Significance
The Hippo-YAP/TAZ pathway plays pivotal roles in controlling tissue growth, and its abnormal regulation is frequently implicated in a variety of cancers. We have identified MAML1/2 as critical regulators for YAP/TAZ nuclear retention and transcription activities. Clinical analysis with specimens of a human cancer patient and a public cancer database reveals pathological association between MAML expression and YAP signature. Furthermore, our results suggest that the cell density governing YAP/TAZ subcellular localization can also function as an important intrinsic mechanism for miR30c-dependent regulation of MAML1 level. Our findings provide mechanistic insights of YAP/TAZ-dependent growth control pathway and tumorigenesis.
Keywords: Hippo signaling, MAML1/2, nuclear localization, TEAD, YAP/TAZ
Abstract
The Hippo pathway plays a pivotal role in tissue homeostasis and tumor suppression. YAP and TAZ are downstream effectors of the Hippo pathway, and their activities are tightly suppressed by phosphorylation-dependent cytoplasmic retention. However, the molecular mechanisms governing YAP/TAZ nuclear localization have not been fully elucidated. Here, we report that Mastermind-like 1 and 2 (MAML1/2) are indispensable for YAP/TAZ nuclear localization and transcriptional activities. Ectopic expression or depletion of MAML1/2 induces nuclear translocation or cytoplasmic retention of YAP/TAZ, respectively. Additionally, mutation of the MAML nuclear localization signal, as well as its YAP/TAZ interacting region, both abolish nuclear localization and transcriptional activity of YAP/TAZ. Importantly, we demonstrate that the level of MAML1 messenger RNA (mRNA) is regulated by microRNA-30c (miR-30c) in a cell-density-dependent manner. In vivo and clinical results suggest that MAML potentiates YAP/TAZ oncogenic function and positively correlates with YAP/TAZ activation in human cancer patients, suggesting pathological relevance in the context of cancer development. Overall, our study not only provides mechanistic insight into the regulation of YAP/TAZ subcellular localization, but it also strongly suggests that the miR30c–MAML–YAP/TAZ axis is a potential therapeutic target for developing novel cancer treatments.
The Hippo pathway is an evolutionarily conserved regulator of organ size that functions by coordinating cell proliferation and apoptosis (1–5). Diverse upstream inputs, such as cell density, mechanical stress, and soluble factors, tightly regulate the Hippo pathway, which, in response to these extracellular signals, controls cell proliferation and organ growth (6–9). Dysregulation of the Hippo pathway induces massive overgrowth in diverse organs due to uncontrolled cell proliferation (10–12). Hippo signaling inactivation and subsequent Yes-associated protein (YAP) activation induce rapid tumor progression in various animal models and are strongly correlated with several types of human cancers such as lung, breast, and liver cancer (10, 13–15). Activation of the Hippo pathway is triggered by a core kinase cassette that comprises the mammalian sterile 20-like kinases (MST1/2), the large tumor suppressor (LATS1/2), and the scaffold protein Salvador1 (SAV1) (16). Sequential phosphorylation and activation of MST1/2 and LATS1/2 by upstream signals such as high cell density result in phosphorylation-dependent cytoplasmic retention of the transcriptional coactivator YAP and its paralogue TAZ (transcriptional coactivator with PDZ-binding motif). This occurs through interaction with 14-3-3, and the retained YAP/TAZ are down-regulated by β-TrCP–mediated proteolytic degradation (6, 9, 17, 18). LATS1/2-mediated phosphorylation of YAP/TAZ can be suppressed by low cell density or cell-proliferative conditions, both of which induce the nuclear translocation of YAP/TAZ. Once in the nucleus, YAP/TAZ interacts with TEA domain transcription factor (TEAD), resulting in the activation of genes involved in cell proliferation, antiapoptosis, and survival (19). Overall, the regulation of YAP/TAZ subcellular localization by a variety of upstream regulators is considered a critical regulatory step in the Hippo pathway (20–22).
In addition to cytoplasmic sequestration of YAP/TAZ through interaction with 14-3-3 (23, 24), a number of studies have reported alternative modulators of YAP/TAZ subcellular localization (3). Merlin, Kibra, and Angiomotin have all been shown to physically bind to and exclude YAP/TAZ from the nucleus (20, 21, 25). Additionally, protein tyrosine phosphatase PTPN14 induces cell-density-dependent nuclear export of YAP, thereby suppressing YAP oncogenic function (26, 27). This inhibitory role of PTPN14 on YAP activity depends on a physical interaction between PTPN14 and YAP1 and is independent of PTPN14’s enzymatic activity (26). More recently, SET1A-mediated monomethylation at the K342 residue of YAP (28), binding of Dishevelled (a positive regulator of canonical Wnt signaling) to phosphorylated YAP (29), and the interaction between TEAD and NES of YAP/TAZ (30) have also been shown to be involved in the nuclear export of YAP. To date, the regulation of cytoplasmic sequestration of YAP/TAZ has been extensively studied; however, the molecular effectors as well as the underlying mechanisms governing nuclear translocation and retention of YAP/TAZ are unclear. In addition, unlike its Drosophila ortholog Yorkie, a functional nuclear localization signal (NLS) sequence for YAP/TAZ has not been reported in mammals, suggesting the existence of a potential novel molecular mediator of YAP/TAZ nuclear localization.
Mastermind-like (MAML) was originally identified as a pivotal coactivator of Notch-dependent transcription, and three distinct MAML genes (MAML1 to 3) are expressed in mammals. In addition to Notch signaling, numerous studies have reported that MAML promotes Wnt, Shh, and nuclear factor-κB signaling target gene transcription by directly interacting with downstream effectors of these pathways (31–33). Therefore, there is strong evidence demonstrating Notch-independent functions of MAML proteins in regulating various biological processes (34). However, despite its importance as a transcriptional coactivator in various signaling pathways, the overall physiological relevance of MAML in homeostasis and disease has not been well studied.
In this study, we identified MAML1 and MAML2, but not MAML3, as potent effectors of the Hippo pathway by promoting YAP/TAZ nuclear localization in a cell-density-dependent manner. We discovered an evolutionarily conserved PPxY-interacting motif within MAML1/2 that physically binds to the WW domain of YAP/TAZ, promoting YAP/TAZ nuclear retention and consequent transcriptional activity. Mutation of a putative MAML NLS sequence repressed YAP/TAZ activity, further demonstrating the importance of MAML1/2 in regulating YAP/TAZ nuclear localization. Importantly, MAML protein abundance is controlled by expression of microRNA-30c (miR-30c) in a cell-density-dependent manner. Furthermore, analysis of various xenograft assays, The Cancer Genome Atlas (TCGA) database, and clinical specimens revealed that high expression of MAML is associated with YAP/TAZ activation. Taken together, our findings not only provide mechanistic insight into YAP/TAZ nuclear localization, but they also propose the miR30c–MAML–YAP/TAZ axis as a potential target for cancer treatment.
Results
MAML1/2 Promote Nuclear Localization and the Transcriptional Activities of YAP/TAZ.
