Significance
Depression is one of the most common, disabling, and expensive of all neuropsychiatric disorders. Emerging evidence implicates circadian rhythm abnormalities in the pathophysiology of depression. In particular, the nucleus accumbens (NAc), a central component of the midbrain dopamine reward circuit, exhibits disturbed circadian rhythms in postmortem brains of depressed patients, as well as in stressed mice exhibiting depression-like (helpless) behavior. Here we provide evidence for a molecular mechanism by which higher levels of the core circadian clock protein CRYPTOCHROME in the NAc may block D1 dopamine receptor activation during the nocturnal active phase of mice, thereby compromising normal daily activation of NAc neurons and leading to helpless behavior. This mechanism suggests a promising target for future antidepressant drugs.
Keywords: Cryptochrome, nucleus accumbens, circadian rhythm, depression
Abstract
The nucleus accumbens (NAc), a central component of the midbrain dopamine reward circuit, exhibits disturbed circadian rhythms in the postmortem brains of depressed patients. We hypothesized that normal mood regulation requires proper circadian timing in the NAc, and that mood disorders are associated with dysfunctions of the NAc cellular circadian clock. In mice exhibiting stress-induced depression-like behavior (helplessness), we found altered circadian clock function and high nighttime expression of the core circadian clock component CRYPTOCHROME (CRY) in the NAc. In the NAc of helpless mice, we found that higher expression of CRY is associated with decreased activation of dopamine 1 receptor-expressing medium spiny neurons (D1R-MSNs). Furthermore, D1R-MSN–specific CRY-knockdown in the NAc reduced susceptibility to stress-induced helplessness and increased NAc neuronal activation at night. Finally, we show that CRY inhibits D1R-induced G protein activation, likely by interacting with the Gs protein. Altered circadian rhythms and CRY expression were also observed in human fibroblasts from major depressive disorder patients. Our data reveal a causal role for CRY in regulating the midbrain dopamine reward system, and provide a mechanistic link between the NAc circadian clock and vulnerability to depression.
A role for circadian clocks in mood disorders has been suggested for decades on the basis of clinical observations. First, circadian rhythm dysregulation is a prominent clinical feature of mood disorders (1, 2). Changes in daily patterns of sleep/wake, energy levels, and appetite are important symptoms of both bipolar disorder and major depressive disorder (MDD). Furthermore, manipulations of light exposure or sleep that affect the circadian clock are now known to affect mood as well (3, 4). Bright light, the primary resetting stimulus for the clock, is an effective antidepressant in both MDD and seasonal affective disorder (5, 6), and sleep deprivation or shifting sleep onset to an earlier time temporarily alleviates depression (7, 8).
A molecular circadian clock has been well characterized in mammalian cells, based on delayed negative feedback in a core transcriptional–translational feedback loop (9). CLOCK/BMAL1 dimers act at E-box elements on DNA to promote transcription of Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2) genes, leading to increases in PER and CRY levels. After delays associated with transcription, translation, dimerization, and nuclear entry, PER/CRY complexes inhibit transcription of their own genes. This leads to declines in PER and CRY levels, thus relieving the inhibition and permitting a new cycle to begin. Aside from their roles in maintaining a functional clock, circadian clock genes also directly or indirectly regulate the expression of many clock-controlled genes critical for neuron physiology and metabolism (10, 11).
Recent studies have directly implicated the molecular circadian clock in the pathogenesis of depression. In postmortem brains from MDD patients, compared to normal control subjects, Li et al. (12) found remarkably weaker daily rhythms of clock gene expression in multiple brain areas, including the dorsolateral prefrontal cortex, anterior cingulate, hippocampus, amygdala, and nucleus accumbens (NAc). Studies in rodent models of depression also implicate circadian rhythm abnormalities in a subset of mood-regulating brain areas, particularly the NAc (13, 14). Some of these animal studies, through genetic manipulation of clock gene expression in specific brain areas, have even provided evidence that circadian clocks play a causal role in mood regulation (15–21).
A potential brain mechanism is suggested by the circadian regulation of dopamine (DA) signaling in the midbrain reward circuit. Several studies have reported circadian transcriptional regulation of tyrosine hydroxylase (22–25) and monoamine oxidase (26), which are enzymes crucial for DA synthesis and deactivation, respectively. In addition, DA release is also modulated by the circadian clock through the DA transporter, which regulates the diurnal variation of DA in the synaptic cleft (27).
Although circadian clock proteins have pervasive effects on gene transcription in the cell nucleus, this is not the only mechanism by which the clock impacts mammalian physiology. In particular, Zhang et al. (28) discovered a novel cytoplasmic action of the circadian clock component CRYPTOCHROME (CRY), in which CRY inhibits both Ca2+ and cAMP signaling by directly binding to and inhibiting the G proteins Gq and Gs, respectively, in the plasma membrane (28, 29). In the liver, for example, CRY rhythmically inhibits glucagon-mediated gluconeogenesis near dawn by interacting with Gs55 coupled to glucagon receptors (29), and it is likely that CRY acts similarly on other Gs protein-coupled receptors expressed in the brain, such as the DA 1 receptor (D1R). This suggests that the molecular circadian clock might regulate DA signaling in the brain’s reward circuit by directly modulating DA receptor activation in the NAc.
The NAc is known to play a crucial role in regulating motivation and reward functions (30–33). The major projecting NAc neurons that regulate these behaviors are inhibitory, GABA-containing neurons, also called medium spiny neurons (MSNs) (34, 35). The MSNs are classified by their DA receptor expression (expressing either D1R or DA 2 receptors [D2R]) and their projections (36–39). Balanced activity of these two neuronal populations facilitates normal behavioral output, while imbalances are implicated in psychiatric and neurological disease (40–43). Previous studies in humans and rodents have shown that activation of the D1R-MSN pathway induces positive reward, whereas activation of the D2R-MSN pathway induces aversion (44). Inhibition of these pathways induces the opposite motivational states (45–48). Also, Fos transcription factors (c-Fos, FosB, ΔFosB) show differential induction patterns in D1R-MSNs vs. D2R-MSNs (49–52), and D1, but not D2, receptor activation is sufficient for such Fos activation (53–55). These data highlight the important role of D1R-MSN activation in mediating resilience to depression and antidepressant action, whereas D2R-MSN activation may mediate susceptibility to depression.
Previously, we found that NAc brain slices from mice susceptible to stress-induced helpless behavior show less circadian rhythmicity and dephased single-cell rhythms compared to those from resilient mice (56). We therefore hypothesized that normal reward circuit function and mood regulation require proper circadian timing in the NAc, and that helpless behavior may be caused by a mistimed increase of the circadian clock component CRY during the nocturnal active phase, thereby inhibiting the normal nightly activation of D1R-MSNs. Here we induce a helpless state in mice by stressful inescapable tail-shock training, and find circadian clock alterations in the NAc of these mice. Helpless mice show higher nighttime CRY expression and reduced D1R-MSN activation compared to resilient and naïve mice. Similar clock dysfunction and changes in CRY expression were observed in cells from male MDD patients. Furthermore, in mice, knocking down CRY specifically in NAc D1R-MSNs reduces susceptibility to stress-induced helplessness and increases NAc neuronal activation at night. Finally, we show that CRY inhibits D1R-induced G protein activation, likely by interacting directly with the Gs protein. Our data reveal a causal role for CRY in the reward system, mediating vulnerability to stress-induced helplessness, and suggest a mechanism involving the action of CRY on the Gs protein in D1R-MSNs of the NAc.
Results
Helpless Behavior Is Associated with Abnormal PER2::LUC Rhythms in Mouse Suprachiasmatic Nucleus and NAc.
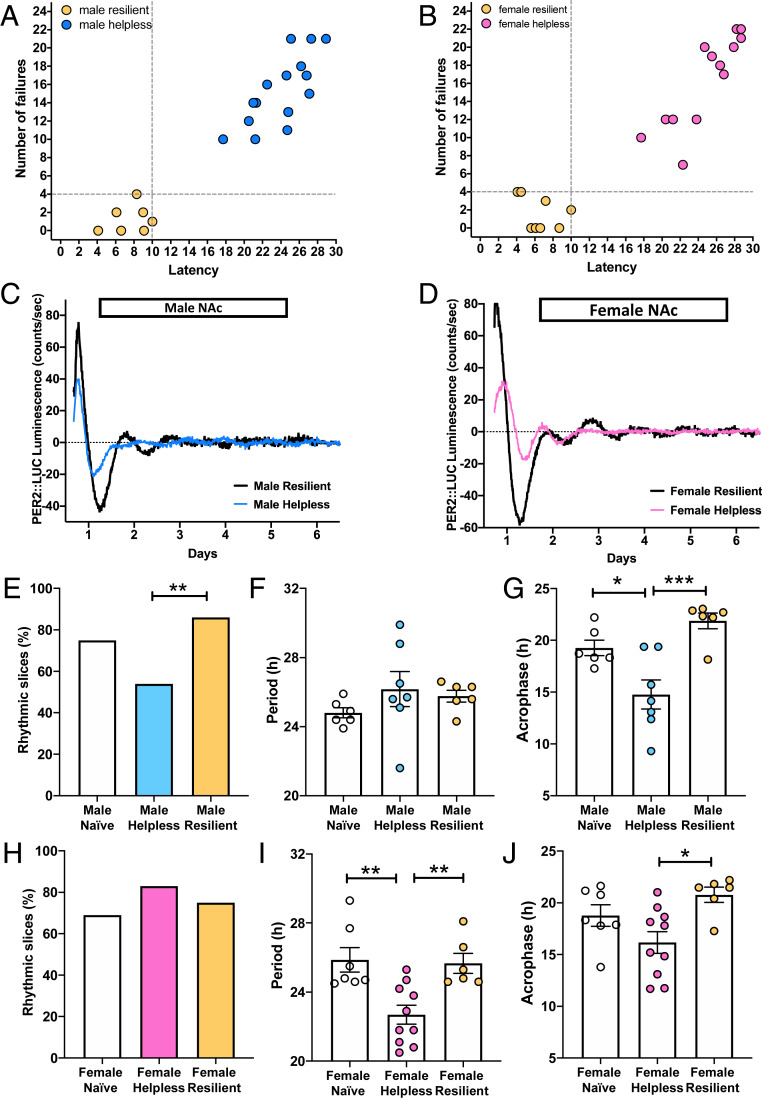
To study circadian clock dysfunction in a mouse model of depression, we first induced “learned helplessness” (LH) in mice harboring the circadian PER2::LUC reporter. This was achieved by subjecting female and male mice to two daily 1-h sessions of inescapable tail shocks (ITS) (57). One day after the second ITS session, mice underwent the tail-suspension test and subsequently the LH paradigm. These are both tests that have been validated extensively as measures of depression-like states in rodents (57, 58). Mice subjected to ITS were classified as resilient by the LH test when they had escape latencies and failures within 2 SDs of those of naïve mice. All mice with greater latency and escape failure values were defined as helpless (Fig. 1 A and B and SI Appendix, Fig. S1 A, B, D, and E). Naïve mice were not subjected to ITS. The tail-suspension test revealed changes in immobility of the animals after the ITS session. Compared to naïve and resilient mice, helpless mice showed significant differences in their total immobility time (SI Appendix, Fig. S1 C and F). Fifteen hours after the LH test, animals were killed, and PER2::LUC rhythms were measured in brain slices containing the suprachiasmatic nucleus (SCN) or the NAc.
