Significance
The balance of proinflammatory and anti-inflammatory cytokines is important for tissue homeostasis. It is well known that excessive production of proinflammatory cytokines promotes tumor development. The human retrovirus human T cell leukemia virus type 1 (HTLV-1) causes both malignant and inflammatory diseases. This study reveals that HTLV-1 changes the immunophenotype of infected cells into that of regulatory T cells (Tregs) and uses the anti-inflammatory cytokine IL-10, but not proinflammatory IL-6, to accelerate the proliferation of infected cells. A viral factor, HTLV-1 bZIP factor, plays a central role in this dysregulation of the cytokine signaling, and consequently triggers oncogenesis of aberrantly differentiated T cells. This is a unique strategy of HTLV-1 to establish persistent infection by hijacking the machinery of Treg differentiation.
Keywords: HTLV-1, HBZ, IL-10, IL-6, JAK/STAT signaling pathway
Abstract
Human T cell leukemia virus type 1 (HTLV-1) is the etiologic agent of a T cell neoplasm and several inflammatory diseases. A viral gene, HTLV-1 bZIP factor (HBZ), induces pathogenic Foxp3-expressing T cells and triggers systemic inflammation and T cell lymphoma in transgenic mice, indicating its significance in HTLV-1–associated diseases. Here we show that, unexpectedly, a proinflammatory cytokine, IL-6, counteracts HBZ-mediated pathogenesis. Loss of IL-6 accelerates inflammation and lymphomagenesis in HBZ transgenic mice. IL-6 innately inhibits regulatory T cell differentiation, suggesting that IL-6 functions as a suppressor against HBZ-associated complications. HBZ up-regulates expression of the immunosuppressive cytokine IL-10. IL-10 promotes T cell proliferation only in the presence of HBZ. As a mechanism of growth promotion by IL-10, HBZ interacts with STAT1 and STAT3 and modulates the IL-10/JAK/STAT signaling pathway. These findings suggest that HTLV-1 promotes the proliferation of infected T cells by hijacking the machinery of regulatory T cell differentiation. IL-10 induced by HBZ likely suppresses the host immune response and concurrently promotes the proliferation of HTLV-1 infected T cells.
Chronic inflammation is known to increase the risk of oncogenesis in some organs (1), and “tumor-promoting inflammation” is recognized as a hallmark of cancer (2). Inflammation innately acts to fight infections and heal wounds; however, excessive and/or prolonged inflammation contributes to the proliferation and survival of malignant cells, induction of genetic instability, angiogenesis, metastasis, and escape from antitumor immunity (3). Proinflammatory cytokines, such as tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β, act as inducers of inflammation and oncogenesis (4). Since IL-6 exerts pleiotropic effects on many types of cells, overproduction of IL-6 triggers a variety of diseases, including cancers and autoimmune diseases (5). Moreover, hyperactivation of the IL-6/JAK/STAT3 pathway is generally associated with a poor prognosis for many types of cancers (6). Thus, blockade of this pathway is considered a promising therapeutic strategy against such diseases (7). In contrast, immunosuppressive cytokines, such as transforming growth factor (TGF)-β and IL-10, have anti-inflammatory functions and suppress the production of proinflammatory cytokines (8, 9). However, immunosuppressive cytokines can also suppress immunity against cancer or against oncogenic pathogens, allowing cancer progression. It is suggested that the effects of the proinflammatory and anti-inflammatory cytokines on cancer development depend on the initial cause of the tumor and the composition of the tumor microenvironment.
Persistent infections are important causes of chronic inflammation leading to malignant diseases; up to 25% of human cancers are thought to be associated with infectious agents (10, 11). Human T cell leukemia virus type 1 (HTLV-1) is a human retrovirus that causes an aggressive malignant disease of CD4+ T cells termed adult T cell leukemia-lymphoma (ATL), along with several chronic inflammatory diseases, including HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP) and uveitis (12, 13). HTLV-1 propagates by cell-to-cell transmission of the virus (de novo infection) and proliferation of infected cells (clonal proliferation) in vivo, and establishes persistent infection (14). Receptors of HTLV-1 include glucose transporter 1, neuropilin 1, and heparan sulfate proteoglycans. Therefore, HTLV-1 can infect many kinds of hematopoietic cells in vitro (15). However, a major immunophenotype of HTLV-1–infected cells, including ATL cells, is CD4+CD25+CCR4+CADM1+ T cells, suggesting that HTLV-1 converts infected CD4+ T cells to this phenotype and induces their clonal expansion.
Among the viral genes encoded in the HTLV-1 provirus, HTLV-1 bZIP factor (HBZ) is thought to be crucial for pathogenesis, since HBZ is constitutively expressed in all HTLV-1–infected subjects, including ATL cases; HBZ accelerates T-cell proliferation (16); and HBZ transgenic (HBZ-Tg) mice demonstrate a similar phenotype to that of HTLV-1–infected individuals: increases in CD4+CD25+CCR4+ cells and development of systemic inflammation and T cell lymphoma (17, 18). In addition, there is a positive correlation between the severity of inflammation and the incidence of T cell lymphoma in this mouse model (19). Thus, the HBZ-Tg mouse is a good animal model for elucidating the molecular mechanisms of HTLV-1–mediated pathogenesis, particularly the impact of inflammatory factors on pathogenesis.
In this study, we show that a proinflammatory cytokine, IL-6, unexpectedly has a suppressive role in the inflammation and lymphomagenesis caused by HBZ. An important mechanism for this pathogenesis is HBZ-induced aberrant differentiation of CD4+ T cells into regulatory T cell (Treg)-like cells. The innate function of IL-6 to block TGF-β–mediated Treg differentiation counteracts HBZ. On the other hand, an immunosuppressive cytokine, IL-10, is up-regulated in HBZ-expressing T cells, and HBZ modulates the IL-10/STAT signaling pathway through interaction with STAT proteins. Our results reveal a unique strategy of HTLV-1 to increase the number of infected cells by hijacking Treg differentiation to promote cell proliferation and evade host immune surveillance.
Results
Loss of IL-6 Unexpectedly Accelerates Inflammation and Lymphomagenesis in HBZ-Tg Mice.