To screen for proteins that promote nuclear localization of YAP/TAZ, the cellular localization of enhanced green fluorescent protein (EGFP)–YAP was examined after cotransfection with various nuclear proteins. Interestingly, ectopic expression of MAML1 clearly induced nuclear localization of YAP, while other nuclear proteins had no effect (Fig. 1A and SI Appendix, Fig. S1A). One exception was TEAD4, which has been shown to form a complex with YAP/TAZ in the nucleus (35). Ectopic expression of MAML1 and TAZ also induced nuclear localization of TAZ (SI Appendix, Fig. S1B). Immunoprecipitation (IP) assay revealed that both endogenous and ectopically expressed MAML1 were able to associate with YAP/TAZ (Fig. 1B and SI Appendix, Fig. S1C). A proximity-ligation assay, which can detect in situ protein interactions with high specificity and sensitivity (36, 37), further confirmed that endogenous MAML1 physically interacts with YAP in the nucleus (Fig. 1C).
Fig. 1.
MAML1/2 promote the nuclear localization and transcriptional activities of YAP/TAZ. (A) Ectopically expressed MAML1 and YAP in HEK293 cells colocalized in the nucleus. (Scale bars, 10 μm.) (B) Co-IP experiments showed that endogenous MAML1 interacts with YAP/TAZ. (C) The fluorescence signal generated by proximity ligation assay demonstrates that endogenous MAML1 and YAP are closely located in the nucleus. (Scale bars, 10 μm.) (D) MAML1 and MAML2, but not MAML3, increased YAP reporter activity in HCT116 cells. (E) Knockdown of MAML1/2 in HEK293A reduced nuclear localization of YAP. The areas marked with white rectangles were magnified and shown in E, Right. (Scale bars, 50 μm.) (F) Knockdown of MAML1/2 in HEK293T cells reduced YAP reporter activity. (G) Quantitative real-time PCR showed that knockdown of MAML1/2 in HeLa cells reduced mRNA levels of Hippo and Notch signaling target genes. (H) A wound-healing assay showed that knockdown of MAML1/2 in HeLa cells reduced migration rate, which was induced by the ectopic expression of YAP. In D and F–H, the statistical analyses represent average values from a representative experiment performed in triplicate. Error bars represent SDs of triplicate assays. **P < 0.01. n.s., not significant. Student’s t test was used for statistical analysis.
MAML was initially discovered to function as a coactivator of Notch signaling, but several reports indicate that MAML may also coactivate other signaling pathways (34). In mammalian cells, there are three distinct members of MAML genes: MAML1, 2, and 3. We found that cotransfection of MAML1 and MAML2, but not MAML3, promoted nuclear localization of EGFP–YAP (SI Appendix, Fig. S1D). Consistently, cotransfection of YAP or TAZ with MAML1 or MAML2, but not MAML3, enhanced TEAD-dependent reporter activity (8xGTIIC-Luc) (SI Appendix, Fig. S1 E and F). Transfection of MAML1 or MAML2, but not MAML3, into HCT116 cells, which express high levels of YAP, also promoted reporter activity (Fig. 1D). In accordance with the above data, knockdown of MAML1 and MAML2 led to relocalization of endogenous YAP from the nucleus to the cytoplasm (Fig. 1E and SI Appendix, Fig. S1G). Additionally, Western blot analysis revealed that knockdown of MAML1/2 dramatically induced YAP in the cytoplasmic fraction, while it reduced YAP in the nuclear fraction (SI Appendix, Fig. S1H). Knockdown of MAML1/2 by short interfering RNA (siRNA) treatment significantly reduced YAP/TAZ-mediated reporter activity in various cell lines (Fig. 1F and SI Appendix, Fig. S1 I–K). Importantly, expression of Notch target genes such as HES1 and NRARP as well as the YAP/TAZ target genes ANKRD1, CTGF, and CYR61 were down-regulated in MAML1/2-depleted cells, thus validating our knockdown approach (Fig. 1G). A wound-healing assay demonstrated that YAP enhanced cell migration, which is consistent with previous reports (38), while the enhanced cell migration was markedly abolished by depletion of MAML1/2 (Fig. 1H). Taken together, these results suggest that MAML1/2 interact with YAP/TAZ and promote their nuclear localization and transcriptional activities.
Nuclear Localization of MAML Is Necessary for the Retention of YAP/TAZ in the Nucleus.
A recent study has shown that YAP contains a conventional nuclear export signal (NES) (28), but it is currently unclear if YAP/TAZ also have an NLS. Previous work suggests that the N-terminal region (1 to 55 amino acids) in Yorkie, a Drosophila homolog of human YAP, has a potential NLS that promotes nuclear import of Yorkie via interaction with importin α1 (39). However, the human homolog of importin α1, KPNA6, did not display a physical interaction with YAP, nor did it induce its nuclear translocation in mammalian cells (39). Upon close inspection, we found that the amino acid sequence of the N-terminal domain of YAP displayed poor homology with that of Yorkie. We also noticed that Arg15, a residue of Yorkie that is critical for binding to importin α1, is not conserved in YAP (SI Appendix, Fig. S2A). Furthermore, an N-terminal truncated form of YAP (YAPΔ58) enhanced reporter activity similar to wild-type YAP, and MAML1-mediated nuclear localization of YAP was unaffected in YAPΔ58 mutants (SI Appendix, Fig. S2 B and C). More recently, it has been shown that the C-terminal domain of TAZ has a nonclassical NLS (30). However, a mutant form of YAP with a mutation in the conserved sequences (hereafter referred to as YAP–mpNLS; SI Appendix, Fig. S2D) induced normal YAP reporter activity. In addition, YAP–mpNLS-mediated reporter activity as well as nuclear localization of YAP–mpNLS were further enhanced by the ectopic expression of MAML1 (SI Appendix, Fig. S2 E and F). Overall, these data suggest that mammalian YAP may lack a functional NLS, or, at the very least, the reported NLS sequences are not necessary for MAML-mediated nuclear localization of YAP.
Because MAML1/2 promote YAP/TAZ nuclear localization, and YAP does not have a distinct NLS, we hypothesized that the NLS of MAML, which has not been characterized, is necessary for YAP nuclear localization. NLS prediction software such as cNLS mapper, PSORTII, and NucPred predicted the 138- to 142-amino-acid stretch (KKTRR) as a putative NLS for MAML1 (Fig. 2A). We mutated the putative NLS of MAML1 (KKTRR to AATLL; hereafter referred to as MAML1–mNLS) and analyzed its subcellular localization in HEK293 cells (Fig. 2B). Notably, MAML1–mNLS was mainly localized in the cytoplasm, and it did not induce nuclear localization of YAP when compared to wild-type MAML1 (Fig. 2B and SI Appendix, Fig. S2G). Consistently, MAML1–mNLS induced much lower reporter activity than wild-type MAML1 in HCT116 colon cancer cells (Fig. 2C). A wound-healing assay revealed reduced cell migration upon depletion of MAML1/2, which was markedly rescued by reintroduction of wild-type MAML1, but not MAML1–mNLS (Fig. 2D). However, our results do not rule out the possibility that MAML1/2 may regulate cell migration through alternative signaling events other than YAP/TAZ. Based on these observations, we hypothesized that MAML1/2 can affect YAP/TAZ nuclear localization by either promoting its transport to the nucleus from the cytoplasm or by enhancing its nuclear retention. Live cell-imaging analysis with HEK293 cells using doxycycline-inducible mCherry–MAML1 and EGFP–YAP showed that MAML1 localized to the nucleus earlier than YAP and that both proteins colocalized in the nucleus only at a later time point (Fig. 2 E, Upper). However, it is still possible that GFP–YAP can be phosphorylated by the Hippo pathway, which may result in slower nuclear translocation of YAP. To address this possibility, nuclear localization of the Hippo-resistant mutant YAP (YAP-5SA) upon inducible expression of mCherry–MAML1 was examined (Fig. 2 E, Lower). Consistently, the YAP-5SA translocated to the nucleus much faster than wild-type YAP upon MAML1 expression. However, we still noticed that nuclear localization of MAML1 was observed earlier than YAP, even in YAP-5SA mutants. While mCherry–MAML1 is strictly present in the nuclei upon doxycycline treatment, the presence of YAP-5SA in the cytoplasm seems to disappear and is enriched in the nucleus upon inducible expression of MAML1. Collectively, our findings suggest that the NLS in MAML1 is essential for the nuclear localization of YAP and that this enhanced nuclear localization is likely caused by nuclear retention. Nonetheless, we cannot completely exclude the possibility that MAML1/2 facilitates the translocation of YAP/TAZ into the nucleus.
