Significance
Our study establishes TiPARP as a turning-off mechanism for a number of important transcription factors, including HIF-1, c-Myc, and estrogen receptor. The study also establishes that TiPARP forms nuclear condensates in an ADP ribosylation-dependent manner, which provides important insights to understand how condensates formation is regulated by protein posttranslational modifications. On the therapeutic front, our study suggests that small molecules that activate TiPARP can be anticancer agents.
Keywords: HIF-1, ADP-ribosylation, nuclear condensates, TiPARP, ubiquitination
Abstract
Precisely controlling the activation of transcription factors is crucial for physiology. After a transcription factor is activated and carries out its transcriptional activity, it also needs to be properly deactivated. Here, we report a deactivation mechanism of HIF-1 and several other oncogenic transcription factors. HIF-1 promotes the transcription of an ADP ribosyltransferase, TiPARP, which serves to deactivate HIF-1. Mechanistically, TiPARP forms distinct nuclear condensates or nuclear bodies in an ADP ribosylation-dependent manner. The TiPARP nuclear bodies recruit both HIF-1α and an E3 ubiquitin ligase HUWE1, which promotes the ubiquitination and degradation of HIF-1α. Similarly, TiPARP promotes the degradation of c-Myc and estrogen receptor. By suppressing HIF-1α and other oncogenic transcription factors, TiPARP exerts strong antitumor effects both in cell culture and in mouse xenograft models. Our work reveals TiPARP as a negative-feedback regulator for multiple oncogenic transcription factors, provides insights into the functions of protein ADP-ribosylation, and suggests activating TiPARP as an anticancer strategy.
ADP-ribosylation is a reversible protein posttranslational modification (PTM) that modifies substrate proteins with a single or multiple ADP-ribosyl groups (1). Intracellular ADP-ribosylation is catalyzed by the ADP-ribosyltransferase diphtheria toxin-like (ARTDs), commonly known as poly-ADP-ribose polymerases (PARPs) (2). Compared to poly-ADP-ribosylation, the function of intracellular mono-ADP-ribosylation is less understood. Nevertheless, it is becoming evident that mono-ADP-ribosylation modulates important signaling pathways, and it has been linked to numerous diseases, including inflammation, diabetes, neurodegeneration, and cancer (3–7). Tetrachlorodibenzo-p-dioxin (TCDD)-inducible poly(ADP-ribose) polymerase (TiPARP) (also known as PARP7 or ARTD14) is a mono-ADP-ribosyltransferase (8). It was first identified as a target gene of aryl hydrocarbon receptor (AHR) in response to TCDD (9, 10). TiPARP regulates transcriptional activity of AHR and liver X receptor via ADP-ribosylation (8, 11). However, the detailed function of TiPARP and its role in modulating transcription was not well understood.
Transcription factors need to be tightly regulated for normal physiology, and many transcription factors are known to be activated only under specific conditions. One paradigm for this is the regulation of hypoxia-inducible factor 1 (HIF-1). HIF-1 is a key regulator of oxygen homeostasis that mediates adaptive responses to changes in oxygenation through transcriptional activation of genes involved in glucose metabolism and cell survival (12, 13). HIF-1 consists of two subunits (α and β) and HIF-1α is hydroxylated and degraded under normoxia conditions, but accumulates under hypoxia conditions to turn on HIF-1. However, whether activated HIF-1 is turned off in the same oxygen-dependent mechanism is not known.
Here, we demonstrate that TiPARP is a target gene of HIF-1 and serves as a negative regulator to deactivate HIF-1. Mechanistically, TiPARP forms distinct nuclear condensates that recruits both HIF-1α and E3 ubiquitin ligases in an ADP-ribosylation-dependent manner. By down-regulating HIF-1 signaling, TiPARP impedes cell growth and metabolic reprogramming in colon and breast cancers. TiPARP similarly regulates other important transcription factors, including c-Myc and estrogen receptor (ER). Our work reveals TiPARP and ADP-ribosylation as a regulatory mechanism that deactivates certain oncogenic transcription factors and provides important insights into the functions of protein ADP-ribosylation.
Results
TiPARP Is a Direct Target Gene of HIF.
TiPARP was previously characterized as a TCDD-responsive gene regulated by AHR (9, 10). We were interested in finding other physiological conditions that could potentially induce TiPARP expression and noticed that AHR is similar to hypoxia-inducible factor 1 α subunit (HIF-1α). The transcriptional activity of both AHR and HIF-1α require their bindings with the coactivator HIF-1β (also known as aryl hydrocarbon receptor nuclear translocator [ARNT]), as well as the recognition of GCGTG core sequence on target genes (14–16). Structurally, they both contain the basic helix–loop–helix (bHLH)-PAS motif that is essential for their heterodimerization with HIF-1β (15, 16). The similarity between AHR and HIF-1α prompted us to investigate the connection between HIF-1 and TiPARP.
We identified a potential hypoxia response element upstream of TiPARP exon 1 (Fig. 1A), implying that HIFs could control TiPARP expression. We examined the mRNA level of TiPARP under hypoxia by real-time quantitative PCR (qRT-PCR), which is a well-established model for studying HIFs function. Indeed, TiPARP mRNA was significantly up-regulated under hypoxia (1% O2) in both HCT116 and MCF-7 cell lines (Fig. 1B). Additionally, TiPARP mRNA level was up-regulated by the treatment of hypoxia-mimetic agents dimethyloxalylglycine (DMOG) and desferrioxamine (DFO) (SI Appendix, Fig. S1 A, Left), or the overexpression of HIF-1α (SI Appendix, Fig. S1 A, Right) or HIF-2α (SI Appendix, Fig. S1B). Western blot analysis of hypoxic cells also showed a significant increase (∼10- to 20-fold) in the endogenous protein level of TiPARP upon treatments of hypoxia or hypoxia-mimetic agents (Fig. 1C and SI Appendix, Fig. S1C). We also examined several other members of the PARP family. The gene expression of other PARPs were not increased under hypoxia (SI Appendix, Fig. S1D).
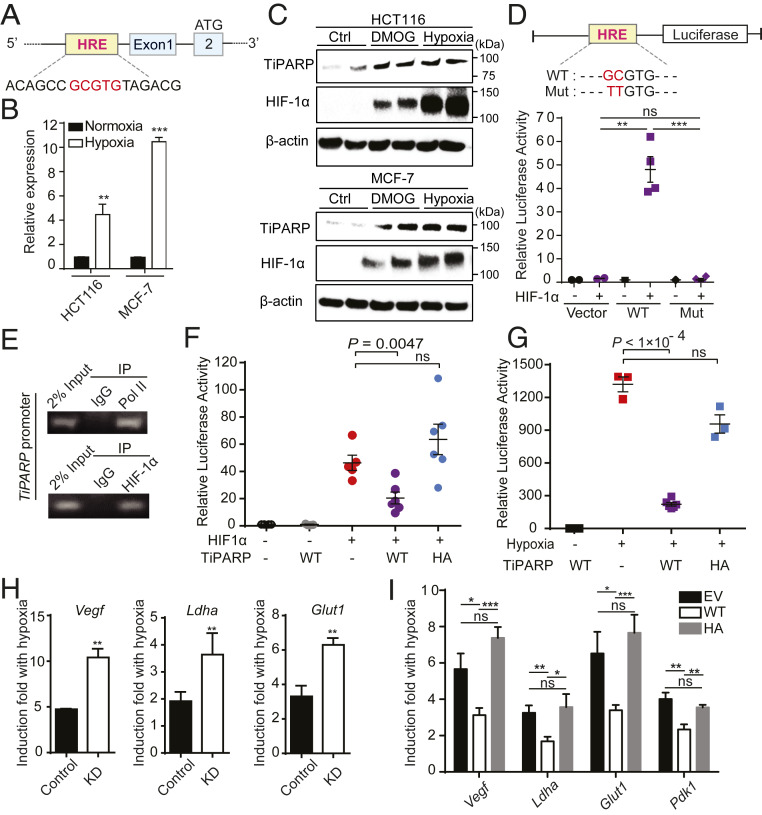
Fig. 1.