Fig. 1.
Helpless mice show altered PER2::LUC circadian rhythms in the NAc. (A and B) Graphs show escape latency and number of escape failures of resilient mice (yellow circles), defined as those showing escape latencies shorter than 10 s and fewer than four escape failures; all other mice were defined as helpless (male: blue circles, female: pink circles). Thresholds are shown as dashed lines. (C) Representative PER2::LUC rhythms of NAc explants from male helpless (blue) and resilient (black) mice. (D) Representative PER2::LUC rhythms of NAc explants from female helpless (pink) and resilient (black) mice. (E and H) Data are shown as percentages of slices that were significantly rhythmic; **P < 0.01, Fisher’s exact test. (F and G) Data show PER2 period and acrophase in the NAc of male mice. Data are shown as means ± SEM; *P < 0.05, ***P < 0.001, one-way ANOVA with Bonferroni posttest. (I and J) Data show PER2 period and acrophase in the NAc of female mice. For I and J, data are shown as in F and G, *P < 0.05, **P < 0.01. Each circle represents one mouse.
For the SCN, the brain’s primary circadian pacemaker controlling rhythmic locomotor activity, the proportion of brain slices showing significant circadian rhythmicity was not affected by LH status (helpless vs. resilient) in either female or male mice (SI Appendix, Fig. S2 A and E), in accordance with our previous results (56). Further analyzing rhythmicity separately in males and females, we found that SCN slices from helpless and resilient female mice showed a shorter PER2 rhythm period compared to naïve female mice (SI Appendix, Fig. S2F). In females, there were no differences in SCN rhythm amplitude or acrophase (time of second PER2 peak) (SI Appendix, Fig. S2 G and H). In helpless male mice, SCN rhythm amplitude was significantly increased compared to naïve and resilient mice (SI Appendix, Fig. S2C). Resilient males also showed a significantly earlier SCN PER2 rhythm acrophase compared to helpless and naïve males, but no differences in period (SI Appendix, Fig. S2 B and D).
For the NAc, a large majority of brain explants from resilient male mice showed rhythmic oscillations (∼80%), whereas significantly fewer NAc explants from helpless male mice (∼50%) were rhythmic (Fig. 1 C and E), confirming our previous results (56). Further analyzing NAc rhythms separately in males and females, we found a shorter PER2 rhythm period in helpless females compared to naïve and resilient females (Fig. 1I), while no such difference was observed in male mice (Fig.1F). No differences in PER2 rhythm amplitude were observed for either female or male mice (SI Appendix, Fig. S3). Interestingly, in both female and male helpless mice, compared to naïve or resilient mice, the acrophase was significantly earlier (Fig. 1 G and J). Overall, these data suggest sex-specific associations between helplessness and circadian rhythm dysfunction in both the SCN and NAc, including reduced rhythmicity and earlier phase of NAc rhythms in helpless mice.
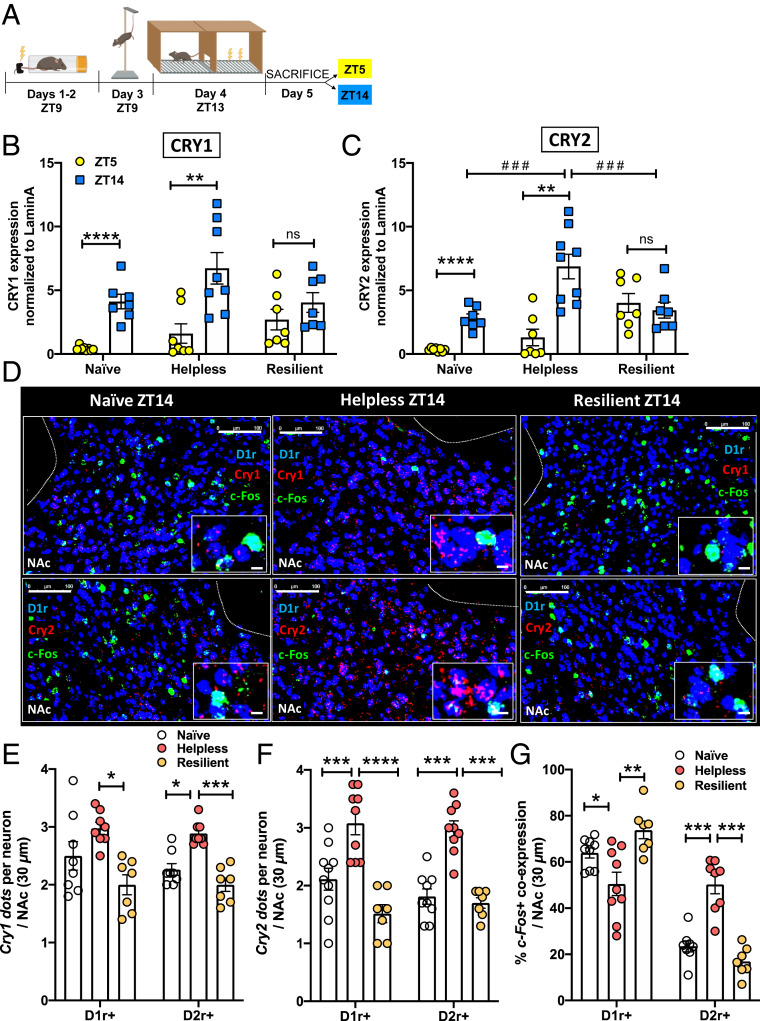
Helpless Mice Show Increased CRY Expression in the NAc at Night.
We next examined whether such altered rhythms in NAc neurons of helpless mice were associated with changes in day vs. night expression patterns of the core circadian clock components CRY1 and CRY2. Levels of CRY1 and CRY2 were measured by western blot in the NAc of naïve, resilient, and helpless mice. One day after the LH test, mice were killed at two different time points: Zeitgeber time 5 (ZT5, daytime, 5 h after lights on) and ZT14 (nighttime, 2 h after lights off). A schematic experimental design is shown in Fig. 2A. Female and male mice showed similar patterns (SI Appendix, Table S1), so CRY expression data for female and male mice are plotted together. We found that naïve and helpless mice showed significant diurnal expression patterns of CRY1 and CRY2, with the highest expression occurring during the night phase, whereas no diurnal variation was detected in resilient mice (Fig. 2 B and C). In addition, helpless mice showed a significant increase in CRY2 expression at ZT14 compared to naïve and resilient mice. Thus, resilient mice showed an altered diurnal pattern of CRY expression in the NAc, with lower expression at the beginning of the active phase.
Fig. 2.
Helpless mice show higher CRY expression and an altered pattern of neuronal activation in the NAc. (A) Timeline of the experimental design: ITS on days 1 to 2 at ZT9, tail suspension test on day 3 at ZT9, LH test on day 4 at ZT13, killing on day 5 at ZT5 or ZT14. Image created with BioRender.com. (B and C) Bar graphs showing CRY1 (B) or CRY2 (C) protein expression in the NAc of naïve, helpless, and resilient mice at ZT5 (light phase, yellow circles) and ZT14 (dark phase, blue squares). Data are shown as means ± SEM. Student t test: **P < 0.01, ****P < 0.0001; two-way ANOVA with Tukey’s multiple comparison posttest: ###P < 0.01, ns = not significant. Column factor: F(1, 15) = 20.42 ***P < 0.001, Time × Column factor: F(2, 23) = 3.53 *P < 0.05 (D) Representative confocal micrographs showing Cry1 (red, Upper) or Cry2 (red, Lower) and c-Fos (green) mRNA expression detected by RNAscope in D1R-MSNs (dark blue, D1r+) at ZT14 in the NAc of naïve, helpless, and resilient mice. c-Fos/D1r+ coexpression is shown in light blue. (Scale bars for each of the three behavioral states: Upper and Lower 100 μm; Insets are shown at higher-magnification, 10 μm.) (E) Bar graph shows Cry1 mRNA expression using semiquantitative scoring of Cry1 dots and clusters per neuron in D1R-MSNs (D1r+) or D2R-MSNs (D2r+). (F) Bar graph shows Cry2 mRNA expression using semiquantitative scoring of Cry1 dots and clusters per neuron in D1R-MSNs (D1r+) or D2R-MSNs (D2r+). (G) Bar graph shows percentage of D1r+ cells or D2r+ cells that were also c-Fos+. For E–G data are shown as means ± SEM for the NAc in a 30-µm section; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Bonferroni posttest. Each circle represents one mouse.