Fig. 2.
Nuclear localization of MAML is essential for the retention of YAP/TAZ in the nucleus. (A) Schematic diagram of human MAML1. The red line indicates a putative NLS sequence, and “KKTRR” was mutated to “AATLL” for inducing loss of NLS function. (B) MAML1-mNLS is present in the cytoplasm, and it does not promote nuclear localization of YAP. (Scale bars, 10 μm.) (C) MAML1–mNLS did not increase YAP reporter activity in HCT116 cells when compared to wild-type (WT) MAML1. Ctrl, control. (D, Left and Center) MAML1–mNLS did not rescue retarded HEK293 cell migration induced by MAML1/2 depletion. (D, Right) Western blot shows normal expression of proteins. (E) Live cell imaging suggests that MAML1 retains YAP in the nucleus, which enhances YAP nuclear localization. (Scale bars, 10 μm.) In C, the statistical analysis represents average values from a representative experiment performed in triplicate. Error bars represent SDs of triplicate assays. In D, Center, the statistical analysis represents average values of three different regions from a representative experiment. Error bars represent SDs of values from three different regions. **P < 0.01. n.s., not significant. Student’s t test was used for statistical analysis.
The WW Domains of YAP/TAZ and the PPxY Motif of MAML1/2 Are Required for the Interaction between YAP/TAZ and MAML1/2.
It is well known that the phosphorylation status of YAP–Ser127 determines the nuclear localization of YAP (40) and that YAP is phosphorylated by LATS1/2 kinase. We tested whether nuclear localization of YAP induced by the absence of LATS1/2 is affected by MAML1/2 depletion. Knockdown of MAML1/2 efficiently redistributed YAP/TAZ to the cytoplasm in wild-type HEK293A cells, while in LATS1/2 knockout (KO) cells, YAP/TAZ was retained in the nucleus (SI Appendix, Fig. S3A). Although it appears that the cytoplasmic staining was somewhat increased, quantification of nuclear YAP did not reveal a significant difference. Despite not observing a significant difference by immunofluorescence (IF) upon MAML depletion in LATS1/2 KO cells, we could see a clear reduction in YAP reporter activity, which is a much more quantitative and reliable analysis compared to IF (SI Appendix, Fig. S3B). We believe that excessive nuclear influx of YAP due to its stabilization via LATS1/2 depletion may override the cytoplasmic localization caused by MAML1/2 depletion. This probably explains why we can only detect a difference with more sensitive methods such as the YAP reporter assay. These data suggest that the unphosphorylated form of YAP can be retained in the nucleus by MAML1. Recently, we suggested that Nemo-like kinase (NLK) functions as a counteracting kinase against LATS1/2 by phosphorylating YAP at Ser-128 (41). We then tested whether the phosphorylation status on YAP may affect its binding affinity to MAML1/2. Co-IP experiments with ectopically expressed phospho-mimic forms of YAP (41) and MAML1 did not show any differences in binding affinities, indicating that the interaction between MAML1/2 and YAP/TAZ may be mediated through other domains rather than those phosphorylated by LATS1/2 or NLK (SI Appendix, Fig. S3C).
As YAP contains WW domains known to be involved in protein–protein interactions (42), we tested mutant constructs for interaction with MAML1 (SI Appendix, Fig. S3D). Strikingly, the MAML1–YAP/TAZ interaction was robustly abolished when the WW domains were mutated in YAP or deleted in TAZ (YAP-mWW or TAZΔWW) (Fig. 3A and SI Appendix, Fig. S3E). Furthermore, nuclear localization and transcriptional activity of YAP-mWW or TAZΔWW were not induced by MAML1 (Fig. 3 B and C and SI Appendix, Fig. S3 F–H). To further confirm the importance of the WW domain, we tested whether nuclear localization of the Hippo signaling insensitive YAP mutant (YAP-5SA and YAP-5SA–mWW [mutation as shown SI Appendix, Fig. S3D was introduced in YAP-5SA]) could be enhanced by ectopic expression of MAML1. Overexpression of MAML1 was able to dramatically enhance the nuclear localization of YAP-5SA, while its effects on YAP-5SA–mWW nuclear localization were minimal (SI Appendix, Fig. S3I). Consistent with these data, overexpression of MAML1 enhanced YAP reporter activity induced by YAP-5SA, whereas YAP-5SA–mWW-induced reporter activity was not enhanced by MAML1 overexpression (SI Appendix, Fig. S3J). These results support our hypotheses that the WW domain of YAP is necessary for the interaction with MAML1 and that nuclear localization of YAP occurs independently of LATS-mediated phosphorylation. Because the WW domain functions as a protein–protein interaction module that binds to a PY motif (PPxY, where x is any amino acid) (43), we attempted to find putative PY motifs in MAML proteins by comparative sequence analysis. Interestingly, we found that MAML1 and MAML2, but not MAML3, contain an evolutionarily conserved PY motif (Fig. 3D). The mutated PY motif (PPxY to PPxA) containing MAML1 (MAML1–PPxA) showed a reduced interaction with YAP in HEK293T cells and did not increase YAP/TAZ reporter activity in HCT116 cells when compared to wild-type MAML1 (Fig. 3 E and F). Furthermore, MAML1–PPxA did not promote nuclear localization of YAP, even though MAML1–PPxA nuclear localization persisted (Fig. 3G and SI Appendix, Fig. S3K). These results are consistent with our data, demonstrating that MAML1/2, but not MAML3, promote nuclear localization and enhance YAP/TAZ reporter activities (Fig. 1D and SI Appendix, Fig. S1 D–F). Overall, these data suggest that the PPxY motif of MAML1/2 is essential for the interaction with and nuclear localization of YAP/TAZ. This notion was further confirmed via wound-healing assay, which showed that the inhibition of cell migration by MAML1/2 knockdown was completely rescued by reinduction of wild-type MAML1, but not by MAML1–PPxA (Fig. 3H). However, our results once again do not rule out the possibility that MAML1/2 may regulate cell migration through alternative signaling events other than YAP/TAZ. Contrary to the effect on YAP/TAZ activities, MAML1–PPxA was able to enhance NICD (Notch Intracellular C-terminal Domain)-mediated reporter activity to a similar degree as wild-type MAML1, demonstrating a Notch-independent effect of MAML1–PPxA in the wound-healing assay shown in Fig. 3H and SI Appendix, Fig. S3L. Collectively, our findings indicate that the interaction between MAML1/2 and YAP/TAZ, which is mediated through the PPxY motif of MAML and the WW domain of YAP/TAZ, is essential for the nuclear localization and transcriptional activity of YAP/TAZ.
