Significance
Pentameric ligand-gated ion channels (pLGICs) are critical for transduction of electrical signals between nerve cells, and highly important for neuropharmacology. Members of this receptor family are also found in prokaryotes, often incorporating additional domains whose roles are largely uncharacterized. Here, we present two structures of a prokaryotic pLGIC with such an extra N-terminal domain. The channel has a gating transition inhibited by Ca2+ binding. Comparison of structures with/without Ca2+ explains its role in the transition and also reveals changes in a cavity known to bind allosteric modulators in other pLGICs, in the interactions between the channel and lipids or between the ligand-binding and N-terminal domains. This illustrates the different ways allosteric modulation can occur in the gating transition of pLGICs.
Keywords: ligand-gated ion channels, structural biology, crystallography, electrophysiology
Abstract
Pentameric ligand-gated ion channels (pLGICs) are allosteric receptors that mediate rapid electrochemical signal transduction in the animal nervous system through the opening of an ion pore upon binding of neurotransmitters. Orthologs have been found and characterized in prokaryotes and they display highly similar structure–function relationships to eukaryotic pLGICs; however, they often encode greater architectural diversity involving additional amino-terminal domains (NTDs). Here we report structural, functional, and normal-mode analysis of two conformational states of a multidomain pLGIC, called DeCLIC, from a Desulfofustis deltaproteobacterium, including a periplasmic NTD fused to the conventional ligand-binding domain (LBD). X-ray structure determination revealed an NTD consisting of two jelly-roll domains interacting across each subunit interface. Binding of Ca2+ at the LBD subunit interface was associated with a closed transmembrane pore, with resolved monovalent cations intracellular to the hydrophobic gate. Accordingly, DeCLIC-injected oocytes conducted currents only upon depletion of extracellular Ca2+; these were insensitive to quaternary ammonium block. Furthermore, DeCLIC crystallized in the absence of Ca2+ with a wide-open pore and remodeled periplasmic domains, including increased contacts between the NTD and classic LBD agonist-binding sites. Functional, structural, and dynamical properties of DeCLIC paralleled those of sTeLIC, a pLGIC from another symbiotic prokaryote. Based on these DeCLIC structures, we would reclassify the previous structure of bacterial ELIC (the first high-resolution structure of a pLGIC) as a “locally closed” conformation. Taken together, structures of DeCLIC in multiple conformations illustrate dramatic conformational state transitions and diverse regulatory mechanisms available to ion channels in pLGICs, particularly involving Ca2+ modulation and periplasmic NTDs.
Pentameric ligand-gated ion channels (pLGICs) play crucial roles in electrochemical signaling in a wide range of organisms (1, 2). In the animal nervous system, binding of neurotransmitters to a receptor favors the open conformation of the internal transmembrane channel, allowing selected ions to permeate the phospholipid bilayer down their electrochemical gradient. Dysfunction of these receptors is linked to major neurodegenerative and psychiatric disorders, such as Alzheimer’s disease, Parkinson’s disease, epilepsy, hyperekplexia, myasthenia gravis, and alcohol dependence (1). Because of their pivotal physiological roles and sensitivity to allosteric modulators, pLGICs are key targets for a variety of common therapeutic agents, such as benzodiazepines, general anesthetics, and barbiturates (3). Since the identification and isolation of the first nicotinic acetylcholine receptor (nAChR) (4), members of this channel family have been subjects of intensive functional and structural studies. Orthologs of the pLGICs have since been found in prokaryotes (5, 6), and provided the first model systems in this family accessible to X-ray structure determination in multiple functional states (7–9). Recent structures of eukaryotic receptors determined by crystallography or cryoelectron microscopy have validated many of these conformational predictions (10–15).
Alongside their value as model systems, distinctive features of prokaryotic pLGICs offer insight into evolutionary and mechanistic diversity. Whereas they generally lack the extended intracellular domain between transmembrane helices M3 and M4 found in eukaryotes (10, 16–19), prokaryotic pLGICs often possess additional N-terminal domains (NTDs) (5, 6) on their extracellular sides. On the basis of sequence homology, pLGIC NTDs include members of the periplasmic binding protein (PBP), calcium channel and chemotaxis receptor (Cache), and methyl-accepting chemotaxis protein N-terminal (MCP-N) families, as well as others that remain to be characterized (5). Based on this, it has been predicted that pLGICs in prokaryotes mediate cell–cell interactions, possibly as chemotaxis or quorum-sensing receptors (5). Indeed, the MCP-N domain is part of the two-component histidine kinase signaling system widespread in bacteria and archaea, and may allow the prokaryotic cell to convert detection of external molecules, such as nutrients or repellents, into flagellar motion. Other families of animal neuronal ion channels, such as the tetrameric AMPA and NMDA receptors (20), also contain large NTDs structurally related to bacterial periplasmic binding proteins fused to their ligand-binding domain (LBDs). In at least some cases, these domains bind allosteric ligands and may play key functional roles (21). However, there is presently no structural information regarding the organization of a full-length pLGIC with a periplasmic NTD, nor how it might modulate the transmembrane domain (TMD) or LBD.
Here we determined X-ray structures of DeCLIC, a pLGIC derived from a symbiotic sulfate-reducing deltaproteobacterium closely related to Desulfofustis glycolicus, including an NTD accounting for about 50% of the total receptor mass. DeCLIC was crystallized in the presence and absence of Ca2+, an apparent effector whose depletion enabled ion conduction in Xenopus oocytes. The resulting structures, determined to 3.55 Å and 3.83 Å resolution, evidenced state-dependent conformational changes in all three domains, including a transition from a nonconducting to wide-open pore. Together with functional and computational data, these structures offer insight into the molecular architecture and accessible states of a full-length prokaryotic pLGIC incorporating a previously uncharacterized NTD.
Results
Structure of a Periplasmic Amino-Terminal Domain in a Full-Length pLGIC.
In pursuit of structural targets to illuminate the diversity of prokaryotic pLGICs fused to periplasmic domains, we performed a BLAST search on sTeLIC, a recently characterized channel expressed in symbiotic bacteria (22). We identified 18 bacterial or archaeal orthologs with over 45% sequence identity to sTeLIC, but with large NTDs fused directly to their LBDs (SI Appendix, Figs. S1 and S2). These NTDs typically encompassed ∼300 residues and accounted for up to 50% of total receptor mass. We focused on one such homolog, DeCLIC. Although the precise physiological role of this protein is unknown, it echoes symbiotic associations of sTeLIC: Desulfofustis grows in close association with sulfide-oxidizing purple sulfur bacteria, with which it forms macroscopic microbial photosynthetic colored aggregates (pink berries) found in some salt marshes (23).