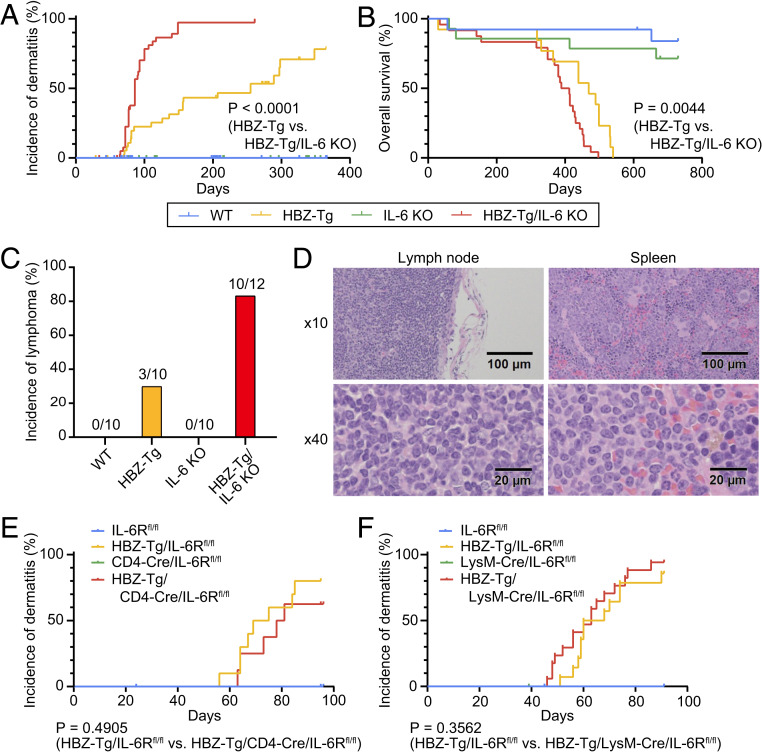
To evaluate the correlation between HBZ-mediated pathogenesis and inflammatory factors, we focused on two proinflammatory cytokines, IFN-γ and IL-6, which are well known for their association with cancer. Indeed, we previously reported that knockout of IFN-γ significantly suppressed the incidence of both dermatitis and T cell lymphomas in HBZ-Tg mice (19). We hypothesized that IL-6 would also contribute to the development of these diseases in HBZ-Tg mice. To assess this possibility, we generated HBZ-Tg/IL-6 KO mice by crossing HBZ-Tg mice with IL-6 KO mice (20) and compared their phenotypes with those of HBZ-Tg mice. As we reported previously (17), ∼45% of HBZ-Tg mice developed dermatitis by age 24 wk (Fig. 1A and Table 1), and histopathological analysis revealed that 30% of them had lymphomas at that age (Fig. 1C and Table 1). Surprisingly, HBZ-Tg/IL-6 KO mice developed dermatitis significantly earlier than HBZ-Tg mice (median, 86 d vs. 255 d; hazard ratio [HR], 0.1726; 95% confidence interval [CI], 0.09454 to 0.3152) (Fig. 1A). In 2 y of observation, the overall survival of HBZ-Tg/IL-6 KO mice was shorter than that of HBZ-Tg mice (median, 400.5 d vs. 469 d; HR, 0.3438; 95% CI, 0.1649 to 0.7169) (Fig. 1B). Moreover, 10 of 12 (83%) HBZ-Tg/IL-6 KO mice developed lymphomas at age 24 wk (Fig. 1 C and D and Table 1). Neither dermatitis nor lymphoma was observed in WT and IL-6 KO mice, and there was no significant difference in overall survival between the parental strains. These results indicate that loss of IL-6 accelerated the development of the inflammation and lymphomagenesis caused by HBZ in vivo.
Fig. 1.
Loss of IL-6 accelerates inflammation and lymphomagenesis in HBZ-Tg mice. (A) Incidence of dermatitis in WT (blue; n = 65), HBZ-Tg (yellow; n = 39), IL-6 KO (green; n = 72), and HBZ-Tg/IL-6 KO (red; n = 42) mice. These mice were observed for 1 y (log-rank test). (B) Overall survival of each strain: WT, n = 13; HBZ-Tg, n = 13; IL-6 KO, n = 14; HBZ-Tg/IL-6 KO, n = 24. These mice were observed for 2 y (log-rank test). (C) Incidence of lymphoma in each strain at age 24 wk. (D) Histopathological analysis of primary lymphoma in lymph node and spleen of an HBZ-Tg/IL-6 KO mouse. (E) Incidence of dermatitis in IL-6Rfl/fl (blue; n = 15), HBZ-Tg/IL-6Rfl/fl (yellow; n = 10), CD4-Cre/IL-6Rfl/fl (green; n = 7), and HBZ-Tg/CD4-Cre/IL-6Rfl/fl (red; n = 8) mice (log-rank test). (F) Incidence of dermatitis in IL-6Rfl/fl (blue; n = 20), HBZ-Tg/IL-6Rfl/fl (yellow; n = 14), LysM-Cre/IL-6Rfl/fl (green; n = 23), and HBZ-Tg/LysM-Cre/IL-6Rfl/fl (red; n = 17) mice (log-rank test).
Table 1.
Histological findings in mice at 24 wk of age
| Mice | Number | Incidence of | Frequency of Foxp3+ cells in lymphoma, % | |||
| Dermatitis, skin, % | Lymphoma, % | |||||
| Spleen | LN | Mean | SD | |||
| WT | 10 | 0 | 0 | 0 | NA | NA |
| HBZ-Tg | 10 | 30 | 30 | 30 | 7.3 | 4.6 |
| IL-6 KO | 10 | 0 | 0 | 0 | NA | NA |
| HBZ-Tg/IL-6 KO | 12 | 41.7 | 66.7 | 83.3 | 38.0 | 10.3 |
LN, lymph node; NA, not applicable.
To understand how the IL-6 signal modulates inflammatory status in HBZ-Tg mice, we next performed conditional knockout of the IL-6 receptor (IL-6R) in this mouse model. IL-6R is known to be expressed on T cells, myeloid cells, and hepatocytes. We crossed IL-6R-flox/flox (IL-6Rfl/fl) mice (21) with CD4-Cre knockin (22) or lysozyme M Cre (LysM-Cre) knockin mice (23) to generate T cell-specific or myeloid cell-specific IL-6R KO mice, respectively. We then crossed the HBZ-Tg mice with each strain to establish HBZ-Tg mice that lack IL-6R specifically in either T cells or myeloid cells. Interestingly, there were no significant differences in the incidence of dermatitis between HBZ-Tg/CD4-Cre/IL-6Rfl/fl mice or HBZ-Tg/LysM-Cre/IL-6Rfl/fl mice and their HBZ-Tg but IL-6R-intact littermates (Fig. 1 E and F).
There are two major modes of IL-6/IL-6R signaling: classic signaling and trans-signaling (24). In classic signaling, IL-6 binds membrane-bound IL-6R with gp130, but only a few types of cells express membrane-bound IL-6R. On the other hand, in trans-signaling, IL-6 forms a complex with soluble IL-6R and targets cells that express only gp130. For example, IL-6/IL-6R trans-signaling is important for the inhibitory effect of IL-6 on the differentiation of Treg cells (25). Since the soluble form of IL-6R was still secreted by other types of cells when we knocked out IL-6R in a single lineage of cells, we could block the classic signaling but could not inhibit the trans-signaling. Although we could not determine the sources of IL-6 and IL-6R affecting the pathogenesis of HBZ, our results suggest that the inhibitory function of IL-6 against HBZ might be exerted via trans-signaling, which is critical in regulating certain types of cells and tissues (24). To understand the mode of action of IL-6R in HBZ-Tg mice, generation and detailed analysis of HBZ-Tg/systemic IL-6R KO mice might be useful. However, in this study, we focus on the characteristics of HBZ-expressing CD4+ T cells in HBZ-Tg and HBZ-Tg/IL-6 KO mice to clarify the mechanisms of accelerated inflammation and T cell lymphoma.
IL-10–Producing CD4+ T Cells Are Increased in HBZ-Tg/IL-6 KO Mice.