TiPARP is a direct target gene and a negative regulator of HIF. (A) A schematic representation of TiPARP promoter. Core sequence of hypoxia-response element (HRE) is highlighted in red. (B) HCT116 and MCF-7 cells were cultured at normoxia or 1% O2 (hypoxia) for 16 h. Expression of TiPARP was analyzed by qRT-PCR. Data are represented as means + SD (n = 3). (C) HCT116 and MCF-7 cells were treated with 1 mM DMOG or incubated at 1% O2 (hypoxia) for 18 h. Endogenous TiPARP and HIF-1α were analyzed by Western blot. (D) Luciferase reporter assay showing that HIF-1 binds to HRE of TiPARP. Luciferase reporter construct used in this experiment contained about 1.2-kb proximal promoter fragment of human Tiparp gene. (Top) Schematic representation of the luciferase reporter construct with WT or mutated (Mut) HRE. (Bottom) HIF-1α transactivation measured by luciferase reporter with WT or mutant HRE from TiPARP promoter. Transfection efficiencies were normalized to cotransfected Renilla-luciferase. Data are represented as means ± SD (n = 2 for the vector control and n = 4 for WT and mutant). (E) ChIP assay assessing the binding of HIF-1α to HRE in endogenous TiPARP promoter. RNA polymerase II (Pol II) was used as a positive control. (F) HIF-1 luciferase reporter activity in HEK 293T cells showing that WT TiPARP, but not inactive H532A mutant (HA), decreases HIF-1 transcriptional activity. Luciferase reporter construct used in this experiment contained three hypoxia response elements from the Pgk-1 gene. Relative luciferase activities were normalized with the cotransfected Renilla-luciferase. Data are represented as means ± SD (n = 6). (G) HIF-1 luciferase reporter activity measured in hypoxic HEK 293T cells transfected with empty vector, Flag-tagged TiPARP WT or H532A mutant. Luciferase reporter construct used in this experiment contained three HREs from the Pgk-1 gene. Data are represented as means ± SD (n = 6 for the hypoxic WT TiPARP samples and n = 3 for other samples). (H) qRT-PCR analysis of HIF-1 target gene induction in response to hypoxia, in HCT116 cells stably expressing control shRNA (Control) or shTiPARP (KD). The ratio of hypoxic to normoxic gene expression is shown. Data are represented as means + SD (n = 3). (I) HCT116 cells were treated with 1 μg/mL doxycycline for 24 h to induce the overexpression of empty vector (EV), Flag-tagged WT TiPARP, or inactive H532A mutant. Cells were then incubated at hypoxia (1% O2) for 16 h. Hypoxic induction of HIF target genes were measured by qRT-PCR. Data are represented as means + SD (n = 6). Statistical analyses were performed using unpaired two-tailed t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
To further confirm TiPARP is a target gene of HIF-1, luciferase reporter assay was performed in HEK 293T cells using constructs with or without the regulatory region of TiPARP. Luciferase reporter constructs containing TiPARP promoter with wild type (WT), but not mutated hypoxia response element (HRE), displayed significantly increased luciferase activity in response to HIF-1α overexpression (Fig. 1D). Moreover, the binding of HIF-1α and RNA polymerase II (Pol II) to TiPARP promoter under hypoxia was validated by chromatin immunoprecipitation-PCR (ChIP-PCR) (Fig. 1E). These results demonstrate that TiPARP is a target gene of HIF-1.
TiPARP Is a Negative Regulator of HIF-1α.
TiPARP has been documented to modulate the activity of transcription factors (8, 11). Thus, we asked whether it also regulates HIF-1 transcriptional activity. Using a luciferase reporter construct, we assessed whether TiPARP affects the transcriptional activity of HIF-1. As a positive control, the luciferase reporter gene was significantly induced by HIF-1α overexpression (Fig. 1F) or hypoxia (Fig. 1G). Cotransfection of TiPARP with HIF-1α resulted in a significant decrease of reporter gene expression (Fig. 1E), indicating that HIF-1α transactivation was inhibited by TiPARP. In addition, TiPARP dramatically repressed the transcriptional activity of endogenous HIF-1α under hypoxia (Fig. 1G). Moreover, the catalytic activity of TiPARP was required for the inhibition, as expressing catalytically inactive H532A mutant TiPARP had little effect on HIF-1α transactivation (Fig. 1 F and G). The activities of WT and H532A mutant of TiPARP were confirmed by Western blot to detect the auto-ADP ribosylation of FLAG-TiPARP immunoprecipitated from HEK 293T cells (SI Appendix, Fig. S1E). Similar inhibitory effect by TiPARP was also observed on HIF-2α (SI Appendix, Fig. S1F).
To further validate this effect in a more physiological setting, we examined the effect of TiPARP knockdown (KD) on HIF-1 target gene expression in HCT116 cells. As expected, the expression of well-characterized HIF-1 target genes, including vascular endothelial growth factor (VEGF), glucose transporter 1 (GLUT1), and lactate dehydrogenase A (LDHA), were elevated under hypoxia, and the activation of these genes was significantly increased by TiPARP KD compared to control KD (Fig. 1H). The knockdown efficiency of TiPARP was 70%, as measured by RT-qPCR (SI Appendix, Fig. S1G). We also examined the mRNA level of HIF-1α target genes in a von Hippel-Lindau (VHL)-mutant clear-cell renal cell carcinoma cell line RCC4 that expresses stabilized HIF-1α (17). As expected, knocking down TiPARP promoted the expression of HIF-1α target genes in RCC4 cells (SI Appendix, Fig. S1 H and I).
We also generated Tet-on-inducible HCT116 stable cells conditionally expressing Flag-tagged TiPARP. Compared with empty vector control, induction of WT TiPARP expression by doxycycline treatment attenuated the activation of HIF-1α target genes in response to hypoxia, while the expression of the inactive H532A mutant had no effect (Fig. 1I). Collectively, all of the data above support the idea that TiPARP functions as a negative regulator of HIF-1.
TiPARP Decreases HIF-1α Protein Level in an ADP-Ribosylation-Dependent Manner.
We next investigated how TiPARP regulates HIF-1. We first examined the interaction between Flag-TiPARP and endogenous HIF-1α. We treated cells with DMOG to increase endogenous HIF-1α protein levels. Both WT Flag-TiPARP and its catalytically inactive mutant H532A were able to bind endogenous HIF-1α (Fig. 2A). In addition, using various HIF-1α truncations for coimmunoprecipitation (co-IP), we found that the bHLH-PAS1 domain of HIF-1α, the conserved structure signature of bHLH-PAS family, was responsible for interacting with TiPARP (SI Appendix, Fig. S2 A and B).
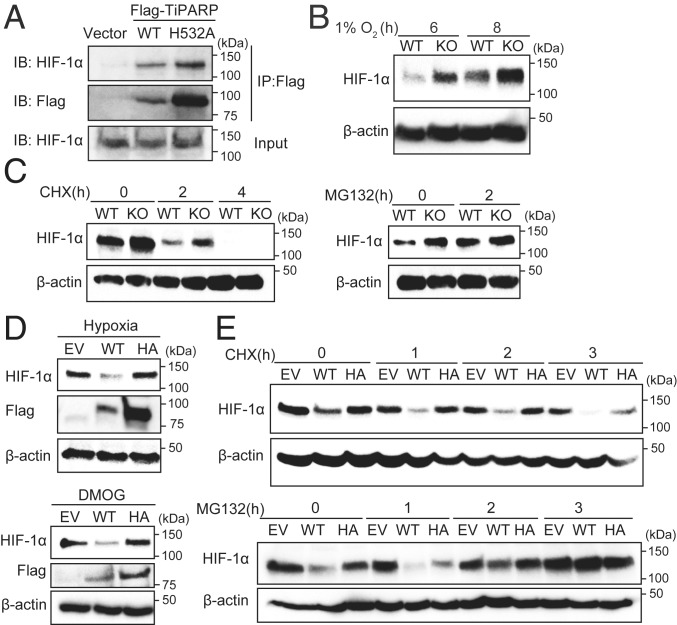
Fig. 2.