Fig. 3.
The WW domains of YAP/TAZ and the PPxY motifs of MAML1/2 are required for the interaction between YAP/TAZ and MAML1/2. (A) YAP-mWW does not interact with MAML1. (B) Nuclear localization of YAP-mWW is not induced by ectopic expression of Flag-MAML1. (Scale bars, 10 μm.) (C) YAP-mWW–mediated reporter activity in HEK293T cells is not further enhanced by MAML1. (D) PPxY motifs in MAML1/2 are highly conserved in various species. (E) MAML1–PPxA shows reduced interaction with YAP. (F) MAML1–PPxA does not enhance YAP reporter activity in HCT116 cells as much as wild-type MAML1. (G) The nuclear localization of YAP was not induced by ectopic expression of MAML1–PPxA. (Scale bars, 10 μm.) (H) MAML1–PPxA did not rescue retarded HEK293 cell migration induced by MAML1/2 depletion. Wound-healing assay and analysis were performed as described in Fig. 2D. In C and F, the statistical analysis represents average values from a representative experiment performed in triplicate. Error bars represent SDs of triplicate assays. **P < 0.01. n.s., not significant. Student’s t test was used for statistical analysis. Ctrl, control; WT, wild type.
MAML1/2 Act as Transcriptional Coactivators by Forming a Trimeric Complex with YAP/TAZ and TEAD.
Since MAML is known to act as a transcriptional coactivator of several signaling pathways (34), we examined whether nuclear MAML could act as a coactivator of YAP/TAZ-mediated transcription. We reasoned that if MAML1/2 are only involved in nuclear retention of YAP/TAZ, MAML1/2 should not enhance the artificial NLS-fused YAP-mediated reporter activity. Contrary to our expectations, depletion of MAML1/2 exhibited significantly reduced reporter activity, despite the fact that NLS-fused YAP did indeed localize to the nucleus (Fig. 4A and SI Appendix, Fig. S4A). Moreover, ectopic expression of MAML1 further increased NLS-fused YAP mediated reporter activity, indicating that MAML1 has additional functions that are independent from YAP/TAZ nuclear retention (Fig. 4B). It is known that the C-terminal domain of MAML acts as a transactivation domain (TAD) to enhance Notch target gene expression via forming a complex with NICD and the transcription factor CSL (34). Interestingly, MAML1ΔTAD, which contains a deletion of the MAML1 Notch TAD (SI Appendix, Fig. S4B), did not further enhance NICD as well as YAP/TAZ-mediated reporter activities (Fig. 4C and SI Appendix, Fig. S4C). These data suggest that MAML proteins function as coactivators by forming a complex with TEAD, which mainly acts as a DNA-binding protein for YAP. IP assay revealed that MAML1 forms a ternary complex together with YAP and TEAD (Fig. 4D). Knockdown of YAP/TAZ and analysis of a YAP mutant (Flag–YAP-S94A that cannot bind TEAD) revealed a reduction in the MAML–TEAD interaction (Fig. 4E and SI Appendix, Fig. S4D), suggesting that MAML–YAP/TAZ–TEAD form a trimeric complex. Through chromatin IP (ChIP) assay, we found that promoter regions of the endogenous YAP/TAZ target genes, ANKRD1 and CTGF, were occupied by MAML1, while the nonspecific gene FAT3 displayed no binding (Fig. 4F). Furthermore, ChIP assay with an anti-V5 antibody after transfection with V5 epitope-tagged MAML1 showed that V5-MAML1 binds to the promoters of YAP target genes, but not to the promoter of the nonspecific gene HBB. Importantly, these binding interactions were significantly reduced by knockdown of TEAD1/3/4 (Fig. 4G). Since we demonstrated that TEAD4 also promoted the localization of YAP (Fig. 1A), we tried to distinguish between the roles of MAML1/2 and TEADs in relation to YAP/TAZ nuclear retention. We analyzed the effect of TEAD4 on the subcellular localization of YAP/TAZ in MAML1/2-depleted cells. Interestingly, ectopic expression of TEAD4 was able to dramatically induce YAP nuclear localization, even after MAML1/2 depletion (SI Appendix, Fig. S4E). Since MAML1/2 does not have a DNA binding domain, we believe that TEADs are required for the localization of MAML1/2 to the promoter region of YAP/TAZ target genes and that the interaction between YAP/TAZ and TEADs is strengthened by MAML1/2 (Fig. 4 E and G and SI Appendix, Figs. S4D and S7). As MAML1/2 bind NICD and YAP/TAZ via different domains, it is possible that these proteins form a large complex in the nucleus and regulate both Hippo and Notch signaling. However, IP data clearly showed that NICD is not present in the YAP–MAML1/2–TEAD protein complex (SI Appendix, Fig. S4F). Consistently, MAML1 enhanced YAP/TAZ-dependent reporter activity, while ectopic expression of NICD had no effect (SI Appendix, Fig. S4G). Consistently, YAP also did not enhance NICD-mediated reporter activity (SI Appendix, Fig. S4H). Overall, these results strongly suggest that MAML1/2 form a complex with YAP and TEAD at the promoter regions of YAP/TAZ target genes in order to enhance their expression.
Fig. 4.
MAML1/2 act as transcriptional coactivators by forming a trimeric complex with YAP/TAZ and TEAD. (A) MAML1/2 KO cell pools exhibited reduced NLS–YAP-mediated reporter activity. (B) NLS–YAP-mediated reporter activity was further enhanced by MAML1 overexpression. (C) MAML1ΔTAD did not induce YAP reporter activity when compared to wild type. (D) Co-IP assay showed that MAML1 interacted with YAP and TEAD1. (E) Depletion of YAP/TAZ reduces interaction between MAML1 and TEAD. (F) ChIP assay showed that MAML1 and YAP co-occupies on the promoters of YAP target genes. FAT3 was used for negative control. (G) ChIP assay showed that TEAD1/3/4 are required for MAML1 to bind the promoters of YAP target genes. HBB was used as a negative control. The statistical analyses represent average values from a representative experiment performed in triplicate. Error bars represent SDs of triplicate assays. *P < 0.05; **P < 0.01. n.s., not significant. Student’s t test was used for statistical analysis. Ctrl, control.
Cell-Density-Dependent Regulation of MAML1 and MAML2.