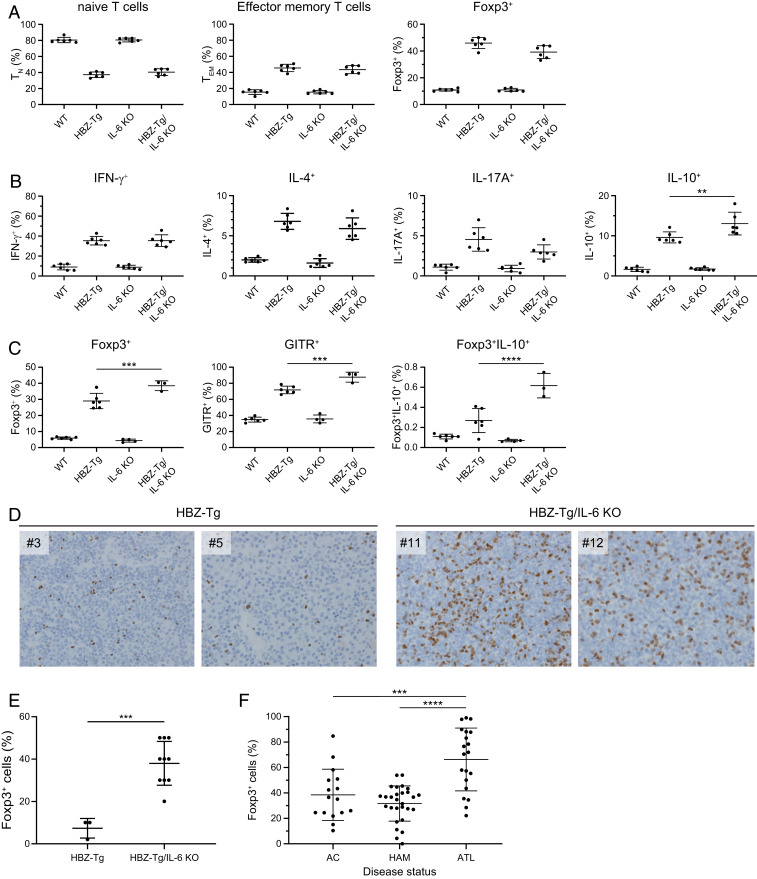
To confirm the immunologic phenotypes of HBZ-Tg/IL-6 KO mice, we next analyzed the T cell subsets and the production of cytokines in CD4+ T cells. Splenocytes from 4-wk-old mice were isolated and subjected to flow cytometry analysis. The percentages of effector memory T cells and CD4+Foxp3+ cells were increased in HBZ-Tg and HBZ-Tg/IL-6 KO mice compared with WT and IL-6 KO mice (Fig. 2A and SI Appendix, Fig. S1). In 28-wk-old mice, the percentage of Foxp3+ T cells was significantly higher in HBZ-Tg/IL-6 KO mice than in HBZ-Tg mice (SI Appendix, Fig. S1).
Fig. 2.
IL-10–producing CD4+ T cells are increased in HBZ-Tg/IL-6 KO mice. (A) Flow cytometry analysis of T cell subsets. Mouse splenocytes were collected from WT, HBZ-Tg, IL-6 KO, and HBZ-Tg/IL-6 KO mice at age 4 wk. Cells were stained with anti-CD4, anti-CD44, and anti-CD62L antibodies for naïve and effector memory T cells, and with anti-Foxp3. (B) Cytokine production by CD4+ T cells from 4-wk-old mice. Splenocytes were stimulated with PMA/ionomycin in the presence of protein transport inhibitor for 5 h and then stained with specific antibodies. (C) Expression of Treg-related molecules in CD4+ cells collected from 16-wk-old mice. Splenocytes were stimulated with PMA/ionomycin in the presence of protein transport inhibitor for 5 h and then stained with specific antibodies. (D) Immunohistochemical analysis for Foxp3 in lymph node and spleen of HBZ-Tg and HBZ-Tg/IL-6 KO mice (original magnification 40×). (E) Percentage of cells that are Foxp3+ in primary lymphomas of HBZ-Tg (n = 3) and HBZ-Tg/IL-6 KO (n = 10) mice. The data were obtained from immunohistochemical analysis. (F) Foxp3 expression in CD4+CADM1+ T cells from HTLV-1–infected subjects. AC, asymptomatic carrier (n = 16); HAM/TSP, HTLV-1–associated myelopathy/tropical spastic paraparesis (n = 28); ATL, adult T cell leukemia/lymphoma (n = 20).
To analyze the expression of cytokines, we stimulated splenocytes in phorbol myristate acetate (PMA) and ionomycin for 5 h and then stained them for intracellular cytokines. The production of IFN-γ, IL-4, IL-17A, and IL-10 was increased in HBZ-Tg and HBZ-Tg/IL-6 KO mice compared with WT and IL-6 KO mice (Fig. 2B). In particular, IL-10–producing cells were more significantly increased in HBZ-Tg/IL-6 KO mice than in HBZ-Tg mice of a young age. On the other hand, the expression of IL-17A was lower in HBZ-Tg/IL-6 KO mice than in HBZ-Tg mice.
To analyze the correlation between Treg markers and IL-10 expression, we stained splenocytes from 16-wk-old mice with antibodies to Foxp3, IL-10, and GITR (a marker of Treg cells). Flow cytometry results showed that the frequencies of CD4+Foxp3+ cells and CD4+GITR+ cells were significantly higher in HBZ-Tg/IL-6KO mice than in HBZ-Tg mice at this age (Fig. 2C). We also confirmed that IL-10–expressing cells in the CD4+Foxp3+ subset were increased in HBZ-Tg/IL-6KO mice compared with HBZ-Tg mice.
Since HBZ induces Foxp3 by activating the TGF-β/Smad signaling pathway (26), these results suggest that knocking out IL-6 in HBZ-Tg mice enhanced the effects of HBZ that drive differentiation of CD4+ T cells toward the Treg-like subset. Interestingly, histopathological analysis demonstrated a higher percentage of Foxp3+ cells in lymphoma tissues from HBZ-Tg/IL-6 KO mice compared with HBZ-Tg mice (Fig. 2 D and E). We also found that Foxp3+ cells were significantly increased in leukemic cells in ATL patients compared with nonleukemic infected cells in HAM/TSP patients and asymptomatic carriers (AC) (Fig. 2F). These findings suggest that Foxp3 expression is associated with oncogenesis by HTLV-1.
Transcriptome Analysis Reveals the Pathways and Genes Involved in Pathogenesis in HBZ-Tg/IL-6 KO Mice.
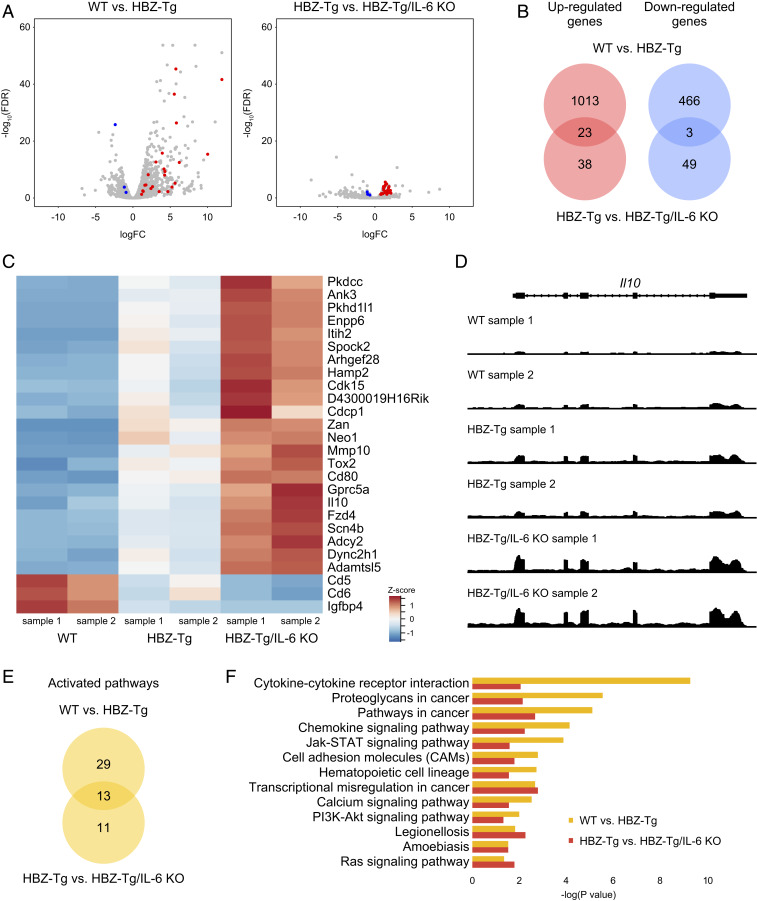
To understand the molecular mechanisms for the acceleration of inflammation and oncogenesis in HBZ-Tg/IL-6 KO mice compared with HBZ-Tg mice, we performed RNA-seq analysis using splenic CD4+ T cells from four strains: WT, HBZ-Tg, IL-6 KO, and HBZ-Tg/IL-6 KO. We hypothesized that HBZ and loss of IL-6 would work cooperatively in promoting inflammation and lymphomagenesis. We first confirmed that the RNA-seq results were compatible with our previously published results obtained by microarray and quantitative RT-PCR (SI Appendix, Fig. S2); many HBZ-target genes, including Ccr4, Trp73, Neo1, Tigit, and Cxcr3 (19, 27, 28), were reproducibly identified. Comparison of HBZ-Tg mice to WT mice showed that 1,035 genes were up-regulated and 469 genes were down-regulated in this study (Fig. 3 A and B).
