ac (Fig. 5G). Thus, inhibition of BRD4 HAT activity by MYC is dependent on its phosphorylation.
In cells, BRD4 partitions into both chromatin-bound and nonchromatin-bound fractions. Chromatin-bound BRD4 is associated with MYC as indicated by the increased levels of BRD4 coimmunoprecipitated with MYC in JQ1-treated MEF cells (SI Appendix, Fig. S4G). This led us to question whether these different forms of BRD4 differ in their regulation of Myc. Surprisingly, BRD4 prebound to histone H3 was severely inhibited in its ability to phosphorylate MYC (SI Appendix, Fig. S4H). Thus, unphosphorylated, but not phosphorylated, Myc inhibits BRD4’s acetylation of histones, while histones bound to BRD4 block its phosphorylation of Myc. These results identify a cross-regulation of BRD4 enzymatic activity by Myc and histones which is modulated as a function of their physical and biochemical status, to achieve fine control of their activities. The net effect is to maintain MYC homeostasis.
ERK1 Interacts with BRD4 and Inhibits Its Kinase Activity.
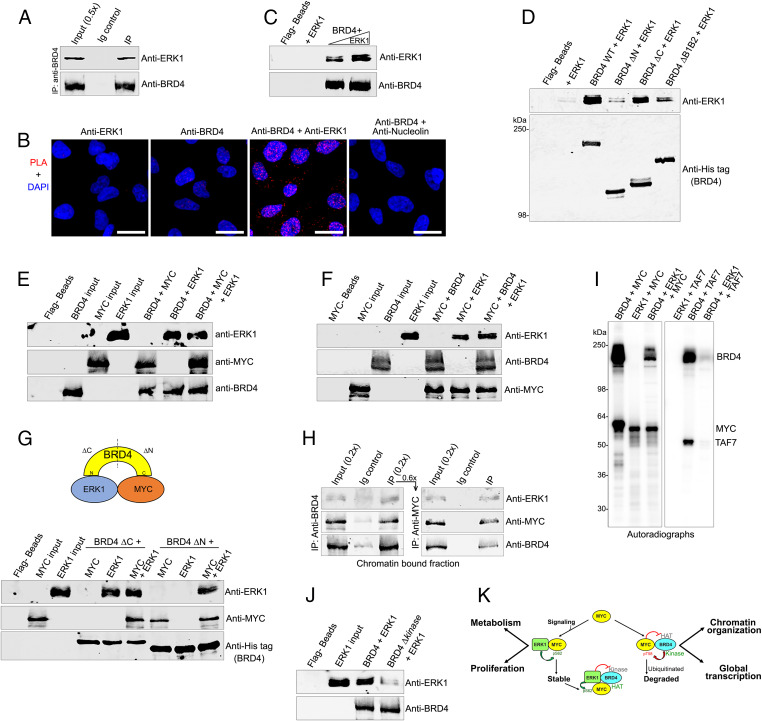
In contrast to BRD4, which phosphorylates MYC at Thr58 leading to MYC degradation, ERK1 phosphorylates MYC Ser62 resulting in MYC stabilization and activation (20). Given the alternative functional consequences of MYC phosphorylation by BRD4 and ERK1, we explored the possibility that BRD4 and ERK1 interact to dynamically regulate MYC levels. Indeed, endogenous BRD4 coimmunoprecipitated ERK1 from HeLa nuclear extracts, indicating that they coexist in a complex (Fig. 6A). PLAs in HeLa cells documented robust BRD4-ERK1 colocalization in situ (Fig. 6B). A direct interaction between BRD4 and ERK1 was demonstrated by in vitro pull-down where recombinant ERK1 was efficiently recovered by Flag-bead bound purified Flag-BRD4 (Fig. 6C).
Fig. 6.