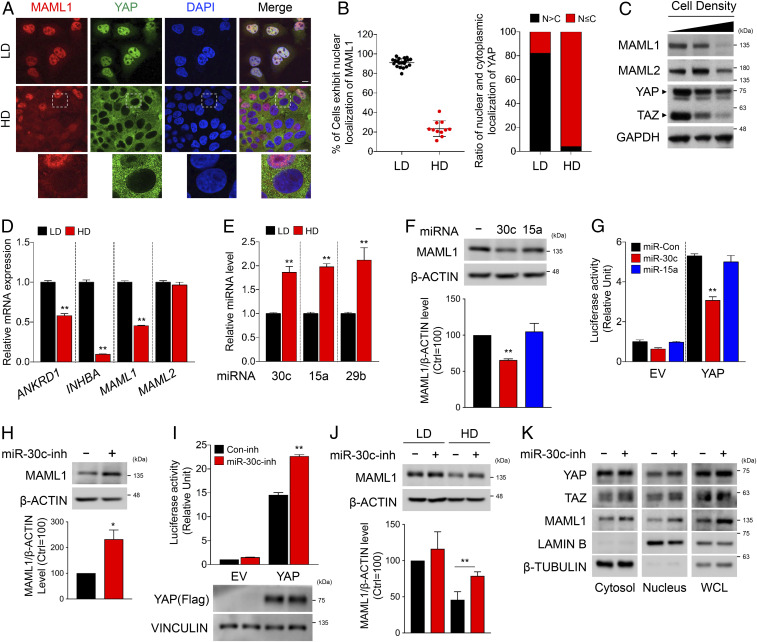
Since the level and nuclear localization of YAP/TAZ are regulated in a cell-density-dependent manner (6) and MAML1/2 bind to YAP/TAZ to promote their nuclear localization, we wondered whether the level of MAML1/2 is also controlled in a cell-density-dependent manner. At low cell density, MAML1, like YAP, displayed high nuclear abundance, and this was substantially reduced at higher cell densities (Fig. 5 A and B). Interestingly, YAP remained in the nucleus to some extent in cells where MAML1 was still present, whereas YAP was predominantly present in the cytoplasm of cells with low levels of MAML1 (Fig. 5 A and B). Western blot analysis further confirmed that YAP/TAZ, as well as MAML1/2, were reduced in a cell-density-dependent manner in various cell lines (Fig. 5C and SI Appendix, Fig. S5A). NICD-mediated Notch reporter activity as well as YAP reporter activity were gradually decreased as cell density increased, further confirming that MAML1/2 levels are reduced at high cell densities (SI Appendix, Fig. S5B).
Fig. 5.
Cell-density-dependent regulation of MAML1 and MAML2 and the involvement of miR-30c in the regulation of MAML1. (A) MAML1 and YAP colocalize in the nucleus or cytoplasm of HEK293 cells in a cell-density-dependent manner. HD, high cell density; LD, low cell density. (Scale bars, 10 μm.) (B) Quantification of MAML1 and YAP cellular localization shown in A. (C) Protein levels of MAML1/2 are reduced in HEK293 cells grown in high density. (D) qPCR assay showed that mRNA levels of MAML1, but not MAML2, were decreased in HEK293A cells grown in high density. (E) The level of miR-30c is increased in cells grown at high density. miR-15a and miR-29b were used for positive controls. (F, Upper) Ectopic expression of miR-30c mimics reduced MAML1 protein levels. (F, Lower) ImageJ was used to calculate the relative level of MAML1. (G) Ectopic expression of miR-30c mimics in HEK293 cells reduced YAP-mediated reporter activity. (H, Upper) Ectopic expression of an inhibitor of miR-30c in HEK293 cells increased MAML1 protein levels. (H, Lower) The relative level of MAML1 was analyzed by ImageJ. (I, Upper) Ectopic expression of miR-30c inhibitor in HEK293 cells increased YAP-mediated reporter activity. (I, Lower) Western blotting shows an equal amount of YAP expression. (J, Upper) Ectopic expression of a miR-30c inhibitor in HEK293 cells increased MAML1 protein level grown in high cell density. (J, Lower) The relative level of MAML1 was analyzed by ImageJ. (K) Ectopic expression of an miR-30c inhibitor in HEK293 cells increased the MAML1 level as well as nuclear localization of YAP and TAZ. The statistical analyses represents the average values from a representative experiment performed in triplicate. Error bars represent SDs of triplicate assays. *P < 0.05; **P < 0.01. Student’s t test was used for statistical analysis.
As both the Hippo pathway and the levels of MAML1/2 are regulated by cell density, we speculated that MAML1/2 levels could be down-regulated by Hippo signaling components that sense and mediate cell-contact inhibition (6, 44). However, knockdown of the core Hippo components NF2, MST1/2, LATS1/2, or YAP did not affect the levels of MAML1/2 (SI Appendix, Fig. S5C). Besides MAML1/2’s roles in nuclear retention and transcriptional activation of YAP, we can imagine that MAML1/2 may regulate AMPK or LATS1/2 activity by an unknown mechanism during active Hippo signaling conditions. We then tested the effect of MAML1/2 depletion on the control of LATS kinase activity and its upstream effector, AMPK (45). However, knockdown of MAML1/2 did not affect the activities of LATS or AMPK, as determined by their phosphorylation levels (SI Appendix, Fig. S5D). Overall, these results suggest that MAML1/2 are controlled by cell-density-related mechanisms that are independent of the Hippo pathway.
To elucidate the molecular mechanisms underlying cell-density-dependent MAML1/2 down-regulation, we examined relative messenger RNA (mRNA) expression at various cell densities. As expected, expression of the YAP/TAZ target genes ANKRD1 and INHBA was reduced at high cell density (Fig. 5D). MAML1 mRNA levels were significantly reduced at high cell density, although the decrease was not as evident as that observed at the protein level. MAML2 mRNA levels were not significantly altered (Fig. 5D). Together, these data point toward posttranscriptional regulation of MAML1/2 at high cell densities. We therefore sought to find specific miRs that can inhibit translation of MAML1/2 mRNAs, since global activation of miR biogenesis has been shown to be induced by cell–cell contact (46–49). Using a reliable prediction program (TargetScanHuman; http://www.targetscan.org/vert_72/), we identified miR-30c as a potential candidate for MAML1. We were not able to identify suitable candidates for MAML2. miR-30c contains a highly conserved region targeting MAML1 mRNA, and the expression of miR-30c was greatly increased at high cell density in a similar manner as miR-15a and miR-29b, both of which have been positively correlated with cell density (46) (Fig. 5E and SI Appendix, Fig. S5E). Consistently, ectopic expression of miR-30c led to a reduction in MAML1 protein levels as well as YAP-mediated reporter activities, while miR-15a, which does not have a seed sequence for targeting MAML1 mRNA, had no effect (Fig. 5 F and G and SI Appendix, Fig. S5 F and G). The decrease in MAML1 resulting from miR-30c treatment was rescued by the addition of an inhibitor of miR-30c, which resulted in increased MAML1 protein levels and YAP reporter activity (Fig. 5 H and I and SI Appendix, Fig. S5H). The reduced MAML1 level observed at high cell density was also restored by treatment with the miR-30c inhibitor (Fig. 5J). In addition, fractionation analysis showed that the levels of nuclear YAP/TAZ were increased by treatment with the miR-30c inhibitor (Fig. 5K and SI Appendix, Fig. S5I). Taken together, these results suggest that MAML1 protein abundance is regulated by both transcriptional and posttranscriptional mechanisms that are directly influenced by cell density. Thus far, we have focused on the role of the miR–MAML axis in mediating cell-density-dependent regulation of YAP/TAZ. We next examined whether the miR–MAML–YAP/TAZ axis can be regulated by other Hippo signaling activation conditions. Although less than the decrease observed by cell density, glucose and serum starvation also reduced the abundance of MAML1/2, whereas actin–cytoskeleton rearrangement had no effect (SI Appendix, Fig. S5 J–L). Consistent with the alteration of MAML1 levels, we found that miR-30c was induced in glucose or serum depletion, while it was not affected by actin rearrangement (SI Appendix, Fig. S5 J–L). Based on these results, we suggest that cell density may be a major physiological factor in regulating miR-30c levels since significantly higher miR-30c expression was induced by high cell density in comparison with other physiological conditions.