The NAc contains two functionally distinct neuronal cell types that control reward and motivational states: D1R-MSNs and D2R-MSNs (36, 37); therefore, we tested whether CRY is differentially expressed in these specific NAc neuronal populations. Because CRY2 expression was significantly higher in helpless mice at ZT14 in the western blot experiments, Cry1 and Cry2 mRNA expression in NAc D1R-MSNs (Fig. 2D) and D2R-MSNs (SI Appendix, Fig. S5A) was tested using the RNAscope assay in naïve, resilient, and helpless mice killed at ZT14. We first designed and validated Cry1 and Cry2 in situ hybridization probes using Cry1-knockout (Cry1−/−) and Cry2-knockout (Cry2−/−) mice. As shown in SI Appendix, Fig. S4, no fluorescent signal was detected for Cry1 in Cry1−/− mice, nor for Cry2 in Cry2−/− mice, compared to WT mice, confirming the specificity of these probes. Quantification of Cry1 and Cry2 expression in D1R- and D2R-MSNs showed that helpless mice, compared to resilient and naïve mice (Cry2 only), displayed a significant increase in Cry1 and Cry2 expression in both neuronal populations in the NAc (Fig. 2 E and F). Female and male mice showed similar patterns as shown in SI Appendix, Table S2. Given previously observed effects of cytoplasmic CRY on G protein-coupled receptors (28, 29), the increase in CRY expression we observed in the NAc at ZT14 could alter the functionality of D1 and/or D2 receptors, and neuronal activation in the NAc reward circuit at the beginning of the active phase.
Helpless Mice Show Altered c-Fos Expression in NAc D1R-MSNs and D2R-MSNs at Night.
Previous studies have shown prominent diurnal variation of extracellular DA levels in the NAc core peaking at ZT13, affecting D1 and D2 receptor activation (27). To test whether the increased CRY expression we observed in helpless mice at ZT14 correlates with changes in D1R- and D2R-MSN neuronal activation in the NAc, we monitored the expression of the immediate-early gene c-Fos, a neuronal activity indicator. Naïve, resilient, and helpless mice were killed at ZT14, and brains were then processed for RNAscope assay to quantify c-Fos expression in a cell-specific manner (Fig. 2D and SI Appendix, Fig. S5A). We found that helpless mice showed greater activation of D2R-MSNs as well as reduced activation of D1R-MSNs compared to resilient and naïve mice (Fig. 2G). Female and male mice showed similar patterns as shown in SI Appendix, Table S2. Thus, altered daily CRY expression in the NAc of helpless mice is associated with an abnormal pattern of activation of D1R- and D2R-MSNs at the beginning of the dark phase. Higher expression of CRY at ZT14 might also affect other downstream signaling pathways of D1 and D2 receptors, such as cyclic AMP response element-binding protein (CREB) activation (59). Using the western blot assay, we found that naïve and helpless mice showed similar daily expression patterns of phospho-CREB (P-CREB), with higher expression at night (ZT14) (SI Appendix, Fig. S5B). However, in resilient mice P-CREB expression is higher during the day (ZT5) (SI Appendix, Fig. S5B), indicating an altered diurnal pattern of DA signaling in the NAc.
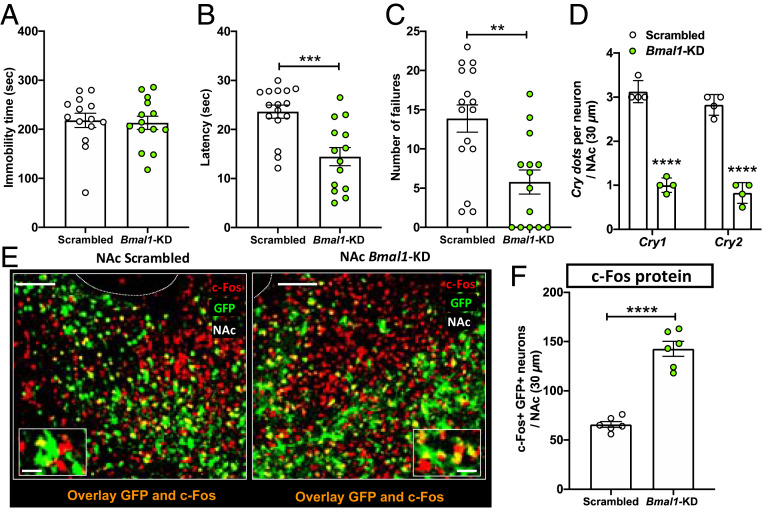
Knockdown of Bmal1 in the NAc Reduces Cry Expression, Increases Neuronal Activation, and Reduces Susceptibility to Helpless Behavior.
In the molecular circadian clock, complexes of BMAL1 and CLOCK promote the transcription of Cry and Per genes. In our previous study we found that Bmal1 knockdown (KD) in the SCN lengthens circadian period and reduces rhythm amplitude (21). We hypothesized that Bmal1 KD in the NAc would alter CRY expression at night, affecting vulnerability to helpless behavior. Mice were injected with either Bmal1-shRNA (Bmal1-KD) or scrambled shRNA (Scrambled) in the NAc. Three weeks after the injection, mice were evaluated using the tail suspension test, followed by ITS training. Prior to stressful ITS training, no differences were observed between Bmal1-KD and control mice in helpless behavior, as reflected by immobility time in the tail suspension test (Fig. 3A), suggesting that Bmal1 KD in the NAc alone does not induce behavioral despair [unlike in the SCN (21)]. Mice were then subjected to 2 d of ITS and then tested for LH. Interestingly, mice injected with Bmal1-shRNA showed a more resilient phenotype, defined by decreased latency and number of failures to escape (Fig. 3 B and C), compared to control mice injected with scrambled shRNA. Immunofluorescence experiments confirmed that Bmal1-shRNA in the NAc reduced BMAL1 protein levels by 60% (SI Appendix, Fig. S6). We then tested whether Bmal1-KD alters Cry expression at ZT14 in the NAc using RNAscope. We found significantly lower Cry1 and Cry2 mRNA expression at ZT14 in the NAc of mice that received Bmal1 shRNA, compared to mice injected with the control virus (Fig. 3D), indicating that Bmal1 down-regulation reduces Cry expression in the dark phase. Because helpless mice showed decreased NAc neuronal activation at ZT14 (Fig. 2G), we examined c-Fos protein expression in mice injected with Bmal1-shRNA and control mice at this time (Fig. 3E). As expected, NAc neurons infected with Bmal1-shRNA showed not only decreased Cry expression but also increased c-Fos expression compared to control (Fig. 3F). These data suggest that Bmal1 KD in the NAc may reduce susceptibility to helpless behavior by reducing CRY-mediated inhibition of neuronal activation.
Fig. 3.
Bmal1 KD in the NAc reduces Cry expression, increases neuronal activation, and reduces helpless behavior in early night. (A) Bar graph shows immobility time, (B) escape latency, and (C) number of escape failures of Bmal1 KD in the NAc (NAc-Bmal1-KD) green circles, compared to Scrambled sequence control (white circles). (D) Bar graph shows semiquantitative scoring of Cry mRNA expression in NAc measured at ZT14 by RNAscope. (Scrambled: males n = 7, females n = 8; Bmal1-KD: male n = 7, females n = 7). (E) Representative confocal micrographs of NAc from control mice injected with virus encoding scrambled control construct (Left) and mice injected with virus encoding Bmal1-shRNA (Right) show transduced cells marked by GFP (green) and c-Fos protein expression marked by immunolabeling (red). Overlays (orange) reveal that most Bmal1-shRNA transduced cells show increased c-Fos expression. (Scale bars: 100 μm; Inset, 20 μm.) (F) Bar graph shows quantification of c-Fos coexpression in GFP+ cells of NAc measured at ZT14 by immunofluorescence. For A–D and F data are shown as means ± SEM; Student t test, **P < 0.01, ***P < 0.001, ****P < 0.0001. Each circle represents one mouse.
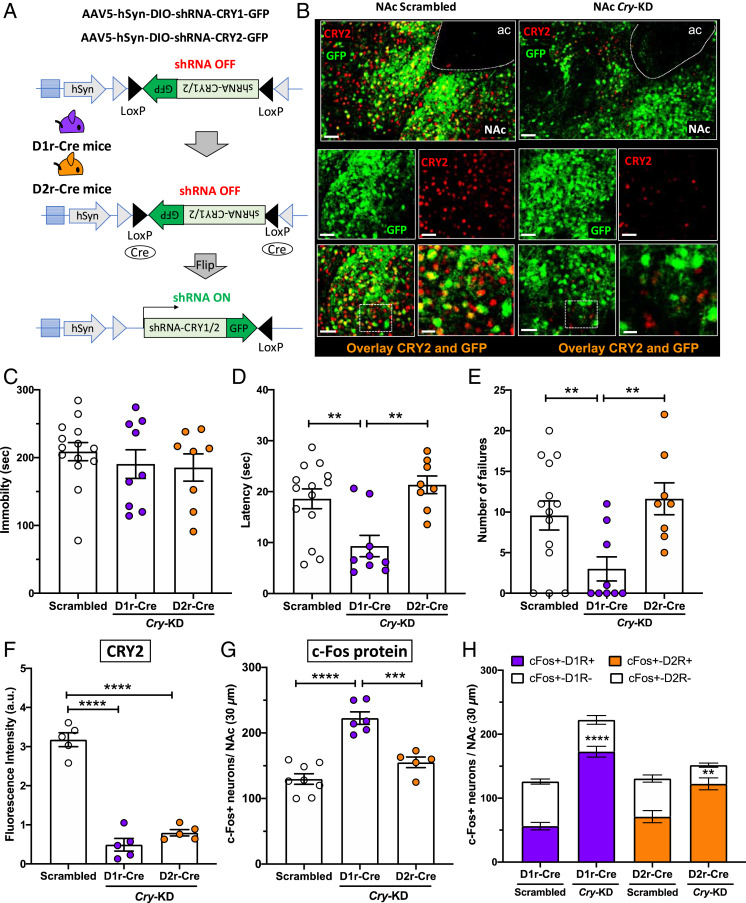
KD of Cry in NAc D1R-MSNs Increases Neuronal Activation and Reduces Susceptibility to Helpless Behavior.