TiPARP promotes the degradation of HIF-1α in a catalytic activity-dependent manner. (A) HEK 293T cells were transfected with empty vector (EV), Flag-tagged WT TiPARP, or the H532A mutant. Cells were treated with 1 mM DMOG to stabilized endogenous HIF-1α protein. Co-IP of Flag-TiPARP and endogenous HIF-1α was detected by Western blot. Protein levels of WT TiPARP in cell lysates were very low due to its self-promoted degradation. (B) Western blot of HIF-1α in HAP-1 WT or TiPARP KO cells after 6 and 8 h of hypoxia (1% O2) treatment. (C) Western blot of HIF-1α in HAP-1 WT or TiPARP KO cells after 6 h of 1 mM DMOG treatment followed by treatment with 50 μM cycloheximide (CHX) (Left) or 20 μM MG132 (Right). (D) HCT116 cells were treated with 1 μg/mL doxycycline for 24 h to induce the expression of empty vector, Flag-tagged TiPARP WT, or inactive H532A mutant (HA). Cells were then incubated at hypoxia (1% O2) (Top) or with DMOG (Bottom) for 6 h. The protein level of HIF-1α was analyzed by Western blot. (E) Western blot of HIF-1α in HCT116 cells expressing empty vector, Flag-tagged WT TiPARP, or H532A mutant treated with DMOG for 6 h followed by treatment with CHX (Top) or MG132 (Bottom).
To understand how TiPARP negatively regulates HIF-1, we examined whether TiPARP affects HIF-1α protein level. We performed Western blot analysis of HIF-1α in control and TiPARP-deficient cell lines under conditions of hypoxia. In HAP-1 TiPARP knockout cells, the level of HIF-1α was higher compared to WT (Fig. 2B and SI Appendix, Fig. S2C). Consistently, knockdown of TiPARP by short interfering RNA (siRNA) or short hairpin RNA (shRNA) in HCT116 also increased the protein levels of HIF-1α under hypoxia (SI Appendix, Fig. S2 D and E).
The protein levels of HIF-1α is tightly regulated by oxygen level. In well-oxygenated environments, HIF-1α is hydroxylated by prolyl hydroxylase domain-containing enzymes (PHDs) and targeted for proteosomal destruction by an E3 ubiquitin ligase, the VHL complex. Under hypoxia or treatments of hypoxia-mimetic agents, the activity of PHD is diminished, and thus HIF-1α degradation is inhibited. To test whether TiPARP inhibits HIF-1α protein level through degradation, we treated cells with the translational inhibitor cycloheximide (CHX) to assess the half-life of HIF-1α after its accumulation induced by hypoxia-mimetic agent DMOG. With CHX treatment, TiPARP knockout still increased HIF-1α protein level, indicating that the regulation of HIF-1α by TiPARP did not depend on protein synthesis (Fig. 2 C, Left). Meanwhile, blocking protein degradation by treatment with MG132 strongly diminished the difference in HIF-1α protein levels in WT and TiPARP knockout/knockdown cells (Fig. 2 C, Right, and SI Appendix, Fig. S2F), suggesting the regulation of HIF-1α by TiPARP depends on proteasomal degradation of HIF-1α.
To investigate the role of TiPARP catalytic activity in HIF-1α inhibition, we overexpressed empty vector (EV), Flag-tagged WT, and the inactive H532A mutant of TiPARP in Tet-on-inducible HCT116 cell line. Only the overexpression of WT TiPARP reduced HIF-1α accumulation upon hypoxia (Fig. 2 D, Top) or DMOG treatments (Fig. 2 D, Bottom). HIF-1α degradation by WT TiPARP was not affected by CHX treatment (Fig. 2 E, Top), but was abolished by MG132 treatment (Fig. 2 E, Bottom). Collectively, our data reveal that, under hypoxia, TiPARP negatively regulates HIF-1α protein level by promoting HIF-1α proteasomal degradation, and this functions is dependent on the catalytic activity of TiPARP.
TiPARP Forms Distinct Nuclear Bodies and Recruits HIF-1α to the Nuclear Bodies.
To understand how TiPARP regulates HIF-1α protein level, we first examined the localization of TiPARP and HIF-1α by confocal imaging. FLAG-tagged WT TiPARP localized to spherical subnuclear structures, whereas catalytically inactive mutant H532A showed a dispersed nuclear distribution (Fig. 3A). We initially hypothesized that TiPARP was recruited to other known nuclear bodies, and thus we analyzed the colocalization of TiPARP with protein markers for known nuclear bodies. No colocalization was observed between TiPARP and coilin, the major component of Cajal bodies (SI Appendix, Fig. S3A). Interestingly, promyelocytic leukemia (PML) protein showed partial colocalization with TiPARP (SI Appendix, Fig. S3 B, Top). However, PML knockdown by shRNA (SI Appendix, Fig. S3C) did not disrupt TiPARP nuclear foci (SI Appendix, Fig. S3 B, Bottom). This result led us to propose that TiPARP could form nuclear bodies or condensates independently.
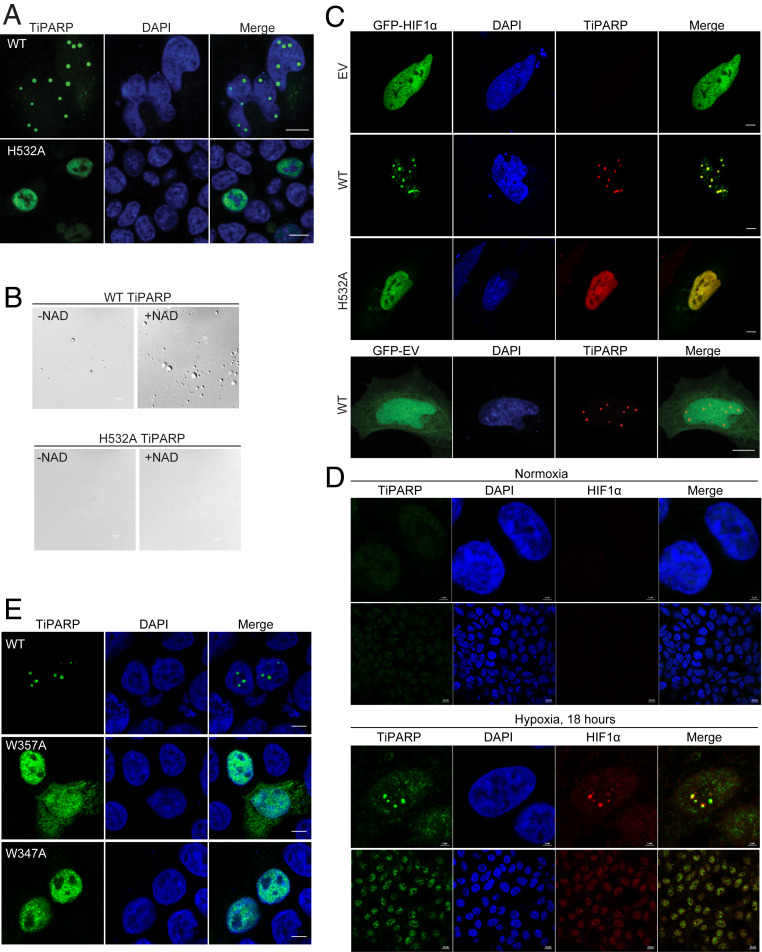
Fig. 3.
TiPARP forms nuclear foci and recruits HIF-1α. (A) HEK 293T cells were transfected with Flag-tagged WT and H532A mutant of TiPARP and analyzed by immunofluorescence microscopy with anti-FLAG antibody (green). Nuclei were stained with DAPI (blue). (B) Purified Flag-tagged WT TiPARP, but not the H532A mutant, formed droplets in the presence of 100 μM NAD+. (Scale bar, 10 μm.) (C, Top) HEK 293T cells were cotransfected with GFP-HIF-1α, as well as empty vector (EV), Flag-tagged WT, or H532A mutant TiPARP. In the Bottom, as a negative control, cells were transfected with GFP empty vector and Flag-TiPARP. (Scale bar, 5 μm.) (D) HEK 293T cells were incubated in normal condition (normoxia) (Top section) or 1% O2 (hypoxia) (Bottom section) for 18 h, followed by fixation and immunofluorescent analysis using anti-TiPARP and anti-HIF1α antibodies. (Scale bar: 2 μm for zoomed images and 10 μm for unzoomed images.) Representative images are shown. (E) HEK 293T cells were transfected with Flag-tagged WT, W347A, or W357A mutant of TiPARP and analyzed by immunofluorescence microscopy with anti-FLAG antibody (green). Nuclei were stained with DAPI (blue). The images are representative of three independent experiments. (Scale bar, 5 μm.)