ERK1 interacts with BRD4 and inhibits its kinase activity. (A) BRD4 associates with ERK1 in vivo. BRD4 was immunoprecipitated from HeLa nuclear extract using anti-BRD4 and immunoblotted with anti-ERK1. (B) BRD4 interacts with ERK1 in situ. PLAs with anti-BRD4 and anti-ERK1 on fixed HeLa cells. PLA with anti-BRD4 and nucleolin as a negative control. PLA, red; DAPI nuclei staining, blue. (Scale bars, 20 µM.) (C) BRD4 interacts directly with ERK1. Recombinant ERK1 (0.1, and 0.2 µg) was pulled down with 0.4 µg recombinant BRD4 immobilized on Flag beads. Beads with or without ERK1 are controls. (D) ERK1 binds the BRD4 N-terminal region. Anti-ERK1 immunoblot of 0.2 µg ERK1 recovered by pull-down with 0.75 µg wild-type His-BRD4-Flag or equimolar amounts of ΔN, ΔC, and ΔB1B2 His-BRD4-Flag mutants on Flag beads (Fig. 1D). Anti-His immunoblot shows BRD4 on beads; beads with or without ERK1 are controls. (E) MYC and ERK1 do not compete for binding to BRD4. ERK1, MYC, and BRD4 immunoblots of pull-down reactions done by incubating BRD4-Flag (0.75 μg) immobilized on beads with MYC (0.3 µg) alone (1:1 molar ratio), with ERK1 (0.2 µg) alone (1:1 molar ratio), or with equimolar amounts of both. Beads alone and the input proteins are controls. (F) BRD4 and ERK1 do not compete for binding to MYC. ERK1, BRD4, and MYC immunoblots of pull-down reactions with equimolar amounts of MYC (0.3 µg) immobilized on agarose beads with ERK1 (0.2 µg) alone, BRD4 (0.75 µg) alone, or both. Beads alone and the input proteins are controls. (G) BRD4, ERK1, and MYC directly interact in a trimeric complex in vitro. (Top) Diagram of BRD4-ERK1-MYC complex. (Bottom) ERK1, BRD4, and MYC immunoblots of pull-down reactions done by incubating either MYC (0.22 µg), ERK1 (0.15 µg), or both with BRD4 ΔC or ΔN proteins (0.3 µg) immobilized on Flag beads. Beads alone and the input proteins are controls. (H) BRD4, ERK1, and MYC exist in a trimeric complex in cellulo. ERK1, BRD4, and MYC immunoblots of BRD4 immunoprecipitated using anti-BRD4 in a primary IP (Left) from chromatin-bound HeLa nuclear protein fraction, subjected to sequential IP with MYC agarose beads (Right). (I) ERK1 inhibits BRD4 kinase activity. Autoradiograph of kinase assays with BRD4 (0.5 µg) or equimolar amounts of ERK1 using 0.5 µg of MYC or TAF7 as substrates. (J) BRD4 Δkinase does not bind ERK1. Anti-ERK1 immunoblot of 0.2 µg ERK1 recovered by pull-down with 0.75 µg wild-type BRD4 or BRD4 Δkinase (lacking aa 502 to 548) on Flag beads. Anti-BRD4 immunoblot shows BRD4. Beads alone and ERK1 input are controls (K). Model of regulatory interactions of MYC, BRD4, and ERK1. BRD4 binding to MYC maintains homeostatic levels of MYC protein and global transcription. MYC stability is regulated by interactions with BRD4 and ERK1 which phosphorylate MYC at Thr58 and Ser62, leading to its degradation and stabilization, respectively. (Right) BRD4 regulates transcription of MYC gene, resulting in increased levels of MYC protein (not shown) and amplified MYC-targeted transcription. MYC inhibits BRD4 HAT activity, limiting chromatin remodeling and transcription. Conversely, BRD4 kinase deregulates MYC protein stability by phosphorylating Thr58. Paradoxically, this enhances pause release while moderating transcriptional amplification. (Left) Signaling through the MAP kinase pathway leads to nuclear localization of ERK1, which phosphorylates MYC at Ser62 and inhibits BRD4 kinase activity, together resulting in stabilized MYC. Phosphorylated MYC is unable to inhibit BRD4 HAT, enabling renewed chromatin remodeling and transcriptional activation.
The interaction domain of BRD4 that binds ERK1 was mapped using the set of BRD4 truncation and deletion mutants (Fig. 1D). ERK1 was efficiently pulled down by full-length BRD4 or the BRD4 mutant lacking the C terminus (ΔC), but not by the ΔN BRD4 protein, lacking the N terminus, mapping the region of ERK1 binding to the N terminus of BRD4. Accordingly, deletion of BRD4 bromodomains substantially reduced ERK1 binding (Fig. 6D). Therefore, ERK1 binds to the N-terminal segment of BRD4 within which the bromodomain regions are important for ERK1 binding.
Since ERK1 binds to the N terminus of BRD4, whereas MYC binds to the C terminus, the ability of BRD4 to simultaneously bind MYC and ERK1 was examined. In pull-down assays where BRD4 was bound on Flag-agarose beads, ERK and MYC both bound to BRD4 (Fig. 6E). The efficiency of binding to BRD4 by either MYC or ERK1 alone or in combination was equivalent, suggesting that they do not compete. Since ERK1 can bind directly to MYC (43), we examined whether BRD4 and ERK1 compete for binding MYC. In pull-down assays where MYC was retained on anti-MYC agarose beads, BRD4 and ERK both bound; efficiency of binding to MYC was equivalent whether each was added alone or in combination (Fig. 6F). Thus, BRD4 and ERK1 do not compete for MYC binding in vitro.