To test the role of CRY in mediating helpless states in mice more directly, we knocked down both Cry1 and Cry2 in either D1R-MSNs or D2R-MSNs selectively. We achieved this by generating Cre-dependent adeno-associated virus (AAV) vectors encoding shRNA to down-regulate Cry1 and Cry2 expression in a cell-specific manner, and by injecting a mixture of both Cry1- and Cry2-specific shRNAs into the NAc of either D1r-Cre or D2r-Cre mice (Fig. 4A). Immunofluorescence staining confirmed down-regulation of CRY2 protein in the NAc of both Cre lines, with significant decreases of CRY2 expression at the injection sites (Fig. 4 B and F). (Unfortunately, there is no available antibody to detect CRY1 protein reliably in immunofluorescence experiments.) Three weeks after the AAV injections, mice underwent tests of helpless behavior. In the tail-suspension test, prior to stressful ITS training, no significant difference was observed in immobility time for D1r-Cre or D2r-Cre mice injected with Cry1 and Cry2 shRNA, compared to mice injected with control virus (Scrambled) (Fig. 4C). One day after the tail-suspension test, mice were subjected to 2 d of ITS and then tested in the LH paradigm. As shown in Fig. 4 D and E, mice with Cry shRNA targeted to D1R-MSNs showed significant decreases in escape latencies and number of escape failures compared to either mice with Cry shRNA targeted to D2R-MSNs or control mice receiving scrambled shRNA.
Fig. 4.
Cry KD in the NAc increases D1R-MSN neuronal activation and reduces helpless behavior in early night. (A) AAV expression constructs encoding GFP and an inhibitory shRNA targeting either Cry1 or Cry2 are shown before and after Cre-mediated recombination. In D1r-Cre and D2r-Cre mice, the NAc was injected with a mixture of two AAVs encoding a GFP reporter and either shRNAs for Cry KD (Cry-KD, one targeting Cry1 and the other targeting Cry2), or scrambled shRNA sequences as a control. (Scrambled D1r-Cre: males n = 3, females n = 4; Cry-KD D1r-Cre: males n = 4, females n = 5; Scrambled D2r-Cre: males n = 3, females n = 4; Cry-KD D2r-Cre: males n = 4, females n = 4). (B) Representative confocal micrographs of NAc from control mice (Left) or mice injected with Cry-shRNA (Right), showing transduced cells marked by GFP (green) and CRY2 protein detected by immunolabeling (red). The overlays show that most cells transduced with Cry-shRNA show reduced CRY2 expression. (Scale bars: 50 μm, except Bottom Right, 20 μm.) (C) Bar graph shows immobility time, (D) escape latency, and (E) number of escape failures of Cry-KD in D1r-Cre (purple circles) or Cry-KD in D2r-Cre (orange circles), compared to Scrambled sequence control (white circles). (F) Bar graph shows quantification of CRY immunofluorescence (arbitrary units), revealing an ∼70% reduction of CRY2 protein levels at ZT14, relative to control mice, in the NAc of D1r-Cre and D2r-Cre mice injected with Cry-shRNA. (G) Bar graph shows c-Fos expression at ZT14 in the NAc of D1r-Cre mice, D2r-Cre mice, relative to controls. For C–G, data are shown as means ± SEM; **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Bonferroni posttests. Each circle represents one mouse. (H) Bar graph shows the proportions of c-Fos+ neurons in D1r-Cre or D2r-Cre mice injected with virus encoding scrambled sequences (Scrambled) or Cry-shRNA (Cry-KD). Each column represents the number of c-Fos+ neurons counted in the NAc in a 30-µm section, and the colored sections show the number of c-Fos+ neurons identified as D1r-MSNs (D1R+, purple) or D2r-MSNs (D2R+, orange) by expression of the GFP reporter. The white sections show the number of c-Fos+ neurons identified as non–D1r-MSNs (D1R−) or non–D2r-MSNs (D2R−) by absence of GFP expression. Data are shown as means ± SEM, n = 4 to 6 mice for each condition. Student t test: c-Fos+D1r+ Scrambled vs. D1rCre Cry-KD ****P < 0.0001; c-Fos+D2r+ Scrambled vs. D2rCre Cry-KD **P < 0.01. For F–H, all cells within the NAc in a 30-µm section were counted.
To test whether CRY down-regulation affects NAc neuronal activation at night, c-Fos expression was evaluated in D1R- and D2R-MSNs at ZT14 in mice receiving Cry1 and Cry2 shRNA. We found that mice injected with Cry shRNA targeted to D1R-MSNs showed greater neuronal activation compared to those injected with Cry shRNA targeted to D2R-MSNs, or to control mice receiving scrambled shRNA (Fig. 4G and SI Appendix, Fig. S7A). Cell-type–specific effects on neuronal activation were tested by quantifying coexpression of c-Fos (marking neuronal activation) and green fluorescent protein (GFP, encoded by Cre-dependent AAV virus, and therefore marking D1-MSNs or D2-MSNs) (SI Appendix, Fig. S7B). This analysis revealed that targeting Cry shRNA to D1R-MSNs activated mostly D1R-MSNs, whereas targeting Cry shRNA to D2R-MSNs activated mostly D2-MSNs (Fig. 4H). These data demonstrate that knocking down Cry1 and Cry2 selectively in D1R-MSNs decreases vulnerability to stress-elicited helpless behavior without altering baseline performance in the tail suspension test, and also suggest a causal role for properly phased rhythmic Cry expression in mediating a normal circadian pattern of D1R-MSN activation in the active phase.
CRYs Interact with Gs Proteins and Inhibit D1 Receptor-Mediated Gs Protein Activation.
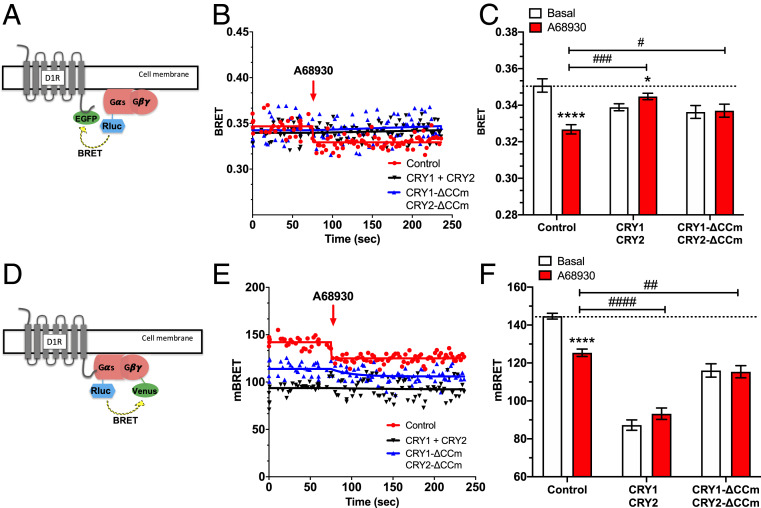
To investigate a possible mechanism by which CRYs alter D1R-MSN activation and vulnerability to helpless behavior, we used a bioluminescence resonance energy transfer (BRET) approach in Chinese hamster ovary (CHO) cells expressing a D1 receptor with its C-terminal fused to GFP (D1R-GFP) and a Gαs subunit fused to Renilla reniformis luciferase (Rluc), as indicated in Fig. 5A. We then used BRET to monitor the interaction between the two fluorescent fusion proteins (D1R-GFP and Gαs-Rluc), in the presence or absence of various CRY protein constructs. Basal and stimulated BRET signal was recorded in cells either coexpressing CRY1 and CRY2, coexpressing the cytoplasmically localized mutants CRY1-ΔCCm and CRY2-ΔCCm (60), or in control cells without CRY constructs. As expected, stimulation of the D1 receptor with the full D1 receptor agonist A68930 induced a conformational change of the D1 receptor-G protein complex, leading to a significant decrease in the detected BRET signal (Fig. 5 B and C). In the presence of CRY1 and CRY2, or the cytoplasmic CRY variants, this response to D1 receptor stimulation was abolished (Fig. 5 B and C). Thus, cytoplasmic CRY blocks D1 agonist-induced conformational rearrangement between the D1 receptor and its G protein, which may affect G protein activation.
Fig. 5.
Cytoplasmic CRYs prevent D1R-mediated Gs protein activation in CHO cells. (A) BRET experiments were performed in CHO cells coexpressing Gαs-RLuc, D1r-EGFP, and either CRYs (CRY1 and CRY2) or their cytoplasm-restricted variants (CRY1-ΔCCm and CRY2-ΔCCm). (B) BRET kinetics measured in the presence or absence (red line and dots) of CRYs (black line and dots) or cytoplasmic CRY variants (blue line and dots). After 80 s of BRET reading, the D1 DA receptor agonist A68930 (10 μM) was applied (red arrow), resulting in a significantly decreased BRET signal due to conformational rearrangement between D1r-EGFP and Gαs-RLuc. The BRET ratio was calculated as the ratio of light emitted by EGFP (530 to 570 nm) to light emitted by RLuc (370 to 470 nm). Results were expressed in mBRET units (change in BRET ratio × 1,000). (C) Bar graph showing BRET signals observed before (white) and after D1 receptor agonist stimulation (red) in the presence of either CRYs or cytoplasmic CRY variants. Data are presented as means ± SEM of four experiments. Student t test: *P < 0.05, ****P < 0.0001; two-way ANOVA with Tukey’s multiple comparison posttest: #P < 0.05, ###P < 0.001. (D) BRET measured in CHO cells coexpressing D1R, Gαs-RLuc, Gγ2-Venus, and either CRYs (CRY1 and CRY2) or their cytoplasm-restricted variants (CRY1-ΔCCm and CRY2-ΔCCm). (E) BRET kinetics measured in the presence or absence of CRYs or cytoplasmic CRY variants. After 80 s of BRET reading, the D1 DA receptor agonist A68930 (10 μM) was applied (red arrow), resulting in a significantly decreased BRET signal due to conformational rearrangement between Gαs-RLuc and Gγ2-Venus. BRET ratio was calculated and reported as above. (F) Bar graph showing BRET signals observed before (white) and after D1 receptor agonist stimulation (red) in the presence of either CRYs or cytoplasmic CRY variants. Data are presented as means ± SEM of four experiments. Student t test: ****P < 0.0001; two-way ANOVA with Tukey’s multiple comparison posttest: ##P < 0.01, ####P < 0.0001.