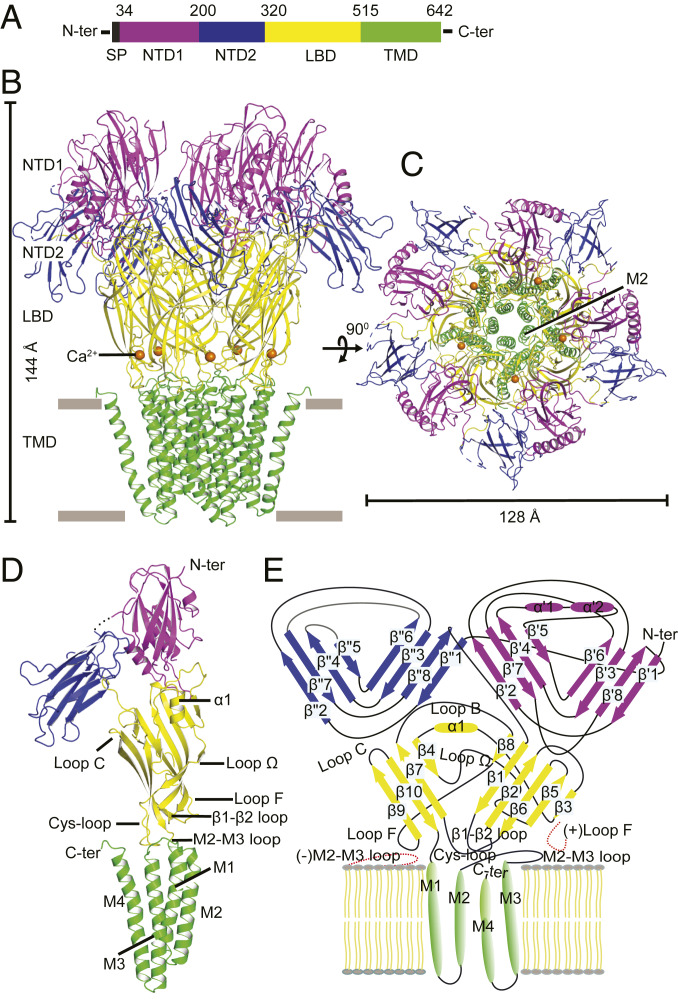
Crystallographic quantities of DeCLIC were produced as fusions with maltose binding protein in C43 Escherichia coli, solubilized, cleaved, and purified in n-dodecyl-β-d-maltoside (DDM) following previously established protocols for other prokaryotic pLGICs (22). Initial hits of crystallogenesis were obtained in the presence of 250-mM Ca2+. The growth of crystals with good diffracting quality required extensive seeding and systematic screening, eventually resulting in an anisotropic dataset with a maximum resolution of 3.55 Å (SI Appendix, Table S1). The initial phases were obtained from molecular replacement; noncrystallographic symmetry averaging aided the subsequent building and refinement of nearly all residues (SI Appendix, Fig. S3 A–F). The resulting structure showed DeCLIC to be a pentameric molecule, ∼144 Å long and 128 Å wide viewed perpendicular to the cell membrane (Fig. 1). Its dimensions and multilayer architecture were reminiscent of NMDA receptors, although the latter are tetrameric oligomers with different topologies. As in other pLGICs, the subunits assembled around a fivefold axis corresponding to the ion pathway, described in detail below; the NTD stood “on top of” the LBD (opposite to the membrane region), forming a fivefold symmetric crown. Adjacent pentamers in the crystal lattice formed parallel linear arrays mediated primarily by contacts between the NTDs (SI Appendix, Fig. S4 A and B). Although the NTD was refined with substantially higher B-factors than the LBD or TMD (SI Appendix, Fig. S5), indicating relatively high flexibility, most residues could be built through iterative refinement. The topology of this domain was further validated by independent crystallization of a soluble NTD fragment (residues 33 to 202), and its structure determined to 1.75 Å using single anomalous diffraction phasing on a SeMet-labeled construct (SI Appendix, Fig. S3G and Table S1). In accordance with secondary structure predictions, the domain contained a majority of β-strands, arranged in two jelly-roll domains, or lobes, connected by a short loop (Fig. 1D). The β-sheets (strands β′1 to β′8) of the first lobe (NTD1, residues 34 to 200) were divided by two α-helices (α′1 and α′2) between β′4 and β′5. This lobe was positioned directly above the LBD of the same subunit, interacting mostly with the LBD β2–β3 and β5–β6 (E) loops (Fig. 1E), and with the second NTD lobe (NTD2, residues 204 to 320) from the principal neighboring subunit (Fig. 1A). The jelly roll of NTD2 (strands β′′1 to β′′8) lacked the α-helices of NTD1, but contained a disulfide bridge between strands β′′6 and β′′7 (SI Appendix, Fig. S3C). This lobe was positioned around the periphery of the upper LBD, interacting primarily with NTD1 and the β2–β3 loop from the complementary LBD (Fig. 1E).
Fig. 1.
Architecture of DeCLIC in high concentration of Ca2+. (A) Domain organization of the full-length DeCLIC receptor. (B) Ribbon representation of the Ca2+-bound structure, viewed from the plane of the membrane, with domains colored as in A. Ca2+ ions are shown as orange spheres; gray lines represent approximate membrane boundaries. (C) View as in B from the periplasmic side. (D) Structure of a single DeCLIC subunit, shown as in B, indicating key structural elements in the LBD and TMD. (E) Topology of a single DeCLIC subunit indicating all secondary structure elements, colored as in A. Proximal loops from neighboring subunits are represented as red dashed lines, and individual lipid tails as olive lines.
The sequence identity between NTD1 and NTD2 was only around 16%, but these lobes exhibited similar topologies and patterns of hydrophobicity (Fig. 1E and SI Appendix, Fig. S6 A and B), with an RMSD of 2.57 Å over 67 Cα atoms. A search for similar structures using the DALI server (24) indicated similar folds in TNF homology domains and several viral capsid VP1 proteins (SI Appendix, Fig. S6C). Proteins that display jelly-roll topology often contain two copies of this domain, especially in viruses (25). However, their relative orientations in DeCLIC appeared to be distinctive, as a DALI search using NTD1+NTD2 from the same subunit or neighboring subunit (domain swapped version) did not detect any hits.
Ca2+-Bound Closed-Pore Structure of the Full-Length Ion Channel.
Beyond the NTD, the DeCLIC LBD and TMD adopted a classic architecture observed in other “Pro-loop” (prokaryotic) or “Cys-loop” (eukaryotic) receptors (6). On the basis of evolutionary relatedness using the ConSurf Server (26), the most conserved regions included the NTD–LBD and LBD–TMD interfaces, as well as the interior of the transmembrane pore (SI Appendix, Fig. S7). In the LBD, each subunit contained 10 β-strands (β1 to β10) in an Ig-like β-sandwich; a single amphipathic helix α1 inserted between β3 and β4 (Figs. 1E and 2A), with its hydrophilic side exposed to the LBD vestibule. Among 18 pLGICs with ≥45% sequence identity to DeCLIC, the most conserved region was the LBD, particularly the β6–β7 (Pro) loop (SI Appendix, Fig. S7). In this region, Q446 formed hydrogen bonds with the backbone of D437 on the opposite side of the Pro-loop hairpin, much like the eponymous Cys-loop linkage in eukaryotic receptors (6) (SI Appendix, Fig. S7C). The Pro-loop conformation was further reinforced by interactions of the strictly conserved N401 (N80 in GLIC) with backbone atoms of I450 (I128). Also in the Pro-loop, D444 contributed to an “electrostatic triad,” previously observed in GLIC and other pLGICs (22), with Q344 in the β1–β2 loop and R513 in the pre-M1 region (SI Appendix, Fig. S8B); this network also involved cation-π stacking between R513 and Loop-F residue W482 (W160 in GLIC). Conserved residues in the upper LBD included an interaction between W359 and P395, which formed the bottom of a vestibular cavity previously identified in ELIC and sTeLIC (22, 27). The β9–β10 (C) loop was one of the least-conserved regions, consistent with idiosyncrasies among agonists specific to different receptor types (SI Appendix, Figs. S2 and S7A).
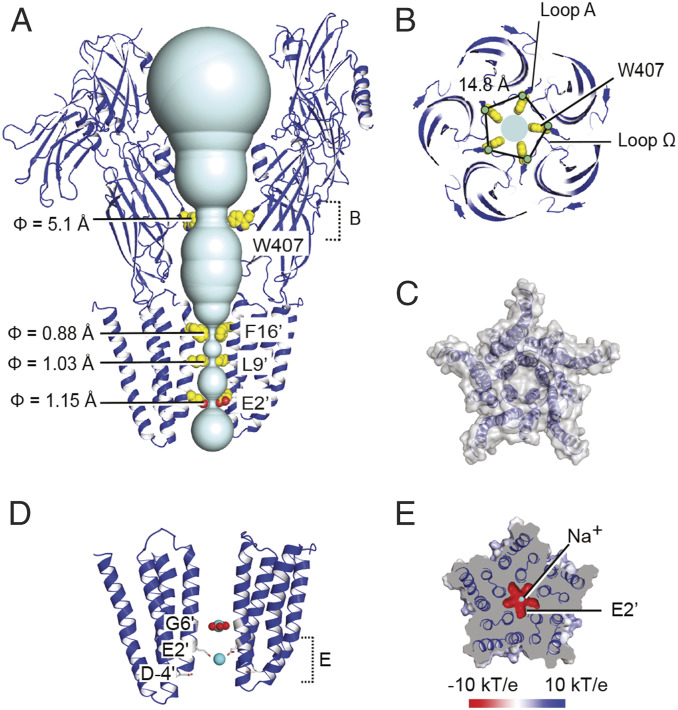
Fig. 2.
Constrictions in the channel pore in the presence Ca2+. (A) View from the membrane as in Fig. 1B of the solvent-accessible volume in the channel (cyan) in the Ca2+-bound conformation. For clarity, only two opposing subunits (blue) are shown; ions and resolved water molecules in the pore are omitted. Side chains for residues forming vestibular and transmembrane constrictions are shown as spheres. (B) View from the periplasm of the LBD vestibular constriction in the Ca2+-bound state, showing distances between Cα atoms of neighboring loop-Ω W407 residues (yellow spheres). (C) View from the periplasm of the TMD surface (gray) in the presence of Ca2+, showing solvent exclusion in the channel pore. (D) View from the membrane of two TMD subunits (blue) in the presence of Ca2+, showing resolved ions (cyan) and waters (red) in the channel pore, and neighboring residues as gray sticks. Dotted bracket indicates cross-sectional slice depicted below. (E) A 20 Å cross section of the Ca2+-bound structure as indicated in D, viewed from the periplasmic side and colored by electrostatic potential according to the scale bar shown below.