Fig. 3.
Expression profiles of splenic CD4+ T cells in each strain. (A) Volcano plots obtained from RNA-seq analysis of splenic CD4+ T cells. Red spots indicate genes that are significantly up-regulated in both a comparison of WT mice (n = 2) and HBZ-Tg mice (n = 2) and in a comparison of HBZ-Tg and HBZ-Tg/IL-6 KO mice (n = 2), while blue spots represent genes that are significantly down-regulated in both experiments (FDR >0.1). (B) Venn diagrams of the overlap of significantly up-regulated and down-regulated genes in WT vs. HBZ-Tg and HBZ-Tg vs. HBZ-Tg/IL-6 KO mice. (C) Heat map of expression profiles for the 26 significant genes (23 up-regulated and 3 down-regulated). (D) Expression tiling of Il10 in each sample. Genomic views of transcription at the Il10 gene locus in WT, HBZ-Tg, and HBZ-Tg/IL-6 KO mice are shown. The y-axis represents the number of reads at the locus, and the maximum read count is set to 500 for all samples. (E) Venn diagrams of significantly up-regulated KEGG pathways in WT vs. HBZ-Tg mice and in HBZ-Tg vs. HBZ-Tg/IL-6 KO mice. (F) Bar plots of significantly (P < 0.05) up-regulated KEGG pathways.
When we looked at the difference between HBZ-Tg mice and HBZ-Tg/IL-6KO mice, there were quite a few significantly up- or down-regulated genes, although the magnitude of their differences was not so great (Fig. 3 A, Right). Among those genes, we focused on 23 up-regulated genes and 3 down-regulated genes for which expression levels in HBZ-Tg/IL-6 KO mice were higher or lower, respectively, than in HBZ-Tg mice (Fig. 3 B and C), since HBZ-Tg/IL-6 KO mice demonstrated enhanced phenotypes of HBZ-Tg mice. A representative genomic view of one differentially regulated gene, the Il10 gene, is shown in Fig. 3D; of the three mouse genotypes, the transcription level of the Il10 gene was the highest in HBZ-Tg/IL-6KO mice, followed by HBZ-Tg mice and WT mice.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tools (29, 30) identified 13 pathways (Fig. 3 E and F) that were activated both in HBZ-Tg mice compared with WT mice and in HBZ-Tg/IL-6 KO mice compared with HBZ-Tg mice. One-half of the pathways were classified as “signal transduction” or “signaling molecules and interaction,” and several cancer-related pathways were also extracted. Among these pathways, the JAK-STAT signaling pathway was up-regulated in HBZ-Tg and HBZ-Tg/IL-6 KO mice. The JAK-STAT signaling pathway is a common pathway downstream of many cytokines, including IL-6, IL-10, IFN-α, and IFN-γ, and is also associated with the growth of many types of cancer (31, 32). Since this pathway is more activated in HBZ-Tg/IL-6 KO mice than in HBZ-Tg mice, we speculated that HBZ triggers activation of the JAK/STAT pathway by IL-10, but not by IL-6.
IL-10 Accelerates the Proliferation of CD4+ T Cells in the Presence of HBZ.
Several recent studies have shown that IL-10 is implicated in pathogenesis by HTLV-1 (33, 34). Our RNA-seq results also suggest that IL-10 is one of the key molecules in HBZ-mediated inflammation and lymphomagenesis. IL-10 is a known anti-inflammatory cytokine that typically protects against various infections and autoimmune diseases (9). For cancer, PEGylated IL-10 has been reported to enhance the activity of CD8+ T cells and lead to tumor regression (35). Interestingly, however, in this study the IL-10 production level was high in HBZ-Tg/IL-6 KO mice, which had a high incidence of lymphoma (Figs. 1C and 2B), suggesting that in HBZ-expressing cells, IL-10 has protumorigenic effects rather than anticancer properties.
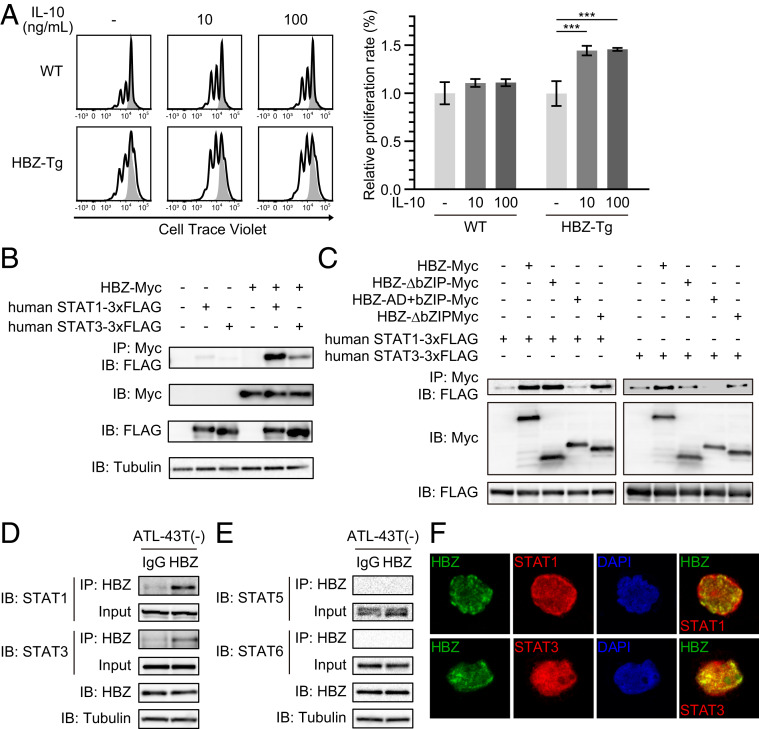
We assessed the influence of IL-10 on cell proliferation using a fluorescence dilution assay. We purified CD4+ T cells from the splenocytes of WT and HBZ-Tg mice, stained the cells with CellTrace Violet, and stimulated them with or without IL-10 in the presence of immobilized anti-CD3 antibody. After 48 h of culture, we analyzed the intensity of CellTrace by flow cytometry. IL-10 did not affect the proliferation of CD4+ T cells from WT mice (Fig. 4A), but on the other hand, it significantly accelerated the proliferation of CD4+ T cells from HBZ-Tg mice. These results indicate that HBZ makes cells sensitive to an IL-10 signaling pathway that promotes proliferation.