To further demonstrate the ability of TiPARP to form nuclear bodies, we expressed and purified FLAG-tagged TiPARP from HEK 293T cells for in vitro studies (SI Appendix, Fig. S3D). To mimic the intracellular environment, 10% PEG was added as a macromolecular crowding agent in physiological buffer (150 mM NaCl and pH 7.5). Without incubating with NAD+, no obvious droplet was observed (Fig. 3 B, Top). After reaction with NAD+, TiPARP spontaneously formed spherical droplets, suggesting that the auto-ADP-ribosylation of TiPARP promotes its condensate formation (Fig. 3 B, Top). Auto-ADP-ribosylation of purified WT TiPARP was confirmed by Western blot using anti–mono-ADP-ribose binding reagent (SI Appendix, Fig. S3E). To further evaluate the role of ADP-ribosylation in droplet formation, catalytic mutant H532A TiPARP was purified and condensate formation was not observed after NAD+ incubation (Fig. 3 B, Bottom), which was consistent with the finding that inactive TiPARP failed to form nuclear bodies in cells (Fig. 3A). Collectively, our data suggests that auto-ADP-ribosylation of TiPARP promoted the formation of condensates in vitro and formation of nuclear bodies in cells.
We next examined the localization of HIF-1α in cells. Without TiPARP expression, GFP-HIF-1α was localized dispersedly in the nucleus and in the cytosol. Interestingly, cotransfection with Flag-TiPARP triggered the translocation of HIF-1α into TiPARP nuclear bodies. However, overexpression of inactive H532A mutant failed to affect HIF-1α localization (Fig. 3 C, Top). As another negative control, TiPARP was co-overexpressed with GFP empty vector and no change in GFP protein localization was observed (Fig. 3 C, Bottom). To assess the physiological relevance of our observation, we used anti-TiPARP antibody to image the formation endogenous TiPARP nuclear bodies during hypoxia by immunofluorescence. HEK 293T cells were incubated at normal condition (Fig. 3 D, Top), or 1% O2 for 18 h (Fig. 3 D, Bottom), 24 h, and 42 h (SI Appendix, Fig. S3F). The specificity of the antibody used was confirmed with overexpressed Flag-tagged TiPARP (SI Appendix, Fig. S3G). In normoxic cells, the endogenous level of TiPARP was very low, and no foci formation was observed. Upon hypoxia, fluorescent signal of endogenous TiPARP elevated and endogenous TiPARP formed multiple nuclear foci, which was consistent with the imaging data obtained with overexpressed TiPARP. More importantly, we observed the localization of endogenous HIF-1α at TiPARP nuclear bodies during hypoxia (Fig. 3 D, Bottom, and SI Appendix, Fig. S3F). Quantitative analysis showed that the number of TiPARP and HIF-1α foci increased over the course of 18 to 42 h of hypoxia (SI Appendix, Fig. S3H). In cells transfected with TiPARP for 24 h, the number of TiPARP bodies was comparable to that of 24 h hypoxia. Collectively, our data demonstrated that TiPARP is able to form nuclear bodies in an ADP-ribosylation-dependent fashion and recruit HIF-1α to the nuclear bodies.
TiPARP Nuclear Body Formation Is Dependent on Its WWE Domain.
PTMs of proteins have been implicated to drive phase separation or condensate formation (18). Given that TiPARP is able to auto-ADP-ribosylate itself, and that inactive mutant TiPARP did not form condensates, we hypothesize that the condensates might be initiated from the noncovalent interaction between ADP-ribosylated TiPARP proteins. Interestingly, some members of the PARP family, including TiPARP, contain a conserved WWE domain (19). WWE domain in PARPs are reported to specifically recognize and bind to ADP-ribose (20), which could potentially modulate the phase separation of automodified TiPARP. To test this hypothesis, we mutated the conserved Trp residues in the WWE domain of TiPARP to Ala (W347A and W357A) and reexamined its cellular distribution. Indeed, neither W347A nor W357A mutant TiPARP was able to form nuclear bodies and both mutants showed diffused distribution (Fig. 3E), similar to the inactive TiPARP H532A mutant (Fig. 3A). We further performed in vitro droplet formation assay with the WWE domain mutant. Consistent with intracellular localization data, mutation of the WWE domain abolished the droplet formation of TiPARP, although it maintained catalytic activity (SI Appendix, Fig. S3 I–L). We thus propose that the WWE domain enables TiPARP to interact with ADP-ribosylated proteins, including itself, thereby initiating the nucleation of subnuclear condensates that help to recruit and degrade HIF-1α.
TiPARP Nuclear Bodies Promote the Ubiquitination of HIF-1α by Recruiting E3 Ubiquitin Ligases.
Ubiquitination is a PTM that tags proteins for degradation by the proteasome. We hypothesized that TiPARP nuclear bodies may promote HIF-1α degradation by promoting its ubiquitination. Indeed, we found that nuclear bodies formed by active TiPARP were surrounded by layers of ubiquitinated proteins (Fig. 4A). In addition, overexpressing WT but not the catalytically inactive TiPARP promoted the ubiquitination of HIF-1α in cells (Fig. 4B).
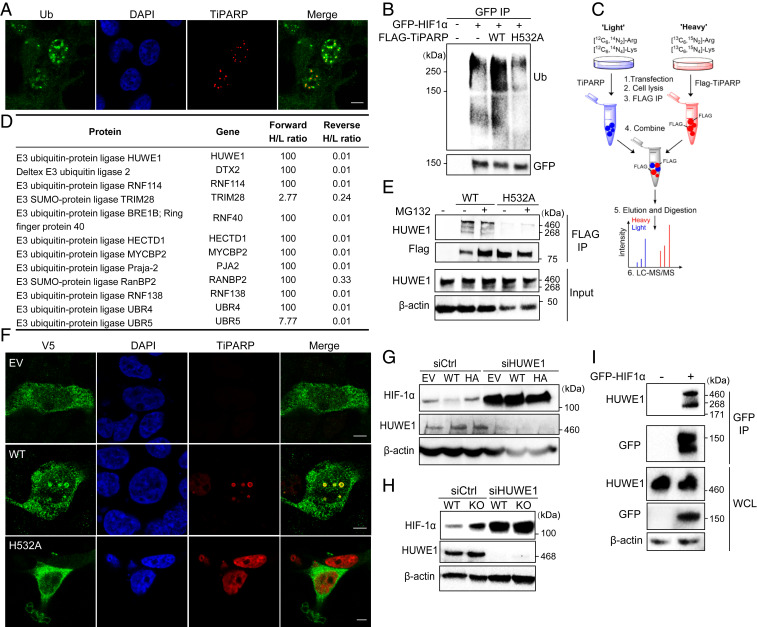
Fig. 4.
TiPARP recruits E3 ubiquitin-protein ligases to degrade HIF-1α. (A) HEK 293T cells transfected with Flag-tagged WT and H532A mutant TiPARP were analyzed by immunofluorescence microscopy with anti-ubiquitin (green) and anti-FLAG (red) antibodies. Nuclei were stained with DAPI (blue). (B) HEK 293T cells cotransfected with GFP-HIF1α and empty vector, Flag-tagged WT, or H532A mutant TiPARP. HIF-1α was immunoprecipitated by anti-GFP affinity resins, and the ubiquitination of HIF-1α was analyzed by Western blot. (C) Scheme showing identification of TiPARP interacting proteins in HEK 293T cells using SILAC. (D) List of E3 ubiquitin ligases from TiPARP interactome and the heavy/light ratios (100 is the maximum in the forward SILAC, and 0.01 is the minimum heavy/light ratio in the reverse SILAC). (E) HEK 293T cells were transfected with empty vector (EV), Flag-tagged WT, or H532A mutant TiPARP overnight, followed by incubation with 10 μM MG132 for 6 h to increase the protein level of WT TiPARP. Flag-tagged TiPARP was immunoprecipitated with anti-FLAG resins, and the co-IP with endogenous HUWE1 was analyzed by Western blot. (F) Colocalization of V5-tagged HUWE1 with Flag-tagged TiPARP WT or H532A mutant in HEK 293T cells was analyzed by immunofluorescence with anti-V5 (green) and anti-FLAG (red) antibodies. (Scale bar, 5 μm.) (G) HUWE1 KD abolished the effect of TiPARP on HIF-1α in HCT116 cells. Cells expressing empty vector (EV), Flag-tagged WT, or H532A mutant (HA) TiPARP and either control or HUWE1 siRNA were incubated in hypoxia for 6 h, and the levels of HIF-1α was detected by Western blot. (H) HUWE1 KD abolished the effect of TiPARP on HIF-1α in HAP-1 cells. HAP-1 WT and TiPARP KO cells expressing control or HUWE1 siRNA were cultured in hypoxia for 6 h. Western blot analysis of cell lysate was performed with indicated antibodies. (I) HEK 293T cells were transfected with empty vector or GFP-HIF1α, followed by IP with anti-GFP affinity resins. Co-IP of GFP-HIF1α and endogenous HUWE1 E3 ubiquitin–protein ligase was detected by Western blot.