Based on examination of the mFo-DFc Fourier difference map, we identified a single strong spherical density at each LBD subunit interface that could be modeled as a Ca2+ ion (Fig. 1 A and B and SI Appendix, Fig. S9 A and B). This site was validated by a 5-Å resolution anomalous dataset from crystals soaked in 200 mM Ba2+ (SI Appendix, Table S1), from which we could unambiguously assign five symmetry-related densities equivalent to the proposed Ca2+ ions (SI Appendix, Fig. S9 C and D). Each ion was coordinated by E347 in the β1–β2 loop, D437 and two backbone carbonyls (P434, P436) in the Pro-loop, and E479 in complementary loop F (SI Appendix, Fig. S9B). The resulting electrostatic interface was further extended by two arginines in the β1–β2 (R345) and M2–M3 (R569) loops, which formed a salt-bridge network with E481 in complementary loop F (SI Appendix, Fig. S8). The only other Ba2+ density evident in DeCLIC was in two of five subunits, at the interface between NTD1 and NTD2 from adjacent chains (SI Appendix, Fig. S9 C and D); we observed no anomalous signal in the ion channel pore.
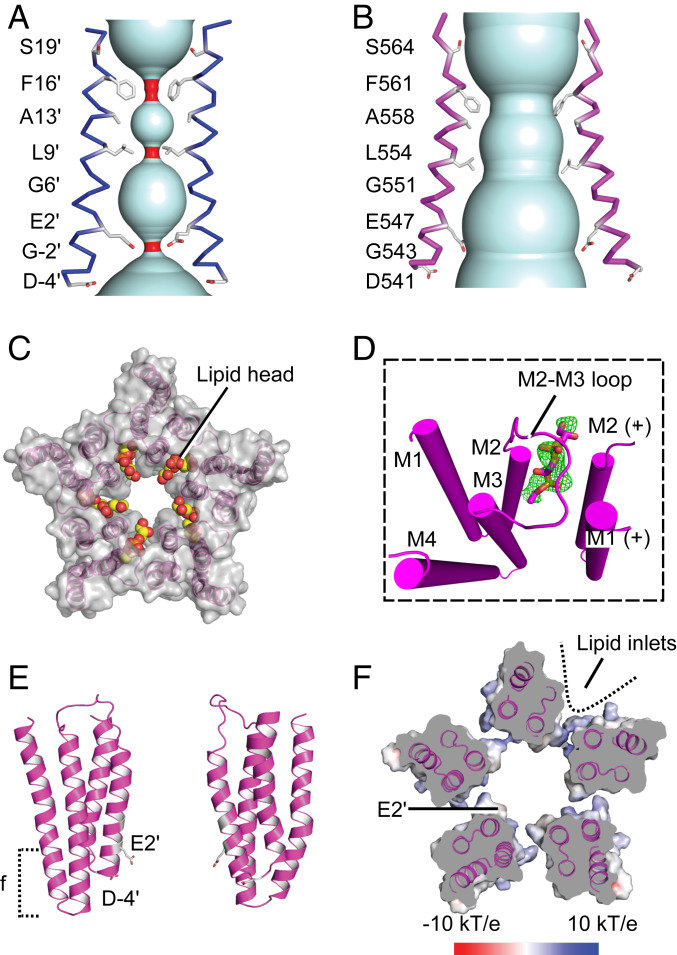
Below the LBD, the transmembrane pore contained two apparent hydrophobic gates. Side chains of M2 residues F561 (F16′) and L554 (L9′) constricted the pore radius to 0.9 Å and 1.0 Å, respectively (Fig. 2A), too small for the passage of a dehydrated Na+ or K+ ion. Below these gates, both mFo-DFc and 2mFo-DFc maps showed a strong central density at the level of G551 (G6′); we modeled this density as Na+, the preponderant monovalent ion in the crystallization buffer, surrounded by five waters in a planar arrangement (Fig. 2D). Further down the DeCLIC pore, an additional, spherical density was evident within a hydrophilic constriction, where the pore radius narrowed to 1.15 Å (Fig. 2A); this density was also modeled as an Na+ ion, coordinated directly by five acidic E547 (E2′) side chains. The cytoplasmic mouth was lined by a second acidic ring of D541 (D-4′) residues (Fig. 2D).
Electrophysiology and Crystallization in the Absence of Ca2+.
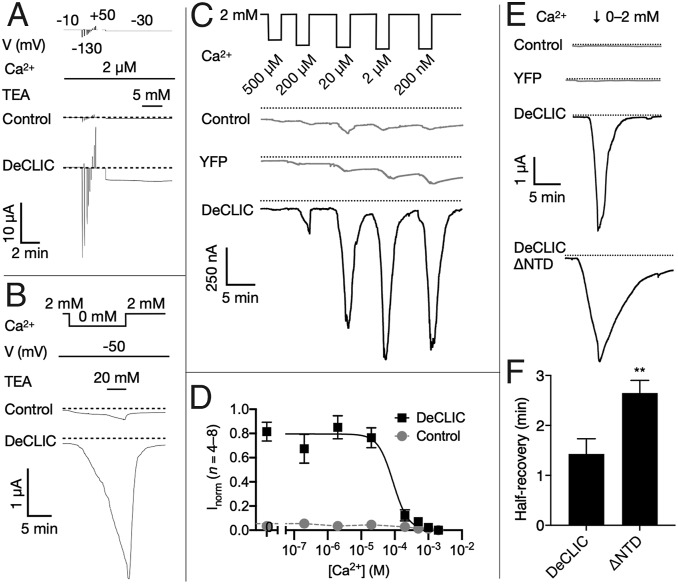
To identify conditions that allow opening of the pore, we injected mRNA encoding DeCLIC into Xenopus laevis oocytes and recorded currents by two-electrode voltage-clamp electrophysiology. We screened a small library of molecules, including sugars, salts, representative amino acids and their derivatives, as well as other pLGIC agonists and modulators, but observed no replicable effects relative to controls (SI Appendix, Table S2). In contrast, depletion of Ca2+ from the oocyte media (reduction from 2 mM to 2 µM) produced currents in DeCLIC-injected cells (Fig. 3A). Under conditions of weak polarization (−30 mV) and elevated pH (8.5) to reduce endogenous leak currents, these currents persisted with no evident desensitization; they were also insensitive to tetraethylammonium (TEA) up to 20 mM (Fig. 3 A and B), consistent with a pore conformation distinct from blocker-sensitive channels in the same family (28). Under most conditions tested, slowly evolving activation (Fig. 3B) and intolerance to prolonged Ca2+ depletion (Fig. 3C) precluded conclusive quantification of steady-state currents, possibly due to slow rates of Ca2+ dissociation or of structural transition to the maximally conducting state. However, based on maximal inward currents evolved at pH 8.5 after 2 min of Ca2+ depletion at −70 mV, concentration-dependent responses were reproducible (n = 4 to 8) with an estimated half-inhibitory concentration (IC50) of 90 µM (Fig. 3 C and D). A DeCLIC construct containing only the LBD and TMD (ΔNTD) produced relatively slow apparent gating, particularly for recovery from Ca2+ depletion (i.e., transition to a nonconducting state) (Fig. 3E). Although this slowed profile excluded direct comparison to the apparent wild-type affinities, we found the time to half-maximal recovery from 0-mM Ca2+ consistently increased nearly twofold in the ΔNTD variant relative to wild-type (Fig. 3F), consistent with a role for the evidently dynamic NTD in gating kinetics.
Fig. 3.
Functional evidence for Ca2+-inhibited DeCLIC currents. (A) Sample traces from two-electrode voltage-clamp electrophysiology in control and DeCLIC-injected Xenopus oocytes, showing equilibration in depleted Ca2+ (2 µM), voltage jumps to identify sustainable currents, then a voltage step (−30 mV) producing inward DeCLIC currents showing no desensitization nor inhibition by TEA (5 mM). Dotted lines represent 0-µA current. Although currents apparently reached a steady state, most oocytes did not tolerate activation by Ca2+ depletion beyond ∼10 min. (B) Sample traces under nonequilibrium conditions (−50 mV, 0 Ca2+), showing slowly developing currents not saturating after 12 min, and not inhibited even by high TEA (20 mM). (C) Sample traces to estimate Ca2+ sensitivity, showing brief (2-min) steps to decreasing Ca2+ concentrations at −70 mV. Maximum DeCLIC currents by this protocol saturated at 2 to 20 µM Ca2+, substantially larger than control or yellow fluorescent protein-expressing cells. (D) Inhibition curves for paired DeCLIC-injected (black squares) and control oocytes (gray circles) according to the protocol in C, normalized to the maximum response in parallel recordings. Solid black line represents nonlinear regression fit to DeCLIC responses, IC50 = 90 µM (n = 4 to 8, R2 = 0.8). (E) Sample traces documenting slowed kinetics of the truncated construct DeCLIC-ΔNTD, showing brief step to 0 mM Ca2+ followed by recovery to baseline in 2 mM Ca2+. Whereas DeCLIC currents decreased to half-maximum within ∼1.5 min, DeCLIC-ΔNTD half-recovery was roughly doubled. (F) Quantification of half-recovery time according to the protocol in E, showing significant slowing in DeCLIC-ΔNTD (n = 7, **P < 0.01).