MAML1/2 Display Oncogenic Properties, and Their Protein Levels Are Positively Correlated with the Expression of YAP/TAZ Target Genes in Cancer.
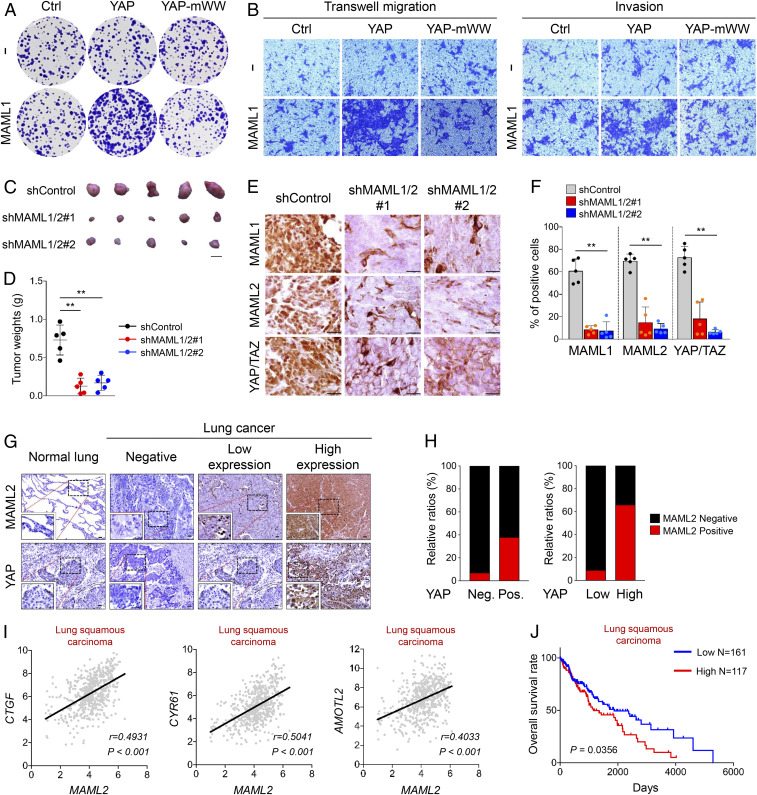
Elevated YAP/TAZ activity and nuclear enrichment of YAP/TAZ have been reported in many types of human cancer such as colon, lung, breast, and liver cancers (10). Given the role of MAML1/2 in promoting YAP/TAZ nuclear localization and activity, we next examined whether MAML1/2 contribute to the oncogenic function of YAP/TAZ. To this end, we first compared the relative expression of MAML1/2 and YAP/TAZ in various colorectal cancer cell lines and found that HCT116 cells express high levels of MAML1/2 and YAP/TAZ (SI Appendix, Fig. S6A). HCT116 cells transfected with either YAP or MAML1 alone exhibited higher growth rates compared to control cells, and cotransfection of YAP and MAML1 led to a synergistic effect (Fig. 3 A–C). Importantly, cotransfection of YAP-mWW and MAML1 did not lead to a synergistic effect on growth rate (SI Appendix, Fig. S6B). To further examine whether MAML1 overexpression cooperates with activated YAP in transformation/tumorigenesis, colony-forming (Fig. 6A and SI Appendix, Fig. S6C), transwell, and invasion assays (Fig. 6B and SI Appendix, Fig. S6D) were performed. In all assays, we obtained similar results to the cell-proliferation assay shown in SI Appendix, Fig. S6B. Moreover, a synergistic effect was observed upon cotransfection of YAP and MAML1, but not with YAP-mWW and MAML1. Taken together, these data strongly suggest that MAML1 promotes YAP-mediated tumorigenesis rather than activating other signaling pathways. To further confirm our hypothesis, we then performed a xenograft assay using HCT116 cells. Strikingly, MAML1/2 depletion by short hairpin RNAs dramatically suppressed xenograft tumor growth (Fig. 6 C and D and SI Appendix, Fig. S6E). Interestingly, immunohistochemistry analysis revealed not only a robust reduction in protein levels, but it also revealed nuclear exclusion of YAP/TAZ in tumors from MAML1/2-depleted HCT116 cells (Fig. 6 E and F). It is thus likely that the in vivo decrease of YAP/TAZ levels may be due to increased cytoplasmic localization of YAP/TAZ, which ultimately leads to phosphorylation by LATS1/2 and proteasomal degradation. To further extend our findings into the pathological condition, we performed immunohistochemical staining of MAML1/2 and YAP using a lung-cancer tissue microarray. MAML1 expression was undetectable in lung tumors, probably due to poor antibody sensitivity or low levels of MAML1 in lung tissue (50, 51). Interestingly, while none of normal lung tissues exhibited positive signals for YAP (0 of 10) and MAML2 (0 of 10), positive expression of MAML2 and YAP was observed in 26.4% (29 of 110) and 62.7% (69 of 110) of lung tumors, respectively (SI Appendix, Fig. S6F). More importantly, a significant positive correlation between MAML2 and YAP expression was observed in lung cancer. In fact, 65.7% (23 of 35) of lung-tumor tissues showed positive MAML2 expression when YAP protein levels were also high (Fig. 6 G and H and SI Appendix, Fig. S6G), whereas 91.2% (31 of 34) of lung tumors with negative MAML2 expression exhibited low YAP levels (Fig. 6 G and H and SI Appendix, Fig. S6G). Overall, these results suggest that high levels of MAML2 promote YAP-mediated tumorigenesis in a substantial proportion of human lung tumors.
Fig. 6.
MAML1/2 display oncogenic activities, and MAML2 protein levels are strongly correlated with the expression of YAP/TAZ target genes in lung cancer patients. (A) MAML1 enhances colony-forming activity of YAP. (B) Migration and invasion ability of HCT116 cells were enhanced upon cotransfection with MAML1 and YAP. (Magnification, x200.) (C and D) Knockdown of MAML1/2 in HCT116 cells shows reduced tumor growth in xenograft experiments. (Scale bar, 1,000 μm [C].) (E and F) MAML1/2-depleted cell-driven tumors exhibit significantly reduced YAP/TAZ levels when compared to control cell driven tumors. (Scale bars, 100 μm.) (F) Graph shows percentage of positive MAML1, MAML2, and YAP/TAZ-stained cells in control and MAML1/2-depleted cell-driven tumors displayed in E. (G and H) The tissue microarray analysis of lung cancer patients showed a positive correlation between MAML2 and YAP protein levels. (G) Histochemical analysis was performed with antibodies indicated on the left. G, Insets show magnification of the small rectangular region. (Scale bars, 20 μm.) (H) Graphs showing correlations between the levels of MAML2 and YAP (raw data were presented in SI Appendix, Fig. S6 F and G). Neg., negative; pos., positive. (I) The mRNA expression datasets of lung squamous carcinoma patients from the TCGA database show a positive correlation between MAML2 and YAP target genes. The correlation coefficient (r) was calculated by Pearman’s linear correlation. Two-tailed P value was used for analyzing statistical significance. (J) Kaplan–Meier plot shows that high expression of MAML2 is associated with poor prognosis of lung cancer patients. Lower 33rd percentile (n = 161) and upper 24th percentile (n = 117) were analyzed. **P < 0.01.