To more directly test the effect of CRY on Gs protein activation in response to D1 receptor agonist binding, we monitored the BRET signal induced by the conformational changes between Gαs and Gγ2 subunits before and after D1 receptor activation. For these experiments, we used the same Gαs-Rluc, but now coexpressed with Gγ2 fused to Venus (Fig. 5D) in CHO cells expressing the D1 receptor, and measured the BRET signal in the presence or absence of CRYs or cytoplasmic CRY variants. As shown in Fig. 5 E and F, the BRET signal was dramatically reduced in the presence of either CRY1 and CRY2, or CRY1-ΔCCm and CRY2-ΔCCm, even before D1 receptor agonist application. Additionally, in the presence of CRY1 and CRY2, or the cytoplasmic CRY variants, the BRET signal induced by D1 receptor activation was abolished (Fig. 5F). Overall, these data indicate that CRY alters Gs protein conformation, likely by interacting directly with the Gαs subunit, and that this interaction inhibits Gs protein activation by the D1 receptor agonist. These observations are consistent with previous evidence of CRY inhibiting Gs protein signaling (29), and reveal that cytoplasmic CRYs inhibit D1 receptor activation by altering the Gαβγ complex.
Fibroblasts from Male Depressed Patients Show Increased Cry1 Expression.
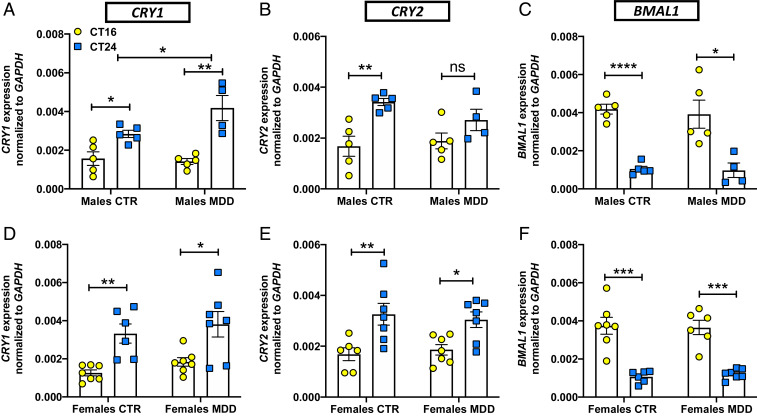
To test for altered circadian rhythmicity or Cry expression in human MDD, we examined fibroblast cell lines that were available from depressed patients. In fibroblasts from patients diagnosed with MDD and from healthy controls, we used qRT-PCR to test clock gene expression at two different time points, circadian time (CT) 16 and 24, after synchronization of circadian rhythms in culture. As expected, Cry expression was robustly CT-dependent in cells from male and female patients and controls (Fig. 6 A, B, D, and E). Bmal1 expression was also robustly rhythmic in cells from male and female patients and controls (Fig. 6 C and F). Cells from male MDD patients showed elevated Cry1 expression levels at CT24 compared to controls (Fig. 6A). Cry expression of fibroblasts from female MDD patients did not differ significantly from controls (Fig. 6 D and E). We also used the PER2::LUC reporter to test whether PER2 rhythms were altered in fibroblasts from MDD patients. No differences were observed in PER2 rhythm period (SI Appendix, Fig. S8A). Interestingly, cells from male MDD patients exhibited an elevated circadian rhythm amplitude compared to either controls or female MDD patients (SI Appendix, Fig. S8B). These data show increased Cry1 expression at dawn and suggest possible circadian rhythm dysfunction in cells from male MDD patients.
Fig. 6.
Fibroblasts from MDD patients show altered Cry1 expression. In fibroblasts from male or female healthy controls (CTR) or patients diagnosed with MDD, mRNA expression of circadian clock genes was measured by qRT-PCR at two circadian phases (CT16, CT24). Bar graphs show levels of CRY1 (A and D), CRY2 (B and E), and BMAL1 (C and F) gene expression at the indicated times. Data are normalized relative to the nonrhythmic control gene GAPDH and presented as means ± SEM of four experiments. Student t test *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; not signifincant. For A: Two-way ANOVA: Interaction F(1, 15) = 4.556, *P = 0.0497. Each circle/square represents one subject.
Discussion
In this study we demonstrate how molecular circadian clock dysfunction in the NAc can increase vulnerability to stress-induced helpless behavior in mice. Previous cellular and molecular studies of circadian clocks have provided tantalizing indications that clock gene defects may be involved in mood disorders (2, 13, 61–64). The NAc, a central component of the midbrain DA reward circuit, exhibits disturbed circadian rhythms in the postmortem brains of depressed patients (12). Furthermore, mice exposed to acute or chronic stress exhibit depression-like behavior and altered circadian rhythms in neurons of the reward circuit (56, 65). Here we demonstrate that the core circadian clock component CRY interacts with G proteins to impair DA D1 receptor activation, alters NAc neuronal activation in a cell type-specific manner predicted to reduce positive reward signaling, and increases vulnerability to stress-induced helpless behavior.
First, using a PER2::LUC reporter gene, we found a sex-specific pattern of altered circadian clock function in NAc slices from helpless mice. In male (but not female) helpless mice, fewer NAc slices were rhythmic compared to resilient mice, confirming our previous finding of less reliable rhythms (56). We found no NAc rhythm amplitude differences in helpless mice, in contrast to a previous study that found increased amplitude after unpredictable chronic mild stress (65), suggesting that acute and chronic stress may affect NAc circadian rhythms differently. We also found that free-running circadian period is shorter in NAc slices from female (but not male) helpless mice. Finally, the most consistent finding was that PER2::LUC rhythms are phased earlier in NAc slices from helpless mice, relative to resilient mice. Overall, these findings indicate that in helpless mice, NAc circadian rhythms are mistimed and (at least in males) less reliable.
Using the same approach, we found a different sex-specific pattern of altered circadian clock function in the SCN of helpless mice. In SCN slices from male (but not female) helpless and naïve mice, PER2::LUC rhythms were phased later relative to resilient mice, in contrast to the earlier phase observed in the NAc of helpless mice. SCN slices from female (but not male) helpless and resilient mice had shorter circadian periods relative to naïve mice, possibly reflecting the effects of ITS exposure, as opposed to a helpless behavioral state. As in our previous study (56), the proportion of SCN slices exhibiting circadian rhythms did not differ among naïve, helpless, and resilient mice, but rhythmic SCN slices from helpless mice did show increased amplitude in the present study. This is in contrast to a recent study finding decreased amplitude in the SCN of mice subjected to unpredictable chronic stress (65). On the other hand, Koresh et al. (66) found increased Per1 and Per2 expression in the SCN after 8 d of exposure to predator scent stress in a posttraumatic stress disorder mouse model. Just as for rhythms in the NAc, these differences among studies may reflect how acute and chronic stress treatments affect SCN circadian rhythms differently.
The disparity in circadian effects we observed in male vs. female mice, or in the NAc vs. SCN, might be related to sex differences in receptors for the stress hormones corticotropin releasing factor (CRF) and glucocorticoids (67, 68). For example, in female rats, stress increases CRF expression in the paraventricular nucleus (69, 70), which receives dense projections from the SCN (71). Greater CRF expression in the hypothalamus of female vs. male rodents can lead to greater activation of the hypothalamic–pituitary–adrenal axis, and greater glucocorticoid release (72–74). In addition, stress down-regulates more NAc genes in female mice than in males, particularly genes regulating nervous system development and function (75). Finally, high levels of CRF and glucocorticoid receptors have also been reported in the NAc (73) but not in the SCN (76), and glucocorticoids are known to shift non-SCN circadian clocks (77, 78). Thus, the increased secretion of glucocorticoids resulting from stressful ITS training could mediate the sex-specific and tissue-specific pattern of clock dysfunctions observed in our helpless mice. Further research is needed to determine the underlying mechanism of sex-specific clock alterations observed in helpless mice.
We also observed dysregulation of CRY expression in the NAc of helpless mice. We found that helpless behavior is associated with higher NAc CRY2 expression at the beginning of the nocturnal active phase (ZT14). In addition, both Cry1 and Cry2 mRNA expression in the NAc at ZT14 was higher in helpless mice compared to naïve and resilient mice. In resilient mice, NAc CRY2 expression was significantly lower at ZT14, with no difference between ZT5 and ZT14, suggesting a difference in NAc circadian rhythm phase between helpless and resilient mice. Indeed, we observed a phase shift of PER2::LUC rhythms in NAc brain slices from helpless mice. This suggests that acute stress induced by ITS alters NAc circadian phase, possibly contributing to susceptibility to helpless behavior.
To determine whether the altered CRY expression we observed in helpless mice is associated with changes in neuronal function, we investigated cell-type–specific activation of NAc MSNs by monitoring the neuronal activity marker c-Fos. In the NAc of helpless mice, we found decreased activation of D1R-MSNs but increased activation of D2R-MSNs, in accordance with previous findings of distinct, opposing roles for NAc MSN subtypes in mediating depression-like behavior in a social-defeat stress paradigm (48). Previous studies have shown that stimulation of D1R-MSNs promotes resilient behavior, whereas stimulation of D2R-MSNs promotes social avoidance after chronic social defeat stress. In addition, repeated restraint stress produces anhedonia through selective attenuation of NAc D1R-MSN excitatory synaptic strength (44). Thus, the important opposing roles of NAc D1R- and D2R-MSNs in regulating reward and motivation is well known (79), but we suggest further that normal circadian activation/inhibition of D1R-MSNs and D2R-MSNs may play a key role in modulating depression phenotypes in mice and humans. In future studies, it would be valuable to characterize the 24-h pattern of neuronal activity in the NAc in normal mice or in mouse models of depression using fiber photometry.