These results suggest that BRD4, ERK1, and MYC form a trimeric complex where MYC and ERK1 bind to the C terminus and N terminus of BRD4, respectively (Figs. 1D and 6D). To validate this, pull-down assays were done with the BRD4ΔC and ΔN proteins immobilized on Flag beads and incubated with either MYC or ERK1 alone or both together (Fig. 6G). BRD4ΔC failed to bind MYC, which binds to the C terminus. However, in the presence of ERK1, which binds the N terminus, BRD4ΔC pulled down both ERK1 and MYC (Fig. 6G). Conversely, BRD4ΔN failed to pull down ERK1 unless MYC was also present (Fig. 6G). These results demonstrate that each protein interacts directly with the other two to form a trimeric complex.
To examine whether a BRD4-MYC-ERK1 trimeric complex exists in cells, sequential immunoprecipitations of endogenous BRD4, MYC, and ERK1 were done (Fig. 6H). Anti-BRD4 antibody immunoprecipitated both ERK1 and MYC (Fig. 6 H, Left). This immunoprecipitate was subjected to a secondary immunoprecipitation with anti-MYC agarose beads; ERK1 and BRD4 were both recovered here, demonstrating the existence of a BRD4-MYC-ERK1 trimeric complex (Fig. 6 H, Right).
The finding of this trimeric complex led to questions of whether 1) ERK1 regulated BRD4 kinase or HAT activities or 2) BRD4 regulated ERK1 kinase activity. ERK1 had no effect on BRD4 HAT activity (SI Appendix, Fig. S5A); BRD4 and ERK1 do not phosphorylate each other (SI Appendix, Fig. S5B). However, ERK1 inhibited BRD4 kinase activity: ERK1 blocked BRD4 phosphorylation of MYC as well as BRD4 autophosphorylation (Fig. 6I and SI Appendix, Fig. S5B). Thus, when both ERK and BRD4 are present in a kinase assay with MYC, MYC was phosphorylated only to the extent of ERK1 phosphorylation (Fig. 6I). Since MYC is a substrate of both BRD4 and ERK1, we validated these conclusions with a BRD4 substrate that is not phosphorylated by ERK1, namely TAF7 (29). While TAF7 was not phosphorylated by ERK1, its phosphorylation by BRD4 was completely abrogated in the presence of ERK1, thus demonstrating that ERK1 is a strong inhibitor of BRD4 kinase activity irrespective of the substrate (Fig. 6I).
BRD4 kinase activity maps to its N-terminal end where ERK1 binds (26) (Fig. 6D). Accordingly, ERK1 binds with less efficiency to the BRD4 Δkinase mutant (Fig. 6I), suggesting that ERK1 targets the 46 aa deleted in the BRD4Δkinase mutant (aa 502 to 548) (Fig. 6J). Thus, ERK1 binding BRD4 at a region critical for its kinase activity inhibits BRD4 kinase activity.
Histones and ERK1 both bind to the N-terminal end of BRD4, with BRD4 bromodomains playing an important role in their interaction (44) (Fig. 4D). We therefore tested if BRD4’s interaction with histones affected its binding to ERK1 or, conversely, if ERK1 reduced BRD4’s binding to histones. In pull-down assays with BRD4ΔC immobilized on beads, ERK1 binding to BRD4 was nearly abrogated in the presence of histone H3, whereas H3 binding to BRD4 was unaffected by ERK1 (SI Appendix, Fig. S5C). These results demonstrate that ERK1 does not bind directly to histone-bound BRD4. We speculate that ERK interacts with chromatin-bound BRD4 through its interaction with Myc.
In conclusion, ERK1 forms a triprotein complex with BRD4 and MYC, to phosphorylate MYC at Ser62 and inhibit BRD4’s phosphorylation of MYCThr58.