Next, we referred to TCGA databases, including lung squamous carcinoma and breast and liver cancers, to determine whether there is a correlation between MAML2 expression and a YAP/TAZ transcriptional signature. These analyses revealed a positive correlation between the level of MAML2 and YAP/TAZ target genes, such as CTGF, CYR61, and AMOTL2, in several types of human cancers (Fig. 6I and SI Appendix, Fig. S6 H and I). However, MAML2 and the Notch target genes HEY1, HES1, and NRARP in human lung cancer were not significantly correlated (SI Appendix, Fig. S6J), suggesting that lung tumors which have a high level of MAML2 are most likely not induced by Notch signaling. Consistently, a high level of MAML2 was associated with worse overall survival in patients with lung squamous carcinoma (Fig. 6J). Thus, elevated MAML2 expression in human cancers is associated with YAP/TAZ activation and reduced survival. Overall, these results not only demonstrate pathological relevance between MAML expression and YAP/TAZ activities in tumorigenesis, they also suggest a potential mechanism underlying YAP/TAZ activation in a variety of human cancers.
Discussion
Regulating nuclear localization of YAP/TAZ is an essential step in the Hippo signaling pathway that controls the expression of genes responsible for maintaining cell proliferation and organ size. The regulation of cytoplasmic YAP/TAZ localization has been almost fully elucidated, but the mechanisms underlying nuclear YAP/TAZ localization are still under intensive investigation (26–30). The reasons for the presence of totally independent mechanisms for regulating nuclear localization of YAP/TAZ are not clear. This may reflect a unique importance of determining nuclear localization of YAP/TAZ or may be simply related to differences in cellular context. In this study, we report that MAML1/2, which are known coactivators of Notch signaling, promote nuclear localization of YAP/TAZ and enhance YAP/TAZ transcriptional activity (SI Appendix, Fig. S7). We show that endogenous MAML1/2 interact with YAP/TAZ through a PPxY motif and WW domain, respectively, and that an NLS in MAML1/2 is essential for the nuclear localization of YAP/TAZ. Xenograft experiments, analysis of human lung cancer tissues, and TCGA data reveal that the levels of MAML2 are strongly correlated with nuclear YAP/TAZ and the expression of YAP/TAZ target genes. Importantly, two recent independent whole genome-wide studies identified highly recurrent YAP1–MAML2 fusions in multiple types of cancers (52, 53). In addition, Gene Set Enrichment Analysis showed that a YAP1-conserved transcriptional signature was the most significant hit in YAP1–MAML2–fusion positive cell lines (52). In both papers, fusions between YAP1 exons encoding the N-terminal domain of YAP1, which has a TEAD binding domain, and the MAML2 exons 2 to 5, which code for the TAD, are the most recurrent. Interestingly, as we identified the NLS for MAML1 (Fig. 2), NLS prediction softwares such as cNLS mapper predicted the 188- to 208-amino-acid stretch, which is encoded by exon2, as a putative NLS for MAML2. These findings substantiate our results in the clinical setting and suggest that blocking the interaction between MAML2 and YAP can be a novel therapeutic target for interruption of cancer progression.
Nuclear localization is highly influenced by protein size. When a protein’s molecular mass is lower than 40 kDa, it can enter the nucleus by passive diffusion (54). YAP’s size is around 75 kDa, so it requires an NLS for nuclear translocation. However, a classical NLS in the YAP amino acid sequence has not yet been discovered, and the mechanism for its nuclear translocation is unknown. Since we found that MAML1/2 interact with YAP/TAZ, and identified a functional NLS in MAML1, we initially hypothesized that MAML1/2 could facilitate the translocation of YAP/TAZ into the nucleus. Although we cannot completely exclude this hypothesis, the following evidence supports the idea that MAML1/2 do not promote YAP/TAZ nuclear translocation; rather, it enhances its retention in the nucleus by forming a complex with TEAD. 1) Live cell-imaging data showed that MAML1 localizes to the nucleus much earlier than YAP (Fig. 2E). 2) MAML1 interacted equally well with various forms of YAP that have differential phosphorylation statuses on YAP–Ser-127 and Ser-128 (SI Appendix, Fig. S3C). If MAML1/2 can translocate YAP to the nucleus, YAP–S127D and YAP–S128A should have a lower affinity to MAML1 since they are mainly localized in the cytoplasm, and this is clearly not the case (41).
It was shown that nuclear YAP interacts with p72, which is a regulatory component of the miR processing complex. YAP sequesters p72, which leads to inhibition of miR biogenesis at low cell density or in cancer (47). Conversely, in high cell density, cytoplasmic YAP cannot interact with nuclear p72, and free p72 promotes miR processing and biogenesis (47). We found that the level of miR-30c, which targets the 3′ untranslated region of MAML1, is increased and inhibits MAML protein synthesis in conditions of high cell density. While the expression of MAML1 is regulated at the transcriptional as well as the posttranscriptional level via miR-30c, the expression of MAML2 seems to be controlled only at the posttranscriptional level in HEK293 cells (Fig. 5). MAML1 is most likely not a target gene of YAP/TAZ, since knockdown of Hippo signaling components did not change the levels of MAML1/2 (SI Appendix, Fig. S5C). Further studies are needed to discover how MAML1 and MAML2 are differentially regulated at the transcriptional and posttranscriptional levels, respectively.
Although our biochemical analyses suggest that MAML1 and MAML2 play similar roles in the promotion of nuclear localization and transcriptional activation of YAP/TAZ, MAML2 seems to play a dominant role in the tissues of lung cancer patients. Tissue microarray data show a strong correlation between MAML2 and YAP levels in lung cancer patients (Fig. 6 G and H). TCGA data show a strong correlation between the mRNA levels of MAML2 and YAP target genes in lung cancer (Fig. 6I). The dominant role of MAML2 might be due to higher expression of MAML2 than MAML1 in lung tissues, which has been shown (50, 51).
We show that MAML1/2 not only promote nuclear localization of YAP/TAZ, they also enhance YAP/TAZ–TEAD-mediated transcriptional activity via forming a functional complex (Fig. 4). The components in the trimeric complex seem to help each other to form nuclear dots, since coexpressed MAML1 and YAP colocalized in the nuclear dots much faster than MAML1 alone (Fig. 2E). The biological significance of colocalization of MAML1 and YAP in nuclear dots is unknown, but it is possible that these dots may contain a specific locus where the trimeric complex can bind and regulate the expression of target genes.
Overall, our findings suggest a role of MAML1/2 in the regulation of Hippo signaling by providing mechanistic insight into the nuclear localization and transcriptional activation of YAP/TAZ (SI Appendix, Fig. S7). Furthermore, they highlight the potential of targeting the miR30c–MAML–YAP/TAZ axis as a novel therapy for the interruption of cancer progression.
Materials and Methods
Cell Culture and Transfection.