Fig. 4.
HBZ modifies the IL-10/JAK/STAT signaling pathway. (A) Fluorescence dilution assay of CD4+ T cells from WT or HBZ-Tg mice. Splenic CD4+ T cells were labeled with 5 μM CellTrace Violet and stimulated by anti-CD3 antibodies with or without IL-10. At 48 h after stimulation, CellTrace Violet was measured by flow cytometry (two-way ANOVA with Turkey’s multiple comparisons). (B) Coimmunoprecipitation of HBZ and human STAT1 or STAT3. The indicated expression vectors were cotransfected into HEK293T cells, and protein interactions were analyzed by immunoprecipitation. (C) Interaction of HBZ mutants with STAT1 or STAT3 was analyzed by immunoprecipitation. (D and E) Interaction between HBZ and STAT proteins in ATL-43T(−). A protein extract of ATL-43T(−) cells was subjected to immunoprecipitation with anti-HBZ antibody or control IgG, and STAT proteins were detected by anti-STAT1, anti-STAT3, anti-STAT5, or anti-STAT6 antibody. (F) Colocalization of HBZ and STAT proteins. HBZ-myc and STAT1 or STAT3-3xFLAG were transfected into Jurkat cells by electroporation. Staining was performed using antibodies against myc (green) and FLAG (red). Nuclei were stained with DAPI (blue).
HBZ Physically Interacts with STAT1 and STAT3.
IL-10 activates the JAK-STAT signaling pathway and signals mainly through STAT1 and STAT3 (36). Since we speculated that the signaling pathway downstream of IL-10 is activated in HBZ-Tg and HBZ-Tg/IL-6KO mice, we first evaluated the expression levels of several genes associated with the JAK/STAT pathway by quantitative RT-PCR (SI Appendix, Fig. S3). There were no genes with whose significantly up-regulated expression in both HBZ-Tg and HBZ-Tg/IL-6KO mice compared with WT and IL-6KO mice, although several genes, such as Il10Ra and Tyk2, were up-regulated in HBZ-Tg mice compared with WT mice.
We next asked whether HBZ physically binds to STAT1 and STAT3, since it is known that HBZ can bind to many transcription factors. To detect any direct interaction between HBZ and STAT proteins, we first used HEK293T cells overexpressing HBZ and STAT proteins. As expected, immunoprecipitation experiments showed that HBZ physically interacted with both STAT1 and STAT3 (Fig. 4B). Interaction of HBZ with both mouse and human STAT proteins was confirmed (SI Appendix, Fig. S4). Experiments using HBZ deletion mutants showed that the central domain of HBZ was critical for its interaction with STAT1 and STAT3 (Fig. 4C).
To assess the interaction of endogenous HBZ and STATs, we next performed immunoprecipitation using the cell line ATL-43T(−), which was derived from an ATL case. We found that endogenous HBZ also bound to STAT1 and STAT3 (Fig. 4D). In contrast, an association between HBZ and other STAT proteins, such as STAT5 and STAT6, was not observed in ATL-43T(−), even though both proteins could be clearly detected in the lysate (Fig. 4E). These results suggest that HBZ interacts preferentially with STAT1 and STAT3 in ATL cells. We could detect the binding between HBZ and STAT1/3 equally, even after ATL-43T(−) was treated with the selective JAK1/2 inhibitor ruxolitinib (SI Appendix, Fig. S5).
Since phosphorylated STAT proteins were not detected in the lysate after the treatment, it has been suggested that their interaction is independent of the phosphorylation of STATs. To see where HBZ interacts with STAT1/3, we immunostained Jurkat cells transfected with HBZ and either STAT1 or STAT3. Although not all HBZ and STAT1/STAT3 colocalized together, we did observe partial colocalization of these proteins in the nucleus (Fig. 4F). The same pattern was obtained using HeLa cells (SI Appendix, Fig. S6), consistent with the potential for intracellular protein–protein interaction. These findings suggest that HBZ might sequester the STAT1 and STAT3 in the nucleus as it does for FoxO3a (37). Recent studies suggest that the subcellular localization of HBZ protein is changed by the disease status: HBZ localizes in the nucleus of ATL cells but in the cytoplasm of infected cells from HAM/TSP patients (38, 39). Nuclear colocalization of HBZ and STAT1/3 might have specific roles in the leukemogenesis of ATL.
HBZ Modulates the JAK-STAT Signaling Triggered by IL-10.
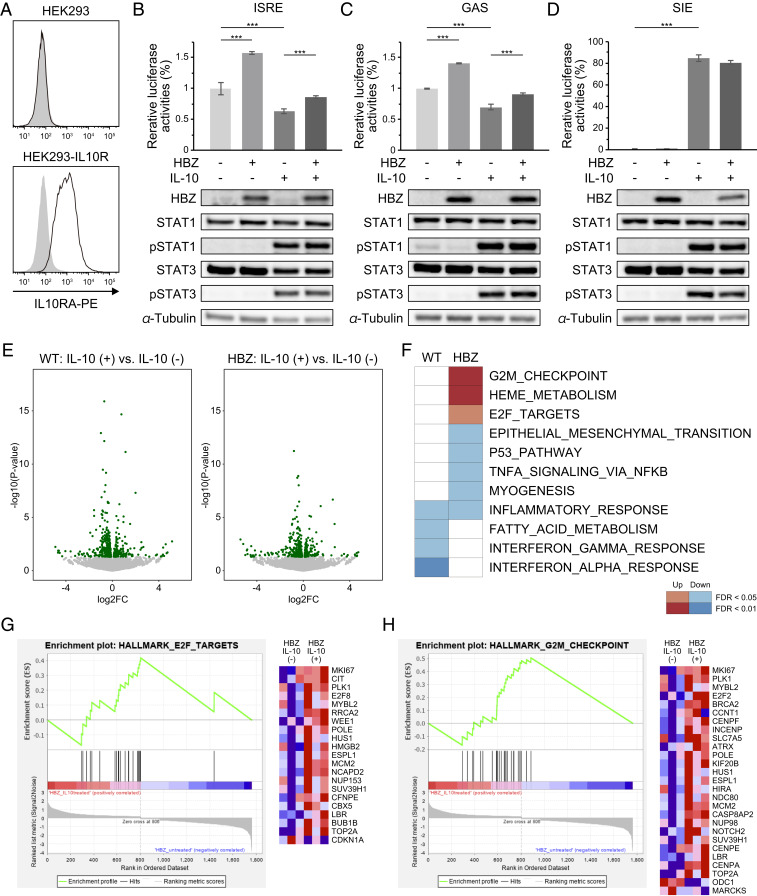
Since HBZ induces IL-10 expression and interacts with its downstream transcription factors STAT1 and STAT3, we expected that HBZ might influence the transcriptional activities of those STAT proteins under stimulation by IL-10. To investigate this possibility, we generated IL-10R–expressing HEK293 cells (Fig. 5A) and evaluated the transcriptional activities of STATs using several reporter constructs. Previous studies have shown that STAT1 and STAT3 form heterodimers or homodimers that can bind to and activate IFN-stimulated response elements (ISREs) (40), the IFN-γ activation site response element (GAS-RE) (41), and the sis-inducible element (SIE) (42). As shown in previous studies (43), IL-10 stimulation decreases the reporter activities of ISRE and GAS-RE (Fig. 5 B and C). We initially expected to find that HBZ would enhance the effects of IL-10 on those elements; however, the results were contrary to our hypothesis: HBZ activated those reporters and canceled the suppression by IL-10. In contrast, IL-10 stimulation increased SIE reporter activity, and HBZ had no effect on this activation (Fig. 5D). In these experiments, immunoblotting showed that IL-10 induced the phosphorylation of STAT1 and STAT3, but HBZ did not influence their phosphorylation status (Fig. 5 B–D). These results suggest that HBZ modifies the effect of IL-10 on each element differently; HBZ activates STAT1 and STAT3 at certain elements (such as ISRE and GAS) for which IL-10 has repressive effects but has no effect on an element (SIE) that IL-10 can activate.