To elucidate the structural basis for Ca2+-dependent gating in DeCLIC, we screened for new crystallization conditions excluding this ion. The best resulting crystals produced an anisotropic dataset with a maximum resolution of 3.83 Å (SI Appendix, Table S1). Similar to the high-Ca2+ condition, individual pentamers in each asymmetric unit aligned in parallel linear arrays, with contact interfaces primarily between the NTDs (SI Appendix, Fig. S4). Molecular replacement was based on the prokaryotic channel sTeLIC (22), and the model again built using fivefold noncrystallographic symmetry. The refined structure was different from that of the Ca2+-bound conformation both at the tertiary and quaternary levels, as described below (Fig. 4 C and D).
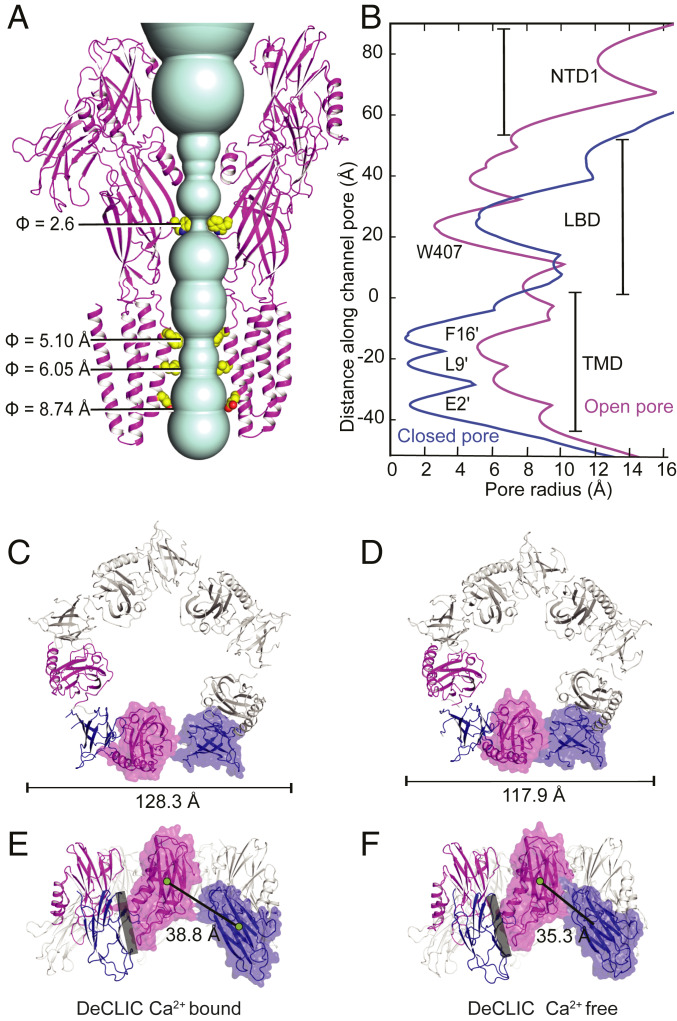
Fig. 4.
Solvent-accessible DeCLIC channel in the absence of Ca2+ and contraction of periplasmic domains. (A) View from the membrane of the channel in the Ca2+-free conformation (magenta). Side chains of residues forming vestibular and transmembrane constrictions are shown as spheres. (B) Pore radius profiles along the channel axis in Ca2+-bound (blue) and Ca2+-free (magenta) states. (C) DeCLIC NTD in the presence of Ca2+, viewed from the periplasmic side. Complete domains from two adjacent subunits are colored by lobe (NTD1, magenta; NTD2, dark blue), with the solvent-accessible surface shown for one subunit. (D) View as in C of the NTD in Ca2+-free DeCLIC. (E) View as in C of the Ca2+-bound DeCLIC NTD, viewed from the membrane plane. Black line indicates center-of-mass distances between two lobes of the same subunit; gray bar indicates the contact interface between principal NTD2 and complementary NTD1 lobes of adjacent subunits. (F) View as in E in Ca2+-free DeCLIC.
Periplasmic Contraction and Transmembrane Expansion in the Absence of Ca2+.
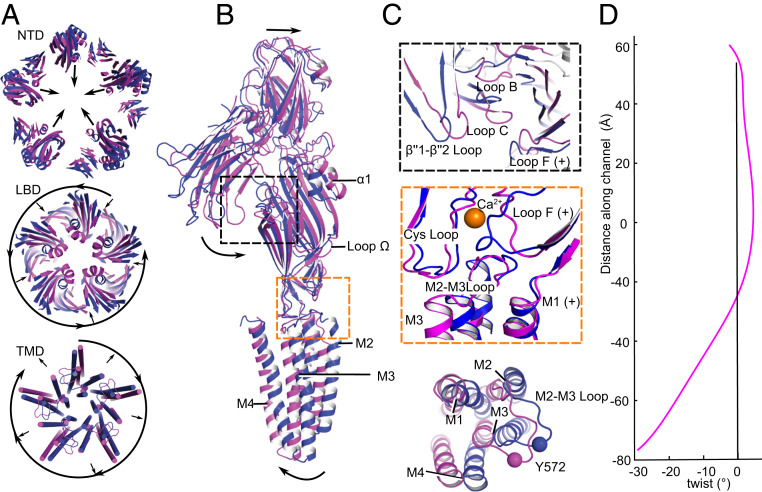
Comparison of DeCLIC structures in the presence and absence of Ca2+ revealed opposing structural transitions in the periplasmic and transmembrane domains. Beginning at the N terminus, the NTD retained its two-lobed jelly-roll fold in the absence of Ca2+, but exhibited an overall radial contraction, decreasing the center-of-mass distances (dCOM) between adjacent NTD2 lobes by over 5 Å (56.8 Å to 51.6 Å) (Fig. 4C and SI Appendix, Fig. S10). Whereas in high Ca2+ the NTD2 lobe interacted primarily with the complementary neighboring subunit, domain contraction in the absence of Ca2+ translated NTD2 inward toward NTD1 of the same subunit, decreasing their dCOM over 3 Å (38.8 Å to 35.3 Å) (Fig. 4 C and D). This motion brought the NTD2 β′′1–β′′2 loop in proximity to LBD loop C (Fig. 5 E and F), near the orthosteric agonist-binding site.
Fig. 5.
Conformational state transitions at the tertiary and quaternary levels. (A) Superposition of DeCLIC structures in the presence (blue) and absence (magenta) of Ca2+, viewed from the periplasmic side, illustrating relative conformational changes in the NTD (Top), LBD (Middle), and TMD (Bottom). Loop regions except the M2–M3 loop are hidden for clarity. Arrows indicate predominant quaternary rearrangements involving radial contraction/expansion or tangential twist. (B) Conformational changes in a single subunit between Ca2+-bound (blue) and -free (magenta) states, aligned by superimposition of the entire pentamer, viewed from the membrane plane. Arrows indicate predominant motions involving contraction of the NTD and LBD, and expansion of the TMD. (C) Details as in B showing remodeling at the NTD2–LBD interface (Top), LBD–TMD interface (Middle), or of a single TMD subunit viewed from the periplasmic side (Bottom). (D) Twist angle (magenta) between Ca2+-bound and -free states in successive z slabs along the linear channel axis. Negative values in the lower channel correspond to a relative clockwise twist of the TMD.
Radial contraction was also evident in the LBD, decreasing the dCOM between adjacent domain-subunits from 28.5 Å to 25.8 Å and increasing buried area at subunit interfaces by 183 Å2 in the Ca2+-free state (SI Appendix, Fig. S10). Below the encroaching NTD2 lobes, the tip of each loop C translated toward the subunit interface by 5.4 Å, making contact with the top of complementary loop F (Fig. 5 B and C). The β7–β8 loop (B) also translated toward the interface, contacting β′7–β′8 of the complementary NTD1 (Fig. 5F). A solvent-accessible cavity beneath loops B and C, shown in other pLGICs to bind orthosteric ligands, was smaller and shallower than in the Ca2+-bound structure (SI Appendix, Fig. S11 A and B).
In the LBD interior, domain contraction was particularly evident in the amphipathic α1 helix, which translated and rotated ∼40° counter-clockwise toward the channel axis (Fig. 5C and SI Appendix, Fig. S12). This motion detached α1 from the β3–β4 (A) loop, established new contacts with the β3 strand and β5–β6 (E) loop, and expanded a vestibular cavity previously implicated in binding of allosteric modulators (SI Appendix, Figs. S11 C and D and S12) (22, 27). Below the α1 helices, an even more constricted ring was formed by the inward-facing β4–β5 loop (Ω22), with the side chains of five W407 residues restricting the vestibular radius to 2.6 Å (Figs. 2B and 4 A and B and SI Appendix, Fig. S13).
In the vicinity of the vacated Ca2+ site, loop F swung inward toward the channel axis by up to 8 Å, away from its ion-mediated intersubunit contacts (Fig. 5 B and C and SI Appendix, Fig. S6). This motion also disrupted the electrostatic network linking E481 in loop F with the β1–β2 (R345) loop of the complementary subunit; instead, E481 accepted a hydrogen bond from Q344 in β1–β2 of the same subunit (SI Appendix, Fig. S8). In turn, the liberated arginine (R345) oriented toward an expanded cleft at the transmembrane subunit interface (SI Appendix, Fig. S8).