HEK293T, HEK293, HEK293A, HEK293A–LATS KO (a gift from Hyun Woo Park, Yonsei University, Seoul, Republic of Korea), HeLa, Huh7, HT29, Caco2, DLD-1, SW480, and HCT116 cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM; Lonza, catalog no. 12-604F; or HyClone, catalog no. SH30243.01) supplemented with 10% fetal bovine serum (Gibco, catalog no. 26140-079) and 1× Antibiotic-Antimyotic (Gibco, catalog no. 15240-062) at 37 °C in a humidified 5% CO2 incubator. The 2-deoxyglucose (25 mM for 4 h) was used for glucose starvation. Serum-depleted DMEM was used for serum starvation. Latrunculin A (0.5 μM for 4 h) was used for F-actin inhibition. To transfect HEK293T cells with plasmids, calcium–phosphate (55) or polyethylenimine (PEI) methods were used. For the PEI method, 10 mg/mL PEI (Sigma, catalog no. 408727) was diluted 1/100 in water (Sigma, catalog no. W4502). Opti-MEM (Gibco, catalog no. 31985-070), plasmids, and PEI were mixed and incubated for 20 min at room temperature. Culture medium was changed with fresh medium before treating transfection mixtures. Lipofectamine 2000 (Invitrogen, catalog no. 11668-019), Lipofectamine 3000 (Invitrogen, catalog no. L3000015), or Turbofect (Thermo Scientific, catalog no. R0531) was used for transforming HEK293, HeLa, or HCT116 according to the manufacturer’s instructions. To transfect HEK293T with siRNA, the calcium–phosphate method was used, and other cell lines were transfected by Lipofectamine RNAiMAX (Invitrogen, catalog no. 13778150). siRNA sequences are listed in SI Appendix, Table S1.
Cloning and Plasmids.
YAP sequence was amplified by using cDNA from HEK293T cells and cloned into pCMV4-Flag (Sigma), pEGFP-C1 (Clontech), and pCS2-HA3 (constructed in our laboratory) expression vectors. Flag-NICD was also cloned as described above. V5-MAML1, Flag–mWW-YAP, and rtTA–2SM2 were purchased from Addgene. mCherry–MAML1 and pBI–mCherry–MAML1 were obtained from Bionics. Flag–MAML1, Flag–MAML2, and Flag–MAML3 were provided by Lizi Wu, University of Florida, Gainesville, FL. The 6×-NRE-Luc and 8×-GTIIC-Luc were provided by Cheol O. Joe and Dae-Sik Lim, KAIST, Daejeon, Korea, respectively. Kun-Liang Guan, University of California San Diego, La Jolla, CA, provided the pRK5–Myc–TEAD4 construct. Flag- or V5-tagged MAML1 mutant constructs (PPxA or mNLS) and EGFP- or Flag-tagged YAP mutant constructs (S127A, S127D, S128A, S128D, mpNLS, 5SA, or 5SA-mWW) were generated by site-directed mutagenesis (56) using KOD-Plus-Neo DNA polymerase (TOYOBO, catalog no. KOD-401). To generate guide RNA (gRNA)–MAML1 and gRNA–MAML2, annealed gRNA sequences which were designed by using the ATUM gRNA design tool (https://www.atum.bio/eCommerce/cas9/input) were inserted into BbsI (NEB, catalog no. R0539S)-digested pSpCas9(BB)-2A-Puro (PX459, Addgene). gRNA sequences are listed in SI Appendix, Table S1.
Detailed descriptions are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the E.h.-J. laboratory for stimulating valuable discussions and Dr. Lizi Wu (University of Florida) for providing the MAML constructs. This study was supported by National Research Foundation of Korea Grants NRF-2016R1E1A1A01943544, 2016R1A5A1010764, 2017M3A9B4062421, and 2020R1A2C3013746 (to E.h.-J.), and NRF-2018R1C1B6002749 (to W.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. F.G.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917969117/-/DCSupplemental.
References
- 1.Pan D., The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu V., Plouffe S. W., Guan K. L., The Hippo pathway in organ development, homeostasis, and regeneration. Curr. Opin. Cell Biol. 49, 99–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misra J. R., Irvine K. D., The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52, 65–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccolo S., Dupont S., Cordenonsi M., The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Kim W., Jho E. H., The history and regulatory mechanism of the Hippo pathway. BMB Rep. 51, 106–118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B. et al., Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont S. et al., Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Yu F. X. et al., Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng Z., Moroishi T., Guan K. L., Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon S., Yeon Park S., Woo Park H., Regulation of the Hippo pathway in cancer biology. Cell. Mol. Life Sci. 75, 2303–2319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moroishi T., Hansen C. G., Guan K. L., The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey K. F., Zhang X., Thomas D. M., The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Johnson R., Halder G., The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F. X., Zhao B., Guan K. L., Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan E. H. et al., The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L., A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C. Y. et al., The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TrCP E3 ligase. J. Biol. Chem. 285, 37159–37169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Yin F. et al., Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J. et al., Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng Z. et al., MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6, 8357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai F. et al., TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J., Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Zhao B. et al., Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W. et al., PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26, 1959–1971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X. et al., PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 32, 1266–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang L. et al., SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell 34, 103–118.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Lee Y. et al., Dishevelled has a YAP nuclear export function in a tumor suppressor context-dependent manner. Nat. Commun. 9, 2301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofler M. et al., Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat. Commun. 9, 4966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alves-Guerra M. C., Ronchini C., Capobianco A. J., Mastermind-like 1 Is a specific coactivator of β-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 67, 8690–8698 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Quaranta R. et al., Maml1 acts cooperatively with Gli proteins to regulate sonic hedgehog signaling pathway. Cell Death Dis. 8, e2942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin B. et al., The mastermind-like 1 (MAML1) co-activator regulates constitutive NF-kappaB signaling and cell survival. J. Biol. Chem. 285, 14356–14365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McElhinny A. S., Li J. L., Wu L., Mastermind-like transcriptional co-activators: Emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 27, 5138–5147 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Qi Y. et al., A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat. Commun. 7, ncomms11840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gullberg M. et al., Cytokine detection by antibody-based proximity ligation. Proc. Natl. Acad. Sci. U.S.A. 101, 8420–8424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söderberg O. et al., Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Overholtzer M. et al., Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S. et al., Importin α1 mediates Yorkie nuclear import via an N-terminal non-canonical nuclear localization signal. J. Biol. Chem. 291, 7926–7937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moroishi T. et al., A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon S. et al., Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 18, 61–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P. et al., Dual function of partitioning-defective 3 in the regulation of YAP phosphorylation and activation. Cell Discov. 2, 16021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H. I., Sudol M., The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishioka N. et al., The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Mo J. S. et al., Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 17, 500–510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang H. W., Wentzel E. A., Mendell J. T., Cell-cell contact globally activates microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 7016–7021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori M. et al., Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 156, 893–906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Y. et al., Deubiquitinase YOD1 potentiates YAP/TAZ activities through enhancing ITCH stability. Proc. Natl. Acad. Sci. U.S.A. 114, 4691–4696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N., Xie C., Lu N., Crosstalk between Hippo signalling and miRNAs in tumour progression. FEBS J. 284, 1045–1055 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Wu L. et al., MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Wu L., Sun T., Kobayashi K., Gao P., Griffin J. D., Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell. Biol. 22, 7688–7700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picco G. et al., Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 10, 2198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekine S. et al., Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J. Clin. Invest. 130, 3827–3832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raices M., D’Angelo M. A., Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 13, 687–699 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Chen C., Okayama H., High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7, 2745–2752 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laible M., Boonrod K., Homemade site directed mutagenesis of whole plasmids. J. Vis. Exp. 27, 1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.