Discussion
MYC is a critical regulator of proliferation and cell growth that affects cell physiology through its interaction with over 300 cellular proteins (11). Among those proteins are ones involved in chromatin structure, DNA damage repair, mitosis, ribosome biogenesis, metabolism, RNA processing, and transcription. Reflecting its broad role in cellular processes, homeostatic levels of MYC are tightly controlled at multiple points of MYC protein biogenesis: transcription, mRNA half-life, and posttranslational regulation of MYC protein turnover (16). Despite the large number of proteins with which MYC interacts, only a handful of kinases regulate its stability. Kinases such as ERK1 and CDK2, phosphorylate MYC at Ser62, leading to its stabilization (19). Phosphorylation of MYC at Thr58 results in its degradation. The only kinase known to phosphorylate MYC at Thr58 was GSK3β, which is largely cytoplasmic, translocating to the nucleus in response to extracellular signaling (21, 45, 46). Thus, GSK3β is unlikely to contribute to homeostatic regulation of MYC. We have discovered that the bromodomain protein, BRD4, binds to MYC and phosphorylates it at Thr58. Since BRD4, unlike GSK3β, is constitutively nuclear, it can contribute to maintaining homeostatic levels of MYC. Furthermore, BRD4 binds ERK1, forming a trimeric complex with MYC. These results identify a regulatory network that maintains homeostatic levels of MYC protein (Fig. 6K).
MYC stability plays a paradoxical role in regulating gene expression. On one hand, MYC protein levels need to be maintained to allow it to function as a general amplifier of transcription (2, 3). This is accomplished in normal cells by regulating both MYC transcription and MYC protein stability by phosphorylation at Ser62 (16). On the other hand, degradation of MYC assembled with the transcription initiation complex is necessary for Pol II pause release and productive elongation at MYC target genes (9). Increased degradation of MYC by phosphorylation at Thr58, reduces MYC levels resulting in reduced global transcription. Thus, dynamically balancing MYC transcript and protein levels through BRD4 HAT and kinase activities is critical to maintain normal patterns of gene expression (Fig. 6K).
BRD4 activates MYC transcription through its HAT and kinase activities. Whereas BRD4 loss can lead to a decrease in MYC transcript abundance, BRD4 overexpression increases MYC transcription but does not alter MYC protein levels (Fig. 2D) (15, 32, 33, 47, 48). The present finding that BRD4 phosphorylation of MYC protein triggers its degradation explains these observations, by demonstrating that BRD4’s differential effects on MYC transcription and protein stability maintain homeostatic MYC protein levels.
Since MYC degradation is necessary for transcriptional pause release (9), we speculate that BRD4-mediated degradation of MYC effects pause release (49, 50) and the accompanying loss of total MYC levels is compensated by BRD4-mediated activation of MYC transcription. Thus, this duality of BRD4 regulation of MYC transcription and protein stability dynamically maintains MYC levels (Fig. 6K). The transcription elongation factor PTEFb, which is recruited and regulated by BRD4 and plays a role in pause release, neither phosphorylates MYC nor influences BRD4 phosphorylation of MYC.
MAPK signaling triggers the translocation of ERK1 from the cytoplasm to the nucleus where it increases MYC half-life both directly through phosphorylation of MYC Ser62, stabilizing it, and indirectly by inhibiting BRD4-mediated MYC degradation. We speculate that in response to upstream MAPK signaling, GSKβ replaces BRD4 in regulating the MYC degradation required for pause release.
While BRD4 contributes to regulation of MYC levels, MYC contributes to regulation of BRD4 function, resulting in a reciprocal relationship. Binding of nonphosphorylated MYC to BRD4, whether free or associated with chromatin, abrogates the BRD4 HAT activity necessary for chromatin remodeling. These findings are consistent with an earlier report that inhibition of MYC degradation blocks histone acetylation at MYC target genes, suggesting MYC turnover is required for HAT activity at these sites (15). Although phosphorylated MYC binds BRD4, it is unable to inhibit BRD4 HAT activity. These interactions are likely to be carefully orchestrated to ensure appropriate levels of MYC under both normal and signaled cellular conditions to avoid inducing tumors.
Dysregulation of MYC levels is associated with over half of all human cancers (24). Understanding its normal regulatory pathways and interactions will be necessary to fully characterize MYC mechanisms of action in cancer. Because of its activation of MYC transcription, BRD4 is being widely investigated as a therapeutic target in MYC-driven cancers. While JQ1-like small molecules reduce MYC transcription but have no effect on MYC stability, MZ1-like molecules which degrade BRD4 have the paradoxical effect of decreasing MYC transcription but increasing MYC stability and levels. Thus, our findings demonstrating BRD4-mediated MYC degradation as well as the reciprocal regulation of BRD4 HAT activity by MYC are likely to have significant translational implications.
Materials and Methods
Details of cell lines, plasmid constructs, and reagents can be found in SI Appendix, as well as all protocols detailing protein purification, mass spectrometry analysis, transient transfections, immunofluorescence, coimmunoprecipitation, in vitro binding assays, PLAs, enzymatic and nucleosome assays, ubiquitination assays, pulse-chase assays, and RNA-seq.