In contrast to the largely contracting motions of the periplasmic NTD and LBD, the TMD exhibited dramatic twisting and outward blooming in Ca2+-free conditions. Based on cumulative rotation between the two states in slices along the channel axis, the TMD twisted clockwise up to −27° upon Ca2+ depletion (Fig. 5D). The M2–M3 loop shifted away from the pore axis by more than 7 Å (Fig. 5 A and C), and distances between adjacent subunits increased by over 5 Å (dCOM 22.0 Å to 27.1 Å) (SI Appendix, Fig. S10). Accordingly, the buried surface area at TMD subunit interfaces decreased by 832 Å2 (SI Appendix, Fig. S10E). Within the pore, we also observed an outward twist and translocation of the M2 helices, which relieved all three constriction gates (Fig. 6 A and B). Compared to the closed state, F16′ and L9′ rotated away from the fivefold axis, expanding to more than 5 Å radius in the upper pore; at the level of E2′, the pore expanded even further to a radius of 9 Å (Fig. 6B). Thus, the Ca2+-free structure is consistent with DeCLIC being in a conducting state in the absence of Ca2+.
Fig. 6.
Pore conformation and lipid interactions in the TMD. (A) View from the membrane of the TMD channel (cyan) in the presence of Ca2+, showing M2 helices of two opposing subunits (blue). Side chains exposed to the pore lumen are shown as sticks, with constrictions of the channel below 1.4 Å in red. (B) View as in A of two opposing M2 helices (magenta) in the Ca2+-free structure. (C) View from the periplasmic side of the TMD surface in the absence of Ca2+, with resolved lipid head-groups as colored spheres. (D) View of two adjacent TMD subunits in the absence of Ca2+ (magenta), with polar heads of l-α-phosphatidylglycerol molecules modeled as sticks. The Fo-Fc omit map (green) is overlaid and contoured at 2.5 σ. (E) View from the membrane of two TMD subunits (blue) in the absence of Ca2+, showing negatively charged residues along channel as sticks. Dotted bracket indicates cross-sectional slice depicted in F. (F) A 20-Å cross section of the Ca2+-bound structure as indicated in E, viewed from the periplasmic side and colored by electrostatic potential according to the scale bar shown below; dotted line indicates an expanded crevice accessible to lipid tails.
Allosteric Transitions and Potential Coupling in TMD and LBD Sites.
Structure determination in Ca2+-bound and -unbound states further revealed remodeling of TMD and LBD sites previously implicated in pLGIC modulation. In the TMD, underneath each M2–M3 loop in the Ca2+-free structure, we observed a strong residual electron density (8 σ in the mFo-DFc difference map), which we modeled as the polar head of a lipid molecule (Fig. 6 C and D and SI Appendix, Fig. S15 A–C). The expanded solvent-accessible surfaces at each subunit interface could accommodate disordered tails of the modeled lipids (Fig. 6F). Each putative lipid site was bounded by the upper M2 and M3 helices of the principal subunit, and by the pre-M1 region and upper M2 helix of the complementary subunit (Fig. 6D and SI Appendix, Fig. S15B). This subunit interface constitutes an apparent general site for lipophilic modulators, penetrating even deeper toward the channel axis than ivermectin cavities in GluCl, GlyRα3, and GlyRα1 (11, 12, 29) (SI Appendix, Fig. S15 D–F).
Remodeling was similarly evident in a cavity facing the LBD vestibule, between helix α1 and loops A and E. An equivalent vestibular cavity in ELIC and sTeLIC was previously shown to bind the allosteric modulators flurazepam and 4-bromocinnamic acid, respectively (22, 27). Indeed, vestibular-site binding was associated with a dramatic redistribution of residue B-factors in sTeLIC, suggesting a dynamical basis for allosteric modulation via this site (22). In DeCLIC, the vestibular cavity was relatively compressed in the Ca2+-bound closed state, but expanded in the Ca2+-free open state, sufficient to accommodate a molecule of similar volume as ELIC/sTeLIC modulators (SI Appendix, Fig. S11 C and D).
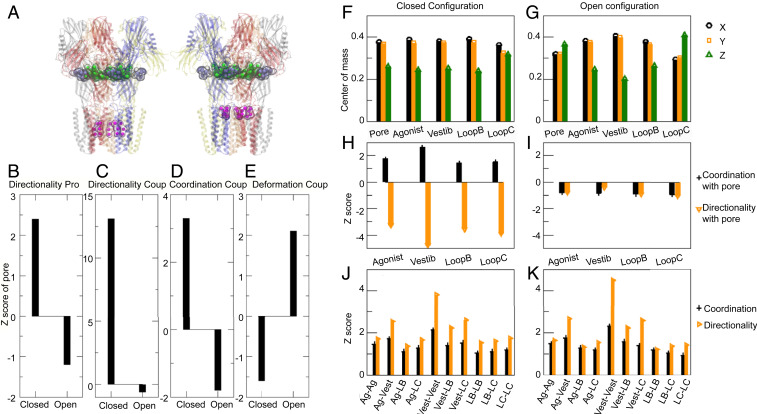
Given the evident relevance of both TMD and LBD sites to allosteric gating transitions in this family, we qualitatively assessed the coupling between DeCLIC domain motions using normal mode analysis. A new method by Bastolla and colleagues (30) has been developed specifically to characterize coupling of ligand-binding sites based on similarities in the direction of motions (codirectionality), fluctuations in interatomic distances (coordination), and how much perturbations in one site modify the structure of another (deformation). Torsional normal mode analysis predicted atomic fluctuations that correlated well with B-factors in both DeCLIC structures (correlation factors of 0.94 and 0.88 for the apparent closed and open states, respectively) (SI Appendix, Fig. S16A), and a limited set of normal modes were sufficient to describe 80% of the torsional component of both apparent gating transitions (18 and 59 normal modes for closed and open pore states, respectively) (SI Appendix, Fig. S16B). Here we examine couplings between the orthosteric site, the vestibular site, and the pore (Fig. 7A).
Fig. 7.
Dynamic coupling between allosteric sites using normal mode analysis averaged over the five chains. (A) Representation of the main functional sites: Pore (magenta), agonist (blue-gray), and vestibular sites (green). Two different definitions of the pore site are represented that give very similar results. (B) The z-score of the directionality profile between atoms of the pore and all other atoms in the protein. (C–E) The z-score of the coupling among atoms of the pore; directionality (C), coordination (D), and deformation (E). Directionality and coordination are larger than expected in the closed conformation and smaller in the open one, while the contrary happens for deformation. (F and G) Squared projections of the movement of the center-of-mass of the functional sites along the three principal axes x (black), y (yellow), and z (green), where the z axis denotes the direction of the pore. The z axis component is the smallest one in the closed conformation (F) but, it is the largest one in the open conformation for the pore (G). (H and I) The z-score of the directionality (yellow) and coordination (black) coupling between the pore and four functional sites: the coordination is positive (higher than expected) in the closed conformation, in particular for the vestibular site, however the directionality is negative (i.e., smaller than expected) (H). In the open conformation the coordination becomes smaller than expected but the directionality is less negative (I). (J and K) The z-score of the directionality and coordination coupling among functional sites. The vestibular site shows the largest couplings among all sites with the agonist site and with itself in both conformations, open and closed.
In the modeled closed-to-open transition associated with Ca2+ depletion, the DeCLIC pore experienced a decrease in their directionality coupling and coordination coupling, and an increase in their deformation coupling (Fig. 7 B–E), which indicates that their dynamical interactions were destabilized in the open form (Fig. 7). Consistently, coordination with the pore was reduced for both orthosteric and vestibular sites. However, the codirectionality and, to a smaller extent, the covariance became less negative. The vestibular site exhibited the largest couplings with other functional sites in both closed and open conformations, and displayed the largest coordination and the most negative codirectionality with the pore in the closed conformation. In the closed-pore conformation, the motions of the functional sites are prevalently directed along the membrane, with smallest component along the pore direction Z (Fig. 7F). In contrast, in the open conformation the motions of the pore and the loop C are prevalently directed along the pore (Fig. 7G) A possible interpretation is that the dynamics of the open-pore atoms accompany ions through the channel, at the price of destabilizing the open-state pore relative to the closed-state one.
Vestibular Constriction and Helix Reorientation in Ca2+-Free DeCLIC.
The lumen of the open-pore LBD was virtually closed by a constriction ring formed by the five side chains of Trp407, projecting toward the C5 axis from loop Ω (SI Appendix, Fig. S13). This kind of constriction ring is also present in sTeLIC (22) and observed recently in the structure of the heteromeric GABAA receptor, where the role of the Trp407 side chain is played by the glycosylation of one of the subunits (15, 31). This constriction is also present in GluCl (4NTV), where five tyrosine residues (Y99) coordinate a central citrate anion, as well as in 5-HT3A-R (4PIR), where five lysine residues (K108) coordinate a central sulfate ion. Strikingly, in the closed-pore conformation of DeCLIC, this constriction ring was absent due to an outward movement of loop Ω (Figs. 4B and 5C).