To find out how TiPARP nuclear bodies could regulate ubiquitination, we performed stable isotope labeling with amino acids in cell culture (SILAC) and affinity-purification mass spectrometry to identify the interactome of TiPARP (Fig. 4C). Both forward and reverse (“heavy” and “light” samples were swapped) SILAC experiments were carried out to increase data reliability. Proteins with a heavy/light ratio (H/L) > 1.5 in forward SILAC, H/L < 0.66 in reverse SILAC, and with at least two unique peptides identified were considered potential TiPARP interacting proteins. In agreement with our hypothesis that TiPARP bodies promote HIF-1α ubiquitination and degradation, most TiPARP-interacting proteins identified are involved in the ubiquitin–proteasome pathway, with several E3 ligases being the top candidates (Fig. 4D and SI Appendix, Table S1). We validated the interaction of TiPARP with several top hits from the SILAC result, including E3 ubiquitin-protein ligase HECT, UBA, and WWE domain containing 1 (HUWE1) (Fig. 4E), DTX2, and RNF114 (SI Appendix, Fig. S4A).
For further analysis, we decided to focus on HUWE1, not only because it showed the highest protein score in the SILAC data, but also because it is one of the E3 ligases that harbors a WWE domain. WWE domains only occur in two functional classes of proteins, ubiquitin ligases and PARPs (19). We thus hypothesize that its WWE domain might modulate its interaction with ADP-ribosylated TiPARP. Consistent with our hypothesis, we found that in HEK 293T cells endogenous HUWE1 coimmunoprecipitated with FLAG-tagged WT TiPARP, while its interaction with the inactive H532A mutant was much weaker (Fig. 4E). These data suggested that the binding of HUWE1 with TiPARP depends on the ADP-ribosylation activity of TiPARP. Furthermore, the overexpression of WT TiPARP relocalized HUWE1 to TiPARP nuclear bodies, which could bring together HIF-1α and HUWE1 to promote HIF-1α ubiquitination (Fig. 4F). Consistent with the co-IP data, overexpressing inactive TiPARP had no effect on the localization of HUWE1 (Fig. 4F). Moreover, we mutated both conserved Try residues in the WWE domain of HUWE1 to alanine (W1617/1619A) and found that mutations in WWE domain disrupted its binding to TiPARP (SI Appendix, Fig. S4B). Together, our data suggest that HUWE1 is recruited to TiPARP nuclear bodies through the recognition of ADP-ribosylation by the WWE domain.
To determine whether HUWE1 was responsible for the degradation of HIF-1α mediated by TiPARP, we examined HIF-1α accumulation in HUWE1 KD cells upon hypoxia or treatment with hypoxia-mimetic agents. Indeed, HUWE1 silencing significantly stabilized the protein level of HIF-1α and diminished the inhibitory effect on HIF-1α by TiPARP under hypoxia or hypoxia-mimetic conditions (Fig. 4 G and H). Co-IP experiment in HEK 293T cells showed that HUWE1 also interacted with HIF-1α (Fig. 4I). Taken together, these results suggest that TiPARP promotes the degradation of nuclear HIF-1α by forming nuclear bodies, which recruit both HIF-1α and its E3 ligase HUWE1 to promote HIF-1α ubiquitination.
Under normal oxygen tension, HIF-1α protein degradation is regulated by O2-dependent prolyl hydroxylation, which targets HIF-1α for ubiquitination by the VHL complex. To examine the effect of TiPARP on HIF1-α hydroxylation and VHL-dependent degradation, we treated cells under normal oxygen tension with the proteasomal inhibitor MG-132 to prevent hydroxylated HIF-1α from being degraded, and assessed the prolyl hydroxylation of HIF-1α. Overexpression of TiPARP did not seem to affect HIF-1α hydroxylation (SI Appendix, Fig. S4C). Thus, we proposed that, during normoxia, HIF-1α is mainly degraded in cytoplasm via the VHL pathway in a TiPARP-independent fashion; under hypoxia, HIF-1α is activated and induces the expression of TiPARP to promote HIF-1α degradation in the nucleus.
Interestingly, the cellular level of TiPARP is also tightly controlled in an auto-ADP-ribosylation and ubiquitination-dependent manner, as catalytically active TiPARP was strongly ubiquitinated and maintained at a fairly low amount, whereas the inactive mutant showed weaker ubiquitination and was expressed at a significantly higher level (SI Appendix, Fig. S4 D and E). This observation suggests that HIF-1 turns on a negative-feedback regulatory mechanism that self-terminates to make sure the TiPARP nuclear bodies do not stay behind to negatively affect HIF-1 activation in the future.
TiPARP Represses the Warburg Effect and Tumorigenesis.
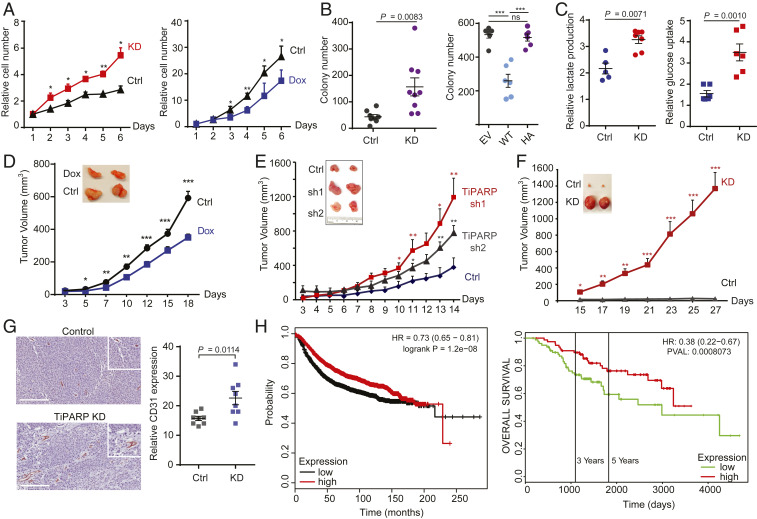
HIF-1α is stabilized or overexpressed in human cancers (13, 21–24) and is crucial for the adaptive responses of tumors to changes in oxygenation by activating the transcription of genes involved in glucose metabolism, angiogenesis, cell survival, and invasion (12, 13, 25, 26). Elevated HIF-1α is strongly correlated with poor patient prognosis and tumor resistance to therapy (27, 28). Given the inhibitory effect of TiPARP on HIF-1α, we hypothesized that TiPARP may also regulate tumorigenesis and tumor growth. We first examined the effect of TiPARP on cancer cell growth by stably knocking down TiPARP or transiently overexpressing TiPARP in Tet-on-inducible HCT116 cells. Compared to control cells, TiPARP KD cells proliferated significantly faster (Fig. 5 A, Left, and SI Appendix, Fig. S5A), while TiPARP overexpressing cells behaved in the opposite way (Fig. 5 A, Right). The growth-promoting effect of TiPARP depletion was also observed in MCF-7 cells (SI Appendix, Fig. S5 B and C). Furthermore, TiPARP KD promoted, while TiPARP overexpression inhibited anchorage-independent growth of HCT116 cells in soft agar (Fig. 5B). Collectively, our data demonstrated that TiPARP suppresses cancer cell growth. TiPARP silencing also enhanced cell migration in various cancer cell lines (SI Appendix, Fig. S5D).
Fig. 5.