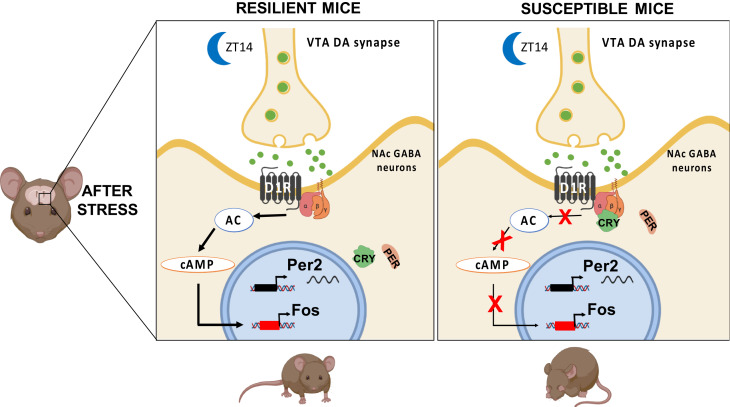
Diurnal variation of phasic DA release at postsynaptic terminals of neurons projecting from the ventral tegmental area (VTA) to the NAc (high at night and low during the day in nocturnally active rodents) (27) implies a parallel diurnal rhythm in postsynaptic receptor activation. D1 receptors, because of their relatively low affinity for DA, are activated by higher levels and phasic release of DA, whereas higher affinity D2 receptors are activated by tonic DA (80, 81). Using a foot-shock avoidance task, Wenzel et al. (82) found that activation of VTA DA neurons enhanced active avoidance in rats, and this was blocked by D1 antagonist injection into the NAc core. In addition, knocking down the DA transporter in the NAc core, which decreases DA reuptake and increases synaptic DA levels, reduces anxiety- and depression-like behaviors (83). As depression is marked by reduced avoidance of aversive stimuli (helplessness), these data imply that dysregulation of NAc D1 receptor activation could increase vulnerability to stress-induced helplessness. We propose that mistimed or weaker NAc circadian rhythmicity leads to excessive levels of CRY in NAc MSNs during the nocturnal active period of mice, and that this reduces D1 receptor activation, thereby compromising the normal daily activation of these cells by DA released from the VTA, and leading to a helpless state (Fig. 7).
Fig. 7.
Proposed mechanism for CRY’s effect on susceptibility to stress-induced depression-like behavior. Our data suggest that stressful ITS training can induce circadian clock dysfunction in the NAc, perhaps a phase shift. This clock dysfunction leads to a change in the 24-h timing of Cry expression such that CRY protein levels are higher in the early night, at the beginning of the active phase. Normal DA reward circuit function and mood regulation depend crucially on MSN activity in the NAc, regulated by synaptic input from dopaminergic neurons in the VTA. During the nocturnal active period of mice, higher levels of CRY may act on G proteins [potentially through inhibition of the adenylate cyclase pathway (28)] in D1R-MSNs of the NAc to block daily activation of D1 receptors, thereby compromising DA reward signaling and leading to depression-like behavior. Individual differences in depression-like behavior may be due, at least in part, to individual differences in prior history of stressful experience during development. Image created with BioRender.com. Adenylate cyclase, AC; cyclic adenosine monophosphate, cAMP.
As a test of this hypothesis, we used a viral-mediated approach to manipulate CRY and the circadian clock in the NAc. First, we showed that knocking down the primary positive circadian transcription factor BMAL1 in the NAc, which reduces Cry expression at night, decreases vulnerability to stress-induced helplessness. In contrast, inducing circadian disruption by repeated 6-h phase advances of the light/dark cycle was recently shown to impair active escape learning (84). Although this may seem paradoxical, a predictable consequence of normal biphasic circadian regulation of behavioral functions is that circadian manipulations may either impair (84) or enhance function (present results), depending on the nature of the circadian phase shift or disruption (85).
Next, we proceeded to test our hypothesis about CRY function in the NAc more directly. CRY1 and CRY2 are both known to be important and to have similar biochemical roles in the critical feedback inhibition of the circadian clock mechanism and in directly modulating G protein function. Because CRY1 is a stronger inhibitor than CRY2 (86), the ratio of CRY1/CRY2 determines circadian period (87), so KD (88) of Cry1 vs. Cry2 have opposite effects on period. CRY1 and CRY2 single knockouts are rhythmic, whereas double knockouts are not (86, 87, 89). Therefore, to both reduce CRY activity and weaken the circadian clock, while also avoiding the confound of opposite period effects observed in single KD, we knocked down both CRYs. Finally, as previous studies have clearly shown that D1R-MSN and D2R-MSN cells in the NAc have distinct or even opposing roles in the DA reward pathway (46–48), we knocked down both CRYs specifically and separately in D1R-MSN and D2R-MSN cells.
Using this cell-type–specific approach targeting both CRYs, we found that down-regulation of CRY in D1R-MSNs reduces vulnerability to stress-induced helplessness, whereas down-regulation of CRY in D2R-MSNs does not. These data are consistent with a previous study showing that the Afterhours mutation of Fbxl3, which reduces Cry1 mRNA [at least in the SCN (90, 91)], reduces depression-like behavior in mice (16). Mood phenotypes have also been reported for Cry knockout mice [reviewed in Vadnie and McClung (20)], but global Cry knockout could affect brain development and many brain regions other than the NAc, so these studies are not comparable to our knockdown of CRY specifically in the NAc.
Given that helpless mice show higher CRY in the NAc and reduced D1R-MSN activation at ZT14, we predicted that D1R-MSN–specific CRY KD would lead to increased D1R-MSN neuronal activation at this time. Indeed, we observed higher D1R-MSN activation at ZT14 when we down-regulated CRY expression selectively in D1R+ neurons; similarly, we observed higher D2R-MSN activation at ZT14 when we down-regulated CRY expression selectively in D2R+ neurons. Using a similar approach, Parekeh and McClung (14) recently reported cell-type–specific actions of the circadian transcription factor NPAS2 in regulating excitatory synaptic activity in D1R-MSNs and cocaine reward-related behavior. These results directly implicate circadian clock components as important regulators of NAc MSN physiology, thereby affecting mood and reward-related behavior.
Circadian clock genes directly or indirectly regulate the transcription of many clock-controlled genes critical for neuron physiology and metabolism (10, 11), but cytoplasmic CRY also interacts with G proteins to modulate receptor-mediated signal transduction pathways (28, 29). Biochemical reconstitution studies have shown previously that CRYs interact directly with Gαs subunits, regulating fasting gluconeogenesis in the liver and inhibiting glucagon-mediated stimulation of cAMP signaling (29). In addition, cytoplasmic CRY has been shown to interact with Gαq subunits to inhibit Ca2+ signaling, whereas no interactions with Gαi were detected (60). These findings suggest a mechanism in which CRY might selectively affects Gs and Gq signaling pathways in the NAc to affect mood. Therefore, we used a BRET approach to test whether CRYs can interact with Gαs to modulate D1 receptor signaling. In CHO cells, we monitored conformational rearrangements at the interfaces between the D1 receptor and Gαs, and between G protein subunits, before and after D1 receptor agonist application. We found that cytoplasmic CRY abolishes D1 agonist-induced conformational reorganization of the D1 receptor–G protein complex. Specifically, we found that CRY stabilizes the heterotrimeric state of the Gs protein and prevents the rearrangement of Gαβγ subunits induced by D1 receptor activation. Thus, CRY inhibits D1R-induced Gs protein activation, likely by interacting directly with the Gs protein; future studies should explore the effects of CRY on D1 receptor signaling in neuronal cells.
Overall, the picture that emerges is that in a helpless state, abnormally high levels of CRY in NAc D1R-MSNs at the beginning of the active phase (ZT14 in mice, early morning in humans) may block D1R-induced Gs protein activation by DA released from presynaptic VTA terminals, thereby compromising neuronal responses to the normal diurnal elevation of DA at this time. Recent studies indicate that novel phosphorylation signaling pathways downstream of the D1 receptor, such as Rap1 or Rho kinase pathways, may play a crucial role in regulating NAc neuronal excitability and emotional behaviors (92, 93). In addition to DA receptors, D1R-MSNs also express M4 cholinergic receptors, dynorphin, and substance P, whereas D2R-MSNs express A2A adenosine receptors, adenosine, enkephalin, and neurotensin. The receptors for all of these neurotransmitters potentially couple to Gs proteins (94), and could also be modulated by CRY. Adenosine, in particular, is well known for balancing neuronal excitability as a neuromodulator (95), so perhaps the increased activation of D2R-MSNs we observed in helpless mice could be related to an effect of CRY on the adenosine receptor pathway as well. However, little is known about how the D2 and A2A receptor signal pathways interact. Future studies should address how CRY modulates different G protein-coupled receptor signal pathways in the NAc and other brain regions.
Our observations of altered circadian rhythm in helpless mice suggest that genetically or epigenetically determined circadian clock defects might also be present in depressed humans; to explore this possibility, we measured Per2-Luc circadian rhythms and clock gene expression in fibroblasts from MDD patients. We found that cells from male MDD patients showed higher rhythm amplitude and higher CRY1 expression at CT24 (dawn) versus control male subjects, whereas no such differences were observed in females. However, cells from female MDD patients had lower amplitude compared to cells from male MDD patients. Possibly, such weaker circadian rhythmicity may contribute to the well-known higher incidence of depression in women (96). Previous studies have found that polymorphisms of CRY1 are associated with a diagnosis of MDD, and variants of CRY2 with bipolar disorder (62–64). These data suggest the possibility that circadian rhythms in human fibroblasts might be a useful biomarker of MDD. More faithful characterization of circadian dysregulation in the brains of MDD patients will require further studies of mouse models in parallel with stem cell-derived neurons from a larger number of MDD and control subjects.
Clinical therapies for depression that alter circadian clocks, such as bright light therapy and sleep deprivation, have proven effective in MDD as well as in seasonal depression (5, 97). Interestingly, the clinically well-characterized antidepressant fluoxetine induces a phase advance in firing rate rhythms of SCN neurons (98). Previous findings of circadian abnormalities in postmortem human brains of depressed patients and in mouse models (99), combined with our results in the present study, support the hypothesis that circadian rhythm disruptions or phase abnormalities in specific neuron types and brain regions may play a key role in the pathophysiology of MDD (2, 13). In a well-validated mouse model of depression, our data reveal a causal role for CRY, a core component of the circadian clock, in regulating the midbrain dopaminergic reward system, which ultimately supports a mechanistic link between the circadian clock and vulnerability to stress-induced helplessness. Thus, targeting circadian rhythms via compounds modulating CRY activity or abundance (100) might be a promising strategy for future antidepressant drugs.