The position of the amphipathic helix α1 at the top of the LBD also differed between open and closed states. In the open state, α1 rotated and translated inward toward the C5 axis. As a consequence, its hydrophobic side detached from the pre-β4 loop and established new contacts with the β3 strand. This reoriented α1 helix position in the Ca2+-free, apparent open state was similar to that observed in both sTeLIC and ELIC (SI Appendix, Fig. S14 A and B). Indeed, the LBD of open-pore DeCLIC aligned closely with those of both sTeLIC (with a similarly wide-open pore) and ELIC, even though the ELIC pore is closed (SI Appendix, Fig. S14 B, D, and F). Comparison to DeCLIC structures in multiple states therefore indicated that current X-ray structures of ELIC may represent a “locally closed” or preactive conformation, with a closed pore but an activated LBD. Consistent with this hypothesis, previous structures of ELIC have shown that its orthosteric site can accommodate agonists, including cysteamine or GABA, without opening the pore in the crystal (27, 32), an observation that has never been clearly explained.
Discussion
Structures of DeCLIC, described here in the presence and absence of Ca2+, provide atomistic details of the molecular architecture and accessible states of a prokaryotic pLGIC incorporating a periplasmic NTD. Despite the presence of this additional domain, the DeCLIC LBD and TMD can readily be compared with other bacterial pLGICs. The first full-length pLGIC characterized by X-ray diffraction, ELIC from the plant pathogen Dickeya dadantii, has been crystallized under numerous conditions but always in a nonconducting state (8). Its pore aligns closely with that of Ca2+-bound DeCLIC, including hydrophobic constrictions at the 16′ and 9′ positions of the pore-lining M2 helix (SI Appendix, Fig. S14F). GLIC from the cyanobacterium Gloeobacter violaceus has been crystallized in apparent open (7, 9), intermediate (33, 34), modulated (35, 36), and resting states (37). The GLIC closed pore is qualitatively similar to nonconducting DeCLIC and ELIC, although somewhat less constricted (SI Appendix, Fig. S14); the open pore of GLIC aligns well with some eukaryotic homologs (11, 12, 16), although both are narrower than the Ca2+-free pore of DeCLIC. Most recently, sTeLIC from a prokaryotic symbiont of the giant tube worm Tevnia jerichonana crystallized under activating conditions with a wide-open pore, similar to that of DeCLIC in the absence of Ca2+ (SI Appendix, Fig. S14). Although we cannot rule out artifactual influence of detergents, crystallographic packing, or other sample conditions, these comparisons were consistent with the nondesensitizing, blocker-insensitive ion conduction of DeCLIC upon Ca2+ depletion in oocytes (Fig. 5). Accordingly, we interpreted our high-Ca2+ DeCLIC structure to represent a closed state, and the Ca2+-free structure a possible open state.
A capacity for ions to enter the DeCLIC pore was evident in the Ca2+-bound structure, which contained two ion sites below the constriction rings, consistent with cation-selective pLGICs. In particular, a planar hydrated ion around the 6′ position was observed in both DeCLIC (Fig. 2D) and the human α4β2 nicotinic receptor (14). The open state of GLIC contains two water pentagons in the same region, albeit with alternative geometry (38). At the cytoplasmic mouth of the pore, ELIC, GLIC, and eukaryotic cationic pLGICs feature a single ring of acidic residues that contribute to selectivity (39); like sTeLIC, DeCLIC exhibited two such rings at the –4′ and 2′ positions, with a hydrophilic constriction at 2′ in the closed state. A dehydrated cation resolved at this cytoplasmic gate in DeCLIC (Fig. 4E) was also observed in recent nAChR structures (40). Ions were not well resolved in the wide-open pore of Ca2+-free DeCLIC, possibly reflecting disruption of coordinated and codirectional motion in this region indicated by normal mode analysis (SI Appendix, Fig. S16C). Insensitivity to quaternary ammonium block has been observed in ELIC and sTeLIC (22, 28, 41) as well as DeCLIC (Fig. 3 A and B), supporting a conservation of conducting states in this subset of prokaryotic channels.
Along with DeCLIC, both ELIC and sTeLIC are also inhibited by external Ca2+ ions (22, 42), indicating some mechanisms of divalent cation modulation could be conserved in this channel family. Structural and functional properties in the presence of Ca2+ indicate DeCLIC should be closed under standard physiological conditions: The periplasmic space is expected to accumulate Ca2+ in excess of 10 mM (43), well above the recorded micromolar IC50 (Fig. 3D). However, local Ca2+ concentrations can be sensitive to precipitation or chelation by anions or other environmental agents, such as sulfate compounds produced by the purple sulfur-bacteria symbiont of Desulfofustis (23), and could transiently drop below the threshold for DeCLIC activation. Notably, several Ca2+-mediated contacts in DeCLIC have previously been implicated in pLGIC gating: An equivalent electrostatic network between the F, β1–β2, and Pro-loops is evident in both ELIC and open GLIC (42, 44). The closest Ca2+ contact in DeCLIC was with E347 in the β1–β2 loop; in GLIC, protonation of the equivalent residue (E35) is thought to bypass classical agonist binding to open the pore (44). ELIC also contains a Ba2+ binding site, involving the F and Pro-loops, adjacent to DeCLIC’s Ca2+ binding site (42).
Remodeling of an extended electrostatic network near the DeCLIC Ca2+ site suggested a pathway for channel gating. Once released from its Ca2+-mediated interactions, loop F in DeCLIC shifted away from the interface toward the subunit interior, replacing intersubunit salt bridges with an intrasubunit hydrogen bond (Q344/E481) and liberating arginines in the β1–β2 and M2–M3 loops to orient toward the expanded TMD subunit interface (R345, R569) (SI Appendix, Fig. S8). This network is at least partially conserved in sTeLIC, where the wide-open structure contains a homologous intrasubunit hydrogen bond (Q25/E161) and orients basic residues in the β1–β2 and M2–M3 loops toward the transmembrane interface (K26, R249). In DeCLIC, this reorganization of basic residues may facilitate the intercalation of lipid head groups, which were resolved in the expanded crevices between subunits in the Ca2+-free state (SI Appendix, Fig. S15C). Intersubunit TMD cavities in pLGICs have been shown to bind various lipophilic potentiators, including alcohol and anesthetics in GLIC (36, 45), and ivermectin in eukaryotic anion channels (11, 12, 46). Lipids themselves act as allosteric effectors of many pLGICs (47), and could contribute to stabilizing DeCLIC in a wide-open state.
Parallel to the inward displacement of loop F, both the LBD and NTD exhibited overall inward contraction upon Ca2+ depletion, manifesting in a hydrophobic occlusion of the periplasmic vestibule by a ring of tryptophans (W407) at the level of loop Ω (Fig. 4 A and B and SI Appendix, Fig. S13). A ring of arginines (R86) similarly occludes the LBD pathway in sTeLIC (22); known structures of ELIC do not feature this constriction, although a ring of Ba2+ ions associate with backbone carbonyls of loop Ω at an equivalent position (42). Among eukaryotic pLGICs, both cation- and anion-selective channels have been crystallized with coordinated anions in this position on the fivefold symmetry axis, also closing the vestibule (10, 48), and recent structures of heteromeric GABAARs show the extracellular domain to be blocked also in this region by large glycans (15, 31, 49). Although its biological relevance is unknown, the occluded LBD vestibule could serve as a reservoir for permeant ions, shielded from the periplasmic or extracellular environments. Ion entry and exit may be regulated by transient opening of the upper occlusion or via routes peripheral to the linear channel pathway. Vestibular constriction could also contribute to an alternating two-gate system analogous to a peristaltic pump, consistent with normal mode analysis of wide-open structures of both sTeLIC (22) and now DeCLIC (SI Appendix, Fig. S16C).
Above the loop Ω occlusion, the linear DeCLIC channel was partially constricted in the Ca2+-free state by inward displacement of the α1 helix, a motif absent or shorter in GLIC and eukaryotic pLGICs, but preserved in ELIC and sTeLIC. Interestingly, a cavity between α1 and the A and E loops forms a vestibular binding site for the allosteric modulator flurazepam in ELIC, or for 4-Bromo cinnamic acid in sTeLIC (22, 27). In the equivalent site of DeCLIC, a molecule of this volume would only be accommodated in the Ca2+-free state (SI Appendix, Fig. S11 C and D). Although several known agonists and modulators failed to produce apparent DeCLIC activation or modulation in oocytes, the α1 (vestibular) cavity could accommodate physiological modulators yet to be identified, or otherwise mediate a conserved mechanism of regulation. Altogether, the remodeling of this cavity between the two forms of DeCLIC rationalizes a posteriori the observation that this vestibular binding site accommodates, in an open-state specific manner, molecules that are modulators in both ELIC and sTeLIC (Table 1 and SI Appendix, Fig. S17).
Table 1.