TiPARP represses Warburg effect and tumorigenesis. (A) TiPARP knockdown (KD) promoted (Left) while doxycycline (Dox)-induced TiPARP overexpression (Right) inhibited the 2D proliferation of HCT116 cells. Relative cell numbers were normalized to that of day 1. Error bars represent SD from three independent experiments. (B, Left) Anchorage-independent growth of HCT116 cells stably expressing shRNA targeting luciferase (Ctrl) or TiPARP (KD) (n = 9). (B, Right) Anchorage-independent growth of HCT116 cells treated with 1 μg/mL doxycycline to induce the overexpression of empty vector (EV), Flag-tagged WT TiPARP (WT), or inactive H532A mutant (HA) (n = 6). Colony numbers in each well of a six-well plate were counted. (C) TiPARP KD increased lactate production (n = 5) and glucose consumption (n = 6) in HCT116 cells cultured in hypoxia for 24 h. Values were normalized to normoxic controls. Data are represented as means ± SD (n = 5 for the control and n = 8 for the KD). (D) Xenograft tumor growth of HCT116 cells with (n = 22) or without (n = 18) Dox-inducible TiPARP expression. (E and F) Xenograft tumor growth of control (Ctrl) and TiPARP KD (sh1 and sh2) HCT116 cells (n = 8) (E) and MCF-7 cells (n = 9) (F). (G) Immunohistochemical analysis of CD31 expression in HCT116 xenografts. Vascular distribution in tumors was quantified by counting CD31-positive microvessels per 20× field (n = 8). (Scale bar, 200 μm.) (H) Kaplan–Meier survival curve of 3,951 (Left) and 157 (Right) breast cancer patients in TCGA, GEO, and EGA databases analyzed using miRpower and PROGgene. Patients were divided into two groups (top and bottom 50% TiPARP expression) based on TiPARP mRNA levels in their tumors. In A–G, results are shown as mean ± SD. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Cancer cells tend to shift from oxidative phosphorylation to aerobic glycolysis (the Warburg effect) to support increased requirement for biosynthesis and adapt to hypoxic microenvironment (29). HIF-1 mediates such metabolic reprogramming through the induction of glycolytic enzymes and glucose transporters (GLUTs) (30). To test whether TiPARP regulates cell growth by modulating metabolic shift to aerobic glycolysis, we measured the lactate production and glucose uptake of cancer cells. Consistent with our data that TiPARP regulates the expression of glycolytic genes through HIF-1α, TiPARP-deficient HCT116 cells consumed more glucose and released more lactate into the media than control knockdown cells under hypoxia (Fig. 5C). TiPARP KD in RCC4 cell line similarly led to increased glucose uptake and lactate secretion (SI Appendix, Fig. S5E). Thus, consistent with the effects of TiPARP on HIF1-α degradation, TiPARP represses the Warburg effect and inhibits cancer cell growth.
Next, we examined whether TiPARP also inhibits tumor growth in vivo. Mice were fed with a doxycycline-containing diet to induce and maintain the expression of TiPARP in Tet-on HCT116 xenografts. TiPARP overexpression resulted in smaller tumors (Fig. 5D). In contrast, HCT116 TiPARP KD xenografts were larger than the control group (Fig. 5E). The tumor-promoting effect of TiPARP KD was even stronger in MCF-7 xenografts (Fig. 5F). mRNAs were isolated from MCF-7 tumor tissues and the expression of HIF-1α targets was quantified by qRT-PCR. As expected, glycolytic genes as well as other HIF-1 target genes were significantly up-regulated in TiPARP KD xenografts (SI Appendix, Fig. S5F). Since we observed an increase in VEGF expression level, we stained tumor tissues for the endothelial marker CD31 to evaluate the angiogenesis in solid tumors. Compared to control group, microvessel density was higher in TiPARP KD tissues (Fig. 5G and SI Appendix, Fig. S5G). Increased HIF-1α protein level and successful knockdown of TiPARP in tumor tissues were confirmed by immunohistochemical staining (SI Appendix, Fig. S5H).
Given the cell line and mouse data, we further checked whether TiPARP expression level correlates with human patient’s prognosis. miRpower and PROGgene analysis (31, 32) showed that lower TiPARP expression strongly correlates with worse patient prognosis (Fig. 5H). Collectively, our data provide strong support that the TiPARP-HIF axis is important for tumor growth in vivo and can be targeted as a potential cancer treatment strategy.
TiPARP Nuclear Bodies Also Promote the Degradation of Estrogen Receptor α and c-MYC.
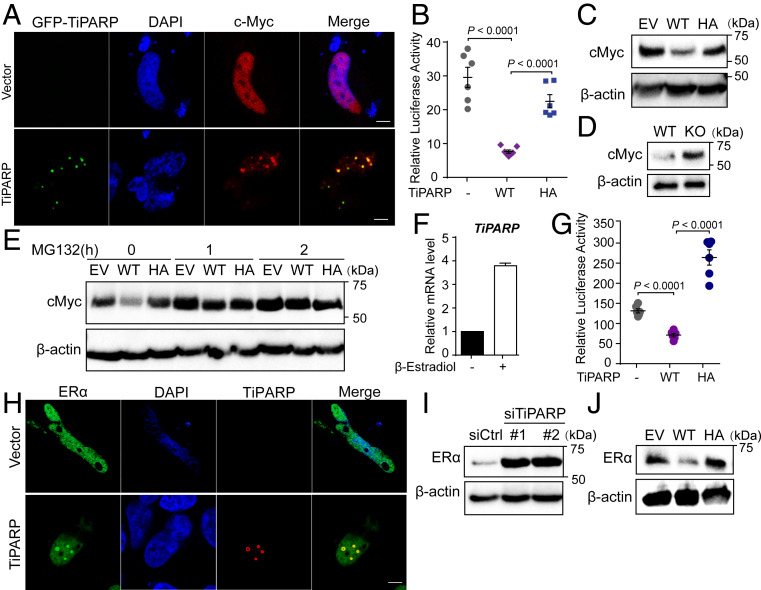
We next asked whether TiPARP mediated protein degradation could also apply to other transcription factors. We first examined other dimeric transcription factors in the bHLH family (33) and found that c-Myc showed TiPARP nuclear bodies localization when cotransfected with TiPARP (Fig. 6A). Moreover, c-Myc transcriptional activity and protein level were also inhibited by overexpressing WT TiPARP in HEK 293T cells (Fig. 6 B and C). Consistently, depletion of TiPARP promoted the accumulation of c-Myc protein (Fig. 6D). Similar to the case of HIF-1α, TiPARP also modulated c-Myc protein through proteasome degradation pathway, as MG132 treatment abolished the effect of TiPARP on c-Myc in HCT116 cells (Fig. 6E).
Fig. 6.
TiPARP inhibits ERα and c-MYC. (A) HEK 293T cells were cotransfected with Flag-cMyc, as well as empty vector (EV) or GFP-TiPARP (green). c-Myc was stained with anti-FLAG (red) antibodies. (Scale bar, 5 μm.) (B) Transcriptional activity of c-Myc was measured using a luciferase reporter with c-Myc binding sites. c-Myc was cotransfected with empty vector, TiPARP WT, or H532A mutant (HA) in HEK 293T cells. Data are represented as means ± SEM (n = 6). (C) Western blot analysis of cMyc in HEK 293T cells expressing empty vector (EV), Flag-tagged WT, or H532A inactive mutant (HA) TiPARP. (D) Western blot analysis of cMyc in WT and TiPARP KO HAP-1 cells. (E) Western blot of cMyc in HCT116 cells expressing empty vector (EV), Flag-tagged WT, or H532A mutant of TiPARP and treated with or without MG132. (F) qRT-PCR analysis of TiPARP mRNA expression in MCF-7 cells treated with DMSO or 10 nM β-estradiol for 24 h. Data are represented as means + SD (n = 8). (G) Transactivation of ERα measured using a luciferase reporter containing estrogen response elements (EREs). HEK 293T cells were transfected with ERE-luciferase, Flag-TiPARP, together with HA-tagged ERα. Twelve hours after transfection, cells were treated with 1 μM β-estradiol for 24 h, followed by luciferase measurement. Data are represented as means ± SEM (n = 6). (H) HeLa cells were cotransfected with HA-tagged ERα and empty vector (EV) or Flag-TiPARP. Colocalization was analyzed by immunofluorescence with anti-HA (green) and anti-FLAG (red) antibodies. Nuclei were stained with DAPI (blue). (Scale bar, 5 μm.) (I) Western blot analysis of endogenous ERα in MCF-7 cells transfected with control or TiPARP siRNA. (J) Western blot of endogenous ERα in HCT116 cells expressing empty vector (EV), Flag-tagged WT, or H532A mutant (HA) of TiPARP.