Materials and Methods
Mice.
Mouse studies were conducted in accordance with regulations of the Institutional Animal Care and Use Committee at University of California, San Diego. Experiments involved male and female adult mice (9- to 16-wk-old), maintained in 12:12 light/dark cycles (12-h light, 12-h dark) with food and water available ad libitum. Complete information is provided in SI Appendix.
Brain Slice Culture and PER2::LUC Measurements.
After all behavioral tests were complete, PER2::LUC mice were anesthetized with isoflurane and killed by cervical dislocation, and organotypic NAc or SCN explants were prepared as described previously (56). Complete information is provided in SI Appendix.
Western Blots.
Brain slices containing NAc were collected from naïve, resilient, and helpless female and male mice 5 h after lights on (ZT5) and 2 h after lights off (ZT14) in the mouse colony. Following collection, brain slices were rinsed briefly in PBS prior to nuclear fractionation, as described previously (101). Nuclear lysates were collected and equilibrated prior to Western blot analysis. Total cell lysates (30 to 50 μg) or nuclear extracts (3 to 5 μg) were separated by SDS/PAGE and transferred to polyvinylidine difluoride membranes. Proteins were detected by standard Western blotting procedures. CRY antibodies anti-Cry1–CT and anti-Cry2–CT were specific to CRY1 and CRY2 cytoplasmic tails, as previously described (102); Lamin-A antibody (L1293) was purchased from Sigma. Western blot quantification was performed using the background subtraction method and ImageJ software (National Institutes of Health).
Immunohistochemistry.
Mice for immunocytochemistry experiments were deeply anesthetized with Xylazine/Ketamine. When no response to a tail/toe pinch was present, mice were transcardially perfused with 1% PBS solution first, followed by 4% paraformaldehyde solution to fix the brain tissue (103). The brain was then removed from the skull and kept in a 30% sucrose solution until use. Frozen brains were sectioned (30 μm) with a standard Leica Cryostat (CM1860). Complete methods are described in SI Appendix.
RNAscope.
RNAscope in situ hybridization (Advanced Cell Diagnostics) for Cry1, Cry2, c-Fos, D1r, and D2r mRNA was performed following the manufacturer’s instructions. Complete RNAscope methods are described in SI Appendix.
Virus Design.
AAV with cDNA encoding Cry1 and Cry2 shRNA were constructed by VectorBuilder. The vector pAAV[Exp]-mU6_TATA-lox-CMV > EGFPTATA-lox: shRNA was used to produce Cre-dependent KD of Cry1 and Cry2. Target sequences used for Cry1 (104) were #13: GCCAAGTGTTTGATAGGAG, and #15: GCGGTTGCCTGTTTCCTGA. Target sequences for Cry2 (104) were #20: GGTTCCTACTGCAATCTCT, and #19: GAATTCGCGTCTGTTTGTA. AAV vectors and sequences for Bmal1 and scrambled shRNA were described previously (21). Viral injections are described in SI Appendix.
Data Analysis.
Statistical analysis was carried out with GraphPad Prism (GraphPad Software). Statistical tests for each study are indicated in the figure legends. Data were checked for normal distribution and homogeneous variance. For normally distributed data, a parametric test was used (one-way ANOVA, two-way ANOVA, or Student’s t test). If the data were not normally distributed, a nonparametric test was used (Mann–Whitney).
Data Availability.
Protocols and data are made available in the main text and in the SI Appendix.
Supplementary Material
Acknowledgments
Human fibroblasts from depressed patients were kindly supplied by Drs. Bruce Cohen (McLean Hospital and Harvard Medical School) and Richard Shelton (Vanderbilt University). G protein constructs for bioluminescence resonance energy transfer were kind gifts of Prof. Bernhard Bettler (University of Basel, Switzerland). Cytoplasmic Cry constructs were gifts of Dr. Mark Montminy (Salk Institute). We thank Dr. Paula Desplats (University of California, San Diego) for use of the confocal microscope. This work was supported by a Veterans Affairs Merit Award (I01 BX001146), a University of California, San Diego Academic Senate Grant (RQ272R), and a Brain and Behavior Research Foundation National Alliance for Research on Schizophrenia and Depression Young Investigator Award (to D.K.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000258117/-/DCSupplemental.
References
- 1.Kronfeld-Schor N., Einat H., Circadian rhythms and depression: Human psychopathology and animal models. Neuropharmacology 62, 101–114 (2012). [DOI] [PubMed] [Google Scholar]
- 2.McCarthy M. J., Welsh D. K., Cellular circadian clocks in mood disorders. J. Biol. Rhythms 27, 339–352 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Karatsoreos I. N., Links between circadian rhythms and psychiatric disease. Front. Behav. Neurosci. 8, 162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteleone P., Maj M., The circadian basis of mood disorders: Recent developments and treatment implications. Eur. Neuropsychopharmacol. 18, 701–711 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Golden R. N., et al. , The efficacy of light therapy in the treatment of mood disorders: A review and meta-analysis of the evidence. Am. J. Psychiatry 162, 662 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Lieverse R., et al. , Bright light in elderly subjects with nonseasonal major depressive disorder: A double blind randomised clinical trial using early morning bright blue light comparing dim red light treatment. Trials 9, 48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J. C., et al. , Rapid and sustained antidepressant response with sleep deprivation and chronotherapy in bipolar disorder. Biol. Psychiatry 66, 298–301 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Boivin D. B., et al. , Complex interaction of the sleep-wake cycle and circadian phase modulates mood in healthy subjects. Arch. Gen. Psychiatry 54, 145–152 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Mohawk J. A., Takahashi J. S., Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 34, 349–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J., Wang H., Liu Y., Shao C., Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 4, e1000193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L., The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J. Z., et al. , Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 110, 9950–9955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan R. W., McClung C. A., Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 20, 49–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parekh P. K., McClung C. A., Circadian mechanisms underlying reward-related neurophysiology and synaptic plasticity. Front. Psychiatry 6, 187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roybal K., et al. , Mania-like behavior induced by disruption of CLOCK. Proc. Natl. Acad. Sci. U.S.A. 104, 6406–6411 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keers R., et al. , Reduced anxiety and depression-like behaviours in the circadian period mutant mouse afterhours. PLoS One 7, e38263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bundel D., Gangarossa G., Biever A., Bonnefont X., Valjent E., Cognitive dysfunction, elevated anxiety, and reduced cocaine response in circadian clock-deficient cryptochrome knockout mice. Front. Behav. Neurosci. 7, 152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer S., et al. , Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur. J. Neurosci. 37, 242–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savalli G., et al. , Anhedonic behavior in cryptochrome 2-deficient mice is paralleled by altered diurnal patterns of amygdala gene expression. Amino Acids 47, 1367–1377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vadnie C. A., McClung C. A., Circadian rhythm disturbances in mood disorders: Insights into the role of the suprachiasmatic nucleus. Neural Plast. 2017, 1504507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landgraf D., et al. , Genetic disruption of circadian rhythms in the suprachiasmatic nucleus causes helplessness, behavioral despair, and anxiety-like behavior in mice. Biol. Psychiatry 80, 827–835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClung C. A., et al. , Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc. Natl. Acad. Sci. U.S.A. 102, 9377–9381 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb I. C., et al. , Diurnal variations in natural and drug reward, mesolimbic tyrosine hydroxylase, and clock gene expression in the male rat. J. Biol. Rhythms 24, 465–476 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Chung S., et al. , Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 157, 858–868 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Logan R. W., et al. , NAD+ cellular redox and SIRT1 regulate the diurnal rhythms of tyrosine hydroxylase and conditioned cocaine reward. Mol. Psychiatry 24, 1668–1684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampp G., et al. , Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr. Biol. 18, 678–683 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Ferris M. J., et al. , Dopamine transporters govern diurnal variation in extracellular dopamine tone. Proc. Natl. Acad. Sci. U.S.A. 111, E2751–E2759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang E. E., et al. , Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatori M., Panda S., CRY links the circadian clock and CREB-mediated gluconeogenesis. Cell Res. 20, 1285–1288 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Shirayama Y., Chaki S., Neurochemistry of the nucleus accumbens and its relevance to depression and antidepressant action in rodents. Curr. Neuropharmacol. 4, 277–291 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nestler E. J., Role of the Brain’s Reward Circuitry in Depression: Transcriptional Mechanisms (Elsevier Inc., ed. 1, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnan V., Nestler E. J., The molecular neurobiology of depression. Nature 455, 894–902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo S. J., Nestler E. J., The brain reward circuitry in mood disorders. Nat. Rev. Neurosci. 14, 609–625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heiman M., et al. , A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobo M. K., Karsten S. L., Gray M., Geschwind D. H., Yang X. W., FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 9, 443–452 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Gerfen C. R., The neostriatal mosaic: Multiple levels of compartmental organization. J. Neural Transm. Suppl. 36, 43–59 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Nicola S. M., The nucleus accumbens as part of a basal ganglia action selection circuit. Psychopharmacology 191, 521–550 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Smith R. J., Lobo M. K., Spencer S., Kalivas P. W., Cocaine-induced adaptations in D1 and D2 accumbens projection neurons (a dichotomy not necessarily synonymous with direct and indirect pathways). Curr. Opin. Neurobiol. 23, 546–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kupchik Y. M., et al. , Coding the direct/indirect pathways by D1 and D2 receptors is not valid for accumbens projections. Nat. Neurosci. 18, 1230–1232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albin R. L., Young A. B., Penney J. B., The functional anatomy of basal ganglia disorders. Trends Neurosci. 12, 366–375 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Maia T. V., Frank M. J., From reinforcement learning models to psychiatric and neurological disorders. Nat. Neurosci. 14, 154–162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kravitz A. V., Kreitzer A. C., Striatal mechanisms underlying movement, reinforcement, and punishment. Physiology 27, 167–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenz J. D., Lobo M. K., Optogenetic insights into striatal function and behavior. Behav. Brain Res. 255, 44–54 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Lim B. K., Huang K. W., Grueter B. A., Rothwell P. E., Malenka R. C., Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature 487, 183–189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlezon W. A. Jr, Thomas M. J., Biological substrates of reward and aversion: A nucleus accumbens activity hypothesis. Neuropharmacology 56 (suppl. 