Data Availability.
RNA-seq data are deposited in the Gene Expression Omnibus database under the accession code GSE147131.
Supplementary Material
Acknowledgments
We thank Drs. Carol Thiele, Louis Staudt, and Avinash Bhandoola for reading the manuscript; laboratory members for discussions; Drs. Qingrong Chen and Anne Gegonne for bioinformatic analyses; and Dr. Ming Zhou for mass spectrometry analysis. This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data are deposited in the Gene Expression Ominibus database under the accession code GSE147131.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919507117/-/DCSupplemental.
References
- 1.Eilers M., Eisenman R. N., Myc’s broad reach. Genes Dev. 22, 2755–2766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nie Z. et al., c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C. Y. et al., Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretones G., Delgado M. D., León J., Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N., The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majello B., Perini G., Myc proteins in cell biology and pathology. Biochim. Biophys. Acta 1849, 467–468 (2015). [DOI] [PubMed] [Google Scholar]
- 7.von Eyss B., Eilers M., Addicted to Myc–But why? Genes Dev. 25, 895–897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahl P. B. et al., c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaenicke L. A. et al., Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol. Cell 61, 54–67 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Baluapuri A. et al., MYC recruits SPT5 to RNA polymerase II to promote processive transcription elongation. Mol. Cell 74, 674–687 e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalkat M. et al., MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell 72, 836–848 e7 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Sammak S., Allen M. D., Hamdani N., Bycroft M., Zinzalla G., The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J. 285, 4165–4180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon S. B., Wood M. A., Cole M. D., The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20, 556–562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang G., Espeseth A., Hazuda D. J., Margolis D. M., c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81, 10914–10923 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaiah B. N. et al., BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 23, 540–548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J., Levens D., Making myc. Curr. Top. Microbiol. Immunol. 302, 1–32 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Dani C. et al., Extreme instability of myc mRNA in normal and transformed human cells. Proc. Natl. Acad. Sci. U.S.A. 81, 7046–7050 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhikary S., Eilers M., Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Farrell A. S., Sears R. C., MYC degradation. Cold Spring Harb. Perspect. Med. 4, a014365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears R. et al., Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welcker M. et al., The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hydbring P. et al., Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc. Natl. Acad. Sci. U.S.A. 107, 58–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang C. V., MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beroukhim R. et al., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S. Y., Chiang C. M., The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282, 13141–13145 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Devaiah B. N. et al., BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 109, 6927–6932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranello L. et al., RNA polymerase II regulates Topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z. et al., Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Devaiah B. N., Singer D. S., Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J. Biol. Chem. 287, 38755–38766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey A., Nishiyama A., Karpova T., McNally J., Ozato K., Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 20, 4899–4909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao R., Nakamura T., Fu Y., Lazar Z., Spector D. L., Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13, 1295–1304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmore J. E. et al., BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertz J. A. et al., Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U.S.A. 108, 16669–16674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S. Y., Lee A. Y., Lai H. T., Zhang H., Chiang C. M., Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell 49, 843–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conacci-Sorrell M., McFerrin L., Eisenman R. N., An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devaiah B. N., Gegonne A., Singer D. S., Bromodomain 4: A cellular Swiss army knife. J. Leukoc. Biol. 100, 679–686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myant K. et al., Serine 62-phosphorylated MYC associates with nuclear lamins and its regulation by CIP2A is essential for regenerative proliferation. Cell Rep. 12, 1019–1031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaiah B. N., Singer D. S., Data from 'Genome wide transcript analysis in Mouse embryonic fibroblasts where endogenous BRD4 is deleted' Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147131. Deposited 17 March 2020.
- 39.Yada M. et al., Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermeking H. et al., Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. U.S.A. 97, 2229–2234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanconato F. et al., Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat. Med. 24, 1599–1610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faiola F. et al., Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol. Cell. Biol. 25, 10220–10234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seth A., Gonzalez F. A., Gupta S., Raden D. L., Davis R. J., Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 267, 24796–24804 (1992). [PubMed] [Google Scholar]
- 44.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K., The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 100, 8758–8763 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory M. A., Qi Y., Hann S. R., Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 278, 51606–51612 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Beurel E., Grieco S. F., Jope R. S., Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lovén J. et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuber J. et al., RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anand P. et al., BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel M. C. et al., BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol. 33, 2497–2507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are deposited in the Gene Expression Omnibus database under the accession code GSE147131.