Summary of the known allosteric binding sites in the LBD of bacterial pLGICs (GLIC, ELIC, sTeLIC, and DeCLIC) with their ligands and PDB ID
| Allosteric site | Loop F | ||||
| pLGIC, State | Pore, PDB | Suborthosteric | Vestibular | Interfacial site | Deep site |
| GLIC | Open | Br-Acetate: 4QH1 | Br-Acetate: 4QH1 | Water network: 6HZW; H+ (E35, T158) | Xenon: 4ZZC |
| Open | 4HFI | Succinate: 6HJZ | Succinate: 6HJZ | ||
| GLIC | Closed | Empty | Empty | Empty | Bromoform: |
| Closed | 4NPQ | MD simulations | |||
| sTeLIC | Open | Empty | 4-Br-cinnamate: 6FLI | Empty | Closed |
| Open | 6FL9 | ||||
| ELIC | Closed | Zopiclone: 4A97 | Flurazepam: 2YOE | Ba2+: 2YN6 | Bromoform: 3ZKR |
| Semiclosed | 2VL0 | Br-Flurazepam: 4A98 | Chlorpromazine: 5LG3 | ||
| Acetylcholine: 3RQW | |||||
| Bromoethanol: 5SXV | |||||
| DeCLIC | Open | Empty | Empty | Empty | Empty |
| Open | 6V4A | ||||
| DeCLIC | Closed | Remodeled/larger | Remodeled/smaller | Ca2+ | Closed |
| Closed | 6V4S | ||||
The periplasmic DeCLIC NTD, a novel feature among reported pLGIC structures, also interacted with functionally relevant regions of the consensus channel in a state-dependent manner. In particular, Ca2+ depletion displaced the NTD2 lobe in each subunit toward loop C, which was itself remodeled toward the complementary subunit interface including NTD1 (Fig. 5 B and C). If activation at the level of the LBD Ca2+ site initiates a conformational wave leading to both pore expansion and NTD remodeling, changes in the NTD could in turn regulate channel kinetics or periplasmic interactions. Alternatively, the NTD could act as a macroscopic agonist, in which reorganization of the outer lobe itself influences the orthosteric site or other elements of the pLGIC gating pathway. Elevated average B-factors of NTD residues in both states (SI Appendix, Fig. S5) suggested this domain was heterogeneous under crystallographic conditions, and could require a binding partner for stabilization. Neither NTD lobe contained any obvious cavity suitable for ligands; however, the overall channel dimensions could enable this domain to interface with the periplasmic peptidoglycan layer, whose effects on channel function have yet to be characterized. Interestingly, two different sugar-binding proteins with jelly-roll topology were retrieved by the Dali search using NTD2 as query (SI Appendix, Fig. S18). It is also suggestive that DeCLIC molecules crystallized in parallel linear arrays, mediated by contacts between their NTDs, and distinct from the packing patterns of most other crystallized pLGICs (SI Appendix, Fig. S4). This phenomenon is reminiscent of 2D lattices formed by GLIC in recent atomic force microscopy experiments (50); in biology, such lattices have been associated with chemotaxis receptors (51). Whether this linear organization of DeCLIC might be reproduced in the cell membrane, or extended to a 2D lattice, remains to be determined.
Combined with previous structures of ELIC and sTeLIC, characterization of DeCLIC in two conformations supports a coherent gating landscape for prokaryotic pLGICs (SI Appendix, Fig. S5). In particular, DeCLIC comparisons offer insight into the annotation of ELIC structures, whose functional states have been controversial. Whereas the nonconducting pore of ELIC is largely superimposable with that of Ca2+-bound DeCLIC, its LBD—including the distinctive α1 helix—is superimposable with that of pore-open DeCLIC or sTeLIC (SI Appendix, Fig. S14 B and D). This correspondence supports the hypothesis that ELIC structures represent a locally closed or preopen state, in which the LBD is activated but the pore is locked in a resting conformation, similar to what has been seen in GLIC (33, 44). Consistent with this hypothesis, X-ray structures of ELIC have shown that both the cysteamine and GABA agonists could bind the agonist binding site of this crystal form without opening the pore (27, 32), as well as positive allosteric modulators in the vestibular cavity (Table 1 and SI Appendix, Fig. S17). Identification in Ca2+-free DeCLIC of a wide-open pore and other features originally observed in sTeLIC also supports the relevance of this structure as a representative open state. Structure-function characterization of DeCLIC thus further illuminates the mechanistic repertoire of prokaryotic pLGICs, enabling annotation of both known and novel structures, and unveiling the role and diversity of accessory domain interactions.
Materials and Methods
Sample Preparation.
Full-length DeCLIC was expressed and purified according to protocols previously optimized for prokaryotic pLGICs (22). Briefly, a gene encoding the predicted processed protein (residues 33 to 642) was codon-optimized and synthesized as a thrombin-cleavable fusion construct with maltose binding protein, and subcloned into vector pET20b. After overnight expression in C43 E. coli, the protein was harvested with the membrane fraction by ultracentrifugation, solubilized in 4% DDM, purified by amylose affinity and size-exclusion chromatography, isolated from its fusion partner by thrombin cleavage and size exclusion, and concentrated to 10 mg/mL, all at 4 °C. The truncated NTD1 sequence (encoding residues 33 to 202) was amplified from the full-length construct and subcloned into vector pRSFDuet as a tobacco etch virus (TEV) protease-cleavable fusion construct with a 14-histidine tag. After overnight expression in BL21 (DE3) Rosetta2 E. coli, the protein was purified by Ni-NTA affinity, isolated from its fusion partner by TEV cleavage and size exclusion, and concentrated to 15 mg/mL, all at 4 °C. SeMet-incorporated NTD1 was expressed in selective media and purified using a similar protocol, with 1 mM DTT added during purification. All proteins were frozen using liquid nitrogen and stored at −80 °C.
Crystallization and Data Collection.
Crystals of DeCLIC were initially grown at 18 °C by 1:1 hanging-drop vapor diffusion and microseeding in a reservoir containing 100 mM Tris pH 7.5, 250 mM CaCl2, 14.5% (wt/vol) PEG-MME 2000 (final pH ∼6.7), and frozen after 1 wk with 25% (vol/vol) ethylene glycol. For the Ba2+-bound dataset, these crystals were soaked in reservoir solution supplemented with 200 mM BaCl2 for 5 to 10 min prior to cryoprotection. For the Ca2+-free dataset, DeCLIC crystals were grown by the same method with 0.3 M NH4-Formate, 0.1 M Tris 7.5, 30% (vol/vol) PEG-MME 500 (final pH ∼7.6), and directly frozen after 3 wk. Crystals of the truncated NTD1, with or without SeMet incorporation, were crystallized similarly with 0.1 M Hepes pH 6.5, 0.7 M NaH2PO4, 0.7 M KH2PO4, and frozen with 30% (vol/vol) glycerol. Diffraction data were collected at Soleil PROXIMA-1 and processed using XDS (52) and CCP4 (53) (SI Appendix, Table S1). Crystals of full-length DeCLIC in the presence or absence of Ca2+ grew in space group P21212 or P21, respectively, with one pentamer in the asymmetric unit. The truncated NTD1 crystallized in space group P61 with two monomers in the asymmetric unit.
Structure Determination and Model Refinement.
Initial phases for the truncated NTD1 were obtained using Phenix (54) from a single anomalous diffraction dataset collected at 0.9791 Å. An initial model was generated using AutoBuild in Phenix, and multiple rounds of model building in Coot (55) and refinement with BUSTER (56) were performed to obtain the final refined model containing residues 33 to 195. For Ca2+-bound DeCLIC, phases were obtained by molecular replacement using a chimeric model containing the extracellular domain of GLIC (PDB ID code 4HFI) (38) and the TMD of ELIC (PDB ID code 2VL0) (57) as a Phaser search model in Phenix (58). The resulting model was refined using Refmac (53) and BUSTER-Refine (56) with alternating cycles of refinement and manual building in Coot (55), with automatic noncrystallographic symmetry restraints applied. Electron density in the NTD became visible after building most of the consensus channel, and was modeled manually with reference to the truncated structure (for NTD1) and backbone trace of PDB ID code 3JQW (for NTD2). For Ca2+-free DeCLIC, a similar approach was taken with sTeLIC (PDB ID code 6FL9) as the search model. Assignments in the vicinity of M3 were further verified by collection of a mercury-edge anomalous dataset from crystals grown with ethylmercury chloride, which was found to label only C596 of six cysteines in each DeCLIC monomer.
Electrophysiology.
Two-electrode voltage-clamp electrophysiology was performed as previously described (59). Briefly, cDNA encoding DeCLIC wild-type or ΔNTD variants was commercially synthesized (Thermo Fisher) and subcloned into the pUNIV oocyte vector (60). DeCLIC and yellow fluorescent protein (YFP) constructs were translated in vitro using T7 RNA polymerase (Thermo Fisher). Commercially isolated X. laevis oocytes were injected with 10 to 30 ng DeCLIC RNA and incubated at 12 °C for 1 to 3 wk, or with 0.5 ng YFP RNA as an injection control. For each recording, control, YFP-, or DeCLIC-injected oocytes were clamped on either side of a parallel recording rig and perfused in 0.5 to 1.0 mL/min running buffer (10 mM Tris pH 8.5, 123 mM NaCl, 10 mM Na+-citrate, 2 mM KCl, 2 mM MgSO4) with varying concentrations of CaCl2. Currents were recorded with an Axon CNS 1440A Digidata system in pCLAMP 10. For Ca2+ sensitivity measurements, peak currents at each concentration were normalized to the maximal response of paired oocytes, and fit to a sigmoidal concentration-response curve using Prism8 (GraphPad).