Fig. 5.
HBZ modulates IL-10/JAK/STAT signaling. (A) Flow cytometry analysis of IL10RA in HEK293-IL10R. Cell were stained by PE-conjugated anti-IL10RA antibody (black line) or isotype control (gray filled); two-tailed unpaired Student’s t test. (B–D, Top) Luciferase assay of ISRE, GAS, and SIE. HEK293-IL10R cells were transfected with HBZ, each reporter, and a reporter plasmid driven by cytomegalovirus immediate-early promoter as an internal control, and then stimulated with IL-10 (100 ng/mL). At 24 h after stimulation, luciferase activities were measured (two-way ANOVA with Turkey’s multiple comparisons). (Bottom) Immunoblotting of HBZ, STAT1, pSTAT1, STAT3, pSTAT3, and tubulin. (E) Volcano plots obtained from RNA-seq analysis of splenic CD4+ T cells of WT or HBZ-Tg (n = 3) cultured with or without IL-10 for 48 h. Green spots indicate differentially expressed genes (P < 0.05) between IL-10–treated and untreated cells. The results from WT and HBZ-Tg mice are shown in the left and right diagrams, respectively. (F) Enriched gene sets modulated by HBZ and IL-10 using GSEA. The top 2,000 differentially expressed genes in IL-10–treated cells compared with untreated cells from WT or HBZ-Tg mice were analyzed. (G and H) GSEA enrichment plots and heat maps of gene expression in CD4+ T cells from HBZ-Tg mice, with or without IL-10 stimulation.
To further investigate the molecules and pathways that are cooperatively regulated by HBZ and IL-10, we carried out transcriptional profiling by RNA-seq. In brief, we isolated CD4+ T cells from WT or HBZ-Tg mice and treated the cells with or without recombinant IL-10. After 48 h of culture, RNAs were extracted from each sample and subjected to analysis (Fig. 5E). Using Gene Set Enrichment Analysis (GSEA) (44, 45), we found that cell cycle-related pathways, such as the G2M checkpoint and E2F targets, were significantly up-regulated by IL-10 in HBZ-Tg mice (Fig. 5 F–H). On the other hand, while IL-10 down-regulated IFN-γ and IFN-α responses in CD4+ T cells from WT mice, these changes were not evident in cells from HBZ-Tg mice (Fig. 5F). Similar results were obtained by the STAT-responsible reporter assays (Fig. 5 B and C). These transcriptional changes suggest that HBZ impedes the immunosuppressive effects of IL-10 in CD4+ T cells and promotes their proliferation, presumably in part through binding to STAT proteins.
Discussion
IL-10 is an immunomodulating cytokine critical for suppressing excessive immune activation and consequent tissue damage (46). Several viruses use the immunosuppressive function of IL-10 to establish persistent infection (47). IL-10 suppresses the antigen-presenting capacity of dendritic cells (DCs) and leads to exhaustion of T cells, which allows viruses to persist (46, 48). In addition, many latent viruses, such as several herpesviruses, encode IL-10 homologs in their genomes, suggesting that induction of cellular/viral IL-10 is an efficient strategy to evade the host immune system. In this study, we show that HTLV-1 uses IL-10 for its persistence in a unique way: HBZ converts infected cells to IL-10–producing Treg-like cells and promotes proliferation of this subset by modulating IL-10/STAT signaling. Moreover, HBZ transactivates the expression of T cell immunoreceptor with Ig and ITIM domains (TIGIT) on T cells, and interaction between TIGIT on T cells and its ligand CD155 on DCs induces IL-10 production from DCs (28). Since increases in Foxp3-expressing cells (Fig. 2 E and F) and plasma levels of IL-10 (34) are correlated with disease status, it appears that autocrine/paracrine activation of the IL-10/STAT pathway and immune suppression by IL-10 are important for the clonal expansion of infected cells and pathogenesis by HTLV-1.
A previous report showed involvement of IL-10 and STAT3 in the proliferation of HTLV-1–immortalized cell lines (33). In this study, HBZ is shown to activate cell cycle-related pathways (Fig. 5 F–H), while apparently inhibiting the suppressive effects of IL-10 on type I IFN immune responses (Fig. 5B). These results suggest that HBZ dysregulates IL-10/STAT signaling and promotes cell expansion in a complex manner. It appears that HBZ-expressing cells tend to become IL-10–producing cells (thus suppressing any immune response against HTLV-1) while perhaps becoming insensitive themselves to the antiproliferative effects of IL-10. Thus, HBZ enables HTLV-1 to establish persistent infection.
It is notable that IL-6 deficiency enhanced inflammation and lymphomagenesis induced by HBZ in vivo. This result was contrary to our initial prediction, since proinflammatory cytokines generally have accelerating effects on chronic inflammation and cancers (32, 49). IL-6 has pleiotropic activities in inflammation, immune reaction, hematopoiesis, and cell differentiation (5); however, it inhibits TGF-β–induced Treg differentiation (50), whereas HBZ induces Foxp3 expression by activating TGF-β/Smad signaling (17, 26). Thus IL-6 may counteract the effects of HBZ on the differentiation of HTLV-1–infected cells. Pathogenesis by HBZ is closely linked to the immunophenotypes of HBZ-expressing cells; HBZ up-regulates a variety of molecules associated with Treg cells, such as Foxp3, CCR4, TIGIT, and IFN-γ, thereby promoting the expansion of these pathogenic Treg-like cells (17–19, 28). Therefore, the disease progression demonstrated by HBZ-Tg/IL-6 KO mice implies that loss of IL-6 accelerates the abnormal differentiation of pathogenic cells by HBZ. Importantly, similar observations in the clinical field have been reported; after treatment with the humanized anti–IL-6R antibody tocilizumab against rheumatoid arthritis, an HTLV-1 carrier developed ATL (51), and HAM/TSP and uveitis were exacerbated in another carrier (52). Our results and those clinical observations alert us to the possibility that blockade of IL-6/IL-6R signaling increases the risk of disease progression in some HTLV-1–infected individuals.
Recently reported results of an integrated genetic analysis show that activating somatic mutations of STAT3 are frequently observed in ATL cases (53, 54). This indicates that STAT3 activation can contribute to leukemogenesis of HTLV-1–infected cells. Importantly, both IL-6 and IL-10 activate mainly STAT3 (55), but only IL-10 up-regulates genes associated with the anti-inflammatory response. These findings indicate that the selective regulation of STAT3 by some factor(s) is important for proinflammatory vs. anti-inflammatory responses. One example of such a molecule is SOCS3. SOCS3 can bind to IL-6R and negatively regulates IL-6-STAT3 signaling, but it cannot bind to IL-10R (56, 57). In addition, loss of SOCS3 changes the character of IL-6 from proinflammatory to anti-inflammatory (57).