Interestingly, we also found that TiPARP expression was induced by the treatment of β-estradiol in MCF-7 cells (Fig. 6F), indicating that TiPARP transcription could also be activated by ER. Increased TiPARP in turn negatively regulated ER transactivation (Fig. 6G) and promoted the translocation of ERα to TiPARP nuclear bodies (Fig. 6H). RNAi knockdown of TiPARP increased the protein level of ERα in MCF-7 cells (Fig. 6I), while overexpressing WT TiPARP had the opposite effect (Fig. 6J), suggesting that TiPARP is also a negative feed-back regulator of ERα.
To evaluate whether TiPARP also promotes the degradation of c-Myc and ERα via HUWE1, we knocked down HUWE1 by siRNA and found that silencing HUWE1 increased the cellular protein abundance of c-Myc and ERα (SI Appendix, Fig. S6A). In addition, we examined the β-subunit of HIF-1 and found that TiPARP overexpression did not change the localization of HIF-1β (SI Appendix, Fig. S6B). Consistent with our hypothesis, since TiPARP did not recruit HIF-1β to nuclear bodies, it had no effect on the protein stability of HIF-1β (SI Appendix, Fig. S6C). We also examined the transcription factor STAT3, and TiPARP did not affect its cellular localization or protein level (SI Appendix, Fig. S6 D–F). These data suggest that the regulation of transcription factors by TiPARP is relatively specific.
Collectively, our data demonstrated that TiPARP nuclear bodies constitute a negative-feedback loop for several oncogenic transcription factors, suggesting that TiPARP may act as a tumor suppressor in many tumors.
Discussion
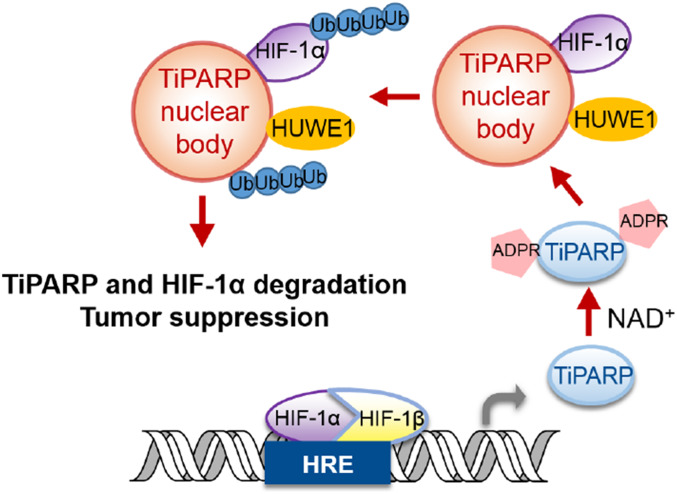
Our study supports a model (Fig. 7) that TiPARP is a target of HIF-1 and acts as a negative-feedback regulator of hypoxic signaling. TiPARP, in an ADP-ribosylation-dependent manner, forms spherical nuclear bodies and recruits HIF-1α, and the E3 ubiquitin ligase HUWE1, leading to the ubiquitination and degradation of HIF-1α. Interestingly, the ability of TiPARP to form nuclear bodies and promote HIF-1α degradation is dependent on its catalytic activity. Although ADP-ribosylation has been reported to modulate transcription (34), our work described here establishes a mechanism via which ADP-ribosylation could regulate condensates formation and transcription. Our data showed that the WWE domain, which can bind ADP-ribosylated proteins, is important for the formation of TiPARP nuclear bodies. However, the detailed mechanism via which WWE domain binding to ADP-ribosylated proteins promotes TiPARP condensates formation requires future investigation.
Fig. 7.
Proposed model depicting the negative-feedback loop regulation of HIF-1α via TiPARP nuclear bodies.
Transcription factors needs to be precisely controlled to avoid undesirable transcriptional activities. Many excellent examples are known regarding how transcription factors are turned on by certain signaling events (35). However, how they are turned off after serving their transcriptional activity is relatively less studied. Our study suggests that turning on TiPARP transcription and forming the TiPARP nuclear bodies could turn off several transcription factors (HIF-1, c-Myc, and ER). Interestingly, TiPARP itself is also degraded by this mechanism, ensuring that there will no excessive TiPARP to negatively impact future activation of the transcription factors. The expression of TiPARP is increased by the corresponding transcription factors in response to hypoxia, dioxin, or estrogen stimulation. The conditional expression of TiPARP, combined with its ability to destruct itself and the transcription factors, forms a precisely controlled negative-feedback loop to make sure that the transcription factors and TiPARP nuclear bodies are tightly controlled.
Our interactome study for TiPARP identified numerous proteins that are associated with ubiquitination, including many E3 ligases. So far, we have only examined the effect of HUWE1 on HIF-1. It is very likely that other E3 ligases are also important for down-regulating the transcription factors. The fact that multiple E3 ligases are associated with TiPARP is a strong indication that TiPARP nuclear bodies could be responsible for the down-regulation of many transcription factors. In a recent screening of TiPARP substrates, proteins involved in “proteasome complex” and “protein polyubiquitination” were enriched as potential TiPARP targets (36). These data support our finding that TiPARP is functionally linked to the ubiquitin–proteasome system.
By inhibiting HIF-1 signaling, TiPARP suppresses the Warburg effect and tumorigenesis in xenograft models of human colon and breast cancers. Thus, TiPARP acts as a tumor suppressor by down-regulating tumor-promoting transcription factors. Recently, in-depth bioinformatics analysis of TiPARP in breast cancer was performed and further confirmed that TiPARP is a prognostic marker and could be considered as a potential therapeutic target for breast cancer (37). Our work suggests that small molecules that increase TiPARP expression or activity may be effective anticancer therapeutics.
Materials and Methods
Reagents.
Dimethyloxallyl glycine and MG132 were purchased from Cayman. CHX, Protease Inhibitor Cocktail Tablets EDTA-free, [13C6,15N2]-l-lysine, [13C6,15N4]-l-arginine, l-lysine, and l-arginine were purchased from Sigma-Aldrich. Lipofectamine RNAiMAX was purchased from Invitrogen. HUWE1 siRNA On-TARGETplus SMARTpools and negative control siRNA were purchased from Dharmacon. Benzonase nuclease lyophilized powder (CAS 9025-65-4) was purchased from Santa Cruz.
Luciferase Reporter Assays.
HEK 293T cells were seeded in 24-well plate and transfected using Fugene 6 (Promega) with 1 μg of the following plasmids as described in the text: pRL Renilla luciferase control reporter vector (Rluc), pGL2-HRE luciferase, pcDNA-HA-HIF1α, pCMV6-flag-TIPARP WT, and pCMV6-flag-TIPARP H532A. After 24 h, cells were lysed, and luciferase activity was determined using the Dual-Luciferase Reporter Assay system (Promega). In hypoxic groups, cells were transfected with Rluc, pGL4-HREx3, and WT or mutant TIPARP. Twelve hours after transfection, cells were switched to hypoxia culture condition (1% O2) for 12 h, followed by lysis and luciferase activity measurement.
Co-IP.
To examine the interaction between FLAG-tagged TIPARP and HA-tagged HIF1α, HEK 293T cells transfected with FLAG-TIPARP and empty vector or HA-HIF1α and cultured overnight. Cells were then collected and lysed in 1% Nonidet P-40 lysis buffer (150 mM NaCl, 25 mM Tris, 1% Nonidet P-40, 10% glycerol, with protease inhibitor mixture freshly supplemented). For each sample, 1 mg of whole-cell lysate (quantified with Bradford regent) was incubated with 10 μL of anti-FLAG M2 affinity gel for 3 h at 4 °C under constant mixing. The resulting affinity gel was washed three times with 1 mL of IP washing buffer (50 mM NaCl, 25 mM Tris, 0.1% Nonidet P-40) and heated in protein loading buffer at 95 °C for 10 min. Western blot was performed to detect the interaction of indicated proteins. Flag- and HA-tagged proteins were detected with HRP-conjugated anti-flag antibody and anti-HA antibody (Santa Cruz), respectively. Endogenous HIF-1β was detected with HIF-1β/ARNT antibody (Cell Signaling; #5537).
Immunofluorescence.