1), 122–132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobo M. K., Nestler E. J., The striatal balancing act in drug addiction: Distinct roles of direct and indirect pathway medium spiny neurons. Front. Neuroanat. 5, 41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freeze B. S., Kravitz A. V., Hammack N., Berke J. D., Kreitzer A. C., Control of basal ganglia output by direct and indirect pathway projection neurons. J. Neurosci. 33, 18531–18539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francis T. C., et al. , Nucleus accumbens medium spiny neuron subtypes mediate depression-related outcomes to social defeat stress. Biol. Psychiatry 77, 212–222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrotti L. I., et al. , Induction of deltaFosB in reward-related brain structures after chronic stress. J. Neurosci. 24, 10594–10602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larson E. B., et al. , Striatal regulation of ΔFosB, FosB, and cFos during cocaine self-administration and withdrawal. J. Neurochem. 115, 112–122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vialou V., et al. , Differential induction of FosB isoforms throughout the brain by fluoxetine and chronic stress. Neuropharmacology 99, 28–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobo M. K., et al. , ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J. Neurosci. 33, 18381–18395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young S. T., Porrino L. J., Iadarola M. J., Cocaine induces striatal c-fos-immunoreactive proteins via dopaminergic D1 receptors. Proc. Natl. Acad. Sci. U.S.A. 88, 1291–1295 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerfen C. R., Keefe K. A., Gauda E. B., D1 and D2 dopamine receptor function in the striatum: Coactivation of D1- and D2-dopamine receptors on separate populations of neurons results in potentiated immediate early gene response in D1-containing neurons. J. Neurosci. 15, 8167–8176 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinberg E. E., et al. , Positive reinforcement mediated by midbrain dopamine neurons requires D1 and D2 receptor activation in the nucleus accumbens. PLoS One 9, e94771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landgraf D., Long J. E., Welsh D. K., Depression-like behaviour in mice is associated with disrupted circadian rhythms in nucleus accumbens and periaqueductal grey. Eur. J. Neurosci. 43, 1309–1320 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Landgraf D., Long J., Der-Avakian A., Streets M., Welsh D. K., Dissociation of learned helplessness and fear conditioning in mice: A mouse model of depression. PLoS One 10, e0125892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castagné V., Moser P., Roux S., Porsolt R. D., Rodent models of depression: Forced swim and tail suspension behavioral despair tests in rats and mice. Curr. Protoc. Neurosci. Chap 8, Unit 8.10A (2011). [DOI] [PubMed] [Google Scholar]
- 59.Nestler E. J., Carlezon W. A. Jr, The mesolimbic dopamine reward circuit in depression. Biol. Psychiatry 59, 1151–1159 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Pongsawakul P., ”Regulation of Second Messenger Pathways by Cryptochrome,” PhD thesis, University of California, San Diego, CA (2013).
- 61.Soria V., et al. , Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 35, 1279–1289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hua P., et al. , Cry1 and Tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J. Affect. Disord. 157, 100–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovanen L., Donner K., Kaunisto M., Partonen T., PRKCDBP (CAVIN3) and CRY2 associate with major depressive disorder. J. Affect. Disord. 207, 136–140 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Buoli M., et al. , The role of clock genes in the etiology of Major Depressive Disorder: Special Section on “Translational and Neuroscience Studies in Affective Disorders”. Section Editor, Maria Nobile MD, PhD. This Section of JAD focuses on the relevance of translational and neuroscience studies in providing a better understanding of the neural basis of affective disorders. The main aim is to briefly summaries relevant research findings in clinical neuroscience with particular regards to specific innovative topics in mood and anxiety disorders. J. Affect. Disord. 234, 351–357 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Logan R. W., et al. , Chronic stress induces brain region-specific alterations of molecular rhythms that correlate with depression-like behavior in mice. Biol. Psychiatry 78, 249–258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koresh O., et al. , The long-term abnormalities in circadian expression of Period 1 and Period 2 genes in response to stress is normalized by agomelatine administered immediately after exposure. Eur. Neuropsychopharmacol. 22, 205–221 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Bangasser D. A., Sex differences in stress-related receptors: “Μicro” differences with “macro” implications for mood and anxiety disorders. Biol. Sex Differ. 4, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howerton A. R., et al. , Sex differences in corticotropin-releasing factor receptor-1 action within the dorsal raphe nucleus in stress responsivity. Biol. Psychiatry 75, 873–883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakase S., Kitayama I., Soya H., Hamanaka K., Nomura J., Increased expression of magnocellular arginine vasopressin mRNA in paraventricular nucleus of stress-induced depression-model rats. Life Sci. 63, 23–31 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Sterrenburg L., et al. , Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One 6, e28128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vrang N., Larsen P. J., Mikkelsen J. D., Direct projection from the suprachiasmatic nucleus to hypophysiotrophic corticotropin-releasing factor immunoreactive cells in the paraventricular nucleus of the hypothalamus demonstrated by means of Phaseolus vulgaris-leucoagglutinin tract tracing. Brain Res. 684, 61–69 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Iwasaki-Sekino A., Mano-Otagiri A., Ohata H., Yamauchi N., Shibasaki T., Gender differences in corticotropin and corticosterone secretion and corticotropin-releasing factor mRNA expression in the paraventricular nucleus of the hypothalamus and the central nucleus of the amygdala in response to footshock stress or psychological stress in rats. Psychoneuroendocrinology 34, 226–237 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Cummings S., Elde R., Ells J., Lindall A., Corticotropin-releasing factor immunoreactivity is widely distributed within the central nervous system of the rat: An immunohistochemical study. J. Neurosci. 3, 1355–1368 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sterrenburg L., et al. , Sex-dependent and differential responses to acute restraint stress of corticotropin-releasing factor-producing neurons in the rat paraventricular nucleus, central amygdala, and bed nucleus of the stria terminalis. J. Neurosci. Res. 90, 179–192 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Hodes G. E., et al. , Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to subchronic variable stress. J. Neurosci. 35, 16362–16376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balsalobre A., et al. , Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Kiessling S., Eichele G., Oster H., Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J. Clin. Invest. 120, 2600–2609 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schibler U., et al. , Clock-Talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 80, 223–232 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Li Z., et al. , Cell-type-specific afferent innervation of the nucleus accumbens core and shell. Front. Neuroanat. 12, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dreyer J. K., Herrik K. F., Berg R. W., Hounsgaard J. D., Influence of phasic and tonic dopamine release on receptor activation. J. Neurosci. 30, 14273–14283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yapo C., et al. , Detection of phasic dopamine by D1 and D2 striatal medium spiny neurons. J. Physiol. 595, 7451–7475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wenzel J. M., et al. , Phasic dopamine signals in the nucleus accumbens that cause active avoidance require endocannabinoid mobilization in the midbrain. Curr. Biol. 28, 1392–1404.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bahi A., Dreyer J.-L., Dopamine transporter (DAT) knockdown in the nucleus accumbens improves anxiety- and depression-related behaviors in adult mice. Behav. Brain Res. 359, 104–115 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Daut R. A., et al. , Circadian misalignment has differential effects on affective behavior following exposure to controllable or uncontrollable stress. Behav. Brain Res. 359, 440–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fernandez F., et al. , Circadian rhythm. Dysrhythmia in the suprachiasmatic nucleus inhibits memory processing. Science 346, 854–857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khan S. K., et al. , Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J. Biol. Chem. 287, 25917–25926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y., Xiong W., Zhang E. E., The ratio of intracellular CRY proteins determines the clock period length. Biochem. Biophys. Res. Commun. 472, 531–538 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Zhang E. E., et al. , A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139, 199–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van der Horst G. T. J., et al. , Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 90.Liu A. C., et al. , Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Godinho S. I. H., et al. , The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Fox M. E., et al. , Dendritic remodeling of D1 neurons by RhoA/Rho-kinase mediates depression-like behavior. Mol. Psychiatry, 10.1038/s41380-018-0211-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang X., et al. , Balance between dopamine and adenosine signals regulates the PKA/Rap1 pathway in striatal medium spiny neurons. Neurochem. Int. 122, 8–18 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Nagai T., Yoshimoto J., Kannon T., Kuroda K., Kaibuchi K., Phosphorylation signals in striatal medium spiny neurons. Trends Pharmacol. Sci. 37, 858–871 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Nam H. W., Bruner R. C., Choi D. S., Adenosine signaling in striatal circuits and alcohol use disorders. Mol. Cells 36, 195–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Labonté B., et al. , Sex-specific transcriptional signatures in human depression. Nat. Med. 23, 1102–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bunney B. G., Bunney W. E., Mechanisms of rapid antidepressant effects of sleep deprivation therapy: Clock genes and circadian rhythms. Biol. Psychiatry 73, 1164–1171 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Sprouse J., Braselton J., Reynolds L., Fluoxetine modulates the circadian biological clock via phase advances of suprachiasmatic nucleus neuronal firing. Biol. Psychiatry 60, 896–899 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Edgar N., McClung C. A., Major depressive disorder: A loss of circadian synchrony? BioEssays 35, 940–944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dong Z., et al. , Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 9, 1556–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lamia K. A., et al. , Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vaughan M., et al. , Phosphorylation of CRY1 serine 71 alters voluntary activity but not circadian rhythms in vivo. J. Biol. Rhythms 34, 401–409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gage G. J., Kipke D. R., Shain W., Whole animal perfusion fixation for rodents. J. Vis. Exp., 3564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ramanathan C., et al. , Cell type-specific functions of period genes revealed by novel adipocyte and hepatocyte circadian clock models. PLoS Genet. 10, e1004244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protocols and data are made available in the main text and in the SI Appendix.