These studies show that activation of STAT3 can be modulated downstream of the cytokine stimulation. In this study, we found that HBZ could activate the STAT-responsive elements ISRE and GAS, which are suppressed by IL-10, but did not affect the SIE, which is activated by IL-10 (Fig. 5 B–D). Thus, HBZ diminishes the repressive capacities of IL-10 on those STAT-responsive elements. RNA-seq results (Fig. 5 E–H) also suggest that HBZ modulates the responsiveness of T cells to IL-10 from immunosuppressive to proliferative phenotypes; however, the target molecules of HBZ and STATs remain unclear. Since HBZ selectively activates the transcription of TGF-β/Smad target genes through recruitment of transcriptional coactivator p300 to the promoter (26), it is possible that similar mechanisms are involved in HBZ-mediated STAT activation. Interestingly, a previous study reported that IL-10, but not IL-6, is crucial for potent STAT3 activation in Foxp3+ Treg cells (58). The Treg cell context might be associated with STAT3 activation by HBZ. Further analysis is needed to clarify the details.
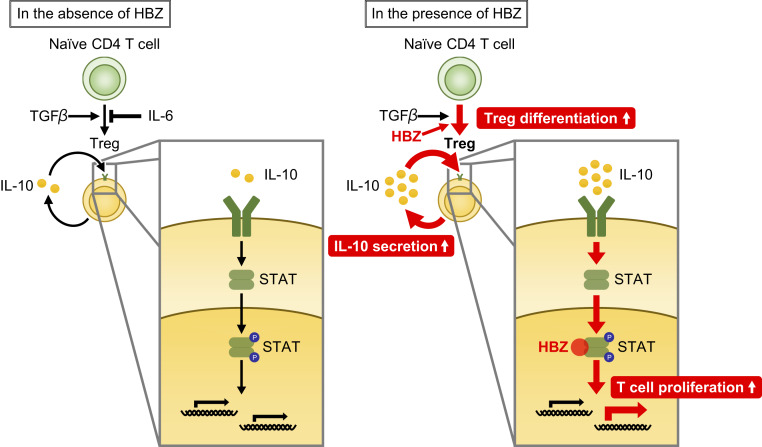
In conclusion, the HTLV-1–encoded viral protein HBZ plays a central role in HTLV-1–mediated pathogenesis by inducing the differentiation of infected cells to cells with Treg-like signatures, increasing IL-10 production, and modulating the IL-10/JAK/STAT signaling to the lymphoproliferative state (Fig. 6). This study demonstrates a previously unknown linkage between HBZ and the immunomodulatory cytokines, and elucidation of the detailed mechanisms can contribute to the development of new strategies for treatment of the refractory diseases induced by HTLV-1.
Fig. 6.
Schema of HBZ-mediated T cell proliferation. HBZ and loss of IL-6 enhances Treg differentiation and IL-10 production. Increased IL-10 activates STAT proteins, and HBZ modulates the IL-10/JAK/STAT signaling toward proinflammatory and proliferative properties.
Materials and Methods
Mice.
C57BL/6J mice were purchased from CLEA. Transgenic mice expressing the spliced form of HBZ driven by the mouse CD4 promoter (HBZ-Tg mice) have been described previously (17). All HBZ-Tg mice were heterozygotes for the transgene. B6.129S2-Il6tm1Kopf/J (IL-6 KO) mice (20) and B6.SJL-Il6ratm1.1Drew/J (21) mice were purchased from The Jackson Laboratory. T cell-specific or myeloid cell-specific IL-6R conditional KO mice were generated by crossing the B6.SJL-Il6ratm1.1Drew/J mice with CD4-Cre knockin mice (kindly provided by Christopher B. Wilson) or lysozyme M-Cre knockin mice (kindly provided by Irmgard Foerster), respectively. All animal experiments carried out in this study were approved by the Animal Research Committee of Kyoto University (approval nos. D13-02, D14-02, D15-02, A10-3, and A17-1).
Cells.
Human embryonic kidney cell lines HEK293 and HEK293T were purchased from RIKEN BioResource Research Center and American Type Culture Collection, respectively, and cultured in Dulbecco’s modified Eagle’s medium (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS) and antibiotics. The Jurkat cell line was provided by S. Sakaguchi, Osaka University. Jurkat and ATL-43T(−) cell lines (59) were grown in RPMI 1640 (Nacalai Tesque) with 10% FBS and antibiotics at 37° in a 5% CO2 atmosphere. The IL10RA expression vector was electroporated into HEK293 cells using the PiggyBac Transposon Vector System (System Biosciences). Stable transfectants were selected in puromycin (1 μg/mL).
Clinical Samples.
Peripheral blood mononuclear cells (PBMCs) were obtained from ATL patients, HAM/TSP patients, and HTLV-1 carriers. All subjects were fully informed of the purpose and procedures of this study, and written consent was obtained from each subject. The PBMCs were collected using Ficoll-Paque PLUS (GE Healthcare). Use of the clinical samples in this research was approved by the Ethics Committee of Kyoto University (approval no. E1649).
RNA Extraction and RNA-Seq.
CD4+ T cells were isolated from splenocytes using anti-mouse CD4 magnetic particles (BD Biosciences). For RNA-seq to analyze the effects of IL-10 on CD4+ T cells (Fig. 5), cells were treated by immobilized anti-CD3 antibodies with or without recombinant IL-10 (Peprotech) for 48 h. RNA was extracted with TRIzol reagent (Invitrogen) and purified with the Direct-zol RNA MiniPrep Kit (Zymo Research). Library preparation and high-throughput sequencing were performed at Macrogen using TruSeq RNA Sample Prep Kit v2 and HiSeq 2000 (Illumina) or GeneWiz using the NEBNext Ultra II Directed RNA library kit (New England BioLabs) and HiSeq X (Illumina). RNA-seq data were mapped to the GRCm38/mm10 using HISAT2 (60). Differently expressed genes were analyzed using HTSEq (61) and edgeR (62) or DESeq2 (63). Volcano plots and heat maps were drawn using ggplot2 (64) and gplots (65). Expression tiling of RNA-seq was visualized using the Integrative Genomics Viewer (66). Functional annotation of gene lists with Gene Ontology terms and KEGG pathways was performed with the DAVID tools (29, 30) and GSEA (44, 45).
Statistical Analysis.
All experiments were biologically and/or technically replicated at least three times, except for the RNA-seq experiment. Statistical analyses were performed using Prism 8 (GraphPad Software). The log-rank (Mantel–Cox) test was used to assess significance in incidence of dermatitis and overall survival as measured by a Kaplan–Meier plot. Statistical significances of two group comparisons were determined using the two-tailed unpaired Student’s t test. Multiple comparisons were performed by one-way or two-way ANOVA with Tukey correction. All data are presented as mean ± SD. The minimum significance level was set at P < 0.05. Asterisks indicate the statistical significance as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Additional information on the study methodology is provided in SI Appendix.
Data Availability.
RNA-seq data have been deposited in the DNA Data Bank of Japan (DDBJ) database for data sharing (accession numbers are DRA009954 and DRA009955).