Cells were seeded in 35-mm glass-bottom dishes (MatTek) and transfected with FLAG-TIPARP, GFP-HIF1α, and V5-HUWE1 in HEK 293T cells overnight. Cells were then rinsed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Fixed cells were washed twice with PBS, permeabilized with 0.1% saponin in PBS, and blocked with 5% bovine serum albumin (BSA) for 30 min at room temperature. Cells were then incubated with indicated antibodies overnight at 4 °C in dark in PBS with 0.1% saponin and 5% BSA. Cells were washed with PBS with 0.1% saponin for three times and incubated with secondary antibody in the dark for 1 h at room temperature. Samples were washed with PBS three times and mounted with Fluoromount-G (Southern Biotech; 0100-01). Samples were imaged with Zeiss LSM880 inverted confocal microscopy or Zeiss LSM710 confocal microscopy. Images were captured at 63×, and about 100 cells per condition per experiment were analyzed. Experiments were repeated three independent times, and representative images are shown in the figures. Images were processed with ZEN and Fiji software. The following primary antibodies are used: mouse anti-FLAG tag antibody at 1:2,000 (Cell Signaling; 14793), rabbit anti-PML protein antibody at 1:1,000 (Abcam; ab179466), rabbit anti-V5 tag at 1:1,000 (Cell Signaling; 13202), rabbit anti-coilin at 1:1,000 (Cell Signaling; 14168), rabbit anti-TiPARP (Abcam; 84664) at 1:50, and mouse anti–HIF-1α (H1alpha67; NB100-105) at 1:50. The secondary antibodies Alexa Fluor 488 goat anti-mouse and cyanine3 goat anti-rabbit were diluted at 1:1,000. For hypoxic imaging, cells were incubated in complete medium at 1% O2, 5% CO2, and 37 °C for various time courses before fixation.
In Vitro Droplet Formation.
To purify TiPARP, HEK 293T cells were transfected with pCMV6-FLAG-TIPARP (WT or H532A inactive mutant) and cultured overnight. After harvesting cells, each group of cells (from four plates of 10-cm dishes) was lysed with 3 mL of Nonidet P-40 lysis buffer and centrifuged at 17,000 rpm for 20 min to obtain clear lysate. Whole-cell lysates were incubated with 50 μL of anti-FLAG affinity resin for 4 h at 4 °C. The resins were then washed three times with lysis buffer, and FLAG-TiPARP was eluted from the resins using 3× FLAG peptide from Millipore Sigma (F4799), followed by buffer exchange and concentration of FLAG-TiPARP to 2 μM. The in vitro assay was prepared with 1 μM FLAG-TiPARP in 20 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM DTT, 25 U (0.1 μL) of benzonase, and 10% PEG-8000. NAD+ was added at last to the samples at 100 μM. Images were captured within 15 min of reaction setup.
SILAC and Mass-Spectrometric Analysis.
HEK 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% dialyzed fetal bovine serum (FBS) and either [13C6,15N2]-l-lysine, [13C6,15N4]-l-arginine (“heavy”) or normal l-lysine, l-arginine (“light”) for six generations. After transient transfection of pCMV-FLAG-TiPARP or pCMV-TiPARP plasmid, heavy and light cells were lysed in 1% Nonidet P-40 lysis buffer separately. Total lysate of 10 mg from either heavy or light cells was subjected for FLAG IP separately. After washing the affinity resin three times with lysis buffer, heavy and light samples were combined and washed two more times with PBS buffer. Resins were resuspended in freshly prepared 8 M urea and incubated at 65° for 10 min. Next, the protein supernatant was reduced with 10 mM DTT at room temperature for 1 h and alkylated by incubation with 40 mM iodoacetamide at room temperature for 1 h. DTT was then added to stop alkylation at room temperature for 1 h. After diluting the urea concentration to 2 M with 50 mM Tris⋅HCl, pH 8.0, and 1 mM CaCl2, 1 μg of trypsin was added and incubated at 37 °C for 18 h. Trifluoroacetic acid (0.1%) was added to quench the trypsin digestion, followed by desalting with a Sep-Pak C18 cartridge. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was performed following previously published methods (38).
Other methods can be found in SI Appendix.
Data and Materials Availability.
Expression vectors, except those obtained from Addgene with material transfer agreement, and other commercial sources are available for not-for-profit researchers upon request. All data are available in the main text or SI Appendix, Materials.
Supplementary Material
Acknowledgments
We thank the Cornell University Biotechnology Resource Center (BRC) Imaging Facility for help with the confocal microscopy. This work was supported in part by funding from Howard Hughes Medical Institute (HHMI), Cornell University, NIH/National Institute of General Medical Sciences (NIGMS) Grant R35GM131808 (to H.L.), and NIH Grant S10OD018516, which supported the BRC Imaging Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921815117/-/DCSupplemental.
References
- 1.Gibson B. A., Kraus W. L., New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Hottiger M. O., Hassa P. O., Lüscher B., Schüler H., Koch-Nolte F., Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Corda D., Di Girolamo M., Functional aspects of protein mono-ADP-ribosylation. EMBO J. 22, 1953–1958 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bütepage M., Eckei L., Verheugd P., Lüscher B., Intracellular mono-ADP-ribosylation in signaling and disease. Cells 4, 569–595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corda D., Di Girolamo M., Mono-ADP-ribosylation: A tool for modulating immune response and cell signaling. Sci. STKE 2002, pe53 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Del Vecchio M., Balducci E., Mono ADP-ribosylation inhibitors prevent inflammatory cytokine release in alveolar epithelial cells. Mol. Cell. Biochem. 310, 77–83 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Scarpa E. S., Fabrizio G., Di Girolamo M., A role of intracellular mono-ADP-ribosylation in cancer biology. FEBS J. 280, 3551–3562 (2013). [DOI] [PubMed] [Google Scholar]
- 8.MacPherson L. et al., 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Q., Induction and superinduction of 2,3,7,8-tetrachlorodibenzo-rho-dioxin-inducible poly(ADP-ribose) polymerase: Role of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator transcription activation domains and a labile transcription repressor. Arch. Biochem. Biophys. 404, 309–316 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Ma Q., Baldwin K. T., Renzelli A. J., McDaniel A., Dong L., TCDD-inducible poly(ADP-ribose) polymerase: A novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 289, 499–506 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Bindesbøll C. et al., TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and co-activates liver X receptors. Biochem. J. 473, 899–910 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Gordan J. D., Simon M. C., Hypoxia-inducible factors: Central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 17, 71–77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza G. L., Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Semenza G. L., Wang G. L., A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang B. H., Rue E., Wang G. L., Roe R., Semenza G. L., Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271, 17771–17778 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L., Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldewijns M. M. et al., VHL and HIF signalling in renal cell carcinogenesis. J. Pathol. 221, 125–138 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Hofweber M., Dormann D., Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L., The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275 (2001). [DOI] [PubMed] [Google Scholar]
- 20.He F. et al., Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 586, 3858–3864 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Semenza G. L., HIF-1 and human disease: One highly involved factor. Genes Dev. 14, 1983–1991 (2000). [PubMed] [Google Scholar]
- 22.Masoud G. N., Li W., HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 5, 378–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong H. et al., Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59, 5830–5835 (1999). [PubMed] [Google Scholar]
- 24.Talks K. L. et al., The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157, 411–421 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan H. E. et al., Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60, 4010–4015 (2000). [PubMed] [Google Scholar]
- 26.Maxwell P. H. et al., Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 94, 8104–8109 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindl M. et al.; Austrian Breast and Colorectal Cancer Study Group , Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin. Cancer Res. 8, 1831–1837 (2002). [PubMed] [Google Scholar]
- 28.Bachtiary B. et al., Overexpression of hypoxia-inducible factor 1alpha indicates diminished response to radiotherapy and unfavorable prognosis in patients receiving radical radiotherapy for cervical cancer. Clin. Cancer Res. 9, 2234–2240 (2003). [PubMed] [Google Scholar]
- 29.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semenza G. L., HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20, 51–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lánczky A. et al., miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160, 439–446 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Goswami C. P., Nakshatri H., PROGgene: Gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinforma. 3, 22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones S., An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus W. L., Lis J. T., PARP goes transcription. Cell 113, 677–683 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Barolo S., Posakony J. W., Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Lu A. Z. et al., Enabling drug discovery for the PARP protein family through the detection of mono-ADP-ribosylation. Biochem. Pharmacol. 167, 97–106 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Cheng L. et al., TCDD-inducible poly-ADP-ribose polymerase (TIPARP), A novel therapeutic target of breast cancer. Cancer Manag. Res. 11, 8991–9004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Cao J., Miller S. P., Jing H., Lin H., Comparative nucleotide-dependent interactome analysis reveals shared and differential properties of KRas4a and KRas4b. ACS Cent. Sci. 4, 71–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.