Normal Mode Analysis.
Dynamical couplings between atoms in four functional sites were computed as previously described (30), adopting the normal modes of the torsional network model (61). Briefly, 1) directionality coupling was defined as the Boltzmann average of the scalar product between the directions of motion of two atoms; 2) coordination coupling was a constant minus the root mean square of the fluctuation of the distance between the two atoms; 3) deformation coupling was defined as the deformation in one atom produced by a perturbation applied to the other with constant amplitude and direction, such that the perturbation is maximized. A z-score was calculated from 1,000 randomly generated pairs of residues having the same distance along the sequence as the considered pair. Full methods are provided in SI Appendix, SI Materials and Methods.
Data Availability.
All crystallographic data have been deposited to the PDB (diffraction data and refined coordinates of the models), under the following ID codes: 6V4S for closed pore conformation of DeCLIC, 6V4A for open pore conformation of DeCLIC, and 6V4B for NTD1 of DeCLIC (residues 34 to 202). All other data, associated protocols, code, and materials in the paper will be available from the authors upon request.
Supplementary Material
Acknowledgments
We thank Pr. J. P. Changeux for initially suggesting that we investigate the architecture of bacterial pLGIC with additional domains, and for discussion and support during the course of this work; the staff of the Synchrotron Soleil (St Aubin, France), especially Dr. P. Legrand, and of the European Synchrotron Radiation Facility (Grenoble, France) for help in data collection; and Sirine Hlioui for help in protein preparation and crystallogenesis in the final stages of this study. R.J.H. and E.L. thank the Swedish Research Council and The Knut and Alice Wallenberg Foundation for support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6V4S, 6V4A, and 6V4B).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922701117/-/DCSupplemental.
References
- 1.Corringer P.-J. et al., Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure 20, 941–956 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Plested A. J. R., Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat. Struct. Mol. Biol. 23, 494–502 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Sauguet L., Shahsavar A., Delarue M., Crystallographic studies of pharmacological sites in pentameric ligand-gated ion channels. Biochim. Biophys. Acta 1850, 511–523 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Changeux J. P., Kasai M., Lee C. Y., Use of a snake venom toxin to characterize the cholinergic receptor protein. Proc. Natl. Acad. Sci. U.S.A. 67, 1241–1247 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasneem A., Iyer L. M., Jakobsson E., Aravind L., Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6, R4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaiteh M., Taly A., Hénin J., Evolution of pentameric ligand-gated ion channels: Pro-loop receptors. PLoS One 11, e0151934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocquet N. et al., X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Hilf R. J. C., Dutzler R., X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hilf R. J. C., Dutzler R., Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hassaine G. et al., X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Hibbs R. E., Gouaux E., Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J., Lü W., Wu S., Cheng Y., Gouaux E., Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller P. S., Aricescu A. R., Crystal structure of a human GABAA receptor. Nature 512, 270–275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales-Perez C. L., Noviello C. M., Hibbs R. E., X-ray structure of the human α4β2 nicotinic receptor. Nature 538, 411–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S. et al., Structure of a human synaptic GABAA receptor. Nature 559, 67–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polovinkin L. et al., Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basak S., Gicheru Y., Rao S., Sansom M. S. P., Chakrapani S., Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritschy J.-M., Harvey R. J., Schwarz G., Gephyrin: Where do we stand, where do we go? Trends Neurosci. 31, 257–264 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Neff R. A. 3rd, Gomez-Varela D., Fernandes C. C., Berg D. K., Postsynaptic scaffolds for nicotinic receptors on neurons. Acta Pharmacol. Sin. 30, 694–701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara P. J. et al., The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron 11, 41–52 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Zhu S., Paoletti P., Allosteric modulators of NMDA receptors: Multiple sites and mechanisms. Curr. Opin. Pharmacol. 20, 14–23 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hu H. et al., Crystal structures of a pentameric ion channel gated by alkaline pH show a widely open pore and identify a cavity for modulation. Proc. Natl. Acad. Sci. U.S.A. 115, E3959–E3968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilbanks E. G. et al., Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh. Environ. Microbiol. 16, 3398–3415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krupovič M., Bamford D. H., Virus evolution: How far does the double β-barrel viral lineage extend? Nat. Rev. Microbiol. 6, 941–948 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Ashkenazy H. et al., ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spurny R. et al., Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. U.S.A. 109, E3028–E3034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Gutierrez G., Grosman C., The atypical cation-conduction and gating properties of ELIC underscore the marked functional versatility of the pentameric ligand-gated ion-channel fold. J. Gen. Physiol. 146, 15–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X., Chen H., Shaffer P. L., Crystal structures of human GlyRα3 bound to ivermectin. Structure 25, 945–950.e2 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Alfayate A., Caceres C. R., Gomes Dos Santos H., Bastolla U., Predicted dynamical couplings of protein residues characterize catalysis, transport and allostery. Bioinformatics 35, 4971–4978 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Phulera S. et al., Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife 7, e39383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Gutierrez G. et al., Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl. Acad. Sci. U.S.A. 109, 6331–6336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prevost M. S. et al., A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat. Struct. Mol. Biol. 19, 642–649 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Gutierrez G., Cuello L. G., Nair S. K., Grosman C., Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 110, 18716–18721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basak S., Schmandt N., Gicheru Y., Chakrapani S., Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel. eLife 6, e23886 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fourati Z. et al., Structural basis for a bimodal allosteric mechanism of general anesthetic modulation in pentameric ligand-gated ion channels. Cell Rep. 23, 993–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Sauguet L. et al., Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. U.S.A. 111, 966–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauguet L. et al., Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 32, 728–741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sine S. M., Wang H.-L., Hansen S., Taylor P., On the origin of ion selectivity in the Cys-loop receptor family. J. Mol. Neurosci. 40, 70–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gharpure A. et al., Agonist selectivity and ion permeation in the α3β4 ganglionic nicotinic receptor. Neuron 104, 501–511.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Gutierrez G., Wang Y., Cymes G. D., Tajkhorshid E., Grosman C., Chasing the open-state structure of pentameric ligand-gated ion channels. J. Gen. Physiol. 149, 1119–1138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann I., Marabelli A., Bertozzi C., Sivilotti L. G., Dutzler R., Inhibition of the prokaryotic pentameric ligand-gated ion channel ELIC by divalent cations. PLoS Biol. 10, e1001429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones H. E., Holland I. B., Campbell A. K., Direct measurement of free Ca(2+) shows different regulation of Ca(2+) between the periplasm and the cytosol of Escherichia coli. Cell Calcium 32, 183–192 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Hu H. et al., Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel. Proc. Natl. Acad. Sci. U.S.A. 115, E12172–E12181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauguet L. et al., Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat. Commun. 4, 1697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X., Chen H., Michelsen K., Schneider S., Shaffer P. L., Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature 526, 277–280 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Baenziger J. E., Corringer P.-J., 3D structure and allosteric modulation of the transmembrane domain of pentameric ligand-gated ion channels. Neuropharmacology 60, 116–125 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Althoff T., Hibbs R. E., Banerjee S., Gouaux E., X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laverty D. et al., Cryo-EM structure of the human α1β3γ2 GABAA receptor in a lipid bilayer. Nature 565, 516–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruan Y. et al., Structural titration of receptor ion channel GLIC gating by HS-AFM. Proc. Natl. Acad. Sci. U.S.A. 115, 10333–10338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briegel A. et al., Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife 3, e02151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winn M. D. et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanc E. et al., Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Bocquet N. et al., A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 (2007). [DOI] [PubMed] [Google Scholar]
- 58.McCoy A. J. et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heusser S. A. et al., Functional characterization of neurotransmitter activation and modulation in a nematode model ligand-gated ion channel. J. Neurochem. 138, 243–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkatachalan S. P. et al., Optimized expression vector for ion channel studies in Xenopus oocytes and mammalian cells using alfalfa mosaic virus. Pflugers Arch. 454, 155–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mendez R., Bastolla U., Torsional network model: Normal modes in torsion angle space better correlate with conformation changes in proteins. Phys. Rev. Lett. 104, 228103 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All crystallographic data have been deposited to the PDB (diffraction data and refined coordinates of the models), under the following ID codes: 6V4S for closed pore conformation of DeCLIC, 6V4A for open pore conformation of DeCLIC, and 6V4B for NTD1 of DeCLIC (residues 34 to 202). All other data, associated protocols, code, and materials in the paper will be available from the authors upon request.