Supplementary Material
Acknowledgments
We thank Dr. Linda Kingsbury for editorial comments and proofreading. We also thank Drs. Shigeo Koyasu and Irmgard Foerster for the lysozyme M-Cre knockin mice, Dr. Christopher B. Wilson for CD4-Cre knockin mice, Dr. Norihiro Takenouchi in Kansai Medical University for HAM/TSP samples, Dr. Koichi Ikuta for pMX-mouse STAT1 and pEFBOS-mouse STAT3, and Dr. Koichi Nakajima for pBS-HA-hSTAT1. This study was supported by the Project for Cancer Research and Therapeutic Evolution (P-CREATE) grants (19cm0106611h0003, to J.-i.Y., and 19cm0106306h0004, to M.M.); the Research Program on Emerging and Re-emerging Infectious Diseases grants (18fk0108027h0003 and 19fk0108088h0001, to J.-i.Y. and M.M.) from the Japan Agency for Medical Research and Development; the Japan Society for the Promotion of Science (JSPS) KAKENHI grants (19H03689, to M.M., and JP17K07166, to J.-i.Y.); the Japan Science and Technology Agency-Mirai Program (JPMJMI18G1 to J.-i.Y.); and the Japan Leukemia Research Fund (to J.-i.Y.). Support for this study was also provided by the JSPS Core-to-Core Program A, Advanced Research Networks.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the DNA Data Bank of Japan (DDBJ) database, https://www.ddbj.nig.ac.jp/index-e.html (accession nos. DRA009954 and DRA009955).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922884117/-/DCSupplemental.
References
- 1.Coussens L. M., Werb Z., Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Colotta F., Allavena P., Sica A., Garlanda C., Mantovani A., Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 30, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Zamarron B. F., Chen W., Dual roles of immune cells and their factors in cancer development and progression. Int. J. Biol. Sci. 7, 651–658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka T., Narazaki M., Kishimoto T., IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6, a016295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S. A., Scheller J., Rose-John S., Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 121, 3375–3383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M. O., Wan Y. Y., Sanjabi S., Robertson A. K., Flavell R. A., Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 24, 99–146 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ouyang W., O’Garra A., IL-10 family cytokines IL-10 and IL-22: From basic science to clinical translation. Immunity 50, 871–891 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Hussain S. P., Harris C. C., Inflammation and cancer: An ancient link with novel potentials. Int. J. Cancer 121, 2373–2380 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Yasunaga J. I., Matsuoka M., Oncogenic spiral by infectious pathogens: Cooperation of multiple factors in cancer development. Cancer Sci. 109, 24–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poiesz B. J. et al., Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 77, 7415–7419 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasunaga J., Matsuoka M., Molecular mechanisms of HTLV-1 infection and pathogenesis. Int. J. Hematol. 94, 435–442 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka M., Jeang K. T., Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Koyanagi Y. et al., In vivo infection of human T-cell leukemia virus type I in non-T cells. Virology 196, 25–33 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Satou Y., Yasunaga J., Yoshida M., Matsuoka M., HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 103, 720–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satou Y. et al., HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 7, e1001274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugata K. et al., HTLV-1 viral factor HBZ induces CCR4 to promote T-cell migration and proliferation. Cancer Res. 76, 5068–5079 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Mitagami Y., Yasunaga J., Kinosada H., Ohshima K., Matsuoka M., Interferon-γ promotes inflammation and development of T-cell lymphoma in HTLV-1 bZIP factor transgenic mice. PLoS Pathog. 11, e1005120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopf M. et al., Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368, 339–342 (1994). [DOI] [PubMed] [Google Scholar]
- 21.McFarland-Mancini M. M. et al., Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 184, 7219–7228 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Lee P. P. et al., A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I., Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S., The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Dominitzki S. et al., Cutting edge: Trans-signaling via the soluble IL-6R abrogates the induction of FoxP3 in naive CD4+CD25 T cells. J. Immunol. 179, 2041–2045 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Zhao T. et al., HTLV-1 bZIP factor enhances TGF-β signaling through p300 coactivator. Blood 118, 1865–1876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawatsuki A., Yasunaga J. I., Mitobe Y., Green P. L., Matsuoka M., HTLV-1 bZIP factor protein targets the Rb/E2F-1 pathway to promote proliferation and apoptosis of primary CD4 (+) T cells. Oncogene 35, 4509–4517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuma K. et al., HTLV-1 bZIP factor impairs anti-viral immunity by inducing co-inhibitory molecule, T cell immunoglobulin and ITIM domain (TIGIT). PLoS Pathog. 12, e1005372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H., Lee H., Herrmann A., Buettner R., Jove R., Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Zaidi M. R., Merlino G., The two faces of interferon-γ in cancer. Clin. Cancer Res. 17, 6118–6124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawada L. et al., IL-10-mediated signals act as a switch for lymphoproliferation in human T-cell leukemia virus type-1 infection by activating the STAT3 and IRF4 pathways. PLoS Pathog. 13, e1006597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagdi H., Demontis M. A., Ramos J. C., Taylor G. P., Switching and loss of cellular cytokine producing capacity characterize in vivo viral infection and malignant transformation in human T-lymphotropic virus type 1 infection. PLoS Pathog. 14, e1006861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naing A. et al., PEGylated IL-10 (Pegilodecakin) induces systemic immune activation, CD8(+) T cell invigoration and polyclonal T cell expansion in cancer patients. Cancer Cell 34, 775–791.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finbloom D. S., Winestock K. D., IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J. Immunol. 155, 1079–1090 (1995). [PubMed] [Google Scholar]
- 37.Tanaka-Nakanishi A., Yasunaga J., Takai K., Matsuoka M., HTLV-1 bZIP factor suppresses apoptosis by attenuating the function of FoxO3a and altering its localization. Cancer Res. 74, 188–200 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Baratella M. et al., Cytoplasmic localization of HTLV-1 HBZ protein: A biomarker of HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). PLoS Negl. Trop. Dis. 11, e0005285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forlani G. et al., HTLV-1 HBZ protein resides exclusively in the cytoplasm of infected cells in asymptomatic carriers and HAM/TSP patients. Front. Microbiol. 10, 819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy D. E., Kessler D. S., Pine R., Reich N., Darnell J. E. Jr., Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 2, 383–393 (1988). [DOI] [PubMed] [Google Scholar]
- 41.Decker T., Kovarik P., Meinke A., GAS elements: A few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17, 121–134 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Wagner B. J., Hayes T. E., Hoban C. J., Cochran B. H., The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 9, 4477–4484 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito S. et al., Interleukin-10 inhibits expression of both interferon alpha- and interferon gamma-induced genes by suppressing tyrosine phosphorylation of STAT1. Blood 93, 1456–1463 (1999). [PubMed] [Google Scholar]
- 44.Mootha V. K. et al., PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rojas J. M., Avia M., Martín V., Sevilla N., IL-10: A multifunctional cytokine in viral infections. J. Immunol. Res. 2017, 6104054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouyang P. et al., IL-10 encoded by viruses: A remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J. Gen. Virol. 95, 245–262 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Blackburn S. D., Wherry E. J., IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 15, 143–146 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Dinarello C. A., Anti-inflammatory agents: Present and future. Cell 140, 935–950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bettelli E. et al., Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Nakamura H. et al., Development of adult T-cell leukemia in a patient with rheumatoid arthritis treated with tocilizumab. Intern. Med. 52, 1983–1986 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Terada Y. et al., Treatment of rheumatoid arthritis with biologics may exacerbate HTLV-1-associated conditions: A case report. Medicine (Baltimore) 96, e6021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kataoka K. et al., Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 47, 1304–1315 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Kataoka K. et al., Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood 131, 215–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong Z., Wen Z., Darnell J. E. Jr., Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Croker B. A. et al., SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4, 540–545 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Yasukawa H. et al., IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 4, 551–556 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Chaudhry A. et al., Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34, 566–578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda S. et al., Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer 109, 559–567 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer, New York, 2016). [Google Scholar]
- 65.Gregory R., et al. , gplots: Various R programming tools for plotting data. https://github.com/talgalili/gplots. Accessed 26 February 2019.
- 66.Robinson J. T. et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the DNA Data Bank of Japan (DDBJ) database for data sharing (accession numbers are DRA009954 and